RNA INTERFERENCE MEDIATED INHIBITION OF GENE EXPRESSION USING SHORT INTERFERING NUCLEIC ACID (siNA) This application is a continuation-in-part of International Patent Application No. PCT/US04/16390, filed May 24, 2004, which is a continuation-in-part of U.S. Patent Application No. 10/826,966, filed April 16, 2004, which is continuation-in-part of U.S. Patent Application No. 10/757,803, filed January 14, 2004, which is a continuation-in- part of U.S. Patent Application No. 10/720,448, filed November 24, 2003, which is a continuation-in-part of U.S. Patent Application No. 10/693,059, filed October 23, 2003, which is a continuation-in-part of U.S. Patent Application No. 10/444,853, filed May 23, 2003, which is a continuation-in-part of International Patent Application No. PCT/US03/05346, filed February 20, 2003, and a continuation-in-part of International Patent Application No. PCT/US03/O5028, filed February 20, 2003, both of which claim the benefit of U.S. Provisional Application No. 60/358,580 filed February 20, 2002, U.S. Provisional Application No. 60/363,124 filed March 11, 2002, U.S. Provisional Application No. 60/386,782 filed June 6, 2002, U.S. Provisional Application No. 60/406,784 filed August 29, 2002, U.S. Provisional Application No. 60/408,378 filed September 5, 2002, U.S. Provisional Application No. 60/409,293 filed September 9, 2002, and U.S. Provisional Application No. 60/440,129 filed January 15, 2003. This application is also a continuation-in-part of International Patent Application No. PCT/US04/13456, filed April 30, 2004, which is a continuation-in-part of U.S. Patent Application No. 10/780,447, filed February 13, 2004, which is a continuation-in-part of U.S. Patent Application No. 10/427,160, filed April 30, 2003, which is a continuation-in- part of International Patent Application No. PCT/US02/15876 filed May 17, 2002, which claims the benefit of U.S. Provisional Application No. 60/292,217, filed May 18, 2001, U.S. Provisional Application No. 60/362,016, filed March 6, 2002, U.S. Provisional Application No. 60/306,883, filed July 20, 2001, and U.S. Provisional Application No. 60/311,865, filed August 13, 2001. This application is also a continuation-in-part of U.S. Patent Application No. 10/727,780 filed December 3, 2003. This application also claims the benefit of U.S. Provisional Application No. 60/543,480, filed February 10, 2004. The instant application claims the benefit of all the listed applications, which are hereby incoφorated by reference herein in their entireties, including the drawings.
Field Of The Invention
The present invention relates to compounds, compositions, and methods for the study, diagnosis, and treatment of traits, diseases and conditions that respond to the modulation of tartget gene expression and/or activity. The present invention is also directed to compounds, compositions, and methods relating to traits, diseases and conditions that respond to the modulation of expression and/or activity of genes involved in gene expression pathways or other cellular processes that mediate the maintenance or development of such traits, diseases and conditions. Specifically, the invention relates to small nucleic acid molecules, such as short interfering nucleic acid (siNA), short interfering RNA (siRNA), double-stranded RNA (dsRNA), micro-RNA (miRNA), and short hairpin RNA (shRNA) molecules capable of mediating RNA interference (RNAi) against target gene expression. Such small nucleic acid molecules are useful, for example, in providing compositions for treatment of traits, diseases and conditions that can respond to modulation of gene expression in a subject or organism. Background Of The Invention
The following is a discussion of relevant art pertaining to RNAi. The discussion is provided only for understanding of the invention that follows. The summary is not an admission that any of the work described below is prior art to the claimed invention.
RNA interference refers to the process of sequence-specific post-transcriptional gene silencing in animals mediated by short interfering RNAs (siRNAs) (Zamore et al,
2000, Cell, 101, 25-33; Fire et al, 1998, Nature, 391, 806; Hamilton et al, 1999,
Science, 286, 950-951; Tin et al, 1999, Nature, 402, 128-129; Sharp, 1999, Genes &
Dev., 13:139-141; and Strauss, 1999, Science, 286, 886). The corresponding process in plants (Heifetz et al, International PCT Publication No. WO 99/61631) is commonly referred to as post-transcriptional gene silencing or RNA silencing and is also referred to as quelling in fungi. The process of post-transcriptional gene silencing is thought to be an evolutionarily-conserved cellular defense mechanism used to prevent the expression of foreign genes and is commonly shared by diverse flora and phyla (Fire et al, 1999,
Trends Genet, 15, 358). Such protection from foreign gene expression may have evolved in response to the production of double-stranded RNAs (dsRNAs) derived from
viral infection or from the random integration of transposon elements into a host genome via a cellular response that specifically destroys homologous single-stranded RNA or viral genomic RNA. The presence of dsRNA in cells triggers the RNAi response through a mechanism that has yet to be fully characterized. This mechanism appears to be different from other known mechanisms involving double stranded RNA-specific ribonucleases, such as the interferon response that results from dsRNA-mediated activation of protein kinase PKR and 2',5'-oligoadenylate synthetase resulting in nonspecific cleavage of mRNA by ribonuclease L (see for example US Patent Nos. 6,107,094; 5,898,031; Clemens et al, 1997, J. Interferon & Cytokine Res., 17, 503-524; Adah et al, 2001, Curr. Med. Chem., 8, 1189).
The presence of long dsRNAs in cells stimulates the activity of a ribonuclease III enzyme referred to as dicer (Bass, 2000, Cell, 101, 235; Zamore et al, 2000, Cell, 101, 25-33; Hammond et al, 2000, Nature, 404, 293). Dicer is involved in the processing of the dsRNA into short pieces of dsRNA known as short interfering RNAs (siRNAs) (Zamore et al, 2000, Cell, 101, 25-33; Bass, 2000, Cell, 101, 235; Berstein et al, 2001, Nature, 409, 363). Short interfering RNAs derived from dicer activity are typically about 21 to about 23 nucleotides in length and comprise about 19 base pair duplexes (Zamore et al, 2000, Cell, 101, 25-33; Elbashir eta , 2001, Genes Dev., 15, 188). Dicer has also been implicated in the excision of 21- and 22-nucleotide small temporal RNAs (stRNAs) from precursor RNA of conserved structure that are implicated in translational control (Hutvagner et al, 2001, Science, 293, 834). The RNAi response also features an endonuclease complex, commonly referred to as an RNA-induced silencing complex (RISC), which mediates cleavage of single-stranded RNA having sequence complementary to the antisense strand of the siRNA duplex. Cleavage of the target RNA takes place in the middle of the region complementary to the antisense strand of the siRNA duplex (Elbashir et al, 2001, Genes Dev., 15, 188).
RNAi has been studied in a variety of systems. Fire et al, 1998, Nature, 391, 806, were the first to observe RNAi in C. elegans. Bahramian and Zarbl, 1999, Molecular and Cellular Biology, 19, 274-283 and Wianny and Goetz, 1999, Nature Cell Biol, 2, 70, describe RNAi mediated by dsRNA in mammalian systems. Hammond et al, 2000,
Nature, 404, 293, describe RNAi in Drosophila cells transfected with dsRNA. Elbashir
et al, 2001, Nature, 411, 494 and Tuschl et al, International PCI' Publication J o. wu 01/75164, describe RNAi induced by introduction of duplexes of synthetic 21 -nucleotide RNAs in cultured mammalian cells including human embryonic kidney and HeLa cells. Recent work in Drosophila embryonic lysates (Elbashir et al, 2001, EMBO J., 20, 6877 and Tuschl et al, International PCT Publication No. WO 01/75164) has revealed certain requirements for siRNA length, structure, chemical composition, and sequence that are essential to mediate efficient RNAi activity. These studies have shown that 21- nucleotide siRNA duplexes are most active when containing 3'-terminal dinucleotide overhangs. Furthermore, complete substitution of one or both siRNA strands with 2'- deoxy (2'-H) or 2'-0-methyl nucleotides abolishes RNAi activity, whereas substitution of the 3 '-terminal siRNA overhang nucleotides with 2'-deoxy nucleotides (2'-H) was shown to be tolerated. Single mismatch sequences in the center of the siRNA duplex were also shown to abolish RNAi activity. In addition, these studies also indicate that the position of the cleavage site in the target RNA is defined by the 5 '-end of the siRNA guide sequence rather than the 3'-end of the guide sequence (Elbashir et al, 2001, EMBO J., 20, 6877). Other studies have indicated that a 5'-phosphate on the target-complementary strand of a siRNA duplex is required for siRNA activity and that ATP is utilized to maintain the 5'-phosphate moiety on the siRNA (Nykanen et al, 2001, Cell, 107, 309).
Studies have shown that replacing the 3 '-terminal nucleotide overhanging segments of a 21-mer siRNA duplex having two-nucleotide 3 '-overhangs with deoxyribonucleotides does not have an adverse effect on RNAi activity. Replacing up to four nucleotides on each end of the siRNA with deoxyribonucleotides has been reported to be well tolerated, whereas complete substitution with deoxyribonucleotides results in no RNAi activity (Elbashir et al, 2001, EMBO J., 20, 6877 and Tuschl et al, International PCT Publication No. WO 01/75164). In addition, Elbashir et al, supra, also report that substitution of siRNA with 2'-0-methyl nucleotides completely abolishes
RNAi activity. Ti et al, International PCT Publication No. WO 00/44914, and Beach et al, International PCT Publication No. WO 01/68836 preliminarily suggest that siRNA may include modifications to either the phosphate-sugar backbone or the nucleoside to include at least one of a nitrogen or sulfur heteroatom, however, neither application postulates to what extent such modifications would be tolerated in siRNA molecules, nor provides any further guidance or examples of such modified siRNA. Kreutzer et al,
Canadian Patent Application No. 2,359,180, also describe certain chemical modifications for use in dsRNA constructs in order to counteract activation of double-stranded RNA- dependent protein kinase PKR, specifically 2'-amino or 2'-0-methyl nucleotides, and nucleotides containing a 2'-0 or 4'-C methylene bridge. However, Kreutzer et al. similarly fails to provide examples or guidance as to what extent these modifications would be tolerated in dsRNA molecules.
Parrish et al, 2000, Molecular Cell, 6, 1077-1087, tested certain chemical modifications targeting the unc-22 gene in C. elegans using long (>25 nt) siRNA transcripts. The authors describe the introduction of thiophosphate residues into these siRNA transcripts by incorporating thiophosphate nucleotide analogs with T7 and T3 RNA polymerase and observed that RNAs with two phosphorothioate modified bases also had substantial decreases in effectiveness as RNAi. Further, Parrish et al. reported that phosphorothioate modification of more than two residues greatly destabilized the RNAs in vitro such that interference activities could not be assayed. Id. at 1081. The authors also tested certain modifications at the 2'-position of the nucleotide sugar in the long siRNA transcripts and found that substituting deoxynucleotides for ribonucleotides produced a substantial decrease in interference activity, especially in the case of Uridine to Thymidine and/or Cytidine to deoxy-Cytidine substitutions. Id. In addition, the authors tested certain base modifications, including substituting, in sense and antisense strands of the siRNA, 4-thiouracil, 5-bromouracil, 5-iodouracil, and 3-(aminoallyl)uracil for uracil, and inosine for guanosine. Whereas 4-thiouracil and 5-bromouracil substitution appeared to be tolerated, Parrish reported that inosine produced a substantial decrease in interference activity when incorporated in either strand. Parrish also reported that incorporation of 5-iodouracil and 3-(aminoallyl)uracil in the antisense strand resulted in a substantial decrease in RNAi activity as well.
The use of longer dsRNA has been described. For example, Beach et al, International PCT Publication No. WO 01/68836, describes specific methods for attenuating gene expression using endogenously-derived dsRNA. Tuschl et al, International PCT Publication No. WO 01/75164, describe a Drosophila in vitro RNAi system and the use of specific siRNA molecules for certain functional genomic and certain therapeutic applications; although Tuschl, 2001, Chem. Biochem., 2, 239-245,
doubts that KJ Ai can be used to cure genetic diseases or viral miection oue to tne danger of activating interferon response. Ti et al, International PCT Publication No. WO 00/44914, describe the use of specific long (141 bp-488 bp) enzymatically synthesized or vector expressed dsRNAs for attenuating the expression of certain target genes. Zernicka-Goetz et al, International PCT Publication No. WO 01/36646, describe certain methods for inhibiting the expression of particular genes in mammalian cells using certain long (550 bp-714 bp), enzymatically synthesized or vector expressed dsRNA molecules. Fire et al, International PCT Publication No. WO 99/32619, describe particular methods for introducing certain long dsRNA molecules into cells for use in inhibiting gene expression in nematodes. Plaetinck et al, International PCT Publication No. WO 00/01846, describe certain methods for identifying specific genes responsible for conferring a particular phenotype in a cell using specific long dsRNA molecules. Mello et al, International PCT Publication No. WO 01/29058, describe the identification of specific genes involved in dsRNA-mediated RNAi. Pachuck et al, International PCT Publication No. WO 00/63364, describe certain long (at least 200 nucleotide) dsRNA constructs. Deschamps Depaillette et al, International PCT Publication No. WO 99/07409, describe specific compositions consisting of particular dsRNA molecules combined with certain anti-viral agents. Waterhouse et al, International PCT Publication No. 99/53050 and 1998, PNAS, 95, 13959-13964, describe certain methods for decreasing the phenotypic expression of a nucleic acid in plant cells using certain dsRNAs. Driscoll et al, International PCT Publication No. WO 01/49844, describe specific DNA expression constructs for use in facilitating gene silencing in targeted organisms.
Others have reported on various RNAi and gene-silencing systems. For example, Parrish et al, 2000, Molecular Cell, 6, 1077-1087, describe specific chemically-modified dsRNA constructs targeting the unc-22 gene of C. elegans. Grossniklaus, International PCT Publication No. WO 01/38551, describes certain methods for regulating polycomb gene expression in plants using certain dsRNAs. Churikov et al, International PCT Publication No. WO 01/42443, describe certain methods for modifying genetic characteristics of an organism using certain dsRNAs. Cogoni et al„ International PCT Publication No. WO 01/53475, describe certain methods for isolating a Neurospora silencing gene and uses thereof. Reed et al, International PCT Publication No. WO
01/68836, describe certain methods for gene silencing in plants. Honer et al., International PCT Publication No. WO 01/70944, describe certain methods of drug screening using transgenic nematodes as Parkinson's Disease models using certain dsRNAs. Deak et al, International PCT Publication No. WO 01/72774, describe certain Drosophila-deήyed gene products that may be related to RNAi in Drosophila. Arndt et al, International PCT Publication No. WO 01/92513 describe certain methods for mediating gene suppression by using factors that enhance RNAi. Tuschl et al, International PCT Publication No. WO 02/44321, describe certain synthetic siRNA constructs. Pachuk et al, International PCT Publication No. WO 00/63364, and Satishchandran et al, International PCT Publication No. WO 01/04313, describe certain methods and compositions for inhibiting the function of certain polynucleotide sequences using certain long (over 250 bp), vector expressed dsRNAs. Echeverri et al, International PCT Publication No. WO 02/38805, describe certain C. elegans genes identified via RNAi. Kreutzer et al, International PCT Publications Nos. WO 02/055692, WO 02/055693, and EP 1144623 B 1 describes certain methods for inhibiting gene expression using dsRNA. Graham et al, International PCT Publications Nos. WO 99/49029 and WO 01/70949, and AU 4037501 describe certain vector expressed siRNA molecules. Fire et al, US 6,506,559, describe certain methods for inhibiting gene expression in vitro using certain long dsRNA (299 bp-1033 bp) constructs that mediate RNAi. Martinez et al, 2002, Cell, 110, 563-574, describe certain single stranded siRNA constructs, including certain 5 '-phosphorylated single stranded siRNAs that mediate RNA interference in Hela cells. Harborth et al, 2003, Antisense & Nucleic Acid Drug Development, 13, 83-105, describe certain chemically and structurally modified siRNA molecules. Chiu and Rana, 2003, RNA, 9, 1034-1048, describe certain chemically and structurally modified siRNA molecules. Woolf et al, International PCT Publication Nos. WO 03/064626 and WO 03/064625 describe certain chemically modified dsRNA constructs.
SUMMARY OF THE INVENTION
This invention relates to compounds, compositions, and methods useful for modulating gene expression using short interfering nucleic acid (siNA) molecules. This invention also relates to compounds, compositions, and methods useful for modulating
the expression and activity of other genes involved in pathways of gene expression and/or activity by RNA interference (RNAi) using small nucleic acid molecules. In particular, the instant invention features small nucleic acid molecules, such as short interfering nucleic acid (siNA), short interfering RNA (siRNA), double-stranded RNA (dsRNA), micro-RNA (miRNA), and short hairpin RNA (shRNA) molecules and methods used to modulate the expression of genes, including gene targets having RNA transcripts referred to by Genbank Accession Numbers shown in Table I.
A siNA of the invention can be unmodified or chemically-modified. A siNA of the instant invention can be chemically synthesized, expressed from a vector or enzymatically synthesized. The instant invention also features various chemically- modified synthetic short interfering nucleic acid (siNA) molecules capable of modulating target gene expression or activity in cells by RNA interference (RNAi). The use of chemically-modified siNA improves various properties of native siNA molecules through increased resistance to nuclease degradation in vivo and or through improved cellular uptake. Further, contrary to earlier published studies, siNA having multiple chemical modifications retains its RNAi activity. The siNA molecules of the instant invention provide useful reagents and methods for a variety of therapeutic, veterinary, diagnostic, target validation, genomic discovery, genetic engineering, and pharmacogenomic applications. In one embodiment, the invention features one or more siNA molecules and methods that independently or in combination modulate the expression of target genes encoding proteins, such as proteins that are associated with the maintenance and/or development of diseases, traits, disorders, and/or conditions as described herein or otherwise known in the art, such as genes encoding sequences comprising those sequences referred to by GenBank Accession Nos. shown in Table I, referred to herein generally as "target". The description below of the various aspects and embodiments of the invention is provided with reference to exemplary target genes referred to herein as gene targets. However, the various aspects and embodiments are also directed to other genes, such as gene homologs, transcript variants, and polymorphisms (e.g., single nucleotide polymoφhism, (SNPs)) associated with certain genes. As such, the various aspects and embodiments are also directed to other genes that are involved in disease,
trait, condition, or disorder related pathways of signal transduction or gene expression that are involved, for example, in the maintenence or development of diseases, traits, conditions, or disorders described herein. These additional genes can be analyzed for target sites using the methods described for exemplary genes herein. Thus, the modulation of other genes and the effects of such modulation of the other genes can be performed, determined, and measured as described herein.
In one embodiment, the invention features a double-stranded short interfering nucleic acid (siNA) molecule that down-regulates expression of a target gene or that directs cleavage of a target RNA, wherein said siNA molecule comprises about 15 to about 28 base pairs.
In one embodiment, the invention features a double stranded short interfering nucleic acid (siNA) molecule that directs cleavage of a target RNA via RNA interference (RNAi), wherem the double stranded siNA molecule comprises a first and a second strand, each strand of the siNA molecule is about 18 to about 28 nucleotides in length, the first strand of the siNA molecule comprises nucleotide sequence having sufficient complementarity to the target RNA for the siNA molecule to direct cleavage of the target RNA via RNA interference, and the second strand of said siNA molecule comprises nucleotide sequence that is complementary to the first strand.
In one embodiment, the invention features a double stranded short interfering nucleic acid (siNA) molecule that directs cleavage of a target RNA via RNA interference (RNAi), wherem the double stranded siNA molecule comprises a first and a second strand, each strand of the siNA molecule is about 18 to about 23 nucleotides in length, the first strand of the siNA molecule comprises nucleotide sequence having sufficient complementarity to the target RNA for the siNA molecule to direct cleavage of the target RNA via RNA interference, and the second strand of said siNA molecule comprises nucleotide sequence that is complementary to the first strand.
In one embodiment, the invention features a chemically synthesized double stranded short interfering nucleic acid (siNA) molecule that directs cleavage of a target
RNA via RNA interference (RNAi), wherein each strand of the siNA molecule is about 18 to about 28 nucleotides in length; and one strand of the siNA molecule comprises
nucleotide sequence having sufficient complementarity to the target RNA for the siNA molecule to direct cleavage of the target RNA via RNA interference.
In one embodiment, the invention features a chemically synthesized double stranded short interfering nucleic acid (siNA) molecule that directs cleavage of a target RNA via RNA interference (RNAi), wherein each strand of the siNA molecule is about 18 to about 23 nucleotides in length; and one strand of the siNA molecule comprises nucleotide sequence having sufficient complementarity to the target RNA for the siNA molecule to direct cleavage of the target RNA via RNA interference.
In one embodiment, the invention features a siNA molecule that down-regulates expression of a target gene or that directs cleavage of a target RNA, for example, wherein the gene comprises protein encoding sequence. In one embodiment, the invention features a siNA molecule that down-regulates expression of a target gene or that directs cleavage of a target RNA, for example, wherein the gene comprises non- coding sequence or encodes sequence of regulatory elements involved in gene expression (e.g., non-coding RNA) .
In one embodiment, a siNA of the invention is used to inhibit the expression of target genes or a target gene family, wherein the genes or gene family sequences share sequence homology. Such homologous sequences can be identified as is known in the art, for example using sequence alignments. siNA molecules can be designed to target such homologous sequences, for example using perfectly complementary sequences or by incoφorating non-canonical base pairs, for example mismatches and/or wobble base pairs, that can provide additional target sequences. In instances where mismatches are identified, non-canonical base pairs (for example, mismatches and/or wobble bases) can be used to generate siNA molecules that target more than one gene sequence. In a non- limiting example, non-canonical base pairs such as UU and CC base pairs are used to generate siNA molecules that are capable of targeting sequences for differing targets that share sequence homology. As such, one advantage of using siNAs of the invention is that a single siNA can be designed to include nucleic acid sequence that is complementary to the nucleotide sequence that is conserved between the homologous genes. In this approach, a single siNA can be used to inhibit expression of more than one gene instead of using more than one siNA molecule to target the different genes.
In one embodiment, a target RNA of the invention is an expressed pseudogene (see for example pseudogene sequences referred to by Genbank Accession Numbers in Table I). As used herein the term "disease related expressed pseudogene" refers to any expressed pseudogene that is associated with a disease, disorder, condition, or trait. In one embodiment, the invention features a siNA molecule having RNAi activity against target RNA (e.g., coding or non-coding RNA), wherein the siNA molecule comprises a sequence complementary to any RNA sequence, such as those sequences having GenBank Accession Nos. shown in Table I. In another embodiment, the invention features a siNA molecule having RNAi activity against target RNA, wherein the siNA molecule comprises a sequence complementary to an RNA having variant encoding sequence, for example other mutant genes not shown in Table I but known in the art to be associated with the maintenance and/or development of diseases, traits, disorders, and/or conditions described herein or otherwise known in the art. Chemical modifications as shown in Table II or otherwise described herein can be applied to any siNA construct of the invention. In another embodiment, a siNA molecule of the invention includes a nucleotide sequence that can interact with nucleotide sequence of a target gene and thereby mediate silencing of gene expression, for example, wherem the siNA mediates regulation of gene expression by cellular processes that modulate the chromatin structure or methylation patterns of the gene and prevent transcription of the gene.
In one embodiment, siNA molecules of the invention are used to down regulate or inhibit the expression of proteins arising from haplotype polymoφhisms that are associated with a disease or condition. Analysis of genes, or protein or RNA levels can be used to identify subjects with such polymoφhisms or those subjects who are at risk of developing traits, conditions, or diseases described herein. These subjects are amenable to treatment, for example, treatment with siNA molecules of the invention and any other composition useful in treating diseases related to gene expression. As such, analysis of protein or RNA levels can be used to determine treatment type and the course of therapy in treating a subject. Monitoring of protein or RNA levels can be used to predict treatment outcome and to determine the efficacy of compounds and compositions that
modulate the level and/or activity of certain proteins associated with a trait, disorder, condition, or disease.
In one embodiment of the invention a siNA molecule comprises an antisense strand comprising a nucleotide sequence that is complementary to a target polynucleotide sequence or a portion thereof. The siNA further comprises a sense strand, wherein said sense strand comprises a nucleotide sequence of a target polynucleotide sequence or a portion thereof, (e.g., about 15 to about 25 or more, or about 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, or 25 or more contiguous nucleotides in a target polynucleotide sequence). In one embodiment, the target polynucleotide sequence is a target DNA. In one embodiment, the target polynucleotide sequence is a target RNA.
In one embodiment, the invention features a siNA molecule comprising a first sequence, for example, the antisense sequence of the siNA construct, complementary to a sequence or portion of sequence comprising sequence represented by GenBank Accession Nos. shown in Table I, and a second sequence, for example a sense sequence, that is complementary to the antisense sequence. Chemical modifications in Table II and described herein can be applied to any siNA construct (e.g., sense or antisenase sequence) of the invention.
In one embodiment of the invention a siNA molecule comprises an antisense strand having about 15 to about 30 (e.g., about 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, or 30) nucleotides, wherein the antisense strand is complementary to a target RNA sequence or a portion thereof, and wherein said siNA further comprises a sense strand having about 15 to about 30 (e.g., about 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, or 30) nucleotides, and wherem said sense strand and said antisense strand are distinct nucleotide sequences where at least about 15 nucleotides in each strand are complementary to the other strand.
In another embodiment of the invention a siNA molecule of the invention comprises an antisense region having about 15 to about 30 (e.g., about 15, 16, 17, 18, 19,
20, 21, 22, 23, 24, 25, 26, 27, 28, 29, or 30) nucleotides, wherein the antisense region is complementary to a target DNA sequence, and wherem said siNA further comprises a sense region having about 15 to about 30 (e.g., about 15, 16, 17, 18, 19, 20, 21, 22, 23,
_ -r, .υ, .1, o, y, or ou) nucleotides, wherem said sense region and said antisense region are comprised in a linear molecule where the sense region comprises at least about 15 nucleotides that are complementary to the antisense region.
In one embodiment, a siNA molecule of the invention has RNAi activity that modulates expression of RNA encoded by one or more genes. Because various genes can share some degree of sequence homology with each other, siNA molecules can be designed to target a class of genes or alternately specific genes (e.g., polymoφhic variants) by selecting sequences that are either shared amongst different gene targets or alternatively that are unique for a specific gene target. Therefore, in one embodiment, the siNA molecule can be designed to target conserved regions of target RNA sequences having homology among several gene variants so as to target a class of genes with one siNA molecule. Accordingly, in one embodiment, the siNA molecule of the invention modulates the expression of one or both gene alleles in a subject. In another embodiment, the siNA molecule can be designed to target a sequence that is unique to a specific target RNA sequence (e.g., a single allele or single nucleotide polymoφhism (SNP)) due to the high degree of specificity that the siNA molecule requires to mediate RNAi activity.
In one embodiment, nucleic acid molecules of the invention that act as mediators of the RNA interference gene silencing response are double-stranded nucleic acid molecules. In another embodiment, the siNA molecules of the invention consist of duplex nucleic acid molecules containing about 15 to about 30 base pairs between oligonucleotides comprising about 15 to about 30 (e.g., about 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, or 30) nucleotides. In yet another embodiment, siNA molecules of the invention comprise duplex nucleic acid molecules with overhanging ends of about 1 to about 3 (e.g., about 1, 2, or 3) nucleotides, for example, about 21- nucleotide duplexes with about 19 base pairs and 3 '-terminal mononucleotide, dinucleotide, or trinucleotide overhangs. In yet another embodiment, siNA molecules of the invention comprise duplex nucleic acid molecules with blunt ends, where both ends are blunt, or alternatively, where one of the ends is blunt. In one embodiment, the invention features one or more chemically-modified siNA constructs having specificity for a target polynucleotide (e.g., RNA or DNA), such as
ωwuuig NΛ sequences reierred to herein by Uenbank Accession number or such RNA sequences referred to herein by Genbank Accession number. In one embodiment, the invention features a RNA based siNA molecule (e.g., a siNA comprising 2'-OH nucleotides) having specificity for target polynucleotides (e.g., RNA or DNA) that includes one or more chemical modifications described herein. Non- limiting examples of such chemical modifications include without limitation phosphorothioate internucleotide linkages, 2'-deoxyribonucleotides, 2'-0-methyl ribonucleotides, 2'-deoxy-2'-fluoro ribonucleotides, "universal base" nucleotides, "acyclic" nucleotides, 5-C-methyl nucleotides, and terminal glyceryl and/or inverted deoxy abasic residue incoφoration. These chemical modifications, when used in various siNA constructs, (e.g., RNA based siNA constructs), are shown to preserve RNAi activity in cells while at the same time, dramatically increasing the serum stability of these compounds. Furthermore, contrary to the data published by Parrish et al, supra, applicant demonstrates that multiple (greater than one) phosphorothioate substitutions are well-tolerated and confer substantial increases in serum stability for modified siNA constructs.
In one embodiment, a siNA molecule of the invention comprises modified nucleotides while maintaining the ability to mediate RNAi. The modified nucleotides can be used to improve in vitro or in vivo characteristics such as stability, activity, and/or bioavailabihty. For example, a siNA molecule of the invention can comprise modified nucleotides as a percentage of the total number of nucleotides present in the siNA molecule. As such, a siNA molecule of the invention can generally comprise about 5% to about 100% modified nucleotides (e.g., about 5%, 10%, 15%, 20%, 25%, 30%, 35%, 40%, 45%, 50%, 55%, 60%, 65%, 70%, 75%, 80%, 85%, 90%, 95% or 100% modified nucleotides). The actual percentage of modified nucleotides present in a given siNA molecule will depend on the total number of nucleotides present in the siNA. If the siNA molecule is single stranded, the percent modification can be based upon the total number of nucleotides present in the single stranded siNA molecules. Likewise, if the siNA molecule is double stranded, the percent modification can be based upon the total number of nucleotides present in the sense strand, antisense strand, or both the sense and antisense strands.
One aspect of the invention features a double-stranded short interfering nucleic acid (siNA) molecule that down-regulates expression of a target gene or that directs cleavage of a target RNA. In one embodiment, the double stranded siNA molecule comprises one or more chemical modifications and each strand of the double-stranded siNA is about 21 nucleotides long. In one embodiment, the double-stranded siNA molecule does not contain any ribonucleotides. In another embodiment, the double- stranded siNA molecule comprises one or more ribonucleotides. In one embodiment, each strand of the double-stranded siNA molecule independently comprises about 15 to about 30 (e.g., about 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, or 30) nucleotides, wherein each strand comprises about 15 to about 30 (e.g., about 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, or 30) nucleotides that are complementary to the nucleotides of the other strand. In one embodiment, one of the strands of the double-stranded siNA molecule comprises a nucleotide sequence that is complementary to a nucleotide sequence or a portion thereof of the gene, and the second strand of the double-stranded siNA molecule comprises a nucleotide sequence substantially similar to the nucleotide sequence of the gene or a portion thereof.
In another embodiment, the invention features a double-stranded short interfering nucleic acid (siNA) molecule that down-regulates expression of a target gene or that directs cleavage of a target RNA, comprising an antisense region, wherein the antisense region comprises a nucleotide sequence that is complementary to a nucleotide sequence of the gene or a portion thereof, and a sense region, wherein the sense region comprises a nucleotide sequence substantially similar to the nucleotide sequence of the gene or a portion thereof. In one embodiment, the antisense region and the sense region independently comprise about 15 to about 30 (e.g. about 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, or 30) nucleotides, wherein the antisense region comprises about 15 to about 30 (e.g. about 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, or 30) nucleotides that are complementary to nucleotides of the sense region.
In another embodiment, the invention features a double-stranded short interfering nucleic acid (siNA) molecule that down-regulates expression of a target gene or that directs cleavage of a target RNA, comprising a sense region and an antisense region, wherein the antisense region comprises a nucleotide sequence that is complementary to a
nucleotide sequence of RNA encoded by the gene or a portion thereof and the sense region comprises a nucleotide sequence that is complementary to the antisense region.
In one embodiment, a siNA molecule of the invention comprises blunt ends, i.e., ends that do not include any overhanging nucleotides. For example, a siNA molecule comprising modifications described herein (e.g., comprising nucleotides having Formulae I-VII or siNA constructs comprising "Stab 00"-"Stab 32" (Table H) or any combination thereof (see Table II)) and/or any length described herein can comprise blunt ends or ends with no overhanging nucleotides.
In one embodiment, any siNA molecule of the invention can comprise one or more blunt ends, i.e. where a blunt end does not have any overhanging nucleotides. In one embodiment, the blunt ended siNA molecule has a number of base pairs equal to the number of nucleotides present in each strand of the siNA molecule. In another embodiment, the siNA molecule comprises one blunt end, for example wherein the 5'- end of the antisense strand and the 3 '-end of the sense strand do not have any overhanging nucleotides. In another example, the siNA molecule comprises one blunt end, for example wherein the 3 '-end of the antisense strand and the 5 '-end of the sense strand do not have any overhanging nucleotides. In another example, a siNA molecule comprises two blunt ends, for example wherein the 3 '-end of the antisense strand and the 5 '-end of the sense strand as well as the 5 '-end of the antisense strand and 3 '-end of the sense strand do not have any overhanging nucleotides. A blunt ended siNA molecule can comprise, for example, from about 15 to about 30 nucleotides (e.g., about 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, or 30 nucleotides). Other nucleotides present in a blunt ended siNA molecule can comprise, for example, mismatches, bulges, loops, or wobble base pairs to modulate the activity of the siNA molecule to mediate RNA interference.
By "blunt ends" is meant symmetric termini or termini of a double stranded siNA molecule having no overhanging nucleotides. The two strands of a double stranded siNA molecule align with each other without over-hanging nucleotides at the termini.
For example, a blunt ended siNA construct comprises terminal nucleotides that are complementary between the sense and antisense regions of the siNA molecule.
In one embodiment, the invention features a double-stranded short interfering nucleic acid (siNA) molecule that down-regulates expression of a target gene or that directs cleavage of a target RNA, wherein the siNA molecule is assembled from two separate oligonucleotide fragments wherein one fragment comprises the sense region and the second fragment comprises the antisense region of the siNA molecule. The sense region can be connected to the antisense region via a linker molecule, such as a polynucleotide linker or a non-nucleotide linker.
In one embodiment, the invention features double-stranded short interfering nucleic acid (siNA) molecule that down-regulates expression of a target gene or that directs cleavage of a target RNA, wherein the siNA molecule comprises about 15 to about 30 (e.g. about 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, or 30) base pairs, and wherein each strand of the siNA molecule comprises one or more chemical modifications. In another embodiment, one of the strands of the double-stranded siNA molecule comprises a nucleotide sequence that is complementary to a nucleotide sequence of a gene or a portion thereof, and the second strand of the double-stranded siNA molecule comprises a nucleotide sequence substantially similar to the nucleotide sequence or a portion thereof of the gene. In another embodiment, one of the strands of the double-stranded siNA molecule comprises a nucleotide sequence that is complementary to a nucleotide sequence of a gene or portion thereof, and the second strand of the double-stranded siNA molecule comprises a nucleotide sequence substantially similar to the nucleotide sequence or portion thereof of the gene. In another embodiment, each strand of the siNA molecule comprises about 15 to about 30 (e.g. about 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, or 30) nucleotides, and each strand comprises at least about 15 to about 30 (e.g. about 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, or 30) nucleotides that are complementary to the nucleotides of the other strand. The gene can comprise, for example, a gene that encodes sequences refeπed to in Table I.
In one embodiment, a siNA molecule of the invention comprises no ribonucleotides. In another embodiment, a siNA molecule of the invention comprises ribonucleotides.
In one embodiment, a siNA molecule of the invention comprises an antisense region comprising a nucleotide sequence that is complementary to a nucleotide sequence of a target gene or a portion thereof, and the siNA further comprises a sense region comprising a nucleotide sequence substantially similar to the nucleotide sequence of the target gene or a portion thereof. In another embodiment, the antisense region and the sense region each comprise about 15 to about 30 (e.g. about 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, or 30) nucleotides and the antisense region comprises at least about 15 to about 30 (e.g. about 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, or 30) nucleotides that are complementary to nucleotides of the sense region. The target gene can comprise, for example, sequence encoding sequences refeπed to in Table I. In another embodiment, the siNA is a double stranded nucleic acid molecule, where each of the two strands of the siNA molecule independently comprise about 15 to about 40 (e.g. about 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 23, 33, 34, 35, 36, 37, 38, 39, or 40) nucleotides, and where one of the strands of the siNA molecule comprises at least about 15 (e.g. about 15, 16, 17, 18, 19, 20, 21, 22, 23, 24 or 25 or more) nucleotides that are complementary to the nucleic acid sequence of the gene or a portion thereof.
In one embodiment, a siNA molecule of the invention comprises a sense region and an antisense region, wherein the antisense region comprises a nucleotide sequence that is complementary to a nucleotide sequence of RNA encoded by a target gene, or a portion thereof, and the sense region comprises a nucleotide sequence that is complementary to the antisense region. In one embodiment, the siNA molecule is assembled from two separate oligonucleotide fragments, wherem one fragment comprises the sense region and the second fragment comprises the antisense region of the siNA molecule. In another embodiment, the sense region is connected to the antisense region via a linker molecule. In another embodiment, the sense region is connected to the antisense region via a linker molecule, such as a nucleotide or non- nucleotide linker. The target gene can comprise, for example, sequence encoding sequences refeπed in to Table I. In one embodiment, the invention features a double-stranded short interfering nucleic acid (siNA) molecule that down-regulates expression of a target gene or that
directs cleavage of a target RNA comprising a sense region and an antisense region, wherein the antisense region comprises a nucleotide sequence that is complementary to a nucleotide sequence of RNA encoded by the target gene or a portion thereof and the sense region comprises a nucleotide sequence that is complementary to the antisense region, and wherein the siNA molecule has one or more modified pyrimidine and/or purine nucleotides. In one embodiment, the pyrimidine nucleotides in the sense region are 2'-0-methyl pyrimidine nucleotides or 2'-deoxy-2'-fluoro pyrimidine nucleotides and the purine nucleotides present in the sense region are 2'-deoxy purine nucleotides. In another embodiment, the pyrimidine nucleotides in the sense region are 2'-deoxy-2'- fluoro pyrimidine nucleotides and the purine nucleotides present in the sense region are 2'-0-methyl purine nucleotides. In another embodiment, the pyrimidine nucleotides in the sense region are 2'-deoxy-2'-fTuoro pyrimidine nucleotides and the purine nucleotides present in the sense region are 2'-deoxy purine nucleotides. In one embodiment, the pyrimidine nucleotides in the antisense region are 2'-deoxy-2'-fluoro pyrimidine nucleotides and the purine nucleotides present in the antisense region are 2 '-O-methyl or 2'-deoxy purine nucleotides. In another embodiment of any of the above-described siNA molecules, any nucleotides present in a non-complementary region of the sense strand (e.g. overhang region) are 2'-deoxy nucleotides.
In one embodiment, the invention features a double-stranded short interfering nucleic acid (siNA) molecule that down-regulates expression of a target gene or that directs cleavage of a target RNA, wherein the siNA molecule is assembled from two separate oligonucleotide fragments wherein one fragment comprises the sense region and the second fragment comprises the antisense region of the siNA molecule, and wherein the fragment comprising the sense region includes a terminal cap moiety at the 5'-end, the 3 '-end, or both of the 5' and 3' ends of the fragment. In one embodiment, the terminal cap moiety is an inverted deoxy abasic moiety or glyceryl moiety. In one embodiment, each of the two fragments of the siNA molecule independently comprise about 15 to about 30 (e.g. about 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, or 30) nucleotides. In another embodiment, each of the two fragments of the siNA molecule independently comprise about 15 to about 40 (e.g. about 15, 16, 17, 18, 19, 20, 21, 22,
23, 24, 25, 26, 27, 28, 29, 30, 31, 23, 33, 34, 35, 36, 37, 38, 39, or 40) nucleotides. In a
non-limiting example, each of the two fragments of the siNA molecule comprise about 21 nucleotides.
In one embodiment, the invention features a siNA molecule comprising at least one modified nucleotide, wherein the modified nucleotide is a 2 '-deoxy-2 '-fluoro nucleotide. The siNA can be, for example, about 15 to about 40 nucleotides in length. In one embodiment, all pyrimidine nucleotides present in the siNA are 2 '-deoxy-2 '-fluoro pyrimidine nucleotides. In one embodiment, the modified nucleotides in the siNA include at least one 2 '-deoxy-2 '-fluoro cytidine or 2 '-deoxy-2 '-fluoro uridine nucleotide. In another embodiment, the modified nucleotides in the siNA include at least one 2'- fluoro cytidine and at least one 2 '-deoxy-2 '-fluoro uridine nucleotides. In one embodiment, all uridine nucleotides present in the siNA are 2 '-deoxy-2 '-fluoro uridine nucleotides. In one embodiment, all cytidine nucleotides present in the siNA are 2'- deoxy-2' -fluoro cytidine nucleotides. In one embodiment, all adenosine nucleotides present in the siNA are 2'-deoxy-2'-fluoro adenosine nucleotides. In one embodiment, all guanosine nucleotides present in the siNA are 2 '-deoxy-2 '-fluoro guanosme nucleotides. The siNA can further comprise at least one modified internucleotidic linkage, such as phosphorothioate linkage. In one embodiment, the 2'-deoxy-2'- fluoronucleotides are present at specifically selected locations in the siNA that are sensitive to cleavage by ribonucleases, such as locations having pyrimidine nucleotides. In one embodiment, the invention features a method of increasing the stability of a siNA molecule against cleavage by ribonucleases comprising introducing at least one modified nucleotide into the siNA molecule, wherein the modified nucleotide is a 2'- deoxy-2' -fluoro nucleotide. In one embodiment, all pyrimidine nucleotides present in the siNA are 2 '-deoxy-2 '-fluoro pyrimidine nucleotides. In one embodiment, the modified nucleotides in the siNA include at least one 2 '-deoxy-2 '-fluoro cytidine or 2'- deoxy-2' -fluoro uridine nucleotide. In another embodiment, the modified nucleotides in the siNA include at least one 2 '-fluoro cytidine and at least one 2 '-deoxy-2 '-fluoro uridine nucleotides. In one embodiment, all uridine nucleotides present in the siNA are 2 '-deoxy-2 '-fluoro uridine nucleotides. In one embodiment, all cytidine nucleotides present in the siNA are 2'-deoxy-2'-fluoro cytidine nucleotides. In one embodiment, all adenosine nucleotides present in the siNA are 2 '-deoxy-2 '-fluoro adenosine nucleotides.
In one embodiment, all guanosine nucleotides present in the siNA are 2 '-deoxy-2 '-fluoro guanosine nucleotides. The siNA can further comprise at least one modified internucleotidic linkage, such as phosphorothioate linkage. In one embodiment, the 2'- deoxy-2'-fluoronucleotides are present at specifically selected locations in the siNA that are sensitive to cleavage by ribonucleases, such as locations having pyrimidine nucleotides.
In one embodiment, the invention features a double-stranded short interfering nucleic acid (siNA) molecule that down-regulates expression of a target gene or that directs cleavage of a target RNA comprising a sense region and an antisense region, wherein the antisense region comprises a nucleotide sequence that is complementary to a nucleotide sequence of RNA encoded by the gene or a portion thereof and the sense region comprises a nucleotide sequence that is complementary to the antisense region, and wherein the purine nucleotides present in the antisense region comprise 2'-deoxy- purine nucleotides. In an alternative embodiment, the purine nucleotides present in the antisense region comprise 2'-0-methyl purine nucleotides. In either of the above embodiments, the antisense region can comprise a phosphorothioate internucleotide linkage at the 3' end of the antisense region. Alternatively, in either of the above embodiments, the antisense region can comprise a glyceryl modification at the 3' end of the antisense region. In another embodiment of any of the above-described siNA molecules, any nucleotides present in a non-complementary region of the antisense strand (e.g. overhang region) are 2'-deoxy nucleotides.
In one embodiment, the antisense region of a siNA molecule of the invention comprises sequence complementary to a portion of a target polynucleotide sequence having sequence unique to a particular disease related allele, such as sequence comprising a single nucleotide polymoφhism (SNP) associated with the disease specific allele. As such, the antisense region of a siNA molecule of the invention can comprise sequence complementary to sequences that are unique to a particular allele to provide specificity in mediating selective RNAi against the disease, condition, or trait related allele. In one embodiment, the invention features a double-stranded short interfering nucleic acid (siNA) molecule that down-regulates expression of a target gene or that
directs cleavage of a target RNA, wherein the siNA molecule is assembled from two separate oligonucleotide fragments wherein one fragment comprises the sense region and the second fragment comprises the antisense region of the siNA molecule. In another embodiment, the siNA molecule is a double stranded nucleic acid molecule, where each strand is about 21 nucleotides long and where about 19 nucleotides of each fragment of the siNA molecule are base-paired to the complementary nucleotides of the other fragment of the siNA molecule, wherein at least two 3' terminal nucleotides of each fragment of the siNA molecule are not base-paired to the nucleotides of the other fragment of the siNA molecule. In another embodiment, the siNA molecule is a double stranded nucleic acid molecule, where each strand is about 19 nucleotide long and where the nucleotides of each fragment of the siNA molecule are base-paired to the complementary nucleotides of the other fragment of the siNA molecule to form at least about 15 (e.g., 15, 16, 17, 18, or 19) base pairs, wherein one or both ends of the siNA molecule are blunt ends. In one embodiment, each of the two 3' terminal nucleotides of each fragment of the siNA molecule is a 2'-deoxy-pyrimidine nucleotide, such as a 2'- deoxy-thymidine. In another embodiment, all nucleotides of each fragment of the siNA molecule are base-paired to the complementary nucleotides of the other fragment of the siNA molecule. In another embodiment, the siNA molecule is a double stranded nucleic acid molecule of about 19 to about 25 base pairs having a sense region and an antisense region, where about 19 nucleotides of the antisense region are base-paired to the nucleotide sequence or a portion thereof of the RNA encoded by the target gene. In another embodiment, about 21 nucleotides of the antisense region are base-paired to the nucleotide sequence or a portion thereof of the RNA encoded by the target gene. In any of the above embodiments, the 5 '-end of the fragment comprising said antisense region can optionally include a phosphate group.
In one embodiment, the invention features a double-stranded short interfering nucleic acid (siNA) molecule that inhibits the expression of a target RNA sequence (e.g., wherein said target RNA sequence is encoded by a gene involved in a pathway of gene expression), wherein the siNA molecule does not contain any ribonucleotides and wherein each strand of the double-stranded siNA molecule is about 15 to about 30 nucleotides. In one embodiment, the siNA molecule is 21 nucleotides in length. Examples of non-ribonucleotide containing siNA constructs are combinations of
stabilization chemistries shown in Table II in any combination of Sense/Antisense chemistries, such as Stab 7/8, Stab 7/11, Stab 8/8, Stab 18/8, Stab 18/11, Stab 12/13, Stab 7/13, Stab 18/13, Stab 7/19, Stab 8/19, Stab 18/19, Stab 7/20, Stab 8/20, Stab 18/20, Stab 7/32, Stab 8/32, or Stab 18/32 (e.g., any siNA having Stab 7, 8, 11, 12, 13, 14, 15, 17, 18, 19, 20, or 32 sense or antisense strands or any combination thereof).
In one embodiment, the invention features a chemically synthesized double stranded RNA molecule that directs cleavage of a target RNA via RNA interference, wherein each, strand of said RNA molecule is about 15 to about 30 nucleotides in length; one strand of the RNA molecule comprises nucleotide sequence having sufficient complementarity to the target RNA for the RNA molecule to direct cleavage of the target RNA via RNA interference; and wherein at least one strand of the RNA molecule optionally comprises one or more chemically modified nucleotides described herein, such as without limitation deoxynucleotides, 2'-0-methyl nucleotides, 2'-deoxy-2'- fluoro nucleotides, 2'-0-methoxyethyl nucleotides etc. In one embodiment, a target RNA of the invention comprises sequence encoding a protein.
In one embodiment, target RNA of the invention comprises non-coding RNA sequence (e.g., miRNA, snRNA siRNA etc.).
In one embodiment, the invention features a medicament comprising a siNA molecule of the invention.
In one embodiment, the invention features an active ingredient comprising a siNA molecule of the invention.
In one embodiment, the invention features the use of a double-stranded short interfering nucleic acid (siNA) molecule to inhibit, down-regulate, or reduce expression of a gene or that directs cleavage of a target RNA, wherein the siNA molecule comprises one or more chemical modifications and each strand of the double-stranded siNA is independently about 15 to about 30 or more (e.g., about 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29 or 30 or more) nucleotides long. In one embodiment, the siNA molecule of the invention is a double stranded nucleic acid molecule comprising one or
more chemical modifications, where each of the two fragments of the siNA molecule independently comprise about 15 to about 40 (e.g. about 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 23, 33, 34, 35, 36, 37, 38, 39, or 40) nucleotides and where one of the strands comprises at least 15 nucleotides that are complementary to nucleotide sequence of target RNA or a portion thereof. In a non-limiting example, each of the two fragments of the siNA molecule comprise about 21 nucleotides. In another embodiment, the siNA molecule is a double stranded nucleic acid molecule comprising one or more chemical modifications, where each strand is about 21 nucleotide long and where about 19 nucleotides of each fragment of the siNA molecule are base-paired to the complementary nucleotides of the other fragment of the siNA molecule, wherein at least two 3' terminal nucleotides of each fragment of the siNA molecule are not base-paired to the nucleotides of the other fragment of the siNA molecule. In another embodiment, the siNA molecule is a double stranded nucleic acid molecule comprising one or more chemical modifications, where each strand is about 19 nucleotide long and where the nucleotides of each fragment of the siNA molecule are base-paired to the complementary nucleotides of the other fragment of the siNA molecule to form at least about 15 (e.g., 15, 16, 17, 18, or 19) base pairs, wherein one or both ends of the siNA molecule are blunt ends. In one embodiment, each of the two 3' terminal nucleotides of each fragment of the siNA molecule is a 2'-deoxy-pyrimidine nucleotide, such as a 2'-deoxy-thymidine. In another embodiment, all nucleotides of each fragment of the siNA molecule are base- paired to the complementary nucleotides of the other fragment of the siNA molecule. In another embodiment, the siNA molecule is a double stranded nucleic acid molecule of about 19 to about 25 base pairs having a sense region and an antisense region and comprising one or more chemical modifications, where about 19 nucleotides of the antisense region are base-paired to the nucleotide sequence or a portion thereof of the RNA encoded by the target gene. In another embodiment, about 21 nucleotides of the antisense region are base-paired to the nucleotide sequence or a portion thereof of the RNA encoded by the target gene. In any of the above embodiments, the 5 '-end of the fragment comprising said antisense region can optionally include a phosphate group. In one embodiment, the invention features the use of a double-stranded short interfering nucleic acid (siNA) molecule that inhibits, down-regulates, or reduces expression of a target gene or that directs cleavage of a target RNA, wherein one of the
strands of the double-stranded siNA molecule is an antisense strand which comprises nucleotide sequence that is complementary to nucleotide sequence of target RNA or a portion thereof, the other strand is a sense strand which comprises nucleotide sequence that is complementary to a nucleotide sequence of the antisense strand and wherein a majority of the pyrimidine nucleotides present in the double-stranded siNA molecule comprises a sugar modification (e.g., 2 '-deoxy-2 '-fluoro, 2'-0-methyl, or 2'-deoxy modifications).
In one embodiment, the invention features a double-stranded short interfering nucleic acid (siNA) molecule that inhibits, down-regulates, or reduces expression of a target gene or that directs cleavage of a target RNA, wherem one of the strands of the double-stranded siNA molecule is an antisense strand which comprises nucleotide sequence that is complementary to nucleotide sequence of target RNA or a portion thereof, wherein the other strand is a sense strand which comprises nucleotide sequence that is complementary to a nucleotide sequence of the antisense strand and wherein a majority of the pyrimidine nucleotides present in the double-stranded siNA molecule comprises a sugar modification (e.g., 2 '-deoxy-2 '-fluoro, 2'-0-methyl, or 2'-deoxy modificatins).
In one embodiment, the invention features a double-stranded short interfering nucleic acid (siNA) molecule that inhibits, down-regulates, or reduces expression of a gene or that directs cleavage of a target RNA, wherem one of the strands of the double- stranded siNA molecule is an antisense strand which comprises nucleotide sequence that is complementary to nucleotide sequence of target RNA that encodes a protein or portion thereof, the other strand is a sense strand which comprises nucleotide sequence that is complementary to a nucleotide sequence of the antisense strand and wherem a majority of the pyrimidine nucleotides present in the double-stranded siNA molecule comprises a sugar modification. In one embodiment, each strand of the siNA molecule comprises about 15 to about 30 or more (e.g., about 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, or 30 or more) nucleotides, wherein each strand comprises at least about 15 nucleotides that are complementary to the nucleotides of the other strand. In one embodiment, the siNA molecule is assembled from two oligonucleotide fragments, wherein one fragment comprises the nucleotide sequence of the antisense strand of the
siNA molecule and a second fragment comprises nucleotide sequence of the sense region of the siNA molecule. In one embodiment, the sense strand is connected to the antisense strand via a linker molecule, such as a polynucleotide linker or a non-nucleotide linker. In a further embodiment, the pyrimidine nucleotides present in the sense strand are 2'- deoxy- 'fluoro pyrimidine nucleotides and the purine nucleotides present in the sense region are 2'-deoxy purine nucleotides. In another embodiment, the pyrimidine nucleotides present in the sense strand are 2 '-deoxy-2 'fluoro pyrimidine nucleotides and the purine nucleotides present in the sense region are 2'-0-methyl purine nucleotides. In still another embodiment, the pyrimidine nucleotides present in the antisense strand are 2'-deoxy-2'-fluoro pyrimidine nucleotides and any purine nucleotides present in the antisense strand are 2'-deoxy purine nucleotides. In another embodiment, the antisense strand comprises one or more 2 '-deoxy-2 '-fluoro pyrimidine nucleotides and one or more '-O-methyl purine nucleotides. In another embodiment, the pyrimidine nucleotides present in the antisense strand are 2 '-deoxy-2 '-fluoro pyrimidine nucleotides and any purine nucleotides present in the antisense strand are 2'-0-methyl purine nucleotides. In a further embodiment the sense strand comprises a 3'-end and a 5'-end, wherein a terminal cap moiety (e.g., an inverted deoxy abasic moiety or inverted deoxy nucleotide moiety such as inverted thymidine) is present at the 5'-end, the 3 '-end, or both of the 5' and 3' ends of the sense strand. In another embodiment, the antisense strand comprises a phosphorothioate internucleotide linkage at the 3' end of the antisense strand. In another embodiment, the antisense strand comprises a glyceryl modification at the 3' end. In another embodiment, the 5 '-end of the antisense strand optionally includes a phosphate group.
In any of the above-described embodiments of a double-stranded short interfering nucleic acid (siNA) molecule that inhibits expression of a target gene or that directs cleavage of a target RNA, wherein a majority of the pyrimidine nucleotides present in the double-stranded siNA molecule comprises a sugar modification, each of the two strands of the siNA molecule can comprise about 15 to about 30 or more (e.g., about 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, or 30 or more) nucleotides. In one embodiment, about 15 to about 30 or more (e.g., about 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, or 30 or more) nucleotides of each strand of the siNA molecule are base-paired to the complementary nucleotides of the other strand of the siNA molecule.
In another embodiment, about 15 to about 30 or more (e.g., about 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, or 30 or more) nucleotides of each strand of the siNA molecule are base-paired to the complementary nucleotides of the other strand of the siNA molecule, wherein at least two 3' terminal nucleotides of each strand of the siNA molecule are not base-paired to the nucleotides of the other strand of the siNA molecule. In another embodiment, each of the two 3' terminal nucleotides of each fragment of the siNA molecule is a 2'-deoxy-pyrimidine, such as 2'-deoxy-thymidine. In one embodiment, each strand of the siNA molecule is base-paired to the complementary nucleotides of the other strand of the siNA molecule. In one embodiment, about 15 to about 30 (e.g., about 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, or 30) nucleotides of the antisense strand are base-paired to the nucleotide sequence of the target RNA or a portion thereof. In one embodiment, about 18 to about 25 (e.g., about 18, 19, 20, 21, 22, 23, 24, or 25) nucleotides of the antisense strand are base-paired to the nucleotide sequence of the target RNA or a portion thereof. In one embodiment, the invention features a double-stranded short interfering nucleic acid (siNA) molecule that inhibits expression of a target gene or that directs cleavage of a target RNA, wherein one of the strands of the double-stranded siNA molecule is an antisense strand which comprises nucleotide sequence that is complementary to nucleotide sequence of target RNA or a portion thereof, the other strand is a sense strand which comprises nucleotide sequence that is complementary to a nucleotide sequence of the antisense strand and wherein a majority of the pyrimidine nucleotides present in the double-stranded siNA molecule comprises a sugar modification, and wherein the 5 '-end of the antisense strand optionally includes a phosphate group. In one embodiment, the invention features a double-stranded short interfering nucleic acid (siNA) molecule that inhibits expression of a target gene or that directs cleavage of a target RNA, wherein one of the strands of the double-stranded siNA molecule is an antisense strand which comprises nucleotide sequence that is complementary to nucleotide sequence of target RNA or a portion thereof, the other strand is a sense strand which comprises nucleotide sequence that is complementary to a nucleotide sequence of the antisense strand and wherem a majority of the pyrimidine
nucleotides present in the double-stranded siNA molecule comprises a sugar modification, and wherem the nucleotide sequence or a portion thereof of the antisense strand is complementary to a nucleotide sequence of the untranslated region or a portion thereof of the target RJSTA. In one embodiment, the invention features a double-stranded short interfering nucleic acid (siNA) molecule that inhibits expression of a target gene or that directs cleavage of a target RNA, wherem one of the strands of the double-stranded siNA molecule is an antisense strand which comprises nucleotide sequence that is complementary to nucleotide sequence of target RNA or a portion thereof, wherein the other strand is a sense strand which comprises nucleotide sequence that is complementary to a nucleotide sequence of the antisense strand, wherein a majority of the pyrimidine nucleotides present in the double-stranded siNA molecule comprises a sugar modification, and wherein the nucleotide sequence of the antisense strand is complementary to a nucleotide sequence of the target RNA or a portion thereof that is present in the target RJSTA.
In one embodiment, the invention features a composition comprising a siNA molecule of the invention in a pharmaceutically acceptable carrier or diluent.
In a non-limiting example, the introduction of chemically-modified nucleotides into nucleic acid molecules provides a powerful tool in overcoming potential limitations of in vivo stability and bioavailability inherent to native RNA molecules that are delivered exogenously. For example, the use of chemically-modified nucleic acid molecules can enable a lower dose of a particular nucleic acid molecule for a given therapeutic effect since chemically-modified nucleic acid molecules tend to have a longer half-life in serum. Furthermore, certain chemical modifications can improve the bioavailability of nucleic acid molecules by targeting particular cells or tissues and/or improving cellular uptake of the nucleic acid molecule. Therefore, even if the activity of a chemically-modified nucleic acid molecule is reduced as compared to a native nucleic acid molecule, for example, when compared to an all-RNA nucleic acid molecule, the overall activity of the modified nucleic acid molecule can be greater than that of the native molecule due to improved stability and/or delivery of the molecule. Unlike native
unmodified siNA, chemically-modified siNA can also minimize the possibility of activating interferon activity in humans.
In any of the embodiments of siNA molecules described herein, the antisense region of a siNA molecule of the invention can comprise a phosphorothioate internucleotide linkage at the 3 '-end of said antisense region. In any of the embodiments of siNA molecules described herein, the antisense region can comprise about one to about five phosphorothioate internucleotide linkages at the 5 '-end of said antisense region. In any of the embodiments of siNA molecules described herein, the 3 '-terminal nucleotide overhangs of a siNA molecule of the invention can comprise ribonucleotides or deoxyribonucleotides that are chemically-modified at a nucleic acid sugar, base, or backbone. In any of the embodiments of siNA molecules described herein, the 3'- terminal nucleotide overhangs can comprise one or more universal base ribonucleotides. In any of the embodiments of siNA molecules described herein, the 3'-terminal nucleotide overhangs can comprise one or more acyclic nucleotides. One embodiment of the invention provides an expression vector comprising a nucleic acid sequence encoding at least one siNA molecule of the invention in a manner that allows expression of the nucleic acid molecule. Another embodiment of the invention provides a mammalian cell comprising such an expression vector. The mammalian cell can be a human cell. The siNA molecule of the expression vector can comprise a sense region and an antisense region. The antisense region can comprise sequence complementary to a RNA or DNA sequence encoding the target and the sense region can comprise sequence complementary to the antisense region. The siNA molecule can comprise two distinct strands having complementary sense and antisense regions. The siNA molecule can comprise a single strand having complementary sense and antisense regions.
In one embodiment, the invention features a chemically-modified short interfering nucleic acid (siNA) molecule capable of mediating RNA interference (RNAi) against a target polynucleotide (e.g., DNA or RNA) inside a cell or reconstituted in vitro system, wherein the chemical modification comprises one or more (e.g., about 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, or more) nucleotides comprising a backbone modified internucleotide linkage having Formula I:

wherein each Rl and R2 is independently any nucleotide, non-nucleotide, or polynucleotide which can be naturally-occurring or chemically-modified, each X and Y is independently O, S, N, alkyl, or substituted alkyl, each Z and W is independently O, S, N, alkyl, substituted alkyl, O-alkyl, S-alkyl, alkaryl, aralkyl, or acetyl and wherein W, X, Y, and Z are optionally not all O. In another embodiment, a backbone modification of the invention comprises a phosphonoacetate and/or thiophosphonoacetate internucleotide linkage (see for example Sheehan et al., 2003, Nucleic Acids Research, 31 , 4109-4118).
The chemically-modified internucleotide linkages having Foπnula I, for example, wherein any Z, W, X, and/or Y independently comprises a sulphur atom, can be present in one or both oligonucleotide strands of the siNA duplex, for example, in the sense strand, the antisense strand, or both strands. The siNA molecules of the invention can comprise one or more (e.g., about 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, or more) chemically- modified internucleotide linkages having Foπnula I at the 3 '-end, the 5'-end, or both of the 3' and 5'-ends of the sense strand, the antisense strand, or both strands. For example, an exemplary siNA molecule of trie invention can comprise about 1 to about 5 or more (e.g., about 1, 2, 3, 4, 5, or more) chemically-modified internucleotide linkages having Formula I at the 5'-end of the sense strand, the antisense strand, or both strands. In another non-limiting example, an exemplary siNA molecule of the invention can comprise one or more (e.g., about 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, or more) pyrimidine nucleotides with chemically-modified internucleotide linkages having Formula I in the sense strand, the antisense strand, or both strands. In yet another non-limiting example, an exemplary siNA molecule of the invention can comprise one or more (e.g., about 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, or more) purine nucleotides with chemically-modified internucleotide linkages having Formula I in the sense strand, the antisense strand, or both strands. In another embodiment, a siNA molecule of the invention having internucleotide linkage(s) of Formula I also comprises a chemically-modified nucleotide or non-nucleotide having any of Formulae I- VII.
In one embodiment, the invention features a chemically-modified short interfering nucleic acid (siNA) molecule capable of mediating RNA interference (RNAi) against a target polynucleotide (e.g., DNA or RNA) inside a cell or reconstituted in vitro system, wherem the chemical modification comprises one or more (e.g., about 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, or more) nucleotides or non-nucleotides having Formula II:
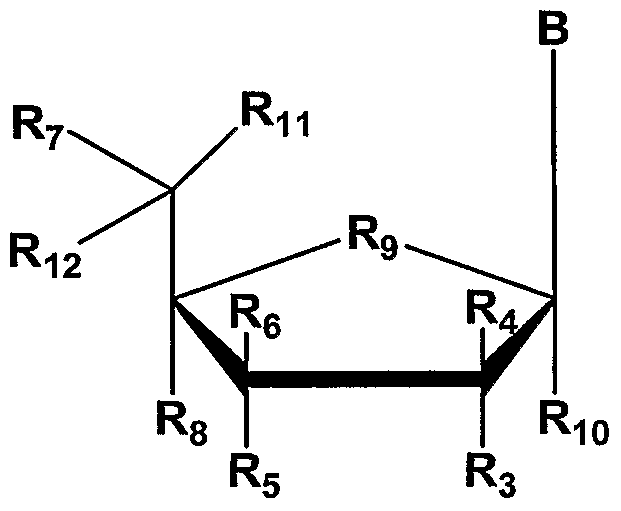
wherem each R3, R4, R5, R6, R7, R8, RIO, Rll and R12 is independently H, OH, alkyl, substituted alkyl, alkaryl or aralkyl, F, Cl, Br, CN, CF3, OCF3, OCN, O-alkyl, S-alkyl, N-alkyl, O-alkenyl, S-alkenyl, N-alkenyl, SO-alkyl, alkyl-OSH, alkyl-OH, O-alkyl-OH, O-alkyl-SH, S-alkyl-OH, S-alkyl-SH, alkyl-S-alkyl, alkyl-O-alkyl, ON02, N02, N3, NH2, aminoalkyl, aminoacid, aminoacyl, ONH2, O-aminoalkyl, O-aminoacid, O- aminoacyl, heterocycloalkyl, heterocycloalkaryl, ammoalkylamino, polyalklylamino, substituted silyl, or group having Formula I or II; R9 is O, S, CH2, S=0, CHF, or CF2, and B is a nucleosidic base such as adenine, guanine, uracil, cytosine, thymine, 2- aminoadenosine, 5-methylcytosine, 2,6-diaminopurine, or any other non-naturally occurring base that can be complementary or non-complementary to target RNA or a non-nucleosidic base such as phenyl, naphthyl, 3-nitropyπole, 5-nitroindole, nebularine, pyridone, pyridinone, or any other non-naturally occurring universal base that can be complementary or non-complementary to target RNA. The chemically-modified nucleotide or non-nucleotide of Formula II can be present in one or both oligonucleotide strands of the siNA duplex, for example in the sense strand, the antisense strand, or both strands. The siNA molecules of the invention can comprise one or more chemically-modified nucleotides or non-nucleotides of Formula II at the 3'-end, the 5'-end, or both of the 3' and 5'-ends of the sense strand, the antisense strand, or both strands. For example, an exemplary siNA molecule of the
invention can comprise about 1 to about 5 or more (e.g., about 1, 2, 3, 4, 5, or more) chemically-modified nucleotides or non-nucleotides of Formula II at the 5'-end of the sense strand, the antisense strand, or both strands. In anther non-limiting example, an exemplary siNA molecule of the invention can comprise about 1 to about 5 or more (e.g., about 1, 2, 3, 4, 5, or more) chemically-modified nucleotides or non-nucleotides of Formula II at the 3 '-end of the sense strand, the antisense strand, or both strands.
In one embodiment, the invention features a chemically-modified short interfering nucleic acid (siNA) molecule capable of mediating RNA interference (RNAi) against a target polynucleotide (e.g., DNA or RNA) inside a cell or reconstituted in vitro system, wherein the chemical modification comprises one or more (e.g., about 1, 2, 3, 4, 5, 6, 7,
8, 9, 10, or more) nucleotides or non-nucleotides having Formula III:

wherem each R3, R4, R5, R6, R7, R8, R10, Rl 1 and R12 is independently H, OH, alkyl, substituted alkyl, alkaryl or aralkyl, F, Cl, Br, CN, CF3, OCF3, OCN, O-alkyl, S-alkyl, N-alkyl, O-alkenyl, S-alkenyl, N-alkenyl, SO-alkyl, alkyl-OSH, alkyl-OH, O-alkyl-OH, O-alkyl-SH, S-alkyl-OH, S-alkyl-SH, alkyl-S-alkyl, alkyl-O-alkyl, ON02, N02, N3, NH2, aminoalkyl, aminoacid, aminoacyl, ONH2, O-aminoalkyl, O-aminoacid, O- aminoacyl, heterocycloalkyl, heterocycloalkaryl, aminoalkylamino, polyalklylamino, substituted silyl, or group having Formula I or II; R9 is O, S, CH2, S=0, CHF, or CF2, and B is a nucleosidic base such as adenine, guanine, uracil, cytosine, thymme, 2- aminoadenosine, 5-mefhylcytosine, 2,6-diaminopurine, or any other non-naturally occurring base that can be employed to be complementary or non-complementary to target RNA or a non-nucleosidic base such as phenyl, naphthyl, 3-nitropyπole, 5- nitroindole, nebularine, pyridone, pyridinone, or any other non-naturally occurring universal base that can be complementary or non-complementary to target RNA.
The chemically-modified nucleotide or non-nucleotide of Formula JJI can be present in one or both oligonucleotide strands of the siNA duplex, for example, in the sense strand, the antisense strand, or both strands. The siNA molecules of the invention can comprise one or more chemically-modified nucleotides or non-nucleotides of Formula III at the 3'-end, the 5'-end, or both of the 3' and 5'-ends of the sense strand, the antisense strand, or both strands. For example, an exemplary siNA molecule of the invention can comprise about 1 to about 5 or more (e.g., about 1, 2, 3, 4, 5, or more) chemically-modified nucleotide(s) or non-nucleotide(s) of Formula III at the 5'-end of the sense strand, the antisense strand, or both strands. In anther non-limiting example, an exemplary siNA molecule of the invention can comprise about 1 to about 5 or more (e.g., about 1, 2, 3, 4, 5, or more) chemically-modified nucleotide or non-nucleotide of Formula III at the 3 '-end of the sense strand, the antisense strand, or both strands.
In another embodiment, a siNA molecule of the invention comprises a nucleotide having Formula II or III, wherein the nucleotide having Formula II or III is in an inverted configuration. For example, the nucleotide having Formula II or III is connected to the siNA construct in a 3'-3', 3'-2', 2'-3', or 5'-5' configuration, such as at the 3'-end, the 5'- end, or both of the 3' and 5'-ends of one or both siNA strands.
In one embodiment, the invention features a chemically-modified short interfering nucleic acid (siNA) molecule capable of mediating RNA interference (RNAi) against a target polynucleotide (e.g., DNA or RNA) inside a cell or reconstituted in vitro system, wherein the chemical modification comprises a 5!-terminal phosphate group having
Formula IV:
Z X — P II — Y w wherein each X and Y is independently O, S, N, alkyl, substituted alkyl, or alkylhalo; wherein each Z and W is independently O, S, N, alkyl, substituted alkyl, O-alkyl, S- alkyl, alkaryl, aralkyl, alkylhalo, or acetyl; and wherein W, X, Y and Z are not all O.
In one embodiment, the invention features a siNA molecule having a 5'-terminal phosphate group having Formula IV on the target-complementary strand, for example, a strand complementary to a target RNA, wherein the siNA molecule comprises an all RNA siNA molecule. In another embodiment, the invention features a siNA molecule having a 5 '-terminal phosphate group having Formula IV on the target-complementary strand wherein the siNA molecule also comprises about 1 to about 3 (e.g., about 1, 2, or 3) nucleotide 3 '-terminal nucleotide overhangs having about 1 to about 4 (e.g., about 1, 2,
3, or 4) deoxyribonucleotides on the 3'-end of one or both strands. In another embodiment, a 5'-terminal phosphate group having Formula IV is present on the target- complementary strand of a siNA molecule of the invention, for example a siNA molecule having chemical modifications having any of Formulae I- VII.
In one embodiment, the invention features a chemically-modified short interfering nucleic acid (siNA) molecule capable of mediating RNA interference (RNAi) against a target polynucleotide (e.g., DNA or RNA) inside a cell or reconstituted in vitro system, wherein the chemical modification comprises one or more phosphorothioate internucleotide linkages. For example, in a non-limiting example, the invention features a chemically-modified short interfering nucleic acid (siNA) having about 1, 2, 3, 4, 5, 6, 7, 8 or more phosphorothioate internucleotide linkages in one siNA strand. In yet another embodiment, the invention features a chemically-modified short interfering nucleic acid (siNA) individually having about 1, 2, 3, 4, 5, 6, 7, 8 or more phosphorothioate internucleotide linkages in both siNA strands. The phosphorothioate internucleotide linkages can be present in one or both oligonucleotide strands of the siNA duplex, for example in the sense strand, the antisense strand, or both strands. The siNA molecules of the invention can comprise one or more phosphorothioate internucleotide linkages at the 3'-end, the 5'-end, or both of the 3'- and 5'-ends of the sense strand, the antisense strand, or both strands. For example, an exemplary siNA molecule of the invention can comprise about 1 to about 5 or more (e.g., about 1, 2, 3, 4, 5, or more) consecutive phosphorothioate internucleotide linkages at the 5 '-end of the sense strand, the antisense strand, or both strands. In another non-limiting example, an exemplary siNA molecule of the invention can comprise one or more (e.g., about 1, 2, 3,
4, 5, 6, 7, 8, 9, 10, or more) pyrimidine phosphorothioate internucleotide linkages in the sense strand, the antisense strand, or both strands. In yet another non-limiting example,
an exemplary siNA molecule of the invention can comprise one or more (e.g., about 1, 2,
3, 4, 5, 6, 7, 8, 9, 10, or more) purine phosphorothioate internucleotide linkages in the sense strand, the antisense strand, or both strands.
In one embodiment, a siNA molecule of the invention is featured, wherein the sense strand comprises one or more, for example, about 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, or more phosphorothioate internucleotide linkages, and/or one or more (e.g., about 1, 2, 3,
4, 5, 6, 7, 8, 9, 10 or more) 2'-deoxy, 2'-0-methyl, 2'-deoxy-2'-fluoro, and/or about one or more (e.g., about 1, 2, 3, 4, 5, 6, 7, 8, 9, 10 or more) universal base modified nucleotides, and optionally a terminal cap molecule at the 3 '-end, the 5 '-end, or both of the 3'- and 5'-ends of the sense strand; and wherein the antisense strand comprises about 1 to about 10 or more, specifically about 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, or more phosphorothioate internucleotide linkages, and or one or more (e.g., about 1, 2, 3, 4, 5, 6, 7, 8, 9, 10 or more) 2'-deoxy, 2'-0-methyl, 2'-deoxy-2'-fluoro, and/or one or more (e.g., about 1, 2, 3, 4, 5, 6, 7, 8, 9, 10 or more) universal base modified nucleotides, and optionally a terminal cap molecule at the 3'-end, the 5'-end, or both of the 3'- and 5'-ends of the antisense strand. In another embodiment, one or more, for example about 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, or more, pyrimidine nucleotides of the sense and/or antisense siNA strand are chemically-modified with 2'-deoxy, 2-O-methyl and/or 2'-deoxy-2'-fluoro nucleotides, with or without one or more, for example about 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, or more, phosphorothioate internucleotide linkages and/or a terminal cap molecule at the 3'- end, the 5'-end, or both of the 3'- and 5'-ends, being present in the same or different strand.
In another embodiment, a siNA molecule of the invention is featured, wherem the sense strand comprises about 1 to about 5, specifically about 1, 2, 3, 4, or 5 phosphorothioate internucleotide linkages, and/or one or more (e.g., about 1, 2, 3, 4, 5, or more) 2'-deoxy, 2'-0-methyl, 2 '-deoxy-2 '-fluoro, and/or one or more (e.g., about 1, 2, 3, 4, 5, or more) universal base modified nucleotides, and optionally a terminal cap molecule at the 3 -end, the 5 '-end, or both of the 3'- and 5 '-ends of the sense strand; and wherein the antisense strand comprises about 1 to ahout 5 or more, specifically about 1, 2, 3, 4, 5, or more phosphorothioate internucleotide linkages, and/or one or more (e.g., about 1, 2, 3, 4, 5, 6, 7, 8, 9, 10 or more) 2'-deoxy, 2'-0-methyl, 2'-deoxy-2'-fluoro,
and/or one or more (e.g., about 1, 2, 3, 4, 5, 6, 7, 8, 9, 10 or more) universal base modified nucleotides, and optionally a terminal cap molecule at the 3 '-end, the 5'-end, or both of the 3'- and 5'-ends of the antisense strand. In another embodiment, one or more, for example about 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, or more, pyrimidine nucleotides of the sense and/or antisense siNA strand are chemically-modified with 2'-deoxy, 2'-0-methyl and/or 2'-deoxy-2'-fluoro nucleotides, with or without about 1 to about 5 or more, for example about 1, 2, 3, 4, 5, or more phosphorothioate internucleotide linkages and/or a terminal cap molecule at the 3 '-end, the 5'-end, or both of the 3'- and 5'-ends, being present in the same or different strand. In one embodiment, a siNA molecule of the invention is featured, wherem the antisense strand comprises one or more, for example, about 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, or more phosphorothioate internucleotide linkages, and or about one or more (e.g., about 1, 2, 3, 4, 5, 6, 7, 8, 9, 10 or more) 2'-deoxy, 2'-0-methyl, 2'-deoxy-2'-fluoro, and/or one or more (e.g., about 1, 2, 3, 4, 5, 6, 7, 8, 9, 10 or more) universal base modified nucleotides, and optionally a terminal cap molecule at the 3'-end, the 5'-end, or both of the 3'- and 5'- ends of the sense strand; and wherem the antisense strand comprises about 1 to about 10 or more, specifically about 1, 2, 3, 4, 5, 6, 7, 8, 9, 10 or more phosphorothioate internucleotide linkages, and or one or more (e.g., about 1, 2, 3, 4, 5, 6, 7, 8, 9, 10 or more) 2'-deoxy, 2'-0-methyl, 2'-deoxy-2'-fluoro, and/or one or more (e.g., about 1, 2, 3, 4, 5, 6, 7, 8, 9, 10 or more) universal base modified nucleotides, and optionally a terminal cap molecule at the 3'-end, the 5'-end, or both of the 3'- and 5'-ends of the antisense strand. In another embodiment, one or more, for example about 1, 2, 3, 4, 5, 6, 7, 8, 9, 10 or more pyrimidine nucleotides of the sense and/or antisense siNA strand are chemically-modified with 2'-deoxy, 2'-0-methyl and/or 2'-deoxy-2'-fluoro nucleotides, with or without one or more, for example, about 1, 2, 3, 4, 5, 6, 7, 8, 9, 10 or more phosphorothioate internucleotide linkages and/or a terminal cap molecule at the 3 '-end, the 5'-end, or both of the 3' and 5'-ends, being present in the same or different strand.
In another embodiment, a siNA molecule of the invention is featured, wherein the antisense strand comprises about 1 to about 5 or more, specifically about 1, 2, 3, 4, 5 or more phosphorothioate internucleotide linkages, and/or one or more (e.g., about 1, 2, 3,
4, 5, 6, 7, 8, 9, 10 or more) 2'-deoxy, 2'-0-methyl, 2'-deoxy-2'-fluoro, and/or one or more
(e.g., about 1, 2, 3, 4, 5, 6, 7, 8, 9, 10 or more) universal base modified nucleotides, and optionally a terminal cap molecule at the 3'-end, the 5'-end, or both of the 3'- and 5'-ends of the sense strand; and wherein the antisense strand comprises about 1 to about 5 or more, specifically about 1, 2, 3, 4, 5 or more phosphorothioate internucleotide linkages, and/or one or more (e.g., about 1, 2, 3, 4, 5, 6, 7, 8, 9, 10 or more) 2'-deoxy, 2'-0-methyl, 2'-deoxy-2'-fluoro, and/or one or more (e.g., about 1, 2, 3, 4, 5, 6, 7, 8, 9, 10 or more) universal base modified nucleotides, and optionally a terminal cap molecule at the 3'- end, the 5 '-end, or both of the 3'- and 5 '-ends of the antisense strand. In another embodiment, one or more, for example about 1, 2, 3, 4, 5, 6, 7, 8, 9, 10 or more pyrimidine nucleotides of the sense and/or antisense siNA strand are chemically- modified with 2'-deoxy, 2'-0-methyl and/or 2'-deoxy-2'-fluoro nucleotides, with or without about 1 to about 5, for example about 1, 2, 3, 4, 5 or more phosphorothioate internucleotide linkages and/or a terminal cap molecule at the 3'-end, the 5'-end, or both of the 3'- and 5'-ends, being present in the same or different strand. In one embodiment, a chemically-modified short interfering nucleic acid (siNA) molecule of the invention comprises about 1 to about 5 or more (specifically about 1, 2, 3, 4, 5 or more) phosphorothioate internucleotide linkages in each strand of the siNA molecule.
In another embodiment, a siNA molecule of the invention comprises 2'-5' internucleotide linkages. The 2'-5' internucleotide linkage(s) can be at the 3'-end, the 5'- end, or both of the 3'- and 5'-ends of one or both siNA sequence strands. In addition, the 2'-5' internucleotide linkage(s) can be present at various other positions within one or both siNA sequence strands, for example, about 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, or more including every internucleotide linkage of a pyrimidine nucleotide in one or both strands of the siNA molecule can comprise a 2'-5' internucleotide linkage, or about 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, or more including every internucleotide linkage of a purine nucleotide in one or both strands of the siNA molecule can comprise a 2'-5' internucleotide linkage.
In another embodiment, a chemically-modified siNA molecule of the invention comprises a duplex having two strands, one or both of which can be chemically- modified, wherein each strand is independently about 15 to about 30 (e.g., about 15, 16,
17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, or 30) nucleotides in length, wherein the
duplex has about 15 to about 30 (e.g., about 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, or 30) base pairs, and wherein the chemical modification comprises a structure having any of Formulae I- VII. For example, an exemplary chemically- modified siNA molecule of the invention comprises a duplex having two strands, one or both of which can be chemically-modified with a chemical modification having any of Formulae I- VII or any combination thereof, wherein each strand consists of about 21 nucleotides, each having a 2-nucleotide 3 '-terminal nucleotide overhang, and wherein the duplex has about 19 base pairs. In another embodiment, a siNA molecule of the invention comprises a single stranded hairpin structure, wherein the siNA is about 36 to about 70 (e.g., about 36, 40, 45, 50, 55, 60, 65, or 70) nucleotides in length having about 15 to about 30 (e.g., about 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, or 30) base pairs, and wherein the siNA can include a chemical modification comprising a structure having any of Formulae I- VII or any combination thereof. For example, an exemplary chemically-modified siNA molecule of the invention comprises a linear oligonucleotide having about 42 to about 50 (e.g., about 42, 43, 44, 45, 46, 47, 48, 49, or 50) nucleotides that is chemically-modified with a chemical modification having any of Formulae I- VII or any combination thereof, wherein the linear oligonucleotide forms a haiφin structure having about 19 to about 21 (e.g., 19, 20, or 21) base pairs and a 2- nucleotide 3 '-terminal nucleotide overhang. In another embodiment, a linear haiφin siNA molecule of the invention contains a stem loop motif, wherein the loop portion of the siNA molecule is biodegradable. For example, a linear haiφin siNA molecule of the invention is designed such that degradation of the loop portion of the siNA molecule in vivo can generate a double-stranded siNA molecule with 3'-terminal overhangs, such as 3 '-terminal nucleotide overhangs comprising about 2 nucleotides. In another embodiment, a siNA molecule of the invention comprises a haiφin structure, wherein the siNA is about 25 to about 50 (e.g., about 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35, 36, 37, 38, 39, 40, 41, 42, 43, 44, 45, 46, 47, 48, 49, or 50) nucleotides in length having about 3 to about 25 (e.g., about 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, or 25) base pairs, and wherein the siNA can include one or more chemical modifications comprising a structure having any of Formulae I- VII or any combination thereof. For example, an exemplary chemically-modified siNA molecule of the invention comprises a linear oligonucleotide having about 25 to about 35 (e.g., about
25, 26, 27, 28, 29, 30, 31, 32, 33, 34, or 35) nucleotides that is chemically-modified with one or more chemical modifications having any of Formulae I- VII or any combination thereof, wherein the linear oligonucleotide forms a haiφin structure having about 3 to about 25 (e.g., about 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, or 25) base pairs and a 5'-terminal phosphate group that can be chemically modified as described herein (for example a 5'-terminal phosphate group having Formula IV). In another embodiment, a linear haiφin siNA molecule of the invention contains a stem loop motif, wherem the loop portion of the siNA molecule is biodegradable. In one embodiment, a linear haiφin siNA molecule of the invention comprises a loop portion comprising a non-nucleotide linker.
In another embodiment, a siNA molecule of the invention comprises an asymmetric haiφin structure, wherein the siNA is about 25 to about 50 (e.g., about 25,
26, 27, 28, 29, 30, 31, 32, 33, 34, 35, 36, 37, 38, 39, 40, 41, 42, 43, 44, 45, 46, 47, 48, 49, or 50) nucleotides in length having about 3 to about 25 (e.g., about 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, or 25) base pairs, and wherem the siNA can include one or more chemical modifications comprising a structure having any of Formulae I- VII or any combination thereof. For example, an exemplary chemically- modified siNA molecule of the invention comprises a linear oligonucleotide having about 25 to about 35 (e.g., about 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, or 35) nucleotides that is chemically-modified with one or more chemical modifications having any of Formulae I- VII or any combination thereof, wherein the linear oligonucleotide forms an asymmetric haiφin structure having about 3 to about 25 (e.g., about 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, or 25) base pairs and a 5'-terminal phosphate group that can be chemically modified as described herein (for example a 5'- terminal phosphate group having Formula IV). In one embodiment, an asymmetric haiφin siNA molecule of the invention contains a stem loop motif, wherein the loop portion of the siNA molecule is biodegradable. In another embodiment, an asymmetric haiφin siNA molecule of the invention comprises a loop portion comprising a non- nucleotide linker. In another embodiment, a siNA molecule of the invention comprises an asymmetric double stranded structure having separate polynucleotide strands comprising
sense and antisense regions, wherein the antisense region is about 1 to about 30 (e.g., about 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, or 30) nucleotides in length, wherein the sense region is about 3 to about 25 (e.g., about 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, or 25) nucleotides in length, wherein the sense region and the antisense region have at least 3 complementary nucleotides, and wherein the siNA can include one or more chemical modifications comprising a structure having any of Formulae I- VII or any combination thereof. For example, an exemplary chemically-modified siNA molecule of the invention comprises an asymmetric double stranded structure having separate polynucleotide strands comprising sense and antisense regions, wherein the antisense region is about 18 to about 23 (e.g., about 18, 19, 20, 21, 22, or 23) nucleotides in length and wherein the sense region is about 3 to about 15 (e.g., about 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, or 15) nucleotides in length., wherem the sense region the antisense region have at least 3 complementary nucleotides, and wherein the siNA can include one or more chemical modifications comprising a structure having any of Formulae I- VII or any combination thereof. In another embodiment, the asymmetric double stranded siNA molecule can also have a 5 '-terminal phosphate group that can be chemically modified as described herein (for example a 5'-terminal phosphate group having Formula IV).
In another embodiment, a siNA molecule of the invention comprises a circular nucleic acid molecule, wherem the siNA is about 38 to about 70 (e.g:, about 38, 40, 45, 50, 55, 60, 65, or 70) nucleotides in length having about 15 to about 30 (e.g., about 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, or 30) base pairs, and wherein the siNA can include a chemical modification, which comprises a structure having any of Formulae I- VII or any combination thereof. For example, an exemplary chemically- modified siNA molecule of the invention comprises a circular oligonucleotide having about 42 to about 50 (e.g., about 42, 43, 44, 45, 46, 47, 48, 49, or 50) nucleotides that is chemically-modified with a chemical modification having any of Formulae I- VII or any combination thereof, wherein the circular oligonucleotide forms a dumbbell shaped structure having about 19 base pairs and 2 loops. In another embodiment, a circular siNA molecule of the invention contains two loop motifs, wherein one or both loop portions of the siNA molecule is biodegradable.
voτ example, a circular si A molecule of the invention is designed such that degradation of the loop portions of the siNA molecule in vivo can generate a double-stranded siNA molecule with 3 '-terminal overhangs, such as 3 '-terminal nucleotide overhangs comprising about 2 nucleotides. In one embodiment, a siNA molecule of the invention comprises at least one (e.g., about 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, or more) abasic moiety, for example a compound having Formula V:
wherein each R3, R4, R5, R6, R7, R8, RIO, Rll, R12, and R13 is independently H, OH, alkyl, substituted alkyl, alkaryl or aralkyl, F, Cl, Br, CN, CF3, OCF3, OCN, O-alkyl, S- alkyl, N-alkyl, O-alkenyl, S-alkenyl, N-alkenyl, SO-alkyl, alkyl-OSH, alkyl-OH, O- alkyl-OH, O-alkyl-SH, S-alkyl-OH, S-alkyl-SH, alkyl-S-alkyl, alkyl-O-alkyl, ON02, N02, N3, NH2, aminoalkyl, aminoacid, aminoacyl, ONH2, 0-aminoalkyl, O-aminoacid, O-aminoacyl, heterocycloalkyl, heterocycloalkaryl, aminoalkylamino, polyalklylamino, substituted silyl, or group having Formula I or II; R9 is O, S, CH2, S=O, CHF, or CF2.
In one embodiment, a siNA molecule of the invention comprises at least one (e.g., about 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, or more) inverted abasic moiety, for example a compound having Formula VI:
wherein each R3, R4, R5, R6, R7, R8, RIO, Rll, R12, and R13 is independently H, OH, alkyl, substituted alkyl, alkaryl or aralkyl, F, Cl, Br, CN, CF3, OCF3, OCN, O-alkyl, S- alkyl, N-alkyl, O-alkenyl, S-alkenyl, N-alkenyl, SO-alkyl, alkyl-OSH, alkyl-OH, O- alkyl-OH, O-alkyl-SH, S-alkyl-OH, S-alkyl-SH, alkyl-S-alkyl, alkyl-O-alkyl, ON02, N02, N3, NH2, aminoalkyl, aminoacid, aminoacyl, ONH2, O-aminoalkyl, O-aminoacid, O-aminoacyl, heterocycloalkyl, heterocycloalkaryl, aminoalkylamino, polyalklylamino, substituted silyl, or group having Formula I or II; R9 is O, S, CH2, S=0, CHF, or CF2, and either R2, R3, R8 or R13 serve as points of attachment to the siNA molecule of the invention. In another embodiment, a siNA molecule of the invention comprises at least one
(e.g., about 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, or more) substituted polyalkyl moieties, for example a compound having Formula VII:
wherein each n is independently an integer from 1 to 12, each Rl, R2 and R3 is independently H, OH, alkyl, substituted alkyl, alkaryl or aralkyl, F, Cl, Br, CTM, CF3, OCF3, OCN, O-alkyl, S-alkyl, N-alkyl, O-alkenyl, S-alkenyl, N-alkenyl, SO-alkyl, alkyl-OSH, alkyl-OH, O-alkyl-OH, O-alkyl-SH, S-alkyl-OH, S-alkyl-SH, alkyl-S-alkyl, alkyl-O-alkyl, ON02, N02, N3, NH2, aminoalkyl, aminoacid, aminoacyl, ONH2, O- aminoalkyl, O-aminoacid, O-aminoacyl, heterocycloalkyl, heterocycloalkaryl, aminoalkylamino, polyalklylamino, substituted silyl, or a group having Formula I, and Rl, R2 or R3 serves as points of attachment to the siNA molecule of the invention.
In another embodiment, a siNA molecule of the invention comprises a compound having Formula VII, wherein Rl and R2 are hydroxyl (OH) groups, n = 1, and R3 comprises O and is the point of attachment to the 3'-end, the 5'-end, or both of trie 3' and 5'-ends of one or both strands of a double-stranded siNA molecule of the invention or to a single-stranded siNA molecule of the invention. This modification is referred to herein as "glyceryl" (for example modification 6 in Figure 10).
In another embodiment, a chemically modified nucleoside or non-nucleoside (e.g. a moiety having any of Formula V, VI or VII) of the invention is at the 3 '-end, the 5' -end, or both of the 3' and 5 '-ends of a siNA molecule of the invention. For example, chemically modified nucleoside or non-nucleoside (e.g., a moiety having Formula V, VI or VII) can be present at the 3'-end, the 5'-end, or both of the 3' and 5'-ends of the antisense strand, the sense strand, or both antisense and sense strands of the siNA molecule. In one embodiment, the chemically modified nucleoside or non-nucleoside (e.g., a moiety having Formula V, VI or VII) is present at the 5'-end and 3'-end of the sense strand and the 3 '-end of the antisense strand of a double stranded siNA molecule of the invention. In one embodiment, the chemically modified nucleoside or non- nucleoside (e.g., a moiety having Formula V, VI or VII) is present at the terminal position of the 5 '-end and 3 '-end of the sense strand and the 3 '-end of the antisense strand of a double stranded siNA molecule of the invention. In one embodiment, the chemically modified nucleoside or non-nucleoside (e.g., a moiety having Formula "V, VI or VII) is present at the two terminal positions of the 5'-end and 3'-end of the sense strand and the 3 '-end of the antisense strand of a double stranded siNA molecule of the invention. In one embodiment, the chemically modified nucleoside or non-nucleoside (e.g., a moiety having Formula V, VI or VII) is present at the penultimate position of the 5 '-end and 3 '-end of the sense strand and the 3 '-end of the antisense strand of a double stranded siNA molecule of the invention. In addition, a moiety having Formula VII can be present at the 3'-end or the 5'-end of a haiφin siNA molecule as described herein.
In another embodiment, a siNA molecule of the invention comprises an abasic residue having Formula V or VI, wherein the abasic residue having Formula VI or VI is connected to the siNA construct in a 3'-3', 3'-2', 2'-3', or 5'-5' configuration, such as at the 3'-end, the 5'-end, or both of the 3' and 5'-ends of one or both siNA strands.
In one embodiment, a siNA molecule of the invention comprises one or more (e.g., about 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, or more) locked nucleic acid (LNA) nucleotides, for example, at the 5'-end, the 3'-end, both of the 5' and 3'-ends, or any combination thereof, of the siNA molecule. In another embodiment, a siNA molecule of the invention comprises one or more
(e.g., about 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, or more) acyclic nucleotides, for example, at the
5'-end, the 3'-end, both of the 5' and 3'-ends, or any combination thereof, of the siNA molecule.
In one embodiment, the invention features a chemically-modified short interfering nucleic acid (siNA) molecule of the invention comprising a sense region, wherein any (e.g., one or more or all) pyrimidine nucleotides present in the sense region are 2'-deoxy- 2'-fluoro pyrimidine nucleotides (e.g., wherein all pyrimidine nucleotides are 2'-deoxy- 2'-fluoro pyrimidine nucleotides or alternately a plurality of pyrimidine nucleotides are 2'-deoxy-2'-fluoro pyrimidine nucleotides), and wherein any (e.g., one or more or all) purine nucleotides present in the sense region are 2'-deoxy purine nucleotides (e.g., wherein all purine nucleotides are 2'-deoxy purine nucleotides or alternately a plurality of purine nucleotides are 2'-deoxy purine nucleotides).
In one embodiment, the invention features a chemically-modified short interfering nucleic acid (siNA) molecule of the invention comprising a sense region, wherein any (e.g., one or more or all) pyrimidine nucleotides present in the sense region are 2'-deoxy- 2'-fluoro pyrimidine nucleotides (e.g., wherem all pyrimidine nucleotides are 2'-deoxy- 2'-fluoro pyrimidine nucleotides or alternately a plurality of pyrimidine nucleotides are 2'-deoxy-2'-fluoro pyrimidine nucleotides), and wherein any (e.g., one or more or all) purine nucleotides present in the sense region are 2'-deoxy purine nucleotides (e.g., wherem all purine nucleotides are 2'-deoxy purine nucleotides or alternately a plurality of purine nucleotides are 2'-deoxy purine nucleotides), wherein any nucleotides comprising a 3 '-terminal nucleotide overhang that are present in said sense region are 2'- deoxy nucleotides.
In one embodiment, the invention features a chemically-modified short interfering nucleic acid (siNA) molecule of the invention comprising a sense region, wherein any (e.g., one or more or all) pyrimidine nucleotides present in the sense region are 2'-deoxy- 2'-fluoro pyrimidine nucleotides (e.g., wherein all pyrimidine nucleotides are 2'-deoxy- 2'-fluoro pyrimidine nucleotides or alternately a plurality of pyrimidine nucleotides are 2'-deoxy-2'-fluoro pyrimidine nucleotides), and wherein any (e.g., one or more or all) purine nucleotides present in the sense region are 2'-0-methyl purine nucleotides (e.g., wherein all purine nucleotides are 2'-0-methyl purine nucleotides or alternately a plurality of purine nucleotides are 2'-0-methyl purine nucleotides).
In one embodiment, the invention features a chemically-modified short interfering nucleic acid (siNA) molecule of the invention comprising a sense region, wherein any (e.g., one or more or all) pyrimidine nucleotides present in the sense region are 2'-deoxy- 2'-fluoro pyrimidine nucleotides (e.g., wherein all pyrimidine nucleotides are 2'-deoxy- 2'-fluoro pyrimidine nucleotides or alternately a plurality of pyrimidine nucleotides are 2'-deoxy-2'-fluoro pyrimidine nucleotides), wherein any (e.g., one or more or all) purine nucleotides present in the sense region are 2'-0-methyl purine nucleotides (e.g., wherein all purine nucleotides are 2'-0-methyl purine nucleotides or alternately a plurality of purine nucleotides are 2'-0-methyl purine nucleotides), and wherem any nucleotides comprising a 3 '-terminal nucleotide overhang that are present in said sense region are 2'- deoxy nucleotides.
In one embodiment, the invention features a chemically-modified short interfering nucleic acid (siNA) molecule of the invention comprising an antisense region, wherein any (e.g., one or more or all) pyrimidine nucleotides present in the antisense region are 2'-deoxy-2'-fluoro pyrimidine nucleotides (e.g., wherein all pyrimidine nucleotides are 2'-deoxy-2'-fluoro pyrimidine nucleotides or alternately a plurality of pyrimidine nucleotides are 2'-deoxy-2'-fluoro pyrimidine nucleotides), and wherem any (e.g., one or more or all) purine nucleotides present in the antisense region are 2'-0-methyl purine nucleotides (e.g., wherem all purine nucleotides are 2'-0-methyl purine nucleotides or alternately a plurality of purine nucleotides are 2'-0-methyl purine nucleotides).
In one embodiment, the invention features a chemically-modified short interfering nucleic acid (siNA) molecule of the invention comprising an antisense region, wherein any (e.g., one or more or all) pyrimidine nucleotides present in the antisense region are 2'-deoxy-2'-fluoro pyrimidine nucleotides (e.g., wherein all pyrimidine nucleotides are 2'-deoxy-2'-fluoro pyrimidine nucleotides or alternately a plurality of pyrimidine nucleotides are 2'-deoxy-2'-fluoro pyrimidine nucleotides), wherein any (e.g., one or more or all) purine nucleotides present in the antisense region are 2' -O-methyl purine nucleotides (e.g., wherem all purine nucleotides are 2'-0-methyl purine nucleotides or alternately a plurality of purine nucleotides are 2'-0-methyl purine nucleotides), and wherein any nucleotides comprising a 3'-terminal nucleotide overhang that are present in said antisense region are 2'-deoxy nucleotides.
In one embodiment, the invention features a chemically-modified short interfering nucleic acid (siNA) molecule of the invention comprising an antisense region, wherein any (e.g., one or more or all) pyrimidine nucleotides present in the antisense region are 2'-deoxy-2'-fluoro pyrimidine nucleotides (e.g., wherein all pyrimidine nucleotides are 2'-deoxy-2'-fluoro pyrimidine nucleotides or alternately a plurality of pyrimidine nucleotides are 2'-deoxy-2'-fluoro pyrimidine nucleotides), and wherein any (e.g., one or more or all) purine nucleotides present in the antisense region are 2'-deoxy purine nucleotides (e.g., wherein all purine nucleotides are 2'-deoxy purine nucleotides or alternately a plurality of purine nucleotides are 2'-deoxy purine nucleotides). In one embodiment, the invention features a chemically-modified short interfering nucleic acid (siNA) molecule of the invention comprising an antisense region, wherem any (e.g., one or more or all) pyrimidine nucleotides present in the antisense region are 2'-deoxy-2'-fiuoro pyrimidine nucleotides (e.g., wherein all pyrimidine nucleotides are 2'-deoxy-2'-fluoro pyrimidine nucleotides or alternately a plurality of pyrimidine nucleotides are 2'-deoxy-2'-fluoro pyrimidine nucleotides), and wherein any (e.g., one or more or all) purine nucleotides present in the antisense region are 2'-0-methyl purine nucleotides (e.g., wherein all purine nucleotides are 2'-0-methyl purine nucleotides or alternately a plurality of purine nucleotides are 2'-0-methyl purine nucleotides).
In one embodiment, the invention features a chemically-modified short interfering nucleic acid (siNA) molecule of the invention capable of mediating RNA interference
(RNAi) against a target polynucleotide (e.g., DNA or RNA) inside a cell or reconstituted in vitro system comprising a sense region, wherein one or more pyrimidine nucleotides present in the sense region are 2'-deoxy-2'-fluoro pyrimidine nucleotides (e.g., wherem all pyrimidine nucleotides are 2'-deoxy-2'-fluoro pyrimidine nucleotides or alternately a plurality of pyrimidine nucleotides are 2'-deoxy-2'-fluoro pyrimidine nucleotides), and one or more purine nucleotides present in the sense region are 2'-deoxy purine nucleotides (e.g., wherem all purine nucleotides are 2'-deoxy purine nucleotides or alternately a plurality of purine nucleotides are 2'-deoxy purine nucleotides), and an antisense region, wherein one or more pyrimidine nucleotides present in the antisense region are 2'-deoxy-2'-fluoro pyrimidine nucleotides (e.g., wherein all pyrimidine nucleotides are 2'-deoxy-2'-fluoro pyrimidine nucleotides or alternately a plurality of
pyrimidine nucleotides are 2'-deoxy-2'-fluoro pyrimidine nucleotides), and one or more purine nucleotides present in the antisense region are 2'-0-methyl purine nucleotides (e.g., wherein all purine nucleotides are 2'-0-methyl purine nucleotides or alternately a plurality of purine nucleotides are 2'-0-methyl purine nucleotides). The sense region and/or the antisense region can have a terminal cap modification, such as any modification described herein or shown in Figure 10, that is optionally present at the 3'- end, the 5'-end, or both of the 3' and 5'-ends of the sense and/or antisense sequence. The sense and/or antisense region can optionally further comprise a 3 '-terminal nucleotide overhang having about 1 to about 4 (e.g., about 1, 2, 3, or 4) 2'-deoxynucleotides. The overhang nucleotides can further comprise one or more (e.g., about 1, 2, 3, 4 or more) phosphorothioate, phosphonoacetate, and/or thiophosphonoacetate internucleotide linkages. Non-limiting examples of these chemically-modified siNAs are shown in Figures 4 and 5 and Table II herein. In any of these described embodiments, the purine nucleotides present in the sense region are alternatively 2'-0-methyl purine nucleotides (e.g., wherein all purine nucleotides are 2'-0-methyl purine nucleotides or alternately a plurality of purine nucleotides are 2'-0-methyl purine nucleotides) and one or more purine nucleotides present in the antisense region are 2'-0-methyl purine nucleotides (e.g., wherein all purine nucleotides are 2'-0-methyl purine nucleotides or alternately a plurality of purine nucleotides are 2'-0-methyl purine nucleotides). Also, in any of these embodiments, one or more purine nucleotides present in the sense region are alternatively purine ribonucleotides (e.g., wherein all purine nucleotides are purine ribonucleotides or alternately a plurality of purine nucleotides are purine ribonucleotides) and any purine nucleotides present in the antisense region are 2'-0-methyl purine nucleotides (e.g., wherem all purine nucleotides are 2'-0-methyl purine nucleotides or alternately a plurality of purine nucleotides are 2'-0-methyl purine nucleotides). Additionally, in any of these embodiments, one or more purine nucleotides present in the sense region and/or present in the antisense region are alternatively selected from the group consisting of 2'-deoxy nucleotides, locked nucleic acid (LNA) nucleotides, 2'- methoxyethyl nucleotides, 4'-thionucleotides, and 2'-0-methyl nucleotides (e.g., wherem all purine nucleotides are selected from the group consisting of 2'-deoxy nucleotides, locked nucleic acid (LNA) nucleotides, 2'-methoxyethyl nucleotides, 4'- thionucleotides, and 2'-0-methyl nucleotides or alternately a plurality of purine
nucleotides are selected from the group consisting of 2 '-deoxy nucleotides, locked nucleic acid (LNA) nucleotides, 2'-methoxyethyl nucleotides, 4'-thionucleotides, and 2'- O-methyl nucleotides).
In another embodiment, any modified nucleotides present in the siNA molecules of the invention, preferably in the antisense strand of the siNA molecules of the invention, but also optionally in the sense and/or both antisense and sense strands, comprise modified nucleotides having properties or characteristics similar to naturally occurring ribonucleotides. For example, the invention features siNA molecules including modified nucleotides having a Northern conformation (e.g., Northern pseudorotation cycle, see for example Saenger, Principles of Nucleic Acid Structure, Springer-Verlag ed., 1984). As such, chemically modified nucleotides present in the siNA molecules of the invention, preferably in the antisense strand of the siNA molecules of the invention, but also optionally in the sense and/or both antisense and sense strands, are resistant to nuclease degradation while at the same time maintaining the capacity to mediate RNAi. Non- limiting examples of nucleotides having a northern configuration include locked nucleic acid (LNA) nucleotides (e.g., 2'-0, 4'-C-methylene-(D-ribofuranosyl) nucleotides); 2'- methoxyethoxy (MOE) nucleotides; 2'-methyl-thio-ethyl, 2 '-deoxy-2 '-fluoro nucleotides, 2 '-deoxy-2 '-chloro nucleotides, 2'-azido nucleotides, and 2'-0-methyl nucleotides. In one embodiment, the sense strand of a double stranded siNA molecule of the invention comprises a terminal cap moiety, (see for example Figure 10) such as an inverted deoxyabaisc moiety, at the 3 '-end, 5 '-end, or both 3' and 5 '-ends of the sense strand.
In one embodiment, the invention features a chemically-modified short interfering nucleic acid molecule (siNA) capable of mediating RNA interference (RNAi) against a target polynucleotide (e.g., DNA or RNA) inside a cell or reconstituted in vitro system, wherein the chemical modification comprises a conjugate covalently attached to the chemically-modified siNA molecule. Non-limiting examples of conjugates contemplated by the invention include conjugates and ligands described in Vargeese et al, USSN 10/427,160, filed April 30, 2003, incoφorated by reference herein in its entirety, including the drawings. In another embodiment, the conjugate is covalently
attached to the chemically-modified siNA molecule via a biodegradable linker. In one embodiment, the conjugate molecule is attached at the 3'-end of either the sense strand, the antisense strand, or both strands of the chemically-modified siNA molecule. In another embodiment, the conjugate molecule is attached at the 5'-end of either the sense strand, the antisense strand, or both strands of the chemically-modified siNA molecule. In yet another embodiment, the conjugate molecule is attached both the 3 '-end and 5'-end of either the sense strand, the antisense strand, or both strands of the chemically- modified siNA molecule, or any combination thereof. In one embodiment, a conjugate molecule of the invention comprises a molecule that facilitates delivery of a chemically- modified siNA molecule into a biological system, such as a cell. In another embodiment, the conjugate molecule attached to the chemically-modified siNA molecule is a polyethylene glycol, human serum albumin, or a ligand for a cellular receptor that can mediate cellular uptake. Examples of specific conjugate molecules contemplated by the instant invention that can be attached to chemically-modified siNA molecules are described in Vargeese et al, U.S. Serial No. 10/201,394, filed July 22, 2002 incoφorated by reference herein. The type of conjugates used and the extent of conjugation of siNA molecules of the invention can be evaluated for improved pharmacokinetic profiles, bioavailability, and/or stability of siNA constructs while at the same time maintaining the ability of the siNA to mediate RNAi activity. As such, one skilled in the art can screen siNA constructs that are modified with various conjugates to determine whether the siNA conjugate complex possesses improved properties while maintaining the ability to mediate RNAi, for example in animal models as are generally known in the art.
In one embodiment, the invention features a short interfering nucleic acid (siNA) molecule of the invention, wherem the siNA further comprises a nucleotide, non- nucleotide, or mixed nucleotide/non-nucleotide linker that joins the sense region of the siNA to the antisense region of the siNA. In one embodiment, a nucleotide linker of the invention can be a linker of > 2 nucleotides in length, for example about 3, 4, 5, 6, 7, 8, 9, or 10 nucleotides in length. In another embodiment, the nucleotide linker can be a nucleic acid aptamer. By "aptamer" or "nucleic acid aptamer" as used herein is meant a nucleic acid molecule that binds specifically to a target molecule wherein the nucleic acid molecule has sequence that comprises a sequence recognized by the target molecule in its natural setting. Alternately, an aptamer can be a nucleic acid molecule that binds to
a target molecule where the target molecule does not naturally bind to a nucleic acid. The target molecule can be any molecule of interest. For example, the aptamer can be used to bind to a ligand-binding domain of a protein, thereby preventing interaction o»f the naturally occurring ligand with the protein. This is a non-limiting example and those in the art will recognize that other embodiments can be readily generated using techniques generally known in the art. (See, for example, Gold et al, 1995, Annu. Rev. Biochem., 64, 763; Brody and Gold, 2000, J. Biotechnol, 74, 5; Sun, 2000, Curr. Opin. Mol. Ther., 2, 100; Kusser, 2000, J. Biotechnol, 74, 27; Hermann and Patel, 200O, Science, 287, 820; and Jayasena, 1999, Clinical Chemistry, 45, 1628.) In yet another embodiment, a non-nucleotide linker of the invention comprises abasic nucleotide, polyether, polyamine, polyamide, peptide, carbohydrate, lipid., polyhydrocarbon, or other polymeric compounds (e.g. polyethylene glycols such as those having between 2 and 100 ethylene glycol units). Specific examples include those described by Seela and Kaiser, Nucleic Acids Res. 1990, S:6353 and Nucleic Acids Re^s. 1987, 75:3113; Cload and Schepartz, J. Am. Chem. Soc. 1991, 113:6324; Richardson and Schepartz, J. Am. Chem. Soc. 1991, 773:5109; Ma et al, Nucleic Acids Res. 1993, 27:2585 and Biochemistry 1993, 32:1751; Durand et al, Nucleic Acids Res. 1990, 75:6353; McCurdy et al, Nucleosides & Nucleotides 1991, 70:287; Jschke et al., Tetrahedron Lett. 1993, 34:301; Ono et al, Biochemistry 1991, 30:9914; Arnold et al., International Publication No. WO 89/02439; Usman et al, International Publication No. WO 95/06731; Dudycz et al, International Publication No. WO 95/11910 and Ferentz and Verdine, J. Am. Chem. Soc. 1991, 773:4000, all hereby incoφorated by reference herein. A "non-nucleotide" further means any group or compound that can be incoφorated into a nucleic acid chain in the place of one or more nucleotide units, including either sugar and/or phosphate substitutions, and allows the remaining bases to exhibit their enzymatic activity. The group or compound can be abasic in that it does not contain a commonly recognized nucleotide base, such as adenosine, guanine, cytosine, uracil or thymine, for example at the Cl position of the sugar.
In one embodiment, the invention features a short interfering nucleic acid (siNA) molecule capable of mediating RNA interference (RNAi) inside a cell or reconstituted in vitro system, wherein one or both strands of the siNA molecule that are assembled fro>m
two separate oligonucleotides do not comprise any ribonucleotides. For example, a siNA molecule can be assembled from a single oligonculeotide where the sense and antisense regions of the siNA comprise separate oligonucleotides that do not have any ribonucleotides (e.g., nucleotides having a 2'-OH group) present in the oligonucleotides. In another example, a siNA molecule can be assembled from a single oligonculeotide where the sense and antisense regions of the siNA are linked or circularized by a nucleotide or non-nucleotide linker as described herein, wherein the oligonucleotide does not have any ribonucleotides (e.g., nucleotides having a 2'-OH group) present in the oligonucleotide. Applicant has suφrisingly found that the presense of ribonucleotides (e.g., nucleotides having a 2'-hydroxyl group) within the siNA molecule is not required or essential to support RNAi activity. As such, in one embodiment, all positions within the siNA can include chemically modified nucleotides and/or non-nucleotides such as nucleotides and or non-nucleotides having Formula I, II, III, JV, V, VI, or VTI or any combination thereof to the extent that the ability of the siNA molecule to support RNAi activity in a cell is maintained.
In one embodiment, a siNA molecule of the invention is a single stranded siNA molecule that mediates RNAi activity in a cell or reconstituted in vitro system comprising a single stranded polynucleotide having complementarity to a target nucleic acid sequence. In another embodiment, the single stranded siNA molecule of the invention comprises a 5 '-terminal phosphate group. In another embodiment, the single stranded siNA molecule of the invention comprises a 5 '-terminal phosphate group and a 3'-terminal phosphate group (e.g., a 2',3'-cyclic phosphate). In another embodiment, the single stranded siNA molecule of the invention comprises about 15 to about 30 (e.g., about 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, or 30) nucleotides. In yet another embodiment, the single stranded siNA molecule of the invention comprises one or more chemically modified nucleotides or non-nucleotides described herein. For example, all the positions within the siNA molecule can include chemically-modified nucleotides such as nucleotides having any of Formulae I- VII, or any combination thereof to the extent that the ability of the siNA molecule to support RNAi activity in a cell is maintained.
In one embodiment, a siNA molecule of the invention is a single stranded siNA molecule that mediates RNAi activity in a cell or reconstituted in vitro system comprising a single stranded polynucleotide having complementarity to a target nucleic acid sequence, wherein one or more pyrimidine nucleotides present in the siNA are 2'- deoxy-2'-fluoro pyrimidine nucleotides (e.g., wherein all pyrimidine nucleotides are 2'- deoxy-2'-fluoro pyrimidine nucleotides or alternately a plurality of pyrimidine nucleotides are 2'-deoxy-2'-fluoro pyrimidine nucleotides), and wherein any purine nucleotides present in the antisense region are 2'-0-methyl purine nucleotides (e.g., wherein all purine nucleotides are 2'-0-methyl purine nucleotides or alternately a plurality of purine nucleotides are 2'-0-methyl purine nucleotides), and a terminal cap modification, such as any modification described herein or shown in Figure 10, that is optionally present at the 3'-end, the 5'-end, or both of the 3' and 5'-ends of the antisense sequence. The siNA optionally further comprises about 1 to about 4 or more (e.g., about 1, 2, 3, 4 or more) terminal 2'-deoxynucleotides at the 3 '-end of the siNA molecule, wherein the terminal nucleotides can further comprise one or more (e.g., 1, 2, 3, 4 or more) phosphorothioate, phosphonoacetate, and/or thiophosphonoacetate internucleotide linkages, and wherein the siNA optionally further comprises a terminal phosphate group, such as a 5 '-terminal phosphate group. In any of these embodiments, any purine nucleotides present in the antisense region are alternatively 2'-deoxy purine nucleotides (e.g., wherein all purine nucleotides are 2'-deoxy purine nucleotides or alternately a plurality of purine nucleotides are 2'-deoxy purine nucleotides). Also, in any of these embodiments, any purine nucleotides present in the siNA (i.e., purine nucleotides present in the sense and/or antisense region) can alternatively be locked nucleic acid (LNA) nucleotides (e.g., wherein all purine nucleotides are LNA nucleotides or alternately a plurality of purine nucleotides are LNA nucleotides). Also, in any of these embodiments, any purine nucleotides present in the siNA are alternatively 2'- methoxyethyl purine nucleotides (e.g., wherein all purine nucleotides are 2'- methoxyethyl purine nucleotides or alternately a plurality of purine nucleotides are 2'- methoxyethyl purine nucleotides). In another embodiment, any modified nucleotides present in the single stranded siNA molecules of the invention comprise modified nucleotides having properties or characteristics similar to naturally occurring ribonucleotides. For example, the invention features siNA molecules including modified
nucleotides having a Northern conformation (e.g., Northern pseudorotation cycle, see for example Saenger, Principles of Nucleic Acid Structure, Springer-Verlag ed., 1984). As such, chemically modified nucleotides present in the single stranded siNA molecules of the invention are preferably resistant to nuclease degradation while at the same time maintaining the capacity to mediate RNAi.
In one embodiment, a siNA molecule of the invention comprises chemically modified nucleotides or non-nucleotides (e.g., having any of Formulae I- VII, such as 2'- deoxy, 2 '-deoxy-2 '-fluoro, or 2'-0-methyl nucleotides) at alternating positions within one or more strands or regions of the siNA molecule. For example, such chemical modifications can be introduced at every other position of a RNA based siNA molecule, starting at either the first or second nucleotide from the 3 '-end or 5 '-end of the siNA. In a non-limiting example, a double stranded siNA molecule of the invention in which each strand of the siNA is 21 nucleotides in length is featured wherein positions 1, 3, 5, 7, 9, 11, 13, 15, 17, 19 and 21 of each strand are chemically modified (e.g., with compounds having any of Formulae 1-VII, such as such as 2 '-deoxy, 2 '-deoxy-2 '-fluoro, or 2'-O- methyl nucleotides). In another non-limiting example, a double stranded siNA molecule of the invention in which each strand of the siNA is 21 nucleotides in length is featured wherein positions 2, 4, 6, 8, 10, 12, 14, 16, 18, and 20 of each strand are chemically modified (e.g., with compounds having any of Formulae 1-VII, such as such as 2'-deoxy, 2 '-deoxy-2 '-fluoro, or 2'-0-methyl nucleotides). Such siNA molecules can further comprise terminal cap moieties and/or backbone modifications as described herein.
In one embodiment, the invention features a method for modulating the expression of a target gene within a cell comprising: (a) synthesizing a siNA molecule of the invention, which can be chemically-modified, wherem one of the siNA strands comprises a sequence complementary to RNA of the target gene; and (b) introducing the siNA molecule into a cell under conditions suitable to modulate the expression of the target gene in the cell.
In one embodiment, the invention features a method for modulating the expression of a target gene within a cell comprising: (a) synthesizing a siNA molecule of the invention, which can be chemically-modified, wherem one of the siNA strands comprises a sequence complementary to RNA of the target gene and wherein the sense
βuαiiu sequence υi cne SUN A compnses a sequence identical or substantially similar to the sequence of the target RNA; and (b) introducing the siNA molecule into a cell under conditions suitable to modulate the expression of the target gene in the cell.
In another embodiment, the invention features a method for modulating the expression of more than one target gene within a cell comprising: (a) synthesizing siNA molecules of the invention, which can be chemically-modified, wherein one of the siNA strands comprises a sequence complementary to RNA of the target genes; and (b) introducing the siNA molecules into a cell under conditions suitable to modulate the expression of the target genes in the cell. In another embodiment, the invention features a method for modulating the expression of two or more target genes within a cell comprising: (a) synthesizing one or more siNA molecules of the invention, which can be chemically-modified, wherein the siNA strands comprise sequences complementary to RNA of the target genes and wherein the sense strand sequences of the siNAs comprise sequences identical or substantially similar to the sequences of the target RNAs; and (b) introducing the siNA molecules into a cell under conditions suitable to modulate the expression of the target genes in the cell.
In another embodiment, the invention features a method for modulating the expression of more than one target gene within a cell comprising: (a) synthesizing a siNA molecule of the invention, which can be chemically-modified, wherem one of the siNA strands comprises a sequence complementary to RNA of the target gene and wherein the sense strand sequence of the siNA comprises a sequence identical or substantially similar to the sequences of the target RNAs; and (b) introducing the siNA molecule into a cell under conditions suitable to modulate the expression of the target genes in the cell.
In one embodiment, siNA molecules of the invention are used as reagents in ex vivo applications. For example, siNA reagents are introduced into tissue or cells that are transplanted into a subject for therapeutic effect. The cells and/or tissue can be derived from an organism or subject that later receives the explant, or can be derived from another organism or subject prior to transplantation. The siNA molecules can be used to
modulate the expression of one or more target genes in the cells or tissue, such that the cells or tissue obtain a desired phenotype or are able to perform a function when transplanted in vivo. In one embodiment, certain target cells from a patient are extracted. These extracted cells are contacted with siNAs targeting a specific nucleotide sequence within the cells under conditions suitable for uptake of the siNAs by these cells (e.g. using delivery reagents such as cationic lipids, liposomes and the like or using techniques such as electroporation to facilitate the delivery of siNAs into cells). The cells are then reintroduced back into the same patient or other patients. In one embodiment, the invention features a method of modulating the expression of a target gene in a tissue explant comprising: (a) synthesizing a siNA molecule of the invention, which can be chemically-modified, wherein one of the siNA strands comprises a sequence complementary to RNA of the target gene; and (b) introducing the siNA molecule into a cell of the tissue explant derived from a particular organism under conditions suitable to modulate the expression of the target gene in the tissue explant. In another embodiment, the method further comprises introducing the tissue explant back into the organism the tissue was derived from or into another orgamsm under conditions suitable to modulate the expression of the target gene in that organism.
In one embodiment, the invention features a method of modulating the expression of a target gene in a tissue explant comprising: (a) synthesizing a siNA molecule of the invention, which can be chemically-modified, wherein one of the siNA strands comprises a sequence complementary to RNA of the target gene and wherem the sense strand sequence of the siNA comprises a sequence identical or substantially similar to the sequence of the target RNA; and (b) introducing the siNA molecule into a cell of the tissue explant derived from a particular organism under conditions suitable to modulate the expression of the gene in the tissue explant. In another embodiment, the method further comprises introducing the tissue explant back into the organism the tissue was derived from or into another organism under conditions suitable to modulate the expression of the target gene in that organism.
In another embodiment, the invention features a method of modulating the expression of more than one target gene in a tissue explant comprising: (a) synthesizing siNA molecules of the invention, which can be chemically-modified, wherein one of the
siNA strands comprises a sequence complementary to RNA of the target genes; and (b) introducing the siNA molecules into a cell of the tissue explant derived from a particular organism under conditions suitable to modulate the expression of the target genes in the tissue explant. In another embodiment, the method further comprises introducing the tissue explant back into the organism the tissue was derived from or into another organism under conditions suitable to modulate the expression of the target genes in that organism.
In one embodiment, the invention features a method of modulating the expression of a target gene in a subject or organism comprising: (a) synthesizing a siNA molecule of the invention, which can be chemically-modified, wherein one of the siNA strands comprises a sequence complementary to RNA of the target gene; and (b) introducing the siNA molecule into the subject or organism under conditions suitable to modulate the expression of the target gene in the subject or organism. The level of protein or RNA can be determined using various methods well-known in the art. In another embodiment, the invention features a method of modulating the expression of more than one target gene in a subject or organism comprising: (a) synthesizing siNA molecules of the invention, which can be chemically-modified, wherein one of the siNA strands comprises a sequence complementary to RNA of the target genes; and (b) introducing the siNA molecules into the subject or organism under conditions suitable to modulate the expression of the target genes in the subject or organism. The level of protein or RNA can be determined as is known in the art.
In one embodiment, the invention features a method for modulating the expression of a target gene within a cell comprising: (a) synthesizing a siNA molecule of the invention, which can be chemically-modified, wherein the siNA comprises a single stranded sequence having complementarity to RNA of the target gene; and (b) introducing the siNA molecule into a cell under conditions suitable to modulate the expression of the target gene in the cell.
In another embodiment, the invention features a method for modulating the expression of more than one target gene within a cell comprising: (a) synthesizing siNA molecules of the invention, which can be chemically-modified, wherein the siNA
comprises a single stranded sequence having complementarity to RNA of the target gene; and (b) contacting the cell in vitro or in vivo with the siNA molecule under conditions suitable to modulate the expression of the target genes in the cell.
In one embodiment, the invention features a method of modulating the expression of a target gene in a tissue explant comprising: (a) synthesizing a siNA molecule of the invention, which- can be chemically-modified, wherein the siNA comprises a single stranded sequence having complementarity to RNA of the gene; and (b) contacting a cell of the tissue explant derived from a particular subject or organism with the siNA molecule under conditions suitable to modulate the expression of the target gene in the tissue explant. In another embodiment, the method further comprises introducing the tissue explant back into the subject or organism the tissue was derived from or into another subject or orgamsm under conditions suitable to modulate the expression of the target gene in that subject or organism.
In another embodiment, the invention features a method of modulating the expression of more than one target gene in a tissue explant comprising: (a) synthesizing siNA molecules of the invention, which can be chemically-modified, wherein the siNA comprises a single stranded sequence having complementarity to RNA of the target gene; and (b) introducing the siNA molecules into a cell of the tissue explant derived from a particular subject or organism under conditions suitable to modulate the expression of the target genes in the tissue explant. In another embodiment, the method further comprises introducing the tissue explant back into the subject or organism the tissue was derived from or into another subject or organism under conditions suitable to modulate the expression of the target genes in that subject or organism.
In one embodiment, the invention features a method of modulating the expression of a target gene in a subject or organism comprising: (a) synthesizing a siNA molecule of the invention, which can be chemically-modified, wherein the siNA comprises a single stranded sequence having complementarity to RNA of the target gene; and (b) introducing the siNA molecule into the subject or organism under conditions suitable to modulate the expression of the target gene in the subject or organism.
In another embodiment, the invention features a method of modulating the expression of more than one target gene in a subject or organism comprising: (a) synthesizing siNA molecules of the invention, which can be chemically-modified, wherein the siNA comprises a single stranded sequence having complementarity to RNA of the target gene; and (b) introducing the siNA molecules into the subject or organism under conditions suitable to modulate the expression of the target genes in the subject or organism.
In one embodiment, the invention features a method of modulating the expression of a target gene in a subject or organism comprising contacting the subject or organism with a siNA molecule of the invention under conditions suitable to modulate the expression of the target gene in the subject or organism.
In one embodiment, the invention features a method for treating or preventing a disease, disorder, trait or condition related to gene expression in a subject or organism comprising contacting the subject or organism with a siNA molecule of the invention under conditions suitable to modulate the expression of the target gene in the subject or organism. The reduction of gene expression and thus reduction in the level of the respective protein/RNA relieves, to some extent, the symptoms of the disease, disorder, trait or condition.
In one embodiment, the invention features a method for treating or preventing cancer in a subject or organism comprising contacting the subject or organism with a siNA molecule of the invention under conditions suitable to modulate the expression of a target gene associated with the maintenance or development of cancer in the subject or organism.
In one embodiment, the invention features a method for treating or preventing a proliferative disease, disorder, trait or condition in a subject or organism comprising contacting the subject or organism with a siNA molecule of the invention under conditions suitable to modulate the expression of a target gene associated with the maintenance or development of the proliferative disease, disorder, trait or condition in the subject or organism.
In one embodiment, the invention features a method for treating or preventing transplant and/or tissue rejection (allograft rejection) in a subject or organism comprising contacting the subject or organism with a siNA molecule of the invention under conditions suitable to modulate the expression of a target gene associated with transplant and/or tissue rejection (allograft rejection) in the subject or organism.
In one embodiment, the invention features a method for treating or preventing an autoimmune disease, disorder, trait or condition in a subject or organism comprising contacting the subject or organism with a siNA molecule of the invention under conditions suitable to modulate the expression of a target gene associated with the maintenance or development of the autoimmune disease, disorder, trait or condition in the subject or orgamsm.
In one embodiment, the invention features a method for treating or preventing an infectious disease, disorder, trait or condition in a subject or organism comprising contacting the subject or organism with a siNA molecule of the invention under conditions suitable to modulate the expression of a target gene associated with the maintenance or development of the infectious disease, disorder, trait or condition in the subject or organism.
In one embodiment, the invention features a method for treating or preventing an age-related disease, disorder, trait or condition in a subject or organism comprising contacting the subject or orgamsm with a siNA molecule of the invention under conditions suitable to modulate the expression of a target gene associated with the maintenance or development of the age-related disease, disorder, trait or condition in the subject or organism.
In one embodiment, the invention features a method for treating or preventing a neurologic or neurodegenerative disease, disorder, trait or condition in a subject or orgamsm comprising contacting the subject or organism with a siNA molecule of the invention under conditions suitable to modulate the expression of a target gene associated with the maintenance or development of the neurologic or neurodegenerative disease, disorder, trait or condition in the subject or organism.
In one embodiment, the invention features a method for treating or preventing a metabolic disease, disorder, trait or condition in a subject or orgamsm comprising contacting the subject or organism with a siNA molecule of the invention under conditions suitable to modulate the expression of a target gene associated with the maintenance or development of the metabolic disease, disorder, trait or condition in the subject or organism.
In one embodiment, the invention features a method for treating or preventing an cardiovascular disease, disorder, trait or condition in a subject or organism comprising contacting the subject or organism with a siNA molecule of the invention under conditions suitable to modulate the expression of a target gene associated with the maintenance or development of the cardiovascular disease, disorder, trait or condition in the subject or organism.
In one embodiment, the invention features a method for treating or preventing a respiratory disease, disorder, trait or condition in a subject or organism comprising contacting the subject or organism with a siNA molecule of the invention under conditions suitable to modulate the expression of a target gene associated with the maintenance or development of the respiratory disease, disorder, trait or condition in the subject or organism.
In one embodiment, the invention features a method for treating or preventing an ocular disease, disorder, trait or condition in a subject or organism comprising contacting the subject or organism with a siNA molecule of the invention under conditions suitable to modulate the expression of a target gene associated with the maintenance or development of the ocular disease, disorder, trait or condition in the subject or orgamsm.
In one embodiment, the invention features a method for treating or preventing a dermatological disease, disorder, trait or condition in a subject or organism comprising contacting the subject or organism with a siNA molecule of the invention under conditions suitable to modulate the expression of a target gene associated with the maintenance or development of the dermatological disease, disorder, trait or condition in the subject or organism.
In one embodiment, the invention features a method for treating or preventing a disease, disorder, trait or condition in a subject or organism comprising contacting the subject or organism with a siNA molecule of the invention under conditions suitable to modulate the expression of a target gene associated with the maintenance or development of the disease, disorder, trait or condition in the subject or organism.
In one embodiment, the invention features a method for treating or preventing a liver disease, disorder, trait or condition in a subject or organism comprising contacting the subject or organism with a siNA molecule of the invention under conditions suitable to modulate the expression of a target gene associated with the maintenance or development of the liver disease, disorder, trait or condition in the subject or orgamsm.
In one embodiment, the invention features a method for treating or preventing a kidney disease, disorder, trait or condition in a subject or organism comprising contacting the subject or organism with a siNA molecule of the invention under conditions suitable to modulate the expression of a target gene associated with the maintenance or development of the kidney disease, disorder, trait or condition in the subject or organism.
In one embodiment, the invention features a method for treating or preventing a bladder disease, disorder, trait or condition in a subject or organism comprising contacting the subject or organism with a siNA molecule of the invention under conditions suitable to modulate the expression of a target gene associated with the maintenance or development of the bladder disease, disorder, trait or condition in the subject or organism.
In another embodiment, the invention features a method of modulating the expression of more than one target gene in a subject or organism comprising contacting the subject or organism with one or more siNA molecules of the invention under conditions suitable to modulate the expression of the genes in the subject or organism.
The siNA molecules of the invention can be designed to down regulate or inhibit target gene expression through RNAi targeting of a variety of RNA molecules. In one embodiment, the siNA molecules of the invention are used to target various RNAs coπesponding to a target gene. Non-limiting examples of such RNAs include messenger
RNA (mRNA), alternate RNA splice variants of target gene(s), post-transcnptionally modified RNA of target gene(s), pre-mRNA of target gene(s), and/or RNA templates. If alternate splicing produces a family of transcripts that are distinguished by usage of appropriate exons, the instant invention can be used to inhibit gene expression through the appropriate exons to specifically inhibit or to distinguish among the functions of gene family members. For example, a protein that contains an alternatively spliced transmembrane domain can be expressed in both membrane bound and secreted forms. Use of the invention to target the exon containing the transmembrane domain can be used to determine the functional consequences of pharmaceutical targeting of membrane bound as opposed to the secreted form of the protein. Non-limiting examples of applications of the invention relating to targeting these RNA molecules include therapeutic pharmaceutical applications, pharmaceutical discovery applications, molecular diagnostic and gene function applications, and gene mapping, for example using single nucleotide polymoφhism mapping with siNA molecules of the invention. Such applications can be implemented using known gene sequences or from partial sequences available from an expressed sequence tag (EST).
In another embodiment, the siNA molecules of the invention are used to target conserved sequences corresponding to a target gene family or target gene families . As such, siNA molecules targeting multiple gene targets can provide increased therapeutic effect. In addition, siNA can be used to characterize pathways of gene function in a variety of applications. For example, the present invention can be used to inhibit the activity of target gene(s) in a pathway to determine the function of uncharacterized gene(s) in gene function analysis, mRNA function analysis, or translational analysis. The invention can be used to determine potential target gene pathways involved in various diseases and conditions toward pharmaceutical development. The invention can be used to understand pathways of gene expression involved in diseases, traits, disorders, and/or conditions described herein or otherwise known in the art.
In one embodiment, siNA molecule(s) and/or methods of the invention are used to down regulate the expression of gene(s) that encode RNA refeπed to by Genbank Accession, for example, target genes encoding RNA sequence(s) refeπed to herein by Genbank Accession number, for example, Genbank Accession Nos. shown in Table I.
In one embodiment, the invention features a method comprising: (a) generating a library of siNA constructs having a predetermined complexity; and (b) assaying the siNA constructs of (a) above, under conditions suitable to determine RNAi target sites within the target RNA sequence. In one embodiment, the siNA molecules of (a) have strands of a fixed length, for example, about 23 nucleotides in length. In another embodiment, the siNA molecules of (a) are of differing length, for example having strands of about 15 to about 30 (e.g., about 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, or 30) nucleotides in length. In one embodiment, the assay can comprise a reconstituted in vitro siNA assay as described herein. In another embodiment, the assay can comprise a cell culture system in which target RNA is expressed. In another embodiment, fragments of target RNA are analyzed for detectable levels of cleavage, for example by gel electrophoresis, northern blot analysis, or RNAse protection assays, to determine the most suitable target site(s) within the target RNA sequence. The target RNA sequence can be obtained as is known in the art, for example, by cloning and/or transcription for in vitro systems, and by cellular expression in in vivo systems.
In one embodiment, the invention features a method comprising: (a) generating a randomized library of siNA constructs having a predetermined complexity, such as of 4N, where N represents the number of base paired nucleotides in each of the siNA construct strands (eg. for a siNA construct having 21 nucleotide sense and antisense strands with 19 base pairs, the complexity would be 419); and (b) assaying the siNA constructs of (a) above, under conditions suitable to determine RNAi target sites within the target RNA sequence. In another embodiment, the siNA molecules of (a) have strands of a fixed length, for example about 23 nucleotides in length. In yet another embodiment, the siNA molecules of (a) are of differing length, for example having strands of about 15 to about 30 (e.g., about 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, or 30) nucleotides in length. In one embodiment, the assay can comprise a reconstituted in vitro siNA assay as described in Example 6 herein. In another embodiment, the assay can comprise a cell culture system in which target RNA is expressed. In another embodiment, fragments of target RNA are analyzed for detectable levels of cleavage, for example, by gel electrophoresis, northern blot analysis, or RNAse protection assays, to determine the most suitable target site(s) within the target target RNA sequence. The target target
RNA sequence can be obtained as is known in the art, for example, by cloning and/or transcription for in vitro systems, and by cellular expression in in vivo systems.
In another embodiment, the invention features a method comprising: (a) analyzing the sequence of a RNA target encoded by a target gene; (b) synthesizing one or more sets of siNA molecules having sequence complementary to one or more regions of the RNA of (a); and (c) assaying the siNA molecules of (b) under conditions suitable to determine RNAi targets within the target RNA sequence. In one embodiment, the siNA molecules of (b) have strands of a fixed length, for example about 23 nucleotides in length. In another embodiment, the siNA molecules of (b) are of differing length, for example having strands of about 15 to about 30 (e.g., about 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, or 30) nucleotides in length. In one embodiment, the assay can comprise a reconstituted in vitro siNA assay as described herein. In another embodiment, the assay can comprise a cell culture system in which target RNA is expressed. Fragments of target RNA are analyzed for detectable levels of cleavage, for example by gel electrophoresis, northern blot analysis, or RNAse protection assays, to determine the most suitable target site(s) within the target RNA sequence. The target RNA sequence can be obtained as is known in the art, for example, by cloning and/or transcription for in vitro systems, and by expression in in vivo systems.
By "target site" is meant a sequence within a target RNA that is "targeted" for cleavage mediated by a siNA construct which contains sequences within its antisense region that are complementary to the target sequence.
By "detectable level of cleavage" is meant cleavage of target RNA (and formation of cleaved product RNAs) to an extent sufficient to discern cleavage products above the background of RNAs produced by random degradation of the target RNA. Production of cleavage products from 1-5% of the target RNA is sufficient to detect above the background for most methods of detection.
In one embodiment, the invention features a composition comprising a siNA molecule of the invention, which can be chemically-modified, in a pharmaceutically acceptable carrier or diluent. In another embodiment, the invention features a pharmaceutical composition comprising siNA molecules of the invention, which can be
chemically-modified, targeting one or more genes in a pharmaceutically acceptable carrier or diluent. In another embodiment, the invention features a method for diagnosing a disease or condition in a subject comprising administering to the subject a composition of the invention under conditions suitable for the diagnosis of the disease or condition in the subject. In another embodiment, the invention features a method for treating or preventing a disease or condition in a subject, comprising administering to the subject a composition of the invention under conditions suitable for the treatment or prevention of the disease or condition in the subject, alone or in conjunction with one or more other therapeutic compounds. In yet another embodiment, the invention features a method for treating or preventing diseases, traits, disorders, and/or conditions in a subject or organism comprising administering to the subject a composition of the invention under conditions suitable for the treatment or prevention of the disease, trait, disorder, and/or condition in the subject or organism.
In another embodiment, the invention features a method for validating a gene target, comprising: (a) synthesizing a siNA molecule of the invention, which can be chemically-modified, wherein one of the siNA strands includes a sequence complementary to RNA of a target gene; (b) introducing the siNA molecule into a cell, tissue, subject, or organism under conditions suitable for modulating expression of the target gene in the cell, tissue, subject, or orgamsm; and (c) determining the function of the target gene by assaying for any phenotypic change in the cell, tissue, subject, or orgamsm.
In another embodiment, the invention features a method for validating a gene target comprising: (a) synthesizing a siNA molecule of the invention, which can be chemically-modified, wherein one of the siNA strands includes a sequence complementary to RNA of a target gene; (b) introducing the siNA molecule into a biological system under conditions suitable for modulating expression of the target gene in the biological system; and (c) determining the function of the gene by assaying for any phenotypic change in the biological system.
By "biological system" is meant, material, in a purified or unpurified form, from biological sources, including but not limited to human or animal, wherein the system comprises the components required for RNAi activity. The term "biological system"
includes, for example, a cell, tissue, subject, or organism, or extract thereof. The term biological system also includes reconstituted RNAi systems that can be used in an in vitro setting.
By "phenotypic change" is meant any detectable change to a cell that occurs in response to contact or treatment with a nucleic acid molecule of the invention (e.g., siNA). Such detectable changes include, but are not limited to, changes in shape, size, proliferation, motility, protein expression or RNA expression or other physical or chemical changes as can be assayed by methods known in the art. The detectable change can also include expression of reporter genes/molecules such as Green Florescent Protein (GFP) or various tags that are used to identify an expressed protein or any other cellular component that can be assayed.
In one embodiment, the invention features a kit containing a siNA molecule of the invention, which can be chemically-modified, that can be used to modulate the expression of a target gene in a biological system, including, for example, in a cell, tissue, subject, or organism. In another embodiment, the invention features a kit containing more than one siNA molecule of the invention, which can be chemically- modified, that can be used to modulate the expression of more than one target gene in a biological system, including, for example, in a cell, tissue, subject, or organism.
In one embodiment, the invention features a cell containing one or more siNA molecules of the invention, which can be chemically-modified. In another embodiment, the cell containing a siNA molecule of the invention is a mammalian cell. In yet another embodiment, the cell containing a siNA molecule of the invention is a human cell.
In one embodiment, the synthesis of a siNA molecule of the invention, which can be chemically-modified, comprises: (a) synthesis of two complementary strands of the siNA molecule; (b) annealing the two complementary strands together under conditions suitable to obtain a double-stranded siNA molecule. In another embodiment, synthesis of the two complementary strands of the siNA molecule is by solid phase oligonucleotide synthesis. In yet another embodiment, synthesis of the two complementary strands of the siNA molecule is by solid phase tandem oligonucleotide synthesis.
In one embodiment, the invention features a method for synthesizing a siNA duplex molecule comprising: (a) synthesizing a first oligonucleotide sequence strand of the siNA molecule, wherein the first oligonucleotide sequence strand comprises a cleavable linker molecule that can be used as a scaffold for the synthesis of the second oligonucleotide sequence strand of the siNA; (b) synthesizing the second oligonucleotide sequence strand of siNA on the scaffold of the first oligonucleotide sequence strand, wherein the second oligonucleotide sequence strand further comprises a chemical moiety than can be used to purify the siNA duplex; (c) cleaving the linker molecule of (a) under conditions suitable for the two siNA oligonucleotide strands to hybridize and form a stable duplex; and (d) purifying the siNA duplex utilizing the chemical moiety of the second oligonucleotide sequence strand. In one embodiment, cleavage of the linker molecule in (c) above takes place during deprotection of the oligonucleotide, for example, under hydrolysis conditions using an alkylamine base such as methylamine. In one embodiment, the method of synthesis comprises solid phase synthesis on a solid support such as controlled pore glass (CPG) or polystyrene, wherein the first sequence of (a) is synthesized on a cleavable linker, such as a succinyl linker, using the solid support as a scaffold. The cleavable linker in (a) used as a scaffold for synthesizing the second strand can comprise similar reactivity as the solid support derivatized linker, such that cleavage of the solid support derivatized linker and the cleavable linker of (a) takes place concomitantly. In another embodiment, the chemical moiety of (b) that can be used to isolate the attached oligonucleotide sequence comprises a trityl group, for example dimethoxytrityl group, which can be employed in a trityl-on synthesis strategy as described herein. In yet another embodiment, the chemical moiety, such as a dimethoxytrityl group, is removed during purification, for example, using acidic conditions.
In a further embodiment, the method for siNA synthesis is a solution phase synthesis or hybrid phase synthesis wherein both strands of the siNA duplex are synthesized in tandem using a cleavable linker attached to the first sequence which acts a scaffold for synthesis of the second sequence. Cleavage of the linker under conditions suitable for hybridization of the separate siNA sequence strands results in formation of the double-stranded siNA molecule.
In another embodiment, the invention features a method for synthesizing a sirs A duplex molecule comprising: (a) synthesizing one oligonucleotide sequence strand of the siNA molecule, wherein the sequence comprises a cleavable linker molecule that can be used as a scaffold for the synthesis of another oligonucleotide sequence; (b) synthesizing a second oligonucleotide sequence having complementarity to the first sequence strand on the scaffold of (a), wherein the second sequence comprises the other strand of the double-stranded siNA molecule and wherein the second sequence further comprises a chemical moiety than can be used to isolate the attached oligonucleotide sequence; (c) purifying the product of (b) utilizing the chemical moiety of the second oligonucleotide sequence strand under conditions suitable for isolating the full-length sequence comprising both siNA oligonucleotide strands connected by the cleavable linker and under conditions suitable for the two siNA oligonucleotide strands to hybridize and form a stable duplex. In one embodiment, cleavage of the linker molecule in (c) above takes place during deprotection of the oligonucleotide, for example, under hydrolysis conditions. In another embodiment, cleavage of the linker molecule in (c) above takes place after deprotection of the oligonucleotide. In another embodiment, the method of synthesis comprises solid phase synthesis on a solid support such as controlled pore glass (CPG) or polystyrene, wherein the first sequence of (a) is synthesized on a cleavable linker, such as a succinyl linker, using the solid support as a scaffold. The cleavable linker in (a) used as a scaffold for synthesizing the second strand can comprise similar reactivity or differing reactivity as the solid support derivatized linker, such that cleavage of the solid support derivatized linker and the cleavable linker of (a) takes place either concomitantly or sequentially. In one embodiment, the chemical moiety of (b) that can be used to isolate the attached oligonucleotide sequence comprises a trityl group, for example a dimethoxytrityl group.
In another embodiment, the invention features a method for making a double- stranded siNA molecule in a single synthetic process comprising: (a) synthesizing an oligonucleotide having a first and a second sequence, wherein the first sequence is complementary to the second sequence, and the first oligonucleotide sequence is linked to the second sequence via a cleavable linker, and wherein a terminal 5'-protecting group, for example, a 5'-0-dimethoxytrityl group (5'-0-DMT) remains on the oligonucleotide having the second sequence; (b) deprotectmg the oligonucleotide whereby the
deprotection results in the cleavage of the linker joining the two oligonucleotide sequences; and (c) purifying the product of (b) under conditions suitable for isolating the double-stranded siNA molecule, for example using a trityl-on synthesis strategy as described herein. In another embodiment, the method of synthesis of siNA molecules of the invention comprises the teachings of Scaringe et al, US Patent Nos. 5,889,136; 6,008,400; and 6,111,086, incoφorated by reference herein in their entirety.
In one embodiment, the invention features siNA constructs that mediate RNAi against a target polynucleotide (e.g., DNA or RNA), wherein the siNA construct comprises one or more chemical modifications, for example, one or more chemical modifications having any of Formulae 1-VII or any combination thereof that increases the nuclease resistance of the siNA construct.
In another embodiment, the invention features a method for generating siNA molecules with increased nuclease resistance comprising (a) introducing nucleotides having any of Formula I- VII or any combination thereof into a siNA molecule, and (b) assaying the siNA molecule of step (a) under conditions suitable for isolating siNA molecules having increased nuclease resistance.
In another embodiment, the invention features a method for generating siNA molecules with improved toxicologic profiles (e.g., have attenuated or no immunstimulatory properties) comprising (a) introducing nucleotides having any of Formula I- VII (e.g., siNA motifs refeπed to in Table II) or any combination thereof into a siNA molecule, and (b) assaying the siNA molecule of step (a) under conditions suitable for isolating siNA molecules having improved toxicologic profiles.
In another embodiment, the invention features a method for generating siNA molecules that do not stimulate an interferon response (e.g., no interferon response or attenuated interferon response) in a cell, subject, or organism, comprising (a) introducing nucleotides having any of Formula I- VII (e.g., siNA motifs refeπed to in Table II) or any combination thereof into a siNA molecule, and (b) assaying the siNA molecule of step (a) under conditions suitable for isolating siNA molecules that do not stimulate an interferon response.
By "improved toxicologic profile", is meant that the chemically modified siNA construct exhibits decreased toxicity in a cell, subject, or orgamsm compared to an unmodified siNA or siNA molecule having fewer modifications or modifications that are less effective in imparting improved toxicology. In a non-limiting example, siNA molecules with improved toxicologic profiles are associated with a decreased or attenuated immunostimulatory response in a cell, subject, or organism compared to an unmodified siNA or siNA molecule having fewer modifications or modifications that are less effective in imparting improved toxicology. In one embodiment, a siNA molecule with an improved toxicological profile comprises no ribonucleotides. In one embodiment, a siNA molecule with an improved toxicological profile comprises less than 5 ribonucleotides (e.g., 1, 2, 3, or 4 ribonucleotides). In one embodiment, a siNA molecule with an improved toxicological profile comprises Stab 7, Stab 8, Stab 11, Stab 12, Stab 13, Stab 16, Stab 17, Stab 18, Stab 19, Stab 20, Stab 23, Stab 24, Stab 25, Stab 26, Stab 27, Stab 28, Stab 29, Stab 30, Stab 31, Stab 32 or any combination thereof (see Table II). In one embodiment, the level of immunostimulatory response associated with a given siNA molecule can be measured as is known in the art, for example by determining the level of PKR/interferon response, proliferation, B-cell activation, and/or cytokine production in assays to quantitate the immunostimulatory response of particular siNA molecules (see, for example, Leifer et al, 2003, J Immunother. 26, 313-9; and U.S. Patent No. 5968909, incoφorated in its entirety by reference).
In one embodiment, the invention features siNA constructs that mediate RNAi against a target polynucleotide (e.g., DNA or RNA), wherein the siNA construct comprises one or more chemical modifications described herein that modulates the binding affinity between the sense and antisense strands of the siNA construct. In another embodiment, the invention features a method for generating siNA molecules with increased binding affinity between the sense and antisense strands of the siNA molecule comprising (a) introducing nucleotides having any of Formula I- VII or any combination thereof into a siNA molecule, and (b) assaying the siNA molecule of step (a) under conditions suitable for isolating siNA molecules having increased binding affinity between the sense and antisense strands of the siNA molecule.
In one embodiment, the invention features siNA constructs that mediate RNAi against a target polynucleotide (e.g., DNA or RNA), wherein the siNA construct comprises one or more chemical modifications described herein that modulates the binding affinity between the antisense strand of the siNA construct and a complementary target RNA sequence within a cell.
In one embodiment, the invention features siNA constructs that mediate RNAi against a target polynucleotide (e.g., DNA or RNA), wherein the siNA construct comprises one or more chemical modifications described herein that modulates the binding affinity between the antisense strand of the siNA construct and a complementary target DNA sequence within a cell.
In another embodiment, the invention features a method for generating siNA molecules with increased binding affinity between the antisense strand of the siNA molecule and a complementary target RNA sequence comprising (a) introducing nucleotides having any of Formula I- VII or any combination thereof into a siNA molecule, and (b) assaying the siNA molecule of step (a) under conditions suitable for isolating siNA molecules having increased binding affinity between the antisense strand of the siNA molecule and a complementary target RNA sequence.
In another embodiment, the invention features a method for generating siNA molecules with increased binding affinity between the antisense strand of the siNA molecule and a complementary target DNA sequence comprising (a) introducing nucleotides having any of Formula I- VII or any combination thereof into a siNA molecule, and (b) assaying the siNA molecule of step (a) under conditions suitable for isolating siNA molecules having increased binding affinity between the antisense strand of the siNA molecule and a complementary target DNA sequence. In one embodiment, the invention features siNA constructs that mediate RNAi against a target polynucleotide (e.g., DNA or RNA), wherein the siNA construct comprises one or more chemical modifications described herein that modulate the polymerase activity of a cellular polymerase capable of generating additional endogenous siNA molecules having sequence homology to the chemically-modified siNA construct.
In another embodiment, the invention features a method for generating siNA molecules capable of mediating increased polymerase activity of a cellular polymerase capable of generating additional endogenous siNA molecules having sequence homology to a chemically-modified siNA molecule comprising (a) introducing nucleotides having any of Formula I- VII or any combination thereof into a siNA molecule, and (b) assaying the siNA molecule of step (a) under conditions suitable for isolating siNA molecules capable of mediating increased polymerase activity of a cellular polymerase capable of generating additional endogenous siNA molecules having sequence homology to the chemically-modified siNA molecule. In one embodiment, the invention features chemically-modified siNA constructs that mediate RNAi against a target polynucleotide (e.g., DNA or RNA) in a cell, wherein the chemical modifications do not significantly effect the interaction of siNA with a target RNA molecule, DNA molecule and/or proteins or other factors that are essential for RNAi in a manner that would decrease the efficacy of RNAi mediated by such siNA constructs.
In another embodiment, the invention features a method for generating siNA molecules with improved RNAi activity against a target polynucleotide (e.g., DNA or RNA) comprising (a) introducing nucleotides having any of Formula I- VII or any combination thereof into a siNA molecule, and (b) assaying the siNA molecule of step (a) under conditions suitable for isolating siNA molecules having improved RNAi activity.
In yet another embodiment, the invention features a method for generating siNA molecules with improved RNAi activity against a target RNA comprising (a) introducing nucleotides having any of Formula I- VII or any combination thereof into a siNA molecule, and (b) assaying the siNA molecule of step (a) under conditions suitable for isolating siNA molecules having improved RNAi activity against the target RNA.
In yet another embodiment, the invention features a method for generating siNA molecules with improved RNAi activity against a target DNA comprising (a) introducing nucleotides having any of Formula I- VII or any combination thereof into a siNA
molecule, and (b) assaying the siNA molecule of step (a) under conditions suitable for isolating siNA molecules having improved RNAi activity against the target DNA.
In one embodiment, the invention features siNA constructs that mediate RNAi against a target polynucleotide (e.g., DNA or RNA), wherein the siNA construct comprises one or more chemical modifications described herein that modulates the cellular uptake of the siNA construct.
In another embodiment, the invention features a method for generating siNA molecules against a target polynucleotide (e.g., DNA or RNA) with improved cellular uptake comprising (a) introducing nucleotides having any of Formula I- VII or any combination thereof into a siNA molecule, and (b) assaying the siNA molecule of step (a) under conditions suitable for isolating siNA molecules having improved cellular uptake.
In one embodiment, the invention features siNA constructs that mediate RNAi against a target polynucleotide (e.g., DNA or RNA), wherein the siNA construct comprises one or more chemical modifications described herein that increases the bioavailability of the siNA construct, for example, by attaching polymeric conjugates such as polyethyleneglycol or equivalent conjugates that improve the pharmacokinetics of the siNA construct, or by attaching conjugates that target specific tissue types or cell types in vivo. Non-limiting examples of such conjugates are described in Vargeese et al, U.S. Serial No. 10/201,394 incoφorated by reference herein.
In one embodiment, the invention features a method for generating siNA molecules of the invention with improved bioavailability comprising (a) introducing a conjugate into the structure of a siNA molecule, and (b) assaying the siNA molecule of step (a) under conditions suitable for isolating siNA molecules having improved bioavailability. Such conjugates can include ligands for cellular receptors, such as peptides derived from naturally occurring protein ligands; protein localization sequences, including cellular ZIP code sequences; antibodies; nucleic acid aptamers; vitamins and other co-factors, such as folate and N-acetylgalactosamine; polymers, such as polyethyleneglycol (PEG); phospholipids; cholesterol; polyamines, such as spermine or spermidine; and others.
In one embodiment, the invention features a double stranded short interfering nucleic acid (siNA) molecule that comprises a first nucleotide sequence complementary to a target RNA sequence or a portion thereof, and a second sequence having complementarity to said first sequence, wherein said second sequence is chemically modified in a manner that it can no longer act as a guide sequence for efficiently mediating RNA interference and/or be recognized by cellular proteins that facilitate RNAi.
In one embodiment, the invention features a double stranded short interfering nucleic acid (siNA) molecule that comprises a first nucleotide sequence complementary to a target RNA sequence or a portion thereof, and a second sequence having complementarity to said first sequence, wherein the second sequence is designed or modified in a manner that prevents its entry into the RNAi pathway as a guide sequence or as a sequence that is complementary to a target nucleic acid (e.g., RNA) sequence. Such design or modifications are expected to enhance the activity of siNA and/or improve the specificity of siNA molecules of the invention. These modifications are also expected to minimize any off-target effects and/or associated toxicity.
In one embodiment, the invention features a double stranded short interfering nucleic acid (siNA) molecule that comprises a first nucleotide sequence complementary to a target RNA sequence or a portion thereof, and a second sequence having complementarity to said first sequence, wherein said second sequence is incapable of acting as a guide sequence for mediating RNA interference.
In one embodiment, the invention features a double stranded short interfering nucleic acid (siNA) molecule that comprises a first nucleotide sequence complementary to a target RNA sequence or a portion thereof, and a second sequence having complementarity to said first sequence, wherein said second sequence does not have a terminal 5'-hydroxyl (5'-OH) or 5'-phosphate group.
In one embodiment, the invention features a double stranded short interfering nucleic acid (siNA) molecule that comprises a first nucleotide sequence complementary to a target RNA sequence or a portion thereof, and a second sequence having complementarity to said first sequence, wherem said second sequence comprises a
terminal cap moiety at the 5 '-end of said second sequence. In one embodiment, the terminal cap moiety comprises an inverted abasic, inverted deoxy abasic, inverted nucleotide moiety, a group shown in Figure 10, an alkyl or cycloalkyl group, a heterocycle, or any other group that prevents RNAi activity in which the second sequence serves as a guide sequence or template for RNAi.
In one embodiment, the invention features a double stranded short interfering nucleic acid (siNA) molecule that comprises a first nucleotide sequence complementary to a target RNA sequence or a portion thereof, and a second sequence having complementarity to said first sequence, wherein said second sequence comprises a terminal cap moiety at the 5 '-end and 3 '-end of said second sequence. In one embodiment, each terminal cap moiety individually comprises an inverted abasic, inverted deoxy abasic, inverted nucleotide moiety, a group shown in Figure 10, an alkyl or cycloalkyl group, a heterocycle, or any other group that prevents RNAi activity in which the second sequence serves as a guide sequence or template for RNAi. In one embodiment, the invention features a method for generating siNA molecules of the invention with improved specificity for down regulating or inhibiting the expression of a target nucleic acid (e.g., a DNA or RNA such as a gene or its coπesponding RNA), comprising (a) introducing one or more chemical modifications into the structure of a siNA molecule, and (b) assaying the siNA molecule of step (a) under conditions suitable for isolating siNA molecules having improved specificity. In another embodiment, the chemical modification used to improve specificity comprises terminal cap modifications at the 5 '-end, 3 '-end, or both 5' and 3 '-ends of the siNA molecule. The terminal cap modifications can comprise, for example, structures shown in Figure 10 (e.g. inverted deoxyabasic moieties) or any other chemical modification that renders a portion of the siNA molecule (e.g. the sense strand) incapable of mediating RNA interference against an off target nucleic acid sequence. In a non-limiting example, a siNA molecule is designed such that only the antisense sequence of the siNA molecule can serve as a guide sequence for RISC mediated degradation of a coπesponding target RNA sequence. This can be accomplished by rendering the sense sequence of the siNA inactive by introducing chemical modifications to the sense strand that preclude recognition of the sense strand as a guide sequence by RNAi machinery. In one
embodiment, such chemical modifications comprise any chemical group at the 5 -end oi the sense strand of the siNA, or any other group that serves to render the sense strand inactive as a guide sequence for mediating RNA interference. These modifications, for example, can result in a molecule where the 5 '-end of the sense strand no longer has a free 5 '-hydroxyl (5'-OH) or a free 5 '-phosphate group (e.g., phosphate, diphosphate, triphosphate, cyclic phosphate etc.). Non-limiting examples of such siNA constructs are described herein, such as "Stab 9/10", "Stab 7/8", "Stab 7/19", "Stab 17/22", "Stab 23/24", "Stab 24/25", and "Stab 24/26" (e.g., any siNA having Stab 7, 9, 17, 23, or 24 sense strands) chemistries and variants thereof (see Table II) wherein the 5 '-end and 3'- end of the sense strand of the siNA do not comprise a hydroxyl group or phosphate group.
In one embodiment, the invention features a method for generating siNA molecules of the invention with improved specificity for down regulating or inhibiting the expression of a target nucleic acid (e.g., a DNA or RNA such as a gene or its coπesponding RNA), comprising introducing one or more chemical modifications into the structure of a siNA molecule that prevent a strand or portion of the siNA molecule from acting as a template or guide sequence for RNAi activity. In one embodiment, the inactive strand or sense region of the siNA molecule is the sense strand or sense region of the siNA molecule, i.e. the strand or region of the siNA that does not have complementarity to the target nucleic acid sequence. In one embodiment, such chemical modifications comprise any chemical group at the 5 '-end of the sense strand or region of the siNA that does not comprise a 5'-hydroxyl (5'-OH) or 5'-phosphate group, or any other group that serves to render the sense strand or sense region inactive as a guide sequence for mediating RNA interference. Non-limiting examples of such siNA constructs are described herein, such as "Stab 9/10", "Stab 7/8", "Stab 7/19", "Stab 17/22", "Stab 23/24", "Stab 24/25", and "Stab 24/26" (e.g., any siNA having Stab 7, 9, 17, 23, or 24 sense strands) chemistries and variants thereof (see Table II) wherein the 5 '-end and 3 '-end of the sense strand of the siNA do not comprise a hydroxyl group or phosphate group. In one embodiment, the invention features a method for screening siNA molecules that are active in mediating RNA interference against a target nucleic acid sequence
comprising (a) generating a plurality of unmodified siNA molecules, (b) screening the siNA molecules of step (a) under conditions suitable for isolating siNA molecules that are active in mediating RNA interference against the target nucleic acid sequence, and (c) introducing chemical modifications (e.g. chemical modifications as described herein or as otherwise known in the art) into the active siNA molecules of (b). In one embodiment, the method further comprises re-screening the chemically modified siNA molecules of step (c) under conditions suitable for isolating chemically modified siNA molecules that are active in mediating RNA interference against the target nucleic acid sequence. In one embodiment, the invention features a method for screening chemically modified siNA molecules that are active in mediating RNA interference against a target nucleic acid sequence comprising (a) generating a plurality of chemically modified siNA molecules (e.g. siNA molecules as described herein or as otherwise known in the art), and (b) screening the siNA molecules of step (a) under conditions suitable for isolating chemically modified siNA molecules that are active in mediating RNA interference against the target nucleic acid sequence.
The term "ligand" refers to any compound or molecule, such as a drug, peptide, hormone, or neurotransmitter, that is capable of interacting with another compound, such as a receptor, either directly or indirectly. The receptor that interacts with a ligand can be present on the surface of a cell or can alternately be an intercellular receptor. Interaction of the ligand with the receptor can result in a biochemical reaction, or can simply be a physical interaction or association.
In another embodiment, the invention features a method for generating siNA molecules of the invention with improved bioavailability comprising (a) introducing an excipient formulation to a siNA molecule, and (b) assaying the siNA molecule of step (a) under conditions suitable for isolating siNA molecules having improved bioavailability. Such excipients include polymers such as cyclodextrins, lipids, cationic lipids, polyamines, phospholipids, nanoparticles, receptors, ligands, and others.
In another embodiment, the invention features a method for generating siNA molecules of the invention with improved bioavailability comprising (a) introducing
nucleotides having any of Formulae I- VII or any combination thereof into a siNA molecule, and (b) assaying the siNA molecule of step (a) under conditions suitable for isolating siNA molecules having improved bioavailability.
In another embodiment, polyethylene glycol (PEG) can be covalently attached to siNA compounds of the present invention. The attached PEG can be any molecular weight, preferably from about 2,000 to about 50,000 daltons (Da).
The present invention can be used alone or as a component of a kit having at least one of the reagents necessary to carry out the in vitro or in vivo introduction of RNA to test samples and/or subjects. For example, prefeπed components of the kit include a siNA molecule of the invention and a vehicle that promotes introduction of the siNA into cells of interest as described herein (e.g., using lipids and other methods of transfection known in the art, see for example Beigelman et al, US 6,395,713). The kit can be used for target validation, such as in determining gene function and/or activity, or in drug optimization, and in drug discovery (see for example Usman et al., USSN 60/402,996). Such a kit can also include instructions to allow a user of the kit to practice the invention.
The term "short interfering nucleic acid", "siNA", "short interfering RNA", "siRNA", "short interfering nucleic acid molecule", "short interfering oligonucleotide molecule", or "chemically-modified short interfering nucleic acid molecule" as used herein refers to any nucleic acid molecule capable of inhibiting or down regulating gene expression or viral replication, for example by mediating RNA interference "RNAi" or gene silencing in a sequence-specific manner; see for example Zamore et al, 2000, Cell, 101, 25-33; Bass, 2001, Nature, 411, 428-429; Elbashir et al, 2001, Nature, 411, 494- 498; and Kreutzer et al, International PCT Publication No. WO 00/44895; Zernicka- Goetz et al, International PCT Publication No. WO 01/36646; Fire, International PCT Publication No. WO 99/32619; Plaetinck et al, International PCT Publication No. WO 00/01846; Mello and Fire, International PCT Publication No. WO 01/29058; Deschamps-Depaillette, International PCT Publication No. WO 99/07409; and Li et al, International PCT Publication No. WO 00/44914; Allshire, 2002, Science, 297, 1818- 1819; Volpe et al, 2002, Science, 297, 1833-1837; Jenuwein, 2002, Science, 297, 2215- 2218; and Hall et al, 2002, Science, 297, 2232-2237; Hutvagner and Zamore, 2002,
Science, 297, 2056-60; McManus et al, 2002, RNA, 8, 842-850; Reinhart et al, 2002,
Gene & Dev., 16, 1616-1626; and Reinhart & Barrel, 2002, Science, 297, 1831). Non limiting examples of siNA molecules of the invention are shown in Figures 4-6 herein. For example the siNA can be a double-stranded polynucleotide molecule comprising self-complementary sense and antisense regions, wherein the antisense region comprises nucleotide sequence that is complementary to nucleotide sequence in a target nucleic acid molecule or a portion thereof and the sense region having nucleotide sequence coπesponding to the target nucleic acid sequence or a portion thereof. The siNA can be assembled from two separate oligonucleotides, where one strand is the sense strand and the other is the antisense strand, wherein the antisense and sense strands are self- complementary (i.e. each strand comprises nucleotide sequence that is complementary to nucleotide sequence in the other strand; such as where the antisense strand and sense strand form a duplex or double stranded structure, for example wherein the double stranded region is about 15 to about 30, e.g., about 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29 or 30 base pairs; the antisense strand comprises nucleotide sequence that is complementary to nucleotide sequence in a target nucleic acid molecule or a portion thereof and the sense strand comprises nucleotide sequence coπesponding to the target nucleic acid sequence or a portion thereof (e.g., about 15 to about 25 or more nucleotides of the siNA molecule are complementary to the target nucleic acid or a portion thereof). Alternatively, the siNA is assembled from a single oligonucleotide, where the self-complementary sense and antisense regions of the siNA are linked by means of a nucleic acid based or non-nucleic acid-based linker(s). The siNA can be a polynucleotide with a duplex, asymmetric duplex, haiφin or asymmetric haiφin secondary structure, having self-complementary sense and antisense regions, wherem the antisense region comprises nucleotide sequence that is complementary to nucleotide sequence in a separate target nucleic acid molecule or a portion thereof and the sense region having nucleotide sequence coπesponding to the target nucleic acid sequence or a portion thereof. The siNA can be a circular single-stranded polynucleotide having two or more loop structures and a stem comprising self-complementary sense and antisense regions, wherem the antisense region comprises nucleotide sequence that is complementary to nucleotide sequence in a target nucleic acid molecule or a portion thereof and the sense region having nucleotide sequence coπesponding to the target nucleic acid sequence or a portion thereof, and wherein the circular polynucleotide can
be processed either in vivo or in vitro to generate an active siNA molecule capable of mediating RNAi. The siNA can also comprise a single stranded polynucleotide having nucleotide sequence complementary to nucleotide sequence in a target nucleic acid molecule or a portion thereof (for example, where such siNA molecule does not require the presence within the siNA molecule of nucleotide sequence coπesponding to the target nucleic acid sequence or a portion thereof), wherein the single stranded polynucleotide can further comprise a terminal phosphate group, such as a 5 '-phosphate (see for example Martinez et al, 2002, Cell, 110, 563-574 and Schwarz et al, 2002, Molecular Cell, 10, 537-568), or 5',3'-diphosphate. In certain embodiments, the siNA molecule of the invention comprises separate sense and antisense sequences or regions, wherein the sense and antisense regions are covalently linked by nucleotide or non- nucleotide linkers molecules as is known in the art, or are alternately non-covalently linked by ionic interactions, hydrogen bonding, van der waals interactions, hydrophobic interactions, and/or stacking interactions. In certain embodiments, the siNA molecules of the invention comprise nucleotide sequence that is complementary to nucleotide sequence of a target gene. In another embodiment, the siNA molecule of the invention interacts with nucleotide sequence of a target gene in a manner that causes inhibition of expression of the target gene. As used herein, siNA molecules need not be limited to those molecules containing only RNA, but further encompasses chemically-modified nucleotides and non-nucleotides. In certain embodiments, the short interfering nucleic acid molecules of the invention lack 2'-hydroxy (2'-OH) containing nucleotides. Applicant describes in certain embodiments short interfering nucleic acids that do not require the presence of nucleotides having a 2'-hydroxy group for mediating RNAi and as such, short interfering nucleic acid molecules of the invention optionally do not include any ribonucleotides (e.g., nucleotides having a 2'-OH group). Such siNA molecules that do not require the presence of ribonucleotides within the siNA molecule to support RNAi can however have an attached linker or linkers or other attached or associated groups, moieties, or chains containing one or more nucleotides with 2'-OH groups. Optionally, siNA molecules can comprise ribonucleotides at about 5, 10, 20, 30, 40, or 50% of the nucleotide positions. The modified short interfering nucleic acid molecules of the invention can also be refeπed to as short interfering modified oligonucleotides "siMON." As used herein, the term siNA is meant to be equivalent to
other terms used to describe nucleic acid molecules that are capable of mediating sequence specific RNAi, for example short interfering RNA (siRNA), double-stranded RNA (dsRNA), micro-RNA (miRNA), short haiφin RNA (shRNA), short interfering oligonucleotide, short interfering nucleic acid, short interfering modified oligonucleotide, chemically-modified siRNA, post-transcriptional gene silencing RNA (ptgsRNA), and others. In addition, as used herein, the term RNAi is meant to be equivalent to other terms used to describe sequence specific RNA interference, such as post transcriptional gene silencing, translational inhibition, or epigenetics. For example, siNA molecules of the invention can be used to epigenetically silence genes at both the post-transcriptional level or the pre-transcriptional level. In a non-limiting example, epigenetic regulation of gene expression by siNA molecules of the invention can result from siNA mediated modification of chromatin structure or methylation pattern to alter gene expression (see, for example, Verdel et al, 2004, Science, 303, 672-676; Pal-Bhadra et al, 2004, Science, 303, 669-672; Allshire, 2002, Science, 297, 1818-1819; Volpe et al, 2002, Science, 297, 1833-1837; Jenuwein, 2002, Science, 297, 2215-2218; and Hall et al, 2002, Science, 297, 2232-2237).
In one embodiment, a siNA molecule of the invention is a duplex forming oligonucleotide "DFO", (see for example Figures 14-15 and Vaish et al., USSN 10/727,780 filed December 3, 2003 and International PCT Application No. US04/16390, filed May 24, 2004).
In one embodiment, a siNA molecule of the invention is a multifunctional siNA,
(see for example Figures 16-21 and Jadhav et al, USSN 60/543,480 filed February 10,
2004 and International PCT Application No. US04/16390, filed May 24, 2004). The multifunctional siNA of the invention can comprise sequence targeting, for example, two regions of target RNA.
By "asymmetric haiφin" as used herein is meant a linear siNA molecule comprising an antisense region, a loop portion that can comprise nucleotides or non- nucleotides, and a sense region that comprises fewer nucleotides than the antisense region to the extent that the sense region has enough complementary nucleotides to base pair with the antisense region and form a duplex with loop. For example, an asymmetric haiφin siNA molecule of the invention can comprise an antisense region having length
sufficient to mediate RNAi in a cell or in vitro system (e.g. about 15 to about 30, or about 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, or 30 nucleotides) and a loop region comprising about 4 to about 12 (e.g., about 4, 5, 6, 7, 8, 9, 10, 11, or 12) nucleotides, and a sense region having about 3 to about 25 (e.g., about 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, or 25) nucleotides that are complementary to the antisense region. The asymmetric haiφin siNA molecule can also comprise a 5 '-terminal phosphate group that can be chemically modified. The loop portion of the asymmetric haiφin siNA molecule can comprise nucleotides, non- nucleotides, linker molecules, or conjugate molecules as described herein. By "asymmetric duplex" as used herein is meant a siNA molecule having two separate strands comprising a sense region and an antisense region, wherem the sense region comprises fewer nucleotides than the antisense region to the extent that the sense region has enough complementary nucleotides to base pair with the antisense region and form a duplex. For example, an asymmetric duplex siNA molecule of the invention can comprise an antisense region having length sufficient to mediate RNAi in a cell or in vitro system (e.g. about 15 to about 30, or about 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, or 30 nucleotides) and a sense region having about 3 to about 25 (e.g., about 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, or 25) nucleotides that are complementary to the antisense region. By "modulate" is meant that the expression of the gene, or level of RNA molecule or equivalent RNA molecules encoding one or more proteins or protein subunits, or activity of one or more proteins or protein subunits is up regulated or down regulated, such that expression, level, or activity is greater than or less than that observed in the absence of the modulator. For example, the term "modulate" can mean "inhibit," but the use of the word "modulate" is not limited to this definition.
By "inhibit", "down-regulate", or "reduce", it is meant that the expression of the gene, or level of RNA molecules or equivalent RNA molecules encoding one or more proteins or protein subunits, or activity of one or more proteins or protein subunits, is reduced below that observed in the absence of the nucleic acid molecules (e.g., siNA) of the invention. In one embodiment, inhibition, down-regulation or reduction with an siNA molecule is below that level observed in the presence of an inactive or attenuated
molecule. In another embodiment, inhibition, down-regulation, or reduction with siNA molecules is below that level observed in the presence of, for example, an siNA molecule with scrambled sequence or with mismatches. In another embodiment, inhibition, down-regulation, or reduction of gene expression with a nucleic acid molecule of the instant invention is greater in the presence of the nucleic acid molecule than in its absence. In one embodiment, inhibition, down regulation, or reduction of gene expression is associated with post transcriptional silencing, such as RNAi mediated cleavage of a target nucleic acid molecule (e.g. RNA) or inhibition of translation. In one embodiment, inhibition, down regulation, or reduction of gene expression is associated with pretranscriptional silencing.
By "gene", or "target gene", is meant a nucleic acid that encodes an RNA, for example, nucleic acid sequences including, but not limited to, structural genes encoding a polypeptide. A gene or target gene can also encode a functional RNA (fRNA) or non- coding RNA (ncRNA), such as small temporal RNA (stRNA), micro RNA (miRNA), small nuclear RNA (snRNA), short interfering RNA (siRNA), small nucleolar RNA (snRNA), ribosomal RNA (rRNA), transfer RNA (fRNA) and precursor RNAs thereof. Such non-coding RNAs can serve as target nucleic acid molecules for siNA mediated RNA interference in modulating the activity of fRNA or ncRNA involved in functional or regulatory cellular processes. Abberant fRNA or ncRNA activity leading to disease can therefore be modulated by siNA molecules of the invention. siNA molecules targeting fRNA and ncRNA can also be used to manipulate or alter the genotype or phenotype of a subject, organism or cell, by intervening in cellular processes such as genetic imprinting, transcription, translation, or nucleic acid processing (e.g., transamination, methylation etc.). The target gene can be a gene derived from a cell, an endogenous gene, a transgene, or exogenous genes such as genes of a pathogen, for example a virus, which is present in the cell after infection thereof. The cell containing the target gene can be derived from or contained in any organism, for example a plant, animal, protozoan, virus, bacterium, or fungus. Non-limiting examples of plants include monocots, dicots, or gymnosperms. Non-limiting examples of animals include vertebrates or invertebrates. Non-limiting examples of fungi include molds or yeasts. For a review, see for example Snyder and Gerstein, 2003, Science, 300, 258-260.
By "non-canonical base pair" is meant any non-Watson Crick base pair, such as mismatches and/or wobble base pairs, including flipped mismatches, single hydrogen bond mismatches, trans-type mismatches, triple base interactions, and quadruple base interactions. Non-limiting examples of such non-canonical base pairs include, but are not limited to, AC reverse Hoogsteen, AC wobble, AU reverse Hoogsteen, GU wobble, AA N7 amino, CC 2-carbonyl-amino(Hl)-N3-amino(H2), GA sheared, UC 4-carbonyl- amino, UU imino-carbonyl, AC reverse wobble, AU Hoogsteen, AU reverse Watson Crick, CG reverse Watson Crick, GC N3-amino-amino N3, AA Nl-amino symmetric, AA N7-amino symmetric, GA N7-N1 amino-carbonyl, GA+ carbonyl-amino N7-N1, GG Nl-carbonyl symmetric, GG N3-amino symmetric, CC carbonyl-amino symmetric, CC N3-amino symmetric, UU 2-carbonyl-imino symmetric, UU 4-carbonyl-imino symmetric, AA amino-N3, AA Nl-amino, AC amino 2-carbonyl, AC N3-amino, AC N7-amino, AU amino-4-carbonyl, AU Nl-imino, AU N3-imino, AU N7-imino, CC carbonyl-amino, GA amino-Nl, GA amino-N7, GA carbonyl-amino, GA N3-amino, GC amino-N3, GC carbonyl-amino, GC N3-amino, GC N7-amino, GG amino-N7, GG carbonyl-imino, GG N7-amino, GU amino-2-carbonyl, GU carbonyl-imino, GU imino- 2-carbonyl, GU N7-imino, psiU imino-2-carbonyl, UC 4-carbonyl-amino, UC imino- carbonyl, UU imino-4-carbonyl, AC C2-H-N3, GA carbonyl-C2-H, UU imino-carbonyl 2 carbonyl-C5-H, AC amino(A) N3(C)-carbonyl, GC imino amino-carbonyl, Gpsi imino-2-carbonyl amino-2- carbonyl, and GU imino amino-2-carbonyl base pairs.
By "target" as used herein is meant, any target protein, peptide, or polypeptide, such as encoded by Genbank Accession Nos. shown in Table I. The term "target" also refers to nucleic acid sequences encoding any protein, peptide, or polypeptide (e.g., DNA and RNA). The term "target" is also meant to include other target encoding sequences, such as other isoforms, mutations, splice variants, and polymoφhisms associated with a given target.
By "homologous sequence" is meant, a nucleotide sequence that is shared by one or more polynucleotide sequences, such as genes, gene transcripts and/or non-coding polynucleotides. For example, a homologous sequence can be a nucleotide sequence that is shared by two or more genes encoding related but different proteins, such as different members of a gene family, different protein epitopes, different protein isoforms or
completely divergent genes, such as a cytokine and its coπesponding receptors. A homologous sequence can be a nucleotide sequence that is shared by two or more non- coding polynucleotides, such as noncoding DNA or RNA, regulatory sequences, introns, and sites of transcriptional control or regulation. Homologous sequences can also include conserved sequence regions shared by more than one polynucleotide sequence. Homology does not need to be perfect homology (e.g., 100%), as partially homologous sequences are also contemplated by the instant invention (e.g., 99%, 98%, 97%, 96%, 95%, 94%, 93%, 92%, 91%, 90%, 89%, 88%, 87%, 86%, 85%, 84%, 83%, 82%, 81%, 80% etc.). By "conserved sequence region" is meant, a nucleotide sequence of one or more regions in a polynucleotide does not vary significantly between generations or from one biological system, subject, or organism to another biological system, subject, or organism. The polynucleotide can include both coding and non-coding DNA and RNA.
By "sense region" is meant a nucleotide sequence of a siNA molecule having complementarity to an antisense region of the siNA molecule. In addition, the sense region of a siNA molecule can comprise a nucleic acid sequence having homology with a target nucleic acid sequence.
By "antisense region" is meant a nucleotide sequence of a siNA molecule having complementarity to a target nucleic acid sequence. In addition, the antisense region of a siNA molecule can optionally comprise a nucleic acid sequence having complementarity to a sense region of the siNA molecule.
By "target nucleic acid" or "target polynucleotide" is meant any nucleic acid sequence whose expression or activity is to be modulated. The target nucleic acid can be DNA or RNA. By "complementarity" is meant that a nucleic acid can form hydrogen bond(s) with another nucleic acid sequence by either traditional Watson-Crick or other non-traditional types. In reference to the nucleic molecules of the present invention, the binding free energy for a nucleic acid molecule with its complementary sequence is sufficient to allow the relevant function of the nucleic acid to proceed, e.g., RNAi activity. Determination of binding free energies for nucleic acid molecules is well known in the
art (see, e.g., Turner et al, 1987, CSH Symp. Quant. Biol. LII pp.123-133; Frier et al, 1986, Proc. Nat. Acad. Sci. USA 83:9373-9377; Turner et al, 1987, J. Am. Chem. Soc. 109:3783-3785). A percent complementarity indicates the percentage of contiguous residues in a nucleic acid molecule that can form hydrogen bonds (e.g., Watson-Crick base pairing) with a second nucleic acid sequence (e.g., 5, 6, 7, 8, 9, or 10 nucleotides out of a total of 10 nucleotides in the first oligonucleotide being based paired to a second nucleic acid sequence having 10 nucleotides represents 50%, 60%, 70%, 80%, 90%, and 100% complementary respectively). "Perfectly complementary" means that all the contiguous residues of a nucleic acid sequence will hydrogen bond with the same number of contiguous residues in a second nucleic acid sequence. In one embodiment, a siNA molecule of the invention comprises about 15 to about 30 or more (e.g., about 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, or 30 or more) nucleotides that are complementary to one or more target nucleic acid molecules or a portion thereof.
In one embodiment, siNA molecules of the invention that down regulate or reduce target gene expression are used for preventing or treating diseases, traits, disorders, and/or conditions in a subject or organism.
By "proliferative disease" or "cancer" as used herein is meant, any disease., condition, trait, genotype or phenotype characterized by unregulated cell growth or replication as is known in the art; including AIDS related cancers such as Kaposi's sarcoma; breast cancers; bone cancers such as Osteosarcoma, Chondrosarcomas, Ewmg's sarcoma, Fibrosarcomas, Giant cell tumors, Adamantinomas, and Chordomas; Brain cancers such as Meningiomas, Glioblastomas, Lower-Grade Astrocytomas, Oligodendrocytomas, Pituitary Tumors, Schwannomas, and Metastatic brain cancers; cancers of the head and neck including various lymphomas such as mantle cell lymphoma, non-Hodgkins lymphoma, adenoma, squamous cell carcinoma, laryngeal carcinoma, gallbladder and bile duct cancers, cancers of the retina such as retinoblastoma, cancers of the esophagus, gastric cancers, multiple myeloma, ovarian cancer, uterine cancer, thyroid cancer, testicular cancer, endometrial cancer, melanoma, colorectal cancer, lung cancer, bladder cancer, prostate cancer, lung cancer (including non-small cell lung carcinoma), pancreatic cancer, sarcomas, Wilms' tumor, cervical cancer, head and neck cancer, skin cancers, nasopharyngeal carcinoma, liposarcoma,
epithelial carcinoma, renal cell carcinoma, gallbladder adeno carcinoma, parotid adenocarcinoma, endometrial sarcoma, multidrug resistant cancers, and leukemias such as acute myelogenous leukemia (AML), chronic myelogenous leukemia (CML), acute lymphocytic leukemia (ALL), and chronic lymphocytic leukemia,; and proliferative diseases and conditions, such as neovascularization associated with tumor angiogenesis, macular degeneration (e.g., wet/dry AMD), comeal neovascularization, diabetic retinopathy, neovascular glaucoma, myopic degeneration and other proliferative diseases and conditions such as restenosis and polycystic kidney disease, and any other cancer or proliferative disease, condition, trait, genotype or phenotype that can respond to the modulation of disease related gene expression in a cell or tissue, alone or in combination with other therapies.
By "inflammatory disease" or "inflammatory condition" as used herein is meant any disease, condition, trait, genotype or phenotype characterized by an inflammatory or allergic process as is known in the art, such as inflammation, acute inflammation, chronic inflammation, respiratory disease, atherosclerosis, restenosis, asthma, allergic rhinitis, atopic dermatitis, septic shock, rheumatoid arthritis, inflammatory bowl disease, inflammotory pelvic disease, pain, ocular inflammatory disease, celiac disease, Leigh Syndrome, Glycerol Kinase Deficiency, Familial eosinophilia (FE), autosomal recessive spastic ataxia, laryngeal inflammatory disease; Tuberculosis, Chronic cholecystitis, Bronchiectasis, Silicosis and other pneumoconioses, and any other inflammatory disease, condition, trait, genotype or phenotype that can respond to the modulation of disease related gene expression in a cell or tissue, alone or in combination with other therapies.
By "autoimmune disease" or "autoimmune condition" as used herein is meant, any disease, condition, trait, genotype or phenotype characterized by autoimmunity as is known in the art, such as multiple sclerosis, diabetes mellitus, lupus, celiac disease, Crohn's disease, ulcerative colitis, Guillain-Baπe syndrome, scleroderms, Goodpasture's syndrome, Wegener's granulomatosis, autoimmune epilepsy, Rasmussen's encephalitis, Primary biliary sclerosis, Sclerosing cholangitis, Autoimmune hepatitis, Addison's disease, Hashimoto's thyroiditis, Fibromyalgia, Menier's syndrome; transplantation rejection (e.g., prevention of allograft rejection) pernicious anemia, rheumatoid arthritis, systemic lupus erythematosus, dermatomyositis, Sjogren's syndrome, lupus
erythematosus, multiple sclerosis, myasthenia gravis, Reiter's syndrome, Grave's disease, and any other autoimmune disease, condition, trait, genotype or phenotype that can respond to the modulation of disease related gene expression in a cell or tissue, alone or in combination with other therapies. By "infectious disease" is meant any disease, condition, trait, genotype or phenotype associated with an infectious agent, such as a virus, bacteria, fungus, prion, or parasite. Non-limiting examples of various viral genes that can be targeted using siNA molecules of the invention include Hepatitis C Virus (HCV, for example Genbank Accession Nos: DI l 168, D50483.1, L38318 and S82227), Hepatitis B Virus (HBV, for example GenBank Accession No. AF100308.1), Human Immunodeficiency Virus type 1 (HIV-1, for example GenBank Accession No. U51188), Human Immunodeficiency Virus type 2 (HIV-2, for example GenBank Accession No. X60667), West Nile Virus (WNV for example GenBank accession No. NC_001563), cytomegalovirus (CMV for example GenBank Accession No. NC_001347), respiratory syncytial virus (RSV for example GenBank Accession No. NC_001781), influenza virus (for example GenBank Accession No. AF037412, rhinovirus (for example, GenBank accession numbers: D00239, X02316, X01087, L24917, M16248, K02121, X01087), papillomavirus (for example GenBank Accession No. NC_001353), Heφes Simplex Virus (HSV for example GenBank Accession No. NC_001345), and other viruses such as HTLV (for example GenBank Accession No. AJ430458). Due to the high sequence variability of many viral genomes, selection of siNA molecules for broad therapeutic applications would likely involve the conserved regions of the viral genome. Nonlimiting examples of conserved regions of the viral genomes include but are not limited to 5 '-Non Coding Regions (NCR), 3'- Non Coding Regions (NCR) and/or internal ribosome entry sites (IRES). siNA molecules designed against conserved regions of various viral genomes will enable efficient inhibition of viral replication in diverse patient populations and may ensure the effectiveness of the siNA molecules against viral quasi species which evolve due to mutations in the non-conserved regions of the viral genome. Non-limiting examples of bacterial infections include Actinomycosis, Anthrax, Aspergillosis, Bacteremia, Bacterial Infections and Mycoses, Bartonella Infections, Botulism, Brucellosis, Burkholderia Infections, Campylobacter Infections, Candidiasis, Cat-Scratch Disease, Chlamydia Infections, Cholera , Clostridium Infections, Coccidioidomycosis,
Cross Infection, Cryptococcosis, Dermatomycoses, Dermatomycoses, Diphtheria, Ehrlichiosis, Escherichia coli Infections, Fasciitis, Necrotizing, Fusobacterium Infections, Gas Gangrene, Gram-Negative Bacterial Infections, Gram-Positive Bacterial Infections, Histoplasmosis, Impetigo, Klebsiella Infections, Legionellosis, Leprosy, Leptospirosis, Listeria Infections, Lyme Disease, Maduromycosis, Melioidosis, Mycobacterium Infections, Mycoplasma Infections, Mycoses, Nocardia Infections, Onychomycosis, Omithosis, Plague, Pneumococcal Infections, Pseudomonas Infections, Q Fever, Rat-Bite Fever, Relapsing Fever, Rheumatic Fever, Rickettsia Infections, Rocky Mountain Spotted Fever, Salmonella Infections, Scarlet Fever, Scrub Typhus, Sepsis, Sexually Transmitted Diseases - Bacterial, Bacterial Skin Diseases, Staphylococcal Infections, Streptococcal Infections, Tetanus, Tick-Borne Diseases, Tuberculosis, Tularemia, Typhoid Fever, Typhus, Epidemic Louse-Bome, Vibrio Infections, Yaws, Yersinia Infections, Zoonoses, and Zygomycosis. Non-limiting examples of fungal infections include Aspergillosis, Blastomycosis, Coccidioidomycosis, Cryptococcosis, Fungal Infections of Fingernails and Toenails, Fungal Sinusitis, Histoplasmosis, Histoplasmosis, Mucomiycosis, Nail Fungal Infection, Paracoccidioidomycosis, Sporotrichosis, Valley Fever (Coccidioidomycosis), and Mold Allergy.
By "neurologic disease" or "neurological disease" is meant any disease, disorder, or condition affecting the central or peripheral nervous system, inlcuding ADHD, AIDS -
Neurological Complications, Absence of the Septum Pellucidum, Acquired Epileptiform
Aphasia, Acute Disseminated Encephalomyelitis, Adrenoleukodystrophy, Agenesis of the Coφus Callosum, Agnosia, Aicardi Syndrome, Alexander Disease, Alpers' Disease,
Alternating Hemiplegia, Alzheimer's Disease, Amyotrophic Lateral Sclerosis, Anencephaly, Aneurysm, Angelman Syndrome, Angiomatosis, Anoxia, Aphasia,
Apraxia, Arachnoid Cysts, Arachnoiditis, Arnold-Chiari Malformation, Arteriovenous
Malformation, Aspartame, Asperger Syndrome, Ataxia Telangiectasia, Ataxia, Attention
Deficit-Hyperactivity Disorder, Autism, Autonomic Dysfunction, Back Pain, Barth
Syndrome, Batten Disease, Behcet's Disease, Bell's Palsy, Benign Essential Blepharospasm, Benign Focal Amyotiophy, Benign Intiacranial Hypertension,
Bernhardt-Roth Syndrome, Binswanger's Disease, Blepharospasm, Bloch-Sulzberger
Syndrome, Brachial Plexus Birth Injuries, Brachial Plexus Injuries, Bradbury-Eggleston
Syndrome, Brain Aneurysm, Brain Injury, Brain and Spinal Tumors, Brown-Sequard Syndrome, Bulbospinal Muscular Atrophy, Canavan Disease, Caφal Tunnel Syndrome, Causalgia, Cavernomas, Cavernous Angioma, Cavernous Malformation, Central Cervical Cord Syndrome, Central Cord Syndrome, Central Pain Syndrome, Cephalic Disorders, Cerebellar Degeneration, Cerebellar Hypoplasia, Cerebral Aneurysm, Cerebral Arteriosclerosis, Cerebral Atrophy, Cerebral Beriberi, Cerebral Gigantism, Cerebral Hypoxia, Cerebral Palsy, Cerebro-Oculo-Facio-Skeletal Syndrome, Charcot- Marie-Tooth Disorder, Chiari Malformation, Chorea, Choreoacanthocytosis, Chronic Inflammatory Demyelinating Polyneuropathy (CIDP), Chronic Orthostatic Intolerance, Chronic Pain, Cockayne Syndrome Type II, Coffin Lowry Syndrome, Coma, including Persistent Vegetative State, Complex Regional Pain Syndrome, Congenital Facial Diplegia, Congenital Myasthenia, Congenital Myopathy, Congenital Vascular Cavernous Malformations, Corticobasal Degeneration, Cranial Arteritis, Craniosynostosis, Creutzfeldt-Jakob Disease, Cumulative Trauma Disorders, Cushing's Syndrome, Cytomegalic Inclusion Body Disease (CIBD), Cytomegalovirus Infection, Dancing Eyes-Dancing Feet Syndrome, Dandy- Walker Syndrome, Dawson Disease, De Morsier's Syndrome, Dejerine-Klumpke Palsy, Dementia - Multi-Infarct, Dementia - Subcortical, Dementia With Lewy Bodies, Dermatomyositis, Developmental Dyspraxia, Devic's Syndrome, Diabetic Neuropathy, Diffuse Sclerosis, Dravet's Syndrome, Dysautonomia, Dysgraphia, Dyslexia, Dysphagia, Dyspraxia, Dystonias, Early Infantile Epileptic Encephalopathy, Empty Sella Syndrome, Encephalitis Lethargica, Encephalitis and Meningitis, Encephaloceles, Encephalopathy, Encephalotrigeminal Angiomatosis, Epilepsy, Erb's Palsy, Erb-Duchenne and Dejerine-Klumpke Palsies, Fabry's Disease, Fahr's Syndrome, Fainting, Familial Dysautonomia, Familial Hemangioma, Familial Idiopathic Basal Ganglia Calcification, Familial Spastic Paralysis, Febrile Seizures (e.g., GEFS and GEFS plus), Fisher Syndrome, Floppy Infant Syndrome, Friedreich's Ataxia, Gaucher's Disease, Gerstmann's Syndrome, Gerstmann-Straussler-Scheinker Disease, Giant Cell Arteritis, Giant Cell Inclusion Disease, Globoid Cell Leukodystrophy, Glossopharyngeal Neuralgia, Guillain-Baπe Syndrome, HTLV-1 Associated Myelopathy, Hallervorden-Spatz Disease, Head Injury, Headache, Hemicrama Continua, Hemifacial Spasm, Hemiplegia Alterans, Hereditary Neuropathies, Hereditary Spastic Paraplegia, Heredopathia Atactica Polyneuritiformis, Heφes Zoster Oticus, Heφes
Zoster, Hirayama Syndrome, Holoprosencephaly, Huntington's Disease, Hydranencephaly, Hydrocephalus - Normal Pressure, Hydrocephalus, Hydromyelia, Hypercortisolism, Hypersomnia, Hypertonia, Hypotonia, Hypoxia, Immune-Mediated Encephalomyelitis, Inclusion Body Myositis, Incontinentia Pigmenti, Infantile Hypotonia, Infantile Phytanic Acid Storage Disease, Infantile Refsum Disease, Infantile Spasms, Inflammatory Myopathy, Intestinal Lipodystrophy, Intiacranial Cysts, Intracranial Hypertension, Isaac's Syndrome, Joubert Syndrome, Kearns-Sayre Syndrome, Kennedy's Disease, Kinsbourne syndrome, Kleine-Levin syndrome, Klippel Feil Syndrome, Klippel-Trenaunay Syndrome (KTS), Kliiver-Bucy Syndrome, Korsakoffs Amnesic Syndrome, Krabbe Disease, Kugelberg-Welander Disease, Kuru, Lambert-Eaton Myasthenic Syndrome, Landau-Kleffher Syndrome, Lateral Femoral Cutaneous Nerve Entrapment, Lateral Medullary Syndrome, Learning Disabilities, Leigh's Disease, Lennox-Gastaut Syndrome, Lesch-Nyhan Syndrome, Leukodystrophy, Levine-Critchley Syndrome, Lewy Body Dementia, Lissencephaly, Locked-In Syndrome, Lou Gehrig's Disease, Lupus - Neurological Sequelae, Lyme Disease - Neurological Complications, Machado-Joseph Disease, Macrencephaly, Megalencephaly, Melkersson-Rosenthal Syndrome, Meningitis, Menkes Disease, Meralgia Paresthetica, Metachromatic Leukodystrophy, Microcephaly, Migraine, Miller Fisher Syndrome, Mini-Strokes, Mitochondrial Myopathies, Mobius Syndrome, Monomelic Amyotiophy, Motor Neuron Diseases, Moyamoya Disease, Mucolipidoses, Mucopolysaccharidoses, Multi-Infarct Dementia, Multifocal Motor Neuropathy, Multiple Sclerosis, Multiple System Atrophy with Orthostatic Hypotension, Multiple System Atrophy, Muscular Dystrophy, Myasthenia - Congenital, Myasthenia Gravis, Myelinoclastic Diffuse Sclerosis, Myoclonic Encephalopathy of Infants, Myoclonus, Myopathy - Congenital, Myopathy - Thyrotoxic, Myopathy, Myotonia Congenita, Myotonia, Narcolepsy, Neuroacanthocytosis, Neurodegeneration with Brain Iron Accumulation, Neurofibromatosis, Neuroleptic Malignant Syndrome, Neurological Complications of AIDS, Neurological Manifestations of Pompe Disease, Neuromyelitis Optica, Neuromyotonia, Neuronal Ceroid Lipofuscinosis, Neuronal Migration Disorders, Neuropathy - Hereditary, Neurosarcoidosis, Neurotoxicity, Nevus Cavernosus, Niemann- Pick Disease, O'Sullivan-McLeod Syndrome, Occipital Neuralgia, Occult Spinal Dysraphism Sequence, Ohtahara Syndrome, Olivopontocerebellar Atrophy, Opsoclonus
Myoclonus, Orthostatic Hypotension, Overuse Syndrome, Pain - Chronic, Paraneoplastic Syndromes, Paresthesia, Parkinson's Disease, Parmyotonia Congenita, Paroxysmal Choreoathetosis, Paroxysmal Hemicrania, Parry-Romberg, Pelizaeus-Merzbacher Disease, Pena Shokeir II Syndrome, Perineural Cysts, Periodic Paralyses, Peripheral Neuropathy, Periventricular Leukomalacia, Persistent Vegetative State, Pervasive Developmental Disorders, Phytanic Acid Storage Disease, Pick's Disease, Piriformis Syndrome, Pituitary Tumors, Polymyositis, Pompe Disease, Porencephaly, Post-Polio Syndrome, Postheφetic Neuralgia, Postinfectious Encephalomyelitis, Postural Hypotension, Postural Orthostatic Tachycardia Syndrome, Postural Tachycardia Syndrome, Primary Lateral Sclerosis, Prion Diseases, Progressive Hemifacial Atrophy, Progressive Locomotor Ataxia, Progressive Multifocal Leukoencephalopathy, Progressive Sclerosing Poliodystrophy, Progressive Supranuclear Palsy, Pseudotumor Cerebri, Pyridoxine Dependent and Pyridoxine Responsive Siezure Disorders, Ramsay Hunt Syndrome Type I, Ramsay Hunt Syndrome Type II, Rasmussen's Encephalitis and other autoimmune epilepsies, Reflex Sympathetic Dystrophy Syndrome, Refsum Disease - Infantile, Refsum Disease, Repetitive Motion Disorders, Repetitive Stress Injuries, Restless Legs Syndrome, Retrovirus-Associated Myelopathy, Rett Syndrome, Reye's Syndrome, Riley-Day Syndrome, SUNCT Headache, Sacral Nerve Root Cysts, Saint Vitus Dance, Salivary Gland Disease, Sandhoff Disease, Schilder's Disease, Schizencephaly, Seizure Disorders, Septo-Optic Dysplasia, Severe Myoclonic Epilepsy of Infancy (SMEI), Shaken Baby Syndrome, Shingles, Shy-Drager Syndrome, Sjogren's Syndrome, Sleep Apnea, Sleeping Sickness, Soto's Syndrome, Spasticity, Spina Bifida, Spinal Cord Infarction, Spinal Cord Injury, Spinal Cord Tumors, Spinal Muscular Atrophy, Spinocerebellar Atrophy, Steele-Richardson-Olszewski Syndrome, Stiff-Person Syndrome, Striatonigral Degeneration, Stroke, Sturge-Weber Syndrome, Subacute Sclerosing Panencephalitis, Subcortical Arteriosclerotic Encephalopathy, Swallowing Disorders, Sydenham Chorea, Syncope, Syphilitic Spinal Sclerosis, Syringohydromyelia, Syringomyelia, Systemic Lupus Erythematosus, Tabes Dorsalis, Tardive Dyskinesia, Tarlov Cysts, Tay-Sachs Disease, Temporal Arteritis, Tethered Spinal Cord Syndrome, Thomsen Disease, Thoracic Outlet Syndrome, Thyrotoxic Myopathy, Tic Douloureux, Todd's Paralysis, Tourette Syndrome, Transient Ischemic Attack, Transmissible Spongiform Encephalopathies, Transverse Myelitis, Traumatic Brain Injury, Tremor,
Trigeminal Neuralgia, Tropical Spastic Paraparesis, Tuberous Sclerosis, Vascular Erectile Tumor, Vasculitis including Temporal Arteritis, Von Economo's Disease, Von Hippel-Lindau disease (VHL), Von Recklinghausen's Disease, Wallenberg's Syndrome, Werdnig-Hoffman Disease, Wernicke-Korsakoff Syndrome, West Syndrome, Whipple's Disease, Williams Syndrome, Wilson's Disease, X-Linked Spinal and Bulbar Muscular Atrophy, and Zellweger Syndrome.
By "respiratory disease" is meant, any disease or condition affecting the respiratory tract, such as asthma, chronic obstructive pulmonary disease or "COPD", allergic rhinitis, sinusitis, pulmonary vasoconstriction, inflammation, allergies, impeded respiration, respiratory distress syndrome, cystic fibrosis, pulmonary hypertension, pulmonary vasoconstriction, emphysema, and any other respiratory disease, condition, trait, genotype or phenotype that can respond to the modulation of disease related gene expression in a cell or tissue, alone or in combination with other therapies.
By "cardiovascular disease" is meant and disease or condition affecting the heart and vasculature, inlcuding but not limited to, coronary heart disease (CHD), cerebrovascular disease (CVD), aortic stenosis, peripheral vascular disease, atherosclerosis, arteriosclerosis, myocardial infarction (heart attack), cerebrovascular diseases (stroke), transient ischaemic attacks (TIA), angina (stable and unstable), atrial fibrillation, aπhythmia, vavular disease, congestive heart failure, hypercholoesterolemia, type I hyperlipoproteinemia, type II hyperlipoproteinemia, type III hyperlipoproteinemia, type IV hyperlipoproteinemia, type V hyperlipoproteinemia, secondary hypertrigliceridemia, and familial lecithin cholesterol acyltransferase deficiency.
By "ocular disease" as used herein is meant, any disease, condition, trait, genotype or phenotype of the eye and related structures as is known in the art, such as Cystoid Macular Edema, Asteroid Hyalosis, Pathological Myopia and Posterior Staphyloma, Toxocariasis (Ocular Larva Migrans), Retinal Vein Occlusion, Posterior Vitreous Detachment, Tractional Retinal Tears, Epiretinal Membrane, Diabetic Retinopathy, Lattice Degeneration, Retinal Vein Occlusion, Retinal Artery Occlusion, Macular Degeneration (e.g., age related macular degeneration such as wet AMD or dry AMD), Toxoplasmosis, Choroidal Melanoma, Acquired Retinoschisis, Hollenhorst Plaque,
Idiopathic Central Serous Chorioretmopathy, Macular Hole, Presumed Ocular
Histoplasmosis Syndrome, Retinal Macroaneursym, Retimtis Pigmentosa, Keunai Detachment, Hypertensive Retmopathy, Retinal Pigment Epithelium (RPE) Detachment, Papillophlebitis, Ocular Ischemic Syndrome, Coats' Disease, Leber's Miliary Aneurysm, Conjunctival Neoplasms, Allergic Conjunctivitis, Vernal Conjunctivitis, Acute Bacterial Conjunctivitis, Allergic Conjunctivitis &Vernal Keratoconjunctivitis, Viral Conjunctivitis, Bacterial Conjunctivitis, Chlamydial & Gonococcal Conjunctivitis, Conjunctival Laceration, Episcleritis, Scleritis, Pingueculitis, Pterygium, Superior Limbic Keratoconjunctivitis (SLK of Theodore), Toxic Conjunctivitis, Conjunctivitis with Pseudomembrane, Giant Papillary Conjunctivitis, Terrien's Marginal Degeneration, Acanthamoeba Keratitis, Fungal Keratitis, Filamentary Keratitis, Bacterial Keratitis, Keratitis Sicca/Dry Eye Syndrome, Bacterial Keratitis, Heφes Simplex Keratitis, Sterile Comeal Infiltrates, Phlyctenulosis, Corneal Abrasion & Recuπent Corneal Erosion, Corneal Foreign Body, Chemical Burs, Epithelial Basement Membrane Dystrophy (EBMD), Thygeson's Superficial Punctate Keratopathy, Comeal Laceration, Salzmann's Nodular Degeneration, Fuchs' Endothelial Dystrophy, Crystalline Lens Subluxation, Ciliary-Block Glaucoma, Primary Open-Angle Glaucoma, Pigment Dispersion Syndrome and Pigmentary Glaucoma, Pseudoexfoliation Syndrom and Pseudoexfoliative Glaucoma, Anterior Uveitis, Primary Open Angle Glaucoma, Uveitic Glaucoma & Glaucomatocyclitic Crisis, Pigment Dispersion Syndrome & Pigmentary Glaucoma, Acute Angle Closure Glaucoma, Anterior Uveitis, Hyphema, Angle Recession Glaucoma, Lens Induced Glaucoma, Pseudoexfoliation Syndrome and Pseudoexfoliative Glaucoma, Axenfeld-Rieger Syndrome, Neovascular Glaucoma, Pars Planitis, Choroidal Rupture, Duane's Retraction Syndrome, Toxic/Nutritional Optic Neuropathy, Abeπant Regeneration of Cranial Nerve III, Intiacranial Mass Lesions, Carotid-Cavernous Sinus Fistula, Anterior Ischemic Optic Neuropathy, Optic Disc Edema & Papilledema, Cranial Nerve III Palsy, Cranial Nerve IV Palsy, Cranial Nerve VI Palsy, Cranial Nerve VII (Facial Nerve) Palsy, Homer's Syndrome, Internuclear Ophthalmoplegia, Optic Nerve Head Hypoplasia, Optic Pit, Tonic Pupil, Optic Nerve Head Drusen, Demyelinating Optic Neuropathy (Optic Neuritis, Retrobulbar Optic Neuritis), Amaurosis Fugax and Transient Ischemic Attack, Pseudotumor Cerebri, Pituitary Adenoma, MoUuscum Contagiosum, Canaliculitis, Verruca and Papilloma, Pediculosis and Pthiriasis, Blepharitis, Hordeolum, Preseptal Cellulitis, Chalazion, Basal Cell Carcinoma, Heφes
Zoster Ophthalmicus, Pediculosis & Phthiriasis, Blow-out Fracture, Chronic Epiphora, Dacryocystitis, Heφes Simplex Blepharitis, Orbital Cellulitis, Senile Entropion, and Squamous Cell Carcinoma.
By "metabolic disease" is meant any disease or condition affecting metabolic pathways as in known in the art. Metabolic disease can result in an abnormal metabolic process, either congenital due to inherited enzyme abnormality (inborn eπors of metabolism) or acquired due to disease of an endocrine organ or failure of a metabolically important organ such as the liver. In one embodiment, metabolic disease includes obesity, insulin resistance, and diabetes (e.g., type I and/or type II diabetes). In one embodiment of the present invention, each sequence of a siNA molecule of the invention is independently about 15 to about 30 nucleotides in length, in specific embodiments about 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, or 30 nucleotides in length. In another embodiment, the siNA duplexes of the invention independently comprise about 15 to about 30 base pairs (e.g., about 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, or 30). In another embodiment, one or more strands of the siNA molecule of the invention independently comprises about 15 to about 30 nucleotides (e.g., about 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, or 30) that are complementary to a target nucleic acid molecule. In yet another embodiment, siNA molecules of the invention comprising haiφin or circular structures are about 35 to about 55 (e.g., about 35, 40, 45, 50 or 55) nucleotides in length, or about 38 to about 44 (e.g., about 38, 39, 40, 41, 42, 43, or 44) nucleotides in length and comprising about 15 to about 25 (e.g., about 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, or 25) base pairs. Exemplary siNA molecules of the invention are shown in Figures 4-5.
As used herein "cell" is used in its usual biological sense, and does not refer to an entire multicellular organism, e.g., specifically does not refer to a human. The cell can be present in an organism, e.g., birds, plants and mammals such as humans, cows, sheep, apes, monkeys, swine, dogs, and cats. The cell can be prokaryotic (e.g., bacterial cell) or eukaryotic (e.g., mammalian or plant cell). The cell can be of somatic or germ line origin, totipotent or pluripotent, dividing or non-dividing. The cell can also be derived from or can comprise a gamete or embryo, a stem cell, or a fully differentiated cell.
The siNA molecules of the invention are added directly, or can be complexed with cationic lipids, packaged within liposomes, or otherwise delivered to target cells or tissues. The nucleic acid or nucleic acid complexes can be locally administered to relevant tissues ex vivo, or in vivo through direct dermal application, transdermal application, or injection, with or without their incoφoration in biopolymers.
In another aspect, the invention provides mammalian cells containing one or more siNA molecules of this invention. The one or more siNA molecules can independently be targeted to the same or different sites.
By "RNA" is meant a molecule comprising at least one ribonucleotide residue. By "ribonucleotide" is meant a nucleotide with a hydroxyl group at the 2' position of a β-D- ribofuranose moiety. The terms include double-stranded RNA, single-stranded RNA, isolated RNA such as partially purified RNA, essentially pure RNA, synthetic RNA, recombinantly produced RNA, as well as altered RNA that differs from naturally occurring RNA by the addition, deletion, substitution and/or alteration of one or more nucleotides. Such alterations can include addition of non-nucleotide material, such as to the end(s) of the siNA or internally, for example at one or more nucleotides of the RNA. Nucleotides in the RNA molecules of the instant invention can also comprise non- standard nucleotides, such as non-naturally occurring nucleotides or chemically synthesized nucleotides or deoxynucleotides. These altered RNAs can be refeπed to as analogs or analogs of naturally-occurring RNA.
By "subject" is meant an organism, which is a donor or recipient of explanted cells or the cells themselves. "Subject" also refers to an organism to which the nucleic acid molecules of the invention can be administered. A subject can be a mammal or mammalian cells, including a human or human cells. The term "phosphorothioate" as used herein refers to an internucleotide linkage having Formula I, wherein Z and/or W comprise a sulfur atom. Hence, the term phosphorothioate refers to both phosphorothioate and phosphorodithioate internucleotide linkages.
The term "phosphonoacetate" as used herein refers to an internucleotide linkage having Formula I, wherein Z and/or W comprise an acetyl or protected acetyl group.
The term "thiophosphonoacetate" as used herein refers to an internucleotide linkage having Formula I, wherein Z comprises an acetyl or protected acetyl group and W comprises a sulfur atom or alternately W comprises an acetyl or protected acetyl group and Z comprises a sulfur atom.
The term "universal base" as used herein refers to nucleotide base analogs that form base pairs with each of the natural DNA/RNA bases with little discrimination between them. Non-limiting examples of universal bases include C-phenyl, C-naphthyl and other aromatic derivatives, inosine, azole carboxamides, and nitroazole derivatives such as 3-nitropyπole, 4-nitroindole, 5-nitroindole, and 6-nitroindole as known in the art (see for example Loakes, 2001, Nucleic Acids Research, 29, 2437-2447).
The term "acyclic nucleotide" as used herein refers to any nucleotide having an acyclic ribose sugar, for example where any of the ribose carbons (Cl, C2, C3, C4, or C5), are independently or in combination absent from the nucleotide.
The nucleic acid molecules of the instant invention, individually, or in combination or in conjunction with other drugs, can be used to for preventing or treating diseases, traits, disorders, and/or conditions described herein or otherwise known in the art, in a subject or organism. For example, the siNA molecules can be administered to a subject or can be administered to other appropriate cells evident to those skilled in the art, individually or in combination with one or more drugs under conditions suitable for the treatment.
In a further embodiment, the siNA molecules can be used in combination with other known treatments to prevent or treat preventing or treating diseases, traits, disorders, and/or conditions described herein or otherwise known in the art, in a subject or organism. For example, the described molecules could be used in combination with one or more known compounds, treatments, or procedures to prevent or treat diseases, traits, disorders, and/or conditions described herein or otherwise known in the art, in a subject or organism.
In one embodiment, the invention features an expression vector comprising a nucleic acid sequence encoding at least one siNA molecule of the invention, in a manner which allows expression of the siNA molecule. For example, the vector can contain sequence(s) encoding both strands of a siNA molecule comprising a duplex. The vector can also contain sequence(s) encoding a single nucleic acid molecule that is self- complementary and thus forms a siNA molecule. Non-limiting examples of such expression vectors are described in Paul et al, 2002, Nature Biotechnology, 19, 505; Miyagishi and Taira, 2002, Nature Biotechnology, 19, 497; Lee et al, 2002, Nature Biotechnology, 19, 500; and Novina et al, 2002, Nature Medicine, advance online publication doi: 10.1038/nm725.
In another embodiment, the invention features a mammalian cell, for example, a human cell, including an expression vector of the invention.
In yet another embodiment, the expression vector of the invention comprises a sequence for a siNA molecule having complementarity to a RNA molecule refeπed to by Genbank Accession numbers, for example Genbank Accession Nos. shown in Table I.
In one embodiment, an expression vector of the invention comprises a nucleic acid sequence encoding two or more siNA molecules, which can be the same or different.
In another aspect of the invention, siNA molecules that interact with target RNA molecules and down-regulate gene encoding target RNA molecules (for example target RNA molecules refeπed to by Genbank Accession numbers herein) are expressed from transcription units inserted into DNA or RNA vectors. The recombinant vectors can be DNA plasmids or viral vectors. siNA expressing viral vectors can be constructed based on, but not limited to, adeno-associated virus, retrovirus, adenovirus, or alphavirus. The recombinant vectors capable of expressing the siNA molecules can be delivered as described herein, and persist in target cells. Alternatively, viral vectors can be used that provide for transient expression of siNA molecules. Such vectors can be repeatedly administered as necessary. Once expressed, the siNA molecules bind and down-regulate gene function or expression via RNA interference (RNAi). Delivery of siNA expressing vectors can be systemic, such as by intravenous or intramuscular administration, by administration to target cells ex-planted from a subject followed by reintroduction into
the subject, or by any other means that would allow for introduction into the desired target cell.
By "vectors" is meant any nucleic acid- and/or viral-based technique used to deliver a desired nucleic acid. Other features and advantages of the invention will be apparent from the following description of the prefeπed embodiments thereof, and from the claims.
BRIEF DESCRIPTION OF THE DRAWINGS
Figure 1 shows a non-limiting example of a scheme for the synthesis of siNA molecules. The complementary siNA sequence strands, stiand 1 and strand 2, are synthesized in tandem and are connected by a cleavable linkage, such as a nucleotide succinate or abasic succinate, which can be the same or different from the cleavable linker used for solid phase synthesis on a solid support. The synthesis can be either solid phase or solution phase, in the example shown, the synthesis is a solid phase synthesis. The synthesis is performed such that a protecting group, such as a dimethoxytrityl group, remains intact on the terminal nucleotide of the tandem oligonucleotide. Upon cleavage and deprotection of the oligonucleotide, the two siNA strands spontaneously hybridize to form a siNA duplex, which allows the purification of the duplex by utilizing the properties of the terminal protecting group, for example by applying a trityl on purification method wherein only duplexes/oligonucleotides with the terminal protecting group are isolated.
Figure 2 shows a MALDI-TOF mass spectrum of a purified siNA duplex synthesized by a method of the invention. The two peaks shown coπespond to the predicted mass of the separate siNA sequence strands. This result demonstrates that the siNA duplex generated from tandem synthesis can be purified as a single entity using a simple trityl-on purification methodology.
Figure 3 shows a non-limiting proposed mechanistic representation of target RNA degradation involved in RNAi. Double-stranded RNA (dsRNA), which is generated by RNA-dependent RNA polymerase (RdRP) from foreign single-stranded RNA, for example viral, transposon, or other exogenous RNA, activates the DICER enzyme that in
turn generates siNA duplexes. Alternately, synthetic or expressed siNA can be introduced directly into a cell by appropriate means. An active siNA complex forms which recognizes a target RNA, resulting in degradation of the target RNA by the RISC endonuclease complex or in the synthesis of additional RNA by RNA-dependent RNA polymerase (RdRP), which can activate DICER and result in additional siNA molecules, thereby amplifying the RNAi response.
Figure 4A-F shows non-limiting examples of chemically-modified siNA constructs of the present invention. In the figure, N stands for any nucleotide (adenosine, guanosine, cytosine, uridine, or optionally thymidine, for example thymidine can be substituted in the overhanging regions designated by parenthesis (N N). Various modifications are shown for the sense and antisense strands of the siNA constructs.
Figure 4A: The sense strand comprises 21 nucleotides wherein the two terminal 3 '-nucleotides are optionally base paired and wherem all nucleotides present are ribonucleotides except for (N N) nucleotides, which can comprise ribonucleotides, deoxynucleotides, universal bases, or other chemical modifications described herein. The antisense strand comprises 21 nucleotides, optionally having a 3 '-terminal glyceryl moiety wherein the two terminal 3 '-nucleotides are optionally complementary to the target RNA sequence, and wherem all nucleotides present are ribonucleotides except for (N N) nucleotides, which can comprise ribonucleotides, deoxynucleotides, universal bases, or other chemical modifications described herein. A modified internucleotide linkage, such as a phosphorothioate, phosphorodithioate or other modified internucleotide linkage as described herein, shown as "s", optionally connects the (N N) nucleotides in the antisense strand.
Figure 4B: The sense strand comprises 21 nucleotides wherein the two terminal 3 '-nucleotides are optionally base paired and wherein all pyrimidine nucleotides that may be present are 2'deoxy-2'-fluoro modified nucleotides and all purine nucleotides that may be present are 2'-0-methyl modified nucleotides except for (N N) nucleotides, which can comprise ribonucleotides, deoxynucleotides, universal bases, or other chemical modifications described herein. The antisense strand comprises 21 nucleotides, optionally having a 3'-terminal glyceryl moiety and wherein the two terminal 3'- nucleotides are optionally complementary to the target RNA sequence, and wherein all
pyrimidine nucleotides that may be present are 2'-deoxy-2'-fluoro modified nucleotides and all purine nucleotides that may be present are 2 '-O-methyl modified nucleotides except for (N N) nucleotides, which can comprise ribonucleotides, deoxynucleotides, universal bases, or other chemical modifications described herein. A modified internucleotide linkage, such as a phosphorothioate, phosphorodithioate or other modified internucleotide linkage as described herein, shown as "s", optionally connects the (N N) nucleotides in the sense and antisense stiand.
Figure 4C: The sense strand comprises 21 nucleotides having 5'- and 3'- terminal cap moieties wherein the two terminal 3 '-nucleotides are optionally base paired and wherein all pyrimidine nucleotides that may be present are 2'-0-methyl or 2'-deoxy-2'- fluoro modified nucleotides except for (N N) nucleotides, which can comprise ribonucleotides, deoxynucleotides, universal bases, or other chemical modifications described herein. The antisense strand comprises 21 nucleotides, optionally having a 3'- ter inal glyceryl moiety and wherein the two terminal 3 '-nucleotides are optionally complementary to the target RNA sequence, and wherein all pyrimidine nucleotides that may be present are 2,-deoxy-2'-fluoro modified nucleotides except for (N N) nucleotides, which can comprise ribonucleotides, deoxynucleotides, universal bases, or other chemical modifications described herein. A modified internucleotide linkage, such as a phosphorothioate, phosphorodithioate or other modified internucleotide linkage as described herein, shown as "s", optionally connects the (N N) nucleotides in the antisense strand.
Figure 4D: The sense strand comprises 21 nucleotides having 5'- and 3'- terminal cap moieties wherein the two terminal 3'-nucleotides are optionally base paired and wherein all pyrimidine nucleotides that may be present are 2'-deoxy-2'-fluoro modified nucleotides except for (N N) nucleotides, which can comprise ribonucleotides, deoxynucleotides, universal bases, or other chemical modifications described herein and wherein and all purine nucleotides that may be present are 2'-deoxy nucleotides. The antisense strand comprises 21 nucleotides, optionally having a 3 '-terminal glyceryl moiety and wherein the two terminal 3 '-nucleotides are optionally complementary to the target RNA sequence, wherein all pyrimidine nucleotides that may be present are 2'- deoxy-2'-fluoro modified nucleotides and all purine nucleotides that may be present are
2'-0-methyl modified nucleotides except for (N N) nucleotides, which can comprise ribonucleotides, deoxynucleotides, universal bases, or other chemical modifications described herein. A modified internucleotide linkage, such as a phosphorothioate, phosphorodithioate or other modified internucleotide linkage as described herein, shown as "s", optionally connects the (N N) nucleotides in the antisense strand.
Figure 4E: The sense strand comprises 21 nucleotides having 5'- and 3'- terminal cap moieties wherein the two terminal 3 '-nucleotides are optionally base paired and wherein all pyrimidine nucleotides that may be present are 2'-deoxy-2'-fluoro modified nucleotides except for (N N) nucleotides, which can comprise ribonucleotides, deoxynucleotides, universal bases, or other chemical modifications described herein. The antisense strand comprises 21 nucleotides, optionally having a 3 '-terminal glyceryl moiety and wherein the two terminal 3 '-nucleotides are optionally complementary to the target RNA sequence, and wherein all pyrimidine nucleotides that may be present are 2'- deoxy-2'-fluoro modified nucleotides and all purine nucleotides that may be present are 2'-0-methyl modified nucleotides except for (N N) nucleotides, which can comprise ribonucleotides, deoxynucleotides, universal bases, or other chemical modifications described herein. A modified internucleotide linkage, such as a phosphorothioate, phosphorodithioate or other modified internucleotide linkage as described herein, shown as "s", optionally connects the (N N) nucleotides in the antisense strand. Figure 4F: The sense strand comprises 21 nucleotides having 5'- and 3'- terminal cap moieties wherein the two terminal 3'-nucleotides are optionally base paired and wherein all pyrimidine nucleotides that may be present are 2'-deoxy-2'-fluoro modified nucleotides except for (N N) nucleotides, which can comprise ribonucleotides, deoxynucleotides, universal bases, or other chemical modifications described herein and wherein and all purine nucleotides that may be present are 2'-deoxy nucleotides. The antisense strand comprises 21 nucleotides, optionally having a 3'-terrninal glyceryl moiety and wherein the two terminal 3 '-nucleotides are optionally complementary to the target RNA sequence, and having one 3 '-terminal phosphorothioate internucleotide linkage and wherem all pyrimidine nucleotides that may be present are 2'-deoxy-2'-fluoro modified nucleotides and all purine nucleotides that may be present are 2'-deoxy nucleotides except for (N N) nucleotides, which can comprise ribonucleotides,
deoxynucleotides, universal bases, or other chemical modifications described herein. A modified internucleotide linkage, such as a phosphorothioate, phosphorodithioate or other modified internucleotide linkage as described herein, shown as "s", optionally connects the (N N) nucleotides in the antisense strand. The antisense strand of constructs A-F comprise sequence complementary to any target nucleic acid sequence of the invention. Furthermore, when a glyceryl moiety (L) is present at the 3 '-end of the antisense strand for any construct shown in Figure 4 A-F, the modified internucleotide linkage is optional.
Figure 5A-F shows non-limiting examples of specific chemically-modified siNA sequences of the invention. A-F applies the chemical modifications described in Figure 4A-F to a target siNA sequence. Such chemical modifications can be applied to any target sequence and/or target polymoφhism sequence.
Figure 6 shows non-limiting examples of different siNA constructs of the invention. The examples shown (constructs 1, 2, and 3) have 19 representative base pairs; however, different embodiments of the invention include any number of base pairs described herein. Bracketed regions represent nucleotide overhangs, for example, comprising about 1, 2, 3, or 4 nucleotides in length, preferably about 2 nucleotides. Constructs 1 and 2 can be used independently for RNAi activity. Construct 2 can comprise a polynucleotide or non-nucleotide linker, which can optionally be designed as a biodegradable linker. In one embodiment, the loop structure shown in construct 2 can comprise a biodegradable linker that results in the formation of construct 1 in vivo and/or in vitro. In another example, construct 3 can be used to generate construct 2 under the same principle wherein a linker is used to generate the active siNA construct 2 in vivo and/or in vitro, which can optionally utilize another biodegradable linker to generate the active siNA construct 1 in vivo and/or in vitro. As such, the stability and/or activity of the sϋSTA constructs can be modulated based on the design of the siNA construct for use in vivo or in vitro and/or in vitro.
Figure 7A-C is a diagrammatic representation of a scheme utilized in generating an expression cassette to generate siNA haiφin constructs.
Figure 7A: A DNA oligomer is synthesized with a S'-restriction site (Rl) sequence followed by a region having sequence identical (sense region of siNA) to a predetermined target sequence, wherein the sense region comprises, for example, about
19, 20, 21, or 22 nucleotides (N) in length, which is followed by a loop sequence of defined sequence (X), comprising, for example, about 3 to about 10 nucleotides.
Figure 7B: The synthetic construct is then extended by DNA polymerase to generate a hairpin structure having self-complementary sequence that will result in a siNA transcript having specificity for a target sequence and having self-complementary sense and antisense regions. Figure 7C: The construct is heated (for example to about 95°C) to linearize the sequence, thus allowing extension of a complementary second DNA strand using a primer to the 3 '-restriction sequence of the first strand. The double-stranded DNA is then inserted into an appropriate vector for expression in cells. The construct can be designed such that a 3 '-terminal nucleotide overhang results from the transcription, for example, by engineering restriction sites and/or utilizing a poly-U termination region as described in Paul et al, 2002, Nature Biotechnology, 29, 505-508.
Figure 8A-C is a diagrammatic representation of a scheme utilized in generating an expression cassette to generate double-stranded siNA constructs.
Figure 8A: A DNA oligomer is synthesized with a 5'-restriction (Rl) site sequence followed by a region having sequence identical (sense region of siNA) to a predetermined target sequence, wherein the sense region comprises, for example, about
19, 20, 21, or 22 nucleotides (N) in length, and which is followed by a 3'-restriction site
(R2) which is adjacent to a loop sequence of defined sequence (X).
Figure 8B: The synthetic construct is then extended by DNA polymerase to generate a haiφin structure having self-complementary sequence.
Figure 8C: The construct is processed by restriction enzymes specific to Rl and R2 to generate a double-stranded DNA which is then inserted into an appropriate vector for expression in cells. The transcription cassette is designed such that a U6 promoter region flanks each side of the dsDNA which generates the separate sense and antisense
strands of the siNA. Poly T termination sequences can be added to the constructs to generate U overhangs in the resulting transcript.
Figure 9A-E is a diagrammatic representation of a method used to determine target sites for siNA mediated RNAi within a particular target nucleic acid sequence, such as messenger RNA.
Figure 9A: A pool of siNA oligonucleotides are synthesized wherem the antisense region of the siNA constructs has complementarity to target sites across the target nucleic acid sequence, and wherein the sense region comprises sequence complementary to the antisense region of the siNA. Figure 9B&C: (Figure 9B) The sequences are pooled and are inserted into vectors such that (Figure 9C) transfection of a vector into cells results in the expression of the siNA.
Figure 9D: Cells are sorted based on phenotypic change that is associated with modulation of the target nucleic acid sequence. Figure 9E: The siNA is isolated from the sorted cells and is sequenced to identify efficacious target sites within the target nucleic acid sequence.
Figure 10 shows non-limiting examples of different stabilization chemistries (1- 10) that can be used, for example, to stabilize the 3 '-end of siNA sequences of the invention, including (1) [3 -3'] -inverted deoxyribose; (2) deoxyribonucleotide; (3) [5'-3']- 3 '-deoxyribonucleotide; (4) [5'-3']-ribonucleotide; (5) [5'-3']-3'-0-methyl ribonucleotide; (6) 3 '-glyceryl; (7) [3 *-5']-3 '-deoxyribonucleotide; (8) [3'-3']-deoxyribonucleotide; (9) [5'- 2']-deoxyribonucleotide; and (10) [5-3']-dideoxyribonucleotide. In addition to modified and unmodified backbone chemistries indicated in the figure, these chemistries can be combined with different backbone modifications as described herein, for example, backbone modifications having Formula I. In addition, the 2'-deoxy nucleotide shown 5' to the terminal modifications shown can be another modified or unmodified nucleotide or non-nucleotide described herein, for example modifications having any of Formulae I- VII or any combination thereof.
Figure 11 shows a non-limiting example of a strategy used to identify chemically modified siNA constructs of the invention that are nuclease resistance while preserving the ability to mediate RNAi activity. Chemical modifications are introduced into the siNA construct based on educated design parameters (e.g. introducing 2'-mofications, base modifications, backbone modifications, terminal cap modifications etc). The modified construct in tested in an appropriate system (e.g. human serum for nuclease resistance, shown, or an animal model for PK/delivery parameters). In parallel, the siNA construct is tested for RNAi activity, for example in a cell culture system such as a luciferase reporter assay). Lead siNA constructs are then identified which possess a particular characteristic while maintaining RNAi activity, and can be further modified and assayed once again. This same approach can be used to identify siNA-conjugate molecules with improved pharmacokinetic profiles, delivery, and RNAi activity.
Figure 12 shows non-limiting examples of phosphorylated siNA molecules of the invention, including linear and duplex constructs and asymmetric derivatives thereof. Figure 13 shows non-limiting examples of chemically modified terminal phosphate groups of the invention.
Figure 14A shows a non-limiting example of methodology used to design self complementary DFO constructs utilizing palidrome and/or repeat nucleic acid sequences that are identified in a target nucleic acid sequence, (i) A palindrome or repeat sequence is identified in a nucleic acid target sequence, (ii) A sequence is designed that is complementary to the target nucleic acid sequence and the palindrome sequence, (iii) An inverse repeat sequence of the non-palindrome/repeat portion of the complementary sequence is appended to the 3 '-end of the complementary sequence to generate a self complementary DFO molecule comprising sequence complementary to the nucleic acid target. (iv) The DFO molecule can self-assemble to form a double stranded oligonucleotide. Figure 14B shows a non-limiting representative example of a duplex forming oligonucleotide sequence. Figure 14C shows a non-limiting example of the self assembly schematic of a representative duplex forming oligonucleotide sequence. Figure 14D shows a non-limiting example of the self assembly schematic of a representative duplex forming oligonucleotide sequence followed by interaction with a target nucleic acid sequence resulting in modulation of gene expression.
Figure 15 shows a non-limiting example of the design of self complementary DFO constructs utilizing palidrome and/or repeat nucleic acid sequences that are incoφorated into the DFO constructs that have sequence complementary to any target nucleic acid sequence of interest. Incoφoration of these palindrome/repeat sequences allow the design of DFO constructs that form duplexes in which each strand is capable of mediating modulation of target gene expression, for example by RNAi. First, the target sequence is identified. A complementary sequence is then generated in which nucleotide or non-nucleotide modifications (shown as X or Y) are introduced into the complementary sequence that generate an artificial palindrome (shown as XYXYXY in the Figure). An inverse repeat of the non-palindrome/repeat complementary sequence is appended to the 3 '-end of the complementary sequence to generate a self complementary DFO comprising sequence complementary to the nucleic acid target. The DFO can self- assemble to form a double stranded oligonucleotide.
Figure 16 shows non-limiting examples of multifunctional siNA molecules of the invention comprising two separate polynucleotide sequences that are each capable of mediating RNAi directed cleavage of differing target nucleic acid sequences. Figure 16A shows a non-limiting example of a multifunctional siNA molecule having a first region that is complementary to a first target nucleic acid sequence (complementary region 1) and a second region that is complementary to a second target nucleic acid sequence (complementary region 2), wherein the first and second complementary regions are situated at the 3 '-ends of each polynucleotide sequence in the multifunctional siNA. The dashed portions of each polynucleotide sequence of the multifunctional siNA construct have complementarity with regard to coπesponding portions of the siNA duplex, but do not have complementarity to the target nucleic acid sequences. Figure 16B shows a non-limiting example of a multifunctional siNA molecule having a first region that is complementary to a first target nucleic acid sequence (complementary region 1) and a second region that is complementary to a second target nucleic acid sequence (complementary region 2), wherein the first and second complementary regions are situated at the 5 '-ends of each polynucleotide sequence in the multifunctional siNA. The dashed portions of each polynucleotide sequence of the multifunctional siNA construct have complementarity with regard to coπesponding portions of the siNA duplex, but do not have complementarity to the target nucleic acid sequences.
Figure 17 shows non-limiting examples of multifunctional siNA molecules of the invention comprising a single polynucleotide sequence comprising distinct regions that are each capable of mediating RNAi directed cleavage of differing target nucleic acid sequences. Figure 17A shows a non-limiting example of a multifunctional siNA molecule having a first region that is complementary to a first target nucleic acid sequence (complementary region 1) and a second region that is complementary to a second target nucleic acid sequence (complementary region 2), wherein the second complementary region is situated at the 3 '-end of the polynucleotide sequence in the multifunctional siNA. The dashed portions of each polynucleotide sequence of the multifunctional siNA construct have complementarity with regard to coπesponding portions of the siNA duplex, but do not have complementarity to the target nucleic acid sequences. Figure 17B shows a non-limiting example of a multifunctional siNA molecule having a first region that is complementary to a first target nucleic acid sequence (complementary region 1) and a second region that is complementary to a second target nucleic acid sequence (complementary region 2), wherein the first complementary region is situated at the 5 '-end of the polynucleotide sequence in the multifunctional siNA. The dashed portions of each polynucleotide sequence of the multifunctional siNA construct have complementarity with regard to coπesponding portions of the siNA duplex, but do not have complementarity to the target nucleic acid sequences. In one embodiment, these multifunctional siNA constructs are processed in vivo or in vitro to generate multifunctional siNA constructs as shown in Figure 16.
Figure 18 shows non-limiting examples of multifunctional siNA molecules of the invention comprising two separate polynucleotide sequences that are each capable of mediating RNAi directed cleavage of differing target nucleic acid sequences and wherein the multifunctional siNA construct further comprises a self complementary, palindrome, or repeat region, thus enabling shorter bifuctional siNA constructs that can mediate RNA interference against differing target nucleic acid sequences. Figure 18A shows a non- limiting example of a multifunctional siNA molecule having a first region that is complementary to a first target nucleic acid sequence (complementary region 1) and a second region that is complementary to a second target nucleic acid sequence (complementary region 2), wherein the first and second complementary regions are situated at the 3 '-ends of each polynucleotide sequence in the multifunctional siNA, and
wherein the first and second complementary regions further comprise a self complementary, palindrome, or repeat region. The dashed portions of each polynucleotide sequence of the multifunctional siNA construct have complementarity with regard to corresponding portions of the siNA duplex, but do not have complementarity to the target nucleic acid sequences. Figure 18B shows a non-limiting example of a multifunctional siNA molecule having a first region that is complementary to a first target nucleic acid sequence (complementary region 1) and a second region that is complementary to a second target nucleic acid sequence (complementary region 2), wherein the first and second complementary regions are situated at the 5 '-ends of each polynucleotide sequence in the multifunctional siNA, and wherein the first and second complementary regions further comprise a self complementary, palindrome, or repeat region. The dashed portions of each polynucleotide sequence of the multifunctional siNA construct have complementarity with regard to coπesponding portions of the siNA duplex, but do not have complementarity to the target nucleic acid sequences. Figure 19 shows non-limiting examples of multifunctional siNA molecules of the invention comprising a single polynucleotide sequence comprising distinct regions that are each capable of mediating RNAi directed cleavage of differing target nucleic acid sequences and wherem the multifunctional siNA construct further comprises a self complementary, palindrome, or repeat region, thus enabling shorter bifuctional siNA constructs that can mediate RNA interference against differing target nucleic acid sequences. Figure 19A shows a non-limiting example of a multifunctional siNA molecule having a first region that is complementary to a first target nucleic acid sequence (complementary region 1) and a second region that is complementary to a second target nucleic acid sequence (complementary region 2), wherein the second complementary region is situated at the 3 '-end of the polynucleotide sequence in the multifunctional siNA, and wherein the first and second complementary regions further comprise a self complementary, palindrome, or repeat region. The dashed portions of each polynucleotide sequence of the multifunctional siNA construct have complementarity with regard to coπesponding portions of the siNA duplex, but do not have complementarity to the target nucleic acid sequences. Figure 19B shows a non- limiting example of a multifunctional siNA molecule having a first region that is complementary to a first target nucleic acid sequence (complementary region 1) and a
second region that is complementary to a second target nucleic acid sequence (complementary region 2), wherein the first complementary region is situated at the 5'- end of the polynucleotide sequence in the multifunctional siNA, and wherein the first and second complementary regions further comprise a self complementary, palindrome, or repeat region. The dashed portions of each polynucleotide sequence of the multifunctional siNA construct have complementarity with regard to coπesponding portions of the siNA duplex, but do not have complementarity to the target nucleic acid sequences. In one embodiment, these multifunctional siNA constructs are processed in vivo or in vitro to generate multifunctional siNA constructs as shown in Figure 18. Figure 20 shows a non-limiting example of how multifunctional siNA molecules of the invention can target two separate target nucleic acid molecules, such as separate RNA molecules encoding differing proteins, for example, a cytokine and its coπesponding receptor, differing viral strains, a virus and a cellular protein involved in viral infection or replication, or differing proteins involved in a common or divergent biologic pathway that is implicated in the maintenance of progression of disease. Each strand of the multifunctional siNA construct comprises a region having complementarity to separate target nucleic acid molecules. The multifunctional siNA molecule is designed such that each strand of the siNA can be utilized by the RISC complex to initiate RNA interference mediated cleavage of its coπesponding target. These design parameters can include destabilization of each end of the siNA construct (see for example Schwarz et al, 2003, Cell, 115, 199-208). Such destabilization can be accomplished for example by using guanosine-cytidine base pairs, alternate base pairs (e.g., wobbles), or destabilizing chemically modified nucleotides at terminal nucleotide positions as is known in the art. Figure 21 shows a non-limiting example of how multifunctional siNA molecules of the invention can target two separate target nucleic acid sequences within the same target nucleic acid molecule, such as alternate coding regions of a RNA, coding and non- coding regions of a RNA, or alternate splice variant regions of a RNA. Each strand of the multifunctional siNA construct comprises a region having complementarity to the separate regions of the target nucleic acid molecule. The multifunctional siNA molecule is designed such that each strand of the siNA can be utilized by the RISC complex to
initiate RNA interference mediated cleavage of its coπesponding target region. These design parameters can include destabilization of each end of the siNA construct (see for example Schwarz et al, 2003, Cell, 115, 199-208). Such destabilization can be accomplished for example by using guanosme-cytidine base pairs, alternate base pairs (e.g., wobbles), or destabilizing chemically modified nucleotides at terminal nucleotide positions as is known in the art.
DETAILED DESCRIPTION OF THE INVENTION
Mechanism of Action of Nucleic Acid Molecules of the Invention
The discussion that follows discusses the proposed mechanism of RNA interference mediated by short interfering RNA as is presently known, and is not meant to be limiting and is not an admission of prior art. Applicant demonstrates herein that chemically-modified short interfering nucleic acids possess similar or improved capacity to mediate RNAi as do siRNA molecules and are expected to possess improved stability and activity in vivo; therefore, this discussion is not meant to be limiting only to siRNA and can be applied to siNA as a whole. By "improved capacity to mediate RNAi" or
"improved RNAi activity" is meant to include RNAi activity measured in vitro and/or in vivo where the RNAi activity is a reflection of both the ability of the siNA to mediate
RNAi and the stability of the siNAs of the invention. In this invention, the product of these activities can be increased in vitro and/or in vivo compared to an all RNA siRNA or a siNA containing a plurality of ribonucleotides. In some cases, the activity or stability of the siNA molecule can be decreased (i.e., less than ten-fold), but the overall activity of the siNA molecule is enhanced in vitro and/or in vivo.
RNA interference refers to the process of sequence specific post-transcriptional gene silencing in animals mediated by short interfering RNAs (siRNAs) (Fire et al, 1998, Nature, 391 , 806). The coπesponding process in plants is commonly refeπed to as post-transcriptional gene silencing or RNA silencing and is also refeπed to as quelling in fungi. The process of post-transcriptional gene silencing is thought to be an evolutionarily-conserved cellular defense mechanism used to prevent the expression of foreign genes which is commonly shared by diverse flora and phyla (Fire et al, 1999, Trends Genet., 15, 358). Such protection from foreign gene expression may have
evolved in response to the production of double-stranded RNAs (dsRNAs) derived from viral infection or the random integration of transposon elements into a host genome via a cellular response that specifically destroys homologous single-stranded RNA or viral genomic RNA. The presence of dsRNA in cells triggers the RNAi response though a mechanism that has yet to be fully characterized. This mechanism appears to be different from the interferon response that results from dsRNA-mediated activation of protein kinase PKR and 2', 5'-oligoadenylate synthetase resulting in non-specific cleavage of mRNA by ribonuclease L.
The presence of long dsRNAs in cells stimulates the activity of a ribonuclease III enzyme referred to as Dicer. Dicer is involved in the processing of the dsRNA into short pieces of dsRNA known as short interfering RNAs (siRNAs) (Berstein et al, 2001, Nature, 409, 363). Short interfering RNAs derived from Dicer activity are typically about 21 to about 23 nucleotides in length and comprise about 19 base pair duplexes. Dicer has also been implicated in the excision of 21- and 22-nucleotide small temporal RNAs (stRNAs) from precursor RNA of conserved structure that are implicated in translational control (Hutvagner et al, 2001, Science, 293, 834). The RNAi response also features an endonuclease complex containing a siRNA, commonly refeπed to as an RNA-induced silencing complex (RISC), which mediates cleavage of single-stranded RNA having sequence homologous to the siRNA. Cleavage of the target RNA takes place in the middle of the region complementary to the guide sequence of the siRNA duplex (Elbashir et al, 2001, Genes Dev., 15, 188). In addition, RNA interference can also involve small RNA (e.g., micro-RNA or miRNA) mediated gene silencing, presumably though cellular mechanisms that regulate chromatm structure and thereby prevent transcription of target gene sequences (see for example Allshire, 2002, Science, 297, 1818-1819; Volpe et al, 2002, Science, 297, 1833-1837; Jenuwein, 2002, Science, 297, 2215-2218; and Hall et al, 2002, Science, 297, 2232-2237). As such, siNA molecules of the invention can be used to mediate gene silencing via interaction with RNA transcripts or alternately by interaction with particular gene sequences, wherein such interaction results in gene silencing either at the transcriptional level or post- transcriptional level.
RNAi has been studied in a variety of systems. Fire et al, 1998, Nature, 391, 806, were the first to observe RNAi in C. elegans. Wianny and Goetz, 1999, Nature Cell Biol, 2, 70, describe RNAi mediated by dsRNA in mouse embryos. Hammond et al, 2000, Nature, 404, 293, describe RNAi in Drosophila cells transfected with dsRNA. Elbashir et al, 2001, Nature, 411, 494, describe RNAi induced by introduction of duplexes of synthetic 21 -nucleotide RNAs in cultured mammalian cells including human embryonic kidney and HeLa cells. Recent work in Drosophila embryonic lysates has revealed certain requirements for siRNA length, structure, chemical composition, and sequence that are essential to mediate efficient RNAi activity. These studies have shown that 21 nucleotide siRNA duplexes are most active when containing two 2-nucleotide 3'- terminal nucleotide overhangs. Furthermore, substitution of one or both siRNA strands with 2'-deoxy or 2'-0-methyl nucleotides abolishes RNAi activity, whereas substitution of 3'-terminal siRNA nucleotides with deoxy nucleotides was shown to be tolerated. Mismatch sequences in the center of the siRNA duplex were also shown to abolish RNAi activity. In addition, these studies also indicate that the position of the cleavage site in the target RNA is defined hy the 5'-end of the siRNA guide sequence rather than the 3'- end (Elbashir et al, 2001, EMBO J., 20, 6877). Other studies have indicated that a 5'- phosphate on the target-complementary strand of a siRNA duplex is required for siRNA activity and that ATP is utilized to maintain the 5'-phosphate moiety on the siRNA (Nykanen et al, 2001, Cell, 107, 309); however, siRNA molecules lacking a 5'- phosphate are active when introduced exogenously, suggesting that 5'-phosphorylation of siRNA constructs may occur in vivo.
Synthesis of Nucleic Acid Molecules
Synthesis of nucleic acids greater than 100 nucleotides in length is difficult using automated methods, and the therapeutic cost of such molecules is prohibitive. In this invention, small nucleic acid motifs ("small" refers to nucleic acid motifs no more than 100 nucleotides in length, preferably no more than 80 nucleotides in length, and most preferably no more than 50 nucleotides in length; e.g., individual siNA oligonucleotide sequences or siNA sequences synthesized in tandem) are preferably used for exogenous delivery. The simple structure of these molecules increases the ability of the nucleic acid
to invade targeted regions of protein and/or RNA structure. Exemplary molecules of the instant invention are chemically synthesized, and others can similarly be synthesized.
Oligonucleotides (e.g., certain modified oligonucleotides or portions of oligonucleotides lacking ribonucleotides) are synthesized using protocols known in the art, for example as described in Caruthers et al, 1992, Methods in Enzymology 211, 3- 19, Thompson et al, International PCT Publication No. WO 99/54459, Wincott et al, 1995, Nucleic Acids Res. 23, 2677-2684, Wincott et al, 1997, Methods Mol. Bio., 74, 59, Brennan et al, 1998, Biotechnol Bioeng., 61, 33-45, and Brennan, U.S. Pat. No. 6,001 ,311. All of these references are incoφorated herein by reference. The synthesis of oligonucleotides makes use of common nucleic acid protecting and coupling groups, such as dimethoxytrityl at the 5'-end, and phosphoramidites at the 3'-end. In a non- limiting example, small scale syntheses are conducted on a 394 Applied Biosystems, Inc. synthesizer using a 0.2 μmol scale protocol with a 2.5 min coupling step for 2'-0- methylated nucleotides and a 45 second coupling step for 2'-deoxy nucleotides or 2'- deoxy-2'-fluoro nucleotides. Table III outlines the amounts and the contact times of the reagents used in the synthesis cycle. Alternatively, syntheses at the 0.2 μmol scale can • be performed on a 96-well plate synthesizer, such as the instrument produced by Protogene (Palo Alto, CA) with minimal modification to the cycle. A 33 -fold excess (60 μL of 0.11 M = 6.6 μmol) of 2'-0-methyl phosphoramidite and a 105-fold excess of S- ethyl tetrazole (60 μL of 0.25 M = 15 μmol) can be used in each coupling cycle of 2'-0- methyl residues relative to polymer-bound 5'-hydroxyl. A 22-fold excess (40 μL of 0.11 M = 4.4 μmol) of deoxy phosphoramidite and a 70-fold excess of S-ethyl tetrazole (40 μL of 0.25 M = 10 μmol) can be used in each coupling cycle of deoxy residues relative to polymer-bound 5'-hydroxyl. Average coupling yields on the 394 Applied Biosystems, Inc. synthesizer, determined by colorimetric quantitation of the trityl fractions, are typically 97.5-99%. Other oligonucleotide synthesis reagents for the 394 Applied Biosystems, Inc. synthesizer include the following: detritylation solution is 3% TCA in methylene chloride (ABI); capping is performed with 16% N-methyl imidazole in THF (ABI) and 10% acetic anhydride/10% 2,6-lutidine in THF (ABI); and oxidation solution is 16.9 mM b., 49 mM pyridine, 9% water in THF (PerSeptive Biosystems, Inc.). Burdick & Jackson Synthesis Grade acetonitrile is used directly from the reagent bottle. S-Ethyltetrazole solution (0.25 M in acetonitrile) is made up from the solid obtained
from American International Chemical, Inc. Alternately, for the introduction of phosphorothioate linkages, Beaucage reagent (3H-l,2-Benzodithiol-3-one 1,1-dioxide, 0.05 M in acetonitrile) is used.
Deprotection of the D]NA-based oligonucleotides is performed as follows: the polymer-bound trityl-on oligoribonucleotide is transfeπed to a 4 mL glass screw top vial and suspended in a solution of 40% aqueous methylamine (1 mL) at 65 °C for 10 minutes. After cooling to -20 °C, the supernatant is removed from the polymer support. The support is washed three times with 1.0 mL of EtOH:MeCN:H20/3:l:l, vortexed and the supernatant is then added to the first supernatant. The combined supernatants, containing the oligoribonucleotide, are dried to a white powder.
The method of synthesis used for RNA including certain siNA molecules of the invention follows the procedure as described in Usman et al, 1987, J. Am. Chem. Soc, 109, 7845; Scaringe et al, 1990, Nucleic Acids Res., 18, 5433; and Wincott et al, 1995, Nucleic Acids Res. 23, 2677-2684 Wincott et al, 1997, Methods Mol. Bio., 74, 59, and makes use of common nucleic acid protecting and coupling groups, such as dimethoxytrityl at the 5 '-end, and phosphoramidites at the 3 '-end. In a non-limiting example, small scale syntheses are conducted on a 394 Applied Biosystems, Inc. synthesizer using a 0.2 μmol scale protocol with a 7.5 min coupling step for alkylsilyl protected nucleotides and a 2.5 min coupling step for 2'-0-methylated nucleotides. Table III outlines the amounts and the contact times of the reagents used in the synthesis cycle. Alternatively, syntheses at the 0.2 μmol scale can be done on a 96-well plate synthesizer, such as the instrument produced by Protogene (Palo Alto, CA) with minimal modification to the cycle. A 33-fold excess (60 μL of 0.11 M = 6.6 μmol) of 2'-0- methyl phosphoramidite and a 75-fold excess of S-ethyl tetrazole (60 μL of 0.25 M = 15 μmol) can be used in each coupling cycle of 2'-0-methyl residues relative to polymer- bound 5'-hydroxyl. A 66-fold excess (120 μL of 0.11 M = 13.2 μmol) of alkylsilyl (ribo) protected phosphoramidite and a 150-fold excess of S-ethyl tetrazole (120 μL of 0.25 M = 30 μmol) can be used in each coupling cycle of ribo residues relative to polymer- bound 5'-hydroxyl. Average coupling yields on the 394 Applied Biosystems, Inc. synthesizer, determined by colorimetric quantitation of the tiityl fractions, are typically 97.5-99%. Other oligonucleotide synthesis reagents for the 394 Applied Biosystems,
Inc. synthesizer include the following: detritylation solution is 3% TCA in methylene chloride (ABI); capping is performed with 16% N-methyl imidazole in THF (ABI) and 10% acetic anhydride/10% 2,6-lutidine in THF (ABI); oxidation solution is 16.9 mM I2, 49 mM pyridine, 9% water in THF (PerSeptive Biosystems, Inc.). Burdick & Jackson Synthesis Grade acetonitrile is used directly from the reagent bottle. S-Ethyltetiazole solution (0.25 M in acetonitrile) is made up from the solid obtained from American International Chemical, Inc. Alternately, for the introduction of phosphorothioate linkages, Beaucage reagent (3H-l,2-Benzodithiol-3-one l,l-dioxide0.05 M in acetonitrile) is used. Deprotection of the RNA is performed using either a two-pot or one-pot protocol.
For the two-pot protocol, the polymer-bound trityl-on oligoribonucleotide is transfeπed to a 4 mL glass screw top vial and suspended in a solution of 40% aq. methylamine (1 mL) at 65 °C for 10 min. After cooling to -20 °C, the supernatant is removed from the polymer support. The support is washed three times with 1.0 mL of EtOH:MeCN:H20/3:l:l, vortexed and the supernatant is then added to the first supernatant. The combined supernatants, containing the oligoribonucleotide, are dried to a white powder. The base deprotected oligoribonucleotide is resuspended in anhydrous TEA/HF/NMP solution (300 μL of a solution of 1.5 mL N-methylpyπolidinone, 750 μL TEA and 1 mL TEA»3HF to provide a 1.4 M HF concentration) and heated to 65 °C. After 1.5 h, the oligomer is quenched with 1.5 M NH4HCO3.
Alternatively, for the one-pot protocol, the polymer-bound trityl-on oligoribonucleotide is transferred to a 4 mL glass screw top vial and suspended in a solution of 33% ethanolic methylamine/DMSO: 1/1 (0.8 mL) at 65 °C for 15 minutes. The vial is brought to room temperature TEA«3HF (0.1 mL) is added and the vial is heated at 65 °C for 15 minutes. The sample is cooled at —20 °C and then quenched with I.5 M NH4HCO3.
For purification of the trityl-on oligomers, the quenched NH4HCO3 solution is loaded onto a C-18 containing cartridge that had been prewashed with acetonitrile followed by 50 mM TEAA. After washing the loaded cartridge with water, the RNA is detritylated with 0.5% TFA for 13 minutes. The cartridge is then washed again with
water, salt exchanged with 1 M NaCl and washed with water again. The oligonucleotide is then eluted with 30% acetonitrile.
The average stepwise coupling yields are typically >98% (Wincott et al, 1995 Nucleic Acids Res. 23, 2677-2684). Those of ordinary skill in the art will recognize that the scale of synthesis can be adapted to be larger or smaller than the example described above including but not limited to 96-well format.
Alternatively, the nucleic acid molecules of the present invention can be synthesized separately and joined together post-synthetically, for example, by ligation (Moore et al, 1992, Science 256, 9923; Draper et al, International PCT publication No. WO 93/23569; Shabarova et al, 1991, Nucleic Acids Research 19, 4247; Bellon et al, 1997, Nucleosides & Nucleotides, 16, 951; Bellon et al, 1997, Bioconjugate Chem. 8, 204), or by hybridization following synthesis and/or deprotection.
The siNA molecules of the invention can also be synthesized via a tandem synthesis methodology as described in Example 1 herein, wherein both siNA strands are synthesized as a single contiguous oligonucleotide fragment or strand separated by a cleavable linker which is subsequently cleaved to provide separate siNA fragments or strands that hybridize and permit purification of the siNA duplex. The linker can be a polynucleotide linker or a non-nucleotide linker. The tandem synthesis of siNA as described herein can he readily adapted to both multiwell/multiplate synthesis platforms such as 96 well or similarly larger multi-well platforms. The tandem synthesis of siNA as described herein can also be readily adapted to large scale synthesis platforms employing batch reactors, synthesis columns and the like.
A siNA molecule can also be assembled from two distinct nucleic acid strands or fragments wherein one fragment includes the sense region and the second fragment includes the antisense region of the RNA molecule.
The nucleic acid molecules of the present invention can be modified extensively to enhance stability by modification with nuclease resistant groups, for example, 2'-amino,
2'-C-allyl, 2'- fluoro, 2'-<9-methyl, 2'-H (for a review see Usman and Cedergren, 1992,
TIBS 17, 34; Usman et al, 1994, Nucleic Acids Symp. Ser. 31, 163). siNA constructs can be purified by gel electrophoresis using general methods or can be purified by high
pressure liquid chromatography (HPLC; see Wincott et al, supra, the totality of which is hereby incoφorated herein by reference) and re-suspended in water.
In another aspect of the invention, siNA molecules of the invention are expressed from transcription units inserted into DNA or RNA vectors. The recombinant vectors can be DNA plasmids or viral vectors. siNA expressing viral vectors can be constructed based on, but not limited to, adeno-associated virus, retrovirus, adenovirus, or alphavirus. The recombinant vectors capable of expressing the siNA molecules can be delivered as described herein, and persist in target cells. Alternatively, viral vectors can be used that provide for transient expression of siNA molecules.
Optimizing Activity of the nucleic acid molecule of the invention.
Chemically synthesizing nucleic acid molecules with modifications (base, sugar and/or phosphate) can prevent their degradation by serum ribonucleases, which can increase their potency (see e.g, Eckstein et al, International Publication No. WO 92/07065; Peπault et al, 1990 Nature 344, 565; Pieken et al, 1991, Science 253, 314; Usman and Cedergren, 1992, Trends in Biochem. Sci. 17, 334; Usman et al, International Publication No. WO 93/15187; and Rossi et al, International Publication No. WO 91/03162; Sproat, U.S. Pat. No. 5,334,711; Gold et al, U.S. Pat. No. 6,300,074; and Burgin et al, supra; all of which are incoφorated by reference herein). All of the above references describe various chemical modifications that can be made to the base, phosphate and/or sugar moieties of the nucleic acid molecules described herein. Modifications that enhance their efficacy in cells, and removal of bases from nucleic acid molecules to shorten oligonucleotide synthesis times and reduce chemical requirements are desired.
There are several examples in the art describing sugar, base and phosphate modifications that can be introduced into nucleic acid molecules with significant enhancement in their nuclease stability and efficacy. For example, oligonucleotides are modified to enhance stability and/or enhance biological activity by modification with nuclease resistant groups, for example, 2'-amino, 2'-C-aHyl, 2'-fluoro, 2'-(?-methyl, 2'-0- allyl, 2'-H, nucleotide base modifications (for a review see Usman and Cedergren, 1992, TIBS. 17, 34; Usman et al, 1994, Nucleic Acids Symp. Ser. 31, 163; Burgin et al, 1996,
Biochemistry, 35, 14090). Sugar modification of nucleic acid molecules have been extensively described in the art (see Eckstein et al, International Publication PCT No. WO 92/07065; Peπault et al. Nature, 1990, 344, 565-568; Pieken et al Science, 1991, 253, 314-317; Usman and Cedergren, Trends in Biochem. Sci., 1992, 17, 334-339; Usman et al. International Publication PCT No. WO 93/15187; Sproat, U.S. Pat. No. 5,334,711 and Beigel an et al, 1995, J. Biol. Chem., 270, 25702; Beigelman et al, International PCT publication No. WO 97/26270; Beigelman et al, U.S. Pat. No. 5,716,824; Usman et al, U.S. Pat. No. 5,627,053; Woolf et al, International PCT Publication No. WO 98/13526; Thompson et al, USSN 60/082,404 which was filed on April 20, 1998; Kaφeisky et al, 1998, Tetrahedron Lett., 39, 1131; Earnshaw and Gait, 1998, Biopolymers (Nucleic Acid Sciences), 48, 39-55; Verma and Eckstein, 1998, Annu. Rev. Biochem., 61, 99-134; and Burlina et al, 1997, Bioorg. Med. Chem., 5, 1999-2010; all of the references are hereby incoφorated in their totality by reference herein). Such publications describe general methods and strategies to determine the location of incoφoration of sugar, base and/or phosphate modifications and the like into nucleic acid molecules without modulating catalysis, and are incoφorated by reference herein. In view of such teachings, similar modifications can be used as described herein to modify the siNA nucleic acid molecules of the instant invention so long as the ability of siNA to promote RNAi is cells is not significantly inhibited. While chemical modification of oligonucleotide internucleotide linkages with phosphorothioate, phosphorodithioate, and/or 5'-methylphosphonate linkages improves stability, excessive modifications can cause some toxicity or decreased activity. Therefore, when designing nucleic acid molecules, the amount of these internucleotide linkages should be minimized. The reduction in the concentration of these linkages should lower toxicity, resulting in increased efficacy and higher specificity of these molecules.
Short interfering nucleic acid (siNA) molecules having chemical modifications that maintain or enhance activity are provided. Such a nucleic acid is also generally more resistant to nucleases than an unmodified nucleic acid. Accordingly, the in vitro and/or in vivo activity should not be significantly lowered. In cases in which modulation is the goal, therapeutic nucleic acid molecules delivered exogenously should optimally be
stable within cells until translation of the target RNA has been modulated long enough to reduce the levels of the undesirable protein. This period of time varies between hours to days depending upon the disease state. Improvements in the chemical synthesis of RNA and DNA (Wincott et al, 1995, Nucleic Acids Res. 23, 2677; Caruthers et al, 1992, Methods in Enzymology 111, 3-19 (incoφorated by reference herein)) have expanded the ability to modify nucleic acid molecules by introducing nucleotide modifications to enhance their nuclease stability, as described above.
In one embodiment, nucleic acid molecules of the invention include one or more (e.g., about 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, or more) G-clamp nucleotides. A G-clamp nucleotide is a modified cytosine analog wherein the modifications confer the ability to hydrogen bond both Watson-Crick and Hoogsteen faces of a complementary guanine within a duplex, see for example Lin and Matteucci, 1998, J. Am. Chem. Soc, 120, 8531- 8532. A single G-clamp analog substitution within an oligonucleotide can result in substantially enhanced helical themial stability and mismatch discrimination when hybridized to complementary oligonucleotides. The inclusion of such nucleotides in nucleic acid molecules of the invention results in both enhanced affinity and specificity to nucleic acid targets, complementary sequences, or template strands. In another embodiment, nucleic acid molecules of the invention include one or more (e.g., about 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, or more) LNA "locked nucleic acid" nucleotides such as a 2', 4'- C methylene bicyclo nucleotide (see for example Wengel et al, International PCT Publication No. WO 00/66604 and WO 99/14226).
In another embodiment, the invention features conjugates and/or complexes of siNA molecules of the invention. Such conjugates and/or complexes can be used to facilitate delivery of siNA molecules into a biological system, such as a cell. The conjugates and complexes provided by the instant invention can impart therapeutic activity by transferring therapeutic compounds across cellular membranes, altering the pharmacokinetics, and/or modulating the localization of nucleic acid molecules of the invention. The present invention encompasses the design and synthesis of novel conjugates and complexes for the delivery of molecules, including, but not limited to, small molecules, lipids, cholesterol, phospholipids, nucleosides, nucleotides, nucleic acids, antibodies, toxins, negatively charged polymers and other polymers, for example
proteins, peptides, hormones, carbohydrates, polyethylene glycols, or polyamines, across cellular membranes. In general, the transporters described are designed to be used either individually or as part of a multi-component system, with or without degradable linkers. These compounds are expected to improve delivery and/or localization of nucleic acid molecules of the invention into a number of cell types originating from different tissues, in the presence or absence of serum (see Sullenger and Cech, U.S. Pat. No. 5,854,038). Conjugates of the molecules described herein can be attached to biologically active molecules via linkers that are biodegradable, such as biodegradable nucleic acid linker molecules. The term "biodegradable linker" as used herein, refers to a nucleic acid or non- nucleic acid linker molecule that is designed as a biodegradable linker to connect one molecule to another molecule, for example, a biologically active molecule to a siNA molecule of the invention or the sense and antisense strands of a siNA molecule of the invention. The biodegradable linker is designed such that its stability can be modulated for a particular puφose, such as delivery to a particular tissue or cell type. The stability of a nucleic acid-based biodegradable linker molecule can be modulated by using various chemistries, for example combinations of ribonucleotides, deoxyribonucleotides, and chemically-modified nucleotides, such as 2'-0-methyl, 2'-fluoro, 2'-amino, 2'-0-amino, 2'-C-allyI, 2'-0-allyl, and other 2'-modified or base modified nucleotides. The biodegradable nucleic acid linker molecule can be a dimer, trimer, tetramer or longer nucleic acid molecule, for example, an oligonucleotide of about 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, or 20 nucleotides in length, or can comprise a single nucleotide with a phosphorus-based linkage, for example, a phosphoramidate or phosphodiester linkage. The biodegradable nucleic acid linker molecule can also comprise nucleic acid backbone, nucleic acid sugar, or nucleic acid base modifications.
The term "biodegradable" as used herein, refers to degradation in a biological system, for example, enzymatic degradation or chemical degradation.
The term "biologically active molecule" as used herein refers to compounds or molecules that are capable of eliciting or modifying a biological response in a system. Non-limiting examples of biologically active siNA molecules either alone or in combination with other molecules contemplated by the instant invention include
therapeutically active molecules such as antibodies, cholesterol, hormones, antivirals, peptides, proteins, chemotherapeutics, small molecules, vitamins, co-factors, nucleosides, nucleotides, oligonucleotides, enzymatic nucleic acids, antisense nucleic acids, triplex forming oligonucleotides, 2,5-A chimeras, siNA, dsRNA, allozymes, aptamers, decoys and analogs thereof. Biologically active molecules of the invention also include molecules capable of modulating the pharmacokinetics and/or pharmacodynamics of other biologically active molecules, for example, lipids and polymers such as polyamines, polyamides, polyethylene glycol and other polyethers.
The term "phospholipid" as used herein, refers to a hydrophobic molecule comprising at least one phosphorus group. For example, a phospholipid can comprise a phosphorus-containing group and saturated or unsaturated alkyl group, optionally substituted with OH, COOH, oxo, amine, or substituted or unsubstituted aryl groups.
Therapeutic nucleic acid molecules (e.g., siNA molecules) delivered exogenously optimally are stable within cells until reverse transcription of the RNA has been modulated long enough to reduce the levels of the RNA transcript. The nucleic acid molecules are resistant to nucleases in order to function as effective intracellular therapeutic agents. Improvements in the chemical synthesis of nucleic acid molecules described in the instant invention and in the art have expanded the ability to modify nucleic acid molecules by introducing nucleotide modifications to enhance their nuclease stability as described above.
In yet another embodiment, siNA molecules having chemical modifications that maintain or enhance enzymatic activity of proteins involved in RNAi are provided. Such nucleic acids are also generally more resistant to nucleases than unmodified nucleic acids. Thus, in vitro and/or in vivo the activity should not be significantly lowered. Use of the nucleic acid-based molecules of the invention will lead to better treatments by affording the possibility of combination therapies (e.g., multiple siNA molecules targeted to different genes; nucleic acid molecules coupled with known small molecule modulators; or intermittent treatment with combinations of molecules, including different motifs and/or other chemical or biological molecules). The treatment of subjects with siNA molecules can also include combinations of different types of
nucleic acid molecules, such as enzymatic nucleic acid molecules (ribozymes), allozymes, antisense, 2,5-A oligoadenylate, decoys, and aptamers.
In another aspect a siNA molecule of the invention comprises one or more 5' and/or a 3'- cap structure, for example, on only the sense siNA strand, the antisense siNA strand, or both siNA strands.
By "cap structure" is meant chemical modifications, which have been incoφorated at either terminus of the oligonucleotide (see, for example, Adamic et al, U.S. Pat. No. 5,998,203, incoφorated by reference herein). These terminal modifications protect the nucleic acid molecule from exonuclease degradation, and may help in delivery and/or localization within a cell. The cap may be present at the 5'-terminus (5'-cap) or at the 3'- terminal (3'-cap) or may be present on both termini. In non-limiting examples, the 5'-cap includes, but is not limited to, glyceryl, inverted deoxy abasic residue (moiety); 4',5'- methylene nucleotide; l-(beta-D-erythrofuranosyl) nucleotide, 4'-thio nucleotide; carbocyclic nucleotide; 1,5-anhydrohexitol nucleotide; L-nucleotides; alpha-nucleotides; modified base nucleotide; phosphorodithioate linkage; t/zreo-pentofuranosyl nucleotide; acyclic 3',4'-seco nucleotide; acyclic 3,4-dihydroxybutyl nucleotide; acyclic 3,5- dihydroxypentyl nucleotide, 3 '-3 '-inverted nucleotide moiety; 3 '-3 '-inverted abasic moiety; 3'-2'-inverted nucleotide moiety; S'^'-inverted abasic moiety; 1,4-butanediol phosphate; 3 '-phosphoramidate; hexylphosphate; aminohexyl phosphate; 3'-phosphate; 3'-phosphorothioate; phosphorodithioate; or bridging or non-bridging methylphosphonate moiety. Non-limiting examples of cap moieties are shown in Figure 10.
Non-limiting examples of the 3 '-cap include, but are not limited to, glyceryl, inverted deoxy abasic residue (moiety), 4', 5'-methylene nucleotide; l-(beta-D- erythrofuranosyl) nucleotide; 4'-thio nucleotide, carbocyclic nucleotide; 5'-amino-alkyl phosphate; l,3-diamino-2-propyl phosphate; 3-aminopropyl phosphate; 6-aminohexyl phosphate; 1,2-aminododecyl phosphate; hydroxypropyl phosphate; 1,5-anhydrohexitol nucleotide; L-nucleotide; alpha-nucleotide; modified base nucleotide; phosphorodithioate; tAreo-pentofuranosyl nucleotide; acyclic 3',4'-seco nucleotide; 3,4- dihydroxybutyl nucleotide; 3,5-dihydroxypentyl nucleotide, 5'-5'-inverted nucleotide moiety; 5'-5'-inverted abasic moiety; 5'-phosρhoramidate; 5'-phosphorothioate; 1,4-
butanediol phosphate; 5'-amino; bridging and or non-bridging 5'-phosphoramidate, phosphorothioate and/or phosphorodithioate, bridging or non bridging methylphosphonate and 5'-mercapto moieties (for more details see Beaucage and Iyer, 1993, Tetrahedron 49, 1925; incoφorated by reference herein). By the term "non-nucleotide" is meant any group or compound which can be incoφorated into a nucleic acid chain in the place of one or more nucleotide units, including either sugar and/or phosphate substitutions, and allows the remaining bases to exhibit their enzymatic activity. The group or compound is abasic in that it does not contain a commonly recognized nucleotide base, such as adenosine, guanine, cytosine, uracil or thymine and therefore lacks a base at the 1 '-position.
An "alkyl" group refers to a saturated aliphatic hydrocarbon, including straight- chain, branched-chain, and cyclic alkyl groups. Preferably, the alkyl group has 1 to 12 carbons. More preferably, it is a lower alkyl of from 1 to 7 carbons, more preferably 1 to 4 carbons. The alkyl group can be substituted or unsubstituted. When substituted the substituted group(s) is preferably, hydroxyl, cyano, alkoxy, =0, =S, N02 or N(CH3)2, amino, or SH. The term also includes alkenyl groups that are unsaturated hydrocarbon groups containing at least one carbon-carbon double bond, including straight-chain, branched-chain, and cyclic groups. Preferably, the alkenyl group has 1 to 12 carbons. More preferably, it is a lower alkenyl of from 1 to 7 carbons, more preferably 1 to 4 carbons. The alkenyl group may be substituted or unsubstituted. When substituted the substituted group(s) is preferably, hydroxyl, cyano, alkoxy, =0, =S, NO2, halogen, N(CH3)2, amino, or SH. The term "alkyl" also includes alkynyl groups that have an unsaturated hydrocarbon group containing at least one carbon-carbon triple bond, including straight-chain, branched-chain, and cyclic groups. Preferably, the alkynyl group has 1 to 12 carbons. More preferably, it is a lower alkynyl of from 1 to 7 carbons, more preferably 1 to 4 carbons. The alkynyl group may be substituted or unsubstituted. When substituted the substituted group(s) is preferably, hydroxyl, cyano, alkoxy, =0, =S, NO2 or N(CH3)2, amino or SH.
Such alkyl groups can also include aryl, alkylaryl, carbocyclic aryl, heterocyclic aryl, amide and ester groups. An "aryl" group refers to an aromatic group that has at
least one ring having a conjugated pi electron system and includes carbocyclic aryl, heterocyclic aryl and biaryl groups, all of which may "be optionally substituted. The prefeπed substituent(s) of aryl groups are halogen, trihialomethyl, hydroxyl, SH, OH, cyano, alkoxy, alkyl, alkenyl, alkynyl, and amino groups. An "alkylaryl" group refers to an alkyl group (as described above) covalently joined to an aryl group (as described above). Carbocyclic aryl groups are groups wherein the ring atoms on the aromatic ring are all carbon atoms. The carbon atoms are optionally substituted. Heterocyclic aryl groups are groups having from 1 to 3 heteroatoms as ring atoms in the aromatic ring and the remainder of the ring atoms are carbon atoms. Suitable heteroatoms include oxygen, sulfur, and nitrogen, and include furanyl, thienyl, pyridyl, pyπolyl, N-lower alkyl pyπolo, pyrimidyl, pyrazinyl, imidazolyl and the like, all optionally substituted. An "amide" refers to an -C(0)-NH-R, where R is either alkyl, aryl, alkylaryl or hydrogen. An "ester" refers to an -C(0)-OR', where R is either alkyl, aryl, alkylaryl or hydrogen.
By "nucleotide" as used herein is as recognized in the art to include natural bases (standard), and modified bases well known in the art. Such bases are generally located at the 1' position of a nucleotide sugar moiety. Nucleotides generally comprise a base, sugar and a phosphate group. The nucleotides can be unmodified or modified at the sugar, phosphate and/or base moiety, (also refeπed to interchangeably as nucleotide analogs, modified nucleotides, non-natural nucleotides, non-standard nucleotides and other; see, for example, Usman and McSwiggen, supra; Eckstein et al, International PCT Publication No. WO 92/07065; Usman et al, International PCT Publication No. WO 93/15187; Uhlman & Peyman, supra, all are hereby incoφorated by reference herein). There are several examples of modified nucleic acid bases known in the art as summarized by Limbach et al, 1994, Nucleic Acids Res. 22, 2183. Some of the non- limiting examples of base modifications that can be introduced into nucleic acid molecules include, inosine, purine, pyridin-4-one, pyridin-2-one, phenyl, pseudouracil, 2, 4, 6-trimethoxy benzene, 3 -methyl uracil, dihydroαridine, naphthyl, aminophenyl, 5-alkylcytidines (e.g., 5-methylcytidine), 5-alkyluxidines (e.g., ribothymidine), 5-halouridine (e.g., 5-bromouridine) or 6-azapyrimidines or 6-alkylpyrimidines (e.g. 6- methyluridine), propyne, and others (Burgin et al, 1996, Biochemistry, 35, 14090; Uhlman & Peyman, supra). By "modified bases" in this aspect is meant nucleotide bases other than adenine, guanine, cytosine and uracil at 1' position or their equivalents.
In one embodiment, the invention features modified siNA molecules, with phosphate backbone modifications comprising one or more phosphorothioate, phosphorodithioate, methylphosphonate, phosphotriester, moφholino, amidate carbamate, carboxymethyl, acetamidate, polyamide, sulfonate, sulfonamide, sulfamate, formacetal, thioformacetal, and/or alkylsilyl, substitutions. For a review of oligonucleotide backbone modifications, see Hunziker and Leumann, 1995, Nucleic Acid Analogues: Synthesis and Properties, in. Modern Synthetic Methods, VCH, 331-417, and Mesmaeker et al, 1994, Novel Backbone Replacements for Oligonucleotides, in Carbohydrate Modifications in Antisense Research, ACS, 24-39. By "abasic" is meant sugar moieties lacking a base or having other chemical groups in place of a base at the 1' position, see for example Adamic et al, U.S. Pat. No. 5,998,203.
By "unmodified nucleoside" is meant one of the bases adenine, cytosine, guanine, thymine, or uracil joined to the T carbon of β-D-ribo-furanose. By "modified nucleoside" is meant any nucleotide base which contains a modification in the chemical structure of an unmodified nucleotide base, sugar and/or phosphate. Non-limiting examples of modified nucleotides are shown by Formulae I- VII and/or other modifications described herein.
In connection with 2'-modified nucleotides as described for the present invention, by "amino" is meant 2'-NH2 or 2'-0 NH2, which can be modified or unmodified. Such modified groups are described, for example, in Eckstein et al, U.S. Pat. No. 5,672,695 and Matulic-Adamic et al, U.S. Pat. No. 6,248,878, which are both incoφorated by reference in their entireties.
Various modifications to nucleic acid siNA structure can be made to enhance the utility of these molecules. Such modifications will enhance shelf-life, half-life in vitro, stability, and ease of introduction of such oligonucleotides to the target site, e.g., to enhance penetration of cellular membranes, and confer the ability to recognize and bind to targeted cells.
Administration of Nucleic Acid Molecules
A siNA molecule of the invention can be adapted for use to prevent or treat diseases, traits, disorders, and/or conditions described herein or otherwise known in the art to be related to gene expression, and/or any other trait, disease, disorder or condition that is related to or will respond to the levels of a target polynucleotide in a cell or tissue, alone or in combination with other therapies. For example, a siNA molecule can comprise a delivery vehicle, including liposomes, for administration to a subject, carriers and diluents and their salts, and/or can be present in pharmaceutically acceptable formulations. Methods for the delivery of nucleic acid molecules are described in Akhtar et al, 1992, Trends Cell Bio., 2, 139; Delivery Strategies for Antisense Oligonucleotide Therapeutics, ed. Akhtar, 1995, Maurer et al, 1999, Mol. Membr. Biol, 16, 129-140; Hofland and Huang, 1999, Handb. Exp. Pharmacol, 137, 165-192; and Lee et al, 2000, ACS Symp. Ser., 752, 184-192, all of which are incoφorated herein by reference. Beigelman et al, U.S. Pat. No. 6,395,713 and Sullivan et al, PCT WO 94/02595 further describe the general methods for delivery of nucleic acid molecules. These protocols can be utilized for the delivery of virtually any nucleic acid molecule. Nucleic acid molecules can be administered to cells by a variety of methods known to those of skill in the art, including, but not restricted to, encapsulation in liposomes, by iontophoresis, or by incoφoration into other vehicles, such as biodegradable polymers, hydrogels, cyclodextrins (see for example Gonzalez et al, 1999, Bioconjugate Chem., 10, 1068-1074; Wang et al. International PCT publication Nos. WO 03/47518 and WO 03/46185), poly(lactic-co-glycolic)acid (PLGA) and PLCA microspheres (see for example US Patent 6,447,796 and US Patent Application Publication No. US 2002130430), biodegradable nanocapsules, and bioadhesive microspheres, or by proteinaceous vectors (O'Hare and Normand, International PCT Publication No. WO 00/53722). Alternatively, the nucleic acid/vehicle combination is locally delivered by direct injection or by use of an infusion pump. Direct injection of the nucleic acid molecules of the invention, whether subcutaneous, intramuscular, or intradermal, can take place using standard needle and syringe methodologies, or by needle-free technologies such as those described in Conry et al, 1999, Clin. Cancer Res., 5, 2330- 2337 and Barry et al, International PCT Publication No. WO 99/31262. The molecules of the instant invention can be used as pharmaceutical agents. Pharmaceutical agents
prevent, modulate the occuπence, or treat (alleviate a symptom to some extent, preferably all of the symptoms) of a disease state in a subject.
In another embodiment, the nucleic acid molecules of the invention can also be formulated or complexed with polyethyleneimine and derivatives thereof, such as polyethyleneimine-polyethyleneglycol-N-acetylgalactosamine (PEI-PEG-GAL) or polyethyleneimine-polyethyleneglycol-tri-N-acetylgalactosamine (PEI-PEG-triGAL) derivatives. In one embodiment, the nucleic acid molecules of the invention are formulated as described in United States Patent Application Publication No. 20030077829, incoφorated by reference herein in its entirety. In one embodiment, a siNA molecule of the invention is complexed with membrane disruptive agents such as those described in U.S. Patent Application Publication No. 20010007666, incoφorated by reference herein in its entirety including the drawings. In another embodiment, the membrane disruptive agent or agents and the siNA molecule are also complexed with a cationic lipid or helper lipid molecule, such as those lipids described in U.S. Patent No. 6,235,310, incoφorated by reference herein in its entirety including the drawings.
In one embodiment, a siNA molecule of the invention is complexed with delivery systems as described in U.S. Patent Application Publication No. 2003077829 and International PCT Publication Nos. WO 00/03683 and WO 02/087541, all incoφorated by reference herein in their entirety including the drawings.
In one embodiment, the nucleic acid molecules of the invention are administered via pulmonary delivery, such as by inhalation of an aerosol or spray dried formulation administered by an inhalation device or nebulizer, providing rapid local uptake of the nucleic acid molecules into relevant pulmonary tissues. Solid particulate compositions containing respirable dry particles of micronized nucleic acid compositions can be prepared by grinding dried or lyophilized nucleic acid compositions, and then passing the micronized composition through, for example, a 400 mesh screen to break up or separate out large agglomerates. A solid particulate composition comprising the nucleic acid compositions of the invention can optionally contain a dispersant which serves to facilitate the formation of an aerosol as well as other therapeutic compounds. A suitable
dispersant is lactose, which can be blended with the nucleic acid compound in any suitable ratio, such as a 1 to 1 ratio by weight.
Aerosols of liquid particles comprising a nucleic acid composition of the invention can be produced by any suitable means, such as with a nebulizer (see for example US 4,501,729). Nebulizers are commercially available devices which transform solutions or suspensions of an active ingredient into a therapeutic aerosol mist either by means of acceleration of a compressed gas, typically air or oxygen, through a naπow venturi orifice or by means of ultrasonic agitation. Suitable formulations for use in nebulizers comprise the active ingredient in a liquid carrier in an amount of up to 40% w/w preferably less than 20% w/w of the formulation. The carrier is typically water or a dilute aqueous alcoholic solution, preferably made isotonic with body fluids by the addition of, for example, sodium chloride or other suitable salts. Optional additives include preservatives if the formulation is not prepared sterile, for example, methyl hydroxybenzoate, anti-oxidants, flavorings, volatile oils, buffering agents and emulsifiers and other formulation surfactants. The aerosols of solid particles comprising the active composition and surfactant can likewise be produced with any solid particulate aerosol generator. Aerosol generators for administering solid particulate therapeutics to a subject produce particles which are respirable, as explained above, and generate a volume of aerosol containing a predetermined metered dose of a therapeutic composition at a rate suitable for human administration. One illustrative type of solid particulate aerosol generator is an insufflator. Suitable formulations for administration by insufflation include finely comminuted powders which can be delivered by means of an insufflator. In the insufflator, the powder, e.g., a metered dose thereof effective to carry out the treatments described herein, is contained in capsules or cartridges, typically made of gelatin or plastic, which are either pierced or opened in situ and the powder delivered by air drawn through the device upon inhalation or by means of a manually-operated pump. The powder employed in the insufflator consists either solely of the active ingredient or of a powder blend comprising the active ingredient, a suitable powder diluent, such as lactose, and an optional surfactant. The active ingredient typically comprises from 0.1 to 100 w/w of the formulation. A second type of illustrative aerosol generator comprises a metered dose inhaler. Metered dose inhalers are pressurized aerosol dispensers, typically containing a suspension or solution formulation of the active ingredient in a liquified
propellant. During use these devices discharge the formulation through a valve adapted to deliver a metered volume to produce a fine particle spray containing the active ingredient. Suitable propellants include certain chlorofluorocarbon compounds, for example, dichlorodifluoromethane, trichlorofluoromethane, dichlorotetrafluoroethane and mixtures thereof. The formulation can additionally contain one or more co-solvents, for example, ethanol, emulsifiers and other formulation surfactants, such as oleic acid or sorbitan trioleate, anti-oxidants and suitable flavoring agents. Other methods for pulmonary delivery are described in, for example US Patent Application No. 20040037780, and US Patent Nos. 6,592,904; 6,582,728; 6,565,885. In one embodiment, the invention features the use of methods to deliver the nucleic acid molecules of the instant invention to the central nervous system and/or peripheral nervous system. Experiments have demonstrated the efficient in vivo uptake of nucleic acids by neurons. As an example of local administration of nucleic acids to nerve cells, Sommer et al, 1998, Antisense Nuc Acid Drug Dev., 8, 75, describe a study in which a 15mer phosphorothioate antisense nucleic acid molecule to c-fos is administered to rats via microinjection into the brain. Antisense molecules labeled with tetramethylrhodamine-isothiocyanate (TRITC) or fluorescein isothiocyanate (FITC) were taken up by exclusively by neurons thirty minutes post-injection. A diffuse cytoplasmic staining and nuclear staining was observed in these cells. As an example of systemic administration of nucleic acid to nerve cells, Epa et al, 2000, Antisense Nuc. Acid Drug Dev., 10, 469, describe an in vivo mouse study in which beta-cyclodextrin-adamantane- oligonucleotide conjugates were used to target the p75 neurotrophin receptor in neuronally differentiated PC 12 cells. Following a two week course of IP administration, pronounced uptake of p75 neurotrophin receptor antisense was observed in dorsal root ganglion (DRG) cells. In addition, a marked and consistent down-regulation of p75 was observed in DRG neurons. Additional approaches to the targeting of nmcleic acid to neurons are described in Broaddus et al, 1998, J. Neurosurg., 88(4), 734; Karle et al, 1997, Eur. J. Pharmocol, 340(2/3), 153; Bannai et al, 1998, Brain Research, 784(1,2), 304; Rajakumar et al, 1997, Synapse, 26(3), 199; Wu-pong et al, 1999, BioPharm, 12(1), 32; Bannai et al, 1998, Brain Res. Protoc, 3(1), 83; Simantov et al, 1996, Neuroscience, 74(1), 39. Nucleic acid molecules of the invention are therefore amenable to delivery to and uptake by cells that express repeat expansion allelic variants for
modulation of RE gene expression. The delivery of nucleic acid molecules of the invention, targeting RE is provided by a variety of different strategies. Traditional approaches to CNS delivery that can be used include, but are not limited to, intrathecal and intracerebroventricular administration, implantation of catheters and pumps, direct injection or perfusion at the site of injury or lesion, injection into the brain arterial system, or by chemical or osmotic opening of the blood-brain barrier. Other approaches can include the use of various transport and carrier systems, for example though the use of conjugates and biodegradable polymers. Furthermore, gene therapy approaches, for example as described in Kaplitt et al, US 6,180,613 and Davidson, WO 04/013280, can be used to express nucleic acid molecules in the CNS.
In one embodiment, nucleic acid molecules of the invention are administered to the central nervous system (CNS) or peripheral nervous system (PNS). Experiments have demonstrated the efficient in vivo uptake of nucleic acids by neurons. As an example of local administration of nucleic acids to nerve cells, Sommer et al, 1998, Antisense Nuc. Acid Drug Dev., 8, 75, describe a study in which a 15mer phosphorothioate antisense nucleic acid molecule to c-fos is administered to rats via microinjection into the brain. Antisense molecules labeled with tetramethylrhodamine-isothiocyanate (TRITC) or fluorescein isothiocyanate (FITC) were taken up by exclusively by neurons thirty minutes post-injection. A diffuse cytoplasmic staining and nuclear staining was observed in these cells. As an example of systemic administration of nucleic acid to nerve cells, Epa et al, 2000, Antisense Nuc. Acid Drug Dev., 10, 469, describe an in vivo mouse study in which beta-cyclodextrin-adamantane-oligonucleotide conjugates were used to target the p75 neurotrophin receptor in neuronally differentiated PC12 cells. Following a two week course of IP administration, pronounced uptake of p75 neurotrophin receptor antisense was observed in dorsal root ganglion (DRG) cells. In addition, a marked and consistent down-regulation of p75 was observed in DRG neurons. Additional approaches to the targeting of nucleic acid to neurons are described in Broaddus et al, 1998, J. Neurosurg, 88(4), 734; Karle et al, 1997, Eur. J. Pharmocol, 340(2/3), 153; Bannai et al, 1998, Brain Research, 784(1,2), 304; Rajakumar et al, 1997, Synapse, 26(3), 199; Wu-pong et al, 1999, BioPharm, 12(1), 32; Bannai et al, 1998, Brain Res. Protoc, 3(1), 83; Simantov et al, 1996, Neuroscience,
74(1), 39. Nucleic acid molecules of the invention are therefore amenable to delivery to and uptake by cells in the CNS and/or PNS.
The delivery of nucleic acid molecules of the invention to the CNS is provided by a variety of different strategies. Traditional approaches to CNS delivery that can be used include, but are not limited to, intrathecal and intracerebroventricular administration, implantation of catheters and pumps, direct injection or perfusion at the site of injury or lesion, injection into the brain arterial system, or by chemical or osmotic opening of the blood-brain barrier. Other approaches can include the use of various transport and carrier systems, for example though the use of conjugates and biodegradable polymers. Furthermore, gene therapy approaches, for example as described in Kaplitt et al, US 6,180,613 and Davidson, WO 04/013280, can be used to express nucleic acid molecules in the CNS.
In one embodiment, delivery systems of the invention include, for example, aqueous and nonaqueous gels, creams, multiple emulsions, microemulsions, liposomes, ointments, aqueous and nonaqueous solutions, lotions, aerosols, hydrocarbon bases and powders, and can contain excipients such as solubilizers, permeation enhancers (e.g., fatty acids, fatty acid esters, fatty alcohols and amino acids), and hydrophilic polymers (e.g., polycarbophil and polyvinylpyrolidone). In one embodiment, the pharmaceutically acceptable carrier is a liposome or a transdermal enhancer. Examples of liposomes which can be used in this invention include the following: (1) CellFectin, 1:1.5 (M M) liposome formulation of the cationic lipid N,NI,NII,NIιI-tetramethyl-N,NI,NII,NIII-tetrapalmit-y- spermine and dioleoyl phosphatidylethanolamine (DOPE) (GIBCO BRL); (2) Cytofectin GSV, 2:1 (M/M) liposome formulation of a cationic lipid and DOPE (Glen Research); (3) DOTAP (N-[l-(2,3-dioleoyloxy)-N,N,N-tri-methyl-ammoniummethylsulfate) (Boehringer Manheim); and (4) Lipofectamine, 3:1 (M/M) liposome formulation of the polycationic lipid DOSPA and the neutral lipid DOPE (GIBCO BRL).
In one embodiment, delivery systems of the invention include patches, tablets, suppositories, pessaries, gels and creams, and can contain excipients such as solubilizers and enhancers (e.g., propylene glycol, bile salts and amino acids), and other vehicles (e.g., polyethylene glycol, fatty acid esters and derivatives, and hydrophilic polymers such as hydroxypropylmethylcellulose and hyaluronic acid).
In one embodiment, siNA molecules of the invention are formulated or complexed with polyethylenimine (e.g., linear or branched PEI) and/or polyethylenimine derivatives, including for example grafted PEIs such as galactose PEI, cholesterol PEI, antibody derivatized PEI, and polyethylene glycol PEI (PEG-PEI) derivatives thereof (see for example Ogris et al, 2001, AAPA PharmSci, 3, 1-11; Furgeson et al., 2003, Bioconjugate Chem., 14, 840-847; Kunath et al., 2002, Phramaceutical Research, 19, 810-817; Choi et al., 2001, Bull. Korean Chem. Soc, 22, 46-52; Bettinger et al., 1999, Bioconjugate Chem., 10, 558-561; Peterson et al., 2002, Bioconjugate Chem., 13 , 845- 854; Erbacher et al., 1999, Journal of Gene Medicine Preprint, 1, 1-18; Godbey et al., 1999., PNAS USA, 96, 5177-5181; Godbey et al., 1999, Journal of Controlled Release, 60, 149-160; Diebold et al., 1999, Journal of Biological Chemistry, 274, 19087-19094; Thomas and Klibanov, 2002, PNAS USA, 99, 14640-14645; and Sagara, US 6,586,524, incoφorated by reference herein.
In one embodiment, a siNA molecule of the invention comprises a bioconjugate, for example a nucleic acid conjugate as described in Vargeese et al., USSN 10/427,160, filed April 30, 2003; US 6,528,631; US 6,335,434; US 6, 235,886; US 6,153,737; US 5,214,136; US 5,138,045, all incoφorated by reference herein.
Thus, the invention features a pharmaceutical composition comprising one or more nucleic acid(s) of the invention in an acceptable carrier, such as a stabilizer, buffer, and the like. The polynucleotides of the invention can be administered (e.g., RNA, DNA or protein) and introduced to a subject by any standard means, with or without stabilizers, buffers, and the like, to form a pharmaceutical composition. When it is desired to use a liposome delivery mechanism, standard protocols for formation of liposomes can be followed. The compositions of the present invention can also be formulated and used as creams, gels, sprays, oils and other suitable compositions for topical, dermal, or transdermal administration as is known in the art.
The present invention also includes pharmaceutically acceptable formulations of the compounds described. These formulations include salts of the above compounds, e.g., acid addition salts, for example, salts of hydrochloric, hydrobromic, acetic acid, and benzene sulfonic acid.
A pharmacological composition or formulation refers to a composition or formulation in a form suitable for administration, e.g., systemic or local administration, into a cell or subject, including for example a human. Suitable forms, in part, depend upon the use or the route of entry, for example oral, tiansdermal, or by injection. Such forms should not prevent the composition or formulation from reaching a target cell (i.e., a cell to which the negatively charged nucleic acid is desirable for delivery). For example, pharmacological compositions injected into the blood stream should be soluble. Other factors are known in the art, and include considerations such as toxicity and forms that prevent the composition or formulation from exerting its effect. In one embodiment, siNA molecules of the invention are administered to a subject by systemic administration in a pharmaceutically acceptable composition or formulation. By "systemic administration" is meant in vivo systemic absoφtion or accumulation of drugs in the blood stream followed by distribution throughout the entire body. Administration routes that lead to systemic absoφtion include, without limitation: intravenous, subcutaneous, intraperitoneal, inhalation, oral, intrapulmonary and intramuscular. Each of these administration routes exposes the siNA molecules of the invention to an accessible diseased tissue. The rate of entry of a drug into the circulation has been shown to be a function of molecular weight or size. The use of a liposome or other drug carrier comprising the compounds of the instant invention can potentially localize the drag, for example, in certain tissue types, such as the tissues of the reticular endothelial system (RES). A liposome formulation that can facilitate the association of drug with the surface of cells, such as, lymphocytes and macrophages is also useful. This approach can provide enhanced delivery of the drug to target cells by taking advantage of the specificity of macrophage and lymphocyte immune recognition of abnormal cells, such as cancer cells.
By "pharmaceutically acceptable formulation" or "pharmaceutically acceptable composition" is meant, a composition or formulation that allows for the effective distribution of the nucleic acid molecules of the instant invention in the physical location most suitable for their desired activity. Non-limiting examples of agents suitable for formulation with the nucleic acid molecules of the instant invention include: P- glycoprotein inhibitors (such as Pluronic P85),; biodegradable polymers, such as poly
(DL-lactide-coglycolide) microspheres for sustained release delivery (Emerich, DF et al, 1999, Cell Transplant, 8, 47-58); and loaded nanoparticles, such as those made of polybutylcyanoacrylate. Other non-limiting examples of delivery strategies for the nucleic acid molecules of the instant invention include material described in Boado et al., 1998, J. Pharm. Sci, 87, 1308-1315; Tyler et al, 1999, FEBS Lett., 421, 280-284; Pardridge et al, 1995, PNAS USA., 92, 5592-5596; Boado, 1995, Adv. Drug Delivery Rev., 15, 73-107; Aldrian-Heπada et al, 1998, Nucleic Acids Res., 26, 4910-4916; and Tyler et al, 1999, PNAS USA., 96, 7053-7058.
The invention also features the use of the composition comprising surface- modified liposomes containing poly (ethylene glycol) lipids (PEG-modified, or long- circulating liposomes or stealth liposomes). These formulations offer a method for increasing the accumulation of drugs in target tissues. This class of drug carriers resists opsonization and elimination by the mononuclear phagocytic system (MPS or RES), thereby enabling longer blood circulation times and enhanced tissue exposure for the encapsulated drug (Lasic et al Chem. Rev. 1995, 95, 2601-2627; Ishiwata et al, Chem. Pharm. Bull. 1995, 43, 1005-1011). Such liposomes have been shown to accumulate selectively in tumors, presumably by extravasation and capture in the neovascularized target tissues (Lasic et al, Science 1995, 267, 1275-1276; Oku et al, 1995, Biochim. Biophys. Ada, 1238, 86-90). The long-circulating liposomes enhance the pharmacokinetics and pharmacodynamics of DNA and RNA, particularly compared to conventional cationic liposomes which are known to accumulate in tissues of the MPS (Liu et al, J. Biol. Chem. 1995, 42, 24864-24870; Choi et al, International PCT Publication No. WO 96/10391; Ansell et al, International PCT Publication No. WO 96/10390; Holland et al, International PCT Publication No. WO 96/10392). Long- circulating liposomes are also likely to protect drags from nuclease degradation to a greater extent compared to cationic liposomes, based on their ability to avoid accumulation in metabolically aggressive MPS tissues such as the liver and spleen.
The present invention also includes compositions prepared for storage or administration that include a pharmaceutically effective amount of the desired compounds in a pharmaceutically acceptable carrier or diluent. Acceptable carriers ox diluents for therapeutic use are well known in the pharmaceutical art, and are described,
for example, in Remington's Pharmaceutical Sciences, Mack Publishing Co. (A.R. Gennaro edit. 1985), hereby incoφorated by reference herein. For example, preservatives, stabilizers, dyes and flavoring agents can be provided. These include sodium benzoate, sorbic acid and esters of >-hydroxybenzoic acid. In addition, antioxidants and suspending agents can be used.
A pharmaceutically effective dose is that dose required to prevent, inhibit the occuπence, or treat (alleviate a symptom to some extent, preferably all of the symptoms) of a disease state. The pharmaceutically effective dose depends on the type of disease, the composition used, the route of administration, the type of mammal being treated, the physical characteristics of the specific mammal under consideration, concurrent medication, and other factors that those skilled in the medical arts will recognize. Generally, an amount between 0.1 mg/kg and 100 mg/kg body weight/day of active ingredients is administered dependent upon potency of the negatively charged polymer.
The nucleic acid molecules of the invention and formulations thereof can be administered orally, topically, parenterally, by inhalation or spray, or rectally in dosage unit formulations containing conventional non-toxic pharmaceutically acceptable carriers, adjuvants and/or vehicles. The term parenteral as used herein includes percutaneous, subcutaneous, intravascular (e.g., intravenous), intramuscular, or intrathecal injection or infusion techniques and the like. In addition, there is provided a pharmaceutical formulation comprising a nucleic acid molecule of the invention and a pharmaceutically acceptable carrier. One or more nucleic acid molecules of the invention can be present in association with one or more non-toxic pharmaceutically acceptable carriers and/or diluents and/or adjuvants, and if desired other active ingredients. The pharmaceutical compositions containing nucleic acid molecules of the invention can be in a form suitable for oral use, for example, as tablets, troches, lozenges, aqueous or oily suspensions, dispersible powders or granules, emulsion, hard or soft capsules, or syrups or elixirs.
Compositions intended for oral use can be prepared according to any method known to the art for the manufacture of pharmaceutical compositions and such compositions can contain one or more such sweetening agents, flavoring agents, coloring agents or preservative agents in order to provide pharmaceutically elegant and palatable
preparations. Tablets contain the active ingredient in admixture with non-toxic pharmaceutically acceptable excipients that are suitable for the manufacture of tablets. These excipients can be, for example, inert diluents; such as calcium carbonate, sodium carbonate, lactose, calcium phosphate or sodium phosphate; granulating and disintegrating agents, for example, com starch, or alginic acid; binding agents, for example starch, gelatin or acacia; and lubricating agents, for example magnesium stearate, stearic acid or talc. The tablets can be uncoated or they can be coated by known techniques. In some cases such coatings can be prepared by known techniques to delay disintegration and absoφtion in the gastrointestinal tract and thereby provide a sustained action over a longer period. For example, a time delay material such as glyceryl monosterate or glyceryl distearate can be employed.
Formulations for oral use can also be presented as hard gelatin capsules wherein the active ingredient is mixed with an inert solid diluent, for example, calcium carbonate, calcium phosphate or kaolin, or as soft gelatin capsules wherein the active ingredient is mixed with water or an oil medium, for example peanut oil, liquid paraffin or olive oil.
Aqueous suspensions contain the active materials in a mixture with excipients suitable for the manufacture of aqueous suspensions. Such excipients are suspending agents, for example sodium carboxymethylcellulose, methylcellulose, hydropropyl- methylcelhilose, sodium alginate, polyvinylpyπolidone, gum tragacanth and gum acacia; dispersing or wetting agents can be a naturally-occurring phosphatide, for example, lecithin, or condensation products of an alkylene oxide with fatty acids, for example polyoxyethylene stearate, or condensation products of ethylene oxide with long chain aliphatic alcohols, for example heptadecaethyleneoxycetanol, or condensation products of ethylene oxide with partial esters derived from fatty acids and a hexitol such as polyoxyethylene sorbitol monooleate, or condensation products of ethylene oxide with partial esters derived from fatty acids and hexitol anhydrides, for example polyethylene sorbitan monooleate. The aqueous suspensions can also contain one or more preservatives, for example ethyl, or n-propyl p-hydroxybenzoate, one or more coloring agents, one or more flavoring agents, and one or more sweetening agents, such as sucrose or saccharin.
Oily suspensions can be formulated by suspending the active ingredients in a vegetable oil, for example arachis oil, olive oil, sesame oil or coconut oil, or in a mineral oil such as liquid paraffin. The oily suspensions can contain a thickening agent, for example beeswax, hard paraffin or cetyl alcohol. Sweetening agents and flavoring agents can be added to provide palatable oral preparations. These compositions can be preserved by the addition of an anti-oxidant such as ascorbic acid
Dispersible powders and granules suitable for preparation of an aqueous suspension by the addition of water provide the active ingredient in admixture with a dispersing or wetting agent, suspending agent and one or more preservatives. Suitable dispersing or wetting agents or suspending agents are exemplified by those already mentioned above. Additional excipients, for example sweetening, flavoring and coloring agents, can also be present.
Pharmaceutical compositions of the invention can also be in the form of oil-in- water emulsions. The oily phase can be a vegetable oil or a mineral oil or mixtures of these. Suitable emulsifying agents can be naturally-occurring gums, for example gum acacia or gum tragacanth, naturally-occurring phosphatides, for example soy bean, lecithin, and esters or partial esters derived from fatty acids and hexitol, anhydrides, for example sorbitan monooleate, and condensation products of the said partial esters with ethylene oxide, for example polyoxyethylene sorbitan monooleate. The emulsions can also contain sweetening and flavoring agents.
Syrups and elixirs can be formulated with sweetening agents, for example glycerol, propylene glycol, sorbitol, glucose or sucrose. Such formulations can also contain a demulcent, a preservative and flavoring and coloring agents. The pharmaceutical compositions can be in the form of a sterile injectable aqueous or oleaginous suspension. This suspension can be formulated according to the known art using those suitable dispersing or wetting agents and suspending agents that have been mentioned above. The sterile injectable preparation can also be a sterile injectable solution or suspension in a non-toxic parentally acceptable diluent or solvent, for example as a solution in 1,3- butanediol. Among the acceptable vehicles and solvents that can be employed are water, Ringer's solution and isotonic sodium chloride solution. In addition, sterile, fixed oils are conventionally employed as a solvent or suspending medium. For this puφose, any
bland fixed oil can be employed including synthetic mono-or diglycerides. In addition, fatty acids such as oleic acid find use in the preparation of injectables.
The nucleic acid molecules of the invention can also be administered in the form of suppositories, e.g., for rectal administration of the drug. These compositions can be prepared by mixing the drag with a suitable non-irritating excipient that is solid at ordinary temperatures but liquid at the rectal temperature and will therefore melt in the rectum to release the drug. Such materials include cocoa butter and polyethylene glycols.
Nucleic acid molecules of the invention can be administered parenterally in a sterile medium. The drug, depending on the vehicle and concentration used, can either be suspended or dissolved in the vehicle. Advantageously, adjuvants such as local anesthetics, preservatives and buffering agents can be dissolved in the vehicle.
Dosage levels of the order of from about 0.1 mg to about 140 mg per kilogram of body weight per day are useful in the treatment of the above-indicated conditions (about 0.5 mg to about 7 g per subject per day). The amount of active ingredient that can be combined with the carrier materials to produce a single dosage form varies depending upon the host treated and the particular mode of administration. Dosage unit forms generally contain between from about 1 mg to about 500 mg of an active ingredient.
It is understood that the specific dose level for any particular subject depends upon a variety of factors including the activity of the specific compound employed, the age, body weight, general health, sex, diet, time of administration, route of administration, and rate of excretion, drag combination and the severity of the particular disease undergoing therapy.
For administration to non-human animals, the composition can also be added to the animal feed or drinking water. It can be convenient to formulate the animal feed and drinking water compositions so that the animal takes in a therapeutically appropriate quantity of the composition along with its diet. It can also be convenient to present the composition as a premix for addition to the feed or drinking water.
The nucleic acid molecules of the present invention can also be administered to a subject in combination with other therapeutic compounds to increase the overall therapeutic effect. The use of multiple compounds to treat an indication can increase the beneficial effects while reducing the presence of side effects. In one embodiment, the invention comprises compositions suitable for administering nucleic acid molecules of the invention to specific cell types. For example, the asialoglycoprotein receptor (ASGPr) (Wu and Wu, 1987, J. Biol. Chem. 262, 4429-4432) is unique to hepatocytes and binds branched galactose-terminal glycoproteins, such as asialoorosomucoid (ASOR). In another example, the folate receptor is overexpressed in many cancer cells. Binding of such glycoproteins, synthetic glycoconjugates, or folates to the receptor takes place with an affinity that strongly depends on the degree of branching of the oligosaccharide chain, for example, triatennary structures are bound with greater affinity than biatenarry or monoatennary chains (Baenziger and Fiete, 1980, Cell, 22, 611-620; Connolly et al, 1982, J. Biol. Chem., 257, 939-945). Lee and Lee, 1987, Glycoconjugate J., 4, 317-328, obtained this high specificity through the use of N-acetyl-D-galactosamine as the carbohydrate moiety, which has higher affinity for the receptor, compared to galactose. This "clustering effect" has also been described for the binding and uptake of mannosyl-terminating glycoproteins or glycoconjugates (Ponpipom et al, 1981, J. Med. Chem., 24, 1388- 1395). The use of galactose, galactosamine, or folate based conjugates to transport exogenous compounds across cell membranes can provide a targeted delivery approach to, for example, the treatment of liver disease, cancers of the liver, or other cancers. The use of bioconjugates can also provide a reduction in the required dose of therapeutic compounds required for treatment. Furthermore, therapeutic bioavailability, pharmacodynamics, and pharmacokinetic parameters can be modulated through the use of nucleic acid bioconjugates of the invention. Non-limiting examples of such bioconjugates are described in Vargeese et al, USSN 10/201,394, filed August 13, 2001; and Matulic-Adamic et al, USSN 60/362,016, filed March 6, 2002.
Alternatively, certain siNA molecules of the instant invention can be expressed within cells from eukaryotic promoters (e.g., Izant and Weintraub, 1985, Science, 229,
345; McGaπy and Lindquist, 1986, Proc. Natl. Acad. Sci., USA 83, 399; Scanlon et al,
1991, Proc. Natl. Acad. Sci. USA, 88, 10591-5; Kashani-Sabet et al, 1992, Antisense Res. Dev., 2, 3-15; Dropulic et al, 1992, J. Virol, 66, 1432-41; Weerasinghe et al, 1991, J. Virol, 65, 5531-4; Ojwang et al, 1992, Proc Natl. Acad. Sci. USA, 89, 10802- 6; Chen et α/., 1992, Nucleic Acids Res., 20, 4581-9; Sarver et al, 1990 Science, 247 ', 1222-1225; Thompson et /., 1995, Nwc/etc ctώs Res., 23, 2259; Good et al, 1997, Gene Therapy, 4, 45. Those skilled in the art realize that any nucleic acid can be expressed in eukaryotic cells from the appropriate DNA/RNA vector. The activity of such nucleic acids can be augmented by their release from the primary transcript by a enzymatic nucleic acid (Draper et al, PCT WO 93/23569, and Sullivan et al, PCT WO 94/02595; Ohkawa et al, 1992, Nucleic Acids Symp. Ser., 27, 15-6; Taira et al, 1991, Nucleic Acids Res., 19, 5125-30; Ventura et al, 1993, Nucleic Acids Res., 21, 3249-55; Chowrira et al, 1994, J. Biol. Chem., 269, 25856.
In another aspect of the invention, RNA molecules of the present invention can be expressed from transcription units (see for example Couture et al, 1996, TIG., 12, 510) inserted into DNA or RNA vectors. The recombinant vectors can be DNA plasmids or viral vectors. siNA expressing viral vectors can be constructed based on, but not limited to, adeno-associated virus, retrovirus, adenovirus, or alphaviras. In another embodiment, pol III based constructs are used to express nucleic acid molecules of the invention (see for example Thompson, U.S. Pats. Nos. 5,902,880 and 6,146,886). The recombinant vectors capable of expressing the siNA molecules can be delivered as described above, and persist in target cells. Alternatively, viral vectors can be used that provide for transient expression of nucleic acid molecules. Such vectors can be repeatedly administered as necessary. Once expressed, the siNA molecule interacts with the target mRNA and generates an RNAi response. Delivery of siNA molecule expressing vectors can be systemic, such as by intravenous or intra-muscular administration, by administration to target cells ex-planted from a subject followed by reintroduction into the subject, or by any other means that would allow for introduction into the desired target cell (for a review see Couture et al, 1996, TIG., 12, 510).
In one aspect the invention features an expression vector comprising a nucleic acid sequence encoding at least one siNA molecule of the instant invention. The expression vector can encode one or both strands of a siNA duplex, or a single self-complementary
strand that self hybridizes into a siNA duplex. The nucleic acid sequences encoding the siNA molecules of the instant invention can be operably linked in a manner that allows expression of the siNA molecule (see for example Paul et al, 2002, Nature Biotechnology, 19, 505; Miyagishi and Taira, 2002, Nature Biotechnology, 19, 497; Lee et al, 2002, Nature Biotechnology, 19, 500; and Novina et al, 2002, Nature Medicine, advance online publication doi:10.1038/nm725).
In another aspect, the invention features an expression vector comprising: a) a transcription initiation region (e.g., eukaryotic pol I, II or III initiation region); b) a transcription termination region (e.g., eukaryotic pol I, II or III termination region); and c) a nucleic acid sequence encoding at least one of the siNA molecules of the instant invention, wherem said sequence is operably linked to said initiation region and said termination region in a manner that allows expression and/or delivery of the siNA molecule. The vector can optionally include an open reading frame (ORF) for a protein operably linked on the 5' side or the 3'-side of the sequence encoding the siNA of the invention; and/or an intron (intervening sequences).
Transcription of the siNA molecule sequences can be driven from a promoter for eukaryotic RNA polymerase I (pol I), RNA polymerase II (pol II), or RNA polymerase III (pol III). Transcripts from pol II or pol III promoters are expressed at high levels in all cells; the levels of a given pol II promoter in a given cell type depends on the nature of the gene regulatory sequences (enhancers, silencers, etc.) present nearby. Prokaryotic RNA polymerase promoters are also used, providing that the prokaryotic RNA polymerase enzyme is expressed in the appropriate cells (Elroy-Stein and Moss, 1990, Proc. Natl. Acad. Sci. USA, 87, 6743-7; Gao and Huang 1993, Nucleic Acids Res., 21, 2867-72; Lieber et al, 1993, Methods Enzymol, 111, 47-66; Zhou et al, 1990, Mol. Cell. Biol, 10, 4529-37). Several investigators have demonstrated that nucleic acid molecules expressed from such promoters can function in mammalian cells (e.g. Kashani-Sabet et al, 1992, Antisense Res. Dev., 2, 3-15; Ojwang et al, 1992, Proc Natl. Acad. Sci. U SA, 89, 10802-6; Chen et al, 1992, Nucleic Acids Res., 20, 4581-9; Yu et al, 1993, Proc. Natl Acad. Sci. USA, 90, 6340-4; L'Huillier et al, 1992, EMBO J., 11, 4411-8; Lisziewicz et al, 1993, Proc. Natl. Acad. Sci. U. S. A, 90, 8000-4; Thompson et al, 1995, Nucleic Acids Res., 23, 2259; Sullenger & Cech, 1993, Science,
262, 1566). More specifically, transcription units such as the ones derived from genes encoding U6 small nuclear (snRNA), transfer RNA (tRNA) and adenovirus VA RNA are useful in generating high concentrations of desired RNA molecules such as siNA in cells (Thompson et al, supra; Couture and Stinchcomb, 1996, supra; Noonberg et al, 1994, Nucleic Acid Res., 22, 2830; Noonberg et al, U.S. Pat. No. 5,624,803; Good et al, 1997, Gene Ther., 4, 45; Beigelman et al, International PCT Publication No. WO 96/18736. The above siNA transcription units can be incoφorated into a variety of vectors for introduction into mammalian cells, including but not restricted to, plasmid DNA vectors, viral DNA vectors (such as adenovirus or adeno-associated virus vectors), or viral RNA vectors (such as retroviral or alphaviras vectors) (for a review see Couture and Stinchcomb, 1996, supra).
In another aspect the invention features an expression vector comprising a nucleic acid sequence encoding at least one of the siNA molecules of the invention in a manner that allows expression of that siNA molecule. The expression vector comprises in one embodiment; a) a transcription initiation region; b) a transcription termination region; and c) a nucleic acid sequence encoding at least one strand of the siNA molecule, wherein the sequence is operably linked to the initiation region and the termination region in a manner that allows expression and/or delivery of the siNA molecule.
In another embodiment the expression vector comprises: a) a transcription initiation region; b) a transcription termination region; c) an open reading frame; and d) a nucleic acid sequence encoding at least one strand of a siNA molecule, wherein the sequence is operably linked to the 3 '-end of the open reading frame and wherein the sequence is operably linked to the initiation region, the open reading frame and the termination region in a manner that allows expression and/or delivery of the siNA molecule. In yet another embodiment, the expression vector comprises: a) a transcription initiation region; b) a transcription termination region; c) an intron; and d) a nucleic acid sequence encoding at least one siNA molecule, wherem the sequence is operably linked to the initiation region, the intron and the termination region in a manner which allows expression and/or delivery of the nucleic acid molecule. In another embodiment, the expression vector comprises: a) a transcription initiation region; b) a transcription termination region; c) an intron; d) an open reading
frame; and e) a nucleic acid sequence encoding at least one strand of a siNA molecule, wherein the sequence is operably linked to the 3 '-end of the open reading frame and wherein the sequence is operably linked to the initiation region, the intron, the open reading frame and the termination region in a manner which allows expression and/or delivery of the siNA molecule.
Expressed pseudogene target biology and biochemistry
Pseudogenes have been considered as nonfunctional sequences of genomic DNA originally derived from functional genes. The assumption follows that all pseudogene mutations are selectively neutral and have equal probability to become fixed in the population. However, pseudogenes that have been suitably investigated often exhibit functional roles, such as gene expression, gene regulation, generation of genetic (e.g., antibody, antigenic, and other) diversity (see for example Balakirev et al, 2003, Annu. Rev. Genet, 37, 123-51). A pseudogene is an evolutionary conserved gene copy that does not produce a functional, full-length protein. The human genome is estimated to contain upwards of 20,000 pseudogenes. Although much effort has been devoted to understanding the function of such pseudogenes, their biological roles remain largely unknown. Some psuedogenes that are expressed have been associated with disease or developmental conditions. For example, Hirotsune et al, 2003, Nature, 423, 91-96, report the role of an expressed pseudogene, specifically regulation of messenger-RNA stability, in a transgene-insertion mouse mutant exhibiting polycystic kidneys and bone deformity. The transgene was integrated into the vicinity of the expressing pseudogene of Makorinl, refeπed to as Makorinl-pl. This insertion attenuated transcription of Makorinl-pl, resulting in destabilization of Makorinl mRNA in trans by way of a cis- acting RNA decay element within the 5' region of Makorinl that is homologous between Makorinl and Makorinl-pl. Either Makorinl or Makorinl-pl tiansgenes could rescue the expressed pseudogene phenotypes. These findings demonstrate a specific regulatory role of an expressed pseudogene, and point to the functional significance of non-coding RNAs (Hirotsune et al, 2003, Nature, 423, 91-96). In a subsequent study, it was determined that 2-3% of human processed pseudogenes are expressed (Yano et al, 2004, J. Mol. Med., 82(7), 414-22). Other reports of expressed pseudogenes are described in, for example Kandouz et al, 2004, Oncogene, 23, 4763-70 (Connexin43); Yoshida et al,
2003, Hum Cell, 16, 65-72 (Makorinl); and Perez Jurado et al, 1998, Hum Mol Genet, 7, 325-34).
Examples:
The following are non-limiting examples showing the selection, isolation, synthesis and activity of nucleic acids of the instant invention.
Example 1 : Tandem synthesis of siNA constructs
Exemplary siNA molecules of the invention are synthesized in tandem using a cleavable linker, for example, a succinyl-based linker. Tandem synthesis as described herein is followed by a one-step purification process that provides RNAi molecules in high yield. This approach is highly amenable to siNA synthesis in support of high throughput RNAi screening, and can be readily adapted to multi-column or multi-well synthesis platforms.
After completing a tandem synthesis of a siNA oligo and its complement in which the 5'-terminal dimethoxytrityl (5'-0-DMT) group remains intact (trityl on synthesis), the oligonucleotides are deprotected as described above. Following deprotection, the siNA sequence strands are allowed to spontaneously hybridize. This hybridization yields a duplex in which one strand has retained the 5'-0-DMT group while the complementary strand comprises a terminal 5'-hydroxyl. The newly formed duplex behaves as a single molecule during routine solid-phase extraction purification (Trityl-On purification) even though only one molecule has a dimethoxytrityl group. Because the strands form a stable duplex, this dimethoxytrityl group (or an equivalent group, such as other trityl groups or other hydrophobic moieties) is all that is required to purify the pair of oligos, for example, by using a C18 cartridge.
Standard phosphoramidite synthesis chemistry is used up to the point of introducing a tandem linker, such as an inverted deoxy abasic succinate or glyceryl succinate linker (see Figure 1) or an equivalent cleavable linker. A non-limiting example of linker coupling conditions that can be used includes a hindered base such as diisopropylethylamine (DIPA) and/or DMAP in the presence of an activator reagent such as Bromotripyπolidinophosphoniumhexaflurorophosphate (PyBrOP). After the linker is
coupled, standard synthesis chemistry is utilized to complete synthesis of the second sequence leaving the terminal the 5'-0-DMT intact. Following synthesis, the resulting oligonucleotide is deprotected according to the procedures described herein and quenched with a suitable buffer, for example with 50mM NaOAc or 1.5M NH4H2CO3. Purification of the siNA duplex can be readily accomplished using solid phase extraction, for example, using a Waters C18 SepPak lg cartridge conditioned with 1 column volume (CV) of acetonitrile, 2 CV H20, and 2 CV 50mM NaOAc. The sample is loaded and then washed with 1 CV H20 or 50mM NaOAc. Failure sequences are eluted with 1 CV 14% ACN (Aqueous with 50mM NaOAc and 50mM NaCl). The column is then washed, for example with 1 CV H20 followed by on-column detritylation, for example by passing 1 CV of 1% aqueous trifluoroacetic acid (TFA) over the column, then adding a second CV of 1% aqueous TFA to the column and allowing to stand for approximately 10 minutes. The remaining TFA solution is removed and the column washed with H20 followed by 1 CV 1M NaCl and additional H20. The siNA duplex product is then eluted, for example, using 1 CV 20% aqueous CAN.
Figure 2 provides an example of MALDI-TOF mass spectrometry analysis of a purified siNA construct in which each peak coπesponds to the calculated mass of an individual siNA strand of the siNA duplex. The same purified siNA provides three peaks when analyzed by capillary gel electrophoresis (CGE), one peak presumably coπesponding to the duplex siNA, and two peaks presumably corresponding to the separate siNA sequence strands. Ion exchange HPLC analysis of the same siNA contract only shows a single peak. Testing of the purified siNA construct using a luciferase reporter assay described below demonstrated the same RNAi activity compared to siNA constructs generated from separately synthesized oligonucleotide sequence strands.
Example 2: Identification of potential siNA target sites in any RNA sequence
The sequence of an RNA target of interest, such as a viral or human mRNA transcript, is screened for target sites, for example by using a computer folding algorithm. In a non-limiting example, the sequence of a gene or RNA gene transcript derived from a database, such as Genbank, is used to generate siNA targets having
complementarity to the target. Such sequences can be obtained from a database, or can be determined experimentally as known in the art. Target sites that are known, for example, those target sites determined to be effective target sites based on studies with other nucleic acid molecules, for example ribozymes or antisense, or those targets known to be associated with a disease or condition such as those sites containing mutations or deletions, can be used to design siNA molecules targeting those sites. Various parameters can be used to determine which sites are the most suitable target sites within the target RNA sequence. These parameters include but are not limited to secondary or tertiary RNA structure, the nucleotide base composition of the target sequence, the degree of homology between various regions of the target sequence, or the relative position of the target sequence within the RNA transcript. Based on these determinations, any number of target sites within the RNA transcript can be chosen to screen siNA molecules for efficacy, for example by using in vitro RNA cleavage assays, cell culture, or animal models. In a non-limiting example, anywhere from 1 to 1000 target sites are chosen within the transcript based on the size of the siNA construct to be used. High throughput screening assays can be developed for screening siNA molecules using methods known in the art, such as with multi-well or multi-plate assays to determine efficient reduction in target gene expression.
Example 3: Selection of siNA molecule target sites in a RNA The following non-limiting steps can be used to carry out the selection of siNAs targeting a given gene sequence or transcript.
1. The target sequence is parsed in silico into a list of all fragments or subsequences of a particular length, for example 23 nucleotide fragments, contained within the target sequence. This step is typically caπied out using a custom Perl script, but commercial sequence analysis programs such as Oligo, MacVector, or the GCG Wisconsin Package can be employed as well.
2. In some instances the siNAs coπespond to more than one target sequence; such would be the case for example in targeting different transcripts of the same gene, targeting different transcripts of more than one gene, or for targeting both the human gene and an animal homolog. In this case, a subsequence list of a particular length is
generated for each of the targets, and then the lists are compared to find matching sequences in each list. The subsequences are then ranked according to the number of target sequences that contain the given subsequence; the goal is to find subsequences that are present in most or all of the target sequences. Alternately, the ranking can identify subsequences that are unique to a target sequence, such as a mutant target sequence. Such an approach would enable the use of siNA to target specifically the mutant sequence and not effect the expression of the normal sequence.
3. In some instances the siNA subsequences are absent in one or more sequences while present in the desired target sequence; such would be the case if the siNA targets a gene with a paralogous family member that is to remain untargeted. As in case 2 above, a subsequence list of a particular length is generated for each of the targets, and then the lists are compared to find sequences that are present in the target gene but are absent in the untargeted paralog.
4. The ranked siNA subsequences can be further analyzed and ranked according to GC content. A preference can be given to sites containing 30-70% GC, with a further preference to sites containing 40-60% GC.
5. The ranked siNA subsequences can be further analyzed and ranked according to self- folding and internal haiφins. Weaker internal folds are prefeπed; strong haiφin structures are to be avoided.
6. The ranked siNA subsequences can be further analyzed and ranked according to whether they have runs of GGG or CCC in the sequence. GGG (or even more Gs) in either strand can make oligonucleotide synthesis problematic and can potentially interfere with RNAi activity, so it is avoided whenever better sequences are available. CCC is searched in the target strand because that will place GGG in the antisense strand.
7. The ranked siNA subsequences can be further analyzed and ranked according to whether they have the dinucleotide UU (uridine dinucleotide) on the 3'-end of the sequence, and/or AA on the 5'-end of the sequence (to yield 3' UU on the antisense sequence). These sequences allow one to design siNA molecules with terminal TT thymidine dinucleotides.
8. Four or five target sites are chosen from the ranked list of subsequences as described above. For example, in subsequences having 23 nucleotides, the right 21 nucleotides of each chosen 23-mer subsequence are then designed and synthesized for the upper (sense) strand of the siNA duplex, while the reverse complement of the left 21 nucleotides of each chosen 23-mer subsequence are then designed and synthesized for the lower (antisense) strand of the siNA duplex. If terminal TT residues are desired for the sequence (as described in paragraph 7), then the two 3' terminal nucleotides of both the sense and antisense strands are replaced by TT prior to synthesizing the oligos.
9. The siNA molecules are screened in an in vitro, cell culture or animal model system to identify the most active siNA molecule or the most prefeπed target site within the target RNA sequence.
10. Other design considerations can be used when selecting target nucleic acid sequences, see, for example, Reynolds et al, 2004, Nature Biotechnology Advanced Online Publication, 1 February 2004, doi:10.1038/nbt936 and Ui-Tei et al., 2004, Nucleic Acids Research, 32, doi:10.1093/nar/gkh247.
In an alternate approach, a pool of siNA constracts specific to a target polynucloetide sequence is used to screen for target sites in cells expressing target RNA. The general strategy used in this approach is shown in Figure 9. Cells expressing target RNA are transfected with the pool of siNA constructs and cells that demonstrate a phenotype associated with target inhibition are sorted. The pool of siNA constracts can be expressed from transcription cassettes inserted into appropriate vectors (see for example Figure 7 and Figure 8). The siNA from cells demonstrating a positive phenotypic change (e.g., decreased proliferation, decreased RNA levels, decreased protein expression), are sequenced to determine the most suitable target site(s) within the target RNA sequence.
Example 4: Targeted siNA design siNA target sites were chosen by analyzing sequences of the target polynucleotide and optionally prioritizing the target sites on the basis of folding (structure of any given sequence analyzed to determine siNA accessibility to the target), by using a library of
siNA molecules as described in Example 3, or alternately by using an in vitro siNA system as described in Example 6 herein. siNA molecules are designed that could bind each target and are optionally individually analyzed by computer folding to assess whether the siNA molecule can interact with the target sequence. Varying the length of the siNA molecules can be chosen to optimize activity. Generally, a sufficient number of complementary nucleotide bases are chosen to bind to, or otherwise interact with, the target RNA, but the degree of complementarity can be modulated to accommodate siNA duplexes or varying length or base composition. By using such methodologies, siNA molecules can be designed to target sites within any known RNA sequence, for example those RNA sequences coπesponding to the any gene transcript.
Chemically modified siNA constructs are designed to provide nuclease stability for systemic administration in vivo and/or improved pharmacokinetic, localization, and delivery properties while preserving the ability to mediate RNAi activity. Chemical modifications as described herein are introduced synthetically using synthetic methods described herein and those generally known in the art. The synthetic siNA constracts are then assayed for nuclease stability in serum and/or cellular/tissue extracts (e.g. liver extracts). The synthetic siNA constructs are also tested in parallel for RNAi activity using an appropriate assay, such as a luciferase reporter assay as described herein or another suitable assay that can quantity RNAi activity. Synthetic siNA constructs that possess both nuclease stability and RNAi activity can be further modified and re- evaluated in stability and activity assays. The chemical modifications of the stabilized active siNA constructs can then be applied to any siNA sequence targeting any chosen RNA and used, for example, in target screening assays to pick lead siNA compounds for therapeutic development (see for example Figure 11).
Example 5: Chemical Synthesis and Purification of siNA siNA molecules can be designed to interact with various sites in the RNA message, for example, target sequences within the RNA sequences described herein. The sequence of one strand of the siNA molecule(s) is complementary to the target site sequences described above. The siNA molecules can be chemically synthesized using methods described herein. Inactive siNA molecules that are used as control sequences can be synthesized by scrambling the sequence of the siNA molecules such that it is not
complementary to the target sequence. Generally, siNA constructs can by synthesized using solid phase oligonucleotide synthesis methods as described herein (see for example Usman et al, US Patent Nos. 5,804,683; 5,831,071; 5,998,203; 6,117,657; 6,353,098; 6,362,323; 6,437,117; 6,469,158; Scaringe et al, US Patent Nos. 6,111,086; 6,008,400; 6, 111 ,086 all incoφorated by reference herein in their entirety).
In a non-limiting example, RNA oligonucleotides are synthesized in a stepwise fashion using the phosphoramidite chemistry as is known in the art. Standard phosphoramidite chemistry involves the use of nucleosides comprising any of 5'-0- dimethoxytrityl, 2'-0-tert-butyldimethylsilyl, 3'-0-2-Cyanoethyl N,N-diisopropylphos- phoroamidite groups, and exocyclic amine protecting groups (e.g. N6-benzoyl adenosine, N4 acetyl cytidine, and N2-isobutyryl guanosine). Alternately, 2'-0-Silyl Ethers can be used in conjunction with acid-labile 2'-0-orthoester protecting groups in the synthesis of RNA as described by Scaringe supra. Differing 2' chemistries can require different protecting groups, for example 2 '-deoxy-2 '-amino nucleosides can utilize N-phthaloyl protection as described by Usman et al, US Patent 5,631,360, incoφorated by reference herein in its entirety).
During solid phase synthesis, each nucleotide is added sequentially (3'- to 5'- direction) to the solid support-bound oligonucleotide. The first nucleoside at the 3 '-end of the chain is covalently attached to a solid support (e.g., controlled pore glass or polystyrene) using various linkers. The nucleotide precursor, a ribonucleoside phosphoramidite, and activator are combined resulting in the coupling of the second nucleoside phosphoramidite onto the 5 '-end of the first nucleoside. The support is then washed and any unreacted 5 '-hydroxyl groups are capped with a capping reagent such as acetic anhydride to yield inactive 5 '-acetyl moieties. The trivalent phosphorus linkage is then oxidized to a more stable phosphate linkage. At the end of the nucleotide addition cycle, the 5'-0-protecting group is cleaved under suitable conditions (e.g., acidic conditions for trityl-based groups and Fluoride for silyl-based groups). The cycle is repeated for each subsequent nucleotide.
Modification of synthesis conditions can be used to optimize coupling efficiency, for example by using differing coupling times, differing reagent/phosphoramidite concentrations, differing contact times, differing solid supports and solid support linker
chemistries depending on the particular chemical composition of the siNA to be synthesized. Deprotection and purification of the siNA can be performed as is generally described in Usman et al, US 5,831,071, US 6,353,098, US 6,437,117, and Bellon et al, US 6,054,576, US 6,162,909, US 6,303,773, or Scaringe supra, incoφorated by reference herein in their entireties. Additionally, deprotection conditions can be modified to provide the best possible yield and purity of siNA constructs. For example, applicant has observed that oligonucleotides comprising 2'-deoxy-2 '-fluoro nucleotides can degrade under inappropriate deprotection conditions. Such oligonucleotides are deprotected using aqueous methylamine at about 35°C for 30 minutes. If the 2'-deoxy- 2'-fluoro containing oligonucleotide also comprises ribonucleotides, after deprotection with aqueous methylamine at about 35°C for 30 minutes, TEA-HF is added and the reaction maintained at about 65°C for an additional 15 minutes.
Example 6: RNAi in vitro assay to assess siNA activity
An in vitro assay that recapitulates RNAi in a cell-free system is used to evaluate siNA constructs targeting target RNA. The assay comprises the system described by Tuschl et al, 1999, Genes and Development, 13, 3191-3197 and Zamore et al, 2000, Cell, 101, 25-33 adapted for use with target RNA. A Drosophila extract derived from syncytial blastoderm is used to reconstitute RNAi activity in vitro. Target RNA is generated via in vitro transcription from an appropriate target expressing plasmid using T7 RNA polymerase or via chemical synthesis as described herein. Sense and antisense siNA strands (for example 20 uM each) are annealed by incubation in buffer (such as 100 mM potassium acetate, 30 mM HEPES-KOH, pH 7.4, 2 mM magnesium acetate) for 1 minute at 90°C followed by 1 hour at 37°C , then diluted in lysis buffer (for example 100 M potassium acetate, 30 M HEPES-KOH at pH 7.4, 2mM magnesium acetate). Annealing can be monitored by gel electrophoresis on an agarose gel in TBE buffer and stained with ethidium bromide. The Drosophila lysate is prepared using zero to two- hour-old embryos from Oregon R flies collected on yeasted molasses agar that are dechorionated and lysed. The lysate is centrifuged and the supernatant isolated. The assay comprises a reaction mixture containing 50% lysate [vol/vol], RNA (10-50 pM final concentration), and 10% [vol/vol] lysis buffer containing siNA (10 nM final concentration). The reaction mixture also contains 10 mM creatme phosphate, 10 ug/ml
creatine phosphokinase, 100 urn GTP, 100 uM UTP, 100 uM CTP, 500 uM ATP, 5 mM DTT, 0.1 U/uL RNasin (Promega), and 100 uM of each amino acid. The final concentration of potassium acetate is adjusted to 100 mM. The reactions are pre- assembled on ice and preincubated at 25° C for 10 minutes before adding RNA, then incubated at 25° C for an additional 60 minutes. Reactions are quenched with 4 volumes of 1.25 x Passive Lysis Buffer (Promega). Target RNA cleavage is assayed by RT-PCR analysis or other methods known in the art and are compared to control reactions in which siNA is omitted from the reaction.
Alternately, internally-labeled target RNA for the assay is prepared by in vitro transcription in the presence of [alpha-32p] CTP, passed over a G50 Sephadex column by spin chromatography and used as target RNA without further purification. Optionally, 32 target RNA is 5'- P-end labeled using T4 polynucleotide kinase enzyme. Assays are performed as described above and target RNA and the specific RNA cleavage products generated by RNAi are visualized on an autoradiograph of a gel. The percentage of cleavage is determined by PHOSPHOR IMAGER® (autoradiography) quantitation of bands representing intact control RNA or RNA from control reactions without siNA and the cleavage products generated by the assay.
In one embodiment, this assay is used to determine target sites in the target RNA target for siNA mediated RNAi cleavage, wherein a plurality of siNA constracts are screened for RNAi mediated cleavage of the target RNA, for example, by analyzing the assay reaction by electrophoresis of labeled target RNA, or by northern blotting, as well as by other methodology well known in the art.
Example 7: Nucleic acid inhibition of target RNA siNA molecules targeted to the human target RNA are designed and synthesized as described above. These nucleic acid molecules can be tested for cleavage activity in vivo, for example, using the following procedure.
Two formats are used to test the efficacy of siNAs of the invention. First, the reagents are tested in cell culture to determine the extent of RNA and protein inhibition. siNA reagents are selected against the target as described herein. RNA inhibition is
measured after delivery of these reagents by a suitable transfection agent to cells. Relative amounts of target RNA are measured versus actin using real-time PCR monitoring of amplification (eg., ABI 7700 TAQMAN® (real-time PCR monitoring of amplification)'). A comparison is made to a mixture of oligonucleotide sequences made to unrelated targets or to a randomized siNA control with the same overall length and chemistry, but randomly substituted at each position. Primary and secondary lead reagents are chosen for the target and optimization performed. After an optimal transfection agent concentration is chosen, a RNA time-course of inhibition is performed with the lead siNA molecule. In addition, a cell-plating format can be used to determine RNA inhibition.
Delivery of siNA to Cells
Cells (e.g., HEKn/HEKa, HeLa, A549, A375 cells) are seeded, for example, at 1x10^ cells per well of a six- well dish in EGM-2 (Bio Whittaker) the day before transfection. siNA (final concentration, for example 20nM) and cationic lipid (e.g., final concentration 2μg/ml) are complexed in EGM basal media (Bio Whittaker) at 37°C for 30 minutes in polystyrene tubes. Following vortexing, the complexed siNA is added to each well and incubated for the times indicated. For initial optimization experiments, cells are seeded, for example, at 1x10^ in 96 well plates and siNA complex added as described. Efficiency of delivery of siNA to cells is determined using a fluorescent siNA complexed with lipid. Cells in 6-well dishes are incubated with siNA for 24 hours, rinsed with PBS and fixed in 2% paraformaldehyde for 15 minutes at room temperature. Uptake of siNA is visualized using a fluorescent microscope.
TAQMAN® ("real-time PCR monitoring of amplification) and Lightcycler quantification of mRNA Total RNA is prepared from cells following siNA delivery, for example, using
Qiagen RNA purification kits for 6-well or Rneasy extraction kits for 96-well assays. For
TAQMAN® analysis (real-time PCR monitoring of amplification), dual-labeled probes are synthesized with the reporter dye, FAM or JOE, covalently linked at the 5'-end and the quencher dye TAMRA conjugated to the 3 '-end. One-step RT-PCR amplifications are performed on, for example, an ABI PRISM 7700 Sequence Detector using 50 μl
reactions consisting of 10 μl total RNA, 100 nM forward primer, 900 nM reverse primer, 100 nM probe, IX TaqMan PCR reaction buffer (PE-Applied Biosystems), 5.5 mM MgCl2, 300 μM each dATP, dCTP, dGTP, and dTTP, 10U RNase Inhibitor (Promega), 1.25U AMPLITAQ GOLD® (DNA polymerase) (PE-Applied Biosystems) and lOU M- MLV Reverse Transcriptase (Promega). The thermal cycling conditions can consist of 30 minutes at 48°C, 10 minutes at 95°C, followed by 40 cycles of 15 seconds at 95°C and 1 minute at 60°C. Quantitation of mRNA levels is determined relative to standards generated from serially diluted total cellular RNA (300, 100, 33, 11 ng/rxn) and normalizing to β-actin or GAPDH mRNA in parallel TAQMAN® reactions (real-time PCR monitoring of amplification). For each gene of interest an upper and lower primer and a fluorescently labeled probe are designed. Real time incoφoration of SYBR Green I dye into a specific PCR product can be measured in glass capillary tubes using a lightcyler. A standard curve is generated for each primer pair using control cKNA. Values are represented as relative expression to GAPDH in each sample.
Western blotting
Nuclear extracts can be prepared using a standard micro preparation technique (see for example Andrews and Faller, 1991, Nucleic Acids Research, 19, 2499). Protein extracts from supernatants are prepared, for example using TCA precipitation. An equal volume of 20% TCA is added to the cell supernatant, incubated on ice for 1 hour and pelleted by centrifugation for 5 minutes. Pellets are washed in acetone, dried and resuspended in water. Cellular protein extracts are run on a 10% Bis-Tris NαPage (nuclear extracts) or 4-12% Tris-Glycine (supernatant extracts) polyacrylamide gel and transfeπed onto nitro-cellulose membranes. Non-specific binding can be blocked by incubation, for example, with 5% non-fat milk for 1 hour followed by primary antibody for 16 hour at 4°C. Following washes, the secondary antibody is applied, for example (1:10,000 dilution) for 1 hour at room temperature and the signal detected with SuperSignal reagent (Pierce).
Example 8: Models useful to evaluate the down-regulation of gene expression
Evaluating the efficacy of siNA molecules of the invention in animal models is an important prerequisite to human clinical trials. Various animal models of cancer,
proliferative, inflammatory, autoimmune, neurologic, ocular, respiratory, metabolic, etc. diseases, conditions, or disorders as are known in the art can be adapted for use for pre- clinical evaluation of the efficacy of nucleic acid compositions of the invetention in modulating target gene expression toward therapeutic, cosmetic, or research use.
Example 9: RNAi mediated inhibition of target gene expression siNA constructs (are tested for efficacy in reducing target RNA expression in cells, (e.g., HEKn/HEKa, HeLa, A549, A375 cells). Cells are plated approximately 24 hours before transfection in 96-well plates at 5,000-7,500 cells/well, 100 μl/well, such that at the time of transfection cells are 70-90% confluent. For transfection, annealed siNAs are mixed with the transfection reagent (Lipofectamine 2000, Invifrogen) in a volume of 50 μl/well and incubated for 20 minutes at room temperature. The siNA transfection mixtures are added to cells to give a final siNA concentration of 25 nM in a volume of 150 μl. Each siNA transfection mixture is added to 3 wells for triplicate siNA treatments. Cells are incubated at 37° for 24 hours in the continued presence of the siNA transfection mixture. At 24 hours, RNA is prepared from each well of treated cells. The supematants with the transfection mixtures are first removed and discarded, then the cells are lysed and RNA prepared from each well. Target gene expression following freatment is evaluated by RT-PCR for the target gene and for a control gene (36B4, an RNA polymerase subunit) for normalization. The triplicate data is averaged and the standard deviations determined for each treatment. Normalized data are graphed and the percent reduction of target mRNA by active siNAs in comparison to their respective inverted control siNAs is determined.
Example 10: Indications
Particular conditions and disease states that can be associated with gene expression modulation include, but are not limited to proliferative, inflammatory, autoimmune, neurologic, ocular, respiratory, metabolic etc. diseases, conditions, or disorders as described herein or otherwise known in the art, and any other diseases, conditions or disorders that are related to or will respond to the levels of a target (e.g., target protein or target polynucleotide) in a cell or tissue, alone or in combination with other therapies.
Those skilled in the art will recognize that other drags such as anti-cancer compounds and therapies can be similarly be readily combined with the nucleic acid molecules of the instant invention (e.g. ribozymes and antisense molecules) and are hence within the scope of the instant invention. Such compounds and therapies are well known in the art. For combination therapy, the nucleic acids of the invention are prepared in one of two ways. First, the agents are physically combined in a preparation of nucleic acid and chemotherapeutic agent, such as a mixture of a nucleic acid of the invention encapsulated in liposomes and ifosfamide in a solution for intravenous administration, wherein both agents are present in a therapeutically effective concentration (e.g., ifosfamide in solution to deliver 1000-1250 mg/m2/day and liposome-associated nucleic acid of the invention in the same solution to deliver 0.1-100 mg/kg/day). Alternatively, the agents are administered separately but simultaneously in their respective effective doses (e.g., 1000-1250 mg/m2/d ifosfamide and 0.1 to 100 mg/kg/day nucleic acid of the invention).
Example 11 : Diagnostic uses
The siNA molecules of the invention can be used in a variety of diagnostic applications, such as in the identification of molecular targets (e.g., RNA) in a variety of applications, for example, in clinical, industrial, environmental, agricultural and/or research settings. Such diagnostic use of siNA molecules involves utilizing reconstituted RNAi systems, for example, using cellular lysates or partially purified cellular lysates. siNA molecules of this invention can be used as diagnostic tools to examine genetic drift and mutations within diseased cells or to detect the presence of endogenous or exogenous, for example viral, RNA in a cell. The close relationship between siNA activity and the stracture of the target RNA allows the detection of mutations in any region of the molecule, which alters the base-pairing and three-dimensional structure of the target RNA. By using multiple siNA molecules described in this invention, one can map nucleotide changes, which are important to RNA structure and function in vitro, as well as in cells and tissues. Cleavage of target RNAs with siNA molecules can be used to inhibit gene expression and define the role of specified gene products in the progression of disease or infection. In this manner, other genetic targets can be defined as important mediators of the disease. These experiments will lead to better treatment of
the disease progression by affording the possibility of combination therapies (e.g., multiple siNA molecules targeted to different genes, siNA molecules coupled with known small molecule inhibitors, or intermittent treatment with combinations siNA molecules and/or other chemical or biological molecules). Other in vitro uses of siNA molecules of this invention are well known in the art, and include detection of the presence of mRNAs associated with a disease, infection, or related condition. Such RNA is detected by determining the presence of a cleavage product after treatment with a siNA using standard methodologies, for example, fluorescence resonance emission transfer (FRET). In a specific example, siNA molecules that cleave only wild-type or mutant forms of the target RNA are used for the assay. The first siNA molecules (i.e., those that cleave only wild-type forms of target RNA) are used to identify wild-type RNA present in the sample and the second siNA molecules (i.e., those that cleave only mutant forms of target RNA) are used to identify mutant RNA in the sample. As reaction controls, synthetic substrates of both wild-type and mutant RNA are cleaved by both siNA molecules to demonstrate the relative siNA efficiencies in the reactions and the afcsence of cleavage of the "non-targeted" RNA species. The cleavage products from the synthetic substrates also serve to generate size markers for the analysis of wild-type and mutant RNAs in the sample population. Thus, each analysis requires two siNA molecules, two substrates and one unknown sample, which is combined into six reactions. The presence of cleavage products is determined using an RNase protection assay so that full-length and cleavage fragments of each RNA can be analyzed in one lane of a polyacrylamide gel. It is not absolutely required to quantify the results to gain insight into the expression of mutant RNAs and putative risk of the desired phenotypic changes in target cells. The expression of mRNA whose protein product is implicated in the development of the phenotype (i.e., disease related or infection related) is adequate to establish risk. If probes of comparable specific activity are used for both transcripts, then a qualitative comparison of RNA levels is adequate and decreases the cost of the initial diagnosis. Higher mutant form to wild-type ratios are coπelated with higher risk whether RNA levels are compared qualitatively or quantitatively.
All patents and publications mentioned in the specification are indicative of the levels of skill of those skilled in the art to which the invention pertains. All references cited in this disclosure are incoφorated by reference to the same extent as if each reference had been incoφorated by reference in its entirety individually. One skilled in the art would readily appreciate that the present invention is well adapted to carry out the objects and obtain the ends and advantages mentioned, as well as those inherent therein. The methods and compositions described herein as presently representative of prefeπed embodiments are exemplary and are not intended as limitations on the scope of the invention. Changes therein and other uses will occur to those skilled in the art, which are encompassed within the spirit of the invention, are defined by the scope of the claims.
It will be readily apparent to one skilled in the art that varying substitutions and modifications can be made to the invention disclosed herein without departing from the scope and spirit of the invention. Thus, such additional embodiments are within the scope of the present invention and the following claims. The present invention teaches one skilled in the art to test various combinations and/or substitutions of chemical modifications described herein toward generating nucleic acid constructs with improved activity for mediating RNAi activity. Such improved activity can comprise improved stability, improved bioavailability, and/or improved activation of cellular responses mediating RNAi. Therefore, the specific embodiments described herein are not limiting and one skilled in the art can readily appreciate that specific combinations of the modifications described herein can be tested without undue experimentation toward identifying siNA molecules with improved RNAi activity.
The invention illustratively described herein suitably can be practiced in the absence of any element or elements, limitation or limitations that are not specifically disclosed herein. Thus, for example, in each instance herein any of the terms "comprising", "consisting essentially of, and "consisting of may be replaced with either of the other two terms. The terms and expressions which have been employed are used as terms of description and not of limitation, and there is no intention that in the use of such terms and expressions of excluding any equivalents of the features shown and described or portions thereof, but it is recognized that various modifications are possible
within the scope of the invention claimed. Thus, it should be understood that although the present invention has been specifically disclosed by prefeπed embodiments, optional features, modification and variation of the concepts herein disclosed may be resorted to by those skilled in the art, and that such modifications and variations are considered to be within the scope of this invention as defined by the description and the appended claims.
In addition, where features or aspects of the invention are described in terms of Markush groups or other grouping of alternatives, those skilled in the art will recognize that the invention is also thereby described in terms of any individual member or subgroup of members of the Markush group or other group.
NMJ300536 Homo sapiens recombination activating gene 2 (RAG2), mRNA NM_000601 Homo sapiens hepatocyte growth factor (hepapoietin A; scatter factor) (HGF NM_000940 Homo sapiens paraoxonase 3 (PON3), mRNA NM_000941 Homo sapiens P450 (cytochrome) oxidoreductase (POR), mRNA NM_000953 Homo sapiens prostaglandin D2 receptor (DP) (PTGDR), mRNA NM_0010011: Homo sapiens intersectin 1 (SH3 domain protein) (ITSN1), transcript variant NM_0010011! Homo sapiens transient receptor potential cation channel, subfamily M, mem NM_0010012! Homo sapiens solute carrier family 2 (facilitated glucose transporter), membe NM_0010013' Homo sapiens trypsin X3 (TRY1), mRNA
NM_0010013: Homo sapiens ATPase, Ca++ transporting, plasma membrane 1 (ATP2B1), t NM_0010013I Homo sapiens similar to ubiquitin-specific protease 17-like protein (LOC4014 NM_0010013: Homo sapiens Kazal type serine protease inhibitor 5-like 2 (SPINK5L2), mRI* NM_00100131 Homo sapiens protein kinase C substrate 80K-H (PRKCSH), transcript variar NM_0010013! Homo sapiens chromosome 10 open reading frame 74 (C10orf74), mRNA NM_0010013: Homo sapiens ATPase, Ca++ transporting, plasma membrane 2 (ATP2B2), t NM_0010013I Homo sapiens cytochrome b5 reductase b5R.2 (CYB5R2), transcript variant NM_0010013' Homo sapiens biogenesis of lysosome-related organelles complex-1 , subunr NM_0010013< Homo sapiens MGC27121 gene (MGC27121), mRNA NM_0010013- Homo sapiens ATPase, Ca++ transporting, plasma membrane 3 (ATP2B3), t NM_0010013< Homo sapiens claudin 20 (CLDN20), mRNA
NM_0010013- Homo sapiens NFKB inhibitor interacting Ras-like 2 (NKIRAS2), transcript va NM_00100131 Homo sapiens CD44 antigen (homing function and Indian blood group syster NM_0010013! Homo sapiens CD44 antigen (homing function and Indian blood group syster NM_0010013! Homo sapiens CD44 antigen (homing function and Indian blood group syster NM_0010013! Homo sapiens CD44 antigen (homing function and Indian blood group syster NM_0010013! Homo sapiens HCG3 gene (HCG3), mRNA
NM_0010013! Homo sapiens LIM domain only 3 (rhombotin-Iike 2) (LM03), transcript variar NM_0010013! Homo sapiens ATPase, Ca++ transporting, plasma membrane 4 (ATP2B4), 1 NM_0010014 Homo sapiens hypothetical protein MGC24381 (MGC24381), mRNA NM_0010014' Homo sapiens similar to Zinc finger protein 208 (LOC163223), mRNA NM_0010014' Homo sapiens family with sequence similarity 26,, member C (FAM26C), mRI NM_0010014' Homo sapiens similar to F-box only protein 2 (LOC342897), mRNA NM_0010014 Homo sapiens zinc finger protein 429 (ZNF429), mRNA NM_0010014' Homo sapiens similar to TBC1 domain family, member 3, centromeric (LOC4 NM_0010014' Homo sapiens similar to TBC1 domain family, member 3, telomeric (MGC44! NM_0010014' Homo sapiens SMAD, mothers against DPP homolog 5 (Drosophila) (SMAD! NM_00100141 Homo sapiens SMAD, mothers against DPP homolog 5 (Drosophila) (SMAD! NM_0010014I Homo sapiens troponin T2, cardiac (TNNT2), transcript variant 2, mRNA NM_0010014: Homo sapiens troponin T2, cardiac (TNNT2), transcript variant 3, mRNA NM_0010014I Homo sapiens troponin T2, cardiac (TNNT2), transcript variant 4, mRNA NM_0010014: Homo sapiens syntaxin 16 (STX16), transcript variant 1 , mRNA NM_0010014: Homo sapiens syntaxin 16 (STX16), transcript variant 3, mRNA NM_0010014: Homo sapiens chemokine (C-C motif) ligand 44ike (CCL4L), mRNA NM_0010014! Homo sapiens similar to RIKEN cDNA 4921524J17 (LOC388272), mRNA NM_0010014: Homo sapiens chemokine (C-C motif) ligand 34ike, centromeric (MGC12815 NM_0010014I Homo sapiens lanosterol synthase (2,3-oxidosqualene-lanosterol cyclase) (L NMJ3010014' Homo sapiens solute carrier family 35, member E4 (SLC35E4), mRNA NM_0010014I Homo sapiens hypothetical protein FLJ11011 (FLJ11011), transcript variant ■ NM_0010014I Homo sapiens hypothetical protein FU11011 (FLJ11011), transcript variant : NM_00100141 Homo sapiens zinc finger, DHHC domain containing 13 (ZDHHC13), transcri NM_00100141 Homo sapiens phosphotriesterase related (PTER), transcript variant 1 , mRN; NM_0010014i Homo sapiens ATPase, Ca++ transporting, type 2C, member 1 (ATP2C1), tπ NM_00100141 Homo sapiens ATPase, Ca++ transporting, type 2C, member 1 (ATP2C1 ), tπ NM_00100141 Homo sapiens ATPase, Ca++ transporting, type 2C, member 1 (ATP2C1), tr; NM_00100151 Homo sapiens synuclein, beta (SNCB), transcript variant 1 , mRNA NM_00100151 Homo sapiens NADH dehydrogenase (ubiquinone) flavoprotein 3, 10kDa (Nt NMJ3010015: Homo sapiens hepatoma-derived growth factor-related protein 2 (HDGF2), tr
NM_00100 Homo sapiens UDP-glucose pyrophosphorylase 2 (UGP2), transcript variant NM_00100 Homo sapiens transgelin (TAGLN), transcript variant 1 , mRNA NM_00100 Homo sapiens RAR-related orphan receptor C (RORC), transcript variant 2, i NM_00100 Homo sapiens transmembrane 6 superfamily member 2 (TM6SF2), transcrip NM_00100 Homo sapiens CD36 antigen (collagen type I receptor, thrombospondin rece| NM_00100 Homo sapiens CD36 antigen (collagen type I receptor, thrombospondin recei NM_00100 Homo sapiens growth factor receptor-bound protein 10 (GRB10), transcript v NM_00100 Homo sapiens growth factor receptor-bound protein 10 (GRB10), transcript v NM_00100 Homo sapiens chromosome 9 open reading frame 103 (C9orf103), mRNA NM_00100 Homo sapiens LEM domain containing 1 (LEMD1), mRNA NM_00100 Homo sapiens germ and embryonic stem cell enriched protein STELLA (STE NM_00100 Homo sapiens growth factor receptor-bound protein 10 (GRB10), transcript v NM_00100 Homo sapiens galactokinase 2 (GALK2), transcript variant 2, mRNA NM_00100 Homo sapiens similar to growth differentiation factor 16 (LOC392255), mR NM_00100 Homo sapiens golgi associated, gamma adaptin ear containing, ARF binding NM_00100 Homo sapiens golgi associated, gamma adaptin ear containing, ARF binding NM_00100 Homo sapiens translocase of inner mitochondrial membrane 50 homolog (ye NM_00100 Homo sapiens phosphodiesterase 9A (PDE9A), transcript variant 2, mRNA NM_00100 Homo sapiens phosphodiesterase 9A (PDE9A), transcript variant 3, mRNA NM_00100 Homo sapiens phosphodiesterase 9A (PDE9A), transcript variant 4, mRNA NM_00100 Homo sapiens phosphodiesterase 9A (PDE9A), transcript variant 5, mRNA NM_00100 Homo sapiens phosphodiesterase 9A (PDE9A), transcript variant 6, mRNA NM_00100 Homo sapiens phosphodiesterase 9A (PDE9A), transcript variant 7, mRNA NM_00100 Homo sapiens phosphodiesterase 9A (PDE9A), transcript variant 8, mRNA NM_00100 Homo sapiens phosphodiesterase 9A (PDE9A), transcript variant 9, mRNA NM_00100 Homo sapiens phosphodiesterase 9A (PDE9A), transcript variant 10, mRNA NM_00100 Homo sapiens phosphodiesterase 9A (PDE9A), transcript variant 11 , mRNA NM_00100 Homo sapiens phosphodiesterase 9A (PDE9A), transcript variant 12, mRNA NM_00100 Homo sapiens phosphodiesterase 9A (PDE9A), transcript variant 13, mRNA NM_00100 Homo sapiens phosphodiesterase 9A (PDE9A), transcript variant 14, mRNA NM_00100 Homo sapiens phosphodiesterase 9A (PDE9A), transcript variant 15, mRNA NM_00100 Homo sapiens phosphodiesterase 9A (PDE9A), transcript variant 16, mRNA NM_00100 Homo sapiens phosphodiesterase 9A (PDE9A), transcript variant 17, mRNA NM_00100 Homo sapiens phosphodiesterase 9A (PDE9A), transcript variant 18, mRNA NM_00100 Homo sapiens phosphodiesterase 9A (PDE9A), transcript variant 19, mRNA NM_00100 Homo sapiens phosphodiesterase 9A (PDE9A), transcript variant 20, mRNA NM_00100 Homo sapiens ATPase, Na+/K+ transporting, alpha 1 polypeptide (ATP1A1), NM_00100 Homo sapiens mediator of RNA polymerase II transcription, subunit 8 homol< NM_00100 Homo sapiens mediator of RNA polymerase II transcription, subunit 8 homoli NM_00100 Homo sapiens mediator of RNA polymerase II transcription, subunit 8 homol< NM_00100 Homo sapiens similar to hypothetical protein 9530023G02 (MGC90512), mR NM_00100 Homo sapiens olfactory receptor, family 9, subfamily A, member 4 (OR9A4), NM_00100 Homo sapiens olfactory receptor, family 9, subfamily A, member 2 (OR9A2), NM_00100 Homo sapiens olfactory receptor, family 2, subfamily A, member 14 (OR2A1 * NM_00100 Homo sapiens hypothetical protein LOC144363 (LOC144363), mRNA NM_00100 Homo sapiens hypothetical protein LOC155054 (LOC155054), mRNA NM_00100 Homo sapiens FLJ16636 protein (FLJ16636), mRNA NM_00100 Homo sapiens hypothetical LOC255349 (bA9F11.1), mRNA NM_00100 Homo sapiens hypothetical protein LOC339745 (LOC339745), mRNA NM_00100 Homo sapiens FLJ 16008 protein (FLJ 16008), mRNA NM_00100 Homo sapiens prostate cancer associated protein 5 (PCANAP5), transcript v NM_00100 Homo sapiens olfactory receptor, family 6, subfamily V, member 1 (OR6V1), NM_00100 Homo sapiens FLJ26175 protein (FLJ26175), mRNA NM_00100 Homo sapiens FLJ41603 protein (FLJ41603), mRNA NM_00100 Homo sapiens FLJ46321 protein (FLJ46321), mRNA NM_00100 Homo sapiens FLJ16518 protein (FLJ16518), mRNA NM 00100 Homo sapiens ribonuclease-like protein 9 (h461), mRNA
316' Homo sapiens FLJ46385 protein (FLJ46385), mRNA
316' Homo sapiens lipocalin 9 (LCN9), mRNA
316" Homo sapiens FLJ46300 protein (FLJ46300), mRNA
316' Homo sapiens FLJ44653 protein (FLJ44653), mRNA
316' Homo sapiens FLJ41423 protein (FLJ41423), mRNA
3161 Homo sapiens FLJ42102 protein (FLJ42102), mRNA
3161 Homo sapiens FLJ45300 protein (FLJ45300), mRNA
)16! Homo sapiens FLJ45530 protein (FLJ45530), mRNA
)16l Homo sapiens similar to HSPC296 (MGC88387), mRNA
316! Homo sapiens FLJ45831 protein (FLJ45831), mRNA
3161 Homo sapiens FLJ45079 protein (FLJ45079), mRNA
3161 Homo sapiens FLJ43870 protein (FLJ43870), mRNA
3161 Homo sapiens FLJ26850 protein (FLJ26850), mRNA
)16l Homo sapiens FLJ35409 protein (FLJ35409), mRNA
)16! Homo sapiens FLJ44005 protein (FLJ44005), mRNA
)16! Homo sapiens hypothetical FLJ42133 (FLJ42133), mRNA
316! Homo sapiens FLJ44790 protein (FLJ44790), mRNA
316! Homo sapiens FLJ45139 protein (FLJ45139), mRNA
316! Homo sapiens FLJ46257 protein (FLJ46257), mRNA
316! Homo sapiens FLJ41993 protein (FLJ41993), mRNA
316! Homo sapiens FLJ42418 protein (FLJ42418), mRNA
316! Homo sapiens FLJ44006 protein (FLJ44006), mRNA
316! Homo sapiens FLJ41821 protein (FLJ41821), mRNA
316! Homo sapiens FLJ43879 protein (FLJ43879), mRNA
316! Homo sapiens FLJ25996 protein (FLJ25996), mRNA
3171 Homo sapiens FLJ45966 protein (FLJ45966), mRNA
3171 Homo sapiens HCV F-transactivated protein 1 (LOC401152), mRNA
3171 Homo sapiens FLJ33360 protein (FLJ33360), mRNA
3171 Homo sapiens FLJ46010 protein (FLJ46010), mRNA
3171 Homo sapiens FLJ44796 protein (FLJ44796), mRNA
3171 Homo sapiens FLJ41649 protein (FLJ41649), mRNA
3171 Homo sapiens FLJ42177 protein (FLJ42177), mRNA
3171 Homo sapiens FLJ45974 protein (FLJ45974), mRNA
1171 Homo sapiens FLJ45537 protein (FLJ45537), mRNA
317' Homo sapiens similar to 4931415M17 protein (LOC401565), mRNA
317' Homo sapiens DNA-damage inducible protein 1 (DDI1), mRNA
317' Homo sapiens lipocalin 10 (LCN10), mRNA
317 Homo sapiens SH3 domain binding glutamic acid-rich protein (SH3BGR), tra
317 Homo sapiens FERM, RhoGEF (ARHGEF) and pleckstrin domain protein 1 (■
317' Homo sapiens nuclear factor of kappa light polypeptide gene enhancer in B-<
317: Homo sapiens chromodomain protein, Y chromosome, 2 related (CDY), mR
3171 Homo sapiens transmembrane protein 1 (TMEM1), transcript variant 2, mRN
317: Homo sapiens Mahlavu hepatocellular carcinoma (HHCM), mRNA
31 : Homo sapiens chromosome 10 open reading frame 130 (C10orf130), mRNA
317: Homo sapiens ATPase, Na+/K+ transporting, alpha 4 polypeptide (ATP1 A4),
317- Homo sapiens constitutive photomorphogenic protein (COP1), transcript vari
317! Homo sapiens BRCC2 mRNA (BRCC2), mRNA
3171 Homo sapiens ATPase, Na+/K+ transporting, beta 1 polypeptide (ATP1 B1), t
31 ! Homo sapiens chromosome 21 open reading frame 24 (C21orf24), mRNA
317! Homo sapiens chromosome 9 open reading frame 105 (C9orf105), mRNA
317! Homo sapiens chromosome 10 open reading frame 55 (C10orf55), mRNA
317! Homo sapiens similar to RIKEN cDNA 1700027J05 gene (MGC33692), mRN
317! Homo sapiens similar to RIKEN cDNA C030006K11 gene (MGC70857), mRt
3181 Homo sapiens olfactory receptor, family 2, subfamily A, member 42 (OR2A42
318! Homo sapiens olfactory receptor, family 2, subfamily T, member 34 (OR2T34
318! Homo sapiens olfactory receptor, family 2, subfamily T, member 27 (OR2T27
318: Homo sapiens olfactory receptor, family 2, subfamily T, member 35 (OR2T3E
NMJ30100 I Homo sapiens similar to RIKEN cDNA A030009B12 gene (MGC21382), mRr NM_00100 ! Homo sapiens inter-alpha (globulin) inhibitor H5 (ITIH5), transcript variant 3, NM 30100 ! Homo sapiens serine/threonine-protein kinase pim-3 (PIM3), mRNA NM_00100 Homo sapiens hypothetical protein MGC14376 (MGC14376), transcript varia NM_00100 Homo sapiens heat shock transcription factor, Y-linked 1 (HSFY1 ), transcript NM_00100 ' Homo sapiens chromosome 14 open reading frame 37 (C14orf37), mRNA NM_00100 ' Homo sapiens hypothetical protein LOC283174 (LOC283174), mRNA NMJ30100 Homo sapiens protein kinase NYD-SP25 (NYD-SP25), transcript variant 2, nr NM_00100 Homo sapiens protein kinase NYD-SP25 (NYD-SP25), transcript variant 3, rr NM_00100 Homo sapiens heat shock transcription factor, Y linked 2 (HSFY2), transcript NM_00100 Homo sapiens heat shock transcription factor, Y linked 2 (HSFY2), transcript NM_00100 l Homo sapiens interferon-induced protein with tetratricopeptide repeats 1 (IFI NM 30100 Homo sapiens variably charged X-C (VCX-C), mRNA NM_00100 Homo sapiens runt-related transcription factor 1 (acute myeloid leukemia 1 ; i NM_00100 Homo sapiens prostate cancer associated protein 5 (PCANAP5), transcript v NM_00100 Homo sapiens tetratricopeptide repeat domain 3 (TTC3), transcript variant 2, NM_00100 Homo sapiens ubiquitin associated and SH3 domain containing, A (UBASH3 NM 30100 Homo sapiens olfactory receptor, family 4, subfamily E, member 2 (OR4E2), NM_00100 Homo sapiens olfactory receptor, family 52, subfamily N, member 1 (OR52N NM_00100 Homo sapiens olfactory receptor, family 2, subfamily G, member 3 (OR2G3), NM_00100 Homo sapiens olfactory receptor, family 2, subfamily G, member 2 (OR2G2), NM_00100 Homo sapiens olfactory receptor, family 52, subfamily J, member 3 (OR52J3 NMJ30100 Homo sapiens olfactory receptor, family 56, subfamily A, member 1 (OR56A' NM_00100 Homo sapiens olfactory receptor, family 5, subfamily BF, member 1 (OR5BF NM_00100 Homo sapiens olfactory receptor, family 5, subfamily AS, member 1 (OR5AS NM_00100 Homo sapiens olfactory receptor OR11-62 (OR11-62), mRNA NM 30100 Homo sapiens mitochondrial tumor suppressor 1 (MTUS1), transcript variant NM 30100 Homo sapiens mitochondrial tumor suppressor 1 (MTUS1), transcript variant NM_00100 Homo sapiens mitochondrial tumor suppressor 1 (MTUS1 ), transcript variant NM_00100 Homo sapiens peroxisome proliferative activated receptor, alpha (PPARA), tr NM_00100 Homo sapiens peroxisome proliferative activated receptor, alpha (PPARA), ti NM_00100 Homo sapiens peroxisome proliferative activated receptor, alpha (PPARA), tt NM 30100 Homo sapiens mitochondrial tumor suppressor 1 (MTUS1), transcript variant NM 30100 Homo sapiens LIM homeobox 8 (LHX8), mRNA NM_00100 Homo sapiens ATP synthase, H+ transporting, mitochondrial F1 complex, alf NM 30100 Homo sapiens KIAA1914 (KIAA1914), transcript variant 1 , mRNA NM_00100 Homo sapiens ATP synthase, H+ transporting, mitochondrial F1 complex, air. NM 30100 Homo sapiens chromosome 9 open reading frame 47 (C9orf47), mRNA NM_00100 Homo sapiens 6-pyruvoyl-tetrahydropterin synthase/dimerization cofactor of I NM_00100 Homo sapiens olfactory receptor, family 10, subfamily G, member 9 (OR10G NM_00100 Homo sapiens olfactory receptor, family 5, subfamily A, member 2 (OR5A2), NM_00100 Homo sapiens olfactory receptor, family 13, subfamily C, member 9 (OR13C! NM 30100 Homo sapiens olfactory receptor, family 2, subfamily W, member 3 (OR2W3] NM_00100 Homo sapiens olfactory receptor, family 7, subfamily G, member 3 (OR7G3), NM 30100 Homo sapiens olfactory receptor, family 11 , subfamily L, member 1 (OR11 L1 NMJ30100 Homo sapiens olfactory receptor, family 5, subfamily , member 2 (OR5W2] NM 30100 Homo sapiens olfactory receptor, family 13, subfamily C, member 3 (OR13C: NM 30100 Homo sapiens olfactory receptor, family 6, subfamily B, member 2 pseudoge NM 30100 Homo sapiens olfactory receptor, family 2, subfamily L, member 8 (OR2L8), i NMJ30100 Homo sapiens olfactory receptor, family 2, subfamily T, member 11 (OR2T1 1 NM_00100 Homo sapiens olfactory receptor, family 4, subfamily D, member 5 (OR4D5), NM_00100 Homo sapiens olfactory receptor, family 5, subfamily AT, member 1 (OR5AT NM 30100 Homo sapiens olfactory receptor, family 5, subfamily D, member 13 (OR5D1 : NM_00100 Homo sapiens olfactory receptor, family 6, subfamily S, member 1 (OR6S1), NM_00100 Homo sapiens family with sequence similarity 13, member C1 (FAM13C1), tr NM_00100 Homo sapiens ATP synthase, H+ transporting, mitochondrial F1 complex, ga NM 00100 Homo sapiens pleckstrin homology domain containing, family A (phosphoino
NM 3010019" Homo sapiens ATP synthase, H+ transporting, mitochondrial F1 complex, de NM 3010019" Homo sapiens arginyltransferase 1 (ATE1), transcript variant 1 , mRNA NMJ3010019' Homo sapiens ATP synthase, H+ transporting, mitochondrial F1 complex, ep NM_0010019! Homo sapiens regeneration associated muscle protease (DKFZP586H2123) NM_0010019! Homo sapiens ubiquitin specific protease 16 (USP16), transcript variant 2, m NM_0010019! Homo sapiens glycoprotein M6B (GPM6B), transcript variant 4, mRNA NM_0010019! Homo sapiens glycoprotein M6B (GPM6B), transcript variant 1 , mRNA NM_0010019! Homo sapiens glycoprotein M6B (GPM6B), transcript variant 2, mRNA NM_0010019! Homo sapiens exosome component 10 (EXOSC10), transcript variant 1 , mRI NM_0010020I Homo sapiens guanosine monophosphate reductase 2 (GMPR2), transcript i NM_0010020I Homo sapiens guanosine monophosphate reductase 2 (GMPR2), transcript > NM_0010020I Homo sapiens guanosine monophosphate reductase 2 (GMPR2), transcript NMJ3010020I Homo sapiens 5'-nucleotidase, cytosolic IB (NT5C1B), transcript variant 1, m NM_0010020 Homo sapiens ATP synthase, H+ transporting, mitochondrial F0 complex, su NM_0010020 Homo sapiens ATP synthase, H+ transporting, mitochondrial F0 complex, su NM_0010020 Homo sapiens host cell factor C1 regulator 1 (XP01 dependant) (HCFC1 R1) NM_0010020 Homo sapiens host cell factor C1 regulator 1 (XP01 dependant) (HCFC1R1) NM 3010020; Homo sapiens phosphofructokinase, liver (PFKL), transcript variant 1 , mRN/* NM_0010020, Homo sapiens claudin 18 (CLDN18), transcript variant 2, mRNA NM_0010020; Homo sapiens ATP synthase, H+ transporting, mitochondrial F0 complex, su NM_0010020; Homo sapiens complement component 4B, centromeric (C4B), mRNA NM_0010020 Homo sapiens ATP synthase, H+ transporting, mitochondrial F0 complex, su NMJ3010020 Homo sapiens hematological and neurological expressed 1 (HN1), transcript NM 3010020 Homo sapiens hematological and neurological expressed 1 (HN1), transcript NM_0010020; Homo sapiens defensin, beta 108 (DEFB108), mRNA NM_0010020; Homo sapiens astacin-like metalloendopeptidase (M12 family) (ASTL), mRN, NM 3010022; Homo sapiens kallikrein 2, prostatic (KLK2), transcript variant 2, mRNA NM_001002Z Homo sapiens kallikrein 2, prostatic (KLK2), transcript variant 3, mRNA NM_0010022; Homo sapiens RAB11 family interacting protein 1 (class I) (RAB11 FIP1), trar NM_0010022; Homo sapiens sodium channel modifier 1 (SCNM1), transcript variant 2, mRI NM_0010022; Homo sapiens serine (or cysteine) proteinase inhibitor, clade A (alpha-1 anti| NM_0010022: Homo sapiens serine (or cysteine) proteinase inhibitor, clade A (alpha-1 anti| NM_001002Z Homo sapiens aftiphilin protein (AFTIPHILIN), transcript variant 3, mRNA NM 001002Z Homo sapiens APC11 anaphase promoting complex subunit 11 homolog (ye NM_0010022- Homo sapiens APC11 anaphase promoting complex subunit 11 homolog (ye NM_0010022' Homo sapiens APC11 anaphase promoting complex subunit 11 homolog (ye NM_0010022< Homo sapiens APC11 anaphase promoting complex subunit 11 homolog (ye NM_0010022' Homo sapiens APC11 anaphase promoting complex subunit 11 homolog (ye NM_0010022' Homo sapiens APC11 anaphase promoting complex subunit 11 homolog (ye NM_0010022! Homo sapiens ADP-ribosylation-like factor 6 interacting protein 4 (ARL6IP4), NM 3010022! Homo sapiens ADP-ribosylation-like factor 6 interacting protein 4 (ARL6IP4), NM_0010022! Homo sapiens SMT3 suppressor of mif two 3 homolog 4 (yeast) (SUM04), rr NM_0010022! Homo sapiens ATP synthase, H+ transporting, mitochondrial F0 complex, su NM_0010022! Homo sapiens acyl-CoA:lysocardiolipin acyltransferase 1 (ALCAT1), transcri| NM_0010022! Homo sapiens ATP synthase, H+ transporting, mitochondrial F0 complex, su NM_0010022! Homo sapiens C1q domain containing 1 (C1QDC1), transcript variant 1 , mRI NMJ30100221 Homo sapiens chromosome 9 open reading frame 58 (C9orf58), transcript VE NM_00100221 Homo sapiens zinc finger, FYVE domain containing 27 (ZFYVE27), transcrip NM 3010022I Homo sapiens zinc finger, FYVE domain containing 27 (ZFYVE27), transcrip NM 3010022I Homo sapiens epithelial stromal interaction 1 (breast) (EPSTI1 ), mRNA NM 3010022I Homo sapiens c-mir, cellular modulator of immune recognition (MIR), transcr NM_0010022l Homo sapiens c-mir, cellular modulator of immune recognition (MIR), transcr NM_0010022l Homo sapiens exosome component 3 (EXOSC3), transcript variant 2, ITIRNJΛ NMJ3010022' Homo sapiens Fc fragment of IgG, low affinity lib, receptor for (CD32) (FCGF NM 3010022' Homo sapiens Fc fragment of IgG, low affinity lib, receptor for (CD32) (FCGF NM_0010022' Homo sapiens Fc fragment of IgG, low affinity lib, receptor for (CD32) (FCGF NM_0010022! Homo sapiens putative NFkB activating protein 373 (FLJ23091), transcript vs
NM_0010022! Homo sapiens flavin containing monooxygenase 3 (FM03), transcript variant NM 3010022! Homo sapiens GATA binding protein 3 (GATA3), transcript variant 1 , mRNA NM_0010022! Homo sapiens golgi autoantigen, golgin subfamily a, 7 (GOLGA7), transcript NM 3010027! Homo sapiens HIRA interacting protein 5 (HIRIP5), transcript variant 2, mRN NM 3010027! Homo sapiens HIRA interacting protein 5 (HIRIP5), transcript variant 3, mRN NM 3010027! Homo sapiens HIRA interacting protein 5 (HIRIP5), transcript variant 4, mRN NM 3010027! Homo sapiens PTPN13-like, Y-linked, centromeric (PRY), mRNA NM_0010027! Homo sapiens chromosome 10 open reading frame 78 (C10orf78), transcript NM 3010027I Homo sapiens basic charge, Y-linked, 2 (BPY2), mRNA NMJ3010027I Homo sapiens basic charge, Y-linked, 2 (BPY2), mRNA NM_ 0010027l Homo sapiens DnaJ (Hsp40) homolog, subfamily B, member 12 (DNAJB12), NM_0010027! Homo sapiens multiple C2-domains with two transmembrane regions 1 (MCI NMJ3010027! Homo sapiens SMC4 structural maintenance of chromosomes 4-like 1 (yeast NM_0010028l Homo sapiens SMC4 structural maintenance of chromosomes 4-like 1 (yeast NM_0010028 Homo sapiens phosphodiesterase 4D interacting protein (myomegalin) (PDE NM_0010028 Homo sapiens phosphodiesterase 4D interacting protein (myomegalin) (PDE NM_0010028 Homo sapiens phosphodiesterase 4D interacting protein (myomegalin) (PDE NM 3010028 Homo sapiens RAB11 family interacting protein 1 (class I) (RAB11 FIP1), trar NM_0010028: Homo sapiens hypothetical protein LOC126208 (LOC126208), mRNA NM 3010028: Homo sapiens phosphatidylinositol (4,5) bisphosphate 5-phosphatase, A (Pll NMJ3010028: Homo sapiens protein kinase, lysine deficient 3 (PRKWNK3), transcript varia NM 3010028- Homo sapiens DKFZp434A0131 protein (DKFZP434A0131), transcript variar NM_0010028' Homo sapiens myosin, light polypeptide 4, alkali; atrial, embryonic (MYL4), tr NM_0010028' Homo sapiens suppressor of hairy wing homolog 4 (Drosophila) (SUHW4), tr NM_0010028- Homo sapiens suppressor of hairy wing homolog 4 (Drosophila) (SUH W4), tr NM 3010028' Homo sapiens suppressor of hairy wing homolog 4 (Drosophila) (SUHW4), tr NM 3010028' Homo sapiens CTF8, chromosome transmission fidelity factor 8 homolog (S. NM_0010028' Homo sapiens lymphocyte antigen 6 complex, locus G5C (LY6G5C), transcri NM_0010028' Homo sapiens lymphocyte antigen 6 complex, locus G5C (LY6G5C), transcri NM 3010028! Homo sapiens annexin A2 (ANXA2), transcript variant 2, mRNA NM_0010028! Homo sapiens annexin A2 (ANXA2), transcript variant 1 , mRNA NM 3010028I Homo sapiens BTB (POZ) domain containing 7 (BTBD7), transcript variant 1 NM_0010028! Homo sapiens Rho guanine nucleotide exchange factor (GEF) 5 (ARHGEF5] NMJ3010028I Homo sapiens chromosome 22 open reading frame 14 (C22orf14), transcript NM_0010028' Homo sapiens chromosome 22 open reading frame 18 (C22orf18), transcript NM_0010028' Homo sapiens chromosome 22 open reading frame 19 (C22orf19), transcript NM_0010028' Homo sapiens chromosome 22 open reading frame 19 (C22orf19), transcript NM__0010028' Homo sapiens chromosome 22 open reading frame 19 (C22orf19), transcript NM_0010028! Homo sapiens chromosome 22 open reading frame 2 (C22orf2), transcript vs NM_0010028! Homo sapiens phosphatidylinositol-3-phosphate/phosphatidylinositol 5-kinas NM_0010029( Homo sapiens hypothetical protein FLJ31052 (FLJ31052), mRNA NM_0010029I Homo sapiens olfactory receptor, family 8, subfamily G, member 1 (OR8G1 P NM_0010029I Homo sapiens X Kell blood group precursor-related, Y-linked 2 (XKRY2), mR NM 3010029I Homo sapiens olfactory receptor, family 8, subfamily K, member 1 (OR8K1), NM 3010029I Homo sapiens KIAA0553 protein (KIAA0553), mRNA NMJ3010029 Homo sapiens cytochrome P450, family 2, subfamily D, polypeptide 7 pseudi NM_0010029 Homo sapiens G protein-coupled receptor 139 (GPR139), mRNA NM_0010029 Homo sapiens chromosome 9 open reading frame 115 (C9orf115), mRNA NM 3010029 Homo sapiens potassium channel tetramerisation domain containing 11 (KC NM 3010029 Homo sapiens insulin growth factor-like family member 2 (IGFL2), mRNA NM 3010029 Homo sapiens H2B histone family, member W, testis-specific (H2BFWT), mF NMJ3010029 Homo sapiens olfactory receptor, family 8, subfamily D, member 1 (OR8D1), NM_0010029 Homo sapiens olfactory receptor, family 8, subfamily D, member 2 (OR8D2), NM 3010029 Homo sapiens hypothetical protein LOC285016 (LOC285016), mRNA NM_0010029: Homo sapiens protein expressed in prostate, ovary, testis, and placenta 8 (P NMJ3010029: Homo sapiens adenylate kinase 3-like 2 (AK3L2), mRNA NMJ3010029; Homo sapiens similar to PM5 (FLJ43542), mRNA
NM_0010029: Homo sapiens insulin growth factor-like family member 4 (IGFL4), rnRNA
NM_0010029: Homo sapiens adaptor-related protein complex 3, sig a 1 subunit (AP3S1),
NM 3010029: Homo sapiens olfactory receptor, family 5, subfamily AP, member 2 (OR5AP
NM 3010029: Homo sapiens TWIST neighbor (TWISTNB), mRNA
NMJ3010033! Homo sapiens Bicaudal D homolog 1 (Drosophila) (BICD1), transcript varian!
NM_0010033! Homo sapiens DKFZp451A211 protein (DKFZp451A211), mRNA
NMJ3010034! Homo sapiens calcium channel, voltage-dependent, alpha 11 subunit (CACN,
NM_0010034- Homo sapiens olfactory receptor, family 56, subfamily A, member 3 (OR56AC
NM_0010036! Homo sapiens protein phosphatase 2, regulatory subunit B, delta isoform (PF
NM_0010036I Homo sapiens similar to hypothetical protein Y97E10AL.1 (DKFZp761P211),
NM_0010036' Homo sapiens chromosome 18 open reading frame 1 (C18orf1), transcript vi
NM_0010036' Homo sapiens chromosome 18 open reading frame 1 (C18orf1), transcript vι
NM 3010036' Homo sapiens leptin receptor (LEPR), transcript variant 2, mRNA
NM_0010036! Homo sapiens leptin receptor (LEPR), transcript variant 3, mRNA
NM_001079 Homo sapiens zeta-chain (TCR) associated protein kinase 70kDa (2AP70), t
NM 001132 Homo sapiens AFG3 ATPase family gene 3-like 1 (yeast) (AFG3L1), mRNA
NMJ301222 Homo sapiens calcium/calmodulin-dependent protein kinase (CaM kinase) II
NM_001369 Homo sapiens dynein, axonemal, heavy polypeptide 5 (DNAH5), mRNA
NM 001376 Homo sapiens dynein, cytoplasmic, heavy polypeptide 1 (DNCH1), mRNA
NM 301378 Homo sapiens dynein, cytoplasmic, intermediate polypeptide 2 (DNCI2), mRI
NM_001410 Homo sapiens EGF-like-domain, multiple 4 (EGFL4), mRNA
NM 001547 Homo sapiens interferon-induced protein with tetratricopeptide repeats 2 (IFI
NM 301556 Homo sapiens inhibitor of kappa light polypeptide gene enhancer in B-cells, I
NM 001636 Homo sapiens solute carrier family 25 (mitochondrial carrier; adenine nucleo
NM_001763 Homo sapiens CD1A antigen, a polypeptide (CD1 A), mRNA
NM_001810 Homo sapiens centromere protein B, 80kDa (CENPB), mRNA
NM 301931 Homo sapiens dihydrolipoamide S-acetyltraπsferase (E2 component of pyruv
NM 001947 Homo sapiens dual specificity phosphatase 7 (DUSP7), mRNA
NMJ301984 Homo sapiens esterase D/formylglutathione hydrolase (ESD), mRNA
NM 001986 Homo sapiens ets variant gene 4 (E1A enhancer binding protein, E1 AF) (EP
NM 302154 Homo sapiens heat shock 70kDa protein 4 (HSPA4), transcript variant 1 , mR
NM_002242 Homo sapiens potassium inwardly-rectifying channel, subfamily J, member 1
NM 302348 Homo sapiens lymphocyte antigen 9 (LY9), mRNA
NM_002399 Homo sapiens Meisl , myeloid ecotropic viral integration site 1 homolog 2 (m
NM 302404 Homo sapiens microfibrillar-associated protein 4 (MFAP4), mRNA
NM_002471 Homo sapiens myosin, heavy polypeptide 6, cardiac muscle, alpha (cardiom;
NM_002498 Homo sapiens NIMA (never in mitosis gene a)-related kinase 3 (NEK3), trans
NM 302523 Homo sapiens neuronal pentraxin II (NPTX2), mRNA
NM 302596 Homo sapiens PCTAIRE protein kinase 3 (PCTK3), transcript variant 3,
NM 302603 Homo sapiens phosphodiesterase 7A (PDE7A), transcript variant 1 , mRNA
NM_002604 Homo sapiens phosphodiesterase 7A (PDE7A), transcript variant 2, mRNA
NM_002605 Homo sapiens phosphodiesterase 8A (PDE8A), transcript variant 1 , mRNA
NM 002679 Homo sapiens postmeiotic segregation increased 2-like 2 (PMS2L2), mRNA
NM 302735 Homo sapiens protein kinase, cAMP-dependent, regulatory, type I, beta (PRI
NM_002746 Homo sapiens mitogen-activated protein kinase 3 (MAPK3), mRNA
NM 302791 Homo sapiens proteasome (prosome, macropain) subunit, alpha type, 6 (PSI
NM 002798 Homo sapiens proteasome (prosome, macropain) subunit, beta type, 6 (PSI
NM 302972 Homo sapiens SET binding factor 1 (SBF1 ), transcript variant 1 , mRNA
NMJ302974 Homo sapiens serine (or cysteine) proteinase inhibitor, clade B (ovalbumin),
NM_002998 Homo sapiens syndecan 2 (heparan sulfate proteoglycan 1, cell surface-asst
NM 303013 Homo sapiens secreted frizzled-related protein 2 (SFRP2), mRNA
NM 303047 Homo sapiens solute carrier family 9 (sodium/hydrogen exchanger), isoform
NM 303106 Homo sapiens SRY (sex determining region Y)-box 2 (SOX2), mRNA
NMJ303111 Homo sapiens Sp3 transcription factor (SP3), mRNA
NM 303179 Homo sapiens synaptophysin (SYP), mRNA
NM 303196 Homo sapiens transcription elongation factor A (Sll), 3 (TCEA3), mRNA
NM 003200 Homo sapiens transcription factor 3 (E2A immunoglobulin enhancer binding ■
NM_003302 Homo sapiens thyroid hormone receptor interactor 6 (TRIP6), mRNA
NM 303415 Homo sapiens zinc finger protein 268 (ZNF268), mRNA
NM 303444 Homo sapiens zinc finger protein 154 (pHZ-92) (ZNF154), mRNA
NM_003502 Homo sapiens axin 1 (AXIN1), transcript variant 1 , mRNA
NM_003517 Homo sapiens histone 2, H2ac (HIST2H2AC), mRNA
NM 303575 Homo sapiens zinc finger protein 282 (ZNF282), mRNA
NM_003598 Homo sapiens TEA domain family member 2 (TEAD2), mRNA
NM 303638 Homo sapiens integrin, alpha 8 (ITGA8), mRNA
NM_003660 Homo sapiens protein tyrosine phosphatase, receptor type, f polypeptide (PT
NMJ303677 Homo sapiens density-regulated protein (DENR), mRNA
NM_003700 Homo sapiens olfactory receptor, family 2, subfamily D, member 2 (OR2D2),
NM 303719 Homo sapiens phosphodiesterase 8B (PDE8B), mRNA
NM_003724 Homo sapiens jerky homolog (mouse) (JRK), mRNA
NM_003741 Homo sapiens chordin (CHRD), transcript variant 1 , mRNA
NM_003817 Homo sapiens a disintegrin and metalloproteinase domain 7 (ADAM7), mRN,
NMJ303818 Homo sapiens CDP-diacylglycerol synthase (phosphatidate cytidylyltransfers
NM_003828 Homo sapiens myotubularin related protein 1 (MTMR1), transcript variant 1 , i
NMJ303845 Homo sapiens dual-specificity tyrosine-(Y)-phosphorylation regulated kinase
NM 303848 Homo sapiens succinate-CoA ligase, GDP-forming, beta subunit (SUCLG2),
NM 303858 Homo sapiens cyclin K (CCNK), mRNA
NM 303898 Homo sapiens synaptojanin 2 (SYNJ2), mRNA
NM 303907 Homo sapiens eukaryotic translation initiation factor 2B, subunit 5 epsilon, 8.
NM 303957 Homo sapiens serine/threonine kinase 29 (STK29), mRNA
NM_003959 Homo sapiens huntingtin interacting protein-1 -related (HIP1R), mRNA
NM_003972 Homo sapiens BTAF1 RNA polymerase II, B-TFIID transcription factor-assoc
NM_004080 Homo sapiens diacylglycerol kinase, beta 90kDa (DGKB), transcript variant 1
NMJ304097 Homo sapiens empty spiracles homolog 1 (Drosophila) (EMX1), mRNA
NM 304118 Homo sapiens forkhead-like 18 (Drosophila) (FKHL18), mRNA
NMJ304136 Homo sapiens iron-responsive element binding protein 2 (IREB2), mRNA
NM 304200 Homo sapiens synaptotagmin VII (SYT7), mRNA
NM 304220 Homo sapiens zinc finger protein 213 (ZNF213), mRNA
NM_004241 Homo sapiens jumonji domain containing 1C (JMJD1C), mRNA
NM_004242 Homo sapiens high mobility group nucleosomal binding domain 3 (HMGN3),
NM_004319 Homo sapiens astrotactin (ASTN), transcript variant 1 , mRNA
NM 304439 Homo sapiens EphA5 (EPHA5), transcript variant 1 , mRNA
NM_004498 Homo sapiens one cut domain, family member 1 (ONECUT1), mRNA
NM_004650 Homo sapiens GS2 gene (DXS1283E), mRNA
NMJ304685 Homo sapiens myotubularin related protein 6 (MTMR6), mRNA
NM_004691 Homo sapiens ATPase, H+ transporting, lysosomal 38kDa, VO subunit d isofi
NM_004764 Homo sapiens piwi-like 1 (Drosophila) (PIWIL1), mRNA
NM_004773 Homo sapiens thyroid hormone receptor interactor 3 (TRIP3), mRNA
NM_004816 Homo sapiens chromosome 9 open reading frame 61 (C9orf61), mRNA
NM_004840 Homo sapiens Rac/Cdc42 guanine nucleotide exchange factor (GEF) 6 (ARF
NM 304884 Homo sapiens putative neuronal cell adhesion molecule (PUNC), mRNA
NMJ304946 Homo sapiens dedicator of cytokinesis 2 (DOCK2), mRNA
NM 304947 Homo sapiens dedicator of cytokinesis 3 (DOCK3), mRNA
NMJ305054 Homo sapiens RAN binding protein 2-like 1 (RANBP2L1), transcript variant 1
NM 305105 Homo sapiens RNA binding motif protein 8A (RBM8A), mRNA
NMJ305126 Homo sapiens nuclear receptor subfamily 1 , group D, member 2 (NR1 D2), rr
NM 305140 Homo sapiens cyclic nucleotide gated channel alpha 2 (CNGA2), mRNA
NM_005144 Homo sapiens hairless homolog (mouse) (HR), transcript variant 1 , mRNA
NM 305153 Homo sapiens ubiquitin specific protease 10 (USP10), mRNA
NM 305202 Homo sapiens collagen, type VIII, alpha 2 (COL8A2), mRNA
NM_005240 Homo sapiens ets variant gene 3 (ETV3), mRNA
NM_005241 Homo sapiens ecotropic viral integration site 1 (EVI1), mRNA
NM 305250 Homo sapiens forkhead box L1 (FOXL1), mRNA
NM_005272 Homo sapiens guanine nucleotide binding protein (G protein), alpha transduc
NM 305278 Homo sapiens glycoprotein M6B (GPM6B), transcript variant 3, mRNA
NM_005349 Homo sapiens recombining binding protein suppressor of hairless (Drosophil
NM_005376 Homo sapiens v-myc myelocytomatosis viral oncogene homolog 1 , lung care
NM 305407 Homo sapiens sal-like 2 (Drosophila) (SALL2), mRNA
NM_005482 Homo sapiens phosphatidylinositol glycan, class K (PIGK), mRNA
NM 305487 Homo sapiens high-mobility group protein 2-like 1 (HMG2L1), mRNA
NM 305533 Homo sapiens interferon-induced protein 35 (IFI35), mRNA
NM 305559 Homo sapiens laminin, alpha 1 (LAMA1), mRNA
NM 305595 Homo sapiens nuclear factor l/A (NFIA), mRNA
NM 305650 Homo sapiens transcription factor 20 (AR1 ) (TCF20), transcript variant 1 , mR
NM_005669 Homo sapiens chromosome 5 open reading frame 18 (C5orf18), mRNA
NM_005680 Homo sapiens TATA box binding protein (TBP)-associated factor, RNA polyn
NMJ305702 Homo sapiens Era G-protein-like 1 (E. coli) (ERAL1), mRNA
NM 305707 Homo sapiens programmed cell death 7 (PDCD7), mRNA
NM 305779 Homo sapiens lipoma HMGIC fusion partner-like 2 (LHFPL2), mRNA
NMJ305788 Homo sapiens HMT1 hnRNP methyltransferase-like 3 (S. cerevisiae) (HRMT
NM_005791 Homo sapiens M-phase phosphoprotein 10 (U3 small nucleolar ribonucleopn
NM 305840 Homo sapiens sprouty homolog 3 (Drosophila) (SPRY3), mRNA
NM_005841 Homo sapiens sprouty homolog 1 , antagonist of FGF signaling (Drosophila) i
NM_005848 Homo sapiens c-myc promoter binding protein (MYCPBP), mRNA
NM_005914 Homo sapiens MCM4 minichromosome maintenance deficient 4 (S. cerevisiε
NM_005942 Homo sapiens molybdenum cofactor synthesis 1 (MOCS1), transcript variant
NM_005943 Homo sapiens molybdenum cofactor synthesis 1 (MOCS1), transcript variant
NM_005946 Homo sapiens metallothionein 1A (functional) (MT1 A), mRNA
NMJ305947 Homo sapiens metallothionein 1 B (functional) (MT1 B), mRNA
NM_005949 Homo sapiens metallothionein 1 F (functional) (MT1 F), mRNA
NM_005964 Homo sapiens myosin, heavy polypeptide 10, non-muscle (MYH10), mRNA
NM_005984 Homo sapiens solute carrier family 25 (mitochondrial carrier; citrate transport
NM_005995 Homo sapiens T-box 10 (TBX10), mRNA
NM_006036 Homo sapiens putative prolyl oligopeptidase (KIAA0436), mRNA
NM_006040 Homo sapiens heparan sulfate (glucosamine) 3-O-sulfotransferase 4 (HS3S1
NM_006062 Homo sapiens SMYD family member 5 (SMYD5), mRNA
NM 306091 Homo sapiens coronin, actin binding protein, 2B (COR02B), mRNA
NM_006108 Homo sapiens spondin 1 , extracellular matrix protein (SPON1), mRNA
NM_006133 Homo sapiens chromosome 11 open reading frame 11 (C11orf11), mRNA
NM_006151 Homo sapiens lactoperoxidase (LPO), mRNA
NM_006154 Homo sapiens neural precursor cell expressed, developmentally down-reguk
NM_006172 Homo sapiens natriuretic peptide precursor A (NPPA), mRNA
NM_006175 Homo sapiens nebulin-related anchoring protein (NRAP), transcript variant 1
NM 306210 Homo sapiens paternally expressed 3 (PEG3), mRNA
NM_006216 Homo sapiens serine (or cysteine) proteinase inhibitor, clade E (nexin, plasrr
NM_006266 Homo sapiens ral guanine nucleotide dissociation stimulator (RALGDS), mRI
NM_006277 Homo sapiens intersectin 2 (ITSN2), transcript variant 1 , mRNA
NM_006452 Homo sapiens phosphoribosylaminoimidazole carboxylase, phosphoribosyla
NMJ306524 Homo sapiens zinc finger protein 138 (clone pHZ-32) (ZNF138), mRNA
NM_006591 Homo sapiens polymerase (DNA-directed), delta 3, accessory subunit (POLE
NM 306617 Homo sapiens nestin (NES), mRNA
NMJ306630 Homo sapiens zinc finger protein 234 (ZNF234), mRNA
NM 306631 Homo sapiens zinc finger protein 266 (ZNF266), mRNA
NM 306635 Homo sapiens zinc finger protein 272 (ZNF272), mRNA
NM_006642 Homo sapiens serologically defined colon cancer antigen 8 (SDCCAG8), mR
NM 306647 Homo sapiens NADPH oxidase activator 1 (NOXA1), mRNA
NM_006673 Homo sapiens AT rich interactive domain 5A (MRF1-like) (ARID5A), transcrif
NM_006714 Homo sapiens sphingomyelin phosphodiesterase, acid-like 3A (SMPDL3A), i
NMJ306722 Homo sapiens microphthalmia-associated transcription factor (MITF), transcr
NM 306742 Homo sapiens protein serine kinase H1 (PSKH1), mRNA
NM_006775 Homo sapiens quaking homolog, KH domain RNA binding (mouse) (QKI), tre
NM_006828 Homo sapiens activating signal cointegrator 1 complex subunit 3 (ASCC3), n
NM 306832 Homo sapiens pleckstrin homology domain containing, family C (with FERM
NM_006857 Homo sapiens putative nucleic acid binding protein RY-1 (RY1), mRNA
NM_006859 Homo sapiens lipoic acid synthetase (LIAS), nuclear gene encoding mitocho
NM_006897 Homo sapiens homeo box C9 (HOXC9), mRNA
NM_006909 Homo sapiens Ras protein-specific guanine nucleotide-releasing factor 2 (R/
NM_006916 Homo sapiens ribulose-5-phosphate-3-epimerase (RPE), transcript variant 2.
NMJ306920 Homo sapiens sodium channel, voltage-gated, type I, alpha (SCN1A), mRN/*
NM 306939 Homo sapiens son of sevenless homolog 2 (Drosophila) (SOS2), mRNA
NM 306955 Homo sapiens zinc finger protein 11 b (KOX 2) (ZNF11 B), mRNA
NM 306956 Homo sapiens zinc finger protein 12 (KOX 3) (ZNF12), mRNA
NM 306959 Homo sapiens zinc finger protein 17 (HPF3, KOX 10) (ZNF17), mRNA
NM_006961 Homo sapiens zinc finger protein 19 (KOX 12) (ZNF19), mRNA
NM 306969 Homo sapiens zinc finger protein 28 (KOX 24) (ZNF28), mRNA
NM 306973 Homo sapiens zinc finger protein 32 (KOX 30) (ZNF32), mRNA
NMJ306974 Homo sapiens zinc finger protein 33a (KOX 31 ) (ZNF33A), mRNA
NM_006996 Homo sapiens solute carrier family 19 (thiamine transporter), member 2 (SLC
NM 307001 Homo sapiens solute carrier family 35, member D2 (SLC35D2), mRNA
NMJ307010 Homo sapiens DEAD (Asp-Glu-Ala-Asp) box polypeptide 52 (DDX52), transc
NM_007041 Homo sapiens arginyltransferase 1 (ATE1), transcript variant 2, mRNA
NM 307078 Homo sapiens LIM domain binding 3 (LDB3), mRNA
NM_007130 Homo sapiens zinc finger protein 41 (ZNF41 ), transcript variant 1 , mRNA
NM 307131 Homo sapiens zinc finger protein 75 (D8C6) (ZNF75), mRNA
NM 307135 Homo sapiens zinc finger protein 79 (pT7) (ZNF79), mRNA
NM 307137 Homo sapiens zinc finger protein 81 (HFZ20) (ZNF81 ), mRNA
NM 307139 Homo sapiens zinc finger protein 92 (HTF12) (ZNF92), mRNA
NM_007149 Homo sapiens zinc finger protein 184 (Kruppel-like) (ZNF184), mRNA
NM_007156 Homo sapiens zinc finger, X-linked, duplicated A (ZXDA), mRNA
NM 307157 Homo sapiens zinc finger, X-linked, duplicated B (ZXDB), mRNA
NM 307162 Homo sapiens transcription factor EB (TFEB), mRNA
NM_007174 Homo sapiens citron (rho-interacting, serine/threonine kinase 21) (CIT), mRr*
NM 307189 Homo sapiens ATP-binding cassette, sub-family F (GCN20), member 2 (ABC
NM 307224 Homo sapiens neurexophilin 4 (NXPH4), mRNA
NM 307225 Homo sapiens neurexophilin 3 (NXPH3), mRNA
NM_007243 Homo sapiens nurim (nuclear envelope membrane protein) (NRM), mRNA
NM_007261 Homo sapiens leukocyte membrane antigen (CMRF-35H), mRNA
NM_007270 Homo sapiens FK506 binding protein 9, 63 kDa (FKBP9), mRNA
NM 307277 Homo sapiens SEC6-like 1 (S. cerevisiae) (SEC6L1), mRNA
NMJ307280 Homo sapiens Opa-interacting protein 5 (OIP5), mRNA
NM_007349 Homo sapiens PAX transcription activation domain interacting protein 1 like (
NM 307356 Homo sapiens laminin, beta 4 (LAMB4), mRNA
NM_012073 Homo sapiens chaperonin containing TCP1 , subunit 5 (epsilon) (CCT5), mRI
NMJ312154 Homo sapiens eukaryotic translation initiation factor 2C, 2 (EIF2C2), mRNA
NM_012156 Homo sapiens erythrocyte membrane protein band 4.1-like 1 (EPB41 L1), trai
NM 312167 Homo sapiens F-box protein 11 (FBX011), transcript variant 3, mRNA
NM_012174 Homo sapiens F-box and WD-40 domain protein 8 (FBXW8), transcript varia
NM 312184 Homo sapiens forkhead box D4 like 1 (FOXD4L1), mRNA
NMJ312212 Homo sapiens leukotriene B4 12-hydroxydehydrogenase (LTB4DH), mRNA
NM_012224 Homo sapiens NIMA (never in mitosis gene a)-related kinase 1 (NEK1 ), mRr^
NM 312232 Homo sapiens polymerase I and transcript release factor (PTRF), mRNA
NM 312235 Homo sapiens SREBP CLEAVAGE-ACTIVATING PROTEIN (SCAP), mRNA
NM_012271 Homo sapiens huntingtin interacting protein B (HYPB), transcript variant 2, rr
NM 312272 Homo sapiens Huntingtin interacting protein C (HYPC), mRNA
NM_012284 Homo sapiens potassium voltage-gated channel, subfamily H (eag-related), i
NM_012292 Homo sapiens minor histocompatibility antigen HA-1 (HA-1), mRNA
NM_012305 Homo sapiens adaptor-related protein complex 2, alpha 2 subunit (AP2A2), r
NM_012309 Homo sapiens SH3 and multiple ankyrin repeat domains 2 (SHANK2), transc
NMJ312315 Homo sapiens kallikrein 9 (KLK9), mRNA
NMJ312335 Homo sapiens myosin IF (MY01 F), mRNA
NM_012363 Homo sapiens olfactory receptor, family 1 , subfamily N, member 1 (OR1 N1),
NM_012364 Homo sapiens olfactory receptor, family 1 , subfamily Q, member 1 (OR1Q1),
NMJ312367 Homo sapiens olfactory receptor, family 2, subfamily B, member 6 (OR2B6),
NM 312374 Homo sapiens olfactory receptor, family 4, subfamily D, member 1 (OR4D1),
NM_012378 Homo sapiens olfactory receptor, family 8, subfamily B, member 8 (OR8B8),
NMJ312393 Homo sapiens phosphoribosylformylglycinamidine synthase (FGAR amidotrε
NM_012398 Homo sapiens phosphatidylinositol-4-phosphate 5-kinase, type I, gamma (PI
NM 312416 Homo sapiens RAN binding protein 6 (RANBP6), mRNA
NMJ312477 Homo sapiens WW domain binding protein 1 (WBP1), mRNA
NMJ312478 Homo sapiens WW domain binding protein 2 (WBP2), mRNA
NM 313304 Homo sapiens zinc finger, DHHC domain containing 1 (ZDHHC1), mRNA
NM 313321 Homo sapiens sorting nexin 8 (SNX8), mRNA
NM 313373 Homo sapiens zinc finger, DHHC domain containing 8 (ZDHHC8), mRNA
NMJ314010 Homo sapiens astrotactin 2 (ASTN2), transcript variant 1 , mRNA
NM 314014 Homo sapiens U5 snRNP-specific protein, 200-KD (U5-200KD), mRNA
NMJ314089 Homo sapiens nucleoporin like 1 (NUPL1), mRNA
NM__014098 Homo sapiens peroxiredoxin 3 (PRDX3), nuclear gene encoding mitochondri
NM__014215 Homo sapiens insulin receptor-related receptor (INSRR), mRNA
NM 314220 Homo sapiens transmembrane 4 superfamily member 1 (TM4SF1), mRNA
NMJ314224 Homo sapiens pepsinogen 5, group I (pepsinogen A) (PGA5), mRNA
NMJ314243 Homo sapiens a disintegrin-like and metalloprotease (reprolysin type) with th
NM__014261 Homo sapiens TIR domain containing adaptor inducing interferon-beta (TRIF
NM_014282 Homo sapiens hyaluronan binding protein 4 (HABP4), mRNA
NM__014284 Homo sapiens neurochondrin (NCDN), mRNA
NM_014290 Homo sapiens tudor domain containing 7 (TDRD7), mRNA
NM_014301 Homo sapiens iron-sulfur cluster assembly enzyme (ISCU), mRNA
NM_014346 Homo sapiens chromosome 22 open reading frame 4 (C22orf4), mRNA
NM__014376 Homo sapiens cytoplasmic FMR1 interacting protein 2 (CYFIP2), mRNA
NMJ314381 Homo sapiens mut homolog 3 (E. coli) (MLH3), mRNA
NM_014389 Homo sapiens proline-, glutamic acid-, leucine-rich protein 1 (PELP1), mRN/
NM_014422 Homo sapiens phosphatidylinositol (4,5) bisphosphate 5-phosphatase, A (Pll
NM_014435 Homo sapiens N-acylsphingosine amidohydrolase (acid ceramidase)-like (AJ
NM 314441 Homo sapiens sialic acid binding Ig-like lectin 9 (SIGLEC9), mRNA
NM 314455 Homo sapiens zinc finger protein 364 (ZNF364), mRNA
NM__014460 Homo sapiens RNA-binding protein pippin (PIPPIN), mRNA
NM 314472 Homo sapiens chromosome 10 open reading frame 28 (C10orf28), mRNA
NM__014494 Homo sapiens trinucleotide repeat containing 6 (TNRC6), mRNA
NM__014507 Homo sapiens ma!onyl-CoA:acyl carrier protein transacylase (malonyltransfe
NM_014508 Homo sapiens apolipoprotein B mRNA editing enzyme, catalytic polypeptide-
NM__014510 Homo sapiens piccolo (presynaptic cytomatrix protein) (PCLO), transcript vai
NM__014562 Homo sapiens orthodenticle homolog 1 (Drosophila) (OTX1), mRNA
NM_014568 Homo sapiens UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylga
NM_014572 Homo sapiens LATS, large tumor suppressor, homolog 2 (Drosophila) (LATS
NM 314573 Homo sapiens hypothetical protein MAC30 (MAC30), mRNA
NM 314594 Homo sapiens zinc finger protein 354C (ZNF354C), mRNA
NM__014602 Homo sapiens phosphoinositide-3-kinase, regulatory subunit 4, p150 (PIK3R
NM__014603 Homo sapiens paraneoplastic antigen (HUMPPA), mRNA
NMJ314607 Homo sapiens UBX domain containing 2 (UBXD2), mRNA
NMJ314608 Homo sapiens cytoplasmic FMR1 interacting protein 1 (CYFIP1), mRNA
NM_014611 Homo sapiens MDN1 , midasin homolog (yeast) (MDN1), mRNA
NMJ314613 Homo sapiens expressed in T-cells and eosinophils in atopic dermatitis (ETE
NM 314614 Homo sapiens proteasome (prosome, macropain) activator subunit 4 (PSME
NM 314615 Homo sapiens KIAA0182 protein (KIAA0182), mRNA
NM 314647 Homo sapiens limkain b1 (LKAP), transcript variant 1 , mRNA
NM 314655 Homo sapiens KIAA0446 gene product (KIAA0446), mRNA
NM J14657 Homo sapiens KIAA0406 gene product (KIAA0406), mRNA
NM_014667 Homo sapiens vestigial like 4 (Drosophila) (VGLL4), mRNA
NM 314691 Homo sapiens aquarius homolog (mouse) (AQR), mRNA
NM_014697 Homo sapiens C-terminal PDZ domain ligand of neuronal nitric oxide synthaj
NM 314701 Homo sapiens KIAA0256 gene product (KIAA0256), mRNA
NM 314756 Homo sapiens KIAA0097 gene product (ch-TOG), mRNA
NM_014798 Homo sapiens pleckstrin homology domain containing, family M (with RUN d
NM_014802 Homo sapiens KIAA0528 gene product (KIAA0528), mRNA
NM_014836 Homo sapiens Rho-related BTB domain containing 1 (RHOBTB1), transcript
NM 314839 Homo sapiens plasticity related gene 1 (LPPR4), mRNA
NM_014850 Homo sapiens SLIT-ROBO Rho GTPase activating protein 2 (SRGAP2), mR
NM 314854 Homo sapiens solute carrier family 35, member E2 (SLC35E2), mRNA
NM 314858 Homo sapiens cerebral protein 11 (HUCEP11), mRNA
NM_014881 Homo sapiens DNA cross-link repair 1 A (PS02 homolog, S. cerevisiae) (DCl
NM_014884 Homo sapiens splicing factor, arginine/serine-rich 14 (SFRS14), mRNA
NM 314919 Homo sapiens Wolf-Hirschhom syndrome candidate 1 (WHSC1), transcript \
NM 314955 Homo sapiens CGI-01 protein (CGI-01), transcript variant 2, mRNA
NM 314957 Homo sapiens KIAA0870 protein (KIAA0870), mRNA
NM 314974 Homo sapiens KIAA0934 (KIAA0934), mRNA
NM_014975 Homo sapiens microtubule associated serine/threonine kinase 1 (MAST1), rr
NM_014982 Homo sapiens pecanex homolog (Drosophila) (PCNX), mRNA
NMJ314989 Homo sapiens regulating synaptic membrane exocytosis 1 (RIMS1), transcrij
NM_014991 Homo sapiens WD repeat and FYVE domain containing 3 (WDFY3), transcri
NMJ314992 Homo sapiens dishevelled associated activator of morphogenesis 1 (DAAM1
NMJ314997 Homo sapiens KIAA0265 protein (KIAA0265), mRNA
NM_015000 Homo sapiens serine/threonine kinase 38 like (STK38L), mRNA
NM_015004 Homo sapiens exosome component 7 (EXOSC7), mRNA
NMJ315008 Homo sapiens KIAA0779 protein (KIAA0779), mRNA
NM_015013 Homo sapiens amine oxidase (flavin containing) domain 2 (AOF2), mRNA
NM 315014 Homo sapiens KIAA0117 protein (KIAA0117), mRNA
NM 315015 Homo sapiens jumonji domain containing 2B (JMJD2B), mRNA
NM_015017 Homo sapiens ubiquitin specific protease 33 (USP33), transcript variant 1 , m
NM_015018 Homo sapiens KIAA1117 (KIAA1117), mRNA
NMJ315022 Homo sapiens PDZ domain containing 3 (PDZK3), transcript variant 2, mRN;
NM_015027 Homo sapiens KIAA0251 protein (KIAA0251 ), mRNA
NM_015029 Homo sapiens processing of precursor 1 , ribonuclease P/MRP subunit (S. cε
NMJ315033 Homo sapiens formin binding protein 1 (FNBP1), mRNA
NM_015035 Homo sapiens zinc fingers and homeoboxes 3 (ZHX3), mRNA
NM 315037 Homo sapiens KIAA0913 (KIAA0913), mRNA
NM_015039 Homo sapiens nicotinamide nucleotide adenylyltransferase 2 (NMNAT2), trai
NM_015040 Homo sapiens phosphatidylinositol-3-phosphate/phosphatidylinositol 5-kinas
NM_015045 Homo sapiens KIAA0261 (KIAA0261 ), mRNA
NM_015047 Homo sapiens KIAA0090 protein (KIAA0090), mRNA
NM 315050 Homo sapiens KIAA0082 (KIAA0082), mRNA
NM_015052 Homo sapiens HECT type E3 ubiquitin ligase (NEDL1 ), mRNA
NM 315055 Homo sapiens SWAP-70 protein (SWAP70), mRNA
NM 315059 Homo sapiens talin 2 (TLN2), mRNA
NM_015061 Homo sapiens jumonji domain containing 2C (JMJD2C), mRNA
NM 315065 Homo sapiens SLAC2-B (SLAC2-B), mRNA
NM_015066 Homo sapiens tripartite motif-containing 35 (TRIM35), transcript variant 1 , ml
NM_015069 Homo sapiens zinc finger protein 423 (ZNF423), mRNA
NM 315076 Homo sapiens cyclin-dependent kinase (CDC2-like) 11 (CDK11), mRNA
NM 315078 Homo sapiens Rho family guanine-nucleotide exchange factor (KIAA0861 ), r
NM 315079 Homo sapiens KIAA1055 protein (KIAA1055), mRNA
NM 315085 Homo sapiens GTPase activating RANGAP domain-like 4 (GARNL4), mRNA
NM_015087 Homo sapiens spastic paraplegia 20, spartin (Troyer syndrome) (SPG20), ml
NM_015089 Homo sapiens p53-associated parkin-like cytoplasmic protein (PARC), mRN;
NM 315091 Homo sapiens K1AA0423 (KIAA0423), mRNA
NM_015094 Homo sapiens hypermethylated in cancer 2 (HIC2), mRNA
NM_015099 Homo sapiens calmodulin binding transcription activator 2 (CAMTA2), mRN/1
NM_015100 Homo sapiens pogo transposable element with ZNF domain (POGZ), transcr
NM 315102 Homo sapiens nephronophthisis 4 (NPHP4), mRNA
NM 315103 Homo sapiens plexin D1 (PLXND1), mRNA
NM 315106 Homo sapiens KIAA0809 protein (SR1SNF2L), mRNA
NMJ315107 Homo sapiens PHD finger protein 8 (PHF8), mRNA
NM_015110 Homo sapiens SMC5 structural maintenance of chromosomes 5-like 1 (yeast
NM 315115 Homo sapiens KIAA0276 protein (KIAA0276), mRNA
NM_015116 Homo sapiens leucine-rich repeats and calponin homology (CH) domain con
NM_015117 Homo sapiens zinc finger CCCH type domain containing 3 (ZC3HDC3), mRt*
NM 315120 Homo sapiens Alstrom syndrome 1 (ALMS1), mRNA
NM 315122 Homo sapiens FCH domain only 1 (FCH01), mRNA
NM_015134 Homo sapiens myosin phosphatase-Rho interacting protein (M-RIP), mRNA
NM 315138 Homo sapiens KIAA0252 (KIAA0252), mRNA
NM 315141 Homo sapiens glycerol-3-phosphate dehydrogenase 1-like (GPD1 L), mRNA
NM_015143 Homo sapiens methionyl aminopeptidase 1 (METAP1), mRNA
NM_015144 Homo sapiens zinc finger, CCHC domain containing 14 (ZCCHC14), mRNA
NM_015150 Homo sapiens raft-linking protein (RAFTLIN), mRNA
NM_015151 Homo sapiens chromosome 21 open reading frame 106 (C21orf106), transci
NM 315157 Homo sapiens pleckstrin homology-like domain, family B, member 1 (PHLDE
NM_015158 Homo sapiens ankyrin repeat domain 15 (ANKRD15), transcript variant 1 , ml
NM 315160 Homo sapiens peptidase (mitochondrial processing) alpha (PMPCA), nucleai
NM_015161 Homo sapiens ADP-ribosylation factor-like 6 interacting protein (ARL6IP), mF
NM 315167 Homo sapiens phosphatidylserine receptor (PTDSR), mRNA
NM 315170 Homo sapiens sulfatase 1 (SULF1 ), mRNA
NM 315171 Homo sapiens exportin 6 (XP06), mRNA
NM_015172 Homo sapiens HBxAg transactivated protein 2 (XTP2), mRNA
NM 315173 Homo sapiens TBC1 (tre-2/USP6, BUB2, cdc16) domain family, member 1 (1
NM_015184 Homo sapiens phospholipase C-like 2 (PLCL2), mRNA
NM 315187 Homo sapiens KIAA0746 protein (KIAA0746), mRNA
NM 315190 Homo sapiens DnaJ (Hsp40) homolog, subfamily C, member 9 (DNAJC9), m
NMJ315191 Homo sapiens salt-inducible serine/threonine kinase 2 (SIK2), mRNA
NM_015198 Homo sapiens cordon-bleu homolog (mouse) (COBL), mRNA
NM 315199 Homo sapiens ankyrin repeat domain 28 (ANKRD28), mRNA
NM_015200 Homo sapiens KIAA0648 protein (KIAA0648), mRNA
NM 315201 Homo sapiens block of proliferation 1 (BOP1), mRNA
NM 315203 Homo sapiens KIAA0460 protein (KIAA0460), mRNA
NM_015210 Homo sapiens KIAA0802 (KIAA0802), mRNA
NM 315213 Homo sapiens RAB6 interacting protein 1 (RAB6IP1), mRNA
NM_015219 Homo sapiens exocyst complex component 7 (EXOC7), mRNA
NM_015221 Homo sapiens dynamin binding protein (DNMBP), mRNA
NM 315229 Homo sapiens KIAA0664 protein (KIAA0664), mRNA
NMJ315234 Homo sapiens G protein-coupled receptor 116 (GPR116), mRNA
NM 315238 Homo sapiens KIBRA protein (KIBRA), mRNA
NM_015243 Homo sapiens Cohen syndrome 1 (COH1), transcript variant 3, mRNA
NM_015245 Homo sapiens ankyrin repeat and sterile alpha motif domain containing 1 (Ar
NM 315246 Homo sapiens mahogunin, ring finger 1 (MGRN1), mRNA
NM 315250 Homo sapiens bicaudal D homolog 2 (Drosophila) (BICD2), mRNA
NMJ315252 Homo sapiens NPF/calponin-like protein (NACSIN), mRNA
NM_015255 Homo sapiens chromosome 6 open reading frame 133 (C6orf133), mRNA
NM 315259 Homo sapiens inducible T-cell co-stimulator ligand (ICOSL), mRNA
NM_015261 Homo sapiens KIAA0056 protein (KIAA0056), mRNA
NM 315263 Homo sapiens rabconnectin-3 (RC3), mRNA
NM 315265 Homo sapiens SATB family member 2 (SATB2), mRNA
NM_015266 Homo sapiens solute carrier family 9 (sodium/hydrogen exchanger), isoform
NM 315268 Homo sapiens DnaJ (Hsp40) homolog, subfamily C, member 13 (DNAJC13),
NM 315274 Homo sapiens mannosidase, alpha, class 2B, member 2 (MAN2B2), mRNA
NM 315275 Homo sapiens KIAA1033 protein (KIAA1033), mRNA
NM 315278 Homo sapiens SAM and SH3 domain containing 1 (SASH1), mRNA
NM 315281 Homo sapiens KIAA1043 protein (KIAA1043), mRNA
NM 315282 Homo sapiens cytoplasmic linker associated protein 1 (CLASP1), mRNA
NM 315284 Homo sapiens KIAA0467 protein (KIAA0467), mRNA
NM_015286 Homo sapiens desmuslin (DMN), transcript variant B, mRNA
NM_015289 Homo sapiens vacuolar protein sorting 39 (yeast) (VPS39), mRNA
NM_015293 Homo sapiens spectrin repeat containing, nuclear envelope 1 (SYNE1), trans
NM_015296 Homo sapiens dedicator of cytokinesis 9 (DOCK9), mRNA
NMJ315305 Homo sapiens KIAA0759 (KIAA0759), mRNA
NM_015308 Homo sapiens formin binding protein 4 (FNBP4), mRNA
NM_015315 Homo sapiens likely ortholog of mouse la related protein (LARP), mRNA
NM_015316 Homo sapiens protein phosphatase 1, regulatory (inhibitor) subunit 13B (PPF
NM_015319 Homo sapiens tensin like C1 domain containing phosphatase (TENC1), trans
NM_015321 Homo sapiens mucoepidermoid carcinoma translocated 1 (MECT1), mRNA
NM 315323 Homo sapiens KIAA0776 (KIAA0776), mRNA
NM_015327 Homo sapiens Est1 p-like protein B (EST1 B), mRNA
NM 315328 Homo sapiens KIAA0828 protein (KIAA0828), mRNA
NM 315329 Homo sapiens KIAA0892 (KIAA0892), mRNA
NM 315330 Homo sapiens KIAA0376 protein (KIAA0376), mRNA
NM 315331 Homo sapiens nicastrin (NCSTN), mRNA
NM 315335 Homo sapiens thyroid hormone receptor associated protein 2 (THRAP2), mF
NM_015336 Homo sapiens zinc finger, DHHC domain containing 17 (ZDHHC17), mRNA
NM 315338 Homo sapiens additional sex combs like 1 (Drosophila) (ASXL1), mRNA
NM_015341 Homo sapiens barren homolog (Drosophila) (BRRN1), mRNA
NM 315342 Homo sapiens KIAA0073 protein (KIAA0073), mRNA
NM_015345 Homo sapiens dishevelled associated activator of morphogenesis 2 (DAAM2
NM_015346 Homo sapiens zinc finger, FYVE domain containing 26 (ZFYVE26), mRNA
NMJ315347 Homo sapiens RIM binding protein 2 (KIAA0318), mRNA
NM_015350 Homo sapiens T-cell activation leucine repeat-rich protein (TA-LRRP), mRN/
NM_015352 Homo sapiens protein O-fucosyltransferase 1 (POFUT1), transcript variant 1 ,
NM 315358 Homo sapiens zinc finger, CW-type with coiled-coil domain 3 (ZCWCC3), mF
NM_015359 Homo sapiens solute carrier family 39 (zinc transporter), member 14 (SLC39
NM_015360 Homo sapiens KIAA0052 (KIAA0052), mRNA
NMJ315374 Homo sapiens unc-84 homolog B (C. elegans) (UNC84B), mRNA
NM_015375 Homo sapiens receptor interacting protein kinase 5 (RIPK5), transcript variar
NM 315378 Homo sapiens vacuolar protein sorting 13D (yeast) (VPS13D), mRNA
NMJ315381 Homo sapiens TAFA protein 5 (TAFA5), mRNA
NM 315382 Homo sapiens HECT domain containing 1 (HECTD1), mRNA
NM_015386 Homo sapiens component of oligomeric golgi complex 4 (COG4), mRNA
NM_015391 Homo sapiens anaphase promoting complex subunit 13 (ANAPC13), mRNA
NM 315395 Homo sapiens DKFZP434B0335 protein (DKFZP434B0335), mRNA
NM_015397 Homo sapiens WD repeat domain 40A (WDR40A), mRNA
NM_015404 Homo sapiens deafness, autosomal recessive 31 (DFNB31), mRNA
NMJ315411 Homo sapiens sulfatase modifying factor 2 (SUMF2), mRNA
NM 315412 Homo sapiens DKFZP434F2021 protein (DKFZP434F2021), mRNA
NMJ315430 Homo sapiens regeneration associated muscle protease (DKFZP586H2123)
NM_015431 Homo sapiens BIA2 (BIA2), mRNA
NM_015433 Homo sapiens hepatocellularcarcinoma-associated antigen HCA557a (DKF2
NM_015436 Homo sapiens ring finger and CHY zinc finger domain containing 1 (RCHY1 )
NMJ315439 Homo sapiens chromosome 6 open reading frame 80 (C6orf80), mRNA
NM_015440 Homo sapiens formyltetrahydrofolate synthetase domain containing 1 (FTHF
NM 315441 Homo sapiens olfactomedin-like 2B (OLFML2B), mRNA
NM 315443 Homo sapiens hypothetical protein LOC284058 (LOC284058), mRNA
NM_015444 Homo sapiens Ras-induced senescence 1 (RIS1), mRNA
NM 315446 Homo sapiens ELYS transcription factor-like protein TMBS62 (ELYS), mRNΛ
NM 315447 Homo sapiens calmodulin regulated spectrin-associated protein 1 (CAMSAP
NM_015448 Homo sapiens deleted in a mouse model of primary ciliary dyskinesia (DPCC
NM 315457 Homo sapiens zinc finger, DHHC domain containing 5 (ZDHHC5), mRNA
NM_015459 Homo sapiens DKFZP564J0863 protein (DKFZP564J0863), mRNA
NM_015460 Homo sapiens myosin VIIA and Rab interacting protein (MYRIP), mRNA
NM 315461 Homo sapiens zinc finger protein 521 (ZNF521), mRNA
NMJ315463 Homo sapiens chromosome 2 open reading frame 32 (C2orf32), mRNA
NM_015464 Homo sapiens sclerostin domain containing 1 (SOSTDC1), mRNA
NM_015465 Homo sapiens gem (nuclear organelle) associated protein 5 (GEMIN5), mRN
NM_015466 Homo sapiens protein tyrosine phosphatase, non-receptor type 23 (PTPN23)
NM 315469 Homo sapiens nipsnap homolog 3A (C. elegans) (NIPSNAP3A), mRNA
NMJ315470 Homo sapiens RAB11 family interacting protein 5 (class I) (RAB11FIP5), mR
NM 315475 Homo sapiens DKFZP564F0522 protein (DKFZP564F0522), mRNA
NM_015476 Homo sapiens chromosome 18 open reading frame 10 (C18orf10), mRNA
NM_015477 Homo sapiens SIN3 homolog A, transcriptional regulator (yeast) (SIN3A), mF
NM_015481 Homo sapiens zinc finger protein 385 (ZNF385), mRNA
NM 315483 Homo sapiens kelch repeat and BTB (POZ) domain containing 2 (KBTBD2),
NM 315488 Homo sapiens myofibrillogenesis regulator 1 (MR-1), mRNA
NM_015503 Homo sapiens SH2-B homolog (SH2B), mRNA
NM_015508 Homo sapiens TCDD-inducible poly(ADP-ribose) polymerase (TIPARP), mRI
NM_015518 Homo sapiens DKFZP434C131 protein (DKFZP434C131 ), mRNA
NM_015522 Homo sapiens dynein 2 light intermediate chain (D2LIC), transcript variant 2,
NM_015529 Homo sapiens monooxygenase, DBH-like 1 (MOXD1), mRNA
NM 315531 Homo sapiens DKFZP586P0123 protein (DKFZP586P0123), mRNA
NM_015532 Homo sapiens glutamate receptor, ionotropic, N-methyl D-aspartate-like 1A (
NMJ315534 Homo sapiens zinc finger, ZZ domain containing 3 (ZZZ3), mRNA
NM 315541 Homo sapiens leucine-rich repeats and immunoglobulin-like domains 1 (LRK
NM 315547 Homo sapiens thioesterase, adipose associated (THEA), transcript variant 1 ,
NMJ315548 Homo sapiens dystonin (DST), transcript variant 1eA, mRNA
NM_015553 Homo sapiens phosphoinositide-binding protein PIP3-E (PIP3-E), mRNA
NM_015555 Homo sapiens zinc finger protein 451 (ZNF451 ), mRNA
NM 315557 Homo sapiens chromodomain helicase DNA binding protein 5 (CHD5), mRN,
NMJ315558 Homo sapiens synovial sarcoma translocation gene on chromosome 18-iike
NM_015560 Homo sapiens optic atrophy 1 (autosomal dominant) (OPA1), nuclear gene e
NM_015565 Homo sapiens zinc finger protein 294 (ZNF294), mRNA
NM_015567 Homo sapiens SLIT and NTRK-like family, member 5 (SLITRK5), mRNA
NM_015568 Homo sapiens protein phosphatase 1 , regulatory (inhibitor) subunit 16B (PPF
NM 315569 Homo sapiens dynamin 3 (DNM3), mRNA
NM_015575 Homo sapiens trinucleotide repeat containing 15 (TNRC15), mRNA
NMJ315576 Homo sapiens CAZ-associated structural protein (CAST), mRNA
NM_015578 Homo sapiens chromosome 19 open reading frame 13 (C19orf13), mRNA
NMJ315585 Homo sapiens chromosome 20 open reading frame 26 (C20orf26), mRNA
NM_015589 Homo sapiens sterile alpha motif domain containing 4 (SAMD4), mRNA
NM 315597 Homo sapiens G-protein signalling modulator 1 (AGS3-like, C. elegans) (GP!
NMJ315600 Homo sapiens chromosome 20 open reading frame 22 (C20orf22), mRNA
NM_015602 Homo sapiens lamina-associated polypeptide 1 B (LAP1 B), mRNA
NM 315604 Homo sapiens WD repeat domain 21 (WDR21), transcript variant 1 , mRNA
NM 315605 Homo sapiens DKFZP566K0524 protein (DKFZP566K0524), mRNA
NM_015608 Homo sapiens chromosome 10 open reading frame 137 (C10orf137), mRNA
NMJ315609 Homo sapiens putative MAPK activating protein PM20.PM21 (DKFZp566C04
NM 315617 Homo sapiens pygopus 1 (PYG01), mRNA
NMJ315627 Homo sapiens LDL receptor adaptor protein (ARH), mRNA
NM_015631 Homo sapiens chromosome 10 open reading frame 61 (C10orf61), mRNA
NM_015633 Homo sapiens FGFR1 oncogene partner 2 (FGFR10P2), mRNA
NM_015634 Homo sapiens KIAA1279 (KIAA1279), mRNA
NM_015635 Homo sapiens DKFZP434C212 protein (DKFZP434C212), mRNA
NM_015639 Homo sapiens GTPase activating RANGAP domain-like 2 pseudogene (GAF
NM_015649 Homo sapiens interferon regulatory factor 2 binding protein 1 (IRF2BP1), mR
NM_015655 Homo sapiens zinc finger protein 337 (ZNF337), mRNA
NM 315659 Homo sapiens DKFZP564M182 protein (DKFZP564M182), mRNA
NMJ315660 Homo sapiens immunity associated protein 2 (HIMAP2), mRNA
NM_015662 Homo sapiens selective LIM binding factor, rat homolog (SLB), mRNA
NM_015666 Homo sapiens GTP binding protein 5 (putative) (GTPBP5), mRNA
NM 315667 Homo sapiens chromosome 9 open reading frame 36 (C9orf36), mRNA
NM_015668 Homo sapiens DKFZP434I092 protein (DKFZP434I092), mRNA
NM_015687 Homo sapiens filamin A interacting protein 1 (FILIP1), mRNA
NM_015690 Homo sapiens serine/threonine kinase 36 (fused homolog, Drosophila) (STK
NM 315691 Homo sapiens KIAA1280 protein (KIAA1280), mRNA
NM_015692 Homo sapiens C3 and PZP-like, alpha-2-macroglobulin domain containing 8
NM 315693 Homo sapiens PDZ domain containing 6 (PDZK6), mRNA
NM 315694 Homo sapiens KIAA1285 protein (KIAA1285), mRNA
NMJ315713 Homo sapiens ribonucleotide reductase M2 B (TP53 inducible) (RRM2B), mF
NM_015723 Homo sapiens intracellular membrane-associated calcium-independent phos
NM_015905 Homo sapiens transcriptional intermediary factor 1 (TIF1), transcript variant 1
NM_015979 Homo sapiens cofactor required for Sp1 transcriptional activation, subunit 3,
NM_016105 Homo sapiens FK506 binding protein 7 (FKBP7), transcript variant 1 , mRNA
NM_016133 Homo sapiens insulin induced gene 2 (INSIG2), mRNA
NM_016320 Homo sapiens nucleoporin 98kDa (NUP98), transcript variant 1 , mRNA
NM_016544 Homo sapiens Ras-associated protein Rap1 (RBJ), mRNA
NM 31 419 Homo sapiens amiloride-sensitive cation channel 5, intestinal (ACCN5), mR
NM_017437 Homo sapiens cleavage and polyadenylation specific factor 2, 100kDa (CPSI
NMJ317440 Homo sapiens nuclear protein double minute 1 (MDM1), mRNA
NM_01 510 Homo sapiens gp25L2 protein (HSGP25L2G), mRNA
NM 317516 Homo sapiens RAB39, member RAS oncogene family (RAB39), mRNA
NM 317519 Homo sapiens AT rich interactive domain 1B (SWH-like) (ARID1B), transcrip
NM_017520 Homo sapiens M-phase phosphoprotein, mppδ (HSMPP8), mRNA
NM_017525 Homo sapiens myotonic dystrophy protein kinase like protein (HSMDPKIN), r
NM 317527 Homo sapiens cDNAfor differentially expressed C016 gene (LY6K), mRNA
NM_017539 Homo sapiens dynein, axonemal, heavy polypeptide 3 (DNAH3), mRNA
NM_017549 Homo sapiens upregulated in colorectal cancer gene 1 (UCC1), mRNA
NM_017550 Homo sapiens KIAA1193 (KIAA1193), mRNA
NM 317553 Homo sapiens homolog of yeast INO80 (INO80), transcript variant 1 , mRNA
NM 317554 Homo sapiens KIAA1268 protein (KIAA1268), mRNA
NM_017556 Homo sapiens filamin-binding LIM protein-1 (FBLP-1), mRNA
NM 317563 Homo sapiens interleukin 17 receptor D (IL17RD), mRNA
NM_017565 Homo sapiens family with sequence similarity 20, member A (FAM20A), mRt
NM_017570 Homo sapiens 5-oxoprolinase (ATP-hydrolysing) (OPLAH), mRNA
NM_017573 Homo sapiens proprotein convertase subtilisin/kexin type 4 (PCSK4), mRNA
NM_017576 Homo sapiens kinesin family member 27 (KIF27), mRNA
NM_017580 Homo sapiens zinc finger, RAN-binding domain containing 1 (ZRANB1), mRI
NM 317602 Homo sapiens hypothetical protein DKFZp761A052 (DKFZp761A052), mRN;
NM 317619 Homo sapiens U11/U12 snRNP 65K protein (FLJ25070), mRNA
NM_017628 Homo sapiens hypothetical protein FLJ20032 (FLJ20032), mRNA
NM 317641 Homo sapiens kinesin family member 21 A (KIF21 A), mRNA
NM_017666 Homo sapiens suppressor of hairy wing homolog 3 (Drosophila) (SUHW3), π
NM_017672 Homo sapiens transient receptor potential cation channel, subfamily M, mem
NM_017725 Homo sapiens hypothetical protein FLJ20249 (FLJ20249), transcript variant :
NM_017747 Homo sapiens ankyrin repeat and KH domain containing 1 (ANKHD1), trans(
NM_017754 Homo sapiens chromosome 6 open reading frame 107 (C6orf107), mRNA
NM 317758 Homo sapiens hypothetical protein FLJ20308 (FLJ20308), mRNA
NM 317771 Homo sapiens PX domain containing serine/threonine kinase (PXK), mRNA
NM_017804 Homo sapiens CTF8, chromosome transmission fidelity factor 8 homolog (S.
NM 317861 Homo sapiens hypothetical protein FLJ20522 (FLJ20522), mRNA
NM 317871 Homo sapiens hypothetical protein FLJ20542 (FLJ20542), mRNA
NM 317879 Homo sapiens hypothetical protein FLJ20557 (FLJ20557), mRNA
NM 317969 Homo sapiens hypothetical protein FLJ10006 (FLJ10006), mRNA
NM_017978 Homo sapiens ankyrin repeat and KH domain containing 1 (ANKHD1), transc
NM_018003 Homo sapiens uveal autoantigen with coiled-coil domains and ankyrin repeal
NMJ318069 Homo sapiens hypothetical protein FLJ10352 (FLJ10352), transcript variant :
NM_018117 Homo sapiens WD repeat domain 11 (WDR11 ), mRNA
NM_018151 Homo sapiens telomere-associated protein RIF1 homolog (Rif1), mRNA
NM_018177 Homo sapiens Nedd4 binding protein 2 (N4BP2), mRNA
NM 318193 Homo sapiens hypothetical protein FLJ10719 (FLJ10719), mRNA
NM_018218 Homo sapiens ubiquitin specific protease 40 (USP40), mRNA
NM_018237 Homo sapiens cell division cycle and apoptosis regulator 1 (CCAR1), mRNA
NM 318284 Homo sapiens guanylate binding protein 3 (GBP3), mRNA
NM 318325 Homo sapiens chromosome 9 open reading frame 72 (C9orf72), transcript vs
NM_018334 Homo sapiens leucine rich repeat neuronal 3 (LRRN3), mRNA
NM 318369 Homo sapiens DEP domain containing 1 B (DEPDC1 B), mRNA
NM 318392 Homo sapiens hypothetical protein FLJ11331 (FLJ11331), mRNA
NMJ318397 Homo sapiens choline dehydrogenase (CHDH), mRNA
NM 318405 Homo sapiens hypothetical protein, clone 2746033 (HSA272196), mRNA
NM_018414 Homo sapiens sialyltransferase 7 ((alpha-N-acetylneuraminyl-2,3-beta-galaci
NM_018420 Homo sapiens solute carrier family 22 (organic cation transporter), member 1
NM_018424 Homo sapiens erythrocyte membrane protein band 4.1 like 4B (EPB41 L4B),
NMJ318429 Homo sapiens B double prime 1 , subunit of RNA polymerase III transcription
NM_018462 Homo sapiens chromosome 3 open reading frame 10 (C3orf10), mRNA
NM 318646 Homo sapiens transient receptor potential cation channel, subfamily V, mem
NM 318689 Homo sapiens KIAA1199 (KIAA1199), mRNA
NM_018703 Homo sapiens retinoblastoma binding protein 6 (RBBP6), transcript variant 2
NM_018704 Homo sapiens hypothetical protein DKFZp547A023 (DKFZp547A023), mRN/
NM_018708 Homo sapiens fem-1 homolog a (C.elegans) (FEM1A), mRNA
NM 318710 Homo sapiens hypothetical protein DKFZp762O076 (DKFZp762O076), mRN
NMJ318711 Homo sapiens hypothetical protein DKFZp761 H039 (DKFZp761 H039), mRN,
NM 318712 Homo sapiens hypothetical protein DKFZp547C176 (DKFZp547C176), mRN,
NM_018714 Homo sapiens component of oligomeric golgi complex 1 (COG1), mRNA
NM 318715 Homo sapiens RCd-like (TD-60), mRNA
NM 318717 Homo sapiens mastermind-like 3 (Drosophila) (MAML3), mRNA
NM_018837 Homo sapiens sulfatase 2 (SULF2), transcript variant 1 , mRNA
NM_018847 Homo sapiens kelch-like 9 (Drosophila) (KLHL9), mRNA
NMJ318981 Homo sapiens DnaJ (Hsp40) homolog, subfamily C, member 10 (DNAJC10),
NM_018987 Homo sapiens sema domain, seven thrombospondin repeats (type 1 and typ
NM 318998 Homo sapiens F-box and WD-40 domain protein 5 (FBXW5), transcript varia
NM 318999 Homo sapiens KIAA1128 protein (KIAA1128), mRNA
NM 319001 Homo sapiens 5-3' exoribonuclease 1 (XRN1), mRNA
NMJ319007 Homo sapiens hypothetical protein FLJ20811 (FLJ20811), mRNA
NM 319010 Homo sapiens keratin 20 (KRT20), mRNA
NM 319015 Homo sapiens chondroitin sulfate glucuronyltransferase (CSGIcA-T), mRNA
NM_019022 Homo sapiens FLJ20793 protein (FLJ20793), mRNA
NM_019026 Homo sapiens putative membrane protein (LOC54499), mRNA
NM_019029 Homo sapiens carboxypeptidase, vitellogenic-like (CPVL), transcript variant i
NM 319030 Homo sapiens DEAH (Asp-Glu-Ala-His) box polypeptide 29 (DHX29), mRNA
NM_019032 Homo sapiens thrombospondin repeat containing 1 (TSRC1), mRNA
NM_019036 Homo sapiens 3-hydroxymethyl-3-methylglutaryl-Coenzyme A lyase-like 1 (H
NM_019051 Homo sapiens mitochondrial ribosomal protein L50 (MRPL50), nuclear gene
NM 319053 Homo sapiens SEC15-like 1 (S. cerevisiae) (SEC15L1 ), mRNA
NM_019055 Homo sapiens roundabout homolog 4, magic roundabout (Drosophila) (ROB'
NM 319065 Homo sapiens EF hand calcium binding protein 2 (EFCBP2), mRNA
NM_019072 Homo sapiens small glutamine-rich tetratricopeptide repeat (TPR)-containing
NM_019075 Homo sapiens UDP glycosyltransferase 1 family, polypeptide A10 (UGT1A1C
NM 319077 Homo sapiens UDP glycosyltransferase 1 family, polypeptide A7 (UGT1A7),
NM 319078 Homo sapiens UDP glycosyltransferase 1 family, polypeptide A5 (UGT1A5),
NM_019085 Homo sapiens F-box and leucine-rich repeat protein 19 (FBXL19), mRNA
NM 319092 Homo sapiens hypothetical protein KIAA1164 (KIAA1164), mRNA
NM 319104 Homo sapiens protein F25965 (F25965), mRNA
NM_019107 Homo sapiens chromosome 19 open reading frame 10 (C19orf10), mRNA
NM 319590 Homo sapiens KIAA1217 (KIAA1217), mRNA
NM 319593 Homo sapiens hypothetical protein KIAA1434 (KIAA1434), mRNA
NM_019594 Homo sapiens leucine rich repeat containing 8 (LRRC8), mRNA
NM_019850 Homo sapiens neuronal guanine nucleotide exchange factor (NGEF), mRNA
NM 320063 Homo sapiens BarH-like 2 (Drosophila) (BARHL2), mRNA
NM 320116 Homo sapiens follistatin-like 5 (FSTL5), mRNA
NM_020124 Homo sapiens interferon, kappa (IFNK), mRNA
NM 320170 Homo sapiens nicalin (LOC56926), mRNA
NM 320172 Homo sapiens SPPL2b (SPPL2B), mRNA
NM_020175 Homo sapiens hypothetical protein from EUROIMAGE 1967720 (LOC56931 )
NM 320192 Homo sapiens chromosome 7 open reading frame 36 (C7orf36), mRNA
NM 320204 Homo sapiens LIM homeobox 9 (LHX9), mRNA
NM_020207 Homo sapiens chromosome 9 open reading frame 102 (C9orf102), mRNA
NM_020209 Homo sapiens src homology 2 domain-containing transforming protein D (Sh
NMJ320210 Homo sapiens sema domain, immunoglobulin domain (Ig), transmembrane c
NM 320211 Homo sapiens RGM domain family, member A (RGMA), mRNA
NM 320212 Homo sapiens hypothetical protein from EUROIMAGE 384293 (LOC56964),
NM 320214 Homo sapiens hypothetical protein from EUROIMAGE 1977056 (LOC56965)
NM_020219 Homo sapiens carcinoembryonic antigen-like 1 (CEAL1), mRNA
NM_020223 Homo sapiens family with sequence similarity 20, member C (FAM20C), mRI
NM_020311 Homo sapiens chemokine orphan receptor 1 (CMKOR1), mRNA
NM 320312 Homo sapiens hypothetical protein DKFZp434K046 (DKFZP434K046), mRN
NMJ320318 Homo sapiens pappalysin 2 (PAPPA2), transcript variant 1 , mRNA
NMJ320319 Homo sapiens ankyrin repeat and MYND domain containing 2 (ANKMY2), ml
NM 320320 Homo sapiens arginyl-tRNA synthetase-like (RARSL), mRNA
NM 320336 Homo sapiens KIAA1219 protein (KIAA1219), mRNA
NM_020338 Homo sapiens retinoic acid induced 17 (RAI17), mRNA
NM 320340 Homo sapiens KIAA1244 (KIAA1244), mRNA
NM 320341 Homo sapiens p21 (CDKN1 A)-activated kinase 7 (PAK7), transcript variant 1 ,
NM 320376 Homo sapiens transport-secretion protein 2.2 (TTS-2.2), mRNA
NM_020378 Homo sapiens K562 cell-derived leucine-zipper-like protein 1 (KLP1 ), mRNA
NM_020383 Homo sapiens X-prolyl aminopeptidase (aminopeptidase P) 1 , soluble (XPNF
NM_020409 Homo sapiens mitochondrial ribosomal protein L47 (MRPL47), nuclear gene
NM_020416 Homo sapiens protein phosphatase 2 (formerly 2A), regulatory subunit B (PF
NM 320417 Homo sapiens T-box 20 (TBX20), mRNA
NM_020420 Homo sapiens deleted in azoospermia 4 (DAZ4), mRNA
NM_020429 Homo sapiens SMAD specific E3 ubiquitin protein ligase 1 (SMURF1), transc
NM_020432 Homo sapiens putative homeodomain transcription factor 2 (PHTF2), mRNA
NMJ320438 Homo sapiens dolichyl pyrophosphate phosphatase 1 (DOLPP1), mRNA
NM_020440 Homo sapiens prostaglandin F2 receptor negative regulator (PTGFRN), mRh
NMJ320447 Homo sapiens chromosome 15 open reading frame 17 (C15orf17), mRNA
NM_020451 Homo sapiens selenoprotein N, 1 (SEPN1), transcript variant 1 , mRNA
NM 320452 Homo sapiens ATPase, Class I, type 8B, member 2 (ATP8B2), mRNA
NM 320453 Homo sapiens ATPase, Class V, type 10D (ATP10D), mRNA
NM 320455 Homo sapiens G protein-coupled receptor 126 (GPR126), mRNA
NM 320456 Homo sapiens chromosome 13 open reading frame 1 (C13orf1), mRNA
NM 320457 Homo sapiens THAP domain containing 11 (THAP11), mRNA
NM 320462 Homo sapiens KIAA1181 protein (KIAA1181), mRNA
NM 320463 Homo sapiens KIAA1387 protein (KIAA1387), mRNA
NM_020468 Homo sapiens sorting nexin 14 (SNX14), transcript variant 2, mRNA
NM_020531 Homo sapiens chromosome 20 open reading frame 3 (C20orf3), mRNA
NM_020532 Homo sapiens reticulon 4 (RTN4), transcript variant 1 , mRNA
NMJ320536 Homo sapiens CSRP2 binding protein (CSRP2BP), transcript variant 1 , mRN
NM 320546 Homo sapiens adenylate cyclase 2 (brain) (ADCY2), mRNA
NM 320631 Homo sapiens putative NFkB activating protein (KIAA0720), transcript varian
NM 320647 Homo sapiens junctophilin 1 (JPH1), mRNA
NM 320693 Homo sapiens Down syndrome cell adhesion molecule like 1 (DSCAML1), m
NM_020695 Homo sapiens transcription elongation factor B polypeptide 3 binding protein
NM 320696 Homo sapiens KIAA1143 protein (KIAA1143), mRNA
NM_020697 Homo sapiens potassium voltage-gated channel, delayed-rectifier, subfamily
NMJ320698 Homo sapiens KIAA1145 protein (KIAA1145), mRNA
NMJ320699 Homo sapiens transcription repressor p66 beta component of the MeCP1 coi
NM 320701 Homo sapiens KIAA1160 protein (KIAA1160), mRNA
NM 320702 Homo sapiens KIAA1161 (KIAA1161), mRNA
NM_020706 Homo sapiens splicing factor, arginine/serine-rich 15 (SFRS15), mRNA
NM_020710 Homo sapiens KIAA1185 protein (KIAA1185), mRNA
NM 320713 Homo sapiens KIAA1196 protein (KIAA1196), mRNA
NM 320714 Homo sapiens zinc finger protein 490 (ZNF490), mRNA
NM_020718 Homo sapiens ubiquitin specific protease 31 (USP31), mRNA
NM 320728 Homo sapiens K1AA1228 protein (KIAA1228), mRNA
NM 320732 Homo sapiens AT rich interactive domain 1 B (SWH-like) (ARID1 B), transcrip
NM 320739 Homo sapiens cell cycle progression 1 (CCPG1), mRNA
NM_020740 Homo sapiens ankyrin repeat and FYVE domain containing 1 (ANKFY1), trar
NM 320742 Homo sapiens neuroligin 4, X-linked (NLGN4X), transcript variant 1 , mRNA
NM_020744 Homo sapiens metastasis associated family, member 3 (MTA3), mRNA
NMJ320745 Homo sapiens alanyl-tRNA synthetase like (AARSL), mRNA
NM 320746 Homo sapiens KIAA1271 protein (KIAA1271), mRNA
NM 320748 Homo sapiens KIAA1287 protein (KIAA1287), mRNA
NMJ320750 Homo sapiens exportin 5 (XP05), mRNA
NM_020751 Homo sapiens component of oligomeric golgi complex 6 (COG6), mRNA
NM 320752 Homo sapiens G protein-coupled receptor 158 (GPR158), mRNA
NM_020753 Homo sapiens CASK interacting protein 2 (CASKIN2), mRNA
NM_020755 Homo sapiens tumor differentially expressed 2 (TDE2), mRNA
NMJ320761 Homo sapiens raptor (raptor), mRNA
NM 320762 Homo sapiens SLIT-ROBO Rho GTPase activating protein 1 (SRGAP1), mR
NM 320765 Homo sapiens retinoblastoma-associated factor 600 (RBAF600), mRNA
NM 320769 Homo sapiens KIAA1318 protein (KIAA1318), mRNA
NM_020771 Homo sapiens HECT domain and ankyrin repeat containing, E3 ubiquitin pro
NM_020772 Homo sapiens 82-kD FMRP Interacting Protein (182-FIP), mRNA
NM 320773 Homo sapiens TBC1 domain family, member 14 (TBC1 D14), mRNA
NM__020774 Homo sapiens mindbomb homolog 1 (Drosophila) (MIB1), mRNA
NM 320775 Homo sapiens mabal (KIAA1324), mRNA
NM 320778 Homo sapiens likely ortholog of mouse myocytic induction/differentiation orig
NM 320779 Homo sapiens WD repeat domain 35 (WDR35), mRNA
NM 320781 Homo sapiens zinc finger protein 398 (ZNF398), transcript variant 2, mRNA
NM 320783 Homo sapiens synaptotagmin IV (SYT4), mRNA
NM_020786 Homo sapiens pyruvate dehydrogenase phosphatase isoenzyme 2 (PDP2), i
NMJ320787 Homo sapiens zinc finger protein 624 (ZNF624), mRNA
NM_020789 Homo sapiens immunoglobulin superfamily, member 9 (IGSF9), mRNA
NMJ320791 Homo sapiens serine/threonine protein kinase TA01 homolog (KIAA1361), n
NM 320792 Homo sapiens KIAA1363 protein (KIAA1363), mRNA
NM 320795 Homo sapiens neuroligin 2 (NLGN2), mRNA
NM 320799 Homo sapiens associated molecule with the SH3 domain of STAM (AMSH) Ii
N J20800 Homo sapiens KIAA1374 protein (KIAA1374), mRNA
NMJ320801 Homo sapiens arrestin domain containing 3 (ARRDC3), mRNA
NM 320803 Homo sapiens kelch-like 8 (Drosophila) (KLHL8), mRNA
NM_ )20804 Homo sapiens protein kinase C and casein kinase substrate in neurons 1 (P;
NM 320808 Homo sapiens signal-induced proliferation-associated 1 like 2 (SIPA1L2), mF
NM 320809 Homo sapiens Rho GTPase activating protein 20 (ARHGAP20), mRNA
NM 320810 Homo sapiens KIAA1393 (KIAA1393), mRNA
NM_020812 Homo sapiens dedicator of cytokinesis 6 (DOCK6), mRNA
NMJ320813 Homo sapiens zinc finger protein 471 (ZNF471), mRNA
NMJ320816 Homo sapiens kinesin family member 17 (KIF17), mRNA
NM 320817 Homo sapiens KIAA1407 protein (KIAA1407), mRNA
NM 320818 Homo sapiens KIAA1409 (KIAA1409), mRNA
NMJ320820 Homo sapiens phosphatidylinositol 3,4,5-trisphosphate-dependent RAC ex
NM_020824 Homo sapiens Rho GTPase activating protein 21 (ARHGAP21), mRNA
NM_020825 Homo sapiens Crm, cramped-like (Drosophila) (CRAMP1 L), mRNA
NMJ320826 Homo sapiens synaptotagmin XIII (SYT13), mRNA
NM_020828 Homo sapiens zinc finger protein 28 homolog (mouse) (ZFP28), mRNA
NM 320832 Homo sapiens KIAA1441 protein (KIAA1441), mRNA
NM 320834 Homo sapiens KIAA1443 (KIAA1443), mRNA
NMJ320839 Homo sapiens WD repeat endosomal protein (KIAA1449), mRNA
NM 320844 Homo sapiens KIAA1456 protein (KIAA1456), mRNA
NM_020845 Homo sapiens phosphatidylinositol transfer protein, membrane-associated 2
NMJ320847 Homo sapiens trinucleotide repeat containing 6 (TNRC6), mRNA
NM 320850 Homo sapiens Ran-binding protein 10 (RANBP10), mRNA
NM 320851 Homo sapiens KIAA1465 protein (KIAA1465), mRNA
NM 320854 Homo sapiens KIAA1468 (KIAA1468), mRNA
NM 320856 Homo sapiens zinc finger protein 537 (ZNF537), mRNA
NM_020858 Homo sapiens sema domain, transmembrane domain (TM), and cytoplasmic
NM_020859 Homo sapiens Shroom-related protein (ShrmL), mRNA
NM_020860 Homo sapiens stromal interaction molecule 2 (STIM2), mRNA
NM_020861 Homo sapiens zinc finger and BTB domain containing 2 (ZBTB2), mRNA
NM_020863 Homo sapiens zinc finger protein 406 (ZNF406), mRNA
NMJ320867 Homo sapiens ubiquitin associated protein 2 (UBAP2), transcript variant 2, nr
NM 320868 Homo sapiens dipeptidylpeptidase 10 (DPP10), mRNA
NMJ320870 Homo sapiens SH3 multiple domains 2 (SH3MD2), mRNA
NM_020871 Homo sapiens leucine-rich repeats and calponin homology (CH) domain con
NM_020873 Homo sapiens leucine rich repeat neuronal 1 (LRRN1), mRNA
NM 320875 Homo sapiens Fraser syndrome 1 (FRAS1), transcript variant 3, mRNA
NM 320880 Homo sapiens zinc finger protein 530 (ZNF530), mRNA
NM 320882 Homo sapiens KIAA1510 protein (KIAA1510), mRNA
NM 320889 Homo sapiens PHD finger protein 12 (PHF12), mRNA
NM 320890 Homo sapiens KIAA1524 protein (KIAA1524), mRNA
NMJ320892 Homo sapiens deltex homolog 2 (Drosophila) (DTX2), mRNA
NM 320895 Homo sapiens KIAA1533 (KIAA1533), mRNA
NMJ320897 Homo sapiens hyperpolarization activated cyclic nucleotide-gated potassium
NM_020899 Homo sapiens zinc finger and BTB domain containing 4 (ZBTB4), mRNA
NM_020914 Homo sapiens chromosome 17 open reading frame 27 (C17orf27), mRNA
NM_020918 Homo sapiens glycerol-3-phosphate acyltransferase, mitochondrial (GPAM),
NM_020922 Homo sapiens protein kinase, lysine deficient 3 (PRKWNK3), transcript varia
NM 320925 Homo sapiens KIAA1573 protein (KIAA1573), mRNA
NM 320926 Homo sapiens BCL6 co-repressor (BCOR), transcript variant 2, mRNA
NM 320927 Homo sapiens KIAA1576 protein (KIAA1576), mRNA
NM_020932 Homo sapiens melanoma antigen, family E, 1 (MAGEE1), mRNA
NMJ320935 Homo sapiens ubiquitin specific protease 37 (USP37), mRNA
NM 320936 Homo sapiens DEAD (Asp-Glu-Ala-Asp) box polypeptide 55 (DDX55), mRN/5
NM 320939 Homo sapiens copine V (CPNE5), mRNA
NM 320944 Homo sapiens glucosidase, beta (bile acid) 2 (GBA2), mRNA
NMJD20947 Homo sapiens KIAA1609 protein (KIAA1609), mRNA
NM_020948 Homo sapiens mesoderm induction early response 1 (MI-ER1), mRNA
NM_020951 Homo sapiens zinc finger protein 529 (ZNF529), mRNA
NM_020952 Homo sapiens transient receptor potential cation channel, subfamily M, mem
NMJ320954 Homo sapiens KIAA1618 (KIAA1618), mRNA
NM 320961 Homo sapiens KIAA1627 protein (KIAA1627), mRNA
NMJ320962 Homo sapiens likely ortholog of mouse neighbor of Punc E11 (NOPE), mRN;
NM 320964 Homo sapiens KIAA1632 protein (KIAA1632), mRNA
NM 320965 Homo sapiens membrane-associated guanylate kinase-related (MAGI-3) (M
NM 320970 Homo sapiens KIAA1641 (KIAA1641 ), mRNA
NMJ320971 Homo sapiens spectrin, beta, non-erythrocytic 4 (SPTBN4), mRNA
NM 321006 Homo sapiens chemokine (C-C motif) ligand 3-like 1 (CCL3L1), mRNA
NM 321009 Homo sapiens ubiquitin C (UBC), mRNA
NM 321035 Homo sapiens KIAA1404 protein (KIAA1404), mRNA
NM_021044 Homo sapiens desert hedgehog homolog (Drosophila) (DHH), mRNA
NM 321045 Homo sapiens zinc finger protein 248 (ZNF248), mRNA
NM 321059 Homo sapiens histone 2, H3c (HIST2H3C), mRNA
NM_021061 Homo sapiens zinc finger protein 647 (ZNF647), mRNA
NM 321072 Homo sapiens hyperpolarization activated cyclic nucleotide-gated potassium
NM 321088 Homo sapiens zinc finger protein 2 (A1-5) (ZNF2), mRNA
NMJ321089 Homo sapiens zinc finger protein 8 (clone HF.18) (ZNF8), mRNA
NM_021116 Homo sapiens adenylate cyclase 1 (brain) (ADCY1), mRNA
NM 321117 Homo sapiens cryptochrome 2 (photolyase-like) (CRY2), mRNA
NM 321143 Homo sapiens zinc finger protein 20 (KOX 13) (ZNF20), mRNA
NMJ321148 Homo sapiens zinc finger protein 273 (ZNF273), mRNA
NM_021149 Homo sapiens coactosin-like 1 (Dictyostelium) (COTL1), mRNA
NM_021164 Homo sapiens splicing factor 4 (SF4), transcript variant b, mRNA
NM 321165 Homo sapiens hypothetical protein from clone 24828 (KIAA1747), mRNA
NM_021180 Homo sapiens transcription factor CP2-like 4 (TFCP2L4), transcript variant 1
NM_021202 Homo sapiens tumor protein p53 inducible nuclear protein 2 (TP53INP2), mF
NM_021217 Homo sapiens zinc finger protein 77 (pT1) (ZNF77), mRNA
NM 321218 Homo sapiens chromosome 9 open reading frame 80 (C9orf80), mRNA
NM 321222 Homo sapiens TcD37 homolog (HTCD37), mRNA
NM 321224 Homo sapiens zinc finger protein 462 (ZNF462), mRNA
NM 321227 Homo sapiens DC2 protein (DC2), mRNA
NM 321228 Homo sapiens serine arginine-rich pre-mRNA splicing factor SR-A1 (SR-A1 ),
NM 321237 Homo sapiens selenoprotein K (SELK), mRNA
NM_021250 Homo sapiens leukocyte Ig-like receptor 9 (LIR9), transcript variant 1 , mRNA
NM_021260 Homo sapiens zinc finger, FYVE domain containing 1 (ZFYVE1), transcript v,
NM_021636 Homo sapiens leucine-rich repeat-containing G protein-coupled receptor 6 (L
NM 321649 Homo sapiens toll-like receptor adaptor molecule 2 (TICAM2), mRNA
NM 321652 Homo sapiens SMA4 (SMA4), mRNA
NMJ321915 Homo sapiens zinc finger protein 69 (Cos5) (ZNF69), mRNA
NMJ321916 Homo sapiens zinc finger protein 70 (Cos17) (ZNF70), mRNA
NM_021936 Homo sapiens pappalysin 2 (PAPPA2), transcript variant 2, mRNA
NM_021937 Homo sapiens elongation factor for selenoprotein translation (SELB), mRNA
NMJ322045 Homo sapiens Mdm2, transformed 3T3 cell double minute 2, p53 binding pro
NM_022075 Homo sapiens LAG1 longevity assurance homolog 2 (S. cerevisiae) (LASS2)
NM_022080 Homo sapiens N-ethylmaleimide-sensitive factor attachment protein, beta (N
NMJ322085 Homo sapiens thioredoxin domain containing 5 (TXNDC5), transcript variant
NM_022092 Homo sapiens CTF18, chromosome transmission fidelity factor 18 homolog (
NM 322106 Homo sapiens chromosome 20 open reading frame 177 (C20orf177), mRNA
NMJ322115 Homo sapiens PR domain containing 15 (PRDM15), mRNA
NM 322138 Homo sapiens SPARC related modular calcium binding 2 (SMOC2), mRNA
NM 322160 Homo sapiens DMRT-like family A1 (DMRTA1), mRNA
NM_022166 Homo sapiens xylosyltransferase I (XYLT1), mRNA
NM 322351 Homo sapiens EF hand calcium binding protein 1 (EFCBP1), mRNA
NM_022475 Homo sapiens hedgehog interacting protein (HHIP), mRNA
NM 322478 Homo sapiens cadherin-like 24 (CDH24), mRNA
NM_022479 Homo sapiens Williams-Beuren syndrome chromosome region 17 (WBSCR1
NMJ322486 Homo sapiens sushi domain containing 1 (SUSD1), mRNA
NM 322491 Homo sapiens likely ortholog of mouse Sds3 (SDS3), mRNA
NMJ322572 Homo sapiens myofibrillogenesis regulator 1 (MR-1), mRNA
NMJ322733 Homo sapiens hypothetical protein AL133206 (LOC64744), mRNA
NM_022742 Homo sapiens hypothetical protein DKFZp434G156 (DKFZP434G156), mRN
NMJ322745 Homo sapiens ATP synthase mitochondrial F1 complex assembly factor 1 (A
NM 322757 Homo sapiens hypothetical protein FLJ12892 (FLJ12892), mRNA
NMJ322824 Homo sapiens F-box and leucine-rich repeat protein 17 (FBXL17), mRNA
NM_022833 Homo sapiens chromosome 9 open reading frame 88 (C9orf88), mRNA
NM 322835 Homo sapiens likely ortholog of mouse common-site lymphoma/leukemia GE
NM 322913 Homo sapiens vasculin (DKFZp761C169), mRNA
NM_023002 Homo sapiens hyaluronan and proteoglycan link protein 4 (HAPLN4), mRNA
NM_023006 Homo sapiens kallikrein 15 (KLK15), transcript variant 1 , mRNA
NM 323939 Homo sapiens hypothetical protein MGC2752 (MGC2752), mRNA
NMJ323943 Homo sapiens hypothetical protein MGC3040 (MGC3040), mRNA
NM 324007 Homo sapiens early B-cell factor (EBF), mRNA
NMJ324019 Homo sapiens neurogenin 2 (NEUROG2), mRNA
NM_024100 Homo sapiens WD repeat domain 18 (WDR18), mRNA
NM 324316 Homo sapiens leukocyte receptor cluster (LRC) member 1 (LENG1), mRNA
NMJ324335 Homo sapiens iroquois homeobox protein 6 (IRX6), mRNA
NM_024336 Homo sapiens iroquois homeobox protein 3 (IRX3), mRNA
NM 324342 Homo sapiens glucocorticoid receptor DNA binding factor 1 (GRLF1), transci
NM_024344 Homo sapiens calpain 3, (p94) (CAPN3), transcript variant 2, mRNA
NM_024420 Homo sapiens phospholipase A2, group IVA (cytosolic, calcium-dependent) i
NM 324493 Homo sapiens zinc finger protein 306 (ZNF306), mRNA
NMJ324496 Homo sapiens chromosome 14 open reading frame 4 (C14orf4), mRNA
NM_024511 Homo sapiens chromosome 4 open reading frame 15 (C4orf15), mRNA
NM_024517 Homo sapiens PHD finger protein 2 (PHF2), transcript variant 2, mRNA
NMJ324553 Homo sapiens hypothetical protein FLJ20097 (FLJ20097), mRNA
NM 324621 Homo sapiens hypothetical protein FLJ12604 (FLJ12604), mRNA
NMJ324625 Homo sapiens zinc finger CCCH type, antiviral 1 (ZC3HAV1), transcript varia
NM 324684 Homo sapiens PTD015 protein (PTD015), mRNA
NM_024742 Homo sapiens armadillo repeat containing 5 (ARMC5), mRNA
NM_024769 Homo sapiens adipocyte-specific adhesion molecule (ASAM), mRNA
NM_024870 Homo sapiens DEP domain containing 2 (DEPDC2), transcript variant 1 , mR
NM 324878 Homo sapiens CGI-72 protein (CGl-72), transcript variant 4, mRNA
NM_024933 Homo sapiens hypothetical protein FLJ12056 (FLJ12056), mRNA
NM 324953 Homo sapiens hypothetical protein FLJ13089 (FLJ13089), mRNA
NM_025169 Homo sapiens zinc finger protein 167 (ZNF167), transcript variant 2, mRNA
NM_025196 Homo sapiens GrpE-like 1 , mitochondrial (E. coli) (GRPEL1), mRNA
NM 325202 Homo sapiens EF hand domain containing 1 (EFHD1), mRNA
NMJ325219 Homo sapiens DnaJ (Hsp40) homolog, subfamily C, member 5 (DNAJC5), m
NM 325224 Homo sapiens BTB (POZ) domain containing 4 (BTBD4), mRNA
NM 325248 Homo sapiens SNAP25-interacting protein (SNIP), mRNA
NM_025252 Homo sapiens Ras association (RalGDS/AF-6) and pleckstrin homology dorr
NMJ325256 Homo sapiens HLA-B associated transcript 8 (BAT8), transcript variant NG3£
NM_030625 Homo sapiens CXXC finger 6 (CXXC6), mRNA
NM_030627 Homo sapiens cytoplasmic polyadenylation element binding protein 4 (CPEB
NM 330628 Homo sapiens KIAA1698 protein (KIAA1698), mRNA
NM__030629 Homo sapiens c-Maf-inducing protein (CMIP), transcript variant Tc-mip, mRN
NMJ330630 Homo sapiens chromosome 17 open reading frame 28 (C17orf28), mRNA
NM 330633 Homo sapiens KIAA1712 (KIAA1712), mRNA
NM 330634 Homo sapiens zinc finger protein 436 (ZNF436), mRNA
NM_030636 Homo sapiens KIAA1706 protein (KIAA1706), mRNA
NM 330637 Homo sapiens DDHD domain containing 1 (DDHD1), mRNA
NM 330639 Homo sapiens KIAA1701 protein (KIAA1 01), mRNA
NM_030640 Homo sapiens dual specificity phosphatase 16 (DUSP16), mRNA
NM_030644 Homo sapiens apolipoprotein L, 3 (APOL3), transcript variant alpha/b, mRN 1
NM 330645 Homo sapiens KIAA1720 protein (KIAA1720), mRNA
NM 330650 Homo sapiens KIAA1715 (KIAA1 15), mRNA
NM_030789 Homo sapiens histocompatibility (minor) 13 (HM13), transcript variant 1 , mRI
NM 330812 Homo sapiens actin like protein (LOC81569), mRNA
NM_030883 Homo sapiens olfactory receptor, family 2, subfamily H, member 1 (OR2H1),
NMJ330906 Homo sapiens serine/threonine kinase 33 (STK33), mRNA
NMJ330919 Homo sapiens chromosome 20 open reading frame 129 (C20orf129), mRNA
NM 330922 Homo sapiens non-imprinted in Prader-Willi/Angelman syndrome 2 (NIPA2),
NM 330923 Homo sapiens hypothetical protein DKFZp566N034 (DKFZP566N034), mRN
NM_030949 Homo sapiens protein phosphatase 1 , regulatory (inhibitor) subunit 14C (PPF
NM_030957 Homo sapiens a disintegrin-like and metalloprotease (reprolysin type) with th
NM 330961 Homo sapiens tripartite motif-containing 56 (TRIM56), mRNA
NM_030962 Homo sapiens Charcot-Marie-Tooth neuropathy 4B2 (autosomal recessive, v
NM 331303 Homo sapiens similar to RIKEN cDNA 4933439B08 gene (MGC33211), mRI*
NM_031444 Homo sapiens chromosome 22 open reading frame 13 (C22orf13), mRNA
NM_031448 Homo sapiens chromosome 19 open reading frame 12 (C19orf12), mRNA
NM_031454 Homo sapiens selenoprotein O (SELO), mRNA
NM_031467 Homo sapiens solute carrier family 4, sodium bicarbonate cotransporter, mer
NM_031490 Homo sapiens peroxisomal Ion protease (LONP), mRNA
NM 331888 Homo sapiens pro-melanin-concentrating hormone-like 2 (PMCHL2), mRNA
NM_031895 Homo sapiens calcium channel, voltage-dependent, gamma subunit 8 (CACI
NM_031912 Homo sapiens synaptotagmin XV (SYT15), transcript variant a, mRNA
NMJ331913 Homo sapiens chr3 synaptotagmin (CHR3SYT), mRNA
NM 331914 Homo sapiens synaptotagmin XlV-like (SYT14L), mRNA
NMJ331935 Homo sapiens hemicentin (FIBL-6), mRNA
NM 332017 Homo sapiens Ser/Thr-like kinase (MGC4796), mRNA
NM_032111 Homo sapiens mitochondrial ribosomal protein L14 (MRPL14), nuclear gene
NM_032119 Homo sapiens monogenic, audiogenic seizure susceptibility 1 homolog (mou
NMJ332123 Homo sapiens kin of IRRE like 2 (Drosophila) (KIRREL2), transcript variant 1
NM 332132 Homo sapiens HORMA domain containing protein (NOHMA), mRNA
NM 332137 Homo sapiens hypothetical protein DKFZp434N1817 (DKFZP434N1817), mF
NMJ332156 Homo sapiens C1q domain containing 1 (C1QDC1), transcript variant 3, mRI
NM_032160 Homo sapiens chromosome 18 open reading frame 4 (C18orf4), mRNA
NM 332165 Homo sapiens hypothetical protein FLJ12303 (FLJ12303), mRNA
NM 332168 Homo sapiens hypothetical protein FLJ12519 (FLJ12519), mRNA
NM_032194 Homo sapiens brix domain containing 1 (BXDC1), mRNA
NM 332195 Homo sapiens SON DNA binding protein (SON), transcript variant b, mRNA
NM_032217 Homo sapiens ankyrin repeat domain 17 (ANKRD17), transcript variant 1 , ml
NM 332222 Homo sapiens hypothetical protein FLJ22374 (FLJ22374), mRNA
NMJ332226 Homo sapiens zinc finger, CCHC domain containing 7 (ZCCHC7), mRNA
NM 332228 Homo sapiens male sterility domain containing 2 (MLSTD2), mRNA
NMJ332230 Homo sapiens hypothetical protein FLJ22789 (FLJ22789), mRNA
NM_032279 Homo sapiens hypothetical protein DKFZp76111011 (DKFZp761 l1011), mRN
NM 332282 Homo sapiens hypothetical protein DKFZp547D155 (DKFZp547D155), mRN
NM_032283 Homo sapiens zinc finger, DHHC domain containing 18 (ZDHHC18), mRNA
NM 332285 Homo sapiens hypothetical protein MGC3207 (MGC3207), mRNA
NM 332286 Homo sapiens hypothetical protein MGC5309 (MGC5309), mRNA
NMJ332422 Homo sapiens G protein-coupled receptor 123 (GPR123), mRNA
NM 332423 Homo sapiens zinc finger protein 528 (ZNF528), mRNA
NM 332425 Homo sapiens KIAA1822 (KIAA1822), mRNA
NM 332427 Homo sapiens mastermind-like 2 (Drosophila) (MAML2), mRNA
NM_032429 Homo sapiens leucine zipper, putative tumor suppressor 2 (LZTS2), mRNA
NM_032430 Homo sapiens KIAA1811 protein (KIAA1811), mRNA
NM_032431 Homo sapiens HRD1 protein (HRD1 ), transcript variant 1 , mRNA
NM 332432 Homo sapiens actin binding LIM protein family, member 2 (ABLIM2), mRNA
NM 332433 Homo sapiens zinc finger protein 333 (ZNF333), mRNA
NM 332434 Homo sapiens zinc finger protein 512 (ZNF512), mRNA
NM_032435 Homo sapiens mixed lineage kinase 4 (KIAA1804), mRNA
NM_032436 Homo sapiens chromosome 13 open reading frame 8 (C13orf8), mRNA
NM 332439 Homo sapiens phytanoyl-CoA hydroxylase interacting protein-like (PHYHIPL]
NM_032440 Homo sapiens ligand-dependent corepressor (MLR2), mRNA
NM_032444 Homo sapiens BTB (POZ) domain containing 12 (BTBD12), mRNA
NM 332448 Homo sapiens KIAA1838 (KIAA1838), mRNA
NM 332452 Homo sapiens junctophilin 4 (JPH4), mRNA
NMJ332458 Homo sapiens PHD finger protein 6 (PHF6), mRNA
NM 332477 Homo sapiens mitochondrial ribosomal protein L41 (MRPL41), nuclear gene
NM_032478 Homo sapiens mitochondrial ribosomal protein L38 (MRPL38), nuclear gene
NM_032479 Homo sapiens mitochondrial ribosomal protein L36 (MRPL36), nuclear gene
NM_032482 Homo sapiens DOT1-like, histone H3 methyltransferase (S. cerevisiae) (DO!
NM 332497 Homo sapiens zinc finger protein 559 (ZNF559), mRNA
NM 332501 Homo sapiens acetyl-Coenzyme A synthetase 2 (AMP forming)-like (ACAS2I
NM_032505 Homo sapiens T-cell activation kelch repeat protein (TA-KRP), mRNA
NM_032506 Homo sapiens KIAA1841 protein (KIAA1841 ), mRNA
NM_032508 Homo sapiens family with sequence similarity 11 , member A (FAM11 A), mRf
NM 332511 Homo sapiens chromosome 6 open reading frame 168 (C6orf168), mRNA
NM 332512 Homo sapiens PDZ domain containing 4 (PDZK4), mRNA
NM 332517 Homo sapiens lysozyme-like 1 (LYZL1), mRNA
NM_032528 Homo sapiens beta-galactoside alpha-2,6-sialyltransferase II (STδGalll), mR
NM 332531 Homo sapiens kin of IRRE like 3 (Drosophila) (KIRREL3), mRNA
NM 332536 Homo sapiens netrin G2 (NTNG2), mRNA
NM 332539 Homo sapiens SLIT and NTRK-like family, member 2 (SLITRK2), mRNA
NM 332550 Homo sapiens KIAA1914 (KIAA1914), transcript variant 2, mRNA
NM_032552 Homo sapiens DAB2 interacting protein (DAB2IP), mRNA
NM_032569 Homo sapiens cytokine-like nuclear factor n-pac (N-PAC), mRNA
NM_032590 Homo sapiens F-box and leucine-rich repeat protein 10 (FBXL10), mRNA
NM 332636 Homo sapiens differential display and activated by p53 (DDA3), mRNA
NM_032869 Homo sapiens chronic myelogenous leukemia tumor antigen 66 (CML66), ml
NM_032870 Homo sapiens chromosome 6 open reading frame 111 (C6orf111 ), mRNA
NM_032947 Homo sapiens putative small membrane protein NID67 (NID67), mRNA
NM_033026 Homo sapiens piccolo (presynaptic cytomatrix protein) (PCLO), transcript vai
NM 333046 Homo sapiens rhotekin (RTKN), mRNA
NM 333052 Homo sapiens DMRT-like family C2 (DMRTC2), mRNA
NM 333053 Homo sapiens DMRT-like family C1 (DMRTC1 ), mRNA
NM_033055 Homo sapiens hippocampus abundant transcript 1 (HIAT1), mRNA
NM 333063 Homo sapiens microtubule-associated protein 6 (MAP6), transcript variant 1 ,
NMJ333064 Homo sapiens ataxia, cerebellar, Cayman type (caytaxin) (ATCAY), mRNA
NM 333067 Homo sapiens DMRT-like family B with proline-rich C-terminal, 1 (DMRTB1),
NMJ333071 Homo sapiens spectrin repeat containing, nuclear envelope 1 (SYNE1), trans
NM 333082 Homo sapiens cytokine induced protein 29 kDa (CIP29), mRNA
NM_033086 Homo sapiens FGD1 family, member 3 (FGD3), mRNA
NM_033088 Homo sapiens family with sequence similarity 40, member A (FAM40A), mRt
NM 333090 Homo sapiens GREB1 protein (GREB1), transcript variant b, mRNA
NMJ333107 Homo sapiens hypothetical protein BC004923 (LOC85865), mRNA
NM 333109 Homo sapiens polyribonucleotide nucleotidyltransferase 1 (PNPT1), mRNA
NM_033112 Homo sapiens chromosome 6 open reading frame 153 (C6orf153), mRNA
NM 333121 Homo sapiens ankyrin repeat domain 13 (ANKRD13), mRNA
NMJ333129 Homo sapiens scratch homolog 2, zinc finger protein (Drosophila) (SCRT2),
NM_033141 Homo sapiens mitogen-activated protein kinase kinase kinase 9 (MAP3K9), i
NM 333160 Homo sapiens DKFZP572C163 protein (DKFZP572C163), mRNA
NM 333161 Homo sapiens surfeit 4 (SURF4), mRNA
NM 333200 Homo sapiens hypothetical protein BC002942 (BC002942), mRNA
NMJ333201 Homo sapiens hypothetical gene BC008967 (BC008967), mRNA
NM 333206 Homo sapiens hypothetical gene FLJ00060 (FLJ00060), mRNA
NM_033253 Homo sapiens 5'-nucleotidase, cytosolic IB (NT5C1 B), transcript variant 2, m
NM 333267 Homo sapiens iroquois homeobox protein 2 (IRX2), mRNA
NM 333271 Homo sapiens BTB (POZ) domain containing 6 (BTBD6), mRNA
NM 333276 Homo sapiens Ku70-binding protein 3 (KUB3), mRNA
NM 333288 Homo sapiens zinc finger protein 160 (ZNF160), transcript variant 1 , mRNA
NM_033364 Homo sapiens chromosome 3 open reading frame 15 (C3orf15), mRNA
NM 333375 Homo sapiens myosin IC (MY01C), mRNA
NMJ333386 Homo sapiens molecule interacting with Rab13 (MIRAB13), mRNA
NM_033387 Homo sapiens chromosome 9 open reading frame 59 (C9orf59), mRNA
NM 333389 Homo sapiens slingshot homolog 2 (Drosophila) (SSH2), mRNA
NM_033392 Homo sapiens mitogen-activated protein kinase 8 interacting protein 3 (MAPI
NM 333393 Homo sapiens KIAA1727 protein (KIAA1727), mRNA
NM 333396 Homo sapiens tankyrase 1 binding protein 1 , 182kDa (TNKS1BP1 ), mRNA
NM 333397 Homo sapiens KIAA1754 (KIAA1754), mRNA
NM 333402 Homo sapiens KIAA1764 protein (KIAA1764), mRNA
NMJ333404 Homo sapiens kinase non-catalytic C-lobe domain (KIND) containing 1 (KND
NM_033405 Homo sapiens peroxisomal proliferator-activated receptor A interacting comp
NM 333407 Homo sapiens dedicator of cytokinesis 7 (D0CK7), mRNA
NM 333425 Homo sapiens DIX domain containing 1 (DIXDC1), mRNA
NM_033426 Homo sapiens KIAA1737 (KIAA1737), mRNA
NM 333429 Homo sapiens calmodulin-like 4 (CALML4), mRNA
NM_033449 Homo sapiens FCH and double SH3 domains 1 (FCHSD1), mRNA
NMJ333450 Homo sapiens ATP-binding cassette, sub-family C (CFTR/MRP), member 10
NM_033452 Homo sapiens tripartite motif-containing 47 (TRIM47), mRNA
NM_033505 Homo sapiens selenoprotein 1, 1 (SELI), mRNA
NMJ333510 Homo sapiens dispatched homolog 2 (Drosophila) (DISP2), mRNA
NM 333512 Homo sapiens TSPY-like 5 (TSPYL5), mRNA
NM_033513 Homo sapiens chromosome 19 open reading frame 20 (C19orf20), mRNA
NM_033520 Homo sapiens chromosome 19 open reading frame 33 (C19orf33), mRNA
NM_033542 Homo sapiens chromosome 20 open reading frame 35 (C20orf35), mRNA
NMJ333548 Homo sapiens similar to ZINC FINGER PROTEIN 257 (BONE MARROW Zll
NM_033553 Homo sapiens guanylate cyclase activator 2A (guanylin) (GUCA2A), mRNA
NM_033557 Homo sapiens similar to putative transmembrane protein; homolog of yeast (
NM_033631 Homo sapiens leucine zipper protein 1 (LUZP1), mRNA
NMJ333647 Homo sapiens helicase (DNA) B (HELB), mRNA
NM_033666 Homo sapiens integrin, beta 1 (fibronectin receptor, beta polypeptide, antigei
NM_033667 Homo sapiens integrin, beta 1 (fibronectin receptor, beta polypeptide, antigei
NM_033668 Homo sapiens integrin, beta 1 (fibronectin receptor, beta polypeptide, antigei
NM_033669 Homo sapiens integrin, beta 1 (fibronectin receptor, beta polypeptide, antigei
NM_052843 Homo sapiens obscurin, cytoskeletal calmodulin and titin-interacting RhoGEF
NM_052846 Homo sapiens elastin microfibril interfacer 3 (EMILIN3), mRNA
NMJ352847 Homo sapiens guanine nucleotide binding protein (G protein), gamma 7 (GN
NM 352849 Homo sapiens hypothetical protein MGC20481 (MGC20481), mRNA
NMJ352850 Homo sapiens growth arrest and DNA-damage-inducible, gamma interacting
NM 352857 Homo sapiens hypothetical protein MGC20398 (MGC20398), mRNA
NM_052864 Homo sapiens TRAF2 binding protein (T2BP), mRNA
NM 352867 Homo sapiens voltage gated channel like 1 (VGCNL1), mRNA
NM_052878 Homo sapiens chromosome 19 open reading frame 36 (C19orf36), mRNA
NM_052892 Homo sapiens polycystic kidney disease 1-like 2 (PKD1 L2), transcript varianl
NM 352896 Homo sapiens CUB and Sushi multiple domains 2 (CSMD2), mRNA
NM 352897 Homo sapiens methyl-CpG binding domain protein 6 (MBD6), mRNA
NM_052899 Homo sapiens G protein-regulated inducer of neurite outgrowth 1 (KIAA1893
NM_052900 Homo sapiens CUB and Sushi multiple domains 3 (CSMD3), transcript variai
NM_052901 Homo sapiens solute carrier family 25 (mitochondrial carrier; phosphate carri
NM_052902 Homo sapiens serine/threonine kinase 11 interacting protein (STK111P), mRI
NM_052903 Homo sapiens tubulin, gamma complex associated protein 5 (TUBGCP5), m
N MJ352904 Homo sapiens KI AA1900 (K1AA1900), mRNA
NM 352905 Homo sapiens formin-like 2 (FMNL2), mRNA
NM_052909 Homo sapiens KIAA1909 protein (KIAA1909), mRNA
NM 352910 Homo sapiens SLIT and NTRK-like family, member 1 (SLITRK1 ), mRNA
NM_052911 Homo sapiens establishment factor-like protein (EF01 ), mRNA
NM 352913 Homo sapiens KIAA1913 (KIAA1913), mRNA
NM 352917 Homo sapiens UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylga
NM_052923 Homo sapiens zinc finger protein 452 (ZNF452), mRNA
NM 352924 Homo sapiens rhophilin, Rho GTPase binding protein 1 (RHPN1), mRNA
NM_052925 Homo sapiens leukocyte receptor cluster (LRC) member 8 (LENG8), mRNA
NM_052926 Homo sapiens paraneoplastic antigen like 5 (PNMA5), mRNA
NM 352928 Homo sapiens SET and MYND domain containing 4 (SMYD4), mRNA
NM 352937 Homo sapiens similar to hypothetical protein FLJ10883 (LOC115294), mRN/
NMJ352964 Homo sapiens mast cell immunoreceptor signal transducer (MIST), mRNA
NM 352965 Homo sapiens chromosome 1 open reading frame 19 (C1orf19), mRNA
NM 353041 Homo sapiens COMM domain containing 7 (COMMD7), mRNA
NM 353044 Homo sapiens serine protease HTRA3 (HTRA3), mRNA
NM 353051 Homo sapiens LYST-interacting protein LIP8 (LIP8), mRNA
NM 353052 Homo sapiens SVAP1 protein (IMAGE3451454), mRNA
NM 353277 Homo sapiens chloride intracellular channel 6 (CLIC6), mRNA
NMJ353279 Homo sapiens chromosome 8 open reading frame 13 (C8orf13), mRNA
NM 353282 Homo sapiens SH2 domain-containing molecule EAT2 (EAT2), mRNA
NM_054104 Homo sapiens olfactory receptor, family 6, subfamily C, member 3 (0R6C3),
NM_054105 Homo sapiens olfactory receptor, family 6, subfamily C, member 2 (OR6C2),
NM_057163 Homo sapiens gonadotropin-releasing hormone (type 2) receptor 2 (GNRHR
NM 358163 Homo sapiens hypothetical protein DT1 P1A10 (DT1 P1A10), mRNA
NMJ358243 Homo sapiens bromodomain containing 4 (BRD4), transcript variant long, mF
NM_080574 Homo sapiens chromosome 20 open reading frame 70 (C20orf70), mRNA
NM_080614 Homo sapiens WAP four-disulfide core domain 3 (WFDC3), transcript varian
NM 380618 Homo sapiens CCCTC-binding factor (zinc finger protein)-like (CTCFL), mRN
NM_080622 Homo sapiens chromosome 20 open reading frame 135 (C20orf135), mRNA
NM_080725 Homo sapiens chromosome 20 open reading frame 139 (C20orf139), mRNA
NM J80747 Homo sapiens keratin protein K6irs (K6IRS2), mRNA
NMJ380751 Homo sapiens transmembrane channel-like 2 (TMC2), mRNA
NM 380753 Homo sapiens WAP four-disulfide core domain 10A (WFDC10A), mRNA
NM_080757 Homo sapiens chromosome 20 open reading frame 127 (C20orf127), mRNA
NM 380764 Homo sapiens suppressor of hairy wing homolog 2 (Drosophila) (SUHW2), n
NM 380827 Homo sapiens WAP four-disulfide core domain 6 (WFDC6), mRNA
NM_080833 Homo sapiens chromosome 20 open reading frame 151 (C20orf151), mRNA
NM_080836 Homo sapiens serine/threonine kinase 35 (STK35), mRNA
NM 380865 Homo sapiens G protein-coupled receptor 62 (GPR62), mRNA
NM_080866 Homo sapiens solute carrier family 22 (organic anion/cation transporter), mei
NM 380868 Homo sapiens ankyrin repeat and SOCS box-containing 17 (ASB17), mRNA
NM 380869 Homo sapiens WAP four-disulfide core domain 12 (WFDC12), mRNA
NMJ380875 Homo sapiens skeletrophin (LOC142678), mRNA
NM_080877 Homo sapiens solute carrier family 34 (sodium phosphate), member 3 (SLCS
NMJ380911 Homo sapiens uracil-DNA glycosylase (UNG), nuclear gene encoding mitoch
NM 380928 Homo sapiens ankyrin repeat and SOCS box-containing 15 (ASB15), mRNA
NM_101395 Homo sapiens dual-specificity tyrosine-(Y)-phosphorylation regulated kinase
NM_130391 Homo sapiens protein tyrosine phosphatase, receptor type, D (PTPRD), tram
NM_130392 Homo sapiens protein tyrosine phosphatase, receptor type, D (PTPRD), tran:
NM_130393 Homo sapiens protein tyrosine phosphatase, receptor type, D (PTPRD), tram
NM_130435 Homo sapiens protein tyrosine phosphatase, receptor type, E (PTPRE), trani
NM_130436 Homo sapiens dual-specificity tyrosine-(Y)-phosphorylation regulated kinase
NM_130437 Homo sapiens dual-specificity tyrosine-(Y)-phosphorylation regulated kinase
NM_130438 Homo sapiens dual-specificity tyrosine-(Y)-phosphorylation regulated kinase
NM_130440 Homo sapiens protein tyrosine phosphatase, receptor type, F (PTPRF), trans
NM_130442 Homo sapiens engulfment and cell motility 1 (ced-12 homolog, C. elegans) (I
NM_130444 Homo sapiens collagen, type XVIII, alpha 1 (COL18A1), transcript variant 3, i
NM_130445 Homo sapiens collagen, type XVIII, alpha 1 (COL18A1), transcript variant 2, i
NM_130465 Homo sapiens F-box protein 23 (FBX023), mRNA
NM_130466 Homo sapiens ubiquitin protein ligase E3B (UBE3B), transcript variant 1 , mR
NM_130470 Homo sapiens MAP-kinase activating death domain (MADD), transcript varia
NM_130471 Homo sapiens MAP-kinase activating death domain (MADD), transcript varia
NM_130472 Homo sapiens MAP-kinase activating death domain (MADD), transcript varia
NM_130473 Homo sapiens MAP-kinase activating death domain (MADD), transcript varia
NM__130474 Homo sapiens MAP-kinase activating death domain (MADD), transcript varia
NM_130475 Homo sapiens MAP-kinase activating death domain (MADD), transcript varia
NM_130476 Homo sapiens MAP-kinase activating death domain (MADD), transcript varia
NM_130760 Homo sapiens mucosal vascular addressin cell adhesion molecule 1 (MADC
NM__130761 Homo sapiens mucosal vascular addressin cell adhesion molecule 1 (MADC
NM_130762 Homo sapiens mucosal vascular addressin cell adhesion molecule 1 (MADC
NM_130766 Homo sapiens skeletal muscle and kidney enriched inositol phosphatase (SH
NM_130771 Homo sapiens osteoclast-associated receptor (OSCAR), transcript variant 3,
NM_130775 Homo sapiens XAGE-5 protein (XAGE-5), mRNA
NM_130776 Homo sapiens G antigen, family D, 4 (GAGED4), transcript variant 2, mRNA
NM_130777 Homo sapiens G antigen, family D, 3 (GAGED3), mRNA
NM_130788 Homo sapiens WW domain containing oxidoreductase (WWOX), transcript v
NM_130790 Homo sapiens WW domain containing oxidoreductase (WWOX), transcript v
NM_130791 Homo sapiens WW domain containing oxidoreductase (WWOX), transcript v
NM_130792 Homo sapiens WW domain containing oxidoreductase (WWOX), transcript v
NM_ 30793 Homo sapiens nucleolar protein family 6 (RNA-associated) (NOL6), transcrip
NM_130794 Homo sapiens cystatin 11 (CST11), transcript variant 1 , mRNA
NM_130797 Homo sapiens dipeptidylpeptidase 6 (DPP6), transcript variant 1 , mRNA
NM_130798 Homo sapiens synaptosomal-associated protein, 23kDa (SNAP23), transcrip
NM_130799 Homo sapiens multiple endocrine neoplasia I (MEN1), transcript variant 2, m
NM_130800 Homo sapiens multiple endocrine neoplasia I (MEN1), transcript variant e1 B,
NM_130801 Homo sapiens multiple endocrine neoplasia I (MEN1), transcript variant e1C,
NM_130802 Homo sapiens multiple endocrine neoplasia I (MEN1), transcript variant e1 D,
NM_130803 Homo sapiens multiple endocrine neoplasia I (MEN1), transcript variant e1E,
NM_130804 Homo sapiens multiple endocrine neoplasia I (MEN1), transcript variant e1F'
NM_130806 Homo sapiens leucine-rich repeat-containing G protein-coupled receptor 8 (L
NM_130807 Homo sapiens MOB1 , Mps One Binder kinase activator-like 2A (yeast) (MOB
NM_130808 Homo sapiens copine IV (CPNE4), mRNA
NM_130809 Homo sapiens hypothetical protein MGC12103 (LOC133619), mRNA
NM_130810 Homo sapiens dyslexia susceptibility 1 candidate 1 (DYX1C1), mRNA
NM_130811 Homo sapiens synaptosomal-associated protein, 25kDa (SNAP25), transcrip
NM_130830 Homo sapiens leucine rich repeat containing 15 (LRRC15), mRNA
NM_130831 Homo sapiens optic atrophy 1 (autosomal dominant) (OPA1), nuclear gene e
NM_130832 Homo sapiens optic atrophy 1 (autosomal dominant) (OPA1), nuclear gene e
NM_130833 Homo sapiens optic atrophy 1 (autosomal dominant) (OPA1), nuclear gene e
NM_130834 Homo sapiens optic atrophy 1 (autosomal dominant) (OPA1), nuclear gene e
NM_130835 Homo sapiens optic atrophy 1 (autosomal dominant) (OPA1), nuclear gene e
NM_130836 Homo sapiens optic atrophy 1 (autosomal dominant) (OPA1), nuclear gene e
NM_130837 Homo sapiens optic atrophy 1 (autosomal dominant) (OPA1), nuclear gene e
NM_130838 Homo sapiens ubiquitin protein ligase E3A (human papilloma virus E6-assoc
NM_130839 Homo sapiens ubiquitin protein ligase E3A (human papilloma virus E6-assoc
NM_130840 Homo sapiens ATPase, H+ transporting, lysosomal V0 subunit a isoform 4 (/
NM_130841 Homo sapiens ATPase, H+ transporting, lysosomal V0 subunit a isoform 4 (/
NM_130842 Homo sapiens protein tyrosine phosphatase, receptor type, N polypeptide 2 |
NM_130843 Homo sapiens protein tyrosine phosphatase, receptor type, N polypeptide 2 |
NM_130844 Homo sapiens WW domain containing oxidoreductase (WWOX), transcript v
NM_130845 Homo sapiens syntrophin, beta 2 (dystrophin-associated protein A1, 59kDa,
NM_130846 Homo sapiens protein tyrosine phosphatase, receptor type, R (PTPRR), tram
NM_130847 Homo sapiens angiomotin like 1 (AMOTL1), mRNA
NM_130848 Homo sapiens dendritic cell nuclear protein 1 (DCNP1), mRNA
NM_130849 Homo sapiens solute carrier family 39 (zinc transporter), member 4 (SLC39A
NM_130850 Homo sapiens bone morphogenetic protein 4 (BMP4), transcript variant 2, m
NM_130851 Homo sapiens bone morphogenetic protein 4 (BMP4), transcript variant 3, m
NM_130852 Homo sapiens palate, lung and nasal epithelium carcinoma associated (PLU
NM_130853 Homo sapiens protein tyrosine phosphatase, receptor type, S (PTPRS), trans
NM_130854 Homo sapiens protein tyrosine phosphatase, receptor type, S (PTPRS), tranj
NM_130855 Homo sapiens protein tyrosine phosphatase, receptor type, S (PTPRS), tran;
NM_130896 Homo sapiens WAP four-disulfide core domain 8 (WFDC8), transcript varian
NM_130897 Homo sapiens dynein, cytoplasmic, light polypeptide 2B (DNCL2B), mRNA
NM_130898 Homo sapiens cAMP responsive element binding protein 3-like 4 (CREB3L4J
NM_130899 Homo sapiens hypothetical protein MGC26988 (MGC26988), mRNA
NM_130900 Homo sapiens retinoic acid early transcript 1 L (RAET1 L), mRNA
NM_130901 Homo sapiens chromosome 15 open reading frame 16 (C15orf16), mRNA
NM_130902 Homo sapiens cytochrome c oxidase subunit Vllb2 (COX7B2), mRNA
NM_130906 Homo sapiens peptidylprolyl isomerase (cyclophilin)-like 3 (PPIL3), transcript
NM_131915 Homo sapiens similar to hypothetical protein DKFZp434K191 (H. sapiens) (L
NM_131916 Homo sapiens peptidylprolyl isomerase (cyclophilin)-like 3 (PPIL3), transcrip!
NM_131917 Homo sapiens Fas (TNFRSF6) associated factor 1 (FAF1), transcript variant
NM_133168 Homo sapiens osteoclast-associated receptor (OSCAR), transcript variant 5,
NM_133169 Homo sapiens osteoclast-associated receptor (OSCAR), transcript variant 4,
NM_133170 Homo sapiens protein tyrosine phosphatase, receptor type, T (PTPRT), trans
NM_133171 Homo sapiens engulfment and cell motility 2 (ced-12 homolog, C. elegans) (I
NM_133172 Homo sapiens amyloid beta (A4) precursor protein-binding, family B, membe
NM_133173 Homo sapiens amyloid beta (A4) precursor protein-binding, family B, membe
NM_133174 Homo sapiens amyloid beta (A4) precursor protein-binding, family B, membe
NM_133175 Homo sapiens amyloid beta (A4) precursor protein-binding, family B, membe
NM_133176 Homo sapiens amyloid beta (A4) precursor protein-binding, family B, membe
NM_133177 Homo sapiens protein tyrosine phosphatase, receptor type, U (PTPRU), tran:
NM_133178 Homo sapiens protein tyrosine phosphatase, receptor type, U (PTPRU), tram
NM_133179 Homo sapiens G antigen, family D, 4 (GAGED4), transcript variant 1 , mRNA
NM_133180 Homo sapiens EPS8-like 1 (EPS8L1), transcript variant 1 , mRNA
NM 33181 Homo sapiens EPS8-like 3 (EPS8L3), transcript variant 2, mRNA
NM_133259 Homo sapiens leucine-rich PPR-motif containing (LRPPRC), mRNA
NM_133261 Homo sapiens PDZ domain protein GIPC3 (GIPC3), mRNA
NM_133262 Homo sapiens ATPase, H+ transporting, lysosomal 13kDa, V1 subunit G isol
NM_133263 Homo sapiens peroxisome proliferative activated receptor, gamma, coactivat
NM_133264 Homo sapiens WIRE protein (WIRE), mRNA
NM_133265 Homo sapiens angiomotin (AMOT), mRNA
NM_133266 Homo sapiens SH3 and multiple ankyrin repeat domains 2 (SHANK2), transc
NM_133267 Homo sapiens homeobox protein GSH-2 (GSH-2), mRNA
NM_133268 Homo sapiens oxysterol binding protein-like 1A (OSBPL1A), transcript varian
NM_133269 Homo sapiens Fc fragment of IgA, receptor for (FCAR), transcript variant 2, r
NM_133271 Homo sapiens Fc fragment of IgA, receptor for (FCAR), transcript variant 3, r
NM_133272 Homo sapiens Fc fragment of IgA, receptor for (FCAR), transcript variant 4, r
NM_133273 Homo sapiens Fc fragment of IgA, receptor for (FCAR), transcript variant 5, r
NM_133274 Homo sapiens Fc fragment of IgA, receptor for (FCAR), transcript variant 6, r
NM_133277 Homo sapiens Fc fragment of IgA, receptor for (FCAR), transcript variant 7, r
NM_133278 Homo sapiens Fc fragment of IgA, receptor for (FCAR), transcript variant 8, r
NM_133279 Homo sapiens Fc fragment of IgA, receptor for (FCAR), transcript variant 9, r
NM_133280 Homo sapiens Fc fragment of IgA, receptor for (FCAR), transcript variant 10,
NM_133282 Homo sapiens RAD1 homolog (S. pombe) (RAD1 ), transcript variant 2, mRN
NM_133325 Homo sapiens PHD finger protein 10 (PHF10), transcript variant 2, mRNA
NM_133326 Homo sapiens ATPase, H+ transporting, lysosomal 13kDa, V1 subunit G isol
NM_133327 Homo sapiens sema domain, transmembrane domain (TM), and cytoplasmic
NM_133328 Homo sapiens death effector domain containing 2 (DEDD2), mRNA
NM_133329 Homo sapiens potassium voltage-gated channel, subfamily G, member 3 (K(
NM_133330 Homo sapiens Wolf-Hirschhom syndrome candidate 1 (WHSC1), transcript \
NM_133331 Homo sapiens Wolf-Hirschhorn syndrome candidate 1 (WHSC1), transcript \
NM 133332 Homo sapiens Wolf-Hirschhorn syndrome candidate 1 (WHSC1), transcript v
NM_133333 Homo sapiens Wolf-Hirschhom syndrome candidate 1 (WHSC1), transcript \
NM 133334 Homo sapiens Wolf-Hirschhorn syndrome candidate 1 (WHSC1), transcript \
NM_133335 Homo sapiens Wolf-Hirschhorn syndrome candidate 1 (WHSC1), transcript \
NM 133336 Homo sapiens Wolf-Hirschhorn syndrome candidate 1 (WHSC1), transcript \
NM_133337 Homo sapiens fer-1-like 3, myoferlin (C. elegans) (FER1L3), transcript variar
NM_133338 Homo sapiens RAD1 homolog (S. pombe) (RAD17), transcript variant 1, mF
NM_133339 Homo sapiens RAD17 homolog (S. pombe) (RAD17), transcript variant 2, mF
NM 133340 Homo sapiens RAD17 homolog (S. pombe) (RAD17), transcript variant 3, mF
NM_133341 Homo sapiens RAD17 homolog (S. pombe) (RAD17), transcript variant 4, mF
NM_133342 Homo sapiens RAD1 homolog (S. pombe) (RAD17), transcript variant 5, mF
NM 133343 Homo sapiens RAD17 homolog (S. pombe) (RAD17), transcript variant 6, mF
NM 133344 Homo sapiens RAD17 homolog (S. pombe) (RAD17), transcript variant 7, mF
NM_133367 Homo sapiens chromosome 6 open reading frame 33 (C6orf33), mRNA
NM 133368 Homo sapiens KIAA1972 protein (KIAA1972), mRNA
NM_1.33370 Homo sapiens splicing factor YT521-B (YT521), mRNA
NM_133371 Homo sapiens myozenin 3 (MYOZ3), mRNA
NM 133373 Homo sapiens phospholipase C, delta 3 (PLCD3), mRNA
NM_133375 Homo sapiens hypothetical protein MGC4562 (MGC4562), mRNA
NM 133376 Homo sapiens integrin, beta 1 (fibronectin receptor, beta polypeptide, antigei
NM_133377 Homo sapiens RAD1 homolog (S. pombe) (RAD1), transcript variant 3, mRN
NM 133378 Homo sapiens titin (TTN), transcript variant N2-A, mRNA
NM 133379 Homo sapiens titin (TTN), transcript variant novex-3, mRNA
NM 133430 Homo sapiens G antigen, family D, 2 (GAGED2), transcript variant 3, mRNA
NM 133431 Homo sapiens G antigen, family D, 2 (GAGED2), transcript variant 2, mRNA
NM_133432 Homo sapiens titin (TTN), transcript variant novex-1 , mRNA
NM_133433 Homo sapiens Nipped-B homolog (Drosophila) (NIPBL), transcript variant A,
NM_133436 Homo sapiens asparagine synthetase (ASNS), transcript variant 1 , mRNA
NM 133437 Homo sapiens titin (TTN), transcript variant novex-2, mRNA
NM 33439 Homo sapiens transcriptional adaptor 2 (ADA2 homolog, yeast)-like (TADA2I
NM_133443 Homo sapiens glutamic pyruvate transaminase (alanine aminotransferase) 2
NM_133444 Homo sapiens zinc finger protein 526 (ZNF526), mRNA
NM 133445 Homo sapiens glutamate receptor, ionotropic, N-methyl-D-aspartate 3A (GRI
NM_133446 Homo sapiens centaurin, gamma-like family, member 1 (CTGLF1), mRNA
NMJ 33448 Homo sapiens KIAA1944 protein (KIAA1944), mRNA
NM 133450 Homo sapiens KIAA1977 protein (KIAA1977), mRNA
NMJ 33452 Homo sapiens RAVER1 (RAVER1), mRNA
NMJ 33455 Homo sapiens EMI domain containing 1 (EMID1), mRNA
NM_133456 Homo sapiens apical protein 2 (APXL2), mRNA
NMJ 33457 Homo sapiens EMI domain containing 2 (EMID2), mRNA
NM 133459 Homo sapiens KIAA1983 protein (FLJ30681), mRNA
NMJ 33462 Homo sapiens tetratricopeptide repeat domain 14 (TTC14), mRNA
NMJ 33466 Homo sapiens zinc finger protein 545 (ZNF545), mRNA
NMJ 33467 Homo sapiens Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxj
NMJ 33468 Homo sapiens BMP-binding endothelial regulator precursor protein (BMPER]
NMJ 33473 Homo sapiens zinc finger protein 431 (ZNF431), mRNA
NMJ 33474 Homo sapiens KIAA1982 protein (KIAA1982), mRNA
NM 133476 Homo sapiens zinc finger protein 384 (ZNF384), mRNA
NMJ 33478 Homo sapiens solute carrier family 4, sodium bicarbonate cotransporter, mer
NM 133479 Homo sapiens solute carrier family 4, sodium bicarbonate cotransporter, mer
NMJ 33480 Homo sapiens transcriptional adaptor 3 (NGG1 homolog, yeast)-like (TADA3
NM 133481 Homo sapiens transcriptional adaptor 3 (NGG1 homolog, yeast)-like (TADA3
NM 133482 Homo sapiens RAD50 homolog (S. cerevisiae) (RAD50), transcript variant 2,
NMJ 33483 Homo sapiens RAC/CDC42 exchange factor (GEFT), transcript variant 2, mF
NMJ 33484 Homo sapiens TRAF family member-associated NFKB activator (TANK), trar
NMJ 33486 Homo sapiens muscleblind-like 3 (Drosophila) (MBNL3), mRNA
NM 133487 Homo sapiens RAD51 homolog (RecA homolog, E. coli) (S. cerevisiae) (RAC
NMJ 33489 Homo sapiens solute carrier family 26, member 10 (SLC26A10), mRNA
NMJ 33490 Homo sapiens potassium voltage-gated channel, subfamily G, member 4 (K(
NMJ 33491 Homo sapiens spermidine/spermine N1 -acetyltransferase 2 (SAT2), mRNA
NMJ 33492 Homo sapiens N-acylsphingosine amidohydrolase (alkaline ceramidase) 3 (A
NMJ33493 Homo sapiens CD109 antigen (Gov platelet alloantigens) (CD109), mRNA
NMJ 33494 Homo sapiens NIMA (never in mitosis gene a)-related kinase 7 (NEK7), mRN
NMJ 33496 Homo sapiens solute carrier family 30 (zinc transporter), member 7 (SLC30A
NMJ 33497 Homo sapiens potassium channel, subfamily V, member 2 (KCNV2), mRNA
NMJ 33498 Homo sapiens sperm acrosome associated 4 (SPACA4), mRNA
NMJ33499 Homo sapiens synapsin I (SYN1), transcript variant lb, mRNA
NMJ 33502 Homo sapiens zinc finger protein 274 (ZNF274), transcript variant ZNF274c,
NMJ 33503 Homo sapiens decorin (DCN), transcript variant A2, mRNA
NMJ 33504 Homo sapiens decorin (DCN), transcript variant B, mRNA
NMJ 33505 Homo sapiens decorin (DCN), transcript variant C, mRNA
NMJ 33506 Homo sapiens decorin (DCN), transcript variant D, mRNA
NMJ 33507 Homo sapiens decorin (DCN), transcript variant E, mRNA
NMJ 33509 Homo sapiens RAD51-like 1 (S. cerevisiae) (RAD51L1), transcript variant 3,
NMJ 33510 Homo sapiens RAD51-like 1 (S. cerevisiae) (RAD51L1), transcript variant 2,
NMJ 33625 Homo sapiens synapsin II (SYN2), transcript variant Ila, mRNA
NMJ 33627 Homo sapiens RAD51-like 3 (S. cerevisiae) (RAD51L3), transcript variant 2,
NMJ 33628 Homo sapiens RAD51-like 3 (S. cerevisiae) (RAD51 L3), transcript variant 3,
NMJ 33629 Homo sapiens RAD51-like 3 (S. cerevisiae) (RAD51L3), transcript variant 4,
NMJ 33630 Homo sapiens RAD51-like 3 (S. cerevisiae) (RAD51L3), transcript variant 5,
NMJ 33631 Homo sapiens roundabout, axon guidance receptor, homolog 1 (Drosophila)
NMJ 33632 Homo sapiens synapsin III (SYN3), transcript variant 11 lb, mRNA
NMJ 33633 Homo sapiens synapsin III (SYN3), transcript variant life, mRNA
NMJ 33634 Homo sapiens protein O-fucosyltransferase 2 (POFUT2), transcript variant 2,
NMJ33635 Homo sapiens protein O-fucosyltransferase 2 (POFUT2), transcript variant 3,
NMJ 33636 Homo sapiens DNA helicase HEL308 (HEL308), mRNA
NMJ 33637 Homo sapiens DEAQ box polypeptide 1 (RNA-dependent ATPase) (DQX1 ), i
NMJ 33638 Homo sapiens a disintegrin-like and metalloprotease (reprolysin type) with th
NMJ 33639 Homo sapiens ras homolog gene family, member V (RHOV), mRNA
NMJ 33640 Homo sapiens surfeit 5 (SURF5), transcript variant b, mRNA
NMJ 33642 Homo sapiens like-glycosyltransferase (LARGE), transcript variant 2, mRNA
NMJ 33644 Homo sapiens GTP binding protein 3 (mitochondrial) (GTPBP3), mRNA
N J 33645 Homo sapiens mitochondrial translation optimization 1 homolog (S. cerevisia
NMJ 33646 Homo sapiens sterile alpha motif and leucine zipper containing kinase AZK (,
NMJ 33650 Homo sapiens spectrin repeat containing, nuclear envelope 1 (SYNE1), trans
NMJ34258 Homo sapiens transducin (beta)-like 1 Y-linked (TBL1 Y), transcript variant 2, i
NMJ 34259 Homo sapiens transducin (beta)-like 1 Y-linked (TBL1 Y), transcript variant 3, i
NMJ 34260 Homo sapiens RAR-related orphan receptor A (RORA), transcript variant 2, r
NMJ 34261 Homo sapiens RAR-related orphan receptor A (RORA), transcript variant 1 , r
NMJ34262 Homo sapiens RAR-related orphan receptor A (RORA), transcript variant 4, r
N J 34263 Homo sapiens solute carrier family 26, member 6 (SLC26A6), transcript varis
NMJ 34264 Homo sapiens WD repeat and SOCS box-containing 1 (WSB1), transcript va
NMJ 34265 Homo sapiens WD repeat and SOCS box-containing 1 (WSB1), transcript va
NMJ 34266 Homo sapiens solute carrier family 26, member 7 (SLC26A7), transcript varic
NMJ 34268 Homo sapiens cytoglobin (CYGB), mRNA
NMJ 34269 Homo sapiens smoothelin (SMTN), transcript variant 2, mRNA
NMJ 34270 Homo sapiens smoothelin (SMTN), transcript variant 1 , mRNA
NMJ 34323 Homo sapiens TAR (HIV) RNA binding protein 2 (TARBP2), transcript varian
NMJ 34324 Homo sapiens TAR (HIV) RNA binding protein 2 (TARBP2), transcript varian
NMJ 34325 Homo sapiens solute carrier family 26, member 9 (SLC26A9), transcript van's
NMJ34421 Homo sapiens hippocalcin-like 1 (HPCAL1), transcript variant 2, mRNA
NMJ 34422 Homo sapiens RAD52 homolog (S. cerevisiae) (RAD52), transcript variant de
NMJ 34423 Homo sapiens RAD52 homolog (S. cerevisiae) (RAD52), transcript variant gj
NMJ 34424 Homo sapiens RAD52 homolog (S. cerevisiae) (RAD52), transcript variant be-
NMJ 34425 Homo sapiens solute carrier family 26 (sulfate transporter), member 1 (SLC2
NMJ 34426 Homo sapiens solute carrier family 26, member 6 (SLC26A6), transcript vari-
NM J 34427 Homo sapiens regulator of G-protein signalling 3 (RGS3), transcript variant 4
NMJ34428 Homo sapiens regulatory factor X, 3 (influences HLA class II expression) (RF
NMJ 34431 Homo sapiens solute carrier organic anion transporter family, member 1 A2 (!
NMJ 34433 Homo sapiens regulatory factor X, 2 (influences HLA class II expression) (RF
NMJ 34434 Homo sapiens RAD54 homolog B (S. cerevisiae) (RAD54B), transcript variar
NMJ 34440 Homo sapiens regulatory factor X-associated ankyrin-containing protein (RF>
NMJ 34441 Homo sapiens relaxin 2 (H2) (RLN2), transcript variant 1 , mRNA
NMJ 34442 Homo sapiens cAMP responsive element binding protein 1 (CREB1 ), transcr
NMJ 34444 Homo sapiens NACHT, leucine rich repeat and PYD containing 4 (NALP4), n
NMJ 34445 Homo sapiens CD99 antigen-like 2 (CD99L2), mRNA
NMJ 34446 Homo sapiens CD99 antigen-like 2 (CD99L2), mRNA
NMJ 34447 Homo sapiens chromosome 19 open reading frame 2 (C19orf2), transcript vs
NMJ 34470 Homo sapiens interleukin 1 receptor accessory protein (IL1RAP), transcript v
NMJ 38270 Homo sapiens alpha thalassemia/mental retardation syndrome X-linked (RAI
NMJ 38271 Homo sapiens alpha thalassemia/mental retardation syndrome X-linked (RAI
NMJ 38272 Homo sapiens chromosome 6 open reading frame 25 (C6orf25), transcript VE
NMJ 38273 Homo sapiens chromosome 6 open reading frame 25 (C6orf25), transcript v£
NMJ 38274 Homo sapiens chromosome 6 open reading frame 25 (C6orf25), transcript VE
NMJ 38275 Homo sapiens chromosome 6 open reading frame 25 (C6orf25), transcript VJ
NMJ 38276 Homo sapiens chromosome 6 open reading frame 25 (C6orf25), transcript VJ
NMJ 38277 Homo sapiens chromosome 6 open reading frame 25 (C6orf25), transcript vs
NMJ 38278 Homo sapiens BCL2/adenovirus E1 B 19kD interacting protein like (BNIPL), r
NMJ 38279 Homo sapiens BCL2/adenovirus E1 B 19kD interacting protein like (BNIPL), r
NMJ 38280 Homo sapiens citrate lyase beta like (CLYBL), transcript variant 1 , mRNA
NMJ 38281 Homo sapiens distal-less homeobox 4 (DLX4), transcript variant 1 , mRNA
NMJ 38282 Homo sapiens ATPase, H+ transporting, lysosomal 13kDa, V1 subunit G isol
NMJ 38283 Homo sapiens cystatin-like 1 (CSTL1), mRNA
NMJ 38284 Homo sapiens interleukin 17D (IL17D), mRNA
NMJ 38285 Homo sapiens nucleoporin 35kDa (NUP35), mRNA
NMJ38286 Homo sapiens hypothetical protein FLJ31526 (LOC148213), mRNA
NMJ 38287 Homo sapiens rhysin 2 (BBAP), mRNA
NMJ 38288 Homo sapiens chromosome 14 open reading frame 147 (C14orf147), mRNA
NMJ 38289 Homo sapiens actin-related protein T1 (ACTRT1), mRNA
NMJ38290 Homo sapiens Rap2-binding protein 9 (RPIB9), mRNA
NMJ 38292 Homo sapiens ataxia telangiectasia mutated (includes complementation groi
NMJ 38293 Homo sapiens ataxia telangiectasia mutated (includes complementation groi
NMJ 38294 Homo sapiens expressed in prostate and testis (PATE), mRNA
NMJ38295 Homo sapiens polycystic kidney disease 1 like 1 (PKD1 L1 ), mRNA
NMJ 38296 Homo sapiens pre T-cell antigen receptor alpha (PTCRA), mRNA
NMJ 38297 Homo sapiens mucin 4, tracheobronchial (MUC4), transcript variant 5, mRN/
NMJ 38298 Homo sapiens mucin 4, tracheobronchial (MUC4), transcript variant 2, mRN/
NMJ 38299 Homo sapiens mucin 4, tracheobronchial (MUC4), transcript variant 3, mRN/
NMJ 38300 Homo sapiens pygopus 2 (PYG02), mRNA
NMJ 38316 Homo sapiens pantothenate kinase 1 (PANK1), transcript variant gamma, ml
NMJ 38317 Homo sapiens potassium channel, subfamily K, member 10 (KCNK10), trans
NMJ 38318 Homo sapiens potassium channel, subfamily K, member 10 (KCNK10), trans
NMJ 38319 Homo sapiens proprotein convertase subtilisin/kexin type 6 (PCSK6), transcr
NMJ 38320 Homo sapiens proprotein convertase subtilisin/kexin type 6 (PCSK6), transcr
NMJ 38321 Homo sapiens proprotein convertase subtilisin/kexin type 6 (PCSK6), transcr
NMJ 38322 Homo sapiens proprotein convertase subtilisin/kexin type 6 (PCSK6), transcr
NMJ 38323 Homo sapiens proprotein convertase subtilisin/kexin type 6 (PCSK6), transcr
NMJ 38324 Homo sapiens proprotein convertase subtilisin/kexin type 6 (PCSK6), transcr
NMJ 38325 Homo sapiens proprotein convertase subtilisin/kexin type 6 (PCSK6), transcr
NMJ 38326 Homo sapiens aminocarboxymuconate semialdehyde decarboxylase (ACMS
NMJ 38327 Homo sapiens trace amine receptor 1 (TRAR1), mRNA
NM 138328 Homo sapiens rhomboid, veinlet-like 4 (Drosophila) (RHBDL4), mRNA
NM 138329 Homo sapiens NACHT, leucine rich repeat and PYD containing 6 (NALP6), n
NM 138330 Homo sapiens TRAFδ-inhibitory zinc finger protein (TIZ), mRNA
NM 138331 Homo sapiens ribonuclease, RNase A family, 8 (RNASE8), mRNA
NM 138333 Homo sapiens chromosome 9 open reading frame 42 (C9orf42), mRNA
NM 138334 Homo sapiens hypothetical transmembrane protein SBBI54 (SBBI54), mRN/1
NMJ 38335 Homo sapiens glucosamine-6-phosphate deaminase 2 (GNPDA2), mRNA
NM 138336 Homo sapiens helicase/primase complex protein (LOC150678), mRNA
NMJ 38337 Homo sapiens myeloid inhibitory C-type lectin-like receptor (MICL), transcript
NMJ 38338 Homo sapiens polymerase (RNA) 111 (DNA directed) polypeptide H (22.9kD) (
NMJ 38340 Homo sapiens abhydrolase domain containing 3 (ABHD3), mRNA
NM 138341 Homo sapiens hypothetical protein BC000282 (LOC89894), mRNA
NM 138342 Homo sapiens hypothetical protein BC008326 (LOC89944), mRNA
NMJ 38343 Homo sapiens kinesin-like 8 (KNSL8), transcript variant 4, mRNA
NM 138344 Homo sapiens chromosome 14 open reading frame 152 (C14orf152), mRNA
NMJ 38346 Homo sapiens hypothetical protein MGC33867 (MGC33867), mRNA
NMJ 38347 Homo sapiens zinc finger protein 551 (ZNF551 ), mRNA
NM 138348 Homo sapiens hypothetical protein BC007706 (LOC90268), mRNA
NMJ 38349 Homo sapiens hypothetical protein BC004507 (LOC90313), mRNA
NMJ 38350 Homo sapiens hypothetical protein MGC33488 (MGC33488), mRNA
NM 138352 Homo sapiens atherin (LOC90378), mRNA
NMJ 38355 Homo sapiens secemin 2 (Ses2), mRNA
NM 138356 Homo sapiens hypothetical protein BC007586 (LOC90525), mRNA
NMJ 38357 Homo sapiens chromosome 10 open reading frame 42 (C10orf42), mRNA
NM 138358 Homo sapiens hypothetical protein BC011833 (LOC90580), mRNA
NMJ 38360 Homo sapiens hypothetical protein BC008134 (LOC90668), mRNA
NM 138361 Homo sapiens leucine rich repeat and sterile alpha motif containing 1 (LRSA
NM 138362 Homo sapiens hypothetical protein BC000919 (LOC90736), mRNA
NMJ 38363 Homo sapiens hypothetical protein BC009518 (LOC90799), mRNA
NMJ 38364 Homo sapiens hypothetical protein BC004337 (LOC90826), mRNA
NMJ 38368 Homo sapiens hypothetical protein BC004895 (LOC91056), mRNA
NMJ 38369 Homo sapiens family with sequence similarity 44, member B (FAM44B), mRt
NMJ 38371 Homo sapiens hypothetical protein MGC16044 (MGC16044), mRNA
NM 138372 Homo sapiens hypothetical protein BC001610 (LOC91661), mRNA
NMJ 38373 Homo sapiens myeloid-associated differentiation marker (MYADM), mRNA
NMJ 38375 Homo sapiens Cdk5 and Abl enzyme substrate 1 (CABLES1), mRNA
NM 138376 Homo sapiens tetratricopeptide repeat domain 5 (TTC5), mRNA
NM 138379 Homo sapiens T-cell immunoglobulin and mucin domain containing 4 (TIMD^
NM 138381 Homo sapiens hypothetical protein BC008322 (MGC15763), mRNA
NM 138383 Homo sapiens hypothetical protein BC002770 (LOC92154), mRNA
NMJ 38384 Homo sapiens shadow of prion protein (Sprn), mRNA
NM 138385 Homo sapiens hypothetical protein BC009331 (LOC92305), mRNA
NMJ 38386 Homo sapiens hypothetical protein BC008207 (LOC92345), mRNA
NM 138387 Homo sapiens glucose 6 phosphatase, catalytic, 3 (G6PC3), mRNA
NMJ 38389 Homo sapiens hypothetical protein BC001096 (LOC92689), mRNA
NM 138390 Homo sapiens hypothetical protein BC008604 (LOC92691), mRNA
NMJ 38391 Homo sapiens chromosome 1 open reading frame 37 (C1orf37), mRNA
NM 138392 Homo sapiens hypothetical protein BC007653 (LOC92799), mRNA
NMJ 38393 Homo sapiens chromosome 19 open reading frame 32 (C19orf32), mRNA
NM 138394 Homo sapiens hypothetical protein BC008217 (LOC92906), mRNA
NMJ 38395 Homo sapiens mitochondrial methionyl-tRNA synthetase (MetRS), mRNA
NMJ 38396 Homo sapiens hypothetical protein BC009489 (LOC92979), mRNA
NMJ 38397 Homo sapiens hypothetical protein BC012317 (LOC93082), mRNA
NMJ 38399 Homo sapiens hypothetical protein BC007772 (LOC93109), mRNA
NM 138401 Homo sapiens hypothetical protein BC011840 (LOC93343), mRNA
NMJ 38402 Homo sapiens hypothetical protein BC004921 (LOC93349), mRNA
NMJ 38403 Homo sapiens myosin light chain 2, precursor lymphocyte-specific (MYLC2P
NMJ 38408 Homo sapiens chromosome 6 open reading frame 51 (C6orf51), mRNA
NMJ 38409 Homo sapiens chromosome 6 open reading frame 117 (C6orf117), mRNA
NMJ 38410 Homo sapiens chemokine-like factor super family 7 (CKLFSF7), transcript va
NMJ 38412 Homo sapiens retinol dehydrogenase 13 (all-trans and 9-cis) (RDH13), mRN
NMJ 38413 Homo sapiens chromosome 10 open reading frame 65 (C10orf65), mRNA
NMJ 38414 Homo sapiens hypothetical protein BC011981 (LOC112869), mRNA
NMJ 38415 Homo sapiens hypothetical protein BC012187 (LOC112885), mRNA
NMJ 38416 Homo sapiens hypothetical protein BC011001 (LOC112937), mRNA
NMJ 38417 Homo sapiens hypothetical protein BC012173 (MGC20419), mRNA
NMJ 38418 Homo sapiens hypothetical protein MGC15416 (MGC15416), mRNA
NMJ 38419 Homo sapiens DUF729 domain containing 1 (DUFD1), mRNA
NMJ 38421 Homo sapiens hypothetical protein BC012010 (LOC113174), mRNA
NMJ 38422 Homo sapiens hypothetical protein BC011824 (L0C113179), mRNA
NMJ 38423 Homo sapiens H63 breast cancer expressed gene (H63), transcript variant 1
NMJ 38424 Homo sapiens kinesin family member 12 (KIF12), mRNA
NMJ 38425 Homo sapiens likely ortholog of mouse gene rich cluster, C10 gene (GRCCt
NMJ 38428 Homo sapiens hypothetical protein BC011880 (LOC113444), mRNA
NMJ 38429 Homo sapiens claudin 15 (CLDN15), mRNA
NMJ 38430 Homo sapiens ADP-ribosylhydrolase like 1 (ADPRHL1 ), transcript variant 1 , i
NMJ 38431 Homo sapiens hypothetical protein BC011982 (LOC113655), mRNA
NMJ 38432 Homo sapiens serine dehydratase-like (SDSL), mRNA
NMJ 38433 Homo sapiens hypothetical protein BC009980 (MGC16635), mRNA
NMJ 38434 Homo sapiens chromosome 7 open reading frame 29 (C7orf29), mRNA
NMJ 38435 Homo sapiens hypothetical protein BC011204 (LOC113828), mRNA
NMJ38436 Homo sapiens hypothetical protein BC013035 (LOC114926), mRNA
NMJ 38437 Homo sapiens GASP2 protein (GASP2), mRNA
NMJ38439 Homo sapiens hypothetical protein BC014089 (LOC114984), mRNA
NMJ 38440 Homo sapiens hypothetical protein BC013767 (LOC114990), mRNA
NMJ 38441 Homo sapiens chromosome 6 open reading frame 150 (C6orf150), mRNA
NMJ 38442 Homo sapiens hypothetical protein BC013949 (LOC115098), mRNA
NMJ 38443 Homo sapiens coiled-coil domain containing 5 (spindle associated) (CCDC5)
NMJ38444 Homo sapiens potassium channel tetramerisation domain containing 12 (KC
NMJ38445 Homo sapiens G protein-coupled receptor 146 (GPR146), mRNA
NMJ 38446 Homo sapiens chromosome 7 open reading frame 30 (C7orf30), mRNA
NMJ 38447 Homo sapiens hypothetical protein BC014000 (LOC115509), mRNA
NMJ 38448 Homo sapiens acylphosphatase 2, muscle type (ACYP2), mRNA
NMJ 38450 Homo sapiens ADP-ribosylation factor-like 11 (ARL11 ), mRNA
NMJ38451 Homo sapiens hypothetical protein BC013151 (LOC115811), mRNA
NMJ 38452 Homo sapiens dehydrogenase/reductase (SDR family) member 1 (DHRS1),
NMJ 38453 Homo sapiens RAB3C, member RAS oncogene family (RAB3C), mRNA
NMJ 38454 Homo sapiens thioredoxin-like 6 (TXNL6), mRNA
NMJ 38455 Homo sapiens collagen triple helix repeat containing 1 (CTHRC1), mRNA
NMJ38456 Homo sapiens hypothetical protein BC012330 (MGC20410), mRNA
NMJ 38457 Homo sapiens forkhead box P4 (FOXP4), mRNA
NMJ 38458 Homo sapiens hypothetical protein BC014022 (LOC116143), mRNA
NMJ 38459 Homo sapiens chromosome 6 open reading frame 68 (C6orf68), mRNA
NMJ 38460 Homo sapiens chemokine-like factor super family 5 (CKLFSF5), transcript va
NMJ 38461 Homo sapiens hypothetical protein BC013113 (LOC116211), mRNA
NMJ 38462 Homo sapiens zinc finger, MYND domain containing 19 (ZMYND19), mRNA
NMJ 38463 Homo sapiens hypothetical protein BC014072 (LOC116238), mRNA
NMJ 38465 Homo sapiens GLI-Kruppel family member GLI4 (GLI4), mRNA
NMJ38467 Homo sapiens hypothetical protein BC009514 (LOC127253), mRNA
NMJ 38468 Homo sapiens amyotrophic lateral sclerosis 2 fluvenile) chromosome region,
NMJ 38471 Homo sapiens hypothetical protein BC007540 (LOC144097), mRNA
NMJ 38473 Homo sapiens Sp1 transcription factor (SP1), mRNA
NMJ 38476 Homo sapiens hypothetical protein MGC5987 (MGC5987), mRNA
NMJ 38477 Homo sapiens congenital dyserythropoietic anemia, type I (CDAN1), mRNA
NMJ38479 Homo sapiens hypothetical protein BC007899 (LOC148898), mRNA
NMJ 38482 Homo sapiens hypothetical protein BC009264 (LOC151534), mRNA
NMJ 38484 Homo sapiens shugoshin-like 1 (S. pombe) (SGOL1), mRNA
NMJ 38487 Homo sapiens hypothetical protein BC007882 (LOC152217), mRNA
NMJ 38492 Homo sapiens hypothetical protein MGC21644 (MGC21644), transcript varia
NMJ 38494 Homo sapiens vav-1 interacting Kruppel-like protein (VIK), transcript variant ■
NMJ 38497 Homo sapiens hypothetical protein BC008050 (LOC158435), mRNA
NMJ 38499 Homo sapiens PWWP domain containing 2 (PWWP2), mRNA
NMJ 38501 Homo sapiens glycoprotein, synaptic 2 (GPSN2), mRNA
NMJ 38551 Homo sapiens thymic stromal lymphopoietin (TSLP), transcript variant 2, mR
NMJ 38553 Homo sapiens B-cell CLL/lymphoma 11A (zinc finger protein) (BCL11A), tran
NMJ 38554 Homo sapiens toll-like receptor 4 (TLR4), transcript variant 1 , mRNA
NMJ 38555 Homo sapiens kinesin family member 23 (KIF23), transcript variant 1 , mRNA
NMJ 38556 Homo sapiens toll-like receptor 4 (TLR4), transcript variant 2, mRNA
NMJ 38557 Homo sapiens toll-like receptor 4 (TLR4), transcript variant 4, mRNA
NMJ 38558 Homo sapiens protein phosphatase 1 , regulatory (inhibitor) subunit 8 (PPP1 F
NMJ38559 Homo sapiens B-cell CLL/lymphoma 11 A (zinc finger protein) (BCL11 A), tran
NMJ 38563 Homo sapiens kallikrein 15 (KLK15), transcript variant 2, mRNA
NMJ 38564 Homo sapiens kallikrein 15 (KLK15), transcript variant 3, mRNA
NMJ 38565 Homo sapiens cortactin (CTTN), transcript variant 2, mRNA
NMJ 38566 Homo sapiens glutaminase 2 (liver, mitochondrial) (GLS2), nuclear gene enc
NMJ 38567 Homo sapiens synaptotagmin VIII (SYT8), mRNA
NMJ 38568 Homo sapiens protein 7 transactivated by hepatitis B virus X antigen (HBxAg
NMJ38569 Homo sapiens chromosome 6 open reading frame 142 (C6orf142), mRNA
NMJ 38570 Homo sapiens hypothetical protein MGC15523 (MGC15523), mRNA
NMJ 38571 Homo sapiens histidine triad nucleotide binding protein 3 (HINT3), mRNA
NMJ 38572 Homo sapiens taube nuss homolog (mouse) (TBN), mRNA
NMJ 38573 Homo sapiens neuregulin 4 (LOC145957), mRNA
NMJ 38574 Homo sapiens PWWP domain containing 1 (PWWP1), mRNA
NMJ 38575 Homo sapiens hypothetical protein MGC5352 (MGC5352), mRNA
NMJ 38576 Homo sapiens B-cell CLL/lymphoma 11 B (zinc finger protein) (BCL11 B), tran
NMJ 38578 Homo sapiens BCL2-like 1 (BCL2L1), nuclear gene encoding mitochondrial
NMJ 38608 Homo sapiens metallophosphoesterase 1 (MPPE1), mRNA
NMJ 38609 Homo sapiens H2A histone family, member Y (H2AFY), transcript variant 1 , r
NMJ38610 Homo sapiens H2A histone family, member Y (H2AFY), transcript variant 3, r
NMJ 38612 Homo sapiens hyaluronan synthase 3 (HAS3), transcript variant 2, mRNA
NMJ 38614 Homo sapiens DEAH (Asp-Glu-Ala-His) box polypeptide 30 (DHX30), transcr
NMJ 38615 Homo sapiens DEAH (Asp-Glu-Ala-His) box polypeptide 30 (DHX30), transcr
NMJ38616 Homo sapiens Rhesus blood group, CcEe antigens (RHCE), transcript variar
NMJ 38617 Homo sapiens Rhesus blood group, CcEe antigens (RHCE), transcript variar
NMJ38618 Homo sapiens Rhesus blood group, CcEe antigens (RHCE), transcript variar
NMJ 38619 Homo sapiens golgi associated, gamma adaptin ear containing, ARF binding
NMJ 38620 Homo sapiens DEAD (Asp-Glu-Ala-Asp) box polypeptide 31 (DDX31 ), transc
NMJ 38621 Homo sapiens BCL2-like 11 (apoptosis facilitator) (BCL2L11 ), transcript varU
NMJ 38622 Homo sapiens BCL2-like 11 (apoptosis facilitator) (BCL2L11 ), transcript vari?
NMJ 38623 Homo sapiens BCL2-like 11 (apoptosis facilitator) (BCL2L11 ), transcript varis
NMJ 38624 Homo sapiens BCL2-like 11 (apoptosis facilitator) (BCL2L11 ), transcript varij
NMJ 38625 Homo sapiens BCL2-like 11 (apoptosis facilitator) (BCL2L11), transcript varij
NMJ 38626 Homo sapiens BCL2-like 11 (apoptosis facilitator) (BCL2L11 ), transcript varic
NMJ 38627 Homo sapiens BCL2-like 11 (apoptosis facilitator) (BCL2L11 ), transcript variϊ
NMJ 38632 Homo sapiens Tara-like protein (HRIHFB2122), transcript variant 2, mRNA
NMJ 38633 Homo sapiens A kinase (PRKA) anchor protein 7 (AKAP7), transcript variant
NMJ 38634 Homo sapiens microseminoprotein, beta- (MSMB), transcript variant PSP57,
NMJ 38635 Homo sapiens H2A histone family, member V (H2AFV), transcript variant 2, i
NMJ 38636 Homo sapiens toll-like receptor 8 (TLR8), transcript variant 2, mRNA
NMJ 38637 Homo sapiens dudulin 2 (TSAP6), mRNA
NMJ 38638 Homo sapiens cofilin 2 (muscle) (CFL2), transcript variant 2, mRNA
NMJ 38639 Homo sapiens BCL2-like 12 (proline rich) (BCL2L12), transcript variant 1 , mF
NMJ 38640 Homo sapiens golgi associated, gamma adaptin ear containing, ARF binding
NMJ 38643 Homo sapiens calcium-binding tyrosine-(Y)-phosphorylation regulated (fibroL
NMJ 38644 Homo sapiens calcium-binding tyrosine-(Y)-phosphorylation regulated (fibroL
NMJ 38687 Homo sapiens phosphatidylinositol-4-phosphate 5-kinase, type II, beta (PIP5
NMJ 38688 Homo sapiens toll-like receptor 9 (TLR9), transcript variant B, mRNA
NMJ 38691 Homo sapiens transmembrane channel-like 1 (TMC1), mRNA
NMJ38693 Homo sapiens Kruppel-like factor 14 (KLF14), mRNA
NMJ 38694 Homo sapiens polycystic kidney and hepatic disease 1 (autosomal recessive
NMJ 38697 Homo sapiens taste receptor, type 1 , member 1 (TAS1 R1 ), transcript variant
NMJ 38698 Homo sapiens prematurely terminated mRNA decay factor-like (LOC91431),
NMJ 38699 Homo sapiens hypothetical protein BC006130 (LOC93622), mRNA
NMJ 38700 Homo sapiens tripartite motif-containing 40 (TRIM40), mRNA
NMJ 38701 Homo sapiens chromosome 7 open reading frame 11 (C7orf11), mRNA
NMJ 38702 Homo sapiens melanoma antigen, family C, 3 (MAGEC3), transcript variant 1
NMJ 38703 Homo sapiens melanoma antigen, family E, 2 (MAGEE2), mRNA
NMJ 38704 Homo sapiens necdin-like 2 (NDNL2), mRNA
NMJ 38705 Homo sapiens calglandulin-like protein (CAGLP), mRNA
NMJ 38706 Homo sapiens beta-1 ,3-N-acetylglucosaminyltransferase protein (IMAGE:49(
NMJ 38707 Homo sapiens B-cell CLL/lymphoma 7B (BCL7B), transcript variant 2, mRN/1
NMJ 38709 Homo sapiens DAB2 interacting protein (DAB2IP), mRNA
NMJ 38711 Homo sapiens peroxisome proliferative activated receptor, gamma (PPARG)
NMJ 38712 Homo sapiens peroxisome proliferative activated receptor, gamma (PPARG)
NMJ 38713 Homo sapiens nuclear factor of activated T-cells 5, tonicity-responsive (NFA"
NMJ 38714 Homo sapiens nuclear factor of activated T-cells 5, tonicity-responsive (NFA"
NMJ 38715 Homo sapiens macrophage scavenger receptor 1 (MSR1), transcript variant
NMJ 38716 Homo sapiens macrophage scavenger receptor 1 (MSR1 ), transcript variant
NMJ 38717 Homo sapiens palmitoyl-protein thioesterase 2 (PPT2), transcript variant 2, π
NMJ 38718 Homo sapiens solute carrier family 26, member 8 (SLC26A8), transcript varis
NMJ 38720 Homo sapiens histone 1 , H2bd (HIST1 H2BD), transcript variant 2, mRNA
NMJ 38722 Homo sapiens BCL2-like 14 (apoptosis facilitator) (BCL2L14), transcript varis
NMJ38723 Homo sapiens BCL2-like 14 (apoptosis facilitator) (BCL2L14), transcript varis
NMJ38724 Homo sapiens BCL2-like 14 (apoptosis facilitator) (BCL2L14), transcript varis
NMJ 38726 Homo sapiens ATP-binding cassette, sub-family C (CFTR/MRP), member 13
NMJ 38727 Homo sapiens suppression of tumorigenicity 7 like (ST7L), transcript variant
NMJ 38728 Homo sapiens suppression of tumorigenicity 7 like (ST7L), transcript variant
NMJ 38729 Homo sapiens suppression of tumorigenicity 7 like (ST7L), transcript variant •
NMJ 38730 Homo sapiens high mobility group nucleosomal binding domain 3 (HMGN3),
NMJ 38731 Homo sapiens mirror-image polydactyly 1 (MIPOL1), mRNA
NMJ 38732 Homo sapiens neurexin 2 (NRXN2), transcript variant alpha-2, mRNA
NMJ 38733 Homo sapiens phosphoglycerate kinase 2 (PGK2), mRNA
NMJ 38734 Homo sapiens neurexin 2 (NRXN2), transcript variant beta, mRNA
NMJ38735 Homo sapiens neurexin 1 (NRXN1), transcript variant beta, mRNA
NMJ 38736 Homo sapiens guanine nucleotide binding protein (G protein), alpha activatin
NMJ 38737 Homo sapiens hephaestin (HEPH), transcript variant 1 , mRNA
NMJ 38738 Homo sapiens SH2 domain containing phosphatase anchor protein 1 (SPAP
NMJ 38739 Homo sapiens SH2 domain containing phosphatase anchor protein 1 (SPAP
NMJ 38740 Homo sapiens NICE-3 protein (NICE-3), mRNA
NMJ 38761 Homo sapiens BCL2-associated X protein (BAX), transcript variant alpha, mF
NMJ 38762 Homo sapiens BCL2-associated X protein (BAX), transcript variant gamma, r
NMJ 38763 Homo sapiens BCL2-associated X protein (BAX), transcript variant delta, mR
NMJ 38764 Homo sapiens BCL2-associated X protein (BAX), transcript variant epsilon, n
NMJ 38765 Homo sapiens BCL2-associated X protein (BAX), transcript variant sigma, m
NMJ 38766 Homo sapiens peptidylglycine alpha-amidating monooxygenase (PAM), trans
NMJ 38768 Homo sapiens myeloma overexpressed gene (in a subset of t(11 ;14) positive
NMJ 38769 Homo sapiens ras homolog gene family, member T2 (RHOT2), mRNA
NMJ 38770 Homo sapiens hypothetical protein BC016861 (LOC90557), mRNA
NMJ 38771 Homo sapiens alpha-1 ,3(6)-mannosylglycoprotein beta-1 ,6-N-acetyl-gIucosa
NMJ 38773 Homo sapiens hypothetical protein BC017169 (LOC91137), mRNA
NMJ 38774 Homo sapiens chromosome 19 open reading frame 22 (C19orf22), mRNA
NMJ 38775 Homo sapiens hypothetical protein BC015183 (LOC91801), mRNA
NMJ 38777 Homo sapiens mitochondrial ribosome recycling factor (MRRF), transcript va
NMJ 38778 Homo sapiens chromosome 9 open reading frame 112 (C9orf112), mRNA
NMJ 38779 Homo sapiens hypothetical protein BC015148 (LOC93081), mRNA
NMJ 38780 Homo sapiens synaptotagmin-like 5 (SYTL5), mRNA
NMJ 38781 Homo sapiens similar to envelope protein (LOC113386), mRNA
NMJ 38783 Homo sapiens zinc finger protein Zip67 (ZIP67), mRNA
NMJ38784 Homo sapiens hypothetical protein BC014341 (LOC116123), mRNA
NMJ 38785 Homo sapiens chromosome 6 open reading frame 72 (C6orf72), mRNA
NMJ 38786 Homo sapiens hypothetical protein BC014339 (LOC116441), mRNA
NMJ 38787 Homo sapiens hypothetical protein BC009561 (LOC119710), mRNA
NMJ 38788 Homo sapiens hypothetical protein BC016153 (LOC120224), mRNA
NMJ38789 Homo sapiens hypothetical protein BC019238 (LOC120379), mRNA
NMJ38790 Homo sapiens hypothetical protein BC015003 (LOC122618), mRNA
NMJ38791 Homo sapiens chromosome 14 open reading frame 148 (C14orf148), mRNA
NMJ 38792 Homo sapiens senescence downregulated leol-Iike (LOC123169), mRNA
NMJ 38793 Homo sapiens ectonucleoside triphosphate diphosphohydrolase 8 (ENTPD8]
NMJ 38794 Homo sapiens lysophospholipase-like 1 (LYPLAL1), mRNA
NMJ 38795 Homo sapiens ADP-ribosylation factor-like 10B (ARL10B), mRNA
NMJ 38796 Homo sapiens hypothetical protein BC014608 (LOC128153), mRNA
NMJ 38797 Homo sapiens hypothetical protein BC014641 (LOC129138), mRNA
NMJ 38798 Homo sapiens hypothetical protein BC018453 (LOC129531), mRNA
NMJ 38799 Homo sapiens O-acyltransferase (membrane bound) domain containing 2 (0
NMJ 38800 Homo sapiens tripartite motif-containing 43 (TRIM43), mRNA
NMJ 38801 Homo sapiens galactose mutarotase (aldose 1-epimerase) (GALM), mRNA
NMJ 38802 Homo sapiens hypothetical protein BC018415 (LOC130617), mRNA
NMJ 38803 Homo sapiens hypothetical protein BC015395 (LOC130940), mRNA
NMJ 38804 Homo sapiens hypothetical protein BC014602 (LOC130951), mRNA
NMJ 38805 Homo sapiens family with sequence similarity 3, member D (FAM3D), mRNA
NMJ 38806 Homo sapiens MOX2 receptor (MOX2R), transcript variant 1 , mRNA
NMJ 38807 Homo sapiens hypothetical protein BC015088 (MGC16471), mRNA
NMJ 38808 Homo sapiens hypothetical protein BC015210 (LOC132200), mRNA
NMJ 38809 Homo sapiens hypothetical protein BC001573 (LOC134147), mRNA
NMJ 38810 Homo sapiens T-cell activation GTPase activating protein (TAGAP), transcrip
N J 38811 Homo sapiens chromosome 7 open reading frame 31 (C7orf31 ), mRNA
NMJ38812 Homo sapiens hypothetical protein BC019250 (LOC143241), mRNA
NMJ 38813 Homo sapiens ATPase, Class I, type 8B, member 3 (ATP8B3), mRNA
NMJ38814 Homo sapiens GS2 like (LOC150379), mRNA
NMJ38815 Homo sapiens hypothetical protein BC018070 (LOC151871), mRNA
NMJ 38817 Homo sapiens solute carrier family 7, (cationic amino acid transporter, y+ sys
NMJ 38818 Homo sapiens chromosome 9 open reading frame 65 (C9orf65), mRNA
NMJ38819 Homo sapiens hypothetical protein BC017868 (LOC159091), mRNA
NMJ 38820 Homo sapiens hypothetical protein MGC2198 (MGC2198), mRNA
NMJ 38821 Homo sapiens peptidylglycine alpha-amidating monooxygenase (PAM), trans
NMJ 38822 Homo sapiens peptidylglycine alpha-amidating monooxygenase (PAM), trans
NMJ 38923 Homo sapiens TAF1 RNA polymerase II, TATA box binding protein (TBP)-as
N J 38924 Homo sapiens guanidinoacetate N-methyltransferase (GAMT), transcript vari
NMJ 38925 Homo sapiens SON DNA binding protein (SON), transcript variant a, mRNA
NMJ 38926 Homo sapiens SON DNA binding protein (SON), transcript variant c, mRNA
NMJ 38927 Homo sapiens SON DNA binding protein (SON), transcript variant f, mRNA
N J 38928 Homo sapiens molybdenum cofactor synthesis 1 (MOCS1 ), transcript variant
NMJ 38929 Homo sapiens diablo homolog (Drosophila) (DIABLO), nuclear gene encodin
NMJ 38930 Homo sapiens diablo homolog (Drosophila) (DIABLO), nuclear gene encodin
NMJ 38931 Homo sapiens B-cell CLL/lymphoma 6 (zinc finger protein 51 ) (BCL6), transc
NMJ 38932 Homo sapiens apobec-1 complementation factor (ACF), transcript variant 2,
NMJ 38933 Homo sapiens apobec-1 complementation factor (ACF), transcript variant 3,
NMJ 38934 Homo sapiens palmitoyl-protein thioesterase 2 (PPT2), transcript variant 3, n
NMJ 38937 Homo sapiens pancreatitis-associated protein (PAP), transcript variant 3, mF
NM 138938 Homo sapiens pancreatitis-associated protein (PAP), transcript variant 2, mF
NMJ 38939 Homo sapiens MOX2 receptor (MOX2R), transcript variant 2, mRNA
NM 138940 Homo sapiens MOX2 receptor (MOX2R), transcript variant 3, mRNA
NMJ 38957 Homo sapiens mitogen-activated protein kinase 1 (MAPK1), transcript varian
NM 138958 Homo sapiens autocrine motility factor receptor (AMFR), transcript variant 2,
NMJ 38959 Homo sapiens vang-like 1 (van gogh, Drosophila) (VANGL1), mRNA
NM 138960 Homo sapiens TGFB-induced factor 2-like, X-linked (TGIF2LX), mRNA
NMJ 38961 Homo sapiens endothelial cell adhesion molecule (ESAM), mRNA
NMJ 38962 Homo sapiens musashi homolog 2 (Drosophila) (MSI2), transcript variant 1 , i
NMJ 38963 Homo sapiens ribosomal protein S4, Y-linked 2 (RPS4Y2), mRNA
NMJ 38964 Homo sapiens G protein-coupled receptor 73 (GPR73), mRNA
NM 138966 Homo sapiens neuropilin (NRP) and tolloid (TLL)-like 1 (NET01), transcript v
NMJ 38967 Homo sapiens secretory carrier membrane protein 5 (SCAMP5), mRNA
NMJ 38969 Homo sapiens retinal short chain dehydrogenase reductase (RDH-E2), mRN
NM 138970 Homo sapiens neurexin 3 (NRXN3), transcript variant beta, mRNA
NMJ 38971 Homo sapiens beta-site APP-cleaving enzyme 1 (BACE1 ), transcript variant
NMJ 38972 Homo sapiens beta-site APP-cleaving enzyme 1 (BACE1), transcript variant
NMJ 38973 Homo sapiens beta-site APP-cleaving enzyme 1 (BACE1), transcript variant
NM 138980 Homo sapiens mitogen-activated protein kinase 10 (MAPK10), transcript vari
NMJ38981 Homo sapiens mitogen-activated protein kinase 10 (MAPK10), transcript vari
NMJ 38982 Homo sapiens mitogen-activated protein kinase 10 (MAPK10), transcript vari
NM 138983 Homo sapiens oligodendrocyte transcription factor 1 (OLIG1), mRNA
NMJ 38991 Homo sapiens beta-site APP-cleaving enzyme 2 (BACE2), transcript variant
NMJ 38992 Homo sapiens beta-site APP-cleaving enzyme 2 (BACE2), transcript variant
NM 138993 Homo sapiens mitogen-activated protein kinase 11 (MAPK11), transcript vari
NMJ 38994 Homo sapiens contactin associated protein-like 4 (CNTNAP4), transcript vari
NMJ 38995 Homo sapiens myosin 1MB (MY03B), mRNA
NMJ 38996 Homo sapiens contactin associated protein-like 5 (CNTNAP5), transcript vari
NMJ 38998 Homo sapiens DEAD (Asp-Glu-Ala-Asp) box polypeptide 39 (DDX39), transc
NMJ 38999 Homo sapiens neuropilin (NRP) and tolloid (TLL)-like 1 (NET01), transcript v
NM 139002 Homo sapiens hemochromatosis (HFE), transcript variant 2, mRNA
NMJ 39003 Homo sapiens hemochromatosis (HFE), transcript variant 3, mRNA
NMJ 39004 Homo sapiens hemochromatosis (HFE), transcript variant 4, mRNA
NMJ 39005 Homo sapiens hemochromatosis (HFE), transcript variant 5, mRNA
NM 139006 Homo sapiens hemochromatosis (HFE), transcript variant 6, mRNA
NMJ 39007 Homo sapiens hemochromatosis (HFE), transcript variant 7, mRNA
NMJ 39008 Homo sapiens hemochromatosis (HFE), transcript variant 8, mRNA
NM 139009 Homo sapiens hemochromatosis (HFE), transcript variant 9, mRNA
NM 139010 Homo sapiens hemochromatosis (HFE), transcript variant 10, mRNA
NMJ 39011 Homo sapiens hemochromatosis (HFE), transcript variant 11 , mRNA
NM 139012 Homo sapiens mitogen-activated protein kinase 14 (MAPK14), transcript vari
NM 139013 Homo sapiens mitogen-activated protein kinase 14 (MAPK14), transcript vari
NMJ39014 Homo sapiens mitogen-activated protein kinase 14 (MAPK14), transcript vari
NM 139015 Homo sapiens signal peptide peptidase 3 (SPPL3), mRNA
NM 139016 Homo sapiens hypothetical gene LOC128439 (LOC128439), mRNA
NMJ 39017 Homo sapiens interleukin 31 receptor A (IL31RA), mRNA
NMJ39018 Homo sapiens NK inhibitory receptor precursor (NKIR), mRNA
NMJ39021 Homo sapiens extracellular signal-regulated kinase 8 (ERK8), mRNA
NMJ 39022 Homo sapiens pan-hematopoietic expression (PHEMX), transcript variant 1,
NMJ 39024 Homo sapiens pan-hematopoietic expression (PHEMX), transcript variant 3,
NM 139025 Homo sapiens a disintegrin-like and metalloprotease (reprolysin type) with th
NM 139026 Homo sapiens a disintegrin-like and metalloprotease (reprolysin type) with th
NM 139027 Homo sapiens a disintegrin-like and metalloprotease (reprolysin type) with th
NMJ 39028 Homo sapiens a disintegrin-like and metalloprotease (reprolysin type) with th
NMJ 39030 Homo sapiens CD151 antigen (CD151), transcript variant 2, mRNA
NMJ 39032 Homo sapiens mitogen-activated protein kinase 7 (MAPK7), transcript varian
NMJ 39033 Homo sapiens mitogen-activated protein kinase 7 (MAPK7), transcript varian
NMJ 39034 Homo sapiens mitogen-activated protein kinase 7 (MAPK7), transcript varian
NMJ 39035 Homo sapiens SWI/SNF related, matrix associated, actin dependent regulat
NMJ 39045 Homo sapiens SWI/SNF related, matrix associated, actin dependent regulate
NMJ 39046 Homo sapiens mitogen-activated protein kinase 8 (MAPK8), transcript varian
NMJ 39047 Homo sapiens mitogen-activated protein kinase 8 (MAPK8), transcript varian
NMJ 39048 Homo sapiens SWI/SNF related, matrix associated, actin dependent regulate
NMJ 39049 Homo sapiens mitogen-activated protein kinase 8 (MAPK8), transcript varian
NMJ 39053 Homo sapiens EPS8-like 3 (EPS8L3), transcript variant 1 , mRNA
NMJ 39054 Homo sapiens a disintegrin-like and metalloprotease (reprolysin type) with th
NMJ 39055 Homo sapiens a disintegrin-like and metalloprotease (reprolysin type) with th
NMJ 39056 Homo sapiens a disintegrin-like and metalloprotease (reprolysin type) with th
NMJ 39057 Homo sapiens a disintegrin-like and metalloprotease (reprolysin type) with th
NMJ 39058 Homo sapiens aristaless related homeobox (ARX), mRNA
NMJ39062 Homo sapiens casein kinase 1 , delta (CSNK1 D), transcript variant 2, mRNA
NMJ 39067 Homo sapiens SWI/SNF related, matrix associated, actin dependent regulate
NMJ 39068 Homo sapiens mitogen-activated protein kinase 9 (MAPK9), transcript varian
NMJ 39069 Homo sapiens mitogen-activated protein kinase 9 (MAPK9), transcript varian
NMJ 39070 Homo sapiens mitogen-activated protein kinase 9 (MAPK9), transcript varian
NMJ 39071 Homo sapiens SWI/SNF related, matrix associated, actin dependent regulate
NMJ 39072 Homo sapiens delta-notch-like EGF repeat-containing transmembrane (DNE
NMJ 39073 Homo sapiens spermatogenesis associated 3 (SPATA3), mRNA
NMJ39074 Homo sapiens defensin, beta 127 (DEFB127), mRNA
NMJ 39075 Homo sapiens two pore segment channel 2 (TPCN2), mRNA
NMJ39076 Homo sapiens hypothetical protein FLJ13614 (FLJ13614), mRNA
NMJ 39078 Homo sapiens mitogen-activated protein kinase-activated protein kinase 5 (l,
NMJ 39118 Homo sapiens YY1 associated protein (YAP), transcript variant 2, mRNA
NMJ 39119 Homo sapiens YY1 associated protein (YAP), transcript variant 3, mRNA
NMJ39120 Homo sapiens YY1 associated protein (YAP), transcript variant 4, mRNA
NMJ39121 Homo sapiens YY1 associated protein (YAP), transcript variant 5, mRNA
NMJ39122 Homo sapiens TAF6 RNA polymerase II, TATA box binding protein (TBP)-as
NMJ39123 Homo sapiens TAF6 RNA polymerase II, TATA box binding protein (TBP)-as
NMJ 39124 Homo sapiens mitogen-activated protein kinase 8 interacting protein 2 (MAPI
NMJ39125 Homo sapiens mannan-binding lectin serine protease 1 (C4/C2 activating coi
NMJ39126 Homo sapiens peptidylprolyl isomerase (cyclophilin)-like 4 (PPIL4), mRNA
NMJ39131 Homo sapiens nucleoporin 98kDa (NUP98), transcript variant 2, mRNA
NMJ39132 Homo sapiens nucleoporin 98kDa (NUP98), transcript variant 4, mRNA
NMJ 39135 Homo sapiens AT rich interactive domain 1A (SWI- like) (ARID1A), transcript
NMJ 39136 Homo sapiens potassium voltage-gated channel, Shaw-related subfamily, mi
NMJ 39137 Homo sapiens potassium voltage-gated channel, Shaw-related subfamily, m<
NMJ39155 Homo sapiens a disintegrin-like and metalloprotease (reprolysin type) with th
NMJ 39156 Homo sapiens adenosine monophosphate deaminase 2 (isoform L) (AMPD2
NMJ39157 Homo sapiens suppression of tumorigenicity 5 (ST5), transcript variant 2, mF
NMJ 39158 Homo sapiens amyotrophic lateral sclerosis 2 (juvenile) chromosome region,
NMJ 39159 Homo sapiens dipeptidylpeptidase 9 (DPP9), mRNA
NMJ 39160 Homo sapiens novel 58.3 KDA protein (LOC91614), mRNA
NMJ 39161 Homo sapiens crumbs homolog 3 (Drosophila) (CRB3), transcript variant 2, r
NMJ39162 Homo sapiens Smith-Magenis syndrome chromosome region, candidate 7 (£
NMJ39163 Homo sapiens amyotrophic lateral sclerosis 2 (juvenile) chromosome region,
NMJ 39164 Homo sapiens START domain containing 4, sterol regulated (STARD4), mRt
NMJ 39165 Homo sapiens retinoic acid early transcript 1 E (RAET1 E), mRNA
NMJ 39166 Homo sapiens striated muscle activator of Rho-dependent signaling (STARS
NMJ 39167 Homo sapiens sarcoglycan zeta (SGCZ), mRNA
NMJ39168 Homo sapiens splicing factor, arginine/serine-rich 12 (SFRS12), mRNA
NMJ 39169 Homo sapiens TruB pseudouridine (psi) synthase homolog 1 (E. coli) (TRUB
NMJ39170 Homo sapiens hypothetical protein AF447587 (LOC146562), mRNA
NMJ 39171 Homo sapiens START domain containing 6 (STARD6), mRNA
NMJ 39172 Homo sapiens MDAC1 (MDAC1), mRNA
NMJ 39173 Homo sapiens CG10806-like (LOC150159), mRNA
NMJ 39174 Homo sapiens testis nuclear RNA-binding protein-like (LOC161931), mRNA
NMJ 39175 Homo sapiens ring finger protein 133 (RNF133), mRNA
NMJ 39176 Homo sapiens NACHT, leucine rich repeat and PYD containing 7 (NALP7), ti
NMJ 39177 Homo sapiens solute carrier family 39 (metal ion transporter), member 11 (SI
NMJ 39178 Homo sapiens prostate cancer antigen-1 (DEPC-1), mRNA
NMJ 39179 Homo sapiens KCCR13L (LOC221955), mRNA
NMJ 39181 Homo sapiens centaurin, delta 2 (CENTD2), transcript variant 1 , mRNA
NMJ 39182 Homo sapiens centaurin, delta 1 (CENTD1), transcript variant 2, mRNA
NMJ 39199 Homo sapiens bromodomain containing 8 (BRD8), transcript variant 2, mRN;
NMJ 39201 Homo sapiens G protein-coupled receptor kinase interactor 2 (GIT2), transcr
NMJ 39202 Homo sapiens megalencephalic leukoencephalopathy with subcortical cysts
NMJ 39204 Homo sapiens EPS8-like 1 (EPS8L1 ), transcript variant 3, mRNA
NMJ 39205 Homo sapiens histone deacetylase 5 (HDAC5), transcript variant 2, mRNA
NMJ 39207 Homo sapiens nucleosome assembly protein 1-like 1 (NAP1L1), transcript vε
NMJ 39208 Homo sapiens mannan-binding lectin serine protease 2 (MASP2), transcript >
NMJ 39209 Homo sapiens G protein-coupled receptor kinase 7 (GRK7), mRNA
NMJ 39211 Homo sapiens homeodomain-only protein (HOP), transcript variant 2, mRNA
NMJ 39212 Homo sapiens homeodomain-only protein (HOP), transcript variant 3, mRNA
NMJ39214 Homo sapiens TGFB-induced factor 2-like, Y-linked (TGIF2LY), mRNA
NMJ39215 Homo sapiens TAF15 RNA polymerase II, TATA box binding protein (TBP)-a
NMJ 39235 Homo sapiens nucleolar protein family 6 (RNA-associated) (NOL6), transcrip
N J 39238 Homo sapiens ADAMTS-like 1 (ADAMTSL1 ), transcript variant 1 , mRNA
NMJ 39239 Homo sapiens T-cell activation NFKB-like protein (TA-NFKBH), mRNA
NMJ 39240 Homo sapiens LOC92346 (LOC92346), mRNA
NMJ 39241 Homo sapiens FGD1 family, member 4 (FGD4), mRNA
NMJ 39242 Homo sapiens methionyl-tRNAformyltransferase, mitochondrial (MtFMT), ml
NMJ 39243 Homo sapiens testis nuclear RNA-binding protein (Tenr), mRNA
NMJ 39244 Homo sapiens syntaxin binding protein 5 (tomosyn) (STXBP5), mRNA
NMJ 39245 Homo sapiens protein phosphatase 1 (formerly 2C)-like (PPM1L), mRNA
NMJ 39246 Homo sapiens chromosome 9 open reading frame 97 (C9orf97), mRNA
NMJ 39247 Homo sapiens adenylate cyclase 4 (ADCY4), mRNA
NMJ 39248 Homo sapiens lipase, member H (LIPH), mRNA
NMJ 39249 Homo sapiens membrane-spanning 4-domains, subfamily A, member 6E (M!
NMJ 39250 Homo sapiens cancer/testis antigen 1 A (CTAG1 A), mRNA
NMJ 39264 Homo sapiens ADAMTS-like 1 (ADAMTSL1), transcript variant 3, mRNA
NMJ 39265 Homo sapiens EH-domain containing 4 (EHD4), mRNA
NMJ 39266 Homo sapiens signal transducer and activator of transcription 1, 91 kDa (STA
NMJ 39267 Homo sapiens START domain containing 7 (STARD7), transcript variant 2, n
NMJ 39273 Homo sapiens cysteinyl-tRNA synthetase (CARS), transcript variant 1 , mRN;
NMJ 39274 Homo sapiens acetyl-Coenzyme A synthetase 2 (ADP forming) (ACAS2), tra
NMJ 39275 Homo sapiens A kinase (PRKA) anchor protein 1 (AKAP1), nuclear gene enc
NMJ 39276 Homo sapiens signal transducer and activator of transcription 3 (acute-phase
NMJ 39277 Homo sapiens kallikrein 7 (chymotryptic, stratum corneum) (KLK7), transcrip
NMJ 39278 Homo sapiens leucine-rich repeat LGI family, member 3 (LGI3), mRNA
NMJ 39279 Homo sapiens multiple coagulation factor deficiency 2 (MCFD2), mRNA
NMJ 39280 Homo sapiens ORM1-like 3 (S. cerevisiae) (ORMDL3), mRNA
NMJ 39281 Homo sapiens WD repeat domain 36 (WDR36), mRNA
NMJ 39282 Homo sapiens paired-like homeobox protein OTEX (OTEX), mRNA
NMJ 39283 Homo sapiens T-cell activation protein phosphatase 2C (TA-PP2C), mRNA
NMJ 39284 Homo sapiens leucine-rich repeat LGI family, member 4 (LGI4), mRNA
NMJ 39285 Homo sapiens growth arrest-specific 2 like 2 (GAS2L2), mRNA
NMJ 39286 Homo sapiens cell division cycle 26 (CDC26), mRNA
NM .139289 Homo sapiens A kinase (PRKA) anchor protein 4 (AKAP4), transcript variant
NM 139290 Homo sapiens angiopoietin 1 (ANGPT1 ), transcript variant 2, mRNA
NM_ 139312 Homo sapiens YME1-like 1 (S. cerevisiae) (YME1 L1), nuclear gene encodinς
NM 139313 Homo sapiens YME1-like 1 (S. cerevisiae) (YME1L1), nuclear gene encoding
NM_ .139314 Homo sapiens angiopoietin-like 4 (ANGPTL4), transcript variant 1 , mRNA
NM 139315 Homo sapiens TAF6 RNA polymerase II, TATA box binding protein (TBP)-as
NM_ .139316 Homo sapiens amphiphysin (Stiff-Man syndrome with breast cancer 128kDa
NM .139317 Homo sapiens baculoviral IAP repeat-containing 7 (livin) (BIRC7), transcript
NM 139318 Homo sapiens potassium voltage-gated channel, subfamily H (eag-related), i
NM .139319 Homo sapiens solute carrier family 17 (sodium-dependent inorganic phosphs
NM .139320 Homo sapiens CHRNA7 (cholinergic receptor, nicotinic, alpha polypeptide 7,
NM .139321 Homo sapiens attractin (ATRN), transcript variant 1 , mRNA
NM .139322 Homo sapiens attractin (ATRN), transcript variant 2, mRNA
NM .139323 Homo sapiens tyrosine 3-monooxygenase/tryptophan 5-monooxygenase acti
NM .139343 Homo sapiens bridging integrator 1 (BIN1), transcript variant 1 , mRNA
NM .139344 Homo sapiens bridging integrator 1 (BIN1), transcript variant 2, mRNA
NM 139345 Homo sapiens bridging integrator 1 (BIN1), transcript variant 3, mRNA
NM .139346 Homo sapiens bridging integrator 1 (B1N1), transcript variant 4, mRNA
NM .139347 Homo sapiens bridging integrator 1 (BIN1), transcript variant 5, mRNA
NM 139348 Homo sapiens bridging integrator 1 (BIN1), transcript variant 6, mRNA
NM .139349 Homo sapiens bridging integrator 1 (BIN1), transcript variant 7, mRNA
NM 139350 Homo sapiens bridging integrator 1 (BIN1), transcript variant 9, mRNA
NM .139351 Homo sapiens bridging integrator 1 (BIN1), transcript variant 10, mRNA
NM .139352 Homo sapiens TATA box binding protein (TBP)-associated factor, RNA polyn
NM 139353 Homo sapiens TATA box binding protein (TBP)-associated factor, RNA polyn
NM 139354 Homo sapiens megakaryocyte-associated tyrosine kinase (MATK), transcript
NM 139355 Homo sapiens megakaryocyte-associated tyrosine kinase (MATK), transcript
NM .144488 Homo sapiens regulator of G-protein signalling 3 (RGS3), transcript variant 6
NM 144489 Homo sapiens regulator of G-protein signalling 3 (RGS3), transcript variant 5
NM .144490 Homo sapiens A kinase (PRKA) anchor protein 11 (AKAP11 ), transcript vari.
NM 144492 Homo sapiens claudin 14 (CLDN14), transcript variant 1 , mRNA
NM 144494 Homo sapiens polyglutamine binding protein 1 (PQBP1), mRNA
NM .144495 Homo sapiens polyglutamine binding protein 1 (PQBP1), mRNA
NM 144497 Homo sapiens A kinase (PRKA) anchor protein (gravin) 12 (AKAP12), transc
NM 144498 Homo sapiens oxysterol binding protein-like 2 (OSBPL2), transcript variant 2
NM 144499 Homo sapiens guanine nucleotide binding protein (G protein), alpha transdue
NM 144501 Homo sapiens F11 receptor (F11 R), transcript variant 2, mRNA
NM .144502 Homo sapiens F11 receptor (F11R), transcript variant 3, mRNA
NM .144503 Homo sapiens F11 receptor (F11 R), transcript variant 4, mRNA
NM 144504 Homo sapiens F11 receptor (F11 R), transcript variant 5, mRNA
NM 144505 Homo sapiens kallikrein 8 (neuropsin/ovasin) (KLK8), transcript variant 2, mF
NM 144506 Homo sapiens kallikrein 8 (neuropsin/ovasin) (KLK8), transcript variant 3, mF
NM_ .144507 Homo sapiens kallikrein 8 (neuropsin/ovasin) (KLK8), transcript variant 4, mF
NM 144508 Homo sapiens AF15q14 protein (AF15Q14), mRNA
NM 144563 Homo sapiens ribose 5-phosphate isomerase A (ribose 5-phosphate epimers
NM 144564 Homo sapiens solute carrier family 39 (zinc transporter), member 3 (SLC39A
NM 144565 Homo sapiens homolog of Drosophila Numb-interacting protein (NIP), mRN/1
NM 44567 Homo sapiens similar to RIKEN cDNA 2610307121 (LOC90806), mRNA
NM 144568 Homo sapiens chromosome 14 open reading frame 9 (C14orf9), mRNA
NM 144569 Homo sapiens hypothetical protein FLJ25348 (FLJ25348), mRNA
NM 144570 Homo sapiens chromosome 16 open reading frame 34 (C16orf34), mRNA
NM_ "144571 Homo sapiens CCR4-NOT transcription complex, subunit 6-like (CNOT6L), n
NM 144573 Homo sapiens nexilin (F actin binding protein) (NEXN), mRNA
NM "144574 Homo sapiens WD repeat domain 20 (WDR20), transcript variant 2, mRNA
NM 144575 Homo sapiens calpain 13 (CAPN13), mRNA
NM_ "144576 Homo sapiens hypothetical protein FLJ32452 (FLJ32452), mRNA
NM_ 144577 Homo sapiens hypothetical protein FLJ32926 (FLJ32926), mRNA
NMJ44578 Homo sapiens chromosome 14 open reading frame 32 (C14orf32), mRNA
NMJ44579 Homo sapiens sideroflexin 5 (SFXN5), mRNA
NMJ44580 Homo sapiens kidney predominant protein NCU-G1 (MGC31963), mRNA
NMJ44581 Homo sapiens chromosome 14 open reading frame 149 (C14orf149), mRNA
NMJ44582 Homo sapiens testis expressed sequence 261 (TEX261), mRNA
NMJ44583 Homo sapiens ATPase, H+ transporting, lysosomal 42kDa, V1 subunit C isot
NMJ44584 Homo sapiens hypothetical protein FLJ30525 (FLJ30525), mRNA
NMJ44585 Homo sapiens solute carrier family 22 (organic anion/cation transporter), mei
NMJ44586 Homo sapiens hypothetical protein MGC29643 (MGC29643), mRNA
NMJ44587 Homo sapiens chromosome 10 open reading frame 87 (C10orf87), mRNA
NMJ44588 Homo sapiens zinc finger, FYVE domain containing 27 (ZFYVE27), transcrip
NMJ44589 Homo sapiens catechol-O-methyltransferase domain containing 1 (COMTD1
NMJ44590 Homo sapiens ankyrin repeat domain 22 (ANKRD22), mRNA
NMJ44591 Homo sapiens chromosome 10 open reading frame 32 (C10orf32), mRNA
NMJ44593 Homo sapiens Ras homolog enriched in brain like 1 (RHEBL1), mRNA
NMJ44594 Homo sapiens hypothetical protein FLJ32942 (FLJ32942), mRNA
NMJ44595 Homo sapiens hypothetical protein FLJ30046 (FLJ30046), mRNA
NMJ44596 Homo sapiens tetratricopeptide repeat domain 8 (TTC8), transcript variant 3,
NMJ44597 Homo sapiens hypothetical protein MGC29937 (MGC29937), mRNA
NMJ44598 Homo sapiens leucine rich repeat containing 28 (LRRC28), mRNA
NMJ44599 Homo sapiens non-imprinted in Prader-Willi/Angelman syndrome 1 (NIPA1 ),
NMJ44600 Homo sapiens hypothetical protein FLJ31153 (FLJ31153), mRNA
NMJ44601 Homo sapiens chemokine-like factor super family 3 (CKLFSF3), transcript va
NMJ44602 Homo sapiens hypothetical protein MGC32905 (MGC33367), mRNA
NMJ44603 Homo sapiens NADPH oxidase organizer 1 (NOX01), transcript variant a, ml
NMJ44604 Homo sapiens hypothetical protein BC001584 (LOC124245), mRNA
NMJ44605 Homo sapiens hypothetical protein FLJ25410 (FLJ25410), mRNA
NMJ44606 Homo sapiens folliculin (BHD), transcript variant 2, mRNA
NMJ44607 Homo sapiens hypothetical protein FLJ32499 (FLJ32499), mRNA
NMJ44608 Homo sapiens hypothetical protein MGC39389 (FLJ32384), mRNA
NMJ44609 Homo sapiens hypothetical protein FLJ31795 (FLJ31795), mRNA
NMJ44610 Homo sapiens hypothetical protein FLJ25006 (FLJ25006), mRNA
NMJ44611 Homo sapiens hypothetical protein MGC32124 (MGC32124), mRNA
NMJ44612 Homo sapiens lipoxygenase homology domains 1 (LOXHD1), mRNA
NMJ44613 Homo sapiens cytochrome c oxidase subunit Vlb, testes-specific (COXVIB2)
NMJ44614 Homo sapiens methyl-CpG binding domain protein 3-like 2 (MBD3L2), mRN/
NMJ44615 Homo sapiens hypothetical protein MGC23244 (MGC23244), mRNA
NMJ44616 Homo sapiens homolog of mouse skeletal muscle sarcoplasmic reticulum pr<
NMJ44617 Homo sapiens heat shock protein, alpha-crystallin-related, B6 (HSPB6), mRr
NMJ44618 Homo sapiens hypothetical protein MGC29891 (MGC29891), mRNA
NMJ44620 Homo sapiens hypothetical protein MGC14816 (MGC14816), mRNA
NMJ44621 Homo sapiens zinc finger and BTB domain containing 8 (ZBTB8), mRNA
NMJ 44622 Homo sapiens hypothetical protein FLJ32934 (FLJ32934), mRNA
NMJ 44623 Homo sapiens hypothetical protein FLJ32784 (FLJ32784), mRNA
NMJ44624 Homo sapiens kinase interacting with leukemia-associated gene (stathmin) (
NMJ44625 Homo sapiens hypothetical protein FLJ32978 (FLJ32978), mRNA
NMJ44626 Homo sapiens hypothetical protein MGC17299 (MGC17299), mRNA
NMJ44627 Homo sapiens SSTK-interacting protein (SSTK-IP), mRNA
NMJ44628 Homo sapiens chromosome 20 open reading frame 140 (C20orf140), mRNA
NMJ44629 Homo sapiens chromosome 2 open reading frame 11 (C2orf11), mRNA
NMJ44631 Homo sapiens zinc finger protein 513 (ZNF513), mRNA
NMJ44632 Homo sapiens hypothetical protein FLJ30294 (FLJ30294), mRNA
NMJ44633 Homo sapiens potassium voltage-gated channel, subfamily H (eag-related), i
NMJ44634 Homo sapiens lysozyme-like 4 (LYZL4), mRNA
NMJ44635 Homo sapiens hypothetical protein MGC21688 (MGC21688), mRNA
NMJ44636 Homo sapiens coiled-coil-helix-coiled-coil-helix domain containing 4 (CHCHC
NMJ44637 Homo sapiens zinc finger, DHHC domain containing 19 (ZDHHC19), mRNA
NMJ 44638 Homo sapiens hypothetical protein MGC29956 (MGC29956), mRNA
NMJ44639 Homo sapiens hypothetical protein FLJ31300 (FLJ31300), mRNA
NMJ 44640 Homo sapiens interleukin 17 receptor E (IL17RE), transcript variant 3, mRN/1
NMJ 44641 Homo sapiens likely ortholog of mouse protein phosphatase 2C eta (FLJ323:
NMJ 44642 Homo sapiens synaptoporin (SYNPR), mRNA
NMJ 44643 Homo sapiens hypothetical protein FLJ30655 (FLJ30655), mRNA
NMJ 44644 Homo sapiens spermatogenesis associated 4 (SPATA4), mRNA
NMJ 44645 Homo sapiens hypothetical protein MGC26744 (MGC26744), mRNA
NMJ 44646 Homo sapiens immunoglobulin J polypeptide, linker protein for immunoglobu
NMJ 44647 Homo sapiens hypothetical protein MGC26610 (MGC26610), mRNA
NMJ44648 Homo sapiens hypothetical protein FLJ32786 (FLJ32786), mRNA
NMJ 44649 Homo sapiens hypothetical protein FLJ33069 (FLJ33069), mRNA
NMJ 44650 Homo sapiens alcohol dehydrogenase, iron containing, 1 (ADHFE1), mRNA
NMJ 44651 Homo sapiens hypothetical protein FLJ25471 (FLJ25471), mRNA
NMJ 44652 Homo sapiens leucine zipper-EF-hand containing transmembrane protein 2 (
NMJ44653 Homo sapiens BTB (POZ) domain containing 14A (BTBD14A), mRNA
NMJ 44654 Homo sapiens chromosome 9 open reading frame 116 (C9orf116), mRNA
NMJ 44657 Homo sapiens hypothetical protein FLJ30678 (FLJ30678), mRNA
NMJ 44658 Homo sapiens dedicator of cytokinesis 11 (DOCK11), mRNA
NMJ 44659 Homo sapiens t-complex 10 (mouse)-like (TCP10L), mRNA
NMJ 44660 Homo sapiens sterile alpha motif domain containing 8 (SAMD8), mRNA
NMJ 44661 Homo sapiens chromosome 10 open reading frame 82 (C10orf82), mRNA
NMJ 44662 Homo sapiens hypothetical protein MGC26605 (MGC26605), mRNA
NMJ 44663 Homo sapiens NOV1 (NOV1), mRNA
NMJ 44664 Homo sapiens hypothetical protein MGC33371 (MGC33371), mRNA
NMJ 44665 Homo sapiens sestrin 3 (SESN3), mRNA
NMJ 44666 Homo sapiens hypothetical protein FLJ32752 (FLJ32752), mRNA
NMJ 44667 Homo sapiens hypothetical protein FLJ32894 (FLJ32894), mRNA
NMJ 44668 Homo sapiens hypothetical protein MGC33630 (MGC33630), mRNA
NMJ 44669 Homo sapiens hypothetical protein FLJ31978 (FLJ31978), mRNA
NMJ 44670 Homo sapiens hypothetical protein FLJ25179 (FLJ25179), mRNA
NMJ 44671 Homo sapiens hypothetical protein FLJ32356 (FLJ32356), mRNA
NMJ 44672 Homo sapiens otoancorin (OTOA), mRNA
NMJ 44673 Homo sapiens chemokine-like factor super family 2 (CKLFSF2), mRNA
NMJ 44674 Homo sapiens hypothetical protein FLJ32871 (FLJ32871), mRNA
NMJ 44675 Homo sapiens hypothetical protein MGC18079 (MGC18079), mRNA
NMJ 44676 Homo sapiens hypothetical protein MGC23911 (MGC23911), mRNA
NMJ 44677 Homo sapiens mannosyl (alpha-1 ,6-)-glycoprotein beta-1 ,6-N-acetyl-glucosa
NMJ 44678 Homo sapiens target of mybl-like 2 (chicken) (TOM1L2), mRNA
NMJ 44679 Homo sapiens hypothetical protein FLJ31528 (FLJ31528), mRNA
NMJ44681 Homo sapiens hypothetical protein FLJ32734 (FLJ32734), mRNA
NMJ 44682 Homo sapiens hypothetical protein FLJ31952 (FLJ31952), mRNA
NMJ 44683 Homo sapiens hypothetical protein MGC23280 (MGC23280), mRNA
NMJ 44684 Homo sapiens zinc finger protein 480 (ZNF480), mRNA
NMJ44685 Homo sapiens homeodomain interacting protein kinase 4 (HIPK4), mRNA
NMJ 44686 Homo sapiens transmembrane channel-like 4 (TMC4), mRNA
NMJ 44687 Homo sapiens NACHT, leucine rich repeat and PYD containing 12 (NALP12)
NMJ 44688 Homo sapiens hypothetical protein FLJ32658 (FLJ32658), mRNA
NMJ 44689 Homo sapiens hypothetical protein FLJ32191 (FLJ32191), mRNA
NMJ 44690 Homo sapiens zinc finger protein 582 (ZNF582), mRNA
NMJ44691 Homo sapiens calpain 12 (CAPN12), mRNA
NMJ44692 Homo sapiens hypothetical protein BC017947 (LOC148137), mRNA
NMJ 44693 Homo sapiens zinc finger protein 558 (ZNF558), mRNA
NMJ 44694 Homo sapiens zinc finger protein 570 (ZNF570), mRNA
NMJ 44695 Homo sapiens hypothetical protein FLJ32421 (FLJ32421), mRNA
NMJ44696 Homo sapiens hypothetical protein FLJ32940 (DKFZp686H1423), transcript -
NMJ44697 Homo sapiens hypothetical protein BC017397 (LOC148523), mRNA
NMJ 44698 Homo sapiens hypothetical protein FLJ25124 (FLJ25124), mRNA
NMJ44699 Homo sapiens ATPase, Na+/K+ transporting, alpha 4 polypeptide (ATP1A4),
NMJ44701 Homo sapiens interleukin-23 receptor (IL23R), mRNA
NMJ 44702 Homo sapiens hypothetical protein FLJ32884 (FLJ32884), mRNA
NMJ44703 Homo sapiens chromosome 20 open reading frame 40 (C20orf40), mRNA
NMJ44704 Homo sapiens hypothetical protein FLJ30473 (FLJ30473), mRNA
NMJ44705 Homo sapiens hypothetical protein MGC27019 (MGC27019), mRNA
NMJ44706 Homo sapiens chromosome 2 open reading frame 15 (C2orf15), mRNA
NMJ 44707 Homo sapiens prominin 2 (PROM2), mRNA
NMJ44709 Homo sapiens hypothetical protein FLJ32312 (FLJ32312), mRNA
NMJ44710 Homo sapiens septin 10 (SEPT10), transcript variant 1 , mRNA
NMJ 44711 Homo sapiens hypothetical protein MGC22679 (MGC22679), mRNA
NMJ 44712 Homo sapiens solute carrier family 23 (nucleobase transporters), member 3 •
NMJ 44713 Homo sapiens hypothetical protein FLJ32954 (FLJ32954), mRNA
NMJ44714 Homo sapiens hypothetical protein MGC27069 (FLJ25449), mRNA
NMJ 44715 Homo sapiens hypothetical protein FLJ25200 (FLJ25200), mRNA
NMJ44716 Homo sapiens hypothetical protein MGC23918 (MGC23918), mRNA
NMJ44717 Homo sapiens hypothetical protein MGC34923 (MGC34923), mRNA
NMJ 44718 Homo sapiens hypothetical protein AY099107 (LOC152185), mRNA
NMJ 44719 Homo sapiens hypothetical protein FLJ25467 (FLJ25467), mRNA
NMJ44720 Homo sapiens multiple coiled-coil GABABR1 -binding protein (MARLIN1), mF
NMJ44721 Homo sapiens THAP domain containing 6 (THAP6), mRNA
NMJ 44722 Homo sapiens KPL2 protein (FLJ23577), transcript variant 2, mRNA
NMJ44723 Homo sapiens hypothetical protein FLJ31121 (FLJ31121), mRNA
NMJ 44724 Homo sapiens MARVEL domain containing 2 (MARVELD2), mRNA
NMJ44725 Homo sapiens hypothetical protein FLJ25439 (FLJ25439), mRNA
NMJ 44726 Homo sapiens hypothetical protein FLJ31951 (FLJ31951), mRNA
NMJ44727 Homo sapiens crystallin, gamma N (CRYGN), mRNA
NMJ44728 Homo sapiens dual specificity phosphatase 10 (DUSP10), transcript variant :
NMJ44729 Homo sapiens dual specificity phosphatase 10 (DUSP10), transcript variant
NMJ44732 Homo sapiens heterogeneous nuclear ribonucleoprotein U-like 1 (HNRPUL1
NMJ44733 Homo sapiens heterogeneous nuclear ribonucleoprotein U-like 1 (HNRPUL1
NMJ 44734 Homo sapiens heterogeneous nuclear ribonucleoprotein U-like 1 (HNRPUL1
NMJ44736 Homo sapiens hypothetical protein PR01853 (PR01853), transcript variant 1
NMJ44765 Homo sapiens epithelial V-like antigen 1 (EVA1), transcript variant 2, mRNA
NMJ 4766 Homo sapiens regulator of G-protein signalling 13 (RGS13), transcript varian
NMJ44767 Homo sapiens A kinase (PRKA) anchor protein 13 (AKAP13), transcript varis
NMJ44769 Homo sapiens forkhead box 11 (FOXI1), transcript variant 2, mRNA
NMJ 44770 Homo sapiens RNA binding motif protein 11 (RBM11), mRNA
NMJ44772 Homo sapiens apolipoprotein A-l binding protein (APOA1BP), mRNA
NMJ 44773 Homo sapiens G protein-coupled receptor 73-like 1 (GPR73L1), mRNA
NMJ44775 Homo sapiens Smith-Magenis syndrome chromosome region, candidate 8 (S
NMJ 44776 Homo sapiens formyltetrahydrofolate dehydrogenase (FTHFD), transcript vai
NMJ 44777 Homo sapiens sciellin (SCEL), transcript variant 2, mRNA
NMJ 44778 Homo sapiens muscleblind-like 2 (Drosophila) (MBNL2), transcript variant 1 ,
NMJ 44779 Homo sapiens FXYD domain containing ion transport regulator 5 (FXYD5), tr
NMJ44780 Homo sapiens degenerative spermatocyte homolog, lipid desaturase (Droso|
NMJ 44781 Homo sapiens programmed cell death 2 (PDCD2), transcript variant 2, mRN;
NMJ44782 Homo sapiens carnitine acetyltransferase (CRAT), transcript variant 3, mRN/
NMJ44947 Homo sapiens kallikrein 11 (KLK11 ), transcript variant 2, mRNA
NMJ 44949 Homo sapiens suppressor of cytokine signaling 5 (SOCS5), transcript varianl
NMJ44956 Homo sapiens protease, serine, 21 (testisin) (PRSS21), transcript variant 2, i
NMJ 44957 Homo sapiens protease, serine, 21 (testisin) (PRSS21), transcript variant 3, i
NMJ44962 Homo sapiens hypothetical protein MGC22776 (MGC22776), mRNA
NMJ 44963 Homo sapiens hypothetical protein FLJ23790 (FLJ23790), mRNA
NMJ 44964 Homo sapiens RNA (guanine-9-) methyltransferase domain containing 3 (RG
NMJ44965 Homo sapiens tetratricopeptide repeat domain 16 (TTC16), mRNA
NMJ44966 Homo sapiens chromosome 9 open reading frame 154 (C9orf154), mRNA
NMJ44967 Homo sapiens hypothetical protein FLJ30058 (FLJ30058), mRNA
NMJ44968 Homo sapiens hypothetical protein FLJ32783 (FLJ32783), mRNA
NMJ44969 Homo sapiens zinc finger, DHHC domain containing 15 (ZDHHC15), mRNA
NMJ44970 Homo sapiens hypothetical protein MGC39350 (MGC39350), mRNA
NMJ 44972 Homo sapiens lactate dehydrogenase A-like 6A (LDHAL6A), mRNA
NMJ 44973 Homo sapiens hypothetical protein MGC24039 (MGC24039), mRNA
NMJ44974 Homo sapiens hypothetical protein FLJ31846 (FLJ31846), mRNA
NMJ44975 Homo sapiens hypothetical protein MGC19764 (MGC19764), mRNA
NMJ44976 Homo sapiens zinc finger protein 564 (ZNF564), mRNA
NMJ44977 Homo sapiens family with sequence similarity 31 , member B (FAM31 B), mRt
NMJ44978 Homo sapiens hypothetical protein FLJ32745 (FLJ32745), mRNA
NMJ44979 Homo sapiens hypothetical protein MGC27016 (MGC27016), mRNA
NMJ44980 Homo sapiens chromosome 6 open reading frame 118 (C6orf118), mRNA
NMJ44981 Homo sapiens hypothetical protein FLJ25059 (FLJ25059), mRNA
NMJ44982 Homo sapiens hypothetical protein MGC23401 (MGC23401 ), mRNA
NMJ44984 Homo sapiens chromosome 10 open reading frame 72 (C10orf72), mRNA
NMJ 44985 Homo sapiens cadherin-like 24 (CDH24), mRNA
NMJ44987 Homo sapiens U2(RNU2) small nuclear RNA auxiliary factor 1-like 3 (U2AF1
NMJ44988 Homo sapiens hypothetical protein MGC19780 (MGC19780), mRNA
NMJ44990 Homo sapiens hypothetical protein FLJ23878 (FLJ23878), mRNA
NMJ44991 Homo sapiens chromosome 21 open reading frame 29 (C21orf29), mRNA
NMJ44992 Homo sapiens hypothetical protein MGC26733 (MGC26733), mRNA
NMJ44994 Homo sapiens ankyrin repeat domain 23 (ANKRD23), mRNA
NMJ44995 Homo sapiens DEAH (Asp-Glu-Ala-Asp/His) box polypeptide 57 (DHX57), trε
NMJ44996 Homo sapiens hypothetical protein DKFZp761 H079 (DKFZp761H079), tran&
NMJ44997 Homo sapiens folliculin (BHD), transcript variant 1 , mRNA
NMJ44998 Homo sapiens stimulated by retinoic acid 13 (STRA13), mRNA
NMJ44999 Homo sapiens hypothetical protein MGC20806 (MGC20806), mRNA
NMJ45000 Homo sapiens hypothetical protein FLJ25422 (FLJ25422), mRNA
NMJ 45001 Homo sapiens serine/threonine kinase 32A (STK32A), mRNA
NMJ45003 Homo sapiens hypothetical protein FLJ31164 (FLJ31164), mRNA
NMJ45004 Homo sapiens a disintegrin and metalloproteinase domain 32 (ADAM32), mF
NMJ45005 Homo sapiens chromosome 9 open reading frame 72 (C9orf72), transcript vs
NMJ45006 Homo sapiens sushi domain containing 3 (SUSD3), mRNA
NMJ45007 Homo sapiens NACHT, leucine rich repeat and PYD containing 11 (NALP11)
NMJ45008 Homo sapiens hypothetical protein FLJ30213 (FLJ30213), mRNA
NMJ45010 Homo sapiens chromosome 10 open reading frame 63 (C10orf63), mRNA
NMJ45011 Homo sapiens zinc finger protein 25 (KOX 19) (ZNF25), mRNA
NMJ45012 Homo sapiens chromosome 10 open reading frame 9 (C10orf9), mRNA
NMJ45013 Homo sapiens hypothetical protein MGC35558 (MGC35558), mRNA
NMJ45014 Homo sapiens hypothetical protein FLJ32915 (FLJ32915), mRNA
NMJ45015 Homo sapiens MAS-related GPR, member F (MRGPRF), mRNA
NMJ45016 Homo sapiens BXMAS2-10 (BXMAS2-10), mRNA
NMJ45017 Homo sapiens IIIG9 protein (FLJ32771), mRNA
NMJ45018 Homo sapiens hypothetical protein FLJ25416 (FLJ25416), mRNA
NMJ45019 Homo sapiens hypothetical protein FLJ30707 (FLJ30707), mRNA
NMJ45020 Homo sapiens hypothetical protein FLJ32743 (FLJ32743), mRNA
NMJ45021 Homo sapiens c-mir, cellular modulator of immune recognition (MIR), transcr
NMJ 45023 Homo sapiens coiled-coil domain containing 7 (CCDC7), mRNA
NMJ45024 Homo sapiens hypothetical protein FLJ31547 (FLJ31547), mRNA
NMJ45025 Homo sapiens chromosome 6 open reading frame 199 (C6orf199), mRNA
NMJ45026 Homo sapiens spermatogenesis associated, serine-rich 1 (SPATS1), mRNA
NMJ45027 Homo sapiens chromosome 6 open reading frame 102 (C6orf102), mRNA
NMJ45028 Homo sapiens chromosome 6 open reading frame 81 (C6orf81), mRNA
NMJ45029 Homo sapiens chromosome 6 open reading frame 136 (C6orf136), mRNA
NMJ45030 Homo sapiens hypothetical protein MGC22793 (MGC22793), mRNA
NMJ 45032 Homo sapiens F-box and leucine-rich repeat protein 13 (FBXL13), mRNA
NMJ 45033 Homo sapiens chromosome 21 open reading frame 100 (C21orf100), mRNA
NMJ 45034 Homo sapiens AF464140 (LOC163590), mRNA
NMJ 45035 Homo sapiens ADMP (ADMP), mRNA
NMJ 45036 Homo sapiens hypothetical protein MGC33887 (MGC33887), mRNA
NMJ 45037 Homo sapiens hypothetical protein MGC15606 (MGC15606), mRNA
NMJ 45038 Homo sapiens CG10958-like (MGC16372), mRNA
NMJ 45039 Homo sapiens hypothetical protein MGC16385 (MGC16385), mRNA
NMJ 45040 Homo sapiens protein kinase C, delta binding protein (PRKCDBP), mRNA
NMJ 45041 Homo sapiens hypothetical protein MGC20235 (MGC20235), mRNA
NMJ 45042 Homo sapiens alpha tubulin-like (MGC16703), mRNA
NMJ 45043 Homo sapiens nei like 2 (E. coli) (NEIL2), mRNA
NMJ 45044 Homo sapiens zinc finger protein 501 (ZNF501), mRNA
NMJ 45045 Homo sapiens hypothetical protein MGC20983 (MGC20983), mRNA
NMJ45046 Homo sapiens calreticulin 3 (CALR3), mRNA
NMJ45047 Homo sapiens oxidored-nitro domain-containing protein (NOR1), transcript vi
NMJ 45048 Homo sapiens hypothetical protein MGC29898 (MGC29898), mRNA
NMJ45049 Homo sapiens hypothetical protein MGC10067 (MGC10067), mRNA
NMJ 45050 Homo sapiens hypothetical protein MGC27434 (MGC27434), mRNA
NMJ45051 Homo sapiens hypothetical protein MGC4734 (MGC4734), mRNA
NMJ 45052 Homo sapiens hypothetical protein MGC23937 similar to CG4798 (MGC239;
NMJ 45053 Homo sapiens hypothetical protein MGC20470 (MGC20470), mRNA
NMJ45054 Homo sapiens hypothetical protein LOC146845 (LOC146845), mRNA
NMJ 45055 Homo sapiens chromosome 18 open reading frame 25 (C18orf25), mRNA
NMJ45056 Homo sapiens thymus expressed gene 3-like (MGC15476), mRNA
NMJ 45057 Homo sapiens CDC42 effector protein (Rho GTPase binding) 5 (CDC42EP5;
NMJ45058 Homo sapiens hypothetical protein MGC7036 (MGC7036), mRNA
NMJ45059 Homo sapiens fucokinase (FUK), mRNA
NMJ 45060 Homo sapiens chromosome 18 open reading frame 24 (C18orf24), mRNA
NMJ45061 Homo sapiens chromosome 13 open reading frame 3 (C13orf3), mRNA
NMJ 45062 Homo sapiens chromosome 6 open reading frame 113 (C6orf113), mRNA
NMJ45063 Homo sapiens chromosome 6 open reading frame 130 (C6orf130), mRNA
NMJ 45064 Homo sapiens SH3 and cysteine rich domain 3 (STAC3), mRNA
NMJ45065 Homo sapiens pellino 3 alpha (MGC35521), mRNA
NMJ 45068 Homo sapiens transient receptor potential cation channel, subfamily V, mem
NMJ45071 Homo sapiens cytokine inducible SH2-containing protein (CISH), transcript v
NMJ 45074 Homo sapiens protease, serine, 25 (PRSS25), nuclear gene encoding mitocl
NMJ45080 Homo sapiens non-SMC (structural maintenance of chromosomes) element
NMJ 45102 Homo sapiens zinc finger protein 95 homolog (mouse) (ZFP95), transcript va
NMJ45109 Homo sapiens mitogen-activated protein kinase kinase 3 (MAP2K3), transcri
NMJ 451 10 Homo sapiens mitogen-activated protein kinase kinase 3 (MAP2K3), transcri
NMJ45111 Homo sapiens hypothetical protein DKFZp727G131 (DKFZp727G131), mRN
NMJ45112 Homo sapiens MAX protein (MAX), transcript variant 2, mRNA
NM J451 13 Homo sapiens MAX protein (MAX), transcript variant 3, mRNA
NMJ45114 Homo sapiens MAX protein (MAX), transcript variant 4, mRNA
NMJ451 15 Homo sapiens zinc finger protein 498 (ZNF498), mRNA
NMJ451 16 Homo sapiens MAX protein (MAX), transcript variant 5, mRNA
NMJ 45117 Homo sapiens neuron navigator 2 (NAV2), transcript variant 2, mRNA
NMJ45119 Homo sapiens praja 1 (PJA1), mRNA
NMJ 45159 Homo sapiens jagged 2 (JAG2), transcript variant 2, mRNA
NMJ45160 Homo sapiens mitogen-activated protein kinase kinase 5 (MAP2K5), transcri
NMJ45161 Homo sapiens mitogen-activated protein kinase kinase 5 (MAP2K5), transcri
NMJ45162 Homo sapiens mitogen-activated protein kinase kinase 5 (MAP2K5), transcri
NMJ45165 Homo sapiens Churchill domain containing 1 (CHURC1), mRNA
NMJ45166 Homo sapiens hypothetical protein KIAA1190 (KIAA1190), mRNA
NMJ45167 Homo sapiens phosphatidylinositol glycan, class M (PIGM), mRNA
NMJ 45168 Homo sapiens NAD(P) dependent steroid dehydrogenase-like (HSPC105), n
NMJ 45169 Homo sapiens chromosome 6 open reading frame 83 (C6orf83), mRNA
NMJ45170 Homo sapiens tetratricopeptide repeat domain 18 (TTC18), mRNA
NMJ45171 Homo sapiens glycoprotein hormone beta 5 (GPHB5), mRNA
NMJ45172 Homo sapiens testis development protein NYD-SP29 (NYD-SP29), mRNA
NMJ45173 Homo sapiens DIRAS family, GTP-binding RAS-like 1 (DIRAS1), mRNA
NMJ 45174 Homo sapiens DnaJ (Hsp40) homolog, subfamily B, member 7 (DNAJB7), m
NMJ 45175 Homo sapiens NSE1 (NSE1), mRNA
NMJ 45176 Homo sapiens solute carrier family 2 (facilitated glucose transporter), membe
NMJ 45177 Homo sapiens dehydrogenase/reductase (SDR family) X-linked (DHRSX), m
NMJ45178 Homo sapiens atonal homolog 7 (Drosophila) (ATOH7), mRNA
NMJ45179 Homo sapiens chromosome 21 open reading frame 93 (C21orf93), mRNA
NMJ 45180 Homo sapiens chromosome 21 open reading frame 94 (C21orf94), mRNA
NMJ 45182 Homo sapiens PYD and CARD domain containing (PYCARD), transcript vari,
NMJ 45183 Homo sapiens PYD and CARD domain containing (PYCARD), transcript vari,
NMJ 45185 Homo sapiens mitogen-activated protein kinase kinase 7 (MAP2K7), mRNA
NMJ45186 Homo sapiens ATP-binding cassette, sub-family C (CFTR/MRP), member 11
NMJ 45187 Homo sapiens ATP-binding cassette, sub-family C (CFTR MRP), member 12
NMJ 45188 Homo sapiens ATP-binding cassette, sub-family C (CFTR/MRP), member 12
NMJ 45189 Homo sapiens ATP-binding cassette, sub-family C (CFTR/MRP), member 12
NMJ 45190 Homo sapiens ATP-binding cassette, sub-family C (CFTR/MRP), member 12
NMJ45196 Homo sapiens lipoyltransferase 1 (LIPT1), transcript variant 2, mRNA
NMJ45197 Homo sapiens lipoyltransferase 1 (LIPT1), transcript variant 3, mRNA
NMJ45198 Homo sapiens lipoyltransferase 1 (LIPT1 ), transcript variant 4, mRNA
NMJ 45199 Homo sapiens lipoyltransferase 1 (LIPT1 ), transcript variant 5, mRNA
NMJ45200 Homo sapiens calcium binding protein 4 (CABP4), mRNA
NMJ45201 Homo sapiens similar to CG3714 gene product (PP3856), mRNA
NMJ45202 Homo sapiens proline-rich acidic protein 1 (PRAP1 ), mRNA
NMJ 45203 Homo sapiens casein kinase 1 , alpha 1 -like (CSNK1 A1 L), mRNA
NMJ 45204 Homo sapiens SUMO/sentrin specific protease family member 8 (SENP8), m
NMJ45205 Homo sapiens HMG2 like (LOC127540), mRNA
NMJ45206 Homo sapiens vesicle transport through interaction with t-SNAREs homolog
NMJ45207 Homo sapiens spermatogenesis associated 5 (SPATA5), mRNA
NMJ 45208 Homo sapiens methyl-CpG binding domain protein 3-like 1 (MBD3L1), mRN/
NMJ 45212 Homo sapiens mitochondrial ribosomal protein L30 (MRPL30), nuclear gene
NMJ 45213 Homo sapiens mitochondrial ribosomal protein L30 (MRPL30), nuclear gene
NMJ45214 Homo sapiens tripartite motif-containing 11 (TRIM11), mRNA
NMJ 45230 Homo sapiens chromosome 7 open reading frame 32 (C7orf32), mRNA
NMJ45231 Homo sapiens chromosome 14 open reading frame 143 (C14orf143), mRNA
NMJ 45232 Homo sapiens LOC90353 (LOC90353), mRNA
NMJ45233 Homo sapiens zinc finger protein 625 (ZNF625), mRNA
NMJ 45234 Homo sapiens chordin-like 1 (CHRDL1), mRNA
NMJ45235 Homo sapiens fibronectin type 3 and ankyrin repeat domains 1 (FANK1 ), mR
NMJ45236 Homo sapiens UDP-GlcNAc:betaGal beta-1 ,3-N-acetylglucosaminyltransfera
NMJ 45237 Homo sapiens similar to RNA polymerase I transcription factor RRN3 (LOC9^
NMJ 45238 Homo sapiens zinc finger protein 31 (KOX 29) (ZNF31), mRNA
NMJ 45239 Homo sapiens similar to lymphocyte antigen 6 complex, locus G5B; G5b prol
NM J45241 Homo sapiens WD repeat domain 31 (WDR31 ), mRNA
NMJ45242 Homo sapiens similar to POSSIBLE GUSTATORY RECEPTOR CLONE PTE
NMJ 45243 Homo sapiens metalloprotease related protein 1 (MPRP-1), mRNA
NMJ45244 Homo sapiens DNA-damage-inducible transcript 4-like (DDIT4L), mRNA
NMJ45245 Homo sapiens ecotropic viral integration site 5-like (EVI5L), mRNA
NMJ 45246 Homo sapiens chromosome 10 open reading frame 4 (C10orf4), transcript vs
NMJ45247 Homo sapiens chromosome 10 open reading frame 78 (C10orf78), transcript
NMJ45248 Homo sapiens LOC122258 (LOC122258), mRNA
NMJ45249 Homo sapiens family with sequence similarity 14, member B (FAM14B), mRr
NMJ45250 Homo sapiens chromosome 14 open reading frame 6 (C14orf6), mRNA
NMJ 45251 Homo sapiens serine/threonine/tyrosine interacting protein (STYX), mRNA
NMJ45252 Homo sapiens similar to common salivary protein 1 (LOC124220), mRNA
NMJ45253 Homo sapiens LOC124402 (LOC124402), mRNA
NMJ 45254 Homo sapiens LOC124491 (LOC124491), mRNA
NMJ 45255 Homo sapiens mitochondrial ribosomal protein L10 (MRPL10), nuclear gene
NMJ 45256 Homo sapiens leucine rich repeat containing 25 (LRRC25), mRNA
NMJ 45257 Homo sapiens LOC126731 (LOC126731 ), mRNA
NMJ45258 Homo sapiens hypothetical protein MGC22773 (MGC22773), mRNA
NMJ45259 Homo sapiens activin A receptor, type IC (ACVR1C), mRNA
NMJ45260 Homo sapiens odd-skipped homolog (Drosophila) (ODD), mRNA
NMJ45261 Homo sapiens homolog of yeast TIM14 (TIM14), transcript variant 1 , mRNA
NMJ45262 Homo sapiens CG9886-like (GLYCTK), mRNA
NMJ 45263 Homo sapiens LOC132671 (LOC132671), mRNA
NMJ45265 Homo sapiens similar to RIKEN cDNA 0610011 N22 (LOC133957), mRNA
NMJ45266 Homo sapiens similar to RIKEN cDNA 2700047N05 (LOC134492), mRNA
NMJ 45267 Homo sapiens chromosome 6 open reading frame 57 (C6orf57), mRNA
NMJ 45268 Homo sapiens LOC136263 (LOC136263), mRNA
NMJ 45269 Homo sapiens similar to CG6405 gene product (LOC137392), mRNA
NMJ45270 Homo sapiens similar to hypothetical protein FLJ13841 (LOC146325), mRN/
NMJ 45271 Homo sapiens similar to hypothetical protein MGC13138 (LOC146542), mRN
NMJ45272 Homo sapiens LOC146853 (LOC146853), mRNA
NMJ45273 Homo sapiens triggering receptor expressed on myeloid cells 4 (TREM4), mF
NMJ45274 Homo sapiens hypothetical protein MGC21518 (MGC21518), mRNA
NMJ 45275 Homo sapiens kinesin light chain 2-like (KLC2L), transcript variant 2, mRNA
NMJ45276 Homo sapiens zinc finger protein 563 (ZNF563), mRNA
NMJ 45277 Homo sapiens hemochromatosis type 2 (juvenile) (HFE2), transcript variant t
NMJ45278 Homo sapiens LOC148823 (LOC148823), mRNA
NMJ 45279 Homo sapiens MOB1 , Mps One Binder kinase activator-like 2C (yeast) (MOE
NMJ 45280 Homo sapiens similar to hepatocellular carcinoma-associated antigen HCA5!
NMJ45282 Homo sapiens similar to CG4995 gene product (LOC153328), mRNA
NMJ45283 Homo sapiens chromosome 9 open reading frame 121 (C9orf121), mRNA
NMJ45284 Homo sapiens similar to hypothetical protein MGC17347 (LOC159090), mRN
NMJ45285 Homo sapiens NK2 transcription factor related, locus 3 (Drosophila) (NKX2-3
NMJ45286 Homo sapiens stomatin (EPB72)-like 3 (STOML3), mRNA
NMJ 45287 Homo sapiens zinc finger protein 519 (ZNF519), mRNA
NMJ 45288 Homo sapiens zinc finger protein 342 (ZNF342), mRNA
NMJ 45291 Homo sapiens zinc finger protein 509 (ZNF509), mRNA
NMJ 45292 Homo sapiens UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylga
NMJ 45293 Homo sapiens similar to hypothetical protein FLJ20897 (LOC196549), mRN/
NMJ45294 Homo sapiens similar to RIKEN cDNA 3230401 M21 [Mus musculus] (LOC19
NMJ 45295 Homo sapiens zinc finger protein 627 (ZNF627), mRNA
NMJ 45296 Homo sapiens immunoglobulin superfamily, member 4C (IGSF4C), mRNA
NMJ 45297 Homo sapiens zinc finger protein 626 (ZNF626), mRNA
NMJ 45298 Homo sapiens apolipoprotein B mRNA editing enzyme, catalytic polypeptide-
NMJ 45299 Homo sapiens similar to Dynein heavy chain at 16F (LOC200383), mRNA
NMJ45300 Homo sapiens LOC200420 (LOC200420), mRNA
NMJ 45301 Homo sapiens similar to CGI-148 protein (LOC201158), mRNA
NMJ 45303 Homo sapiens similar to RIKEN CDNA 2310008M10 (LOC202459), mRNA
NMJ 45304 Homo sapiens chromosome 7 open reading frame 33 (C7orf33), mRNA
NMJ45305 Homo sapiens similar to solute carrier family 25 , member 16 (LOC203427),
NMJ45306 Homo sapiens chromosome 10 open reading frame 35 (C10orf35), mRNA
NMJ45307 Homo sapiens pleckstrin homology domain containing, family K member 1 (F
NMJ 45308 Homo sapiens hypothetical protein BC004224 (LOC220070), mRNA
NMJ45309 Homo sapiens Hypothetical 55.1 kDa protein F09G8.5 in chromosome III (LC
NMJ45310 Homo sapiens zinc finger protein 258 (ZNF258), mRNA
NMJ 45311 Homo sapiens crystallin, zeta (quinone reductase)-like 1 (CRYZL1), transcrip
NMJ 45312 Homo sapiens zinc finger protein 485 (ZNF485), mRNA
NMJ45313 Homo sapiens RasGEF domain family, member 1A (RASGEF1A), mRNA
NMJ45314 Homo sapiens chromosome 10 open reading frame 49 (C10orf49), mRNA
NMJ 45315 Homo sapiens lactation elevated 1 (LACE1), mRNA
NMJ 45316 Homo sapiens chromosome 6 open reading frame 128 (C6orf128), mRNA
NMJ 45320 Homo sapiens oxysterol binding protein-like 3 (OSBPL3), transcript variant 2
NMJ 45321 Homo sapiens oxysterol binding protein-like 3 (OSBPL3), transcript variant 3
NMJ 45322 Homo sapiens oxysterol binding protein-like 3 (OSBPL3), transcript variant 4
NMJ 45323 Homo sapiens oxysterol binding protein-like 3 (OSBPL3), transcript variant 5
NMJ 45324 Homo sapiens oxysterol binding protein-like 3 (OSBPL3), transcript variant 6
NMJ45325 Homo sapiens DNA directed RNA polymerase II polypeptide J-related gene (
NMJ45326 Homo sapiens similar to hypothetical protein FLJ13659 (LOC115648), mRN/
NMJ 45328 Homo sapiens chromosome 21 open reading frame 66 (C21orf66), transcript
NMJ 45330 Homo sapiens mitochondrial ribosomal protein L33 (MRPL33), nuclear gene
NMJ 45331 Homo sapiens mitogen-activated protein kinase kinase kinase 7 (MAP3K7), 1
NMJ 45332 Homo sapiens mitogen-activated protein kinase kinase kinase 7 (MAP3K7), 1
NMJ 45333 Homo sapiens mitogen-activated protein kinase kinase kinase 7 (MAP3K7), 1
NMJ 45341 Homo sapiens programmed cell death 4 (neoplastic transformation inhibitor)
NMJ 45342 Homo sapiens mitogen-activated protein kinase kinase kinase 7 interacting p
NMJ 45343 Homo sapiens apolipoprotein L, 1 (APOL1), transcript variant 2, mRNA
NMJ 45344 Homo sapiens apolipoprotein L, 1 (APOL1), transcript variant 3, mRNA
NMJ 45345 Homo sapiens socius (SOC), mRNA
NMJ 45346 Homo sapiens socius (SOC), mRNA
NMJ 45347 Homo sapiens kringle containing transmembrane protein 2 (KREMEN2), tran
NMJ 45348 Homo sapiens kringle containing transmembrane protein 2 (KREMEN2), tran
NMJ 45349 Homo sapiens scavenger receptor class F, member 1 (SCARF1), transcript \
NMJ 45350 Homo sapiens scavenger receptor class F, member 1 (SCARF1), transcript \
NMJ 45351 Homo sapiens scavenger receptor class F, member 1 (SCARF1), transcript \
NMJ 45352 Homo sapiens scavenger receptor class F, member 1 (SCARF1), transcript \
NMJ 45637 Homo sapiens apolipoprotein L, 2 (APOL2), transcript variant beta, mRNA
NMJ 45638 Homo sapiens oxysterol binding protein-like 5 (OSBPL5), transcript variant 2
NMJ 45639 Homo sapiens apolipoprotein L, 3 (APOL3), transcript variant alpha/c, mRN/1
NMJ 45640 Homo sapiens apolipoprotein L, 3 (APOL3), transcript variant alpha/d, mRN/1
NMJ 45641 Homo sapiens apolipoprotein L, 3 (APOL3), transcript variant beta/a, mRNA
NMJ 45642 Homo sapiens apolipoprotein L, 3 (APOL3), transcript variant beta/b, mRNA
NMJ 45644 Homo sapiens mitochondrial ribosomal protein L35 (MRPL35), nuclear gene
NMJ 45645 Homo sapiens Williams-Beuren Syndrome critical region protein 20 copy B p
NMJ 45646 Homo sapiens DEAH (Asp-Glu-Ala-Asp/His) box polypeptide 57 (DHX57), trε
NMJ 45647 Homo sapiens unknown MGC21654 product (MGC21654), mRNA
NMJ 45648 Homo sapiens solute carrier family 15, member 4 (SLC15A4), mRNA
NMJ 45649 Homo sapiens glucosaminyl (N-acetyl) transferase 2, l-branching enzyme (G
NMJ45650 Homo sapiens mucin 15 (MUC15), mRNA
NMJ 45651 Homo sapiens ligand binding protein RYD5 (RYD5), mRNA
NMJ45652 Homo sapiens WAP four-disulfide core domain 5 (WFDC5), mRNA
NMJ45653 Homo sapiens transcription elongation factor B polypeptide 3C (elongin A3) |
NMJ45654 Homo sapiens RAD52 homolog B (S. cerevisiae) (RAD52B), mRNA
NMJ45655 Homo sapiens glucosaminyl (N-acetyl) transferase 2, l-branching enzyme (G
NMJ45657 Homo sapiens GS homeobox 1 (GSH1), mRNA
NMJ 45658 Homo sapiens sperm equatorial segment protein 1 (SPESP1), mRNA
NMJ45659 Homo sapiens interleukin 27 (IL27), mRNA
NMJ 45660 Homo sapiens apolipoprotein L, 4 (APOL4), transcript variant b, mRNA
NMJ 45662 Homo sapiens SPANX family, member A2 (SPANXA2), mRNA
NMJ 45663 Homo sapiens Dbf4-related factor 1 (DRF1), transcript variant 1 , mRNA
NMJ45664 Homo sapiens SPANX family, member B2 (SPANXB2), mRNA
NMJ45665 Homo sapiens SPANX family, member E (SPANXE), mRNA
NMJ45685 Homo sapiens BRF1 homolog, subunit of RNA polymerase III transcription in
NMJ45686 Homo sapiens mitogen-activated protein kinase kinase kinase kinase 4 (MAF
NMJ45687 Homo sapiens mitogen-activated protein kinase kinase kinase kinase 4 (MAF
NMJ45689 Homo sapiens amyloid beta (A4) precursor protein-binding, family B, membe
NMJ 45690 Homo sapiens tyrosine 3-monooxygenase/tryptophan 5-monooxygenase acti
NMJ45691 Homo sapiens ATP synthase mitochondrial F1 complex assembly factor 2 (A
NMJ45693 Homo sapiens lipin 1 (LPIN1), mRNA
NMJ45695 Homo sapiens diacylglycerol kinase, beta 90kDa (DGKB), transcript variant Σ
NMJ45696 Homo sapiens BRF1 homolog, subunit of RNA polymerase III transcription in
NMJ45697 Homo sapiens cell division cycle associated 1 (CDCA1 ), transcript variant 1 ,
NMJ45698 Homo sapiens acyl-Coenzyme A binding domain containing 5 (ACBD5), mRI
NMJ45699 Homo sapiens apolipoprotein B mRNA editing enzyme, catalytic polypeptide-
NMJ 45701 Homo sapiens cell division cycle associated 4 (CDCA4), transcript variant 2,
NMJ 45702 Homo sapiens tigger transposable element derived 1 (TIGD1), mRNA
NMJ45714 Homo sapiens ataxin 2 related protein (A2LP), transcript variant B, mRNA
NMJ 45715 Homo sapiens tigger transposable element derived 2 (TIGD2), mRNA
NMJ45716 Homo sapiens single stranded DNA binding protein 3 (SSBP3), mRNA
NMJ45719 Homo sapiens tigger transposable element derived 3 (TIGD3), mRNA
NMJ45720 Homo sapiens tigger transposable element derived 4 (TIGD4), mRNA
NMJ45725 Homo sapiens TNF receptor-associated factor 3 (TRAF3), transcript variant '
NMJ45726 Homo sapiens TNF receptor-associated factor 3 (TRAF3), transcript variant .
NMJ45727 Homo sapiens lipoprotein, Lp(a)-like 2 (LPAL2), transcript variant 2, mRNA
NMJ 45728 Homo sapiens desmuslin (DMN), transcript variant A, mRNA
NMJ45729 Homo sapiens mitochondrial ribosomal protein L24 (MRPL24), nuclear gene
NMJ 45730 Homo sapiens adaptor-related protein complex 1 , beta 1 subunit (AP1B1), tπ
NMJ 45731 Homo sapiens synaptogyrin 1 (SYNGR1 ), transcript variant 1 b, mRNA
NMJ 45733 Homo sapiens septin 3 (SEPT3), transcript variant A, mRNA
NMJ 45734 Homo sapiens septin 3 (SEPT3), transcript variant C, mRNA
NMJ45735 Homo sapiens Rho guanine nucleotide exchange factor (GEF) 7 (ARHGEF7]
NMJ45738 Homo sapiens synaptogyrin 1 (SYNGR1), transcript variant 1c, mRNA
NMJ45739 Homo sapiens oxysterol binding protein-like 6 (OSBPL6), transcript variant 2
NMJ45740 Homo sapiens glutathione S-transferase A1 (GSTA1), mRNA
NMJ 45747 Homo sapiens thioredoxin reductase 2 (TXNRD2), nuclear gene encoding ml
NMJ 45748 Homo sapiens thioredoxin reductase 2 (TXNRD2), nuclear gene encoding mi
NMJ45751 Homo sapiens TNF receptor-associated factor 4 (TRAF4), transcript variant i
NMJ45752 Homo sapiens CDP-diacylglycerol-inositol 3-phosphatidyltransferase (phosp
NMJ45753 Homo sapiens pleckstrin homology-like domain, family B, member 2 (PHLDE
NMJ45754 Homo sapiens kinesin family member C2 (KIFC2), mRNA
NMJ45755 Homo sapiens TPR domain containing STI2 (STI2), mRNA
NMJ45756 Homo sapiens zinc finger protein 396 (ZNF396), mRNA
NMJ45759 Homo sapiens TNF receptor-associated factor 5 (TRAF5), transcript variant I
NMJ 45762 Homo sapiens GDNF family receptor alpha 4 (GFRA4), transcript variant 2, r
NMJ 45763 Homo sapiens GDNF family receptor alpha 4 (GFRA4), transcript variant 3, r
NMJ 45764 Homo sapiens microsomal glutathione S-transferase 1 (MGST1 ), transcript v
NMJ45791 Homo sapiens microsomal glutathione S-transferase 1 (MGST1), transcript v
NMJ45792 Homo sapiens microsomal glutathione S-transferase 1 (MGST1), transcript v
NMJ45793 Homo sapiens GDNF family receptor alpha 1 (GFRA1), transcript variant 2, r
NMJ45794 Homo sapiens downstream neighbor of SON (DONSON), transcript variant 2
NMJ 45795 Homo sapiens downstream neighbor of SON (DONSON), transcript variant 3
NMJ 45796 Homo sapiens pogo transposable element with ZNF domain (POGZ), transcr
NMJ45798 Homo sapiens oxysterol binding protein-like 7 (OSBPL7), transcript variant 1
NMJ45799 Homo sapiens septin 6 (SEPT6), transcript variant I, mRNA
NMJ45800 Homo sapiens septin 6 (SEPT6), transcript variant III, mRNA
NMJ 45802 Homo sapiens septin 6 (SEPT6), transcript variant V, mRNA
NMJ45803 Homo sapiens TNF receptor-associated factor 6 (TRAF6), transcript variant '
NMJ 45804 Homo sapiens ankyrin repeat and BTB (POZ) domain containing 2 (ABTB2),
NMJ45805 Homo sapiens ISL2 transcription factor, LIM/homeodomain, (islet-2) (ISL2), r
NMJ45806 Homo sapiens zinc finger protein 511 (ZNF511), mRNA
NMJ45807 Homo sapiens hypothetical protein BC018697 (LOC126147), mRNA
NMJ 45808 Homo sapiens myotrophin (MTPN), mRNA
NMJ 45809 Homo sapiens TL132 protein (LOC220594), mRNA
NMJ45810 Homo sapiens cell division cycle associated 7 (CDCA7), transcript variant 2,
NMJ45811 Homo sapiens calcium channel, voltage-dependent, gamma subunit 5 (CACI
NMJ45812 Homo sapiens programmed cell death 8 (apoptosis-inducing factor) (PDCD8
NMJ 45813 Homo sapiens programmed cell death 8 (apoptosis-inducing factor) (PDCD8
NMJ 45814 Homo sapiens calcium channel, voltage-dependent, gamma subunit 6 (CACI
NMJ 45815 Homo sapiens calcium channel, voltage-dependent, gamma subunit 6 (CACI
NMJ 45818 Homo sapiens component of oligomeric golgi complex 4 (COG4), transcript \
NMJ45858 Homo sapiens crystallin, zeta (quinone reductase)-like 1 (CRYZL1), transcrip
NMJ45859 Homo sapiens programmed cell death 10 (PDCD10), transcript variant 2, mF
NMJ 45860 Homo sapiens programmed cell death 10 (PDCD10), transcript variant 3, mF
NMJ 45861 Homo sapiens EDAR-associated death domain (EDARADD), transcript varia
NMJ 45862 Homo sapiens CHK2 checkpoint homolog (S. pombe) (CHEK2), transcript va
NMJ 45863 Homo sapiens ankyrin repeat and SOCS box-containing 3 (ASB3), transcript
NMJ45864 Homo sapiens kallikrein 3, (prostate specific antigen) (KLK3), transcript varia
NMJ45865 Homo sapiens hypothetical protein FLJ38819 (FLJ38819), mRNA
NM J45867 Homo sapiens leukotriene C4 synthase (LTC4S), transcript variant 1 , mRNA
NMJ 45868 Homo sapiens annexin A11 (ANXA11), transcript variant b, mRNA
NMJ 45869 Homo sapiens annexin A11 (ANXA11), transcript variant c, mRNA
NMJ 45870 Homo sapiens glutathione transferase zeta 1 (maleylacetoacetate isomerase
NMJ 45871 Homo sapiens glutathione transferase zeta 1 (maleylacetoacetate isomerase
NMJ 45872 Homo sapiens ankyrin repeat and SOCS box-containing 4 (ASB4), transcript
NMJ 45886 Homo sapiens leucine-rich repeats and death domain containing (LRDD), tra
NMJ45887 Homo sapiens leucine-rich repeats and death domain containing (LRDD), tra
NMJ 45888 Homo sapiens kallikrein 10 (KLK10), transcript variant 2, mRNA
NMJ 45891 Homo sapiens ataxin 2-binding protein 1 (A2BP1), transcript variant 1 , mRN/
NMJ 45892 Homo sapiens ataxin 2-binding protein 1 (A2BP1), transcript variant 2, mRN/
NMJ 45893 Homo sapiens ataxin 2-binding protein 1 (A2BP1 ), transcript variant 3, mRN/
NMJ45894 Homo sapiens kallikrein 12 (KLK12), transcript variant 2, mRNA
NMJ45895 Homo sapiens kallikrein 12 (KLK12), transcript variant 3, mRNA
NMJ 45896 Homo sapiens prefoldin 5 (PFDN5), transcript variant 2, mRNA
NMJ 45897 Homo sapiens prefoldin 5 (PFDN5), transcript variant 3, mRNA
NMJ 45898 Homo sapiens chemokine (C-C motif) ligand 23 (CCL23), transcript variant C
NMJ 45899 Homo sapiens high mobility group AT-hook 1 (HMGA1), transcript variant 1 , 1
NMJ45901 Homo sapiens high mobility group AT-hook 1 (HMGA1), transcript variant 3, 1
NMJ 45902 Homo sapiens high mobility group AT-hook 1 (HMGA1), transcript variant 4, 1
NMJ 45903 Homo sapiens high mobility group AT-hook 1 (HMGA1), transcript variant 5, i
NMJ 45904 Homo sapiens high mobility group AT-hook 1 (HMGA1), transcript variant 6, i
NMJ45905 Homo sapiens high mobility group AT-hook 1 (HMGA1), transcript variant 7, i
NMJ 45906 Homo sapiens RIO kinase 3 (yeast) (RIOK3), transcript variant 2, mRNA
NMJ 45909 Homo sapiens zinc finger protein 323 (ZNF323), mRNA
NMJ45910 Homo sapiens NIMA (never in mitosis gene a)- related kinase 11 (NEK11), rr
NMJ 45911 Homo sapiens zinc finger protein 23 (KOX 16) (ZNF23), mRNA
NMJ45912 Homo sapiens NFAT activation molecule 1 (NFAM1), mRNA
NMJ 45913 Homo sapiens solute carrier family 5 (iodide transporter), member 8 (SLC5A!
NMJ45914 Homo sapiens zinc finger protein 38 (ZNF38), mRNA
NMJ 45918 Homo sapiens cathepsin L (CTSL), transcript variant 2, mRNA
NMJ46387 Homo sapiens mitochondrial ribosomal protein L4 (MRPL4), nuclear gene en
NMJ46388 Homo sapiens mitochondrial ribosomal protein L4 (MRPL4), nuclear gene en
NMJ46421 Homo sapiens glutathione S-transferase M1 (GSTM1), transcript variant 2, rr
NMJ 47127 Homo sapiens Ellis van Creveld syndrome 2 (limbin) (EVC2), mRNA
NMJ 47128 Homo sapiens zinc and ring finger 2 (ZNRF2), mRNA
NMJ 47129 Homo sapiens hypothetical protein LOC259173 (FLJ36525), transcript variar
NMJ47130 Homo sapiens natural cytotoxicity triggering receptor 3 (NCR3), mRNA
NMJ47131 Homo sapiens gaIactose-1 -phosphate uridylyltransferase (GALT), transcript >
NMJ47132 Homo sapiens galactose-1-phosphate uridylyltransferase (GALT), transcript >
NMJ47133 Homo sapiens nuclear transcription factor, X-box binding 1 (NFX1), transcrip
NMJ 47134 Homo sapiens nuclear transcription factor, X-box binding 1 (NFX1), transcrip
NMJ47147 Homo sapiens blood vessel epicardial substance (BVES), transcript variant E
NMJ47148 Homo sapiens glutathione S-transferase M4 (GSTM4), transcript variant 2, rr
NMJ47149 Homo sapiens glutathione S-transferase M4 (GSTM4), transcript variant 3, rr
NMJ 47150 Homo sapiens A kinase (PRKA) anchor protein 2 (AKAP2), transcript variant
NMJ 47152 Homo sapiens intersectin 2 (ITSN2), transcript variant 2, mRNA
NMJ 47156 Homo sapiens transmembrane protein 23 (TMEM23), mRNA
NMJ 47157 Homo sapiens opioid receptor, sigma 1 (OPRS1), transcript variant 2, mRNA
NMJ 47158 Homo sapiens opioid receptor, sigma 1 (OPRS1), transcript variant 3, mRNA
NMJ47159 Homo sapiens opioid receptor, sigma 1 (OPRS1), transcript variant 4, mRNA
NMJ47160 Homo sapiens opioid receptor, sigma 1 (OPRS1), transcript variant 5, mRNA
NMJ47161 Homo sapiens thioesterase, adipose associated (THEA), transcript variant 2,
NMJ47162 Homo sapiens interleukin 1 1 receptor, alpha (IL11 RA), transcript variant 2, rr
NMJ47164 Homo sapiens ciliary neuro rophic factor receptor (CNTFR), transcript varian
NMJ 47166 Homo sapiens A kinase (PRKA) anchor protein (yotiao) 9 (AKAP9), transcrip
NMJ47168 Homo sapiens chromosome 9 open reading frame 24 (C9orf24), transcript vs
NMJ47169 Homo sapiens chromosome 9 open reading frame 24 (C9orf24), transcript vs
NMJ47171 Homo sapiens A kinase (PRKA) anchor protein (yotiao) 9 (AKAP9), transcrip
NMJ47172 Homo sapiens nudix (nucleoside diphosphate linked moiety X)-type motif 2 (I
NMJ47173 Homo sapiens nudix (nucleoside diphosphate linked moiety X)-type motif 2 (I
NM J47174 Homo sapiens heparan sulfate 6-O-sulfotransferase 2 (HS6ST2), mRNA
NM J47175 Homo sapiens heparan sulfate 6-O-sulfotransferase 2 (HS6ST2), transcript v
NMJ47180 Homo sapiens protein phosphatase 3 (formerly 2B), regulatory subunit B, 191
NMJ47181 Homo sapiens Kv channel interacting protein 4 (KCNIP4), transcript variant 2
NMJ47182 Homo sapiens Kv channel interacting protein 4 (KCNIP4), transcript variant 3
NMJ47183 Homo sapiens Kv channel interacting protein 4 (KCNIP4), transcript variant 4
NMJ 47184 Homo sapiens tumor protein p53 inducible protein 3 (TP53I3), transcript vari;
NMJ47185 Homo sapiens A kinase (PRKA) anchor protein (yotiao) 9 (AKAP9), transcrip
NMJ 47187 Homo sapiens tumor necrosis factor receptor superfamily, member 10b (TNF
NMJ 47188 Homo sapiens F-box protein 22 (FBX022), transcript variant 1 , mRNA
NMJ47189 Homo sapiens hypothetical protein MGC39325 (MGC39325), mRNA
NMJ47190 Homo sapiens LAG1 longevity assurance homolog 5 (S. cerevisiae) (LASS5)
NMJ47191 Homo sapiens matrix metalloproteinase 21 (MMP21), mRNA
NMJ 47192 Homo sapiens diencephalon/mesencephalon homeobox 1 (DMBX1), transcri
NMJ 47193 Homo sapiens GLIS family zinc finger 1 (GLIS1 ), mRNA
NMJ47194 Homo sapiens hypothetical protein MGC35361 (MGC35361), mRNA
NMJ47195 Homo sapiens FLJ35740 protein (FLJ35740), mRNA
NMJ47196 Homo sapiens transmembrane inner ear (TMIE), mRNA
NMJ47197 Homo sapiens WAP four-disulfide core domain 11 (WFDC11), mRNA
NMJ 47198 Homo sapiens WAP four- isulfide core domain 9 (WFDC9), mRNA
NMJ 47199 Homo sapiens G protein-coupled receptor MRGX1 (MRGX1), mRNA
NMJ47200 Homo sapiens chromosome 6 open reading frame 4 (C6orf4), transcript varis
NMJ47202 Homo sapiens chromosome 9 open reading frame 25 (C9orf25), mRNA
NMJ47203 Homo sapiens fibrinogen-like 1 (FGL1), transcript variant 2, mRNA
NMJ47204 Homo sapiens transient receptor potential cation channel, subfamily V, mem
NM J47223 Homo sapiens nuclear receptor coactivator 1 (NCOA1 ), transcript variant 2, r
NMJ 47233 Homo sapiens nuclear receptor coactivator 1 (NCOA1 ), transcript variant 3, r
NMJ 47686 Homo sapiens chromosome 6 open reading frame 4 (C6orf4), transcript varis
NMJ47777 Homo sapiens sorting nexin 15 (SNX15), transcript variant B, mRNA
NMJ 47780 Homo sapiens cathepsin B (CTSB), transcript variant 2, mRNA
NMJ47781 Homo sapiens cathepsin B (CTSB), transcript variant 3, mRNA
NMJ47782 Homo sapiens cathepsin B (CTSB), transcript variant 4, mRNA
NMJ47783 Homo sapiens cathepsin B (CTSB), transcript variant 5, mRNA
NMJ 48169 Homo sapiens F-box protein 17 (FBX017), transcript variant 1 , mRNA
NMJ 48170 Homo sapiens cathepsin C (CTSC), transcript variant 2, mRNA
NMJ 48171 Homo sapiens ubiquitin associated protein 2 (UBAP2), transcript variant 3, rr
NMJ48172 Homo sapiens phosphatidylethanolamine N-methyltransferase (PEMT), nucl<
NMJ 48173 Homo sapiens phosphatidylethanolamine N-methyltransferase (PEMT), nucl<
NMJ48174 Homo sapiens ornithine decarboxylase antizyme inhibitor (OAZIN), transcript
NMJ48175 Homo sapiens peptidylprolyl isomerase (cyclophilin)-like 2 (PPIL2), transcripl
NMJ 48176 Homo sapiens peptidylprolyl isomerase (cyclophilin)-like 2 (PPIL2), transcripl
NMJ48177 Homo sapiens F-box protein 32 (FBX032), transcript variant 2, mRNA
NMJ48178 Homo sapiens chromosome 9 open reading frame 23 (C9orf23), transcript vs
NMJ48179 Homo sapiens chromosome 9 open reading frame 23 (C9orf23), transcript vs
NMJ48414 Homo sapiens ataxin 2 related protein (A2LP), transcript variant C, mRNA
NMJ48415 Homo sapiens ataxin 2 related protein (A2LP), transcript variant D, mRNA
NMJ48416 Homo sapiens ataxin 2 related protein (A2LP), transcript variant E, mRNA
NMJ 48570 Homo sapiens mitochondrial ribosomal protein L27 (MRPL27), nuclear gene
NMJ 48571 Homo sapiens mitochondrial ribosomal protein L27 (MRPL27), nuclear gene
NMJ 48672 Homo sapiens chemokine (C-C motif) ligand 28 (CCL28), transcript variant 2
NMJ 48674 Homo sapiens SMC1 structural maintenance of chromosomes 1-like 2 (yeasl
NMJ48675 Homo sapiens Down syndrome critical region gene 9 (DSCR9), mRNA
NMJ48676 Homo sapiens Down syndrome critical region gene 10 (DSCR10), mRNA
NMJ48842 Homo sapiens Williams-Beuren syndrome chromosome region 16 (WBSCR1
NMJ48886 Homo sapiens Smith-Magenis syndrome chromosome region, candidate 7 (£
NMJ 48887 Homo sapiens mitochondrial ribosomal protein L10 (MRPL10), nuclear gene
NMJ 48888 Homo sapiens chemokine (C-C motif) ligand 25 (CCL25), transcript variant 2
NM J48894 Homo sapiens family with sequence similarity 44, member A (FAM44A), mRt
NMJ48896 Homo sapiens preproneuropeptide B (NPB), mRNA
NMJ48897 Homo sapiens orphan short-chain dehydrogenase / reductase (SDR-O), mRI
NMJ48898 Homo sapiens forkhead box P2 (FOXP2), transcript variant 2, mRNA
NMJ48899 Homo sapiens forkhead box P2 (FOXP2), transcript variant 3, mRNA
NMJ 48900 Homo sapiens forkhead box P2 (FOXP2), transcript variant 4, mRNA
NMJ48901 Homo sapiens tumor necrosis factor receptor superfamily, member 18 (TNFF
NMJ 48902 Homo sapiens tumor necrosis factor receptor superfamily, member 18 (TNFF
NMJ 48903 Homo sapiens GREB1 protein (GREB1), transcript variant c, mRNA
NMJ 48904 Homo sapiens oxysterol binding protein-like 9 (OSBPL9), transcript variant 1
NMJ 48905 Homo sapiens oxysterol binding protein-like 9 (OSBPL9), transcript variant 2
NMJ 48906 Homo sapiens oxysterol binding protein-like 9 (OSBPL9), transcript variant 3
NMJ48907 Homo sapiens oxysterol binding protein-like 9 (OSBPL9), transcript variant 4
NMJ48908 Homo sapiens oxysterol binding protein-like 9 (OSBPL9), transcript variant 5
NMJ48909 Homo sapiens oxysterol binding protein-like 9 (OSBPL9), transcript variant 7
NMJ48910 Homo sapiens toll-interleukin 1 receptor (TIR) domain containing adaptor pro
NMJ 48911 Homo sapiens CHRNA7 (cholinergic receptor, nicotinic, alpha polypeptide 7,
NMJ48912 Homo sapiens Williams Beuren syndrome chromosome region 21 (WBSCRΣ
NMJ 48913 Homo sapiens Williams Beuren syndrome chromosome region 21 (WBSCRΣ
NMJ48914 Homo sapiens Williams Beuren syndrome chromosome region 21 (WBSCR2
NMJ48915 Homo sapiens Williams Beuren syndrome chromosome region 21 (WBSCRΣ
NMJ48916 Homo sapiens Williams Beuren syndrome chromosome region 21 (WBSCRΣ
NMJ48918 Homo sapiens serine hydroxymethyltransferase 1 (soluble) (SHMT1), transcr
NMJ48919 Homo sapiens proteasome (prosome, macropain) subunit, beta type, 8 (large
NMJ48920 Homo sapiens phosphatidylinositol glycan, class Q (PIGQ), transcript variant
NMJ48921 Homo sapiens epsin 2 (EPN2), transcript variant 1 , mRNA
NMJ 48923 Homo sapiens cytochrome b-5 (CYB5), mRNA
NMJ48936 Homo sapiens Williams Beuren syndrome chromosome region 20C (WBSCF
NMJ 48954 Homo sapiens proteasome (prosome, macropain) subunit, beta type, 9 (large
NMJ48955 Homo sapiens sorting nexin 1 (SNX1 ), transcript variant 2, mRNA
NMJ48956 Homo sapiens Williams Beuren syndrome chromosome region 20A (WBSCF
NMJ48957 Homo sapiens tumor necrosis factor receptor superfamily, member 19 (TNFF
NMJ48959 Homo sapiens HUS1 checkpoint homolog b (S. pombe) (HUS1 B), mRNA
NMJ48960 Homo sapiens claudin 19 (CLDN19), mRNA
NMJ 48961 Homo sapiens otospiralin (OTOS), mRNA
NMJ 48962 Homo sapiens oxoeicosanoid (OXE) receptor 1 (OXER1), mRNA
NM J48963 Homo sapiens G protein-coupled receptor, family C, group 6, member A (GP
NMJ48964 Homo sapiens cathepsin E (CTSE), transcript variant 2, mRNA
NMJ 48965 Homo sapiens tumor necrosis factor receptor superfamily, member 25 (TNFF
NMJ48966 Homo sapiens tumor necrosis factor receptor superfamily, member 25 (TNFF
NMJ 48967 Homo sapiens tumor necrosis factor receptor superfamily, member 25 (TNFF
NMJ48968 Homo sapiens tumor necrosis factor receptor superfamily, member 25 (TNFF
NMJ48969 Homo sapiens tumor necrosis factor receptor superfamily, member 25 (TNFF
NMJ48970 Homo sapiens tumor necrosis factor receptor superfamily, member 25 (TNFF
NMJ48971 Homo sapiens tumor necrosis factor receptor superfamily, member 25 (TNFF
NMJ48972 Homo sapiens tumor necrosis factor receptor superfamily, member 25 (TNFF
NMJ48973 Homo sapiens tumor necrosis factor receptor superfamily, member 25 (TNFF
NMJ48974 Homo sapiens tumor necrosis factor receptor superfamily, member 25 (TNFF
NM J48975 Homo sapiens membrane-spanning 4-domains, subfamily A, member 4 (MS'
NMJ48976 Homo sapiens proteasome (prosome, macropain) subunit, alpha type, 1 (PSI
NMJ48977 Homo sapiens pantothenate kinase 1 (PANK1 ), transcript variant alpha, mRC
NMJ48978 Homo sapiens pantothenate kinase 1 (PANK1), transcript variant beta, mRNi
NMJ48979 Homo sapiens cathepsin H (CTSH), transcript variant 2, mRNA
NMJ48980 Homo sapiens Williams Beuren syndrome chromosome region 20C (WBSCF
NMJ49379 Homo sapiens Williams Beuren syndrome chromosome region 20C (WBSCF
NMJ 52132 Homo sapiens proteasome (prosome, macropain) subunit, alpha type, 3 (PSI
NMJ52133 Homo sapiens T-cell activation GTPase activating protein (TAGAP), transcrip
NMJ 52219 Homo sapiens gap junction protein, chi 1 , 31.9kDa (connexin 31.9) (GJC1), r
NMJ 52221 Homo sapiens casein kinase 1 , epsilon (CSNK1 E), transcript variant 1 , mRN
NMJ 52222 Homo sapiens tumor necrosis factor receptor superfamily, member 19-like (7
NMJ 52223 Homo sapiens protein phosphatase, EF hand calcium-binding domain 1 (PPI
NMJ 52224 Homo sapiens protein phosphatase, EF hand calcium-binding domain 1 (PPI
NMJ 52225 Homo sapiens protein phosphatase, EF hand calcium-binding domain 1 (PPI
NMJ 52226 Homo sapiens protein phosphatase, EF hand calcium-binding domain 1 (PPI
NMJ 52227 Homo sapiens sorting nexin 5 (SNX5), transcript variant 1 , mRNA
NMJ 52230 Homo sapiens inositol polyphosphate multikinase (IPMK), mRNA
NMJ 52232 Homo sapiens taste receptor, type 1 , member 2 (TAS1R2), mRNA
NMJ 52233 Homo sapiens sorting nexin 6 (SNX6), transcript variant 2, mRNA
NMJ 52235 Homo sapiens splicing factor, arginine/serine-rich 8 (suppressor-of-white-apr
NMJ 52236 Homo sapiens growth arrest-specific 2 like 1 (GAS2L1), transcript variant 2, i
NMJ 52237 Homo sapiens growth arrest-specific 2 like 1 (GAS2L1), transcript variant 3, i
NMJ 52238 Homo sapiens sorting nexin 7 (SNX7), transcript variant 2, mRNA
NMJ 52240 Homo sapiens p53 target zinc finger protein (WIG1 ), transcript variant 2, mR
NMJ 52243 Homo sapiens CDC42 effector protein (Rho GTPase binding) 1 (CDC42EP1;
NMJ 52244 Homo sapiens sorting nexin 11 (SNX11 ), transcript variant 1 , mRNA
NMJ 52245 Homo sapiens carnitine palmitoyltransferase 1 B (muscle) (CPT1 B), nuclear c.
NMJ 52246 Homo sapiens carnitine palmitoyltransferase 1 B (muscle) (CPT1 B), nuclear f.
NMJ 52247 Homo sapiens carnitine palmitoyltransferase 1 B (muscle) (CPT1 B), nuclear <;
NMJ 52250 Homo sapiens defensin, beta 105 (DEFB105), mRNA
NMJ 52251 Homo sapiens defensin, beta 106 (DEFB106), mRNA
NMJ 52253 Homo sapiens choline kinase beta (CHKB), transcript variant 2, mRNA
NMJ 52255 Homo sapiens proteasome (prosome, macropain) subunit, alpha type, 7 (PSI
NMJ 52257 Homo sapiens KIAA0889 protein (KIAA0889), mRNA
NMJ 52259 Homo sapiens leucine-rich repeat kinase 1 (MGC45866), mRNA
NMJ 52260 Homo sapiens chromosome 15 open reading frame 19 (C15orf19), mRNA
NMJ 52261 Homo sapiens hypothetical protein MGC17943 (MGC17943), mRNA
NMJ 52262 Homo sapiens zinc finger protein 439 (ZNF439), mRNA
NMJ 52263 Homo sapiens tropomyosin 3 (TPM3), mRNA
NMJ 52264 Homo sapiens solute carrier family 39 (zinc transporter), member 13 (SLC39
NMJ 52266 Homo sapiens hypothetical protein MGC32020 (MGC32020), mRNA
NMJ 52267 Homo sapiens hypothetical protein FLJ38628 (FLJ38628), mRNA
NMJ52268 Homo sapiens similar to tRNA synthetase class II (DKFZp727A071 ), mRNA
NMJ 52269 Homo sapiens hypothetical protein FLJ38663 (FLJ38663), mRNA
NMJ 52270 Homo sapiens hypothetical protein FLJ34922 (FLJ34922), mRNA
NMJ 52271 Homo sapiens hypothetical protein FLJ23749 (FLJ23749), mRNA
NMJ 52272 Homo sapiens hypothetical protein MGC29816 (MGC29816), mRNA
NMJ 52274 Homo sapiens hypothetical protein MGC29729 (MGC29729), mRNA
NMJ52275 Homo sapiens hypothetical protein FLJ13946 (FLJ13946), mRNA
NMJ 52277 Homo sapiens dendritic cell-derived ubiquitin-like protein (DC-UbP), mRNA
NMJ 52278 Homo sapiens hypothetical protein MGC23947 (MGC23947), mRNA
NMJ 52279 Homo sapiens zinc finger protein 585B (ZNF585B), mRNA
NMJ 52280 Homo sapiens synaptotagmin XI (SYT11 ), mRNA
NMJ 52281 Homo sapiens NTKL-binding protein 1 (FLJ11752), mRNA
NMJ 52282 Homo sapiens hypothetical protein FLJ23751 (FLJ23751), mRNA
NMJ 52283 Homo sapiens zinc finger protein 62 homolog (mouse) (ZFP62), mRNA
NMJ 52284 Homo sapiens Snf7 homologue associated with Alix 3 (Shax3), mRNA
NMJ 52285 Homo sapiens arrestin domain containing 1 (ARRDC1), mRNA
NMJ 52286 Homo sapiens chromosome 9 open reading frame 111 (C9orf111), mRNA
NMJ 52287 Homo sapiens zinc finger protein 276 homolog (mouse) (ZFP276), mRNA
NMJ 52288 Homo sapiens hypothetical protein MGC13024 (MGC13024), mRNA
NMJ 52289 Homo sapiens zinc finger protein 561 (ZNF561 ), mRNA
NMJ 52290 Homo sapiens hypothetical protein MGC35194 (MGC35194), mRNA
NMJ 52291 Homo sapiens mucin 7, salivary (MUC7), mRNA
NMJ 52292 Homo sapiens RNA (guanine-9-) methyltransferase domain containing 2 (RG
NMJ 52295 Homo sapiens threonyl-tRNA synthetase (TARS), mRNA
NMJ 52296 Homo sapiens ATPase, Na+/K+ transporting, alpha 3 polypeptide (ATP1A3),
NMJ 52298 Homo sapiens nuclear autoantigenic sperm protein (histone-binding) (NASP)
NMJ 52299 Homo sapiens hypothetical protein 384D8_6 (384D8-2), mRNA
NMJ 52300 Homo sapiens DEAD (Asp-Glu-Ala-Asp) box polypeptide 52 (DDX52), transc
NMJ 52301 Homo sapiens PP784 protein (PP784), transcript variant 1 , mRNA
NMJ 52302 Homo sapiens chromosome 20 open reading frame 158 (C20orf158), mRNA
NMJ 52303 Homo sapiens zinc finger protein 554 (ZNF554), mRNA
NMJ 52304 Homo sapiens hypothetical protein MGC45806 (MGC45806), mRNA
NMJ 52305 Homo sapiens x 010 protein (MDS010), mRNA
NMJ 52306 Homo sapiens ubiquitin-like, containing PHD and RING finger domains, 2 (Ul
NMJ 52307 Homo sapiens hypothetical protein FLJ40452 (FLJ40452), mRNA
NMJ 52308 Homo sapiens hypothetical protein MGC24665 (MGC24665), mRNA
NMJ 52309 Homo sapiens phosphoinositide-3-kinase adaptor protein 1 (PIK3AP1), mRN
NMJ52310 Homo sapiens elongation of very long chain fatty acids (FEN1/Elo2, SUR4/EI
NMJ 52311 Homo sapiens hypothetical protein MGC32871 (MGC32871 ), mRNA
NMJ 52312 Homo sapiens glycosyltransferase-like 1 B (GYLTL1 B), mRNA
NMJ 52313 Homo sapiens solute carrier family 36 (proton/amino acid symporter), rπembi
NMJ52314 Homo sapiens hypothetical protein MGC34830 (MGC34830), mRNA
NMJ 52315 Homo sapiens hypothetical protein MGC34290 (MGC34290), mRNA
NMJ 52316 Homo sapiens hypothetical protein FLJ38968 (FLJ38968), mRNA
NMJ 52317 Homo sapiens DEP domain containing 4 (DEPDC4), mRNA
NMJ 52318 Homo sapiens hypothetical protein MGC40397 (MGC40397), mRNA
NMJ 52319 Homo sapiens hypothetical protein MGC35033 (MGC35033), mRNA
NMJ 52320 Homo sapiens hypothetical protein FLJ31295 (FLJ31295), mRNA
NMJ 52321 Homo sapiens hypothetical protein FLJ32115 (FLJ32115), mRNA
NMJ 52322 Homo sapiens BTB (POZ) domain containing 11 (BTBD11 ), mRNA
NMJ 52323 Homo sapiens Spi-C transcription factor (Spi-1/PU.1 related) (SPIC), mRNA
NMJ 52324 Homo sapiens hypothetical protein MGC35169 (MGC35169), mRNA
NMJ 52325 Homo sapiens hypothetical protein MGC40178 (MGC40178), mRNA
NMJ 52326 Homo sapiens ankyrin repeat domain 9 (ANKRD9), mRNA
NMJ 52327 Homo sapiens adenylate kinase 7 (AK7), mRNA
NMJ 52328 Homo sapiens adenylosuccinate synthase like 1 (ADSSL1 ), transcript variant
NMJ 52329 Homo sapiens peptidylprolyl isomerase (cyclophilin) like 5 (PPIL5), transcripl
NMJ 52330 Homo sapiens chromosome 14 open reading frame 31 (C14orf31), mRNA
NMJ 52331 Homo sapiens peroxisomal acyl-CoA thioesterase 2B (PTE2B), mRNA
NMJ 52332 Homo sapiens membrane targeting (tandem) C2 domain containing 1 (MTAC
NMJ 52333 Homo sapiens solute carrier family 25, member 29 (SLC25A29), mRNA
NMJ 52334 Homo sapiens FLJ25005 protein (FLJ25005), mRNA
NMJ 52335 Homo sapiens chromosome 15 open reading frame 27 (C15orf27), mRNA
NMJ52336 Homo sapiens hypothetical protein FLJ32310 (FLJ32310), mRNA
NMJ 52337 Homo sapiens hypothetical protein FLJ32702 (FLJ32702), mRNA
NMJ 52338 Homo sapiens zymogen granule protein 16 (ZG16), mRNA
NMJ 52339 Homo sapiens hypothetical protein MGC26885 (MGC26885), mRNA
NMJ 52340 Homo sapiens hypothetical protein FLJ39075 (FLJ39075), mRNA
NMJ 52341 Homo sapiens progestin and adipoQ receptor family member IV (PAQR4), m
NMJ 52342 Homo sapiens chromodomain protein, Y-like 2 (CDYL2), mRNA
NMJ52343 Homo sapiens hypothetical protein FLJ25414 (FLJ25414), mRNA
NMJ 52344 Homo sapiens hypothetical protein FLJ30656 (FLJ30656), mRNA
NMJ 52345 Homo sapiens hypothetical protein FLJ25555 (FLJ25555), mRNA
NMJ 52346 Homo sapiens hypothetical protein MGC34680 (MGC34680), mRNA
NMJ 52347 Homo sapiens hypothetical protein FLJ40342 (FLJ40342), mRNA
NMJ 52348 Homo sapiens hypothetical protein FLJ33817 (FLJ33817), mRNA
NMJ 52349 Homo sapiens hypothetical protein MGC45562 (MGC45562), mRNA
NMJ 52350 Homo sapiens hypothetical protein MGC40157 (MGC40157), mRNA
NMJ 52351 Homo sapiens solute carrier family 5 (sodium/glucose cotransporter), membe
NMJ 52352 Homo sapiens chromosome 18 open reading frame 19 (C18orf19), mRNA
NMJ 52353 Homo sapiens hypothetical protein MGC33839 (MGC33839), mRNA
NMJ 52354 Homo sapiens zinc finger protein 285 (ZNF285), mRNA
NMJ 52355 Homo sapiens zinc finger protein 441 (ZNF441), mRNA
NMJ 52356 Homo sapiens zinc finger protein 491 (ZNF491), mRNA
NMJ 52357 Homo sapiens zinc finger protein 440 (ZNF440), mRNA
NMJ 52358 Homo sapiens hypothetical protein MGC33947 (MGC33947), mRNA
NMJ 52359 Homo sapiens carnitine palmitoyltransferase 1 C (CPT1 C), mRNA
NMJ 52360 Homo sapiens zinc finger protein 573 (ZNF573), mRNA
NMJ 52361 Homo sapiens hypothetical protein FLJ38944 (FLJ38944), mRNA
NMJ 52362 Homo sapiens hypothetical protein MGC17791 (MGC17791 ), mRNA
NMJ 52363 Homo sapiens hypothetical protein FLJ39369 (FLJ39369), mRNA
NMJ 52365 Homo sapiens hypothetical protein FLJ34633 (FLJ34633), mRNA
NMJ 52366 Homo sapiens hypothetical protein MGC33338 (MGC33338), mRNA
NMJ 52367 Homo sapiens hypothetical protein FLJ38716 (FLJ38716), mRNA
NMJ 52369 Homo sapiens hypothetical protein MGC45474 (MGC45474), mRNA
NMJ 52371 Homo sapiens hypothetical protein MGC26818 (MGC26818), mRNA
NMJ 52372 Homo sapiens myomesin family, member 3 (MYOM3), mRNA
NMJ 52373 Homo sapiens hypothetical protein MGC27466 (MGC27466), mRNA
NMJ 52374 Homo sapiens hypothetical protein FLJ38984 (FLJ38984), mRNA
NMJ 52375 Homo sapiens hypothetical protein FLJ38753 (FLJ38753), mRNA
NMJ 52376 Homo sapiens UBX domain containing 3 (UBXD3), mRNA
NMJ 52377 Homo sapiens hypothetical protein MGC34837 (MGC34837), mRNA
NMJ 52378 Homo sapiens hypothetical protein FLJ31052 (FLJ31052), mRNA
NMJ 52379 Homo sapiens hypothetical protein DKFZp547B1713 (DKFZp547B1713), mF
NMJ 52382 Homo sapiens hypothetical protein FLJ37953 (FLJ37953), mRNA
NMJ 52383 Homo sapiens hypothetical protein MGC42174 (MGC42174), mRNA
NMJ 52384 Homo sapiens Bardet-Biedl syndrome 5 (BBS5), mRNA
NMJ 52385 Homo sapiens hypothetical protein FLJ31438 (FLJ31438), mRNA
NMJ 52386 Homo sapiens sphingosine-1 -phosphate phosphotase 2 (SGPP2), mRNA
NMJ 52387 Homo sapiens hypothetical protein FLJ31322 (FLJ31322), mRNA
NMJ 52388 Homo sapiens amyotrophic lateral sclerosis 2 (juvenile) chromosome region,
NMJ 52389 Homo sapiens hypothetical protein MGC35338 (MGC35338), mRNA
NMJ 52390 Homo sapiens hypothetical protein MGC33926 (MGC33926), mRNA
NMJ52391 Homo sapiens chromosome 2 open reading frame 22 (C2orf22), mRNA
NMJ 52392 Homo sapiens AHA1 , activator of heat shock 90kDa protein ATPase homoloj
NMJ 52393 Homo sapiens kelch repeat and BTB (POZ) domain containing 5 (KBTBD5),
NMJ 52394 Homo sapiens hypothetical protein MGC39662 (MGC39662), mRNA
NMJ 52395 Homo sapiens hypothetical protein FLJ31265 (FLJ31265), mRNA
NMJ 52396 Homo sapiens hypothetical protein MGC24132 (MGC24132), mRNA
NMJ 52397 Homo sapiens hypothetical protein MGC39725 (MGC39725), mRNA
NMJ 52398 Homo sapiens hypothetical protein MGC45416 (MGC45416), mRNA
NMJ 52399 Homo sapiens hypothetical protein FLJ30834 (FLJ30834), mRNA
NMJ 52400 Homo sapiens hypothetical protein FLJ39370 (FLJ39370), mRNA
NMJ 52401 Homo sapiens phosducin-like 2 (PDCL2), mRNA
NMJ 52402 Homo sapiens translocation associated membrane protein 1-like 1 (TRAM1 L
NMJ 52403 Homo sapiens hypothetical protein FLJ39155 (FLJ39155), transcript variant •
NMJ 52404 Homo sapiens hypothetical protein FLJ34658 (FLJ34658), mRNA
NMJ52405 Homo sapiens junction-mediating and regulatory protein (JMY), mRNA
NMJ 52407 Homo sapiens GrpE-like 2, mitochondrial (E. coli) (GRPEL2), mRNA
NMJ 52408 Homo sapiens hypothetical protein FLJ35779 (FLJ35779), mRNA
NMJ 52409 Homo sapiens hypothetical protein FLJ37562 (FLJ37562), mRNA
NMJ 52410 Homo sapiens PARK2 co-regulated (PACRG), mRNA
NMJ52411 Homo sapiens hypothetical protein DKFZp762l137 (DKFZp762l137), mRNA
NMJ 52412 Homo sapiens zinc finger protein 572 (ZNF572), mRNA
NMJ 52413 Homo sapiens hypothetical protein MGC33309 (MGC33309), mRNA
NMJ52414 Homo sapiens basic helix-loop-helix domain containing, class B, 5 (BHLHB5]
NMJ 52415 Homo sapiens hepatocellular carcinoma related protein 1 (FLJ32642), mRN/
NMJ52416 Homo sapiens hypothetical protein MGC40214 (MGC40214), mRNA
NMJ 52417 Homo sapiens hypothetical protein FLJ32370 (FLJ32370), mRNA
NMJ52418 Homo sapiens hypothetical protein FLJ35775 (FLJ35775), mRNA
NMJ52420 Homo sapiens chromosome 9 open reading frame 41 (C9orf41), mRNA
NMJ 52421 Homo sapiens hypothetical protein MGC20262 (MGC20262), mRNA
NMJ 52422 Homo sapiens protein tyrosine phosphatase domain containing 1 (PTPDC1),
NMJ 52423 Homo sapiens hypothetical protein FLJ33516 (FLJ33516), mRNA
NMJ 52424 Homo sapiens hypothetical protein FLJ39827 (FLJ39827), mRNA
NMJ 52425 Homo sapiens hypothetical protein FLJ40249 (FLJ40249), mRNA
NMJ 52427 Homo sapiens cofilin pseudogene 1 (CFLP1), mRNA
NMJ 52428 Homo sapiens FERM and PDZ domain containing 2 (FRMPD2), mRNA
NMJ 52429 Homo sapiens chromosome 10 open reading frame 13 (C10orf13), mRNA
NMJ52430 Homo sapiens hypothetical protein MGC24137 (MGC24137), mRNA
NMJ52431 Homo sapiens piwi-like 4 (Drosophila) (PIWIL4), mRNA
NMJ 52433 Homo sapiens kelch repeat and BTB (POZ) domain containing 3 (KBTBD3),
NMJ 52434 Homo sapiens CWF19-Iike 2, cell cycle control (S. pombe) (CWF19L2), mRr-
NMJ 52435 Homo sapiens hypothetical protein MGC35366 (MGC35366), mRNA
NMJ 52436 Homo sapiens hypothetical protein MGC39497 (MGC39497), mRNA
NMJ 52437 Homo sapiens hypothetical protein DKFZp761 B128 (DKFZp761 B128), mRN;
NMJ 52439 Homo sapiens vitelliform macular dystrophy 2-like 3 (VMD2L3), mRNA
NMJ 52440 Homo sapiens hypothetical protein FLJ32549 (FLJ32549), mRNA
NMJ52441 Homo sapiens F-box and leucine-rich repeat protein 14 (FBXL14), mRNA
NMJ 52442 Homo sapiens RAD9 homolog B (S. cerevisiae) (RAD9B), mRNA
NMJ52443 Homo sapiens retinol dehydrogenase 12 (all-trans and 9-cis) (RDH12), mRN
NMJ 52444 Homo sapiens zinc binding alcohol dehydrogenase, domain containing 1 (ZA
NMJ52445 Homo sapiens chromosome 14 open reading frame 44 (C14orf44), mRNA
NMJ52447 Homo sapiens leucine rich repeat and fibronectin type III domain containing !
NMJ 52448 Homo sapiens hypothetical protein MGC33951 (MGC33951), mRNA
NMJ 52449 Homo sapiens hypothetical protein FLJ33O08 (FLJ33008), mRNA
NMJ 52450 Homo sapiens hypothetical protein MGC26690 (MGC26690), mRNA
NMJ 52451 Homo sapiens GRINL1A complex upstream protein (Gup1), mRNA
NMJ 52453 Homo sapiens hypothetical protein MGC35118 (MGC35118), mRNA
NMJ 52454 Homo sapiens hypothetical protein FLJ31461 (FLJ31461), mRNA
NMJ 52455 Homo sapiens hypothetical protein FLJ35867 (FLJ35867), mRNA
NMJ 52456 Homo sapiens hypothetical protein MGC34647 (MGC34647), mRNA
NMJ 52457 Homo sapiens zinc finger protein 597 (ZN F597), mRNA
NMJ 52458 Homo sapiens hypothetical protein FLJ32130 (FLJ32130), mRNA
NMJ 52459 Homo sapiens hypothetical protein MGC45438 (MGC45438), mRNA
NMJ 52460 Homo sapiens hypothetical protein FLJ31882 (FLJ31882), mRNA
NMJ 52461 Homo sapiens ER to nucleus signalling 1 (ERN1), transcript variant 2, mRN/>
NMJ52462 Homo sapiens transmembrane protein 21A (TMEM21A), mRNA
NMJ 52463 Homo sapiens essential meiotic endonuclease 1 homolog 1 (S. pombe) (EMI
NMJ 52464 Homo sapiens chromosome 17 open reading frame 32 (C17orf32), mRNA
NMJ 52465 Homo sapiens hypothetical protein MGC39650 (MGC39650), mRNA
NMJ 52466 Homo sapiens hypothetical protein FLJ25168 (FLJ25168), mRNA
NMJ 52467 Homo sapiens kelch-like 10 (Drosophila) (KLHL10), mRNA
NMJ 52468 Homo sapiens epidermodysplasia verruciformis 2 (EVER2), mRNA
NMJ 52470 Homo sapiens chromosome 18 open reading frame 23 (C18orf23), mRNA
NMJ 52472 Homo sapiens zinc finger protein 578 (ZNF578), mRNA
NMJ 52473 Homo sapiens hypothetical protein FLJ32214 (FLJ32214), mRNA
NMJ 52474 Homo sapiens chromosome 19 open reading frame 18 (C19orf18), mRNA
NMJ 52475 Homo sapiens hypothetical protein MGC34079 (MGC34079), mRNA
NMJ52476 Homo sapiens zinc finger protein 560 (ZNF560), mRNA
NMJ 52477 Homo sapiens zinc finger protein 565 (ZNF565), mRNA
NMJ 52478 Homo sapiens zinc finger protein 583 (ZNF583), mRNA
NMJ 52479 Homo sapiens hypothetical protein MGC33962 (MGC33962), mRNA
NMJ 52480 Homo sapiens chromosome 19 open reading frame 23 (C19orf23), mRNA
NMJ 52481 Homo sapiens hypothetical protein FLJ25660 (FLJ25660), mRNA
NMJ 52482 Homo sapiens chromosome 19 open reading frame 25 (C19orf25), mRNA
NMJ 52483 Homo sapiens hypothetical protein FLJ25328 (FLJ25328), mRNA
NMJ52484 Homo sapiens zinc finger protein 569 (ZNF569), mRNA
NMJ 52485 Homo sapiens hypothetical protein FLJ25078 (FLJ25078), mRNA
NMJ 52486 Homo sapiens sterile alpha motif domain containing 11 (SAMD11), mRNA
NMJ 52487 Homo sapiens hypothetical protein FLJ31842 (FLJ31842), mRNA
NMJ 52488 Homo sapiens hypothetical protein FLJ32833 (FLJ32833), mRNA
NMJ 52489 Homo sapiens hypothetical protein MGC35130 (MGC35130), mRNA
NMJ52490 Homo sapiens beta 1 ,3-N-acetylgalactosaminyltransferase-ll (MGC39558), n
NMJ 52491 Homo sapiens hypothetical protein FLJ32569 (FLJ32569), mRNA
NMJ 52492 Homo sapiens hypothetical protein FLJ32825 (FLJ32825), mRNA
NMJ 52493 Homo sapiens FLJ25476 protein (FLJ25476), mRNA
NMJ 52494 Homo sapiens hypothetical protein FLJ32785 (FLJ32785), mRNA
NMJ 52495 Homo sapiens hypothetical protein FLJ38993 (FLJ38993), mRNA
NMJ 52496 Homo sapiens hypothetical protein FLJ31434 (FLJ31434), mRNA
NMJ 52497 Homo sapiens hypothetical protein FLJ32206 (FLJ32206), mRNA
NMJ 52498 Homo sapiens hypothetical protein FLJ32000 (FLJ32000), mRNA
NMJ 52499 Homo sapiens hypothetical protein MGC45441 (MGC45441), mRNA
NMJ 52500 Homo sapiens hypothetical protein FLJ33084 (FLJ33084), mRNA
NMJ52501 Homo sapiens interferon-inducible protein X (IFIX), transcript variant a1 , mRI
NMJ 52503 Homo sapiens chromosome 20 open reading frame 132 (C20orf132), transcr
NMJ 52504 Homo sapiens hypothetical protein FLJ25067 (FLJ25067), mRNA
NMJ 52505 Homo sapiens chromosome 21 open reading frame 13 (C21orf13), mRNA
NMJ 52506 Homo sapiens chromosome 21 open reading frame 129 (C21orf129), mRNA
NMJ 52507 Homo sapiens chromosome 21 open reading frame 128 (C21orf128), mRNA
NMJ 52509 Homo sapiens hypothetical protein FLJ31568 (FLJ31568), mRNA
NMJ 52510 Homo sapiens hypothetical protein MGC26710 (MGC26710), mRNA
NMJ 52511 Homo sapiens dual specificity phosphatase 18 (DUSP18), mRNA
NMJ 52512 Homo sapiens hypothetical protein FLJ25421 (FLJ25421), mRNA
NMJ 52515 Homo sapiens hypothetical protein FLJ40629 (FLJ40629), mRNA
NMJ 52516 Homo sapiens copper metabolism (Murrl) domain containing 1 (COMMD1),
NMJ 52517 Homo sapiens hypothetical protein FLJ30990 (FLJ30990), mRNA
NMJ 52519 Homo sapiens hypothetical protein FLJ23861 (FLJ23861), mRNA
NMJ 52520 Homo sapiens zinc finger protein 533 (ZNF533), mRNA
NMJ 52522 Homo sapiens ADP-ribosylation-like factor 6-interacting protein 6 (MGC3386
NMJ 52523 Homo sapiens hypothetical protein FLJ40432 (FLJ40432), mRNA
NMJ 52524 Homo sapiens shugoshin-like 2 (S. pombe) (SGOL2), mRNA
NMJ 52525 Homo sapiens hypothetical protein FLJ25351 (FLJ25351), mRNA
NMJ 52526 Homo sapiens amyotrophic lateral sclerosis 2 (juvenile) chromosome region,
NMJ 52527 Homo sapiens solute carrier family 16 (monocarboxylic acid transporters), mi
NMJ 52528 Homo sapiens WD repeat and SAM domain containing 1 (WDSAM1), mRNA
NMJ 52529 Homo sapiens G protein-coupled receptor 155 (GPR155), mRNA
NMJ52531 Homo sapiens hypothetical protein FLJ35155 (FLJ35155), mRNA
NMJ 52533 Homo sapiens hypothetical protein MGC34728 (MGC34728), mRNA
NMJ 52534 Homo sapiens hypothetical protein FLJ32685 (FLJ32685), mRNA
NMJ 52536 Homo sapiens FYVE, RhoGEF and PH domain containing 5 (FGD5), mRNA
NMJ 52538 Homo sapiens immunoglobulin superfamily, member 11 (IGSF11), mRNA
NMJ 52539 Homo sapiens hypothetical protein FLJ32859 (FLJ32859), mRNA
NMJ 52540 Homo sapiens sed family domain containing 2 (SCFD2), mRNA
NMJ 52542 Homo sapiens hypothetical protein DKFZp761G058 (DKFZp761G058), mRN
NMJ 52543 Homo sapiens hypothetical protein FLJ25371 (FLJ25371), mRNA
NMJ 52544 Homo sapiens hypothetical protein FLJ35725 (FLJ35725), mRNA
NMJ 52545 Homo sapiens RasGEF domain family, member 1 B (RASGEF1 B), mRNA
NMJ 52546 Homo sapiens hypothetical protein FLJ25286 (FLJ25286), mRNA
NMJ 52547 Homo sapiens butyrophilin-like 9 (BTNL9), mRNA
NMJ 52548 Homo sapiens hypothetical protein FLJ25333 (FLJ25333), mRNA
NMJ 52549 Homo sapiens hypothetical protein MGC39633 (MGC39633), mRNA
NMJ 52550 Homo sapiens SH3 domain containing ring finger 2 (SH3RF2), mRNA
NMJ 52551 Homo sapiens chromosome 6 open reading frame 151 (C6orf151), mRNA
NMJ 52552 Homo sapiens sterile alpha motif domain containing 3 (SAMD3), mRNA
NMJ 52553 Homo sapiens IBR domain containing 1 (IBRDC1 ), mRNA
NMJ 52554 Homo sapiens chromosome 6 open reading frame 195 (C6orf195), mRNA
NMJ52556 Homo sapiens hypothetical protein FLJ31818 (FLJ31818), mRNA
NMJ52557 Homo sapiens hypothetical protein FLJ31413 (FLJ31413), mRNA
NMJ 52558 Homo sapiens KIAA1023 protein (KIAA1023), mRNA
NMJ 52559 Homo sapiens Williams Beuren syndrome chromosome region 27 (WBSCRΣ
NMJ 52562 Homo sapiens cell division cycle associated 2 (CDCA2), mRNA
NMJ 52563 Homo sapiens hypothetical protein FLJ10661 (FLJ10661), mRNA
NMJ52564 Homo sapiens Cohen syndrome 1 (COH1), transcript variant 1 , mRNA
NMJ 52565 Homo sapiens ATPase, H+ transporting, lysosomal 38kDa, V0 subunit d isofi
NMJ 52568 Homo sapiens hypothetical protein FLJ25169 (FLJ25169), mRNA
NMJ 52569 Homo sapiens chromosome 9 open reading frame 66 (C9orf66), mRNA
NMJ 52570 Homo sapiens hypothetical protein FLJ31810 (FLJ31810), mRNA
NMJ 52571 Homo sapiens hypothetical protein FLJ36779 (FLJ36779), mRNA
NMJ 52572 Homo sapiens chromosome 9 open reading frame 98 (C9orf98), mRNA
NMJ 52573 Homo sapiens RAS and EF hand domain containing (RASEF), mRNA
NMJ 52574 Homo sapiens chromosome 9 open reading frame 52 (C9orf52), mRNA
NMJ 52577 Homo sapiens hypothetical protein FLJ25735 (FLJ25735), mRNA
NMJ 52578 Homo sapiens fragile X mental retardation 1 neighbor (FMR1 NB), mRNA
NMJ 52579 Homo sapiens hypothetical protein FLJ38564 (FLJ38564), mRNA
NMJ 52581 Homo sapiens motile sperm domain containing 2 (MOSPD2), mRNA
NMJ 52582 Homo sapiens hypothetical protein MGC27O05 (MGC27005), mRNA
NMJ 52583 Homo sapiens hypothetical protein MGC40O53 (MGC40053), mRNA
NMJ 52584 Homo sapiens heat shock transcription factor, Y-linked 1 (HSFY1 ), transcript
NMJ 52585 Homo sapiens RNA binding motif protein, V-linked, family 1 (MGC33094), ml
NMJ 52586 Homo sapiens ubiquitin specific protease 54 (USP54), mRNA
NMJ 52587 Homo sapiens hypothetical protein MGC33948 (MGC33948), mRNA
NMJ 52588 Homo sapiens hypothetical protein DKFZp762A217 (DKFZp762A217), mRN;
NMJ 52589 Homo sapiens hypothetical protein FLJ35821 (FLJ35821), mRNA
NMJ 52590 Homo sapiens hypothetical protein FLJ360O4 (FLJ36004), mRNA
NMJ 52591 Homo sapiens hypothetical protein FLJ35843 (FLJ35843), mRNA
NMJ52592 Homo sapiens chromosome 14 open reading frame 49 (C14orf49), mRNA
NMJ 52594 Homo sapiens sprouty-related, EVH1 domain containing 1 (SPRED1), mRN/
NMJ 52595 Homo sapiens piggyBac transposable element derived 4 (PGBD4), mRNA
NMJ 52596 Homo sapiens hypothetical protein MGC33637 (MGC33637), mRNA
NMJ 52597 Homo sapiens fibrous sheath interacting protein 1 (FSIP1), mRNA
NMJ 52598 Homo sapiens hypothetical protein FLJ35757 (FLJ35757), mRNA
NMJ 52599 Homo sapiens hypothetical protein FLJ35773 (FLJ35773), mRNA
NMJ 52600 Homo sapiens zinc finger protein 579 (ZNF579), mRNA
NMJ 52601 Homo sapiens hypothetical protein FLJ38281 (FLJ38281), mRNA
NMJ 52602 Homo sapiens zinc finger protein 433 (ZNF433), mRNA
NMJ 52603 Homo sapiens zinc finger protein 567 (ZNF567), mRNA
NMJ 52604 Homo sapiens zinc finger protein 383 (ZNF383), mRNA
NMJ 52605 Homo sapiens hypothetical protein FLJ37549 (FLJ37549), mRNA
NMJ 52606 Homo sapiens zinc finger protein 540 (ZNF540), mRNA
NMJ 52607 Homo sapiens hypothetical protein FLJ402O1 (FLJ40201), mRNA
NMJ 52608 Homo sapiens hypothetical protein FLJ35382 (FLJ35382), mRNA
NMJ 52609 Homo sapiens hypothetical protein FLJ32O01 (FLJ32001), mRNA
NMJ 52610 Homo sapiens hypothetical protein FLJ35728 (FLJ35728), mRNA
NMJ 52611 Homo sapiens chromosome 20 open reading frame 75 (C20orf75), mRNA
NMJ 52612 Homo sapiens hypothetical protein FLJ36046 (FLJ36046), mRNA
NMJ52613 Homo sapiens hypothetical protein MGC26816 (MGC26816), mRNA
NMJ52614 Homo sapiens hypothetical protein MGC35154 (MGC35154), mRNA
NMJ 52615 Homo sapiens hypothetical protein FLJ40597 (FLJ40597), mRNA
NMJ 52616 Homo sapiens tripartite motif-containing 42 (TRIM42), mRNA
NMJ 52617 Homo sapiens hypothetical protein FLJ35794 (FLJ35794), mRNA
NMJ 52618 Homo sapiens hypothetical protein FLJ35630 (FLJ35630), mRNA
NMJ 52619 Homo sapiens hypothetical protein MGC45428 (MGC45428), mRNA
NMJ52620 Homo sapiens ring finger protein 129 (RNF129), mRNA
NMJ 52621 Homo sapiens hypothetical protein MGC26963 (MGC26963), mRNA
NMJ 52622 Homo sapiens hypothetical protein FLJ35954 (FLJ35954), mRNA
NMJ 52623 Homo sapiens CDC20-like protein (FLJ37927), mRNA
NMJ 52624 Homo sapiens decapping enzyme hDcp2 (DCP2), mRNA
NMJ52625 Homo sapiens zinc finger protein 366 (ZNF366), mRNA
NMJ 52626 Homo sapiens zinc finger protein 92 (HTF12) (ZNF92), mRNA
NMJ 52628 Homo sapiens hypothetical protein MGC39715 (MGC39715), mRNA
NMJ 52629 Homo sapiens GLIS family zinc finger 3 (GLIS3), mRNA
NMJ 52630 Homo sapiens hypothetical protein MGC26999 (MGC26999), mRNA
N J 52631 Homo sapiens hypothetical protein FLJ35782 (FLJ35782), mRNA
NMJ52632 Homo sapiens chromosome X open reading frame 22 (CXorf22), mRNA
NMJ 52633 Homo sapiens hypothetical protein FLJ34064 (FLJ34064), mRNA
NMJ 52635 Homo sapiens oncoprotein induced transcript 3 (OIT3), mRNA
NMJ 52636 Homo sapiens hypothetical protein FLJ33979 (FLJ33979), mRNA
NMJ 52637 Homo sapiens hypothetical protein MGC17301 (MGC17301), mRNA
NMJ 52638 Homo sapiens hypothetical protein MGC26598 (MGC26598), mRNA
N J 52640 Homo sapiens decapping enzyme hDcpl b (DCP1 B), mRNA
NMJ 52643 Homo sapiens kinase non-catalytic C-lobe domain (KIND) containing 1 (KND
NMJ 52644 Homo sapiens family with sequence similarity 24, member B (FAM24B), mRt
NMJ 52647 Homo sapiens hypothetical protein FLJ32800 (FLJ32800), mRNA
NMJ 52649 Homo sapiens hypothetical protein FLJ34389 (FLJ34389), mRNA
NMJ 52652 Homo sapiens zinc finger protein 553 (ZNF553), mRNA
NMJ 52653 Homo sapiens ubiquitin-conjugating enzyme E2E 2 (UBC4/5 homolog, yeast
NMJ 52654 Homo sapiens hypothetical protein FLJ38607 (FLJ38607), mRNA
NMJ 52655 Homo sapiens zinc finger protein 585A (ZNF585A), transcript variant 1 , mRN
NMJ 52657 Homo sapiens gametogenetin (GGN), transcript variant 1 , mRNA
NMJ 52658 Homo sapiens THAP domain containing 8 (THAP8), mRNA
NMJ 52660 Homo sapiens hypothetical protein MGC34648 (MGC34648), mRNA
NMJ 52662 Homo sapiens hypothetical protein FLJ23867 (FLJ23867), mRNA
NMJ 52663 Homo sapiens Ral GEF with PH domain and SH3 binding motif 2 (RALGPS2
NMJ 52665 Homo sapiens hypothetical protein FLJ40873 (FLJ40873), mRNA
NMJ 52666 Homo sapiens hypothetical protein FLJ40773 (FLJ40773), mRNA
NMJ 52667 Homo sapiens chromosome 20 open reading frame 147 (C20orf147), mRNA
NMJ 52670 Homo sapiens hypothetical protein FLJ25369 (FLJ25369), mRNA
NMJ 52671 Homo sapiens phosphatidylinositol-3-phosphate/phosphatidylinositol 5-kinas
NMJ 52672 Homo sapiens organic solute transporter alpha (OSTalpha), mRNA
NMJ 52673 Homo sapiens mucin 20 (MUC20), mRNA
NMJ 52675 Homo sapiens hypothetical protein FLJ23754 (FLJ23754), mRNA
NMJ 52676 Homo sapiens F-box protein 15 (FBX015), mRNA
NMJ 52677 Homo sapiens zinc finger protein 494 (ZNF494), mRNA
NMJ 52678 Homo sapiens hypothetical protein FLJ34969 (FLJ34969), mRNA
NMJ 52679 Homo sapiens solute carrier family 10 (sodium/bile acid cotransporter family)
NMJ 52680 Homo sapiens hypothetical protein FLJ32028 (FLJ32028), mRNA
NMJ 52681 Homo sapiens hypothetical protein FLJ38482 (FLJ38482), mRNA
NMJ 52682 Homo sapiens hypothetical protein MGC10198 (MGC10198), mRNA
NMJ 52683 Homo sapiens hypothetical protein FLJ33167 (FLJ33167), mRNA
NMJ 52684 Homo sapiens hypothetical protein FLJ39653 (FLJ39653), mRNA
NMJ 52685 Homo sapiens solute carrier family 23 (nucleobase transporters), member 1 •
NMJ 52686 Homo sapiens hypothetical protein MGC29463 (MGC29463), mRNA
NMJ 52687 Homo sapiens hypothetical protein FLJ33641 (FLJ33641), mRNA
NMJ 52688 Homo sapiens KH domain containing, RNA binding, signal transduction asso
NMJ 52689 Homo sapiens hypothetical protein MGC9712 (MGC9712), mRNA
NMJ 52690 Homo sapiens dolichyl-phosphate mannosyltransferase polypeptide 2, regu
NMJ 52692 Homo sapiens core 1 UDP-galactose:N-acetylgalactosamine-alpha-R beta 1
NMJ 52693 Homo sapiens hypothetical protein MGC34827 (MGC34827), mRNA
NMJ 52694 Homo sapiens zinc finger, CCHC domain containing 5 (ZCCHC5), mRNA
NMJ 52695 Homo sapiens hypothetical protein FLJ23614 (FLJ23614), mRNA
NMJ 52696 Homo sapiens homeodomain interacting protein kinase 1 (HIPK1), transcript
NMJ 52697 Homo sapiens hypothetical protein MGC34032 (MGC34032), mRNA
NMJ 52698 Homo sapiens hypothetical protein FLJ38377 (FLJ38377), mRNA
NMJ52699 Homo sapiens SUMOI/sentrin specific protease 5 (SENP5), mRNA
NMJ 52700 Homo sapiens hypothetical protein MGC26597 (MGC26597), mRNA
NMJ 52701 Homo sapiens ATP binding cassette gene, sub-family A (ABC1 ), member 13
NMJ 52702 Homo sapiens chromosome 9 open reading frame 94 (C9orf94), mRNA
NMJ 52704 Homo sapiens hypothetical protein FLJ25477 (FLJ25477), transcript variant •
NMJ 52705 Homo sapiens hypothetical protein MGC9850 (MGC9850), mRNA
NMJ 52706 Homo sapiens hypothetical protein MGC26647 (MGC26647), mRNA
NMJ 52707 Homo sapiens solute carrier family 25 (mitochondrial carrier; Graves disease
NMJ 52710 Homo sapiens chromosome 10 open reading frame 27 (C10orf27), mRNA
NMJ52713 Homo sapiens integral membrane protein 1 (ITM1), mRNA
NMJ 52715 Homo sapiens hypothetical protein MGC10233 (MGC10233), mRNA
NMJ 52716 Homo sapiens hypothetical protein FLJ36874 (FLJ36874), mRNA
NMJ 52717 Homo sapiens hypothetical protein MGC35295 (MGC35295), mRNA
NMJ 52718 Homo sapiens hypothetical protein FLJ32009 (FLJ32009), mRNA
NMJ52719 Homo sapiens testis-specific leucine zipper protein nurit (NURIT), mRNA
NMJ 52720 Homo sapiens NIMA (never in mitosis gene a)-related kinase 3 (NEK3), trans
NMJ 52721 Homo sapiens docking protein 5-like (D0K5L), mRNA
NMJ 52722 Homo sapiens hypothetical protein FLJ25530 (FLJ25530), mRNA
NMJ 52723 Homo sapiens hypothetical protein FLJ38159 (FLJ38159), mRNA
NMJ52724 Homo sapiens Ras suppressor protein 1 (RSU1), transcript variant 2, mRNA
NMJ 52725 Homo sapiens solute carrier family 39 (zinc transporter), member 12 (SLC39
NMJ 52726 Homo sapiens Smhs2 homolog (rat) (FLJ34588), mRNA
NMJ 52727 Homo sapiens copine II (CPNE2), mRNA
NMJ 52728 Homo sapiens chromosome 18 open reading frame 20 (C18orf20), mRNA
NMJ 52729 Homo sapiens 5'-nucleotidase, cytosolic U-like 1 (NT5C2L1), mRNA
NMJ 52730 Homo sapiens chromosome 6 open reading frame 170 (C6orf170), mRNA
NMJ 52731 Homo sapiens chromosome 6 open reading frame 65 (C6orf65), mRNA
NMJ 52732 Homo sapiens chromosome 6 open reading frame 206 (C6orf206), mRNA
NMJ 52733 Homo sapiens BTB (POZ) domain containing 9 (BTBD9), mRNA
NMJ 52734 Homo sapiens chromosome 6 open reading frame 89 (C6orf89), mRNA
NMJ 52735 Homo sapiens zinc finger and BTB domain containing 9 (ZBTB9), mRNA
NMJ 52736 Homo sapiens zinc finger protein 187 (ZNF187), mRNA
NMJ 52737 Homo sapiens hypothetical protein MGC33993 (MGC33993), mRNA
NMJ 52738 Homo sapiens hypothetical protein MGC40222 (MGC40222), mRNA
NMJ 52739 Homo sapiens homeo box A9 (HOXA9), transcript variant 1 , mRNA
NMJ52740 Homo sapiens 3-hydroxyisobutyrate dehydrogenase (HI BADH), mRNA
NMJ 52742 Homo sapiens glypican 2 (cerebroglycan) (GPC2), mRN A
NMJ 52743 Homo sapiens chromosome 7 open reading frame 27 (C7orf27), mRNA
NMJ 52744 Homo sapiens sidekick homolog 1 (chicken) (SDK1), mRNA
NMJ52745 Homo sapiens neurexophilin 1 (NXPH1), mRNA
NMJ52747 Homo sapiens hypothetical protein DKFZp586l1420 (DKFZp586l1420), mRN
NMJ 52748 Homo sapiens hypothetical protein FLJ31340 (FLJ31340), mRNA
NMJ 52749 Homo sapiens hypothetical protein MGC33190 (MGC33190), mRNA
N J 52750 Homo sapiens hypothetical protein FLJ23834 (FLJ23834), mRNA
NMJ 52751 Homo sapiens chromosome 10 open reading frame 30 <C10orf30), mRNA
NMJ 52753 Homo sapiens signal peptide, CUB domain, EGF-like 3 (SCUBE3), mRNA
NMJ 52754 Homo sapiens sema domain, immunoglobulin domain (Ig), short basic doma
NMJ 52755 Homo sapiens hypothetical protein MGC40499 (MGC40499), mRNA
NMJ 52757 Homo sapiens hypothetical protein FLJ30313 (FLJ30313), mRNA
NMJ 52758 Homo sapiens YTH domain family 3 (YTHDF3), mRNA
NMJ52759 Homo sapiens hypothetical protein MGC35140 (MGC35140), mRNA
NMJ 52760 Homo sapiens hypothetical protein FLJ30934 (FLJ30934), mRNA
NMJ 52761 Homo sapiens hypothetical protein FLJ25444 (FLJ25444), mRNA
NMJ 52762 Homo sapiens testis specific, 10 interacting protein (TSGA10IP), mRNA
NMJ 52763 Homo sapiens hypothetical protein MGC26989 (MGC2S989), mRNA
NMJ 52764 Homo sapiens hypothetical protein MGC35212 (MGC35212), mRNA
NMJ 52765 Homo sapiens hypothetical protein MGC33510 (MGC33510), mRNA
NMJ 52766 Homo sapiens hypothetical protein MGC40107 (MGC4O107), mRNA
NMJ 52769 Homo sapiens chromosome 19 open reading frame 26 (C19orf26), mRNA
NMJ 52770 Homo sapiens hypothetical protein MGC35043 (MGC35043), mRNA
NMJ 52771 Homo sapiens chromosome 19 open reading frame 34 (C19orf34), mRNA
NMJ 52772 Homo sapiens hypothetical protein MGC40368 (MGC40368), mRNA
NMJ52773 Homo sapiens hypothetical protein MGC33212 (MGC33212), mRNA
NMJ 52774 Homo sapiens hypothetical protein MGC42090 (MGC42090), mRNA
NMJ 52775 Homo sapiens hypothetical protein MGC33607 (MGC33607), mRNA
NMJ 52776 Homo sapiens hypothetical protein MGC40579 (MGC40579), mRNA
NMJ 52777 Homo sapiens chromosome 14 open reading frame 48 (C14orf48), mRNA
NMJ 52778 Homo sapiens hypothetical protein MGC33302 (MGC33302), mRNA
NMJ 52779 Homo sapiens hypothetical protein MGC26856 (MGC2S856), mRNA
NMJ 52780 Homo sapiens hypothetical protein FLJ14503 (FLJ14503), mRNA
NMJ 52781 Homo sapiens hypothetical protein FLJ32830 (FLJ32830), mRNA
NMJ 52782 Homo sapiens hypothetical protein MGC33329 (MGC33329), mRNA
NMJ52783 Homo sapiens hypothetical protein MGC25181 (MGC25181), mRNA
NMJ 52784 Homo sapiens hypothetical protein MGC39581 (MGC39581), mRNA
N J 52785 Homo sapiens germinal center expressed transcript 2 (GCET2), mRNA
NMJ 52786 Homo sapiens chromosome 9 open reading frame 43 (C9orf43), mRNA
NMJ 52787 Homo sapiens TAK1 -binding protein 3 (TAB3), transcript variant 1 , mRNA
NMJ52788 Homo sapiens E2a-Pbx1 -associated protein (EB-1), transcript variant 1 , mRr
NMJ 52789 Homo sapiens hypothetical protein MGC40405 (MGC4O405), mRNA
NMJ 52791 Homo sapiens zinc finger protein 555 (ZNF555), mRNA.
NMJ 52792 Homo sapiens hypothetical protein FLJ25084 (FLJ25084), mRNA
NMJ 52793 Homo sapiens hypothetical protein Ellsl (Ellsl), mRNA
NMJ 52794 Homo sapiens hypoxia inducible factor 3, alpha subunit (HIF3A), transcript v;
NMJ 52795 Homo sapiens hypoxia inducible factor 3, alpha subunit (HIF3A), transcript vi
NMJ 52796 Homo sapiens hypoxia inducible factor 3, alpha subunit (HIF3A), transcript v;
NMJ 52826 Homo sapiens sorting nexin 1 (SNX1), transcript variant 3, mRNA
NMJ 52827 Homo sapiens sorting nexin 3 (SNX3), transcript variant 2, mRNA
NMJ 52828 Homo sapiens sorting nexin 3 (SNX3), transcript variant 3, mRNA
NMJ52829 Homo sapiens testis derived transcript (3 LIM domains) (TES), transcript vari
NMJ 52830 Homo sapiens angiotensin I converting enzyme (peptidyl-dipeptidase A) 1 (A
NMJ 52831 Homo sapiens angiotensin I converting enzyme (peptidyl-dipeptidase A) 1 (A
NMJ 52832 Homo sapiens Mouse Mammary Turmor Virus Receptor homolog 1 (MTVR1)
NMJ 52834 Homo sapiens transmembrane protein 18 (TMEM18), mRNA
NMJ52835 Homo sapiens casein kinase (LOC149420), mRNA
NMJ 52836 Homo sapiens sorting nexin 16 (SNX16), transcript variant 2, mRNA
NMJ 52837 Homo sapiens sorting nexin 16 (SNX16), transcript variant 3, mRNA
NMJ 52838 Homo sapiens RNA binding motif protein 12 (RBM12), transcript variant 2, m
NMJ 52840 Homo sapiens Hermansky-Pudlak syndrome 4 (HPS4), transcript variant 3, r
NMJ 52841 Homo sapiens Hermansky-Pudlak syndrome 4 (HPS4), transcript variant 2, r
NMJ 52842 Homo sapiens Hermansky-Pudlak syndrome 4 (HPS4), transcript variant 5, r
NMJ 52843 Homo sapiens Hermansky-Pudlak syndrome 4 (HPS4), transcript variant 4, r
NMJ 52850 Homo sapiens phosphatidylinositol glycan, class O (PIGO), transcript variant
NMJ 52851 Homo sapiens membrane-spanning 4-domains, subfamily A, member 6A (M!
NMJ52852 Homo sapiens membrane-spanning 4-domains, subfamily A, member 6A (MJ
NMJ 52854 Homo sapiens tumor necrosis factor receptor superfamily, member 5 (TNFR!
NMJ 52855 Homo sapiens immunoglobulin lambda-like polypeptide 1 (IGLL1), transcript
NMJ 52856 Homo sapiens RNA binding motif protein 10 (RBM10), transcript variant 2, m
NMJ 52857 Homo sapiens Wilms tumor 1 associated protein (WTAP), transcript variant , '
NMJ 52858 Homo sapiens Wilms tumor 1 associated protein (WTAP), transcript variant [
NMJ 52860 Homo sapiens Sp7 transcription factor (SP7), mRNA
NMJ 52862 Homo sapiens actin related protein 2/3 complex, subunit 2, 34kDa (ARPC2),
NMJ 52864 Homo sapiens chromosome 20 open reading frame 58 (C20orf58), mRNA
NMJ 52866 Homo sapiens membrane-spanning 4-domains, subfamily A, member 1 (MS'
NMJ 52867 Homo sapiens membrane-spanning 4-domains, subfamily A, member 1 (MS'
NMJ 52868 Homo sapiens potassium inwardly-rectifying channel, subfamily J, member 4
NMJ 52869 Homo sapiens regucalcin (senescence marker protein-30) (RGN), transcript
NMJ 52870 Homo sapiens abhydrolase domain containing 1 (ABHD1), transcript variant
NMJ 52871 Homo sapiens tumor necrosis factor receptor superfamily, member 6 (TNFR!
NMJ 52872 Homo sapiens tumor necrosis factor receptor superfamily, member 6 (TNFR!
NMJ 52873 Homo sapiens tumor necrosis factor receptor superfamily, member 6 (TNFR!
NMJ 52874 Homo sapiens tumor necrosis factor receptor superfamily, member 6 (TNFR!
NMJ 52875 Homo sapiens tumor necrosis factor receptor superfamily, member 6 (TNFR!
NMJ 52876 Homo sapiens tumor necrosis factor receptor superfamily, member 6 (TNFR!
NMJ 52877 Homo sapiens tumor necrosis factor receptor superfamily, member 6 (TNFR!
NMJ 52878 Homo sapiens v-maf musculoaponeurotic fibrosarcoma oncogene homolog F
NMJ 52879 Homo sapiens diacylglycerol kinase, delta 130kDa (DGKD), transcript varian
NMJ 52880 Homo sapiens PTK7 protein tyrosine kinase 7 (PTK7), transcript variant PTK
NMJ 52881 Homo sapiens PTK7 protein tyrosine kinase 7 (PTK7), transcript variant PTK
NMJ 52882 Homo sapiens PTK7 protein tyrosine kinase 7 (PTK7), transcript variant PTK
NMJ 52883 Homo sapiens PTK7 protein tyrosine kinase 7 (PTK7), transcript variant PTK
NMJ52888 Homo sapiens collagen, type XXII, alpha 1 (COL22A1), mRNA
NMJ 52889 Homo sapiens carbohydrate (chondroitin 4) sulfotransferase 13 (CHST13),
NMJ 52890 Homo sapiens collagen, type XXIV, alpha 1 (COL24A1 ), mRNA
NMJ 52891 Homo sapiens protease, serine, 33 (PRSS33), mRNA
NMJ 52892 Homo sapiens hypothetical protein DKFZp434K1815 (DKFZp434K1815), mF
NMJ52896 Homo sapiens ubiquitin-like, containing PHD and RING finger domains, 2 (Ul
NMJ52897 Homo sapiens chromosome 20 open reading frame 161 (C20orf161), transci
NMJ 52898 Homo sapiens Fer3-like (Drosophila) (FERD3L), mRNA
NMJ52899 Homo sapiens interleukin 4 induced 1 (IL411), transcript variant 1 , mRNA
NMJ 52900 Homo sapiens membrane-associated guanylate kinase-related (MAGI-3) (M/
NMJ 52901 Homo sapiens pyrin-domain containing protein 1 (PYC1), mRNA
NMJ 52902 Homo sapiens putative MAPK activating protein (MGC3794), mRNA
NMJ 52903 Homo sapiens kelch repeat and BTB (POZ) domain containing 6 (KBTBD6),
NMJ 52904 Homo sapiens sperm antigen HCMOGT-1 (HCM0GT-1), mRNA
NMJ 52905 Homo sapiens neural precursor cell expressed, develop entally down-reguk
NMJ 52906 Homo sapiens hypothetical protein DKFZp761 P1121 (DKFZp761 P1121), mF
NMJ 52908 Homo sapiens hypothetical protein FLJ31196 (FLJ31196), mRNA
NMJ 52909 Homo sapiens zinc finger protein 548 (ZNF548), mRNA
NMJ 52910 Homo sapiens diacylglycerol kinase, eta (DGKH), transcript variant 1 , mRNA
NMJ 52911 Homo sapiens polyamine oxidase (exo-N4-amino) (PAOX), transcript variant
NMJ 52912 Homo sapiens mitochondrial translational initiation factor 3 (MTIF3), mRNA
NMJ52913 Homo sapiens hypothetical protein DKFZp761 L1417 (DKFZp761 L1417), mR
NMJ52914 Homo sapiens transcript expressed during hematopoiesis 2 (MGC33894), ml
NMJ 52916 Homo sapiens egf-like module containing, mucin-like, hormone receptor-like
NMJ52917 Homo sapiens egf-like module containing, mucin-like, hormone receptor-like
NMJ52918 Homo sapiens egf-like module containing, mucin-like, hormone receptor-like
NMJ52919 Homo sapiens egf-like module containing, mucin-like, hormone receptor-like
NMJ52920 Homo sapiens egf-like module containing, mucin-like, hormone receptor-like
NMJ52921 Homo sapiens egf-like module containing, mucin-like, hormone receptor-like
NMJ 52924 Homo sapiens abhydrolase domain containing 2 (ABHD2), transcript variant
NMJ52925 Homo sapiens copine I (CPNE1 ), transcript variant 1 , mRNA
NMJ 52926 Homo sapiens copine I (CPNE1), transcript variant 2, mRNA
NMJ 52927 Homo sapiens copine I (CPNE1 ), transcript variant 4, mRNA
NMJ 52928 Homo sapiens copine I (CPNE1 ), transcript variant 5, mRNA
NMJ 52929 Homo sapiens copine I (CPNE1 ), transcript variant 6, mRNA
NMJ 52930 Homo sapiens copine I (CPNE1 ), transcript variant 7, mRNA
NMJ 52931 Homo sapiens copine I (CPNE1 ), transcript variant 8, mRNA
NMJ 52932 Homo sapiens glycosyltransferase AD-017 (AD-017), transcript variant 1 , mF
NMJ 52933 Homo sapiens protein phosphatase, EF hand calcium-binding domain 2 (PPI
NMJ 52934 Homo sapiens protein phosphatase, EF hand calcium-binding domain 2 (PPI
NMJ 52939 Homo sapiens egf-like module containing, mucin-like, hormone receptor-like
NMJ 52942 Homo sapiens tumor necrosis factor receptor superfamily, member 8 (TNFR!
NMJ 52943 Homo sapiens zinc finger protein 268 (ZNF268), transcript variant B, mRNA
NMJ 52945 Homo sapiens developmentally regulated RNA-binding protein 1 (DRB1), mF
NMJ 52988 Homo sapiens SPPL2b (SPPL2B), mRNA
NMJ 52989 Homo sapiens SRY (sex determining region Y)-box 5 (SOX5), transcript varis
NMJ 52990 Homo sapiens peroxisomal, testis specific 1 (PXT1), mRNA
NMJ 52991 Homo sapiens embryonic ectoderm development (EED), transcript variant 2,
NMJ 52992 Homo sapiens POM (POM121 homolog, rat) and ZP3 fusion (POMZP3), tran
NMJ52994 Homo sapiens smooth muscle myosin heavy chain 11 isoform SM1-like (LOC
NMJ 52995 Homo sapiens ovarian zinc finger protein (HOZFP), mRNA
NMJ 52996 Homo sapiens sialyltransferase 7 ((alpha-N-acetylneuraminyl-2,3-beta-galacl
NMJ 52997 Homo sapiens chromosome 4 open reading frame 7 (C4orf7), mRNA
NMJ 52998 Homo sapiens enhancer of zeste homolog 2 (Drosophila) (EZH2), transcript '
NMJ 52999 Homo sapiens six transmembrane epithelial antigen of prostate 2 (STEAP2),
NMJ 53000 Homo sapiens adenomatosis polyposis coli down-regulated 1 (APCDD1), mF
NMJ 53001 Homo sapiens proteasome (prosome, macropain) 26S subunit, ATPase, 4 (F
NMJ 53002 Homo sapiens G protein-coupled receptor 156 (GPR156), mRNA
NMJ 53003 Homo sapiens orofacial cleft 1 candidate 1 (OFCC1), mRNA
NMJ 53005 Homo sapiens RIO kinase 1 (yeast) (R10K1), transcript variant 2, mRNA
NMJ 53006 Homo sapiens N-acetylglutamate synthase (NAGS), mRNA
NMJ 53007 Homo sapiens outer dense fiber of sperm tails 4 (ODF4), mRNA
NMJ 53008 Homo sapiens hypothetical protein FLJ30277 (FLJ30277), m RNA
NMJ 53010 Homo sapiens chromosome 18 open reading frame 16 (C18orf16), mRNA
NMJ 53011 Homo sapiens hypothetical protein FLJ30594 (FLJ30594), m RNA
NMJ 53012 Homo sapiens tumor necrosis factor (ligand) superfamily, member 12 (TNFS
NMJ53013 Homo sapiens hypothetical protein FLJ30596 (FLJ30596), m RNA
NMJ53014 Homo sapiens hypothetical protein FLJ30634 (FLJ30634), m RNA
NMJ 53015 Homo sapiens hypothetical protein FLJ30668 (FLJ30668), mRNA
NMJ 53018 Homo sapiens hypothetical protein FLJ30726 (FLJ30726), mRNA
NMJ 53019 Homo sapiens transmembrane protease, serine 6 (TMPRSS6), mRNA
NMJ 53020 Homo sapiens RNA binding motif protein 24 (RBM24), mRNA
NMJ 53022 Homo sapiens hypothetical protein FLJ31166 (FLJ31166), mRNA
NMJ 53023 Homo sapiens spermatogenesis associated 13 (SPATA13), mRNA
NMJ 53024 Homo sapiens seven transmembrane helix receptor (FLJ31393), mRNA
NMJ53025 Homo sapiens hypothetical protein FLJ31606 (FLJ31606), mRNA
NMJ53026 Homo sapiens prickle-like 1 (Drosophila) (PRICKLE1), mRNA
NMJ53027 Homo sapiens hypothetical protein FLJ31659 (FLJ31659), mRNA
NMJ 53028 Homo sapiens zinc finger protein 75a (ZNF75A), mRNA
NMJ 53029 Homo sapiens Nedd4 binding protein 1 (N4BP1), mRNA
NMJ 53031 Homo sapiens hypothetical protein FLJ32063 (FLJ32063), mRNA
NMJ 53032 Homo sapiens hypothetical protein FLJ32065 (FLJ32065), mRNA
NMJ 53033 Homo sapiens potassium channel tetramerisation domain containing 7 (KCT
NMJ 53034 Homo sapiens zinc finger protein 488 (ZNF488), mRNA
NMJ53035 Homo sapiens hypothetical protein FLJ32112 (FLJ32112), mRNA
NMJ 53036 Homo sapiens chromosome 6 open reading frame 78 (C6orf78), mRNA
NMJ 53038 Homo sapiens hypothetical protein FLJ32447 (FLJ32447), mRNA
NMJ53040 Homo sapiens hypothetical protein FLJ32831 (FLJ32831 ), mRNA
NMJ 53041 Homo sapiens hypothetical protein FLJ32955 (FLJ32955), mRNA
NMJ 53043 Homo sapiens hypothetical protein FLJ37078 (FLJ37078), mRNA
NMJ 53044 Homo sapiens hypothetical protein FLJ35801 (FLJ35801 ), mRNA
NMJ 53045 Homo sapiens chromosome 9 open reading frame 91 (C9orf91), mRNA
NMJ 53046 Homo sapiens tudor domain containing 9 (TDRD9), mRNA
NMJ 53047 Homo sapiens FYN oncogene related to SRC, FGR, YES (FYN), transcript vi
NMJ 53048 Homo sapiens FYN oncogene related to SRC, FGR, YES (FYN), transcript v;
NMJ 53050 Homo sapiens myotubularin related protein 3 (MTMR3), transcript variant 1 , i
NMJ 53051 Homo sapiens myotubularin related protein 3 (MTMR3), transcript variant 2, i
NMJ53181 Homo sapiens neuropilin (NRP) and tolloid (TLL)-like 1 (NET01 ), transcript v
NMJ53182 Homo sapiens MYC induced nuclear antigen (MINA), transcript variant 3, mF
NMJ 53183 Homo sapiens nudix (nucleoside diphosphate linked moiety X)-type motif 10
NMJ53184 Homo sapiens immunoglobulin superfamily, member 4D (IGSF4D), mRNA
NMJ53186 Homo sapiens ankyrin repeat domain 15 (ANKRD15), transcript variant 2, ml
NMJ 53187 Homo sapiens solute carrier family 22 (organic cation transporter), member 1
NMJ53188 Homo sapiens transportin 1 (TNP01), transcript variant 2, mRNA
NMJ 53189 Homo sapiens sperm adhesion molecule 1 (PH-20 hyaluronidase, zona pellu
NMJ53191 Homo sapiens solute carrier family 22 (organic cation transporter), member -
NMJ 53200 Homo sapiens endothelial differentiation-related factor 1 (EDF1), transcript vi
NMJ 53201 Homo sapiens heat shock 70kDa protein 8 (HSPA8), transcript variant 2, mR
NMJ53202 Homo sapiens a disintegrin and metalloproteinase domain 33 (ADAM33), tra
NMJ53204 Homo sapiens chromosome 21 open reading frame 90 (C21orf90), mRNA
NMJ 53206 Homo sapiens adhesion molecule AMIGA (AMIGA), mRNA
NMJ 53207 Homo sapiens AE binding protein 2 (AEBP2), mRNA
NMJ 53208 Homo sapiens hypothetical protein MGC35048 (MGC35048), mRNA
NMJ 53209 Homo sapiens hypothetical protein FLJ37300 (FLJ3730O), mRNA
NMJ 53211 Homo sapiens chromosome 18 open reading frame 17 (C18orf17), mRNA
NMJ 53212 Homo sapiens gap junction protein, beta 4 (connexin 30.3) (GJB4), mRNA
NMJ 53213 Homo sapiens Rho guanine nucleotide exchange factor (GEF) 19 (ARHGEF'
NMJ 53214 Homo sapiens hypothetical protein FLJ37440 (FLJ37440), mRNA
NMJ 53215 Homo sapiens hypothetical protein FLJ38608 (FLJ38608), mRNA
NMJ 53216 Homo sapiens hypothetical protein FLJ25680 (FLJ25680), mRNA
NMJ53217 Homo sapiens hypothetical protein MGC13034 (MGC13034), mRNA
NMJ 53218 Homo sapiens hypothetical protein FLJ38725 (FLJ38725), mRNA
NMJ 53219 Homo sapiens zinc finger protein 524 (ZNF524), mRNA
NMJ 53220 Homo sapiens hypothetical protein MGC35440 (MGC35440), mRNA
NMJ53221 Homo sapiens cartilage intermediate layer protein 2 (CILP2), mRNA
NMJ 53223 Homo sapiens hypothetical protein FLJ36090 (FLJ3609O), mRNA
NMJ 53225 Homo sapiens RPE-spondin (RPESP), mRNA
NMJ 53226 Homo sapiens transmembrane protein 20 (TMEM20), rnRNA
NMJ 53228 Homo sapiens hypothetical protein FLJ38335 (FLJ38335), mRNA
NMJ 53229 Homo sapiens hypothetical protein FLJ33318 (FLJ33318), mRNA
NMJ 53230 Homo sapiens F-box protein 39 (FBX039), mRNA
NMJ 53231 Homo sapiens zinc finger protein 550 (ZNF550), mRNA
NMJ 53232 Homo sapiens CREBBP/EP300 inhibitor 2 (CRI2), mRNA
NMJ 53233 Homo sapiens hypothetical protein FLJ36445 (FLJ36445), mRNA
NMJ 53234 Homo sapiens chromosome 5 open reading frame 11 (C5orf11), mRNA
NMJ 53236 Homo sapiens immune associated nucleotide (hlAN7), mRNA
NMJ 53238 Homo sapiens hypothetical protein MGC22001 (MGC22001), mRNA
NMJ 53239 Homo sapiens hypothetical protein KIAA1924 (KIAA1924), mRNA
NMJ 53240 Homo sapiens nephronophthisis 3 (adolescent) (NPHP3), mRNA
NMJ 53244 Homo sapiens chromosome 10 open reading frame 111 (C1 Oorfl 11 ), mRNA
NMJ 53246 Homo sapiens hypothetical protein MGC45491 (MGC45491), mRNA
NMJ 53247 Homo sapiens solute carrier family 29 (nucleoside transporters), member 4 (■
NMJ53248 Homo sapiens hypothetical protein MGC14276 (MGC14276), mRNA
NMJ 53251 Homo sapiens hypothetical protein FLJ25952 (FLJ25952), mRNA
NMJ 53252 Homo sapiens bromo domain-containing protein disrupted in leukemia (BRO
NMJ 53253 Homo sapiens signal-induced proliferation-associated gene 1 (SIPA1), transt
NMJ53254 Homo sapiens hypothetical protein FLJ36119 (FLJ36119), mRNA
NMJ 53255 Homo sapiens minichromosome maintenance deficient domain containing 1
NMJ 53256 Homo sapiens chromosome 10 open reading frame 47 (C10orf47), mRNA
NMJ 53257 Homo sapiens gonadotropin inducible transcription repressor 1 (GIOT-1), F
NMJ53260 Homo sapiens hypothetical protein FLJ36812 (FLJ36812), mRNA
NMJ 53261 Homo sapiens hypothetical protein FLJ38101 (FLJ38101), mRNA
NMJ 53262 Homo sapiens synaptotagmin XIV (SYT14), mRNA
NMJ 53263 Homo sapiens zinc finger protein 549 (ZNF549), mRNA
NMJ 53264 Homo sapiens hypothetical protein FLJ35880 (FLJ35880), mRNA
NMJ 53265 Homo sapiens hypothetical protein FLJ35827 (FLJ35827), mRNA
NMJ 53266 Homo sapiens hypothetical protein MGC33486 (MGC33486), mRNA
NMJ53267 Homo sapiens MAM domain containing 2 (MAMDC2), mRNA
NMJ 53268 Homo sapiens hypothetical protein FLJ31579 (FLJ31579), mRNA
NMJ 53269 Homo sapiens chromosome 20 open reading frame 96 (C20orf96), mRNA
NMJ53270 Homo sapiens hypothetical protein FLJ34960 (FLJ34960), mRNA
NMJ 53271 Homo sapiens hypothetical protein MGC32065 (MGC32065), mRNA
NMJ 53273 Homo sapiens inositol hexaphosphate kinase 1 (IHPK1), mRNA
NMJ 53274 Homo sapiens vitelliform macular dystrophy 2-like 2 (VMD2L2), mRNA
NMJ 53276 Homo sapiens solute carrier family 22 (organic anion transporter), member 6
NMJ 53277 Homo sapiens solute carrier family 22 (organic anion transporter), member 6
NMJ 53278 Homo sapiens solute carrier family 22 (organic anion transporter), member 6
NMJ 53279 Homo sapiens solute carrier family 22 (organic anion transporter), member 6
NMJ 53280 Homo sapiens ubiquitin-activating enzyme E1 (A1S9T and BN75 temperatuπ
NMJ53281 Homo sapiens hyaluronoglucosaminidase 1 (HYAL1), transcript variant 8, ml
NMJ 53282 Homo sapiens hyaluronoglucosaminidase 1 (HYAL1), transcript variant 2, mf
NMJ 53283 Homo sapiens hyaluronoglucosaminidase 1 (HYAL1), transcript variant 3, ml
NMJ 53284 Homo sapiens hyaluronoglucosaminidase 1 (HYAL1), transcript variant 4, ml
NMJ 53285 Homo sapiens hyaluronoglucosaminidase 1 (HYAL1), transcript variant 5, mf
NMJ 53286 Homo sapiens hyaluronoglucosaminidase 1 (HYAL1), transcript variant 6, mi
NMJ 53289 Homo sapiens defensin, beta 119 (DEFB119), transcript variant 1 , mRNA
NMJ 53290 Homo sapiens family with sequence similarity 10, member A4 (FAM10A4), m
NMJ 53291 Homo sapiens family with sequence similarity 10, member A5 (FAM10A5), m
NMJ 53292 Homo sapiens nitric oxide synthase 2A (inducible, hepatocytes) (NOS2A), trs
NMJ 53320 Homo sapiens solute carrier family 22 (organic anion transporter), member 7
NMJ 53321 Homo sapiens peripheral myelin protein 22 (PMP22), transcript variant 2, mF
NMJ 53322 Homo sapiens peripheral myelin protein 22 (PMP22), transcript variant 3, mF
NMJ 53324 Homo sapiens defensin, beta 123 (DEFB123), mRNA
NMJ53325 Homo sapiens defensin, beta 125 (DEFB125), mRNA
NMJ 53326 Homo sapiens aldo-keto reductase family 1 , member A1 (aldehyde reductas<
NMJ 53328 Homo sapiens retinoblastoma binding protein 9 (RBBP9), transcript variant 2
NMJ 53329 Homo sapiens hypothetical protein MGC10204 (MGC10204), mRNA
NMJ 53330 Homo sapiens DnaJ (Hsp40) homolog, subfamily B, member 8 (DNAJB8), m
NMJ 53331 Homo sapiens potassium channel tetramerisation domain containing 6 (KCT
NMJ 53332 Homo sapiens 3' exoribonuclease (3'HEXO), mRNA
NMJ 53333 Homo sapiens hypothetical protein MGC45400 (MGC45400), mRNA
N J 53334 Homo sapiens scavenger receptor class F, member 2 (SCARF2), transcript \
NMJ 53335 Homo sapiens protein kinase LYK5 (LYK5), mRNA
NMJ53336 Homo sapiens chromosome 10 open reading frame 89 (C10orf89), mRNA
NMJ 53337 Homo sapiens selectin ligand interactor cytoplasmic-1 (SLIC1), mRNA
NMJ53338 Homo sapiens hypothetical protein FLJ90165 (FLJ90165), mRNA
NMJ 53339 Homo sapiens hypothetical protein FLJ90811 (FLJ90811), mRNA
NMJ 53340 Homo sapiens hypothetical protein MGC46534 (MGC46534), mRNA
NMJ 53341 Homo sapiens IBR domain containing 3 (IBRDC3), mRNA
NMJ 53342 Homo sapiens fasting-inducible integral membrane protein TM6P1 (FLJ9002
NMJ 53343 Homo sapiens ectonucleotide pyrophosphatase/phσsphodiesterase 6 (ENPF
NMJ53344 Homo sapiens chromosome 6 open reading frame 141 (C6orf141), rnRNA
NMJ 53345 Homo sapiens hypothetical protein FLJ90586 (FLJ90586), mRNA
NMJ 53346 Homo sapiens chromosome X open reading frame 20 (CXorf20), mRNA
NMJ 53347 Homo sapiens hypothetical protein FLJ90119 (FLJ90119), mRNA
NMJ 53348 Homo sapiens F-box and WD-40 domain protein 8 (FBXW8), transcript varia
NMJ 53350 Homo sapiens F-box and leucine-rich repeat protein 16 (FBXL16), RNA
NMJ 53353 Homo sapiens hypothetical protein MGC27085 (MGC27085), mRNA
NMJ53354 Homo sapiens hypothetical protein MGC33214 (MGC33214), mRNA
NMJ 53355 Homo sapiens T-cell lymphoma breakpoint associated target 1 (TCBA1), mR
NMJ53356 Homo sapiens hypothetical protein MGC34741 (MGC34741), mRNA
NMJ 53357 Homo sapiens solute carrier family 16 (monocarboxylic acid transporters), mi
NMJ 53358 Homo sapiens hypothetical protein FLJ90396 (FLJ90396), mRNA
NMJ 53359 Homo sapiens hypothetical protein MGC24975 (MGC24975), mRNA
NMJ53360 Homo sapiens hypothetical protein FLJ90166 (FLJ90166), mRNA
NMJ 53361 Homo sapiens hypothetical protein MGC42105 (MGC42105), mRNA
NMJ 53362 Homo sapiens protease, serine, 35 (PRSS35), mRNA
NMJ 53363 Homo sapiens hypothetical protein MGC42415 (MGC42415), mRNA
NMJ 53364 Homo sapiens hypothetical protein MGC39520 (MGC39520), mRNA
NMJ 53365 Homo sapiens hypothetical protein FLJ90013 (FLJ90013), mRNA
NMJ53367 Homo sapiens chromosome 10 open reading frame 56 (C10orf56), mRNA
NMJ53368 Homo sapiens connexin40.1 (CX40.1), mRNA
NMJ 53369 Homo sapiens KIAA1919 (KIAA1919), mRNA
NMJ53370 Homo sapiens protease inhibitor 16 (PI16), mRNA
NMJ 53371 Homo sapiens ligand of numb-protein X 2 (LNX2), mRNA
NMJ53373 Homo sapiens hypothetical protein MGC15875 (MGC15875), mRNA
NMJ 53374 Homo sapiens hypothetical protein MGC35274 (MGC35274), mRNA
N J 53375 Homo sapiens placenta-specific 2 (PLAC2), mRNA
NMJ 53376 Homo sapiens hypothetical protein FLJ90575 (FLJ90575), mRNA
NMJ 53377 Homo sapiens leucine-rich repeats and immunoglobulin-like domains 3 (LRK
NMJ53378 Homo sapiens solute carrier family 22 (organic anion/cation transporter), mei
NMJ 53379 Homo sapiens kringle containing transmembrane protein 1 (KREMEN1), tran
NMJ 53380 Homo sapiens zinc finger protein 41 (ZNF41), transcript variant 2, mRNA
NMJ 53381 Homo sapiens pro-melanin-concentrating hormone-like 2 (PMCHL2), mRNA
NMJ 53425 Homo sapiens TNFRSF1 A-associated via death domain (TRADD), transcript
NMJ 53426 Homo sapiens paired-like homeodomain transcription factor 2 (P1TX2), transc
NMJ 53427 Homo sapiens paired-like homeodomain transcription factor 2 (PITX2), transi
N J 53437 Homo sapiens outer dense fiber of sperm tails 2 (ODF2), transcript variant 2,
NMJ 53442 Homo sapiens G protein-coupled receptor 26 (GPR26), mRNA
NMJ 53443 Homo sapiens killer cell immunoglobulin-like receptor, three domains, long c;
NMJ 53444 Homo sapiens olfactory receptor, family 5, subfamily P, member 2 (OR5P2),
NMJ 53445 Homo sapiens olfactory receptor, family 5, subfamily P, member 3 (OR5P3),
NMJ 53446 Homo sapiens UDP-GalNAc:Neu5Acalpha2-3Galbeta-R betal ,4-N-acetylgal;
NMJ 53447 Homo sapiens NACHT, leucine rich repeat and PYD containing 5 (NALP5), n
NMJ 53448 Homo sapiens extraembryonic, spermatogenesis, homeobox 1-like (ESX1 L),
NMJ 53449 Homo sapiens solute carrier family 2 (facilitated glucose transporter), membe
NMJ 53450 Homo sapiens lung cancer metastasis-related protein 1 (LCMR1), mRNA
NMJ 53451 Homo sapiens oral cancer overexpressed 1 (ORAOV1), mRNA
NMJ 53453 Homo sapiens vestigial like 2 (Drosophila) (VGLL2), transcript variant 2, mRf
NMJ 53454 Homo sapiens chromosome 21 open reading frame 86 (C21orf86), mRNA
NMJ 53456 Homo sapiens heparan sulfate 6-O-sulfotransferase 3 (HS6ST3), mRNA
NMJ 53460 Homo sapiens interleukin 17 receptor C (IL17RC), transcript variant 2, mRN/
NMJ 53461 Homo sapiens interleukin 17 receptor C (IL17RC), transcript variant 1 , mRN/
NMJ53462 Homo sapiens interleukin 17 receptor C (IL17RC), transcript variant 4, mRN/
NMJ 53463 Homo sapiens interleukin 17 receptor C (IL17RC), transcript variant 5, mRN/
NMJ53464 Homo sapiens interleukin enhancer binding factor 3, 90kDa (ILF3), transcript
NMJ 53477 Homo sapiens ubiquitously-expressed transcript (UXT), transcript variant 1 , r
NMJ 53478 Homo sapiens chondrosarcoma associated gene 1 (CSAG1 ), transcript varia
NMJ 53479 Homo sapiens chondrosarcoma associated gene 1 (CSAG1 ), transcript varia
NMJ 53480 Homo sapiens interleukin 17 receptor E (IL17RE), transcript variant 1 , mRN
NMJ 53481 Homo sapiens interleukin 17 receptor E (IL17RE), transcript variant 2, mRN/5
NMJ 53482 Homo sapiens interleukin 17 receptor E (IL17RE), transcript variant 4, mRN/1
NMJ 53483 Homo sapiens interleukin 17 receptor E (IL17RE), transcript variant 5, mRN/1
NMJ 53485 Homo sapiens nucleoporin 155kDa (NUP155), transcript variant 1 , mRNA
NMJ 53486 Homo sapiens lactate dehydrogenase D (LDHD), nuclear gene encoding miti
NMJ 53487 Homo sapiens MAM domain containing glycosylphosphatidylinositol anchor 1
NMJ 53488 Homo sapiens melanoma antigen, family A, 2B (MAGEA2B), mRNA
NMJ 53490 Homo sapiens keratin 13 (KRT13), transcript variant 1 , mRNA
NMJ 53497 Homo sapiens mitogen-activated protein kinase kinase kinase 7 interacting p
NMJ 53498 Homo sapiens calcium/calmodulin-dependent protein kinase ID (CAMK1 D), t
NMJ 53499 Homo sapiens calcium/calmodulin-dependent protein kinase kinase 2, beta (
NMJ 53500 Homo sapiens calcium/calmodulin-dependent protein kinase kinase 2, beta (
NMJ53603 Homo sapiens component of oligomeric golgi complex 7 (COG7), mRNA
NMJ 53604 Homo sapiens myocardin (MYOCD), mRNA
NMJ 53606 Homo sapiens hypothetical protein FLJ32796 (FLJ32796), mRNA
NMJ 53607 Homo sapiens adult retina protein (LOC153222), mRNA
NMJ 53608 Homo sapiens hypothetical protein MGC17986 (MGC17986), mRNA
NMJ 53609 Homo sapiens transmembrane protease, serine 6 (TMPRSS6), mRNA
NMJ 53610 Homo sapiens cardiomyopathy associated 5 (CMYA5), mRNA
NMJ 53611 Homo sapiens hypothetical protein MGC20446 (MGC20446), mRNA
NMJ 53612 Homo sapiens heparan sulfate (glucosamine) 3-O-sulfotransferase 5 (HS3S1
NMJ 53613 Homo sapiens PISC domain containing hypothetical protein (LOC254531), rr
NMJ53614 Homo sapiens testis spermatogenesis apoptosis-related protein 6 (TSARG6)
NMJ 53615 Homo sapiens Ral-GDS related protein Rgr (Rgr), mRNA
NMJ 53616 Homo sapiens sema domain, transmembrane domain (TM), and cytoplasmic
NMJ53617 Homo sapiens sema domain, transmembrane domain (TM), and cytoplasmic
NMJ53618 Homo sapiens sema domain, transmembrane domain (TM), and cytoplasmic
NMJ 53619 Homo sapiens sema domain, transmembrane domain (TM), and cytoplasmic
NMJ53620 Homo sapiens homeo box A1 (HOXA1), transcript variant 2, mRNA
NMJ53631 Homo sapiens homeo box A3 (HOXA3), transcript variant 2, mRNA
NMJ 53632 Homo sapiens homeo box A3 (HOXA3), transcript variant 3, mRNA
NMJ 53633 Homo sapiens homeo box C4 (HOXC4), transcript variant 2, mRNA
NMJ 53634 Homo sapiens copine VIII (CPNE8), mRNA
NMJ53635 Homo sapiens copine family member (LOC151835), mRNA
NMJ 53636 Homo sapiens copine VII (CPNE7), transcript variant 1, mRNA
NMJ 53637 Homo sapiens pantothenate kinase 2 (Hallervorden-Spatz syndrome) (PANK
NMJ 53638 Homo sapiens pantothenate kinase 2 (Hallervorden-Spatz syndrome) (PANK
NMJ 53639 Homo sapiens pantothenate kinase 2 (Hallervorden-Spatz syndrome) (PANK
NMJ 53640 Homo sapiens pantothenate kinase 2 (Hallervorden-Spatz syndrome) (PANK
NMJ 53641 Homo sapiens pantothenate kinase 2 (Hallervorden-Spatz syndrome) (PANK
NMJ 53645 Homo sapiens nucleoporin 50kDa (NUP50), transcript variant 3, mRNA
NMJ 53646 Homo sapiens solute carrier family 24 (sodium/potassium/calcium exchangei
NMJ 53647 Homo sapiens solute carrier family 24 (sodium/potassium/calcium exchangei
NMJ 53648 Homo sapiens solute carrier family 24 (sodium/potassium/calcium exchangei
NMJ 53649 Homo sapiens tropomyosin 3 (TPM3), mRNA
NMJ 53675 Homo sapiens forkhead box A2 (FOXA2), transcript variant 2, mRNA
NMJ 53676 Homo sapiens Usher syndrome 1 C (autosomal recessive, severe) (USH1 C),
NMJ 53681 Homo sapiens Down syndrome critical region gene 5 (DSCR5), transcript vai
NMJ 53682 Homo sapiens Down syndrome critical region gene 5 (DSCR5), transcript vai
NMJ 53683 Homo sapiens klotho (KL), transcript variant 2, mRNA
NMJ 53684 Homo sapiens nucleoporin 50kDa (NUP50), transcript variant 1 , mRNA
NMJ 53685 Homo sapiens hypothetical protein DKFZp547D2210 (DKFZp547D2210), mF
NMJ 53686 Homo sapiens transcription factor MLR1 (MLR1), mRNA
NMJ 53687 Homo sapiens hypothetical protein FLJ31051 (FLJ31051), mRNA
NMJ 53688 Homo sapiens zinc finger protein 1 homolog (mouse) (ZFP1), mRNA
NMJ 53689 Homo sapiens hypothetical protein FLJ38973 (FLJ38973), mRNA
NMJ 53690 Homo sapiens family with sequence similarity 43, member A (FAM43A), mRt
NMJ 53691 Homo sapiens hypothetical protein FLJ90036 (FLJ90036), mRNA
NMJ 53692 Homo sapiens hypothetical protein FLJ90724 (FLJ90724), mRNA
NMJ 53693 Homo sapiens homeo box C6 (HOXC6), transcript variant 2, mRNA
NMJ 53694 Homo sapiens synaptonemal complex protein 3 (SYCP3), mRNA
NMJ 53695 Homo sapiens zinc finger protein 367 (ZNF367), mRNA
NMJ 53696 Homo sapiens prostate-specific membrane antigen-like protein (PSMAL/GCF
NMJ 53697 Homo sapiens hypothetical protein DKFZp434D2328 (LOC91526), mRNA
NMJ 53699 Homo sapiens glutathione S-transferase A5 (GSTA5), mRNA
NMJ 53700 Homo sapiens stereocilin (STRC), mRNA
NMJ 53701 Homo sapiens interleukin 12 receptor, beta 1 (IL12RB1), transcript variant 2,
NMJ53702 Homo sapiens hypothetical protein MGC10084 (MGC10084), mRNA
NMJ 53703 Homo sapiens podocan (PODN), mRNA
NMJ 53704 Homo sapiens hypothetical protein MGC26979 (MGC26979), mRNA
NMJ 53705 Homo sapiens KDEL (Lys-Asp-Glu-Leu) containing 2 (KDELC2), mRNA
NMJ 53706 Homo sapiens hypothetical protein MGC33648 (MGC33648), mRNA
NMJ 53707 Homo sapiens chromosome 9 open reading frame 138 (C9orf138), mRNA
NMJ 53708 Homo sapiens hypothetical protein MGC35450 (MGC35450), mRNA
NMJ53709 Homo sapiens hypothetical protein MGC40168 (MGC40168), mRNA
NMJ 53711 Homo sapiens chromosome 6 open reading frame 188 (C6orf188), mRNA
NMJ 53712 Homo sapiens tubulin tyrosine ligase (TTL), mRNA
NMJ 53713 Homo sapiens hypothetical protein MGC46719 (MGC46719), mRNA
NMJ 53714 Homo sapiens chromosome 10 open reading frame 67 (C10orf67), mRNA
NMJ 53715 Homo sapiens homeo box A10 (HOXA10), transcript variant 2, mRNA
NMJ 53716 Homo sapiens heat shock transcription factor, Y linked 2 (HSFY2), transcript
NMJ 53717 Homo sapiens Ellis van Creveld syndrome (EVC), transcript variant 2, mRN 1
NMJ 53718 Homo sapiens nucleoporin 62kDa (NUP62), transcript variant 3, mRNA
NMJ 53719 Homo sapiens nucleoporin 62kDa (NUP62), transcript variant 1 , mRNA
NMJ 53741 Homo sapiens dolichyl-phosphate mannosyltransferase polypeptide 3 (DPIV
NMJ 53742 Homo sapiens cystathionase (cystathionine gamma-lyase) (CTH), transcript •
NMJ 53746 Homo sapiens zinc finger, DHHC domain containing 14 (ZDHHC14), mRNA
NMJ 53747 Homo sapiens phosphatidylinositol glycan, class C (PIGC), transcript variant
NMJ 53748 Homo sapiens potassium voltage-gated channel, Shaw-related subfamily, mi
NMJ53750 Homo sapiens chromosome 21 open reading frame 81 (C21orf81), mRNA
NMJ 53752 Homo sapiens chromosome 21 open reading frame 84 (C21orf84), mRNA
NMJ 53754 Homo sapiens chromosome 21 open reading frame 88 (C21orf88), mRNA
NMJ 53756 Homo sapiens fibronectin type III domain containing 5 (FNDC5), mRNA
NMJ 53757 Homo sapiens nucleosome assembly protein 1 -like 5 (NAP1 L5), mRNA
NMJ 53758 Homo sapiens interleukin 19 (IL19), transcript variant 1 , mRNA
NMJ 53759 Homo sapiens DNA (cytosine-5-)-methyltransferase 3 alpha (DNMT3A), tram
NMJ 53763 Homo sapiens potassium voltage-gated channel, Shaw-related subfamily, mi
NMJ 53764 Homo sapiens potassium inwardly-rectifying channel, subfamily J, member 1
NMJ 53765 Homo sapiens potassium inwardly-rectifying channel, subfamily J, member 1
NMJ 53766 Homo sapiens potassium inwardly-rectifying channel, subfamily J, member 1
NMJ 53767 Homo sapiens potassium inwardly-rectifying channel, subfamily J, member 1
NMJ 53768 Homo sapiens calcium-binding tyrosine-(Y)-phosphorylation regulated (fibroi
NMJ 53769 Homo sapiens calcium-binding tyrosine-(Y)-phosphorylation regulated (fibroi
NMJ 53770 Homo sapiens calcium-binding tyrosine-(Y)-phosphorylation regulated (fibroi
NMJ 53773 Homo sapiens chromosome 21 open reading frame 99 (C21orf99), mRNA
NMJ 53809 Homo sapiens TAF1-like RNA polymerase II, TATA box binding protein (TBP
NMJ 53810 Homo sapiens chromosome 10 open reading frame 46 (C10orf46), mRNA
NMJ 53811 Homo sapiens solute carrier family 38, member 6 (SLC38A6), mRNA
NMJ 53812 Homo sapiens PHD finger protein 13 (PHF13), mRNA
NMJ 53813 Homo sapiens zinc finger protein, multitype 1 (ZFPM1), mRNA
NMJ 53815 Homo sapiens Ras protein-specific guanine nucleotide-releasing factor 1 (R
NMJ53816 Homo sapiens sorting nexin 14 (SNX14), transcript variant 1 , mRNA
NMJ 53818 Homo sapiens peroxisome biogenesis factor 10 (PEX10), transcript variant 1
NMJ 53819 Homo sapiens RAS guanyl releasing protein 2 (calcium and DAG-regulated)
NMJ 53822 Homo sapiens proteasome (prosome, macropain) 26S subunit, non-ATPase,
NMJ 53823 Homo sapiens germ cell associated 1 (GSG1), mRNA
NMJ 53824 Homo sapiens pyrroline-5-carboxylate reductase 1 (PYCR1), transcript variai
NMJ 53825 Homo sapiens soluble liver antigen/liver pancreas antigen (SLA LP), mRNA
NMJ 53826 Homo sapiens membrane cofactor protein (CD46, trophoblast-lymphocyte cr
NMJ 53827 Homo sapiens misshapen/NIK-related kinase (MINK), transcript variant 3, ml
NMJ 53828 Homo sapiens reticulon 4 (RTN4), transcript variant 2, mRNA
NMJ 53831 Homo sapiens PTK2 protein tyrosine kinase 2 (PTK2), transcript variant 1 , m
NMJ 53832 Homo sapiens G protein-coupled receptor 161 (GPR161), mRNA
NMJ 53833 Homo sapiens H1 histone family, member O, oocyte-specific (H1 FOO), mRN
NMJ 53834 Homo sapiens G protein-coupled receptor 112 (GPR112), mRNA
NMJ 53835 Homo sapiens G protein-coupled receptor 113 (GPR113), mRNA
NMJ 53836 Homo sapiens cellular repressor of E1A-stimu!ated genes 2 (CREG2), mRN/
NMJ53837 Homo sapiens G protein-coupled receptor 114 (GPR114), mRNA
NMJ 53838 Homo sapiens G protein-coupled receptor 115 (GPR115), mRNA
NMJ 53839 Homo sapiens G protein-coupled receptor 111 (GPR111 ), mRNA
NMJ 53840 Homo sapiens G protein-coupled receptor 110 (GPR110), mRNA
NMJ 56036 Homo sapiens homeo box B6 (HOXB6), transcript variant 3, mRNA
NMJ 56037 Homo sapiens homeo box B6 (HOXB6), transcript variant 1 , mRNA
NMJ 56038 Homo sapiens colony stimulating factor 3 receptor (granulocyte) (CSF3R), tn
NMJ 56039 Homo sapiens colony stimulating factor 3 receptor (granulocyte) (CSF3R), tn
NMJ 70587 Homo sapiens regulator of G-protein signalling 20 (RGS20), mRNA
NMJ70589 Homo sapiens AF15q14 protein (AF15Q14), mRNA
NMJ 70600 Homo sapiens SH2 domain containing 3C (SH2D3C), mRNA
NMJ 70601 Homo sapiens cytosolic sialic acid 9-O-acetylesterase homolog (CSE-C), mF
NMJ 70602 Homo sapiens RAS guanyl releasing protein 4 (RASGRP4), transcript varian
NMJ 70603 Homo sapiens RAS guanyl releasing protein 4 (RASGRP4), transcript varian
NMJ 70604 Homo sapiens RAS guanyl releasing protein 4 (RASGRP4), transcript varian
NMJ 70605 Homo sapiens InaD-like protein (INADL), transcript variant 1 , mRNA
NMJ 70606 Homo sapiens myeloid/lymphoid or mixed-lineage leukemia 3 (MLL3), mRN/1
NMJ 70607 Homo sapiens transcription factor-like 4 (TCFL4), transcript variant 3, mRNA
NMJ 70609 Homo sapiens cysteine-rich secretory protein 1 (CRISP1), transcript variant .
NMJ 70610 Homo sapiens histone 1 , H2ba (HIST1 H2BA), mRNA
NMJ 70662 Homo sapiens Cas-Br-M (murine) ecotropic retroviral transforming sequence
NMJ 70663 Homo sapiens misshapen/NIK-related kinase (MINK), transcript variant 2, mf
NMJ 70664 Homo sapiens otoancorin (OTOA), mRNA
NMJ 70665 Homo sapiens ATPase, Ca++ transporting, cardiac muscle, slow twitch 2 (Al
NMJ70672 Homo sapiens RAS guanyl releasing protein 3 (calcium and DAG-regulated)
NMJ 70674 Homo sapiens Meisl , myeloid ecotropic viral integration site 1 homolog 2 (m
NMJ 70675 Homo sapiens Meisl , myeloid ecotropic viral integration site 1 homolog 2 (m
NMJ70676 Homo sapiens Meisl , myeloid ecotropic viral integration site 1 homolog 2 (
NMJ70677 Homo sapiens Meisl , myeloid ecotropic viral integration site 1 homolog 2 (m
NMJ 70678 Homo sapiens integrin beta 1 binding protein 3 (ITGB1 BP3), mRNA
NMJ70679 Homo sapiens S-phase kinase-associated protein 1A (p19A) (SKP1A), transi
NMJ 70681 Homo sapiens mitochondrial elongation factor G2 (EFG2), nuclear gene enci
NMJ 70682 Homo sapiens purinergic receptor P2X, ligand-gated ion channel, 2 (P2RX2)
NMJ 70683 Homo sapiens purinergic receptor P2X, ligand-gated ion channel, 2 (P2RX2)
NMJ 70685 Homo sapiens tachykinin 4 (hemokinin) (TAC4), mRNA
NMJ70686 Homo sapiens zinc finger protein 398 (ZNF398), transcript variant 1 , mRNA
NMJ 70691 Homo sapiens mitochondrial elongation factor G2 (EFG2), nuclear gene enc<
NMJ 70692 Homo sapiens RAS protein activator like 2 (RASAL2), transcript variant 2, ml
NMJ 70693 Homo sapiens serum/glucocorticoid regulated kinase 2 (SGK2), transcript va
NMJ 70694 Homo sapiens serine hydrolase-like (SERHL), mRNA
NMJ70695 Homo sapiens TGFB-induced factor (TALE family homeobox) (TGIF), transci
NMJ70696 Homo sapiens aldehyde dehydrogenase 1 family, member A2 (ALDH1 A2), tr
NMJ70697 Homo sapiens aldehyde dehydrogenase 1 family, member A2 (ALDH1 A2), tr
NMJ 70698 Homo sapiens similar to CGI-96 (dJ222E13.2), mRNA
NMJ 70699 Homo sapiens G protein-coupled bile acid receptor 1 (GPBAR1), mRNA
NMJ 70705 Homo sapiens isoprenylcysteine carboxyl methyltransferase (ICMT), transcri|
NMJ 70706 Homo sapiens nicotinamide nucleotide adenylyltransferase 2 (NMNAT2), trai
NMJ 70707 Homo sapiens lamin A/C (LMNA), transcript variant 1 , mRNA
NMJ 70708 Homo sapiens lamin A/C (LMNA), transcript variant 3, mRNA
NMJ 70709 Homo sapiens serum/glucocorticoid regulated kinase-like (SGKL), transcript
NMJ70710 Homo sapiens WD repeat domain 17 (WDR17), transcript variant 1 , mRNA
NMJ 70711 Homo sapiens DAZ associated protein 1 (DAZAP1), transcript variant 1 , mRt
NMJ 70712 Homo sapiens Ras association (RalGDS/AF-6) domain family 1 (RASSF1), ti
NMJ 70713 Homo sapiens Ras association (RalGDS/AF-6) domain family 1 (RASSF1 ), ti
NMJ 70714 Homo sapiens Ras association (RalGDS/AF-6) domain family 1 (RASSF1 ), ti
NMJ 70715 Homo sapiens Ras association (RalGDS/AF-6) domain family 1 (RASSF1 ), ti
NMJ 70716 Homo sapiens Ras association (RalGDS/AF-6) domain family 1 (RASSF1 ), ti
NMJ 70717 Homo sapiens Ras association (RalGDS/AF-6) domain family 1 (RASSF1 ), ti
NMJ 70719 Homo sapiens chromosome 13 open reading frame 23 (C13orf23), transcripl
NMJ 70720 Homo sapiens potassium inwardly-rectifying channel, subfamily J, member 1
NMJ 70721 Homo sapiens musashi homolog 2 (Drosophila) (MSI2), transcript variant 2, i
NMJ 70722 Homo sapiens NOD9 protein (NOD9), transcript variant 2, mRNA
NMJ 70723 Homo sapiens chromodomain protein, Y-linked, 1 (CDY1), transcript variant '
NMJ 70724 Homo sapiens polycystic kidney and hepatic disease 1 (autosomal recessive
NMJ 70725 Homo sapiens piggyBac transposable element derived 2 (PGBD2), mRNA
NMJ 70726 Homo sapiens aldehyde dehydrogenase 4 family, member A1 (ALDH4A1), n
NMJ 70731 Homo sapiens brain-derived neurotrophic factor (BDNF), transcript variant 3,
NMJ 70732 Homo sapiens brain-derived neurotrophic factor (BDNF), transcript variant 2,
NMJ 70733 Homo sapiens brain-derived neurotrophic factor (BDNF), transcript variant 5,
NMJ 70734 Homo sapiens brain-derived neurotrophic factor (BDNF), transcript variant 6,
NMJ 70735 Homo sapiens brain-derived neurotrophic factor (BDNF), transcript variant 1 ,
NMJ 70736 Homo sapiens potassium inwardly-rectifying channel, subfamily J, member 1
NMJ 70737 Homo sapiens potassium inwardly-rectifying channel, subfamily J, member 1
NMJ 70738 Homo sapiens mitochondrial ribosomal protein L11 (MRPL11), nuclear gene
NMJ 70739 Homo sapiens mitochondrial ribosomal protein L11 (MRPL11), nuclear gene
NMJ 70740 Homo sapiens aldehyde dehydrogenase 5 family, member A1 (succinate-ser
NMJ 70741 Homo sapiens potassium inwardly-rectifying channel, subfamily J, member 1
NMJ 70742 Homo sapiens potassium inwardly-rectifying channel, subfamily J, member 1
NMJ 70743 Homo sapiens interleukin 28 receptor, alpha (interferon, lambda receptor) (IL
NMJ 70744 Homo sapiens unc-5 homolog B (C. elegans) (UNC5B), mRNA
NMJ 70745 Homo sapiens histone 1 , H2aa (HIST1 H2AA), mRNA
NMJ 70746 Homo sapiens chromosome 11 open reading frame 31 (C11orf31), mRNA
NMJ 70750 Homo sapiens proteasome (prosome, macropain) 26S subunit, non-ATPase,
NMJ 70751 Homo sapiens chromodomain protein, Y-like (CDYL), transcript variant 2, mF
NMJ 70752 Homo sapiens chromodomain protein, Y-like (CDYL), transcript variant 3, mF
N J 70753 Homo sapiens piggyBac transposable element derived 3 (PGBD3), mRNA
NMJ 70754 Homo sapiens tensin like C1 domain containing phosphatase (TENC1), trans
NMJ 70768 Homo sapiens zinc finger protein 91 homolog (mouse) (ZFP91), transcript va
NMJ 70769 Homo sapiens ring finger protein 39 (RNF39), transcript variant 2, mRNA
NMJ 70770 Homo sapiens ring finger protein 39 (RNF39), transcript variant 3, mRNA
NMJ 70771 Homo sapiens aldehyde dehydrogenase 8 family, member A1 (ALDH8A1), tr
NMJ 70773 Homo sapiens Ras association (RalGDS/AF-6) domain family 2 (RASSF2), ti
NMJ 70774 Homo sapiens Ras association (RalGDS/AF-6) domain family 2 (RASSF2), ti
NMJ 70775 Homo sapiens potassium intermediate/small conductance calcium-activated
NMJ 70776 Homo sapiens G protein-coupled receptor 97 (GPR97), mRNA
NMJ 70780 Homo sapiens MOX2 receptor (MOX2R), transcript variant 4, mRNA
NMJ 70781 Homo sapiens protein phosphatase 1 , regulatory (inhibitor) subunit 11 (PPP1
NMJ 70782 Homo sapiens potassium intermediate/small conductance calcium-activated
NMJ70783 Homo sapiens zinc ribbon domain containing, 1 (ZNRD1), transcript variant ε
NMJ 70784 Homo sapiens McKusick-Kaufman syndrome (MKKS), transcript variant 2, m
NMJ71825 Homo sapiens calcium/calmodulin-dependent protein kinase (CaM kinase) II
NMJ 71827 Homo sapiens CD8 antigen, alpha polypeptide (p32) (CD8A), transcript varia
NMJ 71828 Homo sapiens potassium large conductance calcium-activated channel, subt
NMJ 71829 Homo sapiens potassium large conductance calcium-activated channel, subt
NMJ 71830 Homo sapiens potassium large conductance calcium-activated channel, subt
NMJ 71846 Homo sapiens lactamase, beta (LACTB), nuclear gene encoding mitochondr
NMJ71982 Homo sapiens tripartite motif-containing 35 (TRIM35), transcript variant 2, ml
NMJ 71997 Homo sapiens ubiquitin specific protease 2 (USP2), transcript variant 2, mRt*
NMJ 71998 Homo sapiens RAB39B, member RAS oncogene family (RAB39B), mRNA
NMJ 71999 Homo sapiens sal-like 3 (Drosophila) (SALL3), mRNA
NMJ 72000 Homo sapiens putative membrane protein HE9 (HE9), mRNA
N J 72002 Homo sapiens J-type co-chaperone HSC20 (HSC20), mRNA
NMJ 72003 Homo sapiens COBW domain containing 2 (CBWD2), mRNA
NMJ 72004 Homo sapiens dendritic cell-associated lectin-1 (DCAL1), mRNA
NMJ 72005 Homo sapiens WAP four-disulfide core domain 13 (WFDC13), mRNA
NMJ 72006 Homo sapiens WAP four-disulfide core domain 10B (WFDC10B), transcript \
NMJ 72014 Homo sapiens tumor necrosis factor (ligand) superfamily, member 14 (TNFS
NMJ 72016 Homo sapiens tripartite motif-containing 39 (TRIM39), transcript variant 2, ml
NMJ 72020 Homo sapiens POM121 membrane glycoprotein (rat) (POM121), mRNA
NMJ 72024 Homo sapiens ATP-binding cassette, sub-family C (CFTR/MRP), member 13
NMJ 72025 Homo sapiens ATP-binding cassette, sub-family C (CFTR/MRP), member 13
NMJ 72026 Homo sapiens ATP-binding cassette, sub-family C (CFTR/MRP), member 13
NMJ 72027 Homo sapiens ankyrin repeat and BTB (POZ) domain containing 1 (ABTB1),
NMJ 72028 Homo sapiens ankyrin repeat and BTB (POZ) domain containing 1 (ABTB1),
NMJ72037 Homo sapiens retinol dehydrogenase 10 (all-trans) (RDH10), mRNA
NMJ 72056 Homo sapiens potassium voltage-gated channel, subfamily H (eag-related), i
NMJ 72057 Homo sapiens potassium voltage-gated channel, subfamily H (eag-related), I
NMJ 72058 Homo sapiens eyes absent homolog 1 (Drosophila) (EYA1), transcript varian
NMJ 72059 Homo sapiens eyes absent homolog 1 (Drosophila) (EYA1 ), transcript varian
NMJ 72060 Homo sapiens eyes absent homolog 1 (Drosophila) (EYA1 ), transcript varian
NMJ 72069 Homo sapiens pleckstrin homology domain containing, family H (with MyTH4
NMJ72070 Homo sapiens similar to F10G7.10.p (KIAA2024), mRNA
NMJ 72078 Homo sapiens calcium/calmodulin-dependent protein kinase (CaM kinase) II
NMJ72079 Homo sapiens calcium/calmodulin-dependent protein kinase (CaM kinase) II
NMJ 72080 Homo sapiens calcium/calmodulin-dependent protein kinase (CaM kinase) II
NMJ 72081 Homo sapiens calcium/calmodulin-dependent protein kinase (CaM kinase) II
NMJ 72082 Homo sapiens calcium/calmodulin-dependent protein kinase (CaM kinase) II
NMJ72083 Homo sapiens calcium/calmodulin-dependent protein kinase (CaM kinase) II
NMJ72084 Homo sapiens calcium/calmodulin-dependent protein kinase (CaM kinase) II
NMJ 72087 Homo sapiens tumor necrosis factor (ligand) superfamily, member 13 (TNFS
NMJ 72088 Homo sapiens tumor necrosis factor (ligand) superfamily, member 13 (TNFS
NMJ 72089 Homo sapiens tumor necrosis factor (ligand) superfamily, member 12-membι
NMJ 72095 Homo sapiens cation channel, sperm associated 2 (CATSPER2), transcript \
NMJ 72096 Homo sapiens cation channel, sperm associated 2 (CATSPER2), transcript \
NMJ 72097 Homo sapiens cation channel, sperm associated 2 (CATSPER2), transcript \
NMJ 72098 Homo sapiens eyes absent homolog 3 (Drosophila) (EYA3), transcript varian
NMJ 72099 Homo sapiens CD8 antigen, beta polypeptide 1 (p37) (CD8B1), transcript vai
NMJ 72100 Homo sapiens CD8 antigen, beta polypeptide 1 (p37) (CD8B1), transcript vai
NMJ 72101 Homo sapiens CD8 antigen, beta polypeptide 1 (p37) (CD8B1), transcript vai
NMJ 72102 Homo sapiens CD8 antigen, beta polypeptide 1 (p37) (CD8B1), transcript vai
NMJ 72103 Homo sapiens eyes absent homolog 4 (Drosophila) (EYA4), transcript varian
NMJ 72104 Homo sapiens eyes absent homolog 4 (Drosophila) (EYA4), transcript varian
NMJ 72105 Homo sapiens eyes absent homolog 4 (Drosophila) (EYA4), transcript varian
NMJ 72106 Homo sapiens potassium voltage-gated channel, KQT-like subfamily, membe
NMJ 72107 Homo sapiens potassium voltage-gated channel, KQT-like subfamily, membe
NMJ 72108 Homo sapiens potassium voltage-gated channel, KQT-like subfamily, membe
NMJ 72109 Homo sapiens potassium voltage-gated channel, KQT-like subfamily, membe
NMJ 72110 Homo sapiens eyes absent homolog 2 (Drosophila) (EYA2), transcript varian
NMJ 72111 Homo sapiens eyes absent homolog 2 (Drosophila) (EYA2), transcript varian
NMJ 72112 Homo sapiens eyes absent homolog 2 (Drosophila) (EYA2), transcript varian
NMJ 72113 Homo sapiens eyes absent homolog 2 (Drosophila) (EYA2), transcript varian
NMJ 72115 Homo sapiens calcium/calmodulin-dependent protein kinase (CaM kinase) II
NMJ 72127 Homo sapiens calcium/calmodulin-dependent protein kinase (CaM kinase) II
NMJ 72128 Homo sapiens calcium/calmodulin-dependent protein kinase (CaM kinase) II
NMJ 72130 Homo sapiens potassium voltage-gated channel, shaker-related subfamily, b
NMJ 72131 Homo sapiens WAP four-disulfide core domain 10B (WFDC10B), transcript \
NMJ 72138 Homo sapiens interleukin 28A (interferon, lambda 2) (IL28A), mRNA
NMJ 72139 Homo sapiens interleukin 28B (interferon, lambda 3) (IL28B), mRNA
NMJ72140 Homo sapiens interleukin 29 (interferon, lambda 1) (IL29), mRNA
NMJ 72159 Homo sapiens potassium voltage-gated channel, shaker-related subfamily, b
NMJ 72160 Homo sapiens potassium voltage-gated channel, shaker-related subfamily, b
NMJ 72163 Homo sapiens potassium voltage-gated channel, KQT-like subfamily, membe
NMJ 72164 Homo sapiens nuclear autoantigenic sperm protein (histone-binding) (NASP)
NMJ 72165 Homo sapiens mutS homolog 5 (E. coli) (MSH5), transcript variant 2, mRNA
NMJ 72166 Homo sapiens mutS homolog 5 (E. coli) (MSH5), transcript variant 4, mRNA
NMJ 72167 Homo sapiens NADPH oxidase organizer 1 (NOX01), transcript variant b, ml
NMJ 72168 Homo sapiens NADPH oxidase organizer 1 (NOX01 ), transcript variant c, ml
NMJ 72169 Homo sapiens calcium/calmodulin-dependent protein kinase (CaM kinase) II
NMJ 72170 Homo sapiens calcium/calmodulin-dependent protein kinase (CaM kinase) II
NMJ 72171 Homo sapiens calcium/calmodulin-dependent protein kinase (CaM kinase) II
NMJ 72172 Homo sapiens calcium/calmodulin-dependent protein kinase (CaM kinase) II
NMJ 72173 Homo sapiens calcium/calmodulin-dependent protein kinase (CaM kinase) II
NMJ 72174 Homo sapiens interleukin 15 (IL15), transcript variant 1 , mRNA
NMJ 72175 Homo sapiens interleukin 15 (IL15), transcript variant 2, mRNA
NMJ 72177 Homo sapiens mitochondrial ribosomal protein L42 (MRPL42), nuclear gene
NMJ 72178 Homo sapiens mitochondrial ribosomal protein L42 (MRPL42), nuclear gene
NMJ 72193 Homo sapiens kelch domain containing 1 (KLHDC1), mRNA
N J 72195 Homo sapiens eukaryotic translation initiation factor 2B, subunit 4 delta, 67kl
NMJ 72196 Homo sapiens TFIIA-alpha/beta-like factor (ALF), transcript variant 2, mRNA
NMJ 72197 Homo sapiens advanced glycosylation end product-specific receptor (AGER)
NMJ 72198 Homo sapiens potassium voltage-gated channel, Shal-related subfamily, mei
N J 72199 Homo sapiens adenylate kinase 2 (AK2), transcript variant AK2C, mRNA
NMJ 72200 Homo sapiens interleukin 15 receptor, alpha (IL15RA), transcript variant 2, rr
NMJ 72201 Homo sapiens potassium voltage-gated channel, Isk-related family, member
NMJ 72206 Homo sapiens calcium/calmodulin-dependent protein kinase kinase 1 , alpha
NMJ 72207 Homo sapiens calcium/calmodulin-dependent protein kinase kinase 1 , alpha
NMJ 72208 Homo sapiens TAP binding protein (tapasin) (TAPBP), transcript variant 2, m
NMJ 72209 Homo sapiens TAP binding protein (tapasin) (TAPBP), transcript variant 3, m
NMJ 72210 Homo sapiens colony stimulating factor 1 (macrophage) (CSF1), transcript vι
NMJ 72211 Homo sapiens colony stimulating factor 1 (macrophage) (CSF1), transcript vi
NMJ 72212 Homo sapiens colony stimulating factor 1 (macrophage) (CSF1), transcript v.
NMJ 72213 Homo sapiens CD8 antigen, beta polypeptide 1 (p37) (CD8B1), transcript vai
NMJ 72214 Homo sapiens calcium/calmodulin-dependent protein kinase kinase 2, beta (
NMJ 72215 Homo sapiens calcium/calmodulin-dependent protein kinase kinase 2, beta (
NMJ 72216 Homo sapiens calcium/calmodulin-dependent protein kinase kinase 2, beta (
NMJ 72217 Homo sapiens interleukin 16 (lymphocyte chemoattractant factor) (IL16), trar
NMJ 72218 Homo sapiens sperm associated antigen 1 (SPAG1 ), transcript variant 2, mF
NMJ 72219 Homo sapiens colony stimulating factor 3 (granulocyte) (CSF3), transcript va
NMJ 72220 Homo sapiens colony stimulating factor 3 (granulocyte) (CSF3), transcript va
NMJ 72225 Homo sapiens diencephalon/mesencephalon homeobox 1 (DMBX1), transcri
NMJ 72226 Homo sapiens calcium/calmodulin-dependent protein kinase kinase 2, beta (
NMJ 72229 Homo sapiens kringle containing transmembrane protein 2 (KREMEN2), tran
NMJ 72230 Homo sapiens synovial apoptotis inhibitor 1 , synoviolin (SYVN1 ), transcript v
NMJ 72231 Homo sapiens splicing factor 4 (SF4), transcript variant a, mRNA
NMJ 72232 Homo sapiens ATP-binding cassette, sub-family A (ABC1 ), member 5 (ABC/
NMJ72234 Homo sapiens interleukin 17 receptor B (IL17RB), transcript variant 2, mRN/1
NMJ 72236 Homo sapiens protein O-fucosyltransferase 1 (POFUT1 ), transcript variant 2,
NMJ 72238 Homo sapiens transcription factor AP-2 beta (activating enhancer binding pre
NMJ 72239 Homo sapiens exonuclease GOR (GOR), mRNA
NMJ 72240 Homo sapiens TUWD12 (TUWD12), mRNA
NMJ 72241 Homo sapiens cutaneous T-cell lymphoma-associated antigen 1 (CTAGE1),
NMJ 72242 Homo sapiens sperm associated antigen 6 (SPAG6), transcript variant 2, mF
NMJ 72244 Homo sapiens sarcoglycan, delta (35kDa dystrophin-associated glycoprotein
NMJ 72245 Homo sapiens colony stimulating factor 2 receptor, alpha, low-affinity (granul
NMJ 72246 Homo sapiens colony stimulating factor 2 receptor, alpha, low-affinity (granul
NMJ 72247 Homo sapiens colony stimulating factor 2 receptor, alpha, low-affinity (granul
NMJ 72248 Homo sapiens colony stimulating factor 2 receptor, alpha, low-affinity (granul
NMJ 72249 Homo sapiens colony stimulating factor 2 receptor, alpha, low-affinity (granul
NMJ 72250 Homo sapiens methylmalonic aciduria (cobalamin deficiency) type A (MMAA
NMJ 72251 Homo sapiens mitochondrial ribosomal protein L54 (MRPL54), nuclear gene
NMJ 72311 Homo sapiens stoned B/TFIlA-alpha/beta-like factor (SALF), mRNA
NMJ72312 Homo sapiens sperm associated antigen 8 (SPAG8), transcript variant 2, mF
NMJ 72313 Homo sapiens colony stimulating factor 3 receptor (granulocyte) (CSF3R), tn
NMJ72314 Homo sapiens interleukin 17E (IL17E), transcript variant 2, mRNA
NMJ 72315 Homo sapiens Meisl , myeloid ecotropic viral integration site 1 homolog 2 (m
NMJ 72316 Homo sapiens Meisl , myeloid ecotropic viral integration site 1 homolog 2 (m
NMJ 72318 Homo sapiens potassium voltage-gated channel, subfamily G, member 1 (KC
NMJ 72337 Homo sapiens orthodenticle homolog 2 (Drosophila) (OTX2), transcript variai
NMJ 72341 Homo sapiens presenilin enhancer 2 (PEN2), mRNA
NMJ 72343 Homo sapiens interleukin 17F (IL17F), transcript variant 2, mRNA
NMJ 72344 Homo sapiens potassium voltage-gated channel, subfamily G, member 3 (K(
NMJ 72345 Homo sapiens sperm associated antigen 9 (SPAG9), transcript variant 2, mF
NMJ 72346 Homo sapiens ATP-binding cassette, sub-family A (ABC1 ), member 6 (ABC/
NMJ 72347 Homo sapiens potassium voltage-gated channel, subfamily G, member 4 (KC
NMJ 72348 Homo sapiens interleukin 4 (IL4), transcript variant 2, mRNA
NMJ 72349 Homo sapiens nuclear receptor binding SET domain protein 1 (NSD1), transi
NMJ 72350 Homo sapiens membrane cofactor protein (CD46, trophoblast-lymphocyte cr
NMJ 72351 Homo sapiens membrane cofactor protein (CD46, trophoblast-lymphocyte cr
NMJ 72352 Homo sapiens membrane cofactor protein (CD46, trophoblast-lymphocyte cr
NMJ 72353 Homo sapiens membrane cofactor protein (CD46, trophoblast-lymphocyte cr
NMJ 72354 Homo sapiens membrane cofactor protein (CD46, trophoblast-lymphocyte cr
NMJ 72355 Homo sapiens membrane cofactor protein (CD46, trophoblast-lymphocyte cr
NMJ 72356 Homo sapiens membrane cofactor protein (CD46, trophoblast-lymphocyte cr
NMJ 72357 Homo sapiens membrane cofactor protein (CD46, trophoblast-lymphocyte cr
NMJ 72358 Homo sapiens membrane cofactor protein (CD46, trophoblast-lymphocyte cr
NMJ 72359 Homo sapiens membrane cofactor protein (CD46, trophoblast-lymphocyte cr
NMJ 72360 Homo sapiens membrane cofactor protein (CD46, trophoblast-lymphocyte cr
NMJ 72361 Homo sapiens membrane cofactor protein (CD46, trophoblast-lymphocyte cr
NMJ 72362 Homo sapiens potassium voltage-gated channel, subfamily H (eag-related), i
NMJ 72364 Homo sapiens calcium channel, voltage-dependent, alpha 2/delta subunit 4 (
NMJ72365 Homo sapiens chromosome 14 open reading frame 50 (C14orf50), mRNA
NMJ 72366 Homo sapiens F-box protein 16 (FBX016), mRNA
NMJ 72367 Homo sapiens tumor suppressor candidate 5 (TUSC5), mRNA
NMJ 72369 Homo sapiens complement component 1 , q subcomponent, gamma polypep
NMJ 72370 Homo sapiens D-amino acid oxidase activator (DAOA), mRNA
NMJ 72373 Homo sapiens E74-like factor 1 (ets domain transcription factor) (ELF1), mRI
NMJ 72374 Homo sapiens interleukin 4 induced 1 (IL4I1), transcript variant 2, mRNA
NMJ 72375 Homo sapiens potassium voltage-gated channel, subfamily H (eag-related), I
NMJ 72376 Homo sapiens potassium voltage-gated channel, subfamily H (eag-related), i
NMJ 72377 Homo sapiens cancer/testis antigen 2 (CTAG2), transcript variant 1 , mRNA
N J 72386 Homo sapiens ATP-binding cassette, sub-family A (ABC1 ), member 9 (ABC/
NMJ 72387 Homo sapiens nuclear factor of activated T-cells, cytoplasmic, calcineurin-de
NMJ 72388 Homo sapiens nuclear factor of activated T-cells, cytoplasmic, calcineurin-de
NMJ 72389 Homo sapiens nuclear factor of activated T-cells, cytoplasmic, calcineurin-de
NMJ 72390 Homo sapiens nuclear factor of activated T-cells, cytoplasmic, calcineurin-de
NMJ 73039 Homo sapiens aquaporin 11 (AQP11), mRNA
NMJ73042 Homo sapiens interleukin 18 binding protein (IL18BP), transcript variant A, m
NMJ 73043 Homo sapiens interleukin 18 binding protein (IL18BP), transcript variant B, m
NMJ 73044 Homo sapiens interieukin 18 binding protein (IL18BP), transcript variant D, rr
NMJ 73050 Homo sapiens signal peptide, CUB domain, EGF-like 1 (SCUBE1), mRNA
NMJ 73054 Homo sapiens reelin (RELN), transcript variant 2, mRNA
NMJ 73055 Homo sapiens zonadhesin (ZAN), transcript variant 1 , mRNA
NMJ 73056 Homo sapiens zonadhesin (ZAN), transcript variant 2, mRNA
NMJ 73057 Homo sapiens zonadhesin (ZAN), transcript variant 4, mRNA
N J 73058 Homo sapiens zonadhesin (ZAN), transcript variant 5, mRNA
NMJ 73059 Homo sapiens zonadhesin (ZAN), transcript variant 6, mRNA
NMJ 73060 Homo sapiens calpastatin (CAST), transcript variant 2, mRNA
NMJ 73061 Homo sapiens calpastatin (CAST), transcript variant 3, mRNA
NMJ 73062 Homo sapiens calpastatin (CAST), transcript variant 4, mRNA
NMJ 73064 Homo sapiens interleukin 28 receptor, alpha (interferon, lambda receptor) (IL
NMJ 73065 Homo sapiens interleukin 28 receptor, alpha (interferon, lambda receptor) (IL
NMJ 73073 Homo sapiens solute carrier family 35, member C2 (SLC35C2), transcript va
NMJ 73074 Homo sapiens phosphatidylinositol glycan, class F (PIGF), transcript variant
NMJ 73075 Homo sapiens amyloid beta (A4) precursor protein-binding, family B, membe
NMJ 73076 Homo sapiens ATP-binding cassette, sub-family A (ABC1), member 12 (ABC
NMJ 73077 Homo sapiens carboxypeptidase O (CPO), mRNA
NMJ 73078 Homo sapiens SLIT and NTRK-like family, member 4 (SLITRK4), mRNA
NMJ 73079 Homo sapiens RUN domain containing 1 (RUNDC1), mRNA
NMJ 73080 Homo sapiens small proline rich protein 4 (SPRR4), mRNA
NMJ 73081 Homo sapiens armadillo repeat containing 3 (ARMC3), mRNA
NMJ 73082 Homo sapiens SNF2 histone linker PHD RING helicase (SHPRH), mRNA
NMJ 73083 Homo sapiens TUDOR gene similar (TGS), mRNA
NMJ 73084 Homo sapiens tripartite motif-containing 59 (TRIM59), mRNA
NMJ 73086 Homo sapiens keratin 6E (KRT6E), mRNA
NMJ 73087 Homo sapiens calpain 3, (p94) (CAPN3), transcript variant 3, mRNA
NMJ 73088 Homo sapiens calpain 3, (p94) (CAPN3), transcript variant 4, mRNA
NMJ 73089 Homo sapiens calpain 3, (p94) (CAPN3), transcript variant 5, mRNA
NMJ 73090 Homo sapiens calpain 3, (p94) (CAPN3), transcript variant 6, mRNA
NMJ 73091 Homo sapiens nuclear factor of activated T-cells, cytoplasmic, calcineurin-de
NMJ 73092 Homo sapiens potassium voltage-gated channel, subfamily H (eag-related), i
NMJ 73156 Homo sapiens chromosome 1 open reading frame 16 (C1orf16), transcript VJ
NMJ 73157 Homo sapiens nuclear receptor subfamily 4, group A, member 1 (NR4A1), tn
NMJ 73158 Homo sapiens nuclear receptor subfamily 4, group A, member 1 (NR4A1), tn
NMJ73159 Homo sapiens neuronal PAS domain protein 3 (NPAS3), mRNA
NMJ 73160 Homo sapiens FXYD domain containing ion transport regulator 4 (FXYD4), n
NMJ 73161 Homo sapiens interleukin 1 family, member 10 (theta) (IL1F10), transcript va
NMJ 73162 Homo sapiens potassium voltage-gated channel, subfamily H (eag-related), I
NMJ 73163 Homo sapiens nuclear factor of activated T-cells, cytoplasmic, calcineurin-de
NMJ 73164 Homo sapiens nuclear factor of activated T-cells, cytoplasmic, calcineurin-de
NMJ 73165 Homo sapiens nuclear factor of activated T-cells, cytoplasmic, calcineurin-de
NMJ 73167 Homo sapiens cardiomyopathy associated 4 (CMYA4), mRNA
NMJ 73170 Homo sapiens interleukin 1 family, member 5 (delta) (IL1 F5), transcript variai
NMJ 73171 Homo sapiens nuclear receptor subfamily 4, group A, member 2 (NR4A2), tn
NMJ 73172 Homo sapiens nuclear receptor subfamily 4, group A, member 2 (NR4A2), tn
NMJ73173 Homo sapiens nuclear receptor subfamily 4, group A, member 2 (NR4A2), tn
NMJ 73174 Homo sapiens PTK2B protein tyrosine kinase 2 beta (PTK2B), transcript vari
NMJ73175 Homo sapiens PTK2B protein tyrosine kinase 2 beta (PTK2B), transcript vari
NMJ 73176 Homo sapiens PTK2B protein tyrosine kinase 2 beta (PTK2B), transcript vari
NMJ 73177 Homo sapiens nuclear DNA-binding protein (C1D), transcript variant 2, mRN,
NMJ73178 Homo sapiens interleukin 1 family, member 8 (eta) (IL1F8), transcript variant
NMJ 73179 Homo sapiens solute carrier family 35, member C2 (SLC35C2), transcript va
NMJ73191 Homo sapiens Kv channel interacting protein 2 (KCNIP2), transcript variant 2
NMJ 73192 Homo sapiens Kv channel interacting protein 2 (KCNIP2), transcript variant 3
NMJ 73193 Homo sapiens Kv channel interacting protein 2 (KCNIP2), transcript variant 4
NMJ73194 Homo sapiens Kv channel interacting protein 2 (KCNIP2), transcript variant £
NMJ73195 Homo sapiens Kv channel interacting protein 2 (KCNIP2), transcript variant 6
NMJ73197 Homo sapiens Kv channel interacting protein 2 (KCNIP2), transcript variant 7
NMJ73198 Homo sapiens nuclear receptor subfamily 4, group A, member 3 (NR4A3), tn
NMJ73199 Homo sapiens nuclear receptor subfamily 4, group A, member 3 (NR4A3), tπ
N J 73200 Homo sapiens nuclear receptor subfamily 4, group A, member 3 (NR4A3), tn
NMJ 73201 Homo sapiens ATPase, Ca++ transporting, cardiac muscle, fast twitch 1 (ATI
NMJ73202 Homo sapiens interleukin 1 family, member 7 (zeta) (IL1 F7), transcript variar
NMJ 73203 Homo sapiens interleukin 1 family, member 7 (zeta) (IL1 F7), transcript variar
NMJ 73204 Homo sapiens interleukin 1 family, member 7 (zeta) (IL1 F7), transcript variar
NMJ 73205 Homo sapiens interleukin 1 family, member 7 (zeta) (IL1 F7), transcript variar
NMJ 73206 Homo sapiens protein inhibitor of activated STAT, 2 (PIAS2), transcript variai
NMJ 73207 Homo sapiens TGFB-induced factor (TALE family homeobox) (TGIF), transci
NMJ 73208 Homo sapiens TGFB-induced factor (TALE family homeobox) (TGIF), transci
NMJ 73209 Homo sapiens TGFB-induced factor (TALE family homeobox) (TGIF), transcr
NMJ 73210 Homo sapiens TGFB-induced factor (TALE family homeobox) (TGIF), transci
NMJ73211 Homo sapiens TGFB-induced factor (TALE family homeobox) (TGIF), transci
NMJ 73213 Homo sapiens keratin 23 (histone deacetylase inducible) (KRT23), transcript
NMJ 73214 Homo sapiens nuclear factor of activated T-cells 5, tonicity-responsive (NFA"
NMJ 73215 Homo sapiens nuclear factor of activated T-cells 5, tonicity-responsive (NFA"
NMJ 73216 Homo sapiens sialyitransferase 1 (beta-galactoside alpha-2,6-sialyltransferai
NMJ 73217 Homo sapiens sialyitransferase 1 (beta-galactoside alpha-2,6-sialyltransferai
NMJ 73341 Homo sapiens PHD finger protein 7 (PHF7), transcript variant 2, mRNA
NMJ 73342 Homo sapiens Kv channel interacting protein 2 (KCNIP2), transcript variant S
NMJ 73343 Homo sapiens interleukin 1 receptor, type II (IL1R2), transcript variant 2, mR
NMJ 73344 Homo sapiens sialyitransferase 4A (beta-galactoside alpha-2,3-sialyltransfer
NMJ 73351 Homo sapiens olfactory receptor, family 6, subfamily B, member 3 (OR6B3),
NMJ 73352 Homo sapiens keratin 5b (K5B), mRNA
NMJ 73353 Homo sapiens tryptophan hydroxylase 2 (TPH2), mRNA
NMJ73354 Homo sapiens SNF1-like kinase (SNF1LK), mRNA
NMJ 73355 Homo sapiens uridine phosphorylase 2 (UPP2), mRNA
NMJ 73357 Homo sapiens synovial sarcoma, X breakpoint 6 (SSX6), mRNA
NMJ 73358 Homo sapiens synovial sarcoma, X breakpoint 7 (SSX7), mRNA
NMJ 73360 Homo sapiens spermatogenesis associated 9 (SPATA9), transcript variant 2.
NMJ 73362 Homo sapiens LOC317671 (LOC317671), mRNA
NMJ 73452 Homo sapiens ficolin (collagen/fibrinogen domain containing) 3 (Hakata antig
NMJ 73454 Homo sapiens phosphodiesterase 8A (PDE8A), transcript variant 2, mRNA
NMJ 73455 Homo sapiens phosphodiesterase 8A (PDE8A), transcript variant 3, mRNA
NMJ 73456 Homo sapiens phosphodiesterase 8A (PDE8A), transcript variant 4, mRNA
NMJ 73457 Homo sapiens phosphodiesterase 8A (PDE8A), transcript variant 5, mRNA
NMJ 73459 Homo sapiens interleukin 1 receptor-like 1 (IL1 RL1), transcript variant 3, mRI
NMJ 73460 Homo sapiens defensin, beta 119 (DEFB119), transcript variant 2, mRNA
NMJ 73462 Homo sapiens papilin, proteoglycan-like sulfated glycoprotein (PAPLN), mRI*
NMJ 73463 Homo sapiens hypothetical protein DKFZp761 B107 (DKFZp761 B107), mRN;
NMJ 73464 Homo sapiens i(3)mbt-like 4 (Drosophila) (L3MBTL4), mRNA
NMJ 73465 Homo sapiens collagen, type XXIII, alpha 1 (COL23A1), mRNA
NMJ 73466 Homo sapiens hypothetical protein DKFZp434P055 (DKFZp434P055), mRN;
NMJ 73467 Homo sapiens malonyl-CoA:acyl carrier protein transacylase (malonyltransfe
NMJ 73468 Homo sapiens MOB1 , Mps One Binder kinase activator-like 1 A (yeast) (MOB
NMJ 73469 Homo sapiens hypothetical protein LOC92912 (LOC92912), mRNA
NMJ 73470 Homo sapiens transmembrane protein 32 (TMEM32), mRNA
NMJ 73471 Homo sapiens solute carrier family 25, member 26 (SLC25A26), mRNA
NMJ 73472 Homo sapiens hypothetical protein MGC40179 (MGC40179), mRNA
NMJ 73473 Homo sapiens chromosome 10 open reading frame 104 (C1 Oorfl 04), mRNA
NMJ 73474 Homo sapiens N-terminal asparagine amidase (NTAN1), mRNA
NMJ 73475 Homo sapiens hypothetical protein MGC48972 (MGC48972), mRNA
NMJ 73476 Homo sapiens hypothetical protein FLJ34512 (FLJ34512), mRNA
NMJ73477 Homo sapiens Usher syndrome 1G (autosomal recessive) (USH1G), mRNA
NMJ 73478 Homo sapiens hypothetical protein FLJ40137 (FLJ40137), mRNA
NMJ 73479 Homo sapiens hypothetical protein LOC126248 (LOC126248), mRNA
NMJ 73480 Homo sapiens hypothetical protein LOC126295 (LOC126295), mRNA
NMJ73481 Homo sapiens chromosome 19 open reading frame 21 (C19orf21), mRNA
NMJ 73482 Homo sapiens hypothetical protein FLJ40365 (FLJ40365), mRNA
NMJ 73483 Homo sapiens hypothetical protein FLJ39501 (FLJ39501), mRNA
NMJ 73484 Homo sapiens hypothetical protein FLJ40160 (FLJ40160), mRNA
NMJ 73485 Homo sapiens chromosome 20 open reading frame 17 (C20orf17), mRNA
NMJ 73486 Homo sapiens hypothetical protein FLJ40298 (FLJ40298), mRNA
NMJ 73487 Homo sapiens hypothetical protein LOC132321 (LOC132321), mRNA
NMJ 73488 Homo sapiens solute carrier organic anion transporter family, member 6A1 (!
NMJ 73489 Homo sapiens hypothetical protein FLJ40243 (FLJ40243), mRNA
NMJ 73490 Homo sapiens hypothetical protein LOC134285 (LOC134285), mRNA
NMJ 73491 Homo sapiens LSM11 , U7 small nuclear RNA associated (LSM11 ), mRNA
NMJ 73492 Homo sapiens phosphatidylinositol-4-phosphate 5-kinase-like 1 (PIP5KL1), r
NMJ 73493 Homo sapiens PAS domain containing protein 1 (PASD1), mRNA
NMJ 73494 Homo sapiens hypothetical protein MGC35261 (MGC35261 ), mRNA
NMJ 73495 Homo sapiens hypothetical protein FLJ30296 (FLJ30296), mRNA
NMJ 73496 Homo sapiens membrane protein, palmitoylated 7 (MAGUK p55 subfamily m
NMJ 73497 Homo sapiens HECT domain containing 2 (HECTD2), transcript variant 2, ml
NMJ 73499 Homo sapiens spermatogenesis-related protein 8 (MGC44294), mRNA
NMJ 73500 Homo sapiens tau tubulin kinase 2 (TTBK2), mRNA
NMJ 73501 Homo sapiens hypothetical protein LOC146174 (LOC146174), mRNA
NMJ 73502 Homo sapiens hypothetical protein FLJ90661 (FLJ90661), mRNA
NMJ 73503 Homo sapiens hypothetical protein FLJ25818 (FLJ25818), mRNA
NMJ 73505 Homo sapiens ankyrin repeat domain 29 (ANKRD29), mRNA
NMJ 73506 Homo sapiens hypothetical protein MGC42718 (MGC42718), mRNA
NMJ 73507 Homo sapiens hypothetical protein FLJ37118 (FLJ37118), mRNA
NMJ 73508 Homo sapiens solute carrier family 35, member F3 (SLC35F3), mRNA
NMJ 73509 Homo sapiens hypothetical protein MGC16664 (MGC16664), mRNA
NMJ 73510 Homo sapiens hypothetical protein FLJ33814 (FLJ33814), mRNA
NMJ 73511 Homo sapiens amyotrophic lateral sclerosis 2 (juvenile) chromosome region,
NMJ 73512 Homo sapiens hypothetical protein FLJ39822 (FLJ39822), mRNA
NMJ73513 Homo sapiens hypothetical protein MGC43122 (MGC43122), mRNA
NMJ73514 Homo sapiens hypothetical protein FLJ90709 (FLJ90709), mRNA
NMJ 73515 Homo sapiens membrane associated guanylate kinase interacting protein-lik
NMJ 73516 Homo sapiens poly(A)-specific ribonuclease (PARN)-like domain containing '
NMJ 73517 Homo sapiens vitamin K epoxide reductase complex, subunit 1-like 1 (VKOR
NMJ 73518 Homo sapiens hypothetical protein FLJ25692 (FLJ25692), mRNA
NMJ 73519 Homo sapiens hypothetical protein MGC34646 (MGC34646), mRNA
NMJ 73521 Homo sapiens chromosome 9 open reading frame 84 (C9orf84), mRNA
NMJ 73522 Homo sapiens hypothetical protein FLJ36576 (FLJ36576), mRNA
NMJ 73523 Homo sapiens melanoma antigen, family B, 6 (MAGEB6), mRNA
NMJ 73524 Homo sapiens chromosome 10 open reading frame 64 (C10orf64), mRNA
NMJ 73525 Homo sapiens hypothetical protein MGC34805 (MGC34805), mRNA
NMJ 73526 Homo sapiens chromosome 14 open reading frame 54 (C14orf54), mRNA
NMJ 73527 Homo sapiens hypothetical protein FLJ38964 (FLJ38964), mRNA
NMJ 73528 Homo sapiens chromosome 15 open reading frame 26 (C15orf26), mRNA
NMJ 73529 Homo sapiens hypothetical protein MGC33382 (MGC33382), mRNA
NMJ 73530 Homo sapiens zinc finger protein 610 (ZNF610), mRNA
NMJ 73531 Homo sapiens zinc finger protein 100 (ZNF100), mRNA
NMJ 73532 Homo sapiens hypothetical protein FLJ35838 (FLJ35838), mRNA
NMJ 73533 Homo sapiens tudor domain containing 5 (TDRD5), mRNA
NMJ 73535 Homo sapiens C-type (calcium dependent, carbohydrate-recognition domain
NMJ 73536 Homo sapiens gamma-aminobutyric acid (GABA) A receptor, gamma 1 (GAE
NMJ 73537 Homo sapiens GTF2I repeat domain containing 2 (GTF2IRD2), mRNA
NMJ 73538 Homo sapiens hypothetical protein FLJ35802 (FLJ35802), mRNA
NMJ 73539 Homo sapiens zinc finger protein 596 (ZNF596), mRNA
NMJ 73540 Homo sapiens fucosyltransferase 11 (alpha (1 ,3) fucosyltransferase) (FUT11
NMJ 73541 Homo sapiens chromosome 10 open reading frame 91 (C10orf91), mRNA
NMJ 73542 Homo sapiens hypothetical protein LOC196463 (LOC196463), mRNA
NMJ 73543 Homo sapiens hypothetical protein FLJ32844 (FLJ32844), mRNA
NMJ 73544 Homo sapiens B-cell novel protein 1 (BCNP1), mRNA
NMJ 73545 Homo sapiens chromosome 2 open reading frame 13 (C2orf13), mRNA
NMJ 73546 Homo sapiens hypothetical protein MGC35097 (MGC35097), mRNA
NMJ 73547 Homo sapiens hypothetical protein LOC201292 (LOC201292), mRNA
NMJ 73548 Homo sapiens zinc finger protein 584 (ZNF584), mRNA
NMJ 73549 Homo sapiens hypothetical protein FLJ39553 (FLJ39553), mRNA
NMJ 73550 Homo sapiens chromosome 9 open reading frame 93 (C9orf93), mRNA
NMJ 73551 Homo sapiens sterile alpha motif domain containing 6 (SAMD6), mRNA
NMJ 73552 Homo sapiens hypothetical protein MGC33365 (MGC33365), mRNA
NMJ 73553 Homo sapiens hypothetical protein FLJ25801 (FLJ25801), mRNA
NMJ 73554 Homo sapiens chromosome 10 open reading frame 107 (C1 Oorfl 07), mRNA
NMJ 73555 Homo sapiens trypsin domain containing 1 (TYSND1), mRNA
NMJ 73556 Homo sapiens hypothetical protein MGC34732 (MGC34732), mRNA
NMJ 73557 Homo sapiens ring finger protein 152 (RNF152), mRNA
NMJ 73558 Homo sapiens FGD1 family, member 2 (FGD2), mRNA
NMJ 73559 Homo sapiens hypothetical protein FLJ25791 (FLJ25791), mRNA
NMJ 73560 Homo sapiens regulatory factor X domain containing 1 (RFXDC1), mRNA
NMJ 73561 Homo sapiens unc-5 homolog C (C. elegans)-like (UNC5CL), mRNA
NMJ 73562 Homo sapiens chromosome 6 open reading frame 69 (C6orf69), mRNA
NMJ73563 Homo sapiens chromosome 6 open reading frame 146 (C6orf146), mRNA
NMJ 73564 Homo sapiens hypothetical protein FLJ37538 (FLJ37538), mRNA
NMJ 73565 Homo sapiens hypothetical protein LOC222967 (LOC222967), mRNA
NMJ 73566 Homo sapiens hypothetical protein MGC50372 (MGC50372), mRNA
NMJ 73567 Homo sapiens abhydrolase domain containing 7 (ABHD7), mRNA
NMJ 73568 Homo sapiens uromodulin-like 1 (UMODL1), mRNA
NMJ 73570 Homo sapiens zinc finger, DHHC domain containing 23 (ZDHHC23), mRNA
NMJ 73571 Homo sapiens hypothetical protein LOC255313 (LOC255313), mRNA
NMJ 73572 Homo sapiens chromosome 10 open reading frame 93 (C10orf93), mRNA
NMJ 73573 Homo sapiens hypothetical protein MGC35138 (MGC35138), mRNA
NMJ 73574 Homo sapiens hypothetical protein MGC33414 (MGC33414), mRNA
NMJ 73575 Homo sapiens serine/threonine kinase 32C (STK32C), mRNA
NMJ 73576 Homo sapiens chromosome 10 open reading frame 48 (C10orf48), mRNA
NMJ 73578 Homo sapiens hypothetical protein FLJ90834 (FLJ90834), mRNA
NMJ 73579 Homo sapiens hypothetical protein FLJ40224 (FLJ40224), mRNA
NMJ 73580 Homo sapiens hypothetical protein FLJ39058 (FLJ39058), mRNA
NMJ 73581 Homo sapiens hypothetical protein FLJ90231 (FLJ90231), mRNA
NMJ 73582 Homo sapiens phosphoglucomutase 2-like 1 (PGM2L1), mRNA
NMJ 73583 Homo sapiens hypothetical protein FLJ33790 (FLJ33790), mRNA
NMJ 73584 Homo sapiens hypothetical protein MGC45840 (MGC45840), mRNA
NMJ 73586 Homo sapiens hypothetical protein MGC34821 (MGC34821), mRNA
NMJ 73587 Homo sapiens REST corepressor 2 (RCOR2), mRNA
NMJ 73588 Homo sapiens hypothetical protein FLJ37794 (FLJ37794), mRNA
NMJ 73589 Homo sapiens hypothetical protein FLJ35709 (FLJ35709), mRNA
NMJ 73590 Homo sapiens hypothetical protein FLJ36102 (FLJ36102), mRNA
NMJ 73591 Homo sapiens hypothetical protein FLJ90579 (FLJ90579), mRNA
NMJ 73593 Homo sapiens beta 1 ,4-N-acetylgalactosaminyltransferase-transferase-lll (B'
NMJ 73596 Homo sapiens solute carrier family 39 (metal ion transporter), member 5 (SD
NMJ 73597 Homo sapiens hypothetical protein FLJ37587 (FLJ37587), mRNA
NMJ 73598 Homo sapiens kinase suppressor of Ras-2 (KSR2), mRNA
NMJ73599 Homo sapiens hypothetical protein FLJ40126 (FLJ40126), mRNA
NMJ 73605 Homo sapiens potassium channel regulator (KCNRG), mRNA
NMJ73607 Homo sapiens chromosome 14 open reading frame 24 (C14orf24), mRNA
NMJ73608 Homo sapiens chromosome 14 open reading frame 80 (C14orf80), mRNA
NMJ 73609 Homo sapiens chromosome 15 open reading frame 21 (C15orf21), mRNA
NMJ 73610 Homo sapiens hypothetical protein FLJ33768 (FLJ33768), mRNA
NMJ 73611 Homo sapiens hypothetical protein FLJ38426 (FLJ38426), mRNA
NMJ 73613 Homo sapiens hypothetical protein FLJ35785 (FLJ35785), mRNA
NMJ73614 Homo sapiens hypothetical protein LOC283820 (LOC283820), mRNA
NMJ 73616 Homo sapiens hypothetical protein FLJ35894 (FLJ35894), mRNA
NMJ 73617 Homo sapiens hypothetical protein FLJ36701 (FLJ36701), mRNA
NMJ 73618 Homo sapiens hypothetical protein FLJ90652 (FLJ90652), mRNA
NMJ 73619 Homo sapiens hypothetical protein MGC34761 (MGC34761), mRNA
NMJ 73620 Homo sapiens hypothetical protein FLJ23825 (FLJ23825), mRNA
NMJ 73621 Homo sapiens hypothetical protein FLJ34790 (FLJ34790), mRNA
NMJ 73622 Homo sapiens hypothetical protein FLJ36674 (FLJ36674), mRNA
NMJ 73623 Homo sapiens hypothetical protein FLJ35808 (FLJ35808), mRNA
NMJ 73624 Homo sapiens hypothetical protein FLJ40504 (FLJ40504), mRNA
NMJ 73625 Homo sapiens hypothetical protein FLJ39647 (FLJ39647), mRNA
NMJ 73626 Homo sapiens solute carrier family 26, member 11 (SLC26A11 ), mRNA
NMJ 73627 Homo sapiens hypothetical protein FLJ35220 (FLJ35220), mRNA
NMJ 73628 Homo sapiens hypothetical protein FLJ40457 (FLJ40457), mRNA
NMJ73629 Homo sapiens chromosome 18 open reading frame 26 (C18orf26), mRNA
NMJ 73630 Homo sapiens rotatin (RTTN), mRNA
NMJ 73631 Homo sapiens zinc finger protein 547 (ZNF547), mRNA
NMJ 73632 Homo sapiens hypothetical protein FLJ38288 (FLJ38288), mRNA
NMJ 73633 Homo sapiens hypothetical protein FLJ90805 (FLJ90805), mRNA
NMJ 73635 Homo sapiens hypothetical protein FLJ40235 (FLJ40235), mRNA
NMJ73636 Homo sapiens chromosome 19 open reading frame 14 (C19orf14), mRNA
NMJ 73637 Homo sapiens hypothetical protein MGC34725 (MGC34725), mRNA
NMJ 73638 Homo sapiens hypothetical protein MGC8902 (MGC8902), mRNA
NMJ 73639 Homo sapiens hypothetical protein FLJ35976 (FLJ35976), mRNA
NMJ 73640 Homo sapiens likely ortholog of mouse roof plate-specific spondin (R-spondii
NMJ 73641 Homo sapiens hypothetical protein FLJ33655 (FLJ33655), mRNA
NMJ 73642 Homo sapiens hypothetical protein MGC47816 (MGC47816), mRNA
NMJ 73643 Homo sapiens hypothetical protein DKFZp547G0215 (DKFZp547G0215), mF
NMJ 73644 Homo sapiens hypothetical protein FLJ33860 (FLJ33860), mRNA
NMJ 73645 Homo sapiens hypothetical protein FLJ37357 (FLJ37357), mRNA
NMJ 73646 Homo sapiens hypothetical protein FLJ39660 (FLJ39660), mRNA
NMJ 73647 Homo sapiens ring finger protein 149 (RNF149), mRNA
NMJ 73648 Homo sapiens hypothetical protein FLJ39502 (FLJ39502), mRNA
NMJ 73649 Homo sapiens hypothetical protein FLJ40172 (FLJ40172), mRNA
NMJ 73650 Homo sapiens DnaJ (Hsp40) homolog, subfamily C, member 5 gamma (DN
NMJ 73651 Homo sapiens fibrous sheath interacting protein 2 (FSIP2), mRNA
NMJ 73652 Homo sapiens hypothetical protein MGC34824 (MGC34824), mRNA
NMJ 73653 Homo sapiens solute carrier family 9 (sodium/hydrogen exchanger), isoform
NMJ 73654 Homo sapiens hypothetical protein MGC34132 (MGC34132), mRNA
NMJ 73656 Homo sapiens zinc finger protein 619 (ZNF619), mRNA
NMJ 73657 Homo sapiens hypothetical protein FLJ31139 (FLJ31139), mRNA
NMJ 73658 Homo sapiens hypothetical protein FLJ36870 (FLJ36870), mRNA
NMJ 73659 Homo sapiens hypothetical protein MGC29784 (MGC29784), mRNA
NMJ 73660 Homo sapiens hypothetical protein FLJ33718 (FLJ33718), mRNA
NMJ 73661 Homo sapiens hypothetical protein FLJ35424 (FLJ35424), mRNA
NMJ 73662 Homo sapiens hypothetical protein LOC285533 (LOC285533), mRNA
NMJ 73663 Homo sapiens NY-REN-7 antigen (NY-REN-7), mRNA
NMJ 73664 Homo sapiens ADP-ribosylation factor-like 10A (ARL10A), mRNA
NMJ 73665 Homo sapiens hypothetical protein MGC34713 (MGC34713), mRNA
NMJ 73666 Homo sapiens hypothetical protein FLJ33977 (FLJ33977), mRNA
NMJ 73667 Homo sapiens hypothetical protein FLJ37543 (FLJ37543), mRNA
NMJ 73669 Homo sapiens hypothetical protein FLJ34047 (FLJ34047), mRNA
NMJ 73670 Homo sapiens RGM domain family, member B (RGMB), mRNA
NMJ 73671 Homo sapiens hypothetical protein FLJ37396 (FLJ37396), mRNA
NMJ 73672 Homo sapiens peptidylprolyl isomerase (cyclophilin)-like 6 (PPIL6), mRNA
NMJ 73673 Homo sapiens hypothetical protein FLJ34503 (FLJ34503), mRNA
NMJ 73674 Homo sapiens discoidin, CUB and LCCL domain containing 1 (DCBLD1), mF
NMJ 73675 Homo sapiens hypothetical protein FLJ33708 (FLJ33708), mRNA
NMJ 73676 Homo sapiens patatin-like phospholipase domain containing 1 (PNPLA1), ml
NMJ 73677 Homo sapiens hypothetical protein FLJ40852 (FLJ40852), mRNA
NMJ 73678 Homo sapiens hypothetical protein FLJ40722 (FLJ40722), mRNA
NMJ 73680 Homo sapiens hypothetical protein MGC33584 (MGC33584), mRNA
NMJ 73682 Homo sapiens hypothetical protein FLJ40288 (FLJ40288), mRNA
NMJ 73683 Homo sapiens chromosome 8 open reading frame 21 (C8orf21), mRNA
NMJ 73685 Homo sapiens hypothetical protein FLJ32440 (FLJ32440), mRNA
NMJ 73687 Homo sapiens hypothetical protein FLJ37131 (FLJ37131), mRNA
NMJ 73688 Homo sapiens hypothetical protein FLJ39630 (FLJ39630), mRNA
NMJ 73689 Homo sapiens crumbs homolog 2 (Drosophila) (CRB2), mRNA
NMJ 73690 Homo sapiens chromosome 9 open reading frame 126 (C9orf126), mRNA
NMJ 73691 Homo sapiens chromosome 9 open reading frame 75 (C9orf75), mRNA
NMJ 73694 Homo sapiens ATPase, Class VI, type 11C (ATP11C), mRNA
NMJ 73695 Homo sapiens hypothetical protein FLJ36601 (FLJ36601), mRNA
NMJ 73698 Homo sapiens hypothetical protein FLJ37659 (FLJ37659), mRNA
NMJ 73699 Homo sapiens hypothetical protein MGC33889 (MGC33889), mRNA
NMJ 73700 Homo sapiens BCL6 co-repressor-like 2 (BCORL2), mRNA
NMJ 73701 Homo sapiens tryptophanyl-tRNA synthetase (WARS), transcript variant 2, rr
NMJ 73728 Homo sapiens Rho guanine nucleotide exchange factor (GEF) 15 (ARHGEF1
NMJ 73791 Homo sapiens PDZ domain containing 8 (PDZK8), mRNA
NMJ 73793 Homo sapiens hypothetical protein LOC128977 (LOC128977), mRNA
NMJ 73794 Homo sapiens FUN14 domain containing 1 (FUNDC1), mRNA
NMJ 73795 Homo sapiens hypothetical protein FLJ32096 (FLJ32096), mRNA
NMJ 73797 Homo sapiens PAP associated domain containing 4 (PAPD4), mRNA
NMJ 73798 Homo sapiens zinc finger, CCHC domain containing 12 (ZCCHC12), mRNA
NMJ 73799 Homo sapiens hypothetical protein FLJ39873 (FLJ39873), mRNA
N J 73800 Homo sapiens laeverin (FLJ90650), mRNA
NMJ 73801 Homo sapiens hypothetical protein FLJ36198 (FLJ36198), mRNA
NMJ 73802 Homo sapiens hypothetical protein MGC50559 (MGC50559), mRNA
NMJ 73803 Homo sapiens hypothetical protein FLJ39599 (FLJ39599), mRNA
NMJ 73804 Homo sapiens hypothetical protein MGC30208 (MGC30208), mRNA
N J 73805 Homo sapiens hypothetical protein FLJ38723 (FLJ38723), mRNA
NMJ 73806 Homo sapiens hypothetical protein MGC50721 (MGC50721), mRNA
NMJ 73807 Homo sapiens hypothetical protein MGC33370 (MGC33370), mRNA
NMJ 73808 Homo sapiens neuronal growth regulator 1 (NEGR1), mRNA
NMJ 73809 Homo sapiens biogenesis of lysosome-related organelles complex-1 , subunr
NMJ 73810 Homo sapiens hypothetical protein MGC29649 (MGC29649), mRNA
NMJ 73811 Homo sapiens hypothetical protein FLJ32675 (FLJ32675), mRNA
NMJ 73812 Homo sapiens hypothetical protein FLJ32949 (FLJ32949), mRNA
N J 73813 Homo sapiens hypothetical protein FLJ34154 (FLJ34154), mRNA
NMJ 73815 Homo sapiens hypothetical protein FLJ37464 (FLJ37464), mRNA
NMJ 73821 Homo sapiens hypothetical protein FLJ33590 (FLJ33590), mRNA
NMJ 73822 Homo sapiens hypothetical protein MGC39518 (MGC39518), mRNA
NMJ 73824 Homo sapiens hypothetical protein MGC26717 (MGC26717), mRNA
NMJ 73825 Homo sapiens RAB, member of RAS oncogene family-like 3 (RABL3), mRN/
NMJ 73826 Homo sapiens hypothetical protein DKFZp313N0621 (DKFZp313N0621), mF
NMJ 73827 Homo sapiens hypothetical protein FLJ38991 (FLJ38991), mRNA
NMJ 73828 Homo sapiens chromosome 5 open reading frame 16 (C5orf16), mRNA
NMJ 73829 Homo sapiens hypothetical protein FLJ36754 (FLJ36754), mRNA
NMJ 73830 Homo sapiens chromosome 6 open reading frame 182 (C6orf182), mRNA
NMJ 73831 Homo sapiens hypothetical protein LOC286075 (LOC286075), mRNA
NMJ 73832 Homo sapiens hypothetical protein FLJ38705 (FLJ38705), mRNA
NMJ 73833 Homo sapiens hypothetical protein MGC45780 (MGC45780), mRNA
NMJ73834 Homo sapiens hypothetical protein MGC21416 (MGC21416), mRNA
NMJ 73841 Homo sapiens interleukin 1 receptor antagonist (IL1RN), transcript variant 2,
NMJ 73842 Homo sapiens interleukin 1 receptor antagonist (IL1 RN), transcript variant 1 ,
NMJ 73843 Homo sapiens interleukin 1 receptor antagonist (IL1 RN), transcript variant 4,
NMJ 73844 Homo sapiens mucosa associated lymphoid tissue lymphoma translocation c
NMJ73846 Homo sapiens chromosome 14 open reading frame 8 (C14orf8), mRNA
NMJ 73847 Homo sapiens sperm acrosome associated 3 (SPACA3), mRNA
NMJ 73848 Homo sapiens hypothetical protein LOC138046 (LOC138046), mRNA
NMJ 73849 Homo sapiens goosecoid (GSC), mRNA
NMJ 73850 Homo sapiens serine (or cysteine) proteinase inhibitor, clade A (alpha-1 anti|
NMJ 73851 Homo sapiens solute carrier family 30 (zinc transporter), member 8 (SLC30A
NMJ 73852 Homo sapiens keratinocyte associated protein 2 (KRTCAP2), mRNA
NMJ 73853 Homo sapiens keratinocyte associated protein 3 (KRTCAP3), mRNA
NMJ73854 Homo sapiens solute carrier family 41 , member 1 (SLC41A1), mRNA
NMJ 73855 Homo sapiens morn (LOC283385), mRNA
NMJ73856 Homo sapiens vomeronasal 1 receptor 2 (VN1 R2), mRNA
NMJ 73857 Homo sapiens vomeronasal 1 receptor 4 (VN1R4), mRNA
NMJ 73858 Homo sapiens vomeronasal 1 receptor 5 (VN1R5), mRNA
NMJ 73859 Homo sapiens breast cancer and salivary gland expression gene (BASE), ml
NMJ 73860 Homo sapiens homeo box C12 (HOXC12), mRNA
NMJ 73872 Homo sapiens chloride channel 3 (CLCN3), transcript variant e, mRNA
NMJ 74855 Homo sapiens isocitrate dehydrogenase 3 (NAD+) beta (IDH3B), nuclear ger
NMJ 74856 Homo sapiens isocitrate dehydrogenase 3 (NAD+) beta (IDH3B), nuclear ger
NMJ 74858 Homo sapiens adenylate kinase 5 (AK5), transcript variant 1 , mRNA
NMJ 74869 Homo sapiens isocitrate dehydrogenase 3 (NAD+) gamma (IDH3G), nuclear
NMJ 74871 Homo sapiens proteasome (prosome, macropain) 26S subunit, non-ATPase,
NMJ 74872 Homo sapiens purinergic receptor P2X, ligand-gated ion channel, 2 (P2RX2)
NMJ 74873 Homo sapiens purinergic receptor P2X, ligand-gated ion channel, 2 (P2RX2)
NMJ 74878 Homo sapiens Usher syndrome 3A (USH3A), transcript variant 1 , mRNA
NMJ 74880 Homo sapiens Usher syndrome 3A (USH3A), transcript variant 3, mRNA
NMJ 74881 Homo sapiens crumbs homolog 3 (Drosophila) (CRB3), transcript variant 3, r
NMJ 74882 Homo sapiens crumbs homolog 3 (Drosophila) (CRB3), transcript variant 1 , r
NMJ 74886 Homo sapiens TGFB-induced factor (TALE family homeobox) (TGIF), transci
NMJ 74887 Homo sapiens intraflagellar transport protein IFT20 (LOC90410), mRNA
NMJ 74889 Homo sapiens hypothetical protein LOC91942 (LOC91942), mRNA
NMJ 74890 Homo sapiens AN1 , ubiquitin-like, homolog (Xenopus laevis) (ANUBL1), mR
NMJ74891 Homo sapiens chromosome 14 open reading frame 79 (C14orf79), mRNA
NMJ 74892 Homo sapiens triggering receptor expressed on myeloid cells 5 (TREM5), mF
NMJ 74896 Homo sapiens hypothetical protein MGC24133 (MGC24133), mRNA
NMJ 74897 Homo sapiens bactericidal/permeability-increasing protein-like 3 (BPIL3), mF
NMJ 74898 Homo sapiens hypothetical protein LOC129530 (LOC129530), mRNA
NMJ 74899 Homo sapiens F-box protein 36 (FBX036), mRNA
NMJ 74900 Homo sapiens zinc finger protein 42 (ZFP42), mRNA
NMJ 74901 Homo sapiens family with sequence similarity 9, member C (FAM9C), mRNA
NMJ 74902 Homo sapiens hypothetical protein LOC143458 (L0C143458), mRNA
NMJ74905 Homo sapiens hypothetical protein LOC147965 (LOC147965), mRNA
NMJ 74906 Homo sapiens hypothetical protein MGC39724 (MGC39724), mRNA
NMJ 74907 Homo sapiens protein phosphatase 4, regulatory subunit 2 (PPP4R2), mRN/
NMJ 74908 Homo sapiens chromosome 3 open reading frame 6 (C3orf6), transcript varis
NMJ 74909 Homo sapiens hypothetical protein MGC23909 (MGC23909), mRNA
NMJ 74910 Homo sapiens t-complex-associated-testis-expressed 3 (TCTE3), mRNA
NMJ 74911 Homo sapiens breast cancer membrane protein 101 (NSE2), mRNA
NMJ74912 Homo sapiens hypothetical protein FLJ31204 (FLJ31204), mRNA
NMJ74913 Homo sapiens chromosome 14 open reading frame 21 (C14orf21), mRNA
NMJ74914 Homo sapiens hypothetical protein LOC167127 (LOC167127), mRNA
NMJ 74916 Homo sapiens ubiquitin protein ligase E3 component n-recognin 1 (UBR1), n
NMJ 74917 Homo sapiens hypothetical protein LOC197322 (LOC197322), mRNA
NMJ 74918 Homo sapiens hypothetical protein LOC199675 (LOC199675), mRNA
NMJ 74920 Homo sapiens hypothetical protein LOC201191 (LOC201191), mRNA
NMJ 74921 Homo sapiens hypothetical protein LOC201895 (LOC201895), mRNA
NMJ 74922 Homo sapiens aarF domain containing kinase 5 (ADCK5), mRNA
NMJ 74923 Homo sapiens hypothetical protein MGC31967 (MGC31967), mRNA
NMJ 74924 Homo sapiens hypothetical protein LOC204474 (LOC204474), mRNA
NMJ 74925 Homo sapiens hypothetical protein LOC205251 (LOC205251), mRNA
NMJ 74926 Homo sapiens hypothetical protein MGC17839 (MGC17839), mRNA
NMJ 74927 Homo sapiens spergen-1 (SPAS1), mRNA
NMJ74928 Homo sapiens hypothetical protein LOC221143 (LOC221143), mRNA
NMJ 74930 Homo sapiens postmeiotic segregation increased 2-like 5 (PMS2L5), mRNA
NMJ 74931 Homo sapiens hypothetical protein FLJ38348 (FLJ38348), mRNA
NMJ 74932 Homo sapiens bactericidal/permeability-increasing protein-like 2 (BPIL2), mF
NMJ 74933 Homo sapiens phytanoyl-CoA dioxygenase domain containing 1 (PHYHD1),
NMJ 74934 Homo sapiens sodium channel, voltage-gated, type IV, beta (SCN4B), mRN/
NMJ 74936 Homo sapiens proprotein convertase subtilisin/kexin type 9 (PCSK9), mRNA
NMJ 74937 Homo sapiens transcription elongation regulator 1-like (TCERG1L), mRNA
NMJ 74938 Homo sapiens FERM domain containing 3 (FRMD3), mRNA
NMJ 74939 Homo sapiens hypothetical protein MGC39681 (MGC39681), mRNA
NMJ 74940 Homo sapiens hypothetical protein LOC283232 (LOC283232), mRNA
NMJ 74941 Homo sapiens scavenger receptor cysteine-rich type 1 protein M160 (M160),
NMJ 74942 Homo sapiens growth arrest-specific 2 like 3 (GAS2L3), mRNA
NMJ 74943 Homo sapiens hypothetical protein FLJ25976 (FLJ25976), mRNA
NMJ74944 Homo sapiens chromosome 14 open reading frame 20 (C14orf20), mRNA
NMJ 74945 Homo sapiens zinc finger protein 575 (ZNF575), mRNA
NMJ 74947 Homo sapiens chromosome 19 open reading frame 30 (C19orf30), mRNA
NMJ 74950 Homo sapiens hypothetical protein FLJ30435 (FLJ30435), mRNA
NMJ 74951 Homo sapiens family with sequence similarity 9, member A (FAM9A), mRNA
NMJ 74952 Homo sapiens hypothetical protein MGC46496 (MGC46496), mRNA
NMJ 74953 Homo sapiens ATPase, Ca++ transporting, ubiquitous (ATP2A3), transcript v
NMJ 74954 Homo sapiens ATPase, Ca++ transporting, ubiquitous (ATP2A3), transcript \
NMJ 74955 Homo sapiens ATPase, Ca++ transporting, ubiquitous (ATP2A3), transcript v
NMJ 74956 Homo sapiens ATPase, Ca++ transporting, ubiquitous (ATP2A3), transcript v
NMJ 74957 Homo sapiens ATPase, Ca++ transporting, ubiquitous (ATP2A3), transcript v
N J 74958 Homo sapiens ATPase, Ca++ transporting, ubiquitous (ATP2A3), transcript \
NMJ 74959 Homo sapiens hypothetical protein LOC136306 (LOC136306), mRNA
NMJ 74961 Homo sapiens synovial sarcoma, X breakpoint 8 (SSX8), mRNA
NMJ 74962 Homo sapiens synovial sarcoma, X breakpoint 9 (SSX9), mRNA
NMJ 74963 Homo sapiens sialyitransferase 6 (N-acetyllacosaminide alpha 2,3-sialyltrans
NMJ 74964 Homo sapiens sialyitransferase 6 (N-acetyllacosaminide alpha 2,3-sialyltrans
NMJ 74965 Homo sapiens sialyitransferase 6 (N-acetyllacosaminide alpha 2,3-sialyltrans
NMJ 74966 Homo sapiens sialyitransferase 6 (N-acetyllacosaminide alpha 2,3-sialyltrans
NMJ 74967 Homo sapiens sialyitransferase 6 (N-acetyllacosaminide alpha 2,3-sialyltrans
N J 74968 Homo sapiens sialyitransferase 6 (N-acetyllacosaminide alpha 2,3-sialyltrans
NMJ 74969 Homo sapiens sialyitransferase 6 (N-acetyllacosaminide alpha 2,3-sialyltrans
NMJ 74970 Homo sapiens sialyitransferase 6 (N-acetyllacosaminide alpha 2,3-sialyltrans
NMJ 74971 Homo sapiens sialyitransferase 6 (N-acetyllacosaminide alpha 2,3-sialyltrans
N J 74972 Homo sapiens sialyitransferase 6 (N-acetyllacosaminide alpha 2,3-sialyltrans
NMJ 74975 Homo sapiens SEC14-like 3 (S. cerevisiae) (SEC14L3), mRNA
NMJ 74976 Homo sapiens zinc finger, DHHC domain containing 22 (ZDHHC22), mRNA
NMJ 74977 Homo sapiens SEC14-like 4 (S. cerevisiae) (SEC14L4), mRNA
NMJ74978 Homo sapiens chromosome 14 open reading frame 39 (C14orf39), mRNA
NMJ 74980 Homo sapiens vomeronasal 1 receptor 3 (VN1R3), mRNA
NMJ 74981 Homo sapiens ankyrin repeat domain 21 (ANKRD21), mRNA
NMJ 74983 Homo sapiens chromosome 19 open reading frame 28 (C19orf28), mRNA
NMJ 75038 Homo sapiens contactin 1 (CNTN1), transcript variant 2, mRNA
NMJ 75039 Homo sapiens sialyitransferase 7D ((alpha-N-acetylneuraminyl-2,3-beta-gala
NMJ 75040 Homo sapiens sialyitransferase 7D ((alpha-N-acetylneuraminyl-2,3-beta-gala
NMJ 75047 Homo sapiens paired immunoglobin-like type 2 receptor beta (PILRB), transc
NMJ 75052 Homo sapiens sialyitransferase 8D (alpha-2, 8-polysialyltransferase) (SIAT8I
NMJ 75053 Homo sapiens keratin 6 irs4 (K6IRS4), mRNA
NMJ 75054 Homo sapiens histone 4, H4 (HIST4H4), mRNA
NMJ 75055 Homo sapiens histone 3, H2bb (HIST3H2BB), mRNA
NMJ 75056 Homo sapiens hypothetical protein LOC131368 (LOC131368), mRNA
NMJ 75057 Homo sapiens trace amine receptor 3 (TRAR3), mRNA
NMJ75058 Homo sapiens hypothetical protein LOC144100 (LOC144100), mRNA
NMJ 75060 Homo sapiens chromosome 14 open reading frame 27 (C14orf27), mRNA
NMJ75061 Homo sapiens juxtaposed with another zinc finger gene 1 (JAZF1), mRNA
NMJ 75062 Homo sapiens RasGEF domain family, member 1 C (RASGEF1 C), mRNA
NMJ 75063 Homo sapiens hypothetical protein LOC284361 (LOC284361), transcript vari
NMJ 75064 Homo sapiens Williams Beuren syndrome chromosome region 19 (WBSCR1
NMJ 75065 Homo sapiens histone 2, H2ab (HIST2H2AB), mRNA
NMJ 75066 Homo sapiens DEAD (Asp-Glu-Ala-Asp) box polypeptide 51 (DDX51), mRN/1
N J 75067 Homo sapiens trace amine receptor 4 (TRAR4), mRNA
NMJ 75068 Homo sapiens keratin 6 irs3 (K6IRS3), mRNA
NMJ 75069 Homo sapiens aprataxin (APTX), transcript variant 2, mRNA
N J 75071 Homo sapiens aprataxin (APTX), transcript variant 5, mRNA
NMJ 75072 Homo sapiens aprataxin (APTX), transcript variant 3, mRNA
NMJ 75073 Homo sapiens aprataxin (APTX), transcript variant 1 , mRNA
NMJ 75075 Homo sapiens hypothetical protein INM01 (INM01), mRNA
NMJ 75077 Homo sapiens Src-like-adaptor 2 (SLA2), transcript variant 2, mRNA
NMJ 75078 Homo sapiens keratin 1 B (KRT1 B), mRNA
NMJ 75080 Homo sapiens purinergic receptor P2X, ligand-gated ion channel, 5 (P2RX5)
NMJ75081 Homo sapiens purinergic receptor P2X, ligand-gated ion channel, 5 (P2RX5)
NMJ 75085 Homo sapiens phosphoribosylglycinamide formyltransferase, phosphoribosyl
NMJ 75566 Homo sapiens contactin 5 (CNTN5), transcript variant 2, mRNA
NMJ 75567 Homo sapiens purinergic receptor P2X, ligand-gated ion channel, 4 (P2RX4)
NMJ 75568 Homo sapiens purinergic receptor P2X, ligand-gated ion channel, 4 (P2RX4)
NMJ 75569 Homo sapiens Xg blood group (pseudoautosomal boundary-divided on the X
NMJ 75571 Homo sapiens human immune associated nucleotide 6 (hlAN6), mRNA
NMJ 75573 Homo sapiens adhesion regulating molecule 1 (ADRM1), transcript variant 2,
NMJ 75575 Homo sapiens WFIKKN-related protein (WFIKKNRP), mRNA
NMJ 75605 Homo sapiens tetratricopeptide repeat domain 10 (TTC10), transcript variant
NMJ 75607 Homo sapiens contactin 4 (CNTN4), transcript variant 1 , mRNA
NMJ 75609 Homo sapiens ADP-ribosylation factor GTPase activating protein 1 (ARFGAF
N J 75610 Homo sapiens tight junction protein 1 (zona occludens 1 ) (TJP1 ), transcript v
NMJ 75611 Homo sapiens glutamate receptor, ionotropic, kainate 1 (GRIK1 ), transcript v
NMJ 75612 Homo sapiens contactin 4 (CNTN4), transcript variant 2, mRNA
NMJ 75613 Homo sapiens contactin 4 (CNTN4), transcript variant 3, mRNA
NMJ 75614 Homo sapiens NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 11 ,
NMJ 75616 Homo sapiens FIS (FIS), mRNA
NMJ 75617 Homo sapiens metallothionein 1E (functional) (MT1E), mRNA
NMJ 75619 Homo sapiens zygote arrest 1 (ZAR1), mRNA
NMJ 75622 Homo sapiens metallothionein U (MTU), mRNA
NMJ 75623 Homo sapiens RAB3A interacting protein (rabin3) (RAB3IP), transcript variar
NMJ 75624 Homo sapiens RAB3A interacting protein (rabin3) (RAB3IP), transcript variar
NMJ 75625 Homo sapiens RAB3A interacting protein (rabin3) (RAB3IP), transcript variar
NMJ 75626 Homo sapiens RAB3A interacting protein (rabin3) (RAB3IP), transcript variar
NMJ 75627 Homo sapiens RAB3A interacting protein (rabin3) (RAB3IP), transcript variar
NMJ 75629 Homo sapiens DNA (cytosine-5-)-methyltransferase 3 alpha (DNMT3A), tram
NMJ 75630 Homo sapiens DNA (cytosine-5-)-methyltransferase 3 alpha (DNMT3A), trans
NMJ 75634 Homo sapiens core-binding factor, runt domain, alpha subunit 2; translocatee
NMJ 75635 Homo sapiens core-binding factor, runt domain, alpha subunit 2; translocatec
NMJ 75636 Homo sapiens core-binding factor, runt domain, alpha subunit 2; translocatee
NMJ 75698 Homo sapiens synovial sarcoma, X breakpoint 2 (SSX2), transcript variant 2,
N J 75709 Homo sapiens chromobox homolog 7 (CBX7), mRNA
N J 75711 Homo sapiens synovial sarcoma, X breakpoint 3 (SSX3), transcript variant 2,
NMJ 75719 Homo sapiens thyroid peroxidase (TPO), transcript variant 2, mRNA
NMJ 75720 Homo sapiens thyroid peroxidase (TPO), transcript variant 3, mRNA
NMJ 75721 Homo sapiens thyroid peroxidase (TPO), transcript variant 4, mRNA
NMJ 75722 Homo sapiens thyroid peroxidase (TPO), transcript variant 5, mRNA
N J 75723 Homo sapiens synovial sarcoma, X breakpoint 5 (SSX5), transcript variant 2,
NMJ75724 Homo sapiens interleukin 5 receptor, alpha (IL5RA), transcript variant 2, mRI
NMJ 75725 Homo sapiens interleukin 5 receptor, alpha (IL5RA), transcript variant 3, mRI
NMJ 75726 Homo sapiens interleukin 5 receptor, alpha (IL5RA), transcript variant 4, mRI
NMJ75727 Homo sapiens interleukin 5 receptor, alpha (IL5RA), transcript variant 5, mRI
NMJ 75728 Homo sapiens interleukin 5 receptor, alpha (IL5RA), transcript variant 6, mRI
NMJ 75729 Homo sapiens synovial sarcoma, X breakpoint 4 (SSX4), transcript variant 2,
NMJ 75733 Homo sapiens synaptotagmin IX (SYT9), mRNA
NMJ 75734 Homo sapiens hypothetical protein LOC201243 (LOC201243), mRNA
NMJ 75735 Homo sapiens lysozyme-like (LYG2), mRNA
NMJ 75736 Homo sapiens formin-Iike 3 (FMNL3), transcript variant 1 , mRNA
NMJ 75737 Homo sapiens klotho beta like (LOC152831), mRNA
N J 75738 Homo sapiens RAB37, member RAS oncogene family (RAB37), mRNA
NMJ 75739 Homo sapiens serine (or cysteine) proteinase inhibitor, clade A (alpha-1 anti|
NMJ 75741 Homo sapiens nuclear protein in testis (NUT), mRNA
NMJ 75742 Homo sapiens melanoma antigen, family A, 2 (MAGEA2), transcript variant 2
NMJ 75743 Homo sapiens melanoma antigen, family A, 2 (MAGEA2), transcript variant 3
NMJ 75744 Homo sapiens ras homolog gene family, member C (RHOC), mRNA
NMJ 75745 Homo sapiens cancer/testis antigen 3 (CTAG3), transcript variant 1 , mRNA
NMJ 75747 Homo sapiens oligodendrocyte transcription factor 3 (OLIG3), mRNA
NMJ75748 Homo sapiens chromosome 14 open reading frame 130 (C14orf130), mRNA
NMJ 75767 Homo sapiens interleukin 6 signal transducer (gp130, oncostatin M receptor)
NMJ 75768 Homo sapiens glutamate receptor, ionotropic, kainate 2 (GRIK2), transcript v
NMJ 75834 Homo sapiens keratin 6L (KRT6L), mRNA
NMJ 75839 Homo sapiens spermine oxidase (SMOX), transcript variant 1 , mRNA
NMJ 75840 Homo sapiens spermine oxidase (SMOX), transcript variant 2, mRNA
NMJ 75841 Homo sapiens spermine oxidase (SMOX), transcript variant 3, mRNA
NMJ 75842 Homo sapiens spermine oxidase (SMOX), transcript variant 4, mRNA
NMJ 75847 Homo sapiens polypyrimidine tract binding protein 1 (PTBP1 ), transcript varij
NMJ75848 Homo sapiens DNA (cytosine-5-)-methyltransferase 3 beta (DNMT3B), transi
NMJ 75849 Homo sapiens DNA (cytosine-5-)-methyltransferase 3 beta (DNMT3B), transi
NMJ 75850 Homo sapiens DNA (cytosine-5-)-methyltransferase 3 beta (DNMT3B), transi
NMJ 75851 Homo sapiens a disintegrin-like and metalloprotease (reprolysin type) with th
NMJ75852 Homo sapiens taxilin (DKFZp451 J0118), mRNA
NMJ 75854 Homo sapiens PABP1 -dependent poly A-specific ribonuclease subunit PANS
NMJ 75857 Homo sapiens keratin associated protein 8-1 (KRTAP8-1), mRNA
NMJ 75858 Homo sapiens keratin associated protein 11-1 (KRTAP11-1), mRNA
NMJ 75859 Homo sapiens CTP synthase II (CTPS2), transcript variant 2, mRNA
NMJ75861 Homo sapiens ARG99 protein (ARG99), mRNA
NMJ 75862 Homo sapiens CD86 antigen (CD28 antigen ligand 2, B7-2 antigen) (CD86),
NMJ75863 Homo sapiens AT rich interactive domain 1B (SWH-like) (ARID1B), transcrip
NMJ 75864 Homo sapiens core-binding factor, runt domain, alpha subunit 2; translocatee
NMJ 75865 Homo sapiens ELYS transcription factor-like protein TMBS62 (ELYS), mRNA
NMJ 75866 Homo sapiens kinase interacting with leukemia-associated gene (stathmin) (
NMJ 75867 Homo sapiens DNA (cytosine-5-)-methyltransferase 3-like (DNMT3L), transci
NMJ 75868 Homo sapiens melanoma antigen, family A, 6 (MAGEA6), transcript variant 2
NMJ 75870 Homo sapiens hypothetical protein LOC90925 (LOC90925), mRNA
NMJ 75871 Homo sapiens hypothetical protein FLJ35119 (FLJ35119), mRNA
NMJ 75872 Homo sapiens FLJ38451 protein (FLJ38451), mRNA
NMJ 75873 Homo sapiens hypothetical protein LOC134548 (LOC134548), mRNA
NMJ 75874 Homo sapiens hypothetical protein MGC47869 (MGC47869), mRNA
NMJ 75875 Homo sapiens sine oculis homeobox homolog 5 (Drosophila) (SIX5), mRNA
NMJ 75876 Homo sapiens exocyst complex component 8 (EXOC8), mRNA
NMJ 75877 Homo sapiens hypothetical protein MGC35023 (MGC35023), mRNA
NMJ 75878 Homo sapiens hypothetical protein MGC57211 (MGC57211 ), mRNA
NMJ 75881 Homo sapiens hypothetical protein MGC48986 (MGC48986), mRNA
NMJ 75882 Homo sapiens intramembrane protease 5 (IMP5), mRNA
NMJ 75884 Homo sapiens hypothetical protein FLJ36031 (FLJ36031), mRNA
NMJ 75885 Homo sapiens hypothetical protein MGC33846 (MGC33846), mRNA
NMJ 75886 Homo sapiens phosphoribosyl pyrophosphate synthetase 1-like 1 (PRPS1L1
NMJ 75887 Homo sapiens hypothetical protein LOC222171 (LOC222171), mRNA
NMJ 75892 Homo sapiens hypothetical protein FLJ37266 (FLJ37266), mRNA
NMJ 75895 Homo sapiens hypothetical protein FLJ25590 (FLJ25590), mRNA
NMJ 75898 Homo sapiens hypothetical protein LOC283687 (LOC283687), mRNA
NMJ 75900 Homo sapiens hypothetical protein FLJ35681 (FLJ35681 ), mRNA
NMJ 75901 Homo sapiens hypothetical protein LOC283932 (LOC283932), mRNA
NMJ 75902 Homo sapiens hypothetical protein FLJ22222 (FLJ22222), mRNA
NMJ 75903 Homo sapiens hypothetical protein LOC284033 (LOC284033), mRNA
NMJ75904 Homo sapiens hypothetical protein FLJ40121 (FLJ40121), mRNA
NMJ 75906 Homo sapiens hypothetical protein MGC33608 (MGC33608), mRNA
NMJ 75907 Homo sapiens zinc binding alcohol dehydrogenase, domain containing 2 (ZA
NMJ 75908 Homo sapiens hypothetical protein LOC284296 (LOC284296), mRNA
NMJ 75910 Homo sapiens zinc finger protein 493 (ZNF493), mRNA
NMJ 75911 Homo sapiens hypothetical protein MGC40047 (MGC40047), mRNA
NMJ 75913 Homo sapiens junctophilin 2 (JPH2), transcript variant 2, mRNA
NMJ 75918 Homo sapiens hypothetical protein FLJ34443 (FLJ34443), mRNA
NMJ 75920 Homo sapiens hypothetical protein FLJ39485 (FLJ39485), mRNA
NMJ 75921 Homo sapiens hypothetical protein LOC285636 (LOC285636), mRNA
NMJ 75922 Homo sapiens hypothetical protein MGC35308 (MGC35308), mRNA
NMJ 75923 Homo sapiens hypothetical protein MGC42630 (MGC42630), mRNA
NMJ 75924 Homo sapiens immunoglobulin-like domain containing receptor 1 (ILDR1), m
NMJ75929 Homo sapiens fibroblast growth factor 14 (FGF14), transcript variant 2, mRN
NMJ 75931 Homo sapiens core-binding factor, runt domain, alpha subunit 2; translocatec
NMJ 75932 Homo sapiens proteasome (prosome, macropain) 26S subunit, non-ATPase,
NMJ 75940 Homo sapiens dual oxidase 1 (DUOX1), transcript variant 2, mRNA
NMJ 76071 Homo sapiens purinergic receptor P2Y, G-protein coupled, 2 (P2RY2), transi
NMJ 76072 Homo sapiens purinergic receptor P2Y, G-protein coupled, 2 (P2RY2), transt
NMJ 76081 Homo sapiens DNA (cytosine-5-)-methyltransferase 2 (DNMT2), transcript vε
NM J 76083 Homo sapiens DNA (cytosine-5-)-methyltransferase 2 (DNMT2), transcript vε
NMJ 76084 Homo sapiens DNA (cytosine-5-)-methyltransferase 2 (DNMT2), transcript vε
NMJ 76085 Homo sapiens DNA (cytosine-5-)-methyltransferase 2 (DNMT2), transcript vε
NMJ 76086 Homo sapiens DNA (cytosine-5-)-methyltransferase 2 (DNMT2), transcript vε
NMJ 76095 Homo sapiens CDK5 regulatory subunit associated protein 3 (CDK5RAP3), t
NMJ 76096 Homo sapiens CDK5 regulatory subunit associated protein 3 (CDK5RAP3), t
NMJ 76782 Homo sapiens hyporthetical protein MGC27169 (MGC27169), mRNA
NMJ 76783 Homo sapiens proteasome (prosome, macropain) activator subunit 1 (PA28 ι
NMJ 76786 Homo sapiens interleukin 9 receptor (IL9R), transcript variant 2, mRNA
NMJ 76787 Homo sapiens phosphatidylinositol glycan, class N (PIGN), transcript variant
NMJ 76789 Homo sapiens myotubularin related protein 1 (MTMR1), transcript variant 2, i
NMJ 76791 Homo sapiens chromosome 20 open reading frame 65 (C20orf65), mRNA
NMJ 76792 Homo sapiens mitochondrial ribosomal protein L43 (MRPL43), nuclear gene
NMJ 76793 Homo sapiens mitochondrial ribosomal protein L43 (MRPL43), nuclear gene
NMJ 76794 Homo sapiens mitochondrial ribosomal protein L43 (MRPL43), nuclear gene
NMJ 76795 Homo sapiens v-Ha-ras Harvey rat sarcoma viral oncogene homolog (HRAS]
NMJ 76796 Homo sapiens pyrimidinergic receptor P2Y, G-protein coupled, 6 (P2RY6), tr
NMJ 76797 Homo sapiens pyrimidinergic receptor P2Y, G-protein coupled, 6 (P2RY6), tr
NMJ 76798 Homo sapiens pyrimidinergic receptor P2Y, G-protein coupled, 6 (P2RY6), tr
NMJ 76799 Homo sapiens integrin-linked kinase-associated serine/threonine phosphatas
NMJ 76800 Homo sapiens PRP4 pre-mRNA processing factor 4 homolog B (yeast) (PRF
NMJ 76801 Homo sapiens adducin 1 (alpha) (ADD1), transcript variant 4, mRNA
NMJ 76805 Homo sapiens mitochondrial ribosomal protein S11 (MRPS11 ), nuclear gene
NMJ 76806 Homo sapiens molybdenum cofactor synthesis 2 (MOCS2), transcript variant
NMJ 76810 Homo sapiens NACHT, leucine rich repeat and PYD containing 13 (NALP13)
NMJ 76811 Homo sapiens NACHT, leucine rich repeat and PYD containing 8 (NALP8), n
NMJ 76812 Homo sapiens chromosome 20 open reading frame 178 (C20orf178), mRNA
NMJ 76813 Homo sapiens breast cancer membrane protein 11 (BCMP11 ), mRNA
NMJ 76814 Homo sapiens hypothetical protein LOC168850 (LOC168850), mRNA
NMJ 76815 Homo sapiens hypothetical protein LOC200895 (LOC200895), mRNA
NMJ 76816 Homo sapiens Kenae (KENAE), mRNA
NMJ 76817 Homo sapiens taste receptor, type 2, member 38 (TAS2R38), mRNA
NMJ 76818 Homo sapiens hypothetical protein 15E1.2 (15E1.2), mRNA
NMJ 76820 Homo sapiens NACHT, leucine rich repeat and PYD containing 9 (NALP9), n
NMJ 76821 Homo sapiens NACHT, leucine rich repeat and PYD containing 10 (NALP10)
NMJ76822 Homo sapiens NACHT, leucine rich repeat and PYD containing 14 (NALP14)
NMJ 76823 Homo sapiens S100 calcium binding protein A15 (S100A15), mRNA
NMJ 76824 Homo sapiens Bardet-Biedl syndrome 7 (BBS7), transcript variant 1 , mRNA
NMJ 76825 Homo sapiens sulfotransferase family, cytosolic, 1C, member 1 (SULT1C1),
NMJ 76826 Homo sapiens ilvB (bacterial acetolactate synthase)-like (ILVBL), transcript v
NMJ 76853 Homo sapiens C-terminal modulator protein (CTMP), transcript variant 2, mR
NMJ 76863 Homo sapiens proteasome (prosome, macropain) activator subunit 3 (PA28
NMJ 76866 Homo sapiens inorganic pyrophosphatase 2 (PPA2), transcript variant 3, mR
NMJ 76867 Homo sapiens inorganic pyrophosphatase 2 (PPA2), transcript variant 4, mR
NMJ 76869 Homo sapiens inorganic pyrophosphatase 2 (PPA2), transcript variant 1 , mR
NMJ 76870 Homo sapiens metallothionein 1 K (MT1 K), mRNA
NMJ 76871 Homo sapiens PDZ and LIM domain 2 (mystique) (PDLIM2), transcript variar
NMJ 76874 Homo sapiens sulfotransferase family 4A, member 1 (SULT4A1 ), transcript v
NMJ 76875 Homo sapiens cholecystokinin B receptor (CCKBR), mRNA
NMJ 76876 Homo sapiens purinergic receptor P2Y, G-protein coupled, 12 (P2RY12), trai
NMJ 76877 Homo sapiens InaD-like protein (INADL), transcript variant 2, mRNA
NMJ 76878 Homo sapiens InaD-like protein (INADL), transcript variant 4, mRNA
NMJ 76880 Homo sapiens TR4 orphan receptor associated protein TRA16 (TRA16), mR
NMJ 76881 Homo sapiens taste receptor, type 2, member 39 (TAS2R39), mRNA
NMJ 76882 Homo sapiens taste receptor, type 2, member 40 (TAS2R40), mRNA
NMJ 76883 Homo sapiens taste receptor, type 2, member 41 (TAS2R41), mRNA
NMJ 76884 Homo sapiens taste receptor, type 2, member 43 (TAS2R43), mRNA
NMJ 76885 Homo sapiens taste receptor, type 2, member 44 (TAS2R44), mRNA
NMJ 76886 Homo sapiens taste receptor, type 2, member 45 (TAS2R45), mRNA
NMJ 76887 Homo sapiens taste receptor, type 2, member 46 (TAS2R46), mRNA
NMJ 76888 Homo sapiens taste receptor, type 2, member 48 (TAS2R48), mRNA
NMJ 76889 Homo sapiens taste receptor, type 2, member 49 (TAS2R49), mRNA
NMJ 76890 Homo sapiens taste receptor, type 2, member 50 (TAS2R50), mRNA
NMJ 76891 Homo sapiens interferon epsilon 1 (IFNE1), mRNA
NMJ 76894 Homo sapiens purinergic receptor P2Y, G-protein coupled, 13 (P2RY13), trai
NMJ 76895 Homo sapiens phosphatidic acid phosphatase type 2A (PPAP2A), transcript
NMJ 77398 Homo sapiens LIM homeobox transcription factor 1 , alpha (LMX1A), transcri|
NMJ 77399 Homo sapiens LIM homeobox transcription factor 1 , alpha (LMX1 A), transcri|
NMJ 77400 Homo sapiens NK6 transcription factor related, locus 2 (Drosophila) (NKX6-2
NMJ77402 Homo sapiens synaptotagmin II (SYT2), mRNA
NMJ 77403 Homo sapiens RAB7B, member RAS oncogene family (RAB7B), mRNA
NMJ 77404 Homo sapiens melanoma antigen, family B, 1 (MAGEB1), transcript variant 2
NMJ 77405 Homo sapiens cat eye syndrome chromosome region, candidate 1 (CECR1 ).
NMJ 77414 Homo sapiens phosphatidic acid phosphatase type 2B (PPAP2B), transcript
NMJ 77415 Homo sapiens melanoma antigen, family B, 1 (MAGEB1), transcript variant 3
NMJ 77417 Homo sapiens kinesin light chain 2-like (KLC2L), transcript variant 1 , mRNA
NMJ 77422 Homo sapiens eukaryotic translation initiation factor 2C, 3 (EIF2C3), transcri|
NMJ 77423 Homo sapiens protein tyrosine phosphatase, receptor type, f polypeptide (PT
NMJ 77424 Homo sapiens syntaxin 12 (STX12), mRNA
NMJ 77427 Homo sapiens purinergic receptor P2X, ligand-gated ion channel, 7 (P2RX7)
NMJ 77433 Homo sapiens melanoma antigen, family D, 2 (MAGED2), transcript variant _
NMJ77434 Homo sapiens FtsJ homolog 1 (E. coli) (FTSJ1), transcript variant 2, mRNA
NMJ 77435 Homo sapiens peroxisome proliferative activated receptor, delta (PPARD), tn
NMJ77436 Homo sapiens CSE1 chromosome segregation 1-like (yeast) (CSE1 L), transi
NMJ 77437 Homo sapiens taste receptor, type 2, member 60 (TAS2R60), mRNA
NMJ 77438 Homo sapiens Dicerl , Dcr-1 homolog (Drosophila) (DICER1), transcript variε
NMJ 77439 Homo sapiens FtsJ homolog 1 (E. coli) (FTSJ1), transcript variant 3, mRNA
NMJ 77441 Homo sapiens hypothetical protein MGC3123 (MGC3123), mRNA
NMJ 77442 Homo sapiens FtsJ homolog 2 (E. coli) (FTSJ2), transcript variant 2, mRNA
NMJ 77444 Homo sapiens PTPRF interacting protein, binding protein 1 (liprin beta 1) (PF
NMJ 77452 Homo sapiens trafficking protein particle complex 6B (TRAPPC6B), mRNA
NMJ 77453 Homo sapiens progestin and adipoQ receptor family member III (PAQR3), m
NMJ 77454 Homo sapiens KIAA1946 (KIAA1946), mRNA
NMJ77455 Homo sapiens class II bHLH protein MIST1 (MIST1), mRNA
NMJ 77456 Homo sapiens melanoma antigen, family C, 3 (MAGEC3), transcript variant .
NMJ 77457 Homo sapiens Ly-6 neurotoxin-like protein 1 (LYNX1), transcript variant 3, m
NMJ 77458 Homo sapiens secreted Ly6/uPAR related protein 2 (SLURP2), mRNA
NMJ 77476 Homo sapiens Ly-6 neurotoxin-like protein 1 (LYNX1), transcript variant 4, m
NMJ 77477 Homo sapiens Ly-6 neurotoxin-like protein 1 (LYNX1), transcript variant 5, m
NMJ 77478 Homo sapiens ferritin mitochondrial (FTMT), mRNA
NMJ 77483 Homo sapiens glycosylphosphatidylinositol specific phospholipase D1 (GPLC
NMJ 77524 Homo sapiens mesoderm specific transcript homolog (mouse) (MEST), trans
NMJ 77525 Homo sapiens mesoderm specific transcript homolog (mouse) (MEST), trans
NMJ 77526 Homo sapiens phosphatidic acid phosphatase type 2C (PPAP2C), transcript
NMJ77528 Homo sapiens sulfotransferase family, cytosolic, 1A, phenol-preferring, mem
NMJ 77529 Homo sapiens sulfotransferase family, cytosolic, 1 A, phenol-preferring, mem
NMJ 77530 Homo sapiens sulfotransferase family, cytosolic, 1A, phenol-preferring, mem
NMJ 77531 Homo sapiens polycystic kidney and hepatic disease 1 (autosomal recessive
NMJ 77532 Homo sapiens Ras association (RalGDS/AF-6) domain family 6 (RASSF6), ti
NMJ 77533 Homo sapiens nudix (nucleoside diphosphate linked moiety X)-type motif 14
NMJ77534 Homo sapiens sulfotransferase family, cytosolic, 1A, phenol-preferring, mem
NMJ 77535 Homo sapiens melanoma antigen, family D, 4 (MAGED4), transcript variant i
NMJ 77536 Homo sapiens sulfotransferase family, cytosolic, 1A, phenol-preferring, mem
NMJ 77537 Homo sapiens melanoma antigen, family D, 4 (MAGED4), transcript variant
NMJ 77538 Homo sapiens cytochrome P450, family 20, subfamily A, polypeptide 1 (CYP
NMJ 77539 Homo sapiens taste receptor, type 1 , member 1 (TAS1 R1 ), transcript variant
NMJ 77540 Homo sapiens taste receptor, type 1 , member 1 (TAS1 R1 ), transcript variant
NMJ 77541 Homo sapiens taste receptor, type 1 , member 1 (TAS1 R1 ), transcript variant
NMJ 77542 Homo sapiens small nuclear ribonucleoprotein D2 polypeptide 16.5kDa (SNF
NMJ 77543 Homo sapiens phosphatidic acid phosphatase type 2C (PPAP2C), transcript
NMJ 77549 Homo sapiens M8 protein (LOC149830), mRNA
NMJ 77550 Homo sapiens solute carrier family 13 (sodium-dependent citrate transporter
NMJ 77551 Homo sapiens G protein-coupled receptor 109A (GPR109A), mRNA
NMJ 77552 Homo sapiens sulfotransferase family, cytosolic, 1A, phenol-preferring, mem
NMJ 77553 Homo sapiens growth arrest-specific 2 (GAS2), transcript variant 2, mRNA
NMJ 77554 Homo sapiens acid phosphatase 1 , soluble (ACP1 ), transcript variant 1 , mRI1
NMJ 77555 Homo sapiens trophinin (TRO), transcript variant 1 , mRNA
NMJ 77556 Homo sapiens trophinin (TRO), transcript variant 2, mRNA
NMJ 77557 Homo sapiens trophinin (TRO), transcript variant 5, mRNA
NMJ 77558 Homo sapiens trophinin (TRO), transcript variant 4, mRNA
NMJ 77559 Homo sapiens casein kinase 2, alpha 1 polypeptide (CSNK2A1), transcript v;
NMJ 77560 Homo sapiens casein kinase 2, alpha 1 polypeptide (CSNK2A1), transcript vi
NMJ 77924 Homo sapiens N-acylsphingosine amidohydrolase (acid ceramidase) 1 (ASA
NMJ 77925 Homo sapiens H2A histone family, member J (H2AFJ), transcript variant 2, n
NMJ 77926 Homo sapiens CSRP2 binding protein (CSRP2BP), transcript variant 2, mRN
NMJ 77937 Homo sapiens golgi phosphoprotein 2 (GOLPH2), transcript variant 2, mRNA
NMJ 77938 Homo sapiens hypoxia-inducible factor prolyl 4-hydroxylase (PH-4), mRNA
NMJ 77939 Homo sapiens hypoxia-inducible factor prolyl 4-hydroxylase (PH-4), transcrip
NMJ 77947 Homo sapiens armadillo repeat containing, X-linked 3 (ARMCX3), transcript '
NMJ 77948 Homo sapiens armadillo repeat containing, X-linked 3 (ARMCX3), transcript *
NMJ 77949 Homo sapiens armadillo repeat containing, X-linked 2 (ARMCX2), mRNA
NMJ77951 Homo sapiens protein phosphatase 1A (formerly 2C), magnesium-dependen
NMJ 77952 Homo sapiens protein phosphatase 1A (formerly 2C), magnesium-dependen
NMJ 77953 Homo sapiens dynein, cytoplasmic, light polypeptide 2A (DNCL2A), transcrip
NMJ 77954 Homo sapiens dynein, cytoplasmic, light polypeptide 2A (DNCL2A), transcrip
NMJ 77959 Homo sapiens docking protein 5 (DOK5), transcript variant 2, mRNA
NMJ77963 Homo sapiens synaptotagmin XII (SYT12), mRNA
NMJ 77964 Homo sapiens hypothetical protein LOC130576 (LOC130576), mRNA
N M J 77965 Homo sapiens hypothetical protein LOC157657 (LOC157657), mRNA
NMJ77966 Homo sapiens hypothetical protein DKFZp667B1218 (DKFZp667B1218), mF
NMJ 77967 Homo sapiens phosphoglycerate dehydrogenase like 1 (PHGDHL1), mRNA
NMJ 77968 Homo sapiens protein phosphatase 1 B (formerly 2C), magnesium-dependen
NMJ 77969 Homo sapiens protein phosphatase 1 B (formerly 2C), magnesium-dependen
NMJ 77972 Homo sapiens tubby homolog (mouse) (TUB), transcript variant 2, mRNA
NMJ 77973 Homo sapiens sulfotransferase family, cytosolic, 2B, member 1 (SULT2B1), 1
NMJ 77974 Homo sapiens H63 breast cancer expressed gene (H63), transcript variant 2
NMJ 77976 Homo sapiens ADP-ribosylation factor-like 6 (ARL6), transcript variant 2, mR
NMJ 77977 Homo sapiens huntingtin-associated protein 1 (neuroan 1) (HAP1), transcripl
NMJ 77978 Homo sapiens chordin (CHRD), transcript variant 2, mRNA
NMJ 77979 Homo sapiens chordin (CHRD), transcript variant 3, mRNA
NMJ 77980 Homo sapiens cadherin-like 26 (CDH26), transcript variant a, mRNA
NMJ 77983 Homo sapiens protein phosphatase 1 G (formerly 2C), magnesium-dependen
NMJ 77985 Homo sapiens ADP-ribosylation factor-like 5 (ARL5), transcript variant 2, mR
NMJ 77986 Homo sapiens desmoglein 4 (DSG4), mRNA
NMJ 77987 Homo sapiens tubulin, beta 8 (TUBB8-pending), mRNA
NMJ 77988 Homo sapiens mitochondrial ribosomal protein L47 (MRPL47), nuclear gene
NMJ 77989 Homo sapiens actin-like 6A (ACTL6A), transcript variant 2, mRNA
NMJ 77990 Homo sapiens p21 (CDKN1 A)-activated kinase 7 (PAK7), transcript variant 2,
NMJ 77991 Homo sapiens dual specificity phosphatase-like 15 (DUSP15), transcript vari
NMJ 77995 Homo sapiens protein tyrosine phosphatase domain containing 1 (PTPDC1),
NMJ77996 Homo sapiens erythrocyte membrane protein band 4.1-like 1 (EPB41L1), trai
NMJ 77998 Homo sapiens otopetrin 1 (OTOP1), mRNA
NMJ 77999 Homo sapiens ankyrin repeat and SOCS box-containing 6 (ASB6), transcript
NMJ 78000 Homo sapiens protein phosphatase 2A, regulatory subunit B' (PR 53) (PPP2I
NMJ 78001 Homo sapiens protein phosphatase 2A, regulatory subunit B' (PR 53) (PPP2I
NMJ 78002 Homo sapiens protein phosphatase 2A, regulatory subunit B' (PR 53) (PPP2I
NMJ78003 Homo sapiens protein phosphatase 2A, regulatory subunit B' (PR 53) (PPP2I
NMJ 78004 Homo sapiens proline rich membrane anchor 1 (PRIMA1), transcript variant I
NMJ 78006 Homo sapiens START domain containing 13 (STARD13), transcript variant a
NMJ 78007 Homo sapiens START domain containing 13 (STARD13), transcript variant b
NMJ 78008 Homo sapiens START domain containing 13 (STARD13), transcript variant d
NMJ78009 Homo sapiens diacylglycerol kinase, eta (DGKH), transcript variant 2, mRNA
NMJ78010 Homo sapiens SRY (sex determining region Y)-box 5 (SOX5), transcript varis
NMJ 78011 Homo sapiens leucine rich repeat transmembrane neuronal 3 (LRRTM3), mF
NMJ 78012 Homo sapiens tubulin, beta polypeptide paralog (MGC8685), mRNA
NMJ78013 Homo sapiens proline rich membrane anchor 1 (PRIMA1), mRNA
NMJ78014 Homo sapiens beta 5-tubulin (OK/SW-cl.56), mRNA
NMJ 78019 Homo sapiens cation channel, sperm associated 3 (CATSPER3), mRNA
NMJ 78025 Homo sapiens gamma-glutamyltransferase-like 3 (GGTL3), transcript variant
NMJ 78026 Homo sapiens gamma-glutamyltransferase-like 3 (GGTL3), transcript variant
NMJ 78031 Homo sapiens heat shock 70kDa protein 5 (glucose-regulated protein, 78kD;
NMJ 78033 Homo sapiens cytochrome P450, family 4, subfamily X, polypeptide 1 (CYP4
NMJ 78034 Homo sapiens phospholipase A2, group IVD (cytosolic) (PLA2G4D), mRNA
NMJ 78037 Homo sapiens Rab6-interacting protein 2 (ELKS), transcript variant beta, mR
NMJ 78038 Homo sapiens Rabδ-interacting protein 2 (ELKS), transcript variant gamma,
NMJ 78039 Homo sapiens Rabδ-interacting protein 2 (ELKS), transcript variant delta, mF
NMJ 78040 Homo sapiens Rab6-interacting protein 2 (ELKS), transcript variant epsilon, r
NMJ 78042 Homo sapiens actin-like 6A (ACTL6A), transcript variant 3, mRNA
NMJ78043 Homo sapiens FLJ10378 protein (FLJ10378), transcript variant 2, mRNA
NMJ 78044 Homo sapiens hypothetical protein MGC5178 (MGC5178), transcript variant
NMJ 78120 Homo sapiens distal-less homeo box 1 (DLX1), mRNA
NMJ 78121 Homo sapiens HBV pre-s2 binding protein 1 (SBP1), mRNA
NMJ 78122 Homo sapiens hypothetical protein LOC90529 (LOC90529), mRNA
NMJ 78123 Homo sapiens SEC14 and spectrin domains 1 (SESTD1), mRNA
NMJ 78124 Homo sapiens hypothetical protein LOC91966 (LOC91966), mRNA
NMJ78125 Homo sapiens tripartite motif-containing 50A (TRIM50A), mRNA
NMJ 78126 Homo sapiens hypothetical protein LOC162427 (LOC162427), mRNA
NMJ 78127 Homo sapiens angiopoietin-like 5 (ANGPTL5), mRNA
NMJ 78128 Homo sapiens similar to delta 5 fatty acid desaturase (LOC283985), mRNA
NMJ 78129 Homo sapiens purinergic receptor P2Y, G-protein coupled, 8 (P2RY8), mRN;
NMJ 78130 Homo sapiens thioredoxin domain containing 6 (TXNDC6), mRNA
NMJ 78134 Homo sapiens cytochrome P450, family 4, subfamily Z, polypeptide 1 (CYP4.
NMJ78135 Homo sapiens short-chain dehydrogenase/reductase 9 (SCDR9), mRNA
NMJ 78136 Homo sapiens polymerase (DNA-directed), delta interacting protein 3 (POLD
NMJ 78138 Homo sapiens LIM homeobox 3 (LHX3), transcript variant 1 , mRNA
NMJ 78140 Homo sapiens PDZ domain containing 3 (PDZK3), transcript variant 1 , mRN/
NMJ 78145 Homo sapiens Ras association (RalGDS/AF-6) domain family 4 (RASSF4), ti
NMJ 78148 Homo sapiens solute carrier family 35, member B2 (SLC35B2), mRNA
NMJ78150 Homo sapiens F-box protein, helicase, 18 (FBX018), transcript variant 2, mF
NMJ 78151 Homo sapiens doublecortex; lissencephaly, X-linked (doublecortin) (DCX), tn
NMJ 78152 Homo sapiens doublecortex; lissencephaly, X-linked (doublecortin) (DCX), tn
NMJ 78153 Homo sapiens doublecortex; lissencephaly, X-linked (doublecortin) (DCX), tn
NMJ 78154 Homo sapiens fucosyltransferase 8 (alpha (1 ,6) fucosyltransferase) (FUT8), I
NMJ 78155 Homo sapiens fucosyltransferase 8 (alpha (1 ,6) fucosyltransferase) (FUT8), 1
NMJ 78156 Homo sapiens fucosyltransferase 8 (alpha (1 ,6) fucosyltransferase) (FUT8), 1
N 78157 Homo sapiens fucosyltransferase 8 (alpha (1 ,6) fucosyltransferase) (FUT8), 1
NMJ78159 Homo sapiens hypothetical protein FLJ12949 (FLJ12949), transcript variant :
NM 78160 Homo sapiens otopetrin 2 (OTOP2), mRNA
NMJ78161 Homo sapiens pancreas specific transcription factor, 1a (PTF1A), mRNA
NMJ78167 Homo sapiens zinc finger protein 598 (ZNF598), mRNA
NMJ78168 Homo sapiens olfactory receptor, family 10, subfamily A, member 5 (OR10A5
N 78169 Homo sapiens Ras association (RalGDS/AF-6) domain family 3 (RASSF3), n
NMJ78170 Homo sapiens NIMA (never in mitosis gene a)- related kinase 8 (NEK8), mRI
NMJ 78171 Homo sapiens gasdermin 1 (GSDM1 ), mRNA
NMJ78172 Homo sapiens high density lipoprotein-binding protein (LOC338328), mRNA
NMJ 78173 Homo sapiens hypothetical protein LOC339834 (LOC339834), mRNA
NMJ 78174 Homo sapiens triggering receptor expressed on myeloid cells-like 1 (TREML
NMJ 78175 Homo sapiens lipoma HMGIC fusion partner-like 1 (LHFPL1 ), mRNA
NMJ 78176 Homo sapiens monoacylglycerol O-acyltransferase 3 (MOGAT3), mRNA
NMJ 78177 Homo sapiens nicotinamide nucleotide adenylyltransferase 3 (NMNAT3), mF
NMJ 78181 Homo sapiens CUB domain-containing protein 1 (CDCP1), transcript variant
NMJ78190 Homo sapiens ATPase inhibitory factor 1 (ATPIF1), nuclear gene encoding n
NMJ78191 Homo sapiens ATPase inhibitory factor 1 (ATPIF1), nuclear gene encoding n
NMJ 78221 Homo sapiens APG4 autophagy 4 homolog C (S. cerevisiae) (APG4C), trans
NMJ 78225 Homo sapiens F-box and WD-40 domain protein 5 (FBXW5), transcript varia
NMJ78226 Homo sapiens F-box and WD-40 domain protein 5 (FBXW5), transcript varia
NMJ 78228 Homo sapiens killer cell immunoglobulin-like receptor, two domains, short cy
NMJ 78229 Homo sapiens IQ motif containing GTPase activating protein 3 (IQGAP3), ml
NMJ 78230 Homo sapiens cyclophilin-LC (COAS2), mRNA
NMJ78231 Homo sapiens amyotrophic lateral sclerosis 2 (juvenile) chromosome region,
NMJ 78232 Homo sapiens hyaluronan and proteoglycan link protein 3 (HAPLN3), mRNA
NMJ 78233 Homo sapiens otopetrin 3 (OTOP3), mRNA
NMJ 78234 Homo sapiens tumor suppressor candidate 3 (TUSC3), transcript variant 2, n
NMJ 78237 Homo sapiens SEC3-like 1 (S. cerevisiae) (SEC3L1), transcript variant 2, mF
NMJ 78238 Homo sapiens paired immunoglobin-like type 2 receptor beta (PILRB), transc
NMJ 78270 Homo sapiens APG4 autophagy 4 homolog A (S. cerevisiae) (APG4A), trans
NMJ 78271 Homo sapiens APG4 autophagy 4 homolog A (S. cerevisiae) (APG4A), trans
NMJ 78272 Homo sapiens paired immunoglobin-like type 2 receptor alpha (PILRA), trans
NMJ 78273 Homo sapiens paired immunoglobin-like type 2 receptor alpha (PILRA), trans
NMJ78275 Homo sapiens eEF1A2 binding protein (DKFZp434B1231), mRNA
NMJ 78276 Homo sapiens chromosome 5 open reading frame 12 (C5orf12), mRNA
N J 78311 Homo sapiens gamma-glutamyltransferase-like activity 4 (GGTLA4), transcrii
NMJ78312 Homo sapiens gamma-glutamyltransferase-like activity 4 (GGTLA4), transcrii
NMJ78313 Homo sapiens spectrin, beta, non-erythrocytic 1 (SPTBN1), transcript variant
NMJ78314 Homo sapiens hypothetical protein FLJ39378 (FLJ39378), mRNA
NMJ 78324 Homo sapiens serine palmitoyltransferase, long chain base subunit 1 (SPTLI
NMJ 78326 Homo sapiens APG4 autophagy 4 homolog B (S. cerevisiae) (APG4B), trans
NMJ 78329 Homo sapiens chemokine (C-C motif) receptor 3 (CCR3), transcript variant 2
NMJ 78331 Homo sapiens gonadotropin-releasing hormone 2 (GNRH2), transcript variar
NMJ 78332 Homo sapiens gonadotropin-releasing hormone 2 (GNRH2), transcript variar
NMJ 78335 Homo sapiens chromosome 3 open reading frame 6 (C3orf6), transcript varis
NMJ 78336 Homo sapiens mitochondrial ribosomal protein L52 (MRPL52), nuclear gene
NMJ 78338 Homo sapiens AP20 region protein (APRG1 ), transcript variant A, mRNA
NMJ 78339 Homo sapiens AP20 region protein (APRG1), transcript variant B, mRNA
NMJ 78340 Homo sapiens AP20 region protein (APRG1), transcript variant C, mRNA
NMJ 78341 Homo sapiens AP20 region protein (APRG1), transcript variant D, mRNA
N J 78342 Homo sapiens AP20 region protein (APRG1 ), transcript variant E, mR A
NMJ 78343 Homo sapiens AP20 region protein (APRG1 ), transcript variant F, mRNA
NMJ 78344 Homo sapiens AP20 region protein (APRG1 ), transcript variant G, mRNA
NMJ 78348 Homo sapiens late envelope protein 1 (LEP1), mRNA
NMJ 78349 Homo sapiens small proline rich-like (epidermal differentiation complex) 2A (•
NMJ 78351 Homo sapiens late envelope protein 3 (LEP3), mRNA
NMJ 78352 Homo sapiens late envelope protein 4 (LEP4), mRNA
NMJ 78353 Homo sapiens late envelope protein 5 (LEP5), mRNA
NMJ 78354 Homo sapiens late envelope protein 6 (LEP6), mRNA
NMJ 78356 Homo sapiens small proline rich-like (epidermal differentiation complex) 4A (•
NMJ 78422 Homo sapiens membrane progestin receptor alpha (MPRA), mRNA
NMJ 78423 Homo sapiens histone deacetylase 9 (HDAC9), transcript variant 4, mRNA
NMJ 78424 Homo sapiens SRY (sex determining region Y)-box 30 (SOX30), transcript vε
NMJ 78425 Homo sapiens histone deacetylase 9 (HDAC9), transcript variant 5, mRNA
NMJ 78426 Homo sapiens aryl hydrocarbon receptor nuclear translocator (ARNT), transc
NMJ 78427 Homo sapiens aryl hydrocarbon receptor nuclear translocator (ARNT), transc
NMJ 78428 Homo sapiens late envelope protein 9 (LEP9), mRNA
NMJ 78429 Homo sapiens late envelope protein 11 (LEP11), mRNA
NMJ 78430 Homo sapiens small proline rich-like (epidermal differentiation complex) 1A (•
NMJ 78431 Homo sapiens late envelope protein 13 (LEP13), mRNA
NMJ 78432 Homo sapiens cell cycle related kinase (CCRK), transcript variant 1 , mRNA
NMJ 78433 Homo sapiens late envelope protein 14 (LEP14), mRNA
NMJ 78434 Homo sapiens small proline rich-like (epidermal differentiation complex) 3A (■
NMJ 78435 Homo sapiens late envelope protein 17 (LEP17), mRNA
NMJ 78438 Homo sapiens small proline rich-like (epidermal differentiation complex) 5A (•
NMJ 78439 Homo sapiens germ cell-less homolog 1 (Drosophila) (GCL), mRNA
NMJ 78441 Homo sapiens zinc finger, FYVE domain containing 1 (ZFYVE1), transcript v;
NMJ 78443 Homo sapiens UNC-112 related protein 2 (URP2), transcript variant URP2LF
NMJ 78445 Homo sapiens chemokine (C-C motif) receptor-like 1 (CCRL1), transcript var
NMJ 78448 Homo sapiens chromosome 9 open reading frame 140 (C9orf140), mRNA
NMJ 78449 Homo sapiens tuberoinfundibular 39 residue protein precursor (TIP39), mRN
NMJ 78450 Homo sapiens hypothetical protein MGC48332 (MGC48332), mRNA
NMJ 78451 Homo sapiens zinc finger, MYND domain containing 17 (ZMYND17), mRNA
NMJ78452 Homo sapiens similar to RIKEN cDNA 4930457P18 (LOC123872), mRNA
NMJ 78453 Homo sapiens hypothetical protein MGC52282 (MGC52282), mRNA
NMJ 78454 Homo sapiens hypothetical protein MGC54289 (MGC54289), mRNA
NMJ 78456 Homo sapiens chromosome 20 open reading frame 85 (C20orf85), mRNA
NMJ 78460 Homo sapiens protein tyrosine phosphatase, non-receptor type substrate 1 — Ii
NMJ 78463 Homo sapiens chromosome 20 open reading frame 166 (C20orf166), mRNA
NMJ 78465 Homo sapiens TSPY-like 3 (TSPYL3), mRNA
NMJ78466 Homo sapiens chromosome 20 open reading frame 71 (C20orf71), mRNA
NMJ 78467 Homo sapiens high-mobility group (nonhistone chromosomal) protein 4-like (
NMJ 78468 Homo sapiens chromosome 20 open reading frame 128 (C20orf128), mRNA
NMJ 78470 Homo sapiens WD repeat domain 40B (WDR40B), mRNA
NMJ 78471 Homo sapiens G protein-coupled receptor 119 (GPR119), mRNA
NMJ 78472 Homo sapiens chromosome 20 open reading frame 53 (C20orf53), mRNA
NMJ78477 Homo sapiens chromosome 20 open reading frame 179 (C20orf179), mRNA
NMJ 78483 Homo sapiens chromosome 20 open reading frame 79 (C20orf79), mRNA
NMJ 78491 Homo sapiens R3H domain (binds single-stranded nucleic acids) containing-
NMJ78493 Homo sapiens hypothetical protein LOC147111 (LOC147111), mRNA
NMJ78494 Homo sapiens hypothetical protein FLJ40125 (FLJ40125), mRNA
NMJ 78495 Homo sapiens KIAA1754-like (KIAA1754L), mRNA
NMJ78496 Homo sapiens similar to BcDNA:GH11415 gene product (LOC151963), mRN
NMJ 78497 Homo sapiens hypothetical protein FLJ23657 (FLJ23657), mRNA
NMJ 78498 Homo sapiens hypothetical protein MGC52019 (MGC52019), mRNA
NMJ 78499 Homo sapiens hypothetical protein MGC39827 (MGC39827), mRNA
NMJ 78500 Homo sapiens phosphatase, orphan 1 (PHOSPH01), mRNA
NMJ 78502 Homo sapiens deltex 3 homolog (Drosophila) (DTX3), mRNA
NMJ 78504 Homo sapiens hypothetical protein FLJ40427 (FLJ40427), mRNA
NMJ 78505 Homo sapiens transmembrane protein 26 (TMEM26), mRNA
NMJ 78507 Homo sapiens NS5ATP13TP2 protein (NS5ATP13TP2), m RNA
NMJ 78508 Homo sapiens hypothetical protein MGC57858 (MGC57858), mRNA
NMJ 78509 Homo sapiens syntaxin binding protein 4 (STXBP4), mRNA
NMJ 78510 Homo sapiens ankyrin repeat and kinase domain containing 1 (ANKK1), mRI
NMJ 78514 Homo sapiens hypothetical protein LOC283487 (LOC283487), mRNA
NMJ 78516 Homo sapiens hypothetical protein LOC283849 (LOC283849), mRNA
NMJ 78517 Homo sapiens phosphatidylinositol glycan, class W (PIGW), mRNA
NMJ78518 Homo sapiens hypothetical protein FLJ36878 (FLJ36878), mRNA
NMJ 78519 Homo sapiens hypothetical protein FLJ39421 (FLJ39421), mRNA
NMJ78520 Homo sapiens hypothetical protein FLJ38792 (FLJ38792), mRNA
NMJ 78523 Homo sapiens zinc finger protein 616 (ZNF616), mRNA
NMJ 78525 Homo sapiens hypothetical protein MGC33407 (MGC33407), mRNA
NMJ 78527 Homo sapiens hypothetical protein MGC43026 (MGC43026), mRNA
NMJ 78530 Homo sapiens hypothetical protein FLJ38379 (FLJ38379), mRNA
NMJ 78532 Homo sapiens hypothetical protein LOC285671 (LOC285671 ), mRNA
NMJ78536 Homo sapiens lipocalin 12 (LCN12), mRNA
NMJ78537 Homo sapiens beta1,4-N-acetylgalactosaminyltransferases IV (Beta4GalNAc
NMJ 78538 Homo sapiens hypothetical protein LOC338799 (LOC338799), mRNA
N J 78539 Homo sapiens TAFA2 protein (TAFA2), mRNA
NMJ 78540 Homo sapiens hypothetical protein MGC48915 (MGC48915), mRNA
NMJ78542 Homo sapiens hypothetical protein DKFZp762C2414 (DKFZp762C2414), mF
NMJ 78543 Homo sapiens ectonucleotide pyrophosphatase/phosphodiesterase 7 (ENPF
NMJ 78544 Homo sapiens zinc finger protein 546 (ZNF546), mRNA
NMJ 78545 Homo sapiens hypothetical protein LOC339456 (LOC339456), mRNA
N J 78546 Homo sapiens hypothetical protein LOC339483 (LOC339483), mRNA
NMJ 78547 Homo sapiens archease (ARCH), mRNA
NMJ 78548 Homo sapiens adaptor-related protein complex 2, epsilon subunit (AP2E), ml
NMJ 78549 Homo sapiens hypothetical protein MGC42493 (MGC42493), mRNA
NMJ 78550 Homo sapiens hypothetical protein MGC48998 (MGC48998), mRNA
NMJ 78552 Homo sapiens hypothetical protein MGC35206 (MGC35206), mRNA
NMJ 78553 Homo sapiens hypothetical protein MGC44505 (MGC44505), mRNA
NMJ 78554 Homo sapiens kyphoscoliosis peptidase (KY), mRNA
NMJ 78555 Homo sapiens hypothetical protein FLJ25770 (FLJ25770), mRNA
NMJ 78556 Homo sapiens hypothetical protein FLJ36180 (FLJ36180), mRNA
NMJ 78557 Homo sapiens hypothetical protein FLJ37478 (FLJ37478), mRNA
NMJ 78558 Homo sapiens hypothetical protein FLJ90430 (FLJ90430), mRNA
NMJ 78559 Homo sapiens ATP-binding cassette, sub-family B (MDR/TAP), member 5 (A
NMJ78562 Homo sapiens hypothetical protein MGC50844 (MGC50844), mRNA
NMJ 78563 Homo sapiens hypothetical protein LOC340351 (LOC340351), mRNA
NMJ 78564 Homo sapiens hypothetical protein LOC340371 (LOC340371), mRNA
NMJ 78565 Homo sapiens hypothetical protein MGC35555 (MGC35555), mRNA
NMJ 78566 Homo sapiens zinc finger, DHHC domain containing 21 (ZDHHC21), mRNA
NMJ 78568 Homo sapiens reticulon 4 receptor-like 1 (RTN4RL1), mRNA
NMJ 78569 Homo sapiens CEI protein (CEI), mRNA
N J 78570 Homo sapiens reticulon 4 receptor-like 2 (RTN4RL2), mRNA
NMJ78571 Homo sapiens hypothetical protein MGC51025 (MGC51025), mRNA
NMJ 78578 Homo sapiens proteasome (prosome, macropain) inhibitor subunit 1 (PI31) (
NMJ 78579 Homo sapiens proteasome (prosome, macropain) inhibitor subunit 1 (PI31) (
NMJ 78580 Homo sapiens histocompatibility (minor) 13 (HM13), transcript variant 2, mRt
NMJ 78581 Homo sapiens histocompatibility (minor) 13 (HM13), transcript variant 3, mRI
NMJ78582 Homo sapiens histocompatibility (minor) 13 (HM13), transcript variant 4, mRt
NMJ 78583 Homo sapiens WD repeat and FYVE domain containing 3 (WDFY3), transcri
NMJ 78584 Homo sapiens septin 10 (SEPT10), transcript variant 2, mRNA
NMJ 78585 Homo sapiens WD repeat and FYVE domain containing 3 (WDFY3), transcri
NMJ 78586 Homo sapiens protein phosphatase 2, regulatory subunit B (B56), gamma isc
NMJ 78587 Homo sapiens protein phosphatase 2, regulatory subunit B (B56), gamma isc
NMJ 78588 Homo sapiens protein phosphatase 2, regulatory subunit B (B56), gamma isc
NMJ 78812 Homo sapiens LYRIC/3D3 (LYRIC), mRNA
NMJ 78813 Homo sapiens A-kinase anchoring protein 28 (AKAP28), mRNA
NMJ 78814 Homo sapiens adaptor-related protein complex 1 , sigma 3 subunit (AP1S3),
NMJ 78815 Homo sapiens ADP-ribosylation factor-like 8 (ARL8), mRNA
NMJ 78816 Homo sapiens cancer susceptibility candidate 2 (CASC2), mRNA
NMJ78817 Homo sapiens chromosome 21 open reading frame 61 (C21orf61), transcript
NMJ 78818 Homo sapiens chemokine-like factor super family 4 (CKLFSF4), transcript va
NMJ 78819 Homo sapiens putative lysophosphatidic acid acyltransferase (DKFZp586M1
NMJ 78820 Homo sapiens F-box protein 27 (FBX027), mRNA
NMJ 78821 Homo sapiens hypothetical protein FLJ25955 (FLJ25955), mRNA
NMJ 78822 Homo sapiens immunoglobulin superfamily, member 10 (IGSF10), mRNA
NMJ 78823 Homo sapiens chromosome 6 open reading frame 165 (C6orf165), mRNA
NMJ 78824 Homo sapiens hypothetical protein FLJ33620 (FLJ33620), mRNA
NMJ 78826 Homo sapiens transmembrane protein 16D (TMEM16D), mRNA
NMJ 78827 Homo sapiens hypothetical protein FLJ35834 (FLJ35834), mRNA
NMJ 78828 Homo sapiens chromosome 9 open reading frame 79 (C9orf79), mRNA
NMJ 78829 Homo sapiens chromosome 7 open reading frame 34 (C7orf34), mRNA
NMJ 78830 Homo sapiens hypothetical protein FLJ36888 (FLJ36888), mRNA
NMJ 78831 Homo sapiens opposite strand transcription unit to STAG3 (GATS), mRNA
NMJ 78832 Homo sapiens chromosome 10 open reading frame 83 (C10orf83), mRNA
NMJ 78833 Homo sapiens hypothetical protein BC009732 (LOC133308), mRNA
NMJ 78834 Homo sapiens layilin (LOC143903), mRNA
NMJ 78835 Homo sapiens hypothetical protein LOC152485 (LOC152485), mRNA
NMJ78836 Homo sapiens similar to CG12314 gene product (LOC201164), mRNA
NMJ 78837 Homo sapiens similar to hypothetical testis protein from macaque (LOC3529
NMJ 78838 Homo sapiens hypothetical protein LOC90768 (MGC45800), mRNA
NMJ 78839 Homo sapiens leucine rich repeat transmembrane neuronal 1 (LRRTM1), mF
NMJ 78840 Homo sapiens hypothetical protein MGC24047 (MGC24047), mRNA
NMJ 78841 Homo sapiens ring finger protein 166 (RNF166), mRNA
NMJ 78842 Homo sapiens LAG1 longevity assurance homolog 3 (S. cerevisiae) (LASS3)
NMJ 78844 Homo sapiens NOD3 protein (NOD3), mRNA
NMJ 78849 Homo sapiens hepatocyte nuclear factor 4, alpha (HNF4A), transcript variant
NMJ 78850 Homo sapiens hepatocyte nuclear factor 4, alpha (HNF4A), transcript variant
NMJ 78857 Homo sapiens retinitis pigmentosa 1 -like 1 (RP1L1), mRNA
NMJ 78858 Homo sapiens sideroflexin 2 (SFXN2), mRNA
NMJ 78859 Homo sapiens organic solute transporter beta (OSTbeta), mRNA
NMJ 78860 Homo sapiens seizure related 6 homolog (mouse) (SEZ6), mRNA
NMJ 78861 Homo sapiens zinc finger protein 183-like 1 (ZNF183L1), mRNA
NMJ78862 Homo sapiens source of immunodominant MHC-associated peptides (SIMP)
NMJ 8863 Homo sapiens potassium channel tetra erisation domain containing 13 (KC
NMJ 78864 Homo sapiens HLH-PAS transcription factor NXF (NXF), mRNA
NMJ 78865 Homo sapiens tumor differentially expressed 2-like (TDE2L), mRNA
NMJ 78867 Homo sapiens sideroflexin 4 (SFXN4), transcript variant 2, mRNA
NMJ 78868 Homo sapiens chemokine-like factor super family 8 (CKLFSF8), mRNA
NMJ80699 Homo sapiens U11/U12 snRNP 35K (U1SNRNPBP), transcript variant 3, mR
NMJ 80703 Homo sapiens U11/U12 snRNP 35K (U1SNRNPBP), transcript variant 4, mR
NMJ 80976 Homo sapiens protein phosphatase 2, regulatory subunit B (B56), delta isofo
NMJ 80977 Homo sapiens protein phosphatase 2, regulatory subunit B (B56), delta isofo
NMJ 80981 Homo sapiens mitochondrial ribosomal protein L52 (MRPL52), nuclear gene
NMJ 80982 Homo sapiens mitochondrial ribosomal protein L52 (MRPL52), nuclear gene
NMJ 80989 Homo sapiens intimal thickness-related receptor (ITR), mRNA
NMJ 80990 Homo sapiens ligand-gated ion channel subunit (LGICZ), mRNA
NMJ 80991 Homo sapiens solute carrier organic anion transporter family, member 4C1 (!
NMJ81041 Homo sapiens polybromo 1 (PB1), mRNA
NMJ81042 Homo sapiens polybromo 1 (PB1), mRNA
NMJ 81050 Homo sapiens axin 1 (AXIN1), transcript variant 2, mRNA
NMJ 81054 Homo sapiens hypoxia-inducible factor 1 , alpha subunit (basic helix-loop-heli
NMJ 81076 Homo sapiens golgin-67 (GOLGIN-67), transcript variant 2, mRNA
NMJ81077 Homo sapiens golgin-67 (GOLGIN-67), transcript variant 3, mRNA
NMJ 81078 Homo sapiens interleukin 21 receptor (IL21 R), transcript variant 2, mRNA
NMJ 81079 Homo sapiens interleukin 21 receptor (IL21 R), transcript variant 3, mRNA
NMJ 81093 Homo sapiens ezrin-binding partner PACE-1 (PACE-1), transcript variant 2, r
NMJ 81265 Homo sapiens WD repeat domain 17 (WDR17), transcript variant 2, mRNA
NMJ 81268 Homo sapiens chemokine-like factor super family 1 (CKLFSF1), transcript va
NMJ 81269 Homo sapiens chemokine-like factor super family 1 (CKLFSF1 ), transcript va
NMJ 81270 Homo sapiens chemokine-like factor super family 1 (CKLFSF1), transcript va
NMJ 81271 Homo sapiens chemokine-like factor super family 1 (CKLFSF1), transcript va
N J 81272 Homo sapiens chemokine-like factor super family 1 (CKLFSF1 ), transcript va
NMJ 81283 Homo sapiens chemokine-like factor super family 1 (CKLFSF1), transcript va
NMJ 81285 Homo sapiens chemokine-like factor super family 1 (CKLFSF1), transcript va
NMJ 81286 Homo sapiens chemokine-like factor super family 1 (CKLFSF1), transcript va
NMJ 81287 Homo sapiens chemokine-like factor super family 1 (CKLFSF1), transcript va
N J 81288 Homo sapiens chemokine-like factor super family 1 (CKLFSF1 ), transcript va
NMJ 81289 Homo sapiens chemokine-like factor super family 1 (CKLFSF1 ), transcript va
NMJ81290 Homo sapiens chemokine-like factor super family 1 (CKLFSF1), transcript va
NMJ 81291 Homo sapiens WD repeat domain 20 ( DR20), transcript variant 1 , mRNA
NMJ 81292 Homo sapiens chemokine-like factor super family 1 (CKLFSF1), transcript va
NMJ81293 Homo sapiens chemokine-like factor super family 1 (CKLFSF1), transcript va
NMJ 81294 Homo sapiens chemokine-like factor super family 1 (CKLFSF1), transcript va
NMJ 81295 Homo sapiens chemokine-like factor super family 1 (CKLFSF1 ), transcript va
NMJ 81296 Homo sapiens chemokine-like factor super family 1 (CKLFSF1), transcript va
NMJ 81297 Homo sapiens chemokine-like factor super family 1 (CKLFSF1), transcript va
NMJ81298 Homo sapiens chemokine-like factor super family 1 (CKLFSF1), transcript va
NMJ81299 Homo sapiens chemokine-like factor super family 1 (CKLFSF1), transcript va
NMJ 81300 Homo sapiens chemokine-like factor super family 1 (CKLFSF1), transcript va
NMJ 81301 Homo sapiens chemokine-like factor super family 1 (CKLFSF1), transcript va
NMJ 81302 Homo sapiens WD repeat domain 20 (WDR20), transcript variant 4, mRNA
NMJ 81304 Homo sapiens mitochondrial ribosomal protein L52 (MRPL52), nuclear gene
NMJ81305 Homo sapiens mitochondrial ribosomal protein L52 (MRPL52), nuclear gene
NMJ 81306 Homo sapiens mitochondrial ribosomal protein L52 (MRPL52), nuclear gene
NMJ81307 Homo sapiens mitochondrial ribosomal protein L52 (MRPL52), nuclear gene
NMJ 81308 Homo sapiens WD repeat domain 20 (WDR20), transcript variant 3, mRNA
NMJ 81309 Homo sapiens interleukin 22 receptor, alpha 2 (IL22RA2), transcript variant 5
NMJ81310 Homo sapiens interleukin 22 receptor, alpha 2 (IL22RA2), transcript variant S
NMJ 81311 Homo sapiens tafazzin (cardiomyopathy, dilated 3A (X-linked); endocardial fi
NMJ 81312 Homo sapiens tafazzin (cardiomyopathy, dilated 3A (X-linked); endocardial fi
NMJ81313 Homo sapiens tafazzin (cardiomyopathy, dilated 3A (X-linked); endocardial fi
NMJ 81314 Homo sapiens tafazzin (cardiomyopathy, dilated 3A (X-linked); endocardial fi
NMJ 81332 Homo sapiens neuroligin 4, X-linked (N LGN4X), transcript variant 2, mRNA
NMJ 81333 Homo sapiens Rho GTPase activating protein 8 (ARHGAP8), transcript varia
NMJ 81334 Homo sapiens Rho GTPase activating protein 8 (ARHGAP8), transcript varia
NMJ 81335 Homo sapiens Rho GTPase activating protein 8 (ARHGAP8), transcript varia
NMJ 81336 Homo sapiens LEM domain containing 2 (LEMD2), mRNA
NMJ 81337 Homo sapiens kidney associated antigen 1 (KAAG1), mRNA
NMJ 81339 Homo sapiens interleukin 24 (IL24), transcript variant 2, mRNA
NMJ 81340 Homo sapiens WD repeat domain 21 (WDR21 ), transcript variant 2, mRNA
NMJ 81341 Homo sapiens WD repeat domain 21 (WDR21), transcript variant 3, mRNA
NMJ 81342 Homo sapiens FK506 binding protein 7 (FKBP7), transcript variant 2, mRNA
NMJ 81349 Homo sapiens SMAD specific E3 ubiquitin protein ligase 1 (SMURF1), transc
NMJ 81351 Homo sapiens neural cell adhesion molecule 1 (NCAM1), mRNA
NMJ 81353 Homo sapiens inhibitor of DNA binding , dominant negative helix-loop-helix
NMJ 81354 Homo sapiens oxidation resistance 1 (OXR1), mRNA
NM 181355 Homo sapiens frequently rearranged in advanced T-cell lymphomas (FRAT1
NMJ 81356 Homo sapiens suppressor of Ty 3 homolog (S. cerevisiae) (SUPT3H), mRNA
NMJ 81357 Homo sapiens WD repeat domain 23 (WDR23), transcript variant 2, mRNA
NM 181358 Homo sapiens homeodomain interacting protein kinase 1 (HIPK1), transcript
NMJ 81359 Homo sapiens interleukin 6 receptor (IL6R), transcript variant 2, mRNA
NM 181361 Homo sapiens potassium large conductance calcium-activated channel, subt
NM 181425 Homo sapiens Friedreich ataxia (FRDA), nuclear gene encoding mitochondri
NMJ 81427 Homo sapiens GA binding protein transcription factor, beta subunit 2, 47kDa
NMJ81428 Homo sapiens thymosin-like 6 (TMSL6), mRNA
NM 181429 Homo sapiens candidate taste receptor hT2R55 (hT2R55), mRNA
NMJ 81430 Homo sapiens forkhead box K2 (FOXK2), transcript variant 2, mRNA
NMJ81431 Homo sapiens forkhead box K2 (FOXK2), transcript variant 3, mRNA
NM 181435 Homo sapiens C1q and tumor necrosis factor related protein 3 (C1QTNF3), r
NMJ81441 Homo sapiens mitochondrial ribosomal protein L55 (MRPL55), nuclear gene
NM 181442 Homo sapiens activity-dependent neuroprotector (ADNP), transcript variant 2
NMJ81443 Homo sapiens BTB (POZ) domain containing 3 (BTBD3), transcript variant 2
NMJ81446 Homo sapiens follicle stimulating hormone receptor (FSHR), transcript variar
NM 181449 Homo sapiens immune receptor expressed on myeloid cells 2 (IREM2), mRiN
NMJ81453 Homo sapiens GRIP and coiled-coil domain containing 2 (GCC2), transcript >
NM 181454 Homo sapiens mitochondrial ribosomal protein L55 (MRPL55), nuclear gene
NMJ 81455 Homo sapiens mitochondrial ribosomal protein L55 (MRPL55), nuclear gene
NM 181456 Homo sapiens mitochondrial ribosomal protein L55 (MRPL55), nuclear gene
NMJ81457 Homo sapiens paired box gene 3 (Waardenburg syndrome 1 ) (PAX3), transc
NMJ81458 Homo sapiens paired box gene 3 (Waardenburg syndrome 1 ) (PAX3), transc
NM 181459 Homo sapiens paired box gene 3 (Waardenburg syndrome 1 ) (PAX3), transc
NMJ81460 Homo sapiens paired box gene 3 (Waardenburg syndrome 1 ) (PAX3), transc
NM 181461 Homo sapiens paired box gene 3 (Waardenburg syndrome 1 ) (PAX3), transc
NMJ81462 Homo sapiens mitochondrial ribosomal protein L55 (MRPL55), nuclear gene
NM 181463 Homo sapiens mitochondrial ribosomal protein L55 (MRPL55), nuclear gene
NMJ81464 Homo sapiens mitochondrial ribosomal protein L55 (MRPL55), nuclear gene
NM 181465 Homo sapiens mitochondrial ribosomal protein L55 (MRPL55), nuclear gene
NM 181466 Homo sapiens integrin beta 4 binding protein (ITGB4BP), transcript variant 4
NMJ81467 Homo sapiens integrin beta 4 binding protein (ITGB4BP), transcript variant 5
NMJ 81468 Homo sapiens integrin beta 4 binding protein (ITGB4BP), transcript variant 2
NM 181469 Homo sapiens integrin beta 4 binding protein (ITGB4BP), transcript variant 3
NM 181471 Homo sapiens replication factor C (activator 1 ) 2, 40kDa (RFC2), transcript v
NMJ 81472 Homo sapiens chemokine-like factor super family 7 (CKLFSF7), transcript va
NMJ81481 Homo sapiens chromosome 18 open reading frame 1 (C18orf1), transcript VΪ
NM 181482 Homo sapiens chromosome 18 open reading frame 1 (C18orf1 ), transcript VE
NMJ 81483 Homo sapiens chromosome 18 open reading frame 1 (C18orf1), transcript VJ
NMJ 81484 Homo sapiens KIAA1847 (KIAA1847), transcript variant 2, mRNA
NM 181485 Homo sapiens KIAA1847 (KIAA1847), transcript variant 3, mRNA
NMJ 81486 Homo sapiens T-box 5 (TBX5), transcript variant 4, mRNA
NM 181489 Homo sapiens zinc finger protein 445 (ZNF445), mRNA
NM 181491 Homo sapiens surfeit 5 (SURF5), transcript variant c, mRNA
NMJ 81492 Homo sapiens transcription factor 20 (AR1 ) (TCF20), transcript variant 2, mF
NM 181493 Homo sapiens inosine triphosphatase (nucleoside triphosphate pyrophospha
NM 181500 Homo sapiens cut-like 1 , CCAAT displacement protein (Drosophila) (CUTL1)
NM 181501 Homo sapiens integrin, alpha 1 (ITGA1), mRNA
NM 181502 Homo sapiens serine protease inhibitor-like, with Kunitz and WAP domains 1
NM 181503 Homo sapiens exosome component 8 (EXOSC8), mRNA
NM 181504 Homo sapiens phosphoinositide-3-kinase, regulatory subunit, polypeptide 1 (
NMJ 81505 Homo sapiens protein phosphatase 1 , regulatory (inhibitor) subunit 1B (dopa
NM 181506 Homo sapiens synleurin (SLRN), mRNA
NM 181507 Homo sapiens Hermansky-Pudlak syndrome 5 (HPS5), transcript variant 1 , r
NMJ 81508 Homo sapiens Hermansky-Pudlak syndrome 5 (HPS5), transcript variant 3, r
NMJ 81509 Homo sapiens microtubule-associated protein 1 light chain 3 alpha (MAP1 LC
NMJ 81510 Homo sapiens WAP four-disulfide core domain 8 (WFDC8), transcript varian
NMJ81512 Homo sapiens mitochondrial ribosomal protein L21 (MRPL21), nuclear gene
NMJ 81513 Homo sapiens mitochondrial ribosomal protein L21 (MRPL21), nuclear gene
NMJ81514 Homo sapiens mitochondrial ribosomal protein L21 (MRPL21), nuclear gene
NMJ 81515 Homo sapiens mitochondrial ribosomal protein L21 (MRPL21), nuclear gene
NMJ81519 Homo sapiens synaptotagmin XV (SYT15), transcript variant b, mRNA
NMJ 81521 Homo sapiens chemokine-like factor super family 4 (CKLFSF4), transcript va
NMJ 81522 Homo sapiens WAP four-disulfide core domain 3 (WFDC3), transcript varian
NMJ 81523 Homo sapiens phosphoinositide-3-kinase, regulatory subunit, polypeptide 1 (
NMJ 81524 Homo sapiens phosphoinositide-3-kinase, regulatory subunit, polypeptide 1 (
NMJ 81525 Homo sapiens WAP four-disulfide core domain 3 (WFDC3), transcript varian
NMJ 81526 Homo sapiens myosin, light polypeptide 9, regulatory (MYL9), transcript variε
NMJ81527 Homo sapiens Nl -acetyltransferase 5 (ARD1 homolog, S. cerevisiae) (NAT5),
NMJ81528 Homo sapiens N -acetyltransferase 5 (ARD1 homolog, S. cerevisiae) (NAT5),
NMJ81530 Homo sapiens WAP four-disulfide core domain 3 (WFDC3), transcript varian
NMJ81531 Homo sapiens butyrophilin, subfamily 2, member A2 (BTN2A2), transcript va
NMJ81532 Homo sapiens ES cell expressed Ras (ERAS), mRNA
NMJ 81533 Homo sapiens chromosome 14 open reading frame 29 (C14orf29), transcripl
NMJ 81534 Homo sapiens keratin 25A (KRT25A), mRNA
NMJ 81535 Homo sapiens keratin 25D (KRT25D), mRNA
NMJ81536 Homo sapiens polycystic kidney disease 1-like 3 (PKD1 L3), mRNA
NMJ 81537 Homo sapiens keratin 25C (KRT25C), mRNA
NMJ81538 Homo sapiens gap junction protein, epsilon 1 , 29kDa (GJE1), mRNA
NMJ 81539 Homo sapiens keratin 25B (KRT25B), mRNA
NMJ 81552 Homo sapiens cut-like 1 , CCAAT displacement protein (Drosophila) (CUTL1)
NMJ 81553 Homo sapiens chemokine-like factor superfamily 3 (CKLFSF3), transcript va
NMJ 81554 Homo sapiens chemokine-like factor super family 3 (CKLFSF3), transcript va
NMJ 81555 Homo sapiens chemokine-like factor super family 3 (CKLFSF3), transcript va
NMJ81558 Homo sapiens replication factor C (activator 1) 3, 38kDa (RFC3), transcript v
NMJ 81571 Homo sapiens cAMP responsive element modulator (CREM), transcript varia
NMJ 81572 Homo sapiens regulator of G-protein signalling like 1 (RGSL1), mRNA
NMJ 81573 Homo sapiens replication factor C (activator 1 ) 4, 37kDa (RFC4), transcript v
NMJ 81575 Homo sapiens ancient ubiquitous protein 1 (AUP1), transcript variant 2, mR
NMJ81576 Homo sapiens ancient ubiquitous protein 1 (AUP1), transcript variant 3, mRh
NMJ 81578 Homo sapiens replication factor C (activator 1) 5, 36.5kDa (RFC5), transcript
NMJ81581 Homo sapiens protein similar to E.coli yhdg and R. capsulatus nifR3 (PP35),
NMJ 81597 Homo sapiens uridine phosphorylase 1 (UPP1), transcript variant 2, mRNA
NMJ 81598 Homo sapiens spastic paraplegia 3A (autosomal dominant) (SPG3A), mRNA
NMJ 81599 Homo sapiens keratin associated protein 13-1 (KRTAP13-1), mRNA
NMJ 81600 Homo sapiens keratin associated protein 13-4 (KRTAP13-4), mRNA
NMJ 81602 Homo sapiens keratin associated protein 6-1 (KRTAP6-1), mRNA
NMJ 81604 Homo sapiens keratin associated protein 6-2 (KRTAP6-2), mRNA
NMJ81605 Homo sapiens keratin associated protein 6-3 (KRTAP6-3), mRNA
NMJ 81607 Homo sapiens keratin associated protein 19-1 (KRTAP19-1), mRNA
NMJ 81608 Homo sapiens keratin associated protein 19-2 (KRTAP19-2), mRNA
NMJ81609 Homo sapiens keratin associated protein 19-3 (KRTAP19-3), mRNA
NMJ81610 Homo sapiens keratin associated protein 19-4 (KRTAP19-4), mRNA
NMJ 81611 Homo sapiens keratin associated protein 19-5 (KRTAP19-5), mRNA
NMJ81612 Homo sapiens keratin associated protein 19-6 (KRTAP19-6), mRNA
NMJ81614 Homo sapiens keratin associated protein 19-7 (KRTAP19-7), mRNA
NMJ81615 Homo sapiens keratin associated protein 20-1 (KRTAP20-1), mRNA
NMJ81616 Homo sapiens keratin associated protein 20-2 (KRTAP20-2), mRNA
NMJ81617 Homo sapiens keratin associated protein 21-2 (KRTAP21-2), mRNA
NMJ 81618 Homo sapiens chemokine-like factor super family 5 (CKLFSF5), transcript va
NMJ81619 Homo sapiens keratin associated protein 21-1 (KRTAP21-1), mRNA
NMJ81620 Homo sapiens keratin associated protein 22-1 (KRTAP22-1), mRNA
NMJ 81621 Homo sapiens keratin associated protein 13-2 (KRTAP13-2), nuclear gene e
NMJ 81622 Homo sapiens keratin associated protein 13-3 (KRTAP13-3), mRNA
NMJ81623 Homo sapiens keratin associated protein 15-1 (KRTAP15-1), mRNA
NMJ 81624 Homo sapiens keratin associated protein 23-1 (KRTAP23-1), mRNA
NMJ 81640 Homo sapiens chemokine-like factor (CKLF), transcript variant 2, mRNA
NMJ81641 Homo sapiens chemokine-like factor (CKLF), transcript variant 4, mRNA
NMJ 81642 Homo sapiens serine protease inhibitor, Kunitz type 1 (SPINT1), transcript vε
NMJ 81643 Homo sapiens hypothetical protein LOC128344 (LOC128344), mRNA
NMJ81644 Homo sapiens hypothetical protein DKFZp761N1114 (DKFZp761N1114), mF
NMJ 81645 Homo sapiens hypothetical protein FLJ25393 (FLJ25393), mRNA
NMJ 81646 Homo sapiens hypothetical protein FLJ32110 (FLJ32110), mRNA
NMJ 81647 Homo sapiens hypothetical protein LOC285398 (LOC285398), mRNA
NMJ 81651 Homo sapiens peroxiredoxin 5 (PRDX5), nuclear gene encoding mitochondri
NMJ 81652 Homo sapiens peroxiredoxin 5 (PRDX5), nuclear gene encoding mitochondri
NMJ 81654 Homo sapiens complexin 4 (CPLX4), mRNA
NMJ81655 Homo sapiens hypothetical protein LOC284018 (LOC284018), transcript vari
NMJ 81656 Homo sapiens hypothetical protein LOC284018 (LOC284018), transcript vari
NMJ 81657 Homo sapiens leukotriene B4 receptor (LTB4R), mRNA
NMJ81659 Homo sapiens nuclear receptor coactivator 3 (NCOA3), transcript variant 1 , r
NMJ81661 Homo sapiens Cohen syndrome 1 (COH1), transcript variant 4, mRNA
NMJ 81670 Homo sapiens E2a-Pbx1 -associated protein (EB-1), transcript variant 2, mRt
NMJ81671 Homo sapiens phosphatidylinositol transfer protein, cytoplasmic 1 (PITPNC1
NMJ 81672 Homo sapiens O-linked N-acetylglucosamine (GlcNAc) transferase (UDP-N-i
NMJ 81673 Homo sapiens O-linked N-acetylglucosamine (GlcNAc) transferase (UDP-N-i
NMJ81674 Homo sapiens protein phosphatase 2 (formerly 2A), regulatory subunit B (PF
NMJ 81675 Homo sapiens protein phosphatase 2 (formerly 2A), regulatory subunit B (PF
NMJ 81676 Homo sapiens protein phosphatase 2 (formerly 2A), regulatory subunit B (PF
NMJ 81677 Homo sapiens protein phosphatase 2 (formerly 2A), regulatory subunit B (PF
NMJ81678 Homo sapiens protein phosphatase 2 (formerly 2A), regulatory subunit B (PF
NMJ 81679 Homo sapiens NFS1 nitrogen fixation 1 (S. cerevisiae) (NFS1), nuclear gene
NMJ 81684 Homo sapiens keratin associated protein 12-2 (KRTAP12-2), mRNA
NMJ 81686 Homo sapiens keratin associated protein 12-1 (KRTAP12-1), mRNA
NMJ81688 Homo sapiens keratin associated protein 10-10 (KRTAP10-10), mRNA
NMJ 81689 Homo sapiens neuronatin (NNAT), transcript variant 2, mRNA
NMJ 81690 Homo sapiens v-akt murine thymoma viral oncogene homolog 3 (protein kinε
NMJ81696 Homo sapiens peroxiredoxin 1 (PRDX1), transcript variant 2, mRNA
NMJ 81697 Homo sapiens peroxiredoxin 1 (PRDX1), transcript variant 3, mRNA
NMJ 81698 Homo sapiens chromosome 10 open reading frame 9 (C10orf9), mRNA
NMJ 81699 Homo sapiens protein phosphatase 2 (formerly 2A), regulatory subunit A (PF
NMJ 81701 Homo sapiens quiescin Q6-like 1 (QSCN6L1), mRNA
NMJ 81702 Homo sapiens GTP binding protein overexpressed in skeletal muscle (GEM)
NMJ 81703 Homo sapiens gap junction protein, alpha 5, 40kDa (connexin 40) (GJA5), trc
NMJ 81704 Homo sapiens B melanoma antigen family, member 4 (BAGE4), mRNA
NMJ 81705 Homo sapiens hypothetical protein LOC90624 (LOC90624), mRNA
NMJ 81706 Homo sapiens zinc finger, CSL domain containing 3 (ZCSL3), mRNA
NMJ 81707 Homo sapiens hypothetical protein LOC124773 (LOC124773), mRNA
NMJ81708 Homo sapiens hypothetical protein LOC144233 (LOC144233), mRNA
NMJ81709 Homo sapiens hypothetical protein LOC144347 (LOC144347), mRNA
NMJ 81710 Homo sapiens zinc and ring finger 4 (ZNRF4), mRNA
NMJ 81711 Homo sapiens GRP1 (general receptor for phosphoinositides 1 )-associated s
NMJ81712 Homo sapiens hypothetical protein LOC163782 (LOC163782), mRNA
NMJ 81713 Homo sapiens UBX domain containing 4 (UBXD4), mRNA
NMJ81714 Homo sapiens chromosome 6 open reading frame 152 (C6orf152), mRNA
NMJ 81715 Homo sapiens transducer of regulated cAMP response element-binding proti
NMJ 81716 Homo sapiens nuclear protein p30 (p30), mRNA
NMJ 81717 Homo sapiens HLA complex group 27 (HCG27), mRNA
NMJ 81718 Homo sapiens hypothetical protein LOC253982 (LOC253982), mRNA
NMJ 81719 Homo sapiens hypothetical protein LOC255104 (LOC255104), mRNA
NM 181720 Homo sapiens hypothetical protein LOC257106 (LOC257106), mRNA
NMJ 81721 Homo sapiens forkhead box R1 (FOXR1), mRNA
NM 181722 Homo sapiens hypothetical protein LOC285908 (LOC285908), mRNA
NM 181723 Homo sapiens hypothetical protein LOC286097 (LOC286097), mRNA
NM 181724 Homo sapiens hypothetical protein LOC338773 (LOC338773), mRNA
NMJ81725 Homo sapiens hypothetical protein FLJ 12760 (FLJ 12760), mRNA
NM 181726 Homo sapiens low density lipoprotein receptor-related protein binding proteir
NMJ 81727 Homo sapiens spermatogenesis associated 12 (SPATA12), mRNA
NM 181733 Homo sapiens component of oligomeric golgi complex 5 (COG5), transcript \
NMJ 81737 Homo sapiens peroxiredoxin 2 (PRDX2), nuclear gene encoding mitochondri
NM 181738 Homo sapiens peroxiredoxin 2 (PRDX2), nuclear gene encoding mitochondri
NMJ 81739 Homo sapiens WINS1 protein with Drosophila Lines (Lin) homologous doma
NMJ 81740 Homo sapiens WINS1 protein with Drosophila Lines (Lin) homologous doma
NM 181741 Homo sapiens origin recognition complex, subunit 4-like (yeast) (ORC4L), trε
NMJ 81742 Homo sapiens origin recognition complex, subunit 4-like (yeast) (ORC4L), trε
NM 181744 Homo sapiens opsin 5 (OPN5), mRNA
NMJ81745 Homo sapiens G protein-coupled receptor 120 (GPR120), mRNA
NM 181746 Homo sapiens LAG1 longevity assurance homolog 2 (S. cerevisiae) (LASS2)
NMJ 81747 Homo sapiens origin recognition complex, subunit 5-like (yeast) (ORC5L), trε
NMJ 81755 Homo sapiens hydroxysteroid (11 -beta) dehydrogenase 1 (HSD11 B1), transc
NMJ 81756 Homo sapiens zinc finger protein 233 (ZNF233), mRNA
NM 181762 Homo sapiens ubiquitin-conjugating enzyme E2A (RAD6 homolog) (UBE2A),
NMJ 81773 Homo sapiens ARP10 protein (ARP10), mRNA
NMJ 81774 Homo sapiens solute carrier family 36 (proton/amino acid symporter), membi
NM 181775 Homo sapiens hypothetical protein DKFZp434G0625 (DKFZp434G0625), mF
NMJ 81776 Homo sapiens solute carrier family 36 (proton/amino acid symporter), membi
NM 181777 Homo sapiens ubiquitin-conjugating enzyme E2A (RAD6 homolog) (UBE2A),
NMJ 81780 Homo sapiens B and T lymphocyte associated (BTLA), mRNA
NM 181781 Homo sapiens hypothetical protein FLJ20403 (FLJ20403), transcript variant :
NMJ 81782 Homo sapiens nuclear receptor coactivator 7 (NCOA7), mRNA
NM 181783 Homo sapiens SMILE protein (SMILE), mRNA
NMJ81784 Homo sapiens sprouty-related, EVH1 domain containing 2 (SPRED2), mRN/
NM 181785 Homo sapiens hypothetical protein LOC283537 (LOC283537), mRNA
NMJ 81786 Homo sapiens GLI-Kruppel family member HKR1 (HKR1), mRNA
NM 181787 Homo sapiens hypothetical protein LOC286148 (LOC286148), mRNA
NMJ 81788 Homo sapiens HANP1 (LOC341567), mRNA
NM 181789 Homo sapiens collomin (COLM), mRNA
NMJ 81790 Homo sapiens G protein-coupled receptor 142 (GPR142), mRNA
NM 181791 Homo sapiens G protein-coupled receptor 141 (GPR141), mRNA
NMJ 81794 Homo sapiens protein kinase (cAMP-dependent, catalytic) inhibitor beta (PKI
NM 181795 Homo sapiens protein kinase (cAMP-dependent, catalytic) inhibitor beta (PKI
NM 181797 Homo sapiens potassium voltage-gated channel, KQT-like subfamily, membe
NM 181798 Homo sapiens potassium voltage-gated channel, KQT-like subfamily, memb«
NMJ 81799 Homo sapiens ubiquitin-conjugating enzyme E2C (UBE2C), transcript varian
NM 181800 Homo sapiens ubiquitin-conjugating enzyme E2C (UBE2C), transcript varian'
NM 181801 Homo sapiens ubiquitin-conjugating enzyme E2C (UBE2C), transcript varian
NM 181802 Homo sapiens ubiquitin-conjugating enzyme E2C (UBE2C), transcript varian
NM 181803 Homo sapiens ubiquitin-conjugating enzyme E2C (UBE2C), transcript varian
NM 181804 Homo sapiens protein kinase (cAMP-dependent, catalytic) inhibitor gamma (I
NMJ 81805 Homo sapiens protein kinase (cAMP-dependent, catalytic) inhibitor gamma (I
NM 181806 Homo sapiens 2-aminoadipic 6-semialdehyde dehydrogenase (NRPS998), n
NM 181807 Homo sapiens doublecortin domain containing 1 (DCDC1), mRNA
NMJ81808 Homo sapiens polymerase (DNA directed) nu (POLN), mRNA
NM 181809 Homo sapiens bone morphogenetic protein 8a (BMP8A), mRNA
NM 181814 Homo sapiens chromosome 14 open reading frame 29 (C14orf29), transcript
NM 181825 Homo sapiens neurofibromin 2 (bilateral acoustic neuroma) (NF2), transcript
NM 181826 Homo sapiens neurofibromin 2 (bilateral acoustic neuroma) (NF2), transcript
NMJ 81827 Homo sapiens neurofibromin 2 (bilateral acoustic neuroma) (NF2), transcript
NMJ 81828 Homo sapiens neurofibromin 2 (bilateral acoustic neuroma) (NF2), transcript
NMJ 81829 Homo sapiens neurofibromin 2 (bilateral acoustic neuroma) (NF2), transcript
NMJ 81830 Homo sapiens neurofibromin 2 (bilateral acoustic neuroma) (NF2), transcript
NMJ 81831 Homo sapiens neurofibromin 2 (bilateral acoustic neuroma) (NF2), transcript
NMJ 81832 Homo sapiens neurofibromin 2 (bilateral acoustic neuroma) (NF2), transcript
NMJ 81833 Homo sapiens neurofibromin 2 (bilateral acoustic neuroma) (NF2), transcript
NMJ 81834 Homo sapiens neurofibromin 2 (bilateral acoustic neuroma) (NF2), transcript
NMJ 81835 Homo sapiens neurofibromin 2 (bilateral acoustic neuroma) (NF2), transcript
NMJ 81836 Homo sapiens CGI-109 protein (CGI-109), mRNA
NMJ 81837 Homo sapiens origin recognition complex, subunit 3-like (yeast) (ORC3L), trε
NMJ 81838 Homo sapiens ubiquitin-conjugating enzyme E2D 2 (UBC4/5 homolog, yeast
NMJ 81839 Homo sapiens protein kinase (cAMP-dependent, catalytic) inhibitor alpha (PF
NMJ 81840 Homo sapiens TWIK-related spinal cord K+ channel (TRIK), mRNA
NMJ 81841 Homo sapiens transmembrane channel-like 3 (TMC3), mRNA
NMJ 81842 Homo sapiens zinc finger and BTB domain containing 12 (ZBTB12), mRNA
NMJ 81843 Homo sapiens nudix (nucleoside diphosphate linked moiety X)-type motif 8 (I
NMJ 81844 Homo sapiens B-cell CLL/lymphoma 6, member B (zinc finger protein) (BCLf
NMJ 81846 Homo sapiens GLI-Kruppel family member HKR2 (HKR2), mRNA
NMJ 81847 Homo sapiens amphoterin induced gene 2 (AMIG02), mRNA
NMJ 81861 Homo sapiens apoptotic protease activating factor (APAF1 ), transcript varian
NMJ 81862 Homo sapiens brain acyl-CoA hydrolase (BACH), transcript variant hBACHa/
NMJ 81863 Homo sapiens brain acyl-CoA hydrolase (BACH), transcript variant hBACHa/
NMJ 81864 Homo sapiens brain acyl-CoA hydrolase (BACH), transcript variant hBACHb,
NMJ 81865 Homo sapiens brain acyl-CoA hydrolase (BACH), transcript variant hBACHc,
NMJ 81866 Homo sapiens brain acyl-CoA hydrolase (BACH), transcript variant hBACHd,
NMJ 81868 Homo sapiens apoptotic protease activating factor (APAF1 ), transcript varian
NMJ 81869 Homo sapiens apoptotic protease activating factor (APAF1 ), transcript varian
NMJ 81870 Homo sapiens dishevelled, dsh homolog 1 (Drosophila) (DVL1), transcript va
NMJ 81871 Homo sapiens pyruvate kinase, liver and RBC (PKLR), nuclear gene encodir
NMJ 81872 Homo sapiens doublesex and mab-3 related transcription factor 2 (DMRT2),
NMJ 81873 Homo sapiens cisplatin resistance associated (CRA), mRNA
NMJ 81874 Homo sapiens glutamate receptor, metabotropic 7 (GRM7), transcript variant
NMJ 81875 Homo sapiens glutamate receptor, metabotropic 7 (GRM7), transcript variant
NMJ 81876 Homo sapiens protein phosphatase 2 (formerly 2A), regulatory subunit B (PF
NMJ 81877 Homo sapiens zinc finger protein 29 (ZFP29), mRNA
NMJ 81879 Homo sapiens leukocyte Ig-like receptor 9 (LIR9), transcript variant 3, mRNA
NMJ 81880 Homo sapiens variable charge, Y-linked 1 B (VCY1 B), mRNA
NMJ 81882 Homo sapiens periaxin (PRX), mRNA
NMJ 81885 Homo sapiens G protein-coupled receptor 100 (GPR100), mRNA
NMJ 81886 Homo sapiens ubiquitin-conjugating enzyme E2D 3 (UBC4/5 homolog, yeast
NMJ 81887 Homo sapiens ubiquitin-conjugating enzyme E2D 3 (UBC4/5 homolog, yeast
NMJ 81888 Homo sapiens ubiquitin-conjugating enzyme E2D 3 (UBC4/5 homolog, yeast
NMJ 81889 Homo sapiens ubiquitin-conjugating enzyme E2D 3 (UBC4/5 homolog, yeast
NMJ 81890 Homo sapiens ubiquitin-conjugating enzyme E2D 3 (UBC4/5 homolog, yeast
NMJ 81891 Homo sapiens ubiquitin-conjugating enzyme E2D 3 (UBC4/5 homolog, yeast
NMJ 81892 Homo sapiens ubiquitin-conjugating enzyme E2D 3 (UBC4/5 homolog, yeast
NMJ81893 Homo sapiens ubiquitin-conjugating enzyme E2D 3 (UBC4/5 homolog, yeast
NMJ 81894 Homo sapiens glutamate receptor, ionotrophic, AMPA 3 (GRIA3), transcript \
NMJ 81897 Homo sapiens protein phosphatase 2 (formerly 2A), regulatory subunit B", al
NMJ 81900 Homo sapiens START domain containing 5 (STARD5), transcript variant 1 , n
NMJ 81985 Homo sapiens leukocyte Ig-like receptor 9 (LIR9), transcript variant 2, mRNA
NMJ 81986 Homo sapiens leukocyte Ig-like receptor 9 (LIR9), transcript variant 4, mRNA
NMJ 82314 Homo sapiens cytosolic ovarian carcinoma antigen 1 (COVA1 ), transcript vai
NMJ 82398 Homo sapiens ribosomal protein S6 kinase, 90kDa, polypeptide 5 (RPS6KAE
NMJ 82470 Homo sapiens pyruvate kinase, muscle (PKM2), transcript variant 2, mRNA
NMJ 82471 Homo sapiens pyruvate kinase, muscle (PKM2), transcript variant 3, mRNA
NMJ 82472 Homo sapiens EphA5 (EPHA5), transcript variant 2, mRNA
NMJ 82476 Homo sapiens coenzyme Q6 homolog (yeast) (C0Q6), transcript variant 1 , n
NMJ 82477 Homo sapiens gametogenetin (GGN), transcript variant 2, mRNA
NMJ 82480 Homo sapiens coenzyme Q6 homolog (yeast) (C0Q6), transcript variant 2, n
NMJ 82481 Homo sapiens B melanoma antigen family, member 3 (BAGE3), mRNA
NMJ 82482 Homo sapiens B melanoma antigen family, member 2 (BAGE2), mRNA
NMJ 82483 Homo sapiens NSFL1 (p97) cofactor (p47) (NSFL1C), transcript variant 3, m
NMJ 82484 Homo sapiens B melanoma antigen family, member 5 (BAGE5), mRNA
NMJ 82485 Homo sapiens cytoplasmic polyadenylation element binding protein 2 (CPEB
NMJ 82486 Homo sapiens C1q and tumor necrosis factor related protein 6 (C1QTNF6), t
NMJ 82487 Homo sapiens olfactomedin-like 2A (OLFML2A), mRNA
NMJ 82488 Homo sapiens ubiquitin specific protease 12 (USP12), mRNA
NMJ 82489 Homo sapiens STRA8 (LOC346673), mRNA
NMJ 82490 Homo sapiens zinc finger protein 227 (ZNF227), mRNA
NMJ 82491 Homo sapiens hypothetical protein LOC90637 (LOC90637), mRNA
NMJ 82492 Homo sapiens hypothetical protein DKFZp434O0213 (DKFZp434O0213), mF
NMJ 82493 Homo sapiens myosin light chain kinase (MLCK) (LOC91807), mRNA
NMJ 82494 Homo sapiens family with sequence similarity 26, member A (FAM26A), mRt
NMJ 82495 Homo sapiens hypothetical protein FLJ25224 (FLJ25224), mRNA
NMJ 82496 Homo sapiens hypothetical protein FLJ40089 (FLJ40089), mRNA
NMJ 82497 Homo sapiens type I hair keratin KA36 (KA36), mRNA
NMJ 82498 Homo sapiens hypothetical protein MGC51082 (MGC51082), mRNA
NMJ 82499 Homo sapiens hypothetical protein DKFZp434M202 (DKFZp434M202), mRN
NMJ 82500 Homo sapiens hypothetical protein FLJ25143 (FLJ25143), mRNA
NMJ 82501 Homo sapiens hypothetical protein MGC61716 (MGC61716), mRNA
NMJ 82502 Homo sapiens hypothetical protein DKFZp686L1818 (DKFZp686L1818), mR
NMJ 82503 Homo sapiens deaminase domain containing 1 (DEADC1), mRNA
NMJ 82504 Homo sapiens Williams-Beuren syndrome critical region 28 (WBSCR28), mF
NMJ 82505 Homo sapiens chromosome 9 open reading frame 85 (C9orf85), transcript vs
NMJ 82506 Homo sapiens hypothetical protein FLJ32965 (FLJ32965), mRNA
NMJ82507 Homo sapiens hypothetical protein LOC144501 (LOC144501), mRNA
NMJ 82508 Homo sapiens hypothetical protein FLJ40919 (FLJ40919), mRNA
NMJ82509 Homo sapiens thrombospondin, type I, domain containing 3 (THSD3), transc
NMJ 82510 Homo sapiens hypothetical protein FLJ32252 (FLJ32252), mRNA
NMJ 82511 Homo sapiens cerebellin 2 precursor (CBLN2), mRNA
NMJ 82513 Homo sapiens kinetochore protein Spc24 (Spc24), mRNA
NMJ 82516 Homo sapiens hypothetical protein FLJ32011 (FLJ32011), mRNA
NMJ 82517 Homo sapiens hypothetical protein MGC52423 (MGC52423), mRNA
N M J 82518 Homo sapiens hypothetical protein LOC149469 (LOC149469), mRNA
NMJ 82519 Homo sapiens chromosome 20 open reading frame 186 (C20orf186), mRNA
NMJ 82520 Homo sapiens chromosome 22 open reading frame 15 (C22orf15), mRNA
NMJ 82521 Homo sapiens zinc finger, SWIM domain containing 2 (ZSWIM2), mRNA
NMJ 82522 Homo sapiens TAFA4 protein (TAFA4), mRNA
NMJ 82523 Homo sapiens hypothetical protein MGC61571 (MGC61571 ), mRNA
NMJ 82524 Homo sapiens zinc finger protein 595 (ZNF595), mRNA
NMJ 82525 Homo sapiens hypothetical protein FLJ32770 (FLJ32770), mRNA
NMJ 82526 Homo sapiens hypothetical protein FLJ33387 (FLJ33387), mRNA
NMJ 82527 Homo sapiens calcium binding protein 7 (CABP7), mRNA
NMJ 82528 Homo sapiens complement component 1 , q subcomponent-like 2 (C1QL2), r
NMJ 82529 Homo sapiens THAP domain containing 5 (THAP5), mRNA
NMJ 82530 Homo sapiens hypothetical protein FLJ25056 (FLJ25056), mRNA
NMJ 82531 Homo sapiens hypothetical protein FLJ31875 (FLJ31875), mRNA
NMJ 82532 Homo sapiens hypothetical protein LOC199964 (LOC199964), mRNA
NMJ 82533 Homo sapiens hypothetical protein FLJ31031 (FLJ31031), mRNA
NMJ 82534 Homo sapiens hypothetical protein FLJ23703 (FLJ23703), mRNA
NMJ 82535 Homo sapiens hypothetical protein LOC200261 (LOC200261), mRNA
NMJ 82536 Homo sapiens down-regulated in gastric cancer GDDR (GDDR), mRNA
NMJ 82537 Homo sapiens 5-hydroxytryptamine (serotonin) receptor 3 family member D (
NMJ 82538 Homo sapiens hypothetical protein MGC29671 (MGC29671), mRNA
NMJ 82539 Homo sapiens hypothetical protein MGC33600 (MGC33600), mRNA
NMJ 82541 Homo sapiens transmembrane protein 31 (TMEM31), mRNA
NMJ82543 Homo sapiens nucleolar protein (NOL1/NOP2/sun) and PUA domains 1 (NOI
NMJ 82546 Homo sapiens hypothetical protein MGC33530 (MGC33530), mRNA
NMJ 82547 Homo sapiens putative NFkB activating protein HNLF (HNLF), mRNA
NMJ 82548 Homo sapiens hypothetical protein MGC33835 (MGC33835), mRNA
NMJ 82549 Homo sapiens major histocompatibility complex, class II, DQ beta 2 (HLA-DC
NMJ 82551 Homo sapiens acyl-CoA:lysocardiolipin acyltransferase 1 (ALCAT1), transcrii
NMJ 82552 Homo sapiens hypothetical protein MGC43690 (MGC43690), mRNA
NMJ 82553 Homo sapiens hypothetical protein MGC50896 (MGC50896), mRNA
NMJ 82554 Homo sapiens chromosome 10 open reading frame 53 (C10orf53), mRNA
NMJ 82556 Homo sapiens hypothetical protein LOC283130 (LOC283130), mRNA
NMJ 82557 Homo sapiens B-cell CLL/lymphoma 9-like (BCL9L), mRNA
NMJ 82558 Homo sapiens hypothetical protein FLJ33810 (FLJ33810), mRNA
NMJ 82559 Homo sapiens hypothetical protein MGC57341 (MGC57341), mRNA
NMJ 82560 Homo sapiens hypothetical protein FLJ25773 (FLJ25773), mRNA
NMJ 82561 Homo sapiens hypothetical protein FLJ36144 (FLJ36144), mRNA
NMJ 82562 Homo sapiens hypothetical protein FLJ39743 (FLJ39743), mRNA
NMJ 82563 Homo sapiens hypothetical protein MGC21830 (MGC21830), mRNA
NMJ 82564 Homo sapiens hypothetical protein FLJ40319 (FLJ40319), mRNA
NMJ 82565 Homo sapiens hypothetical protein MGC29814 (MGC29814), mRNA
NMJ82566 Homo sapiens secretory protein LOC284013 (LOC284013), mRNA
NMJ82568 Homo sapiens hypothetical protein FLJ36492 (FLJ36492), mRNA
NMJ82569 Homo sapiens hypothetical protein FLJ37451 (FLJ37451), mRNA
NMJ82570 Homo sapiens hypothetical protein FLJ25715 (FLJ25715), mRNA
NMJ 82572 Homo sapiens zinc finger and SCAN domain containing 1 (ZSCAN1 ), mRNA
NMJ 82573 Homo sapiens hypothetical protein FLJ30469 (FLJ30469), mRNA
NMJ 82574 Homo sapiens hypothetical protein FLJ36070 (FLJ36070), mRNA
NMJ 82575 Homo sapiens hypothetical protein MGC34799 (MGC34799), mRNA
NMJ 82577 Homo sapiens chromosome 19 open reading frame 19 (C19orf19), mRNA
NMJ 82578 Homo sapiens hypothetical protein FLJ37964 (FLJ37964), mRNA
NMJ 82579 Homo sapiens hypothetical protein FLJ40343 (FLJ40343), mRNA
NMJ 82580 Homo sapiens cytochrome b-561 domain containing 1 (CYB561 D1), mRNA
NMJ 82581 Homo sapiens hypothetical protein LOC284680 (LOC284680), mRNA
NMJ 82583 Homo sapiens hypothetical protein FLJ38374 (FLJ38374), mRNA
NMJ 82584 Homo sapiens hypothetical protein FLJ33706 (FLJ33706), mRNA
NMJ 82585 Homo sapiens hypothetical protein DKFZp451 M2119 (DKFZp451 M2119), ml
NMJ 82586 Homo sapiens hypothetical protein FLJ33534 (FLJ33534), mRNA
NMJ 82587 Homo sapiens chromosome 2 open reading frame 21 (C2orf21 ), mRNA
NMJ 82589 Homo sapiens 5-hydroxytryptamine (serotonin) receptor 3, family member E
NMJ 82590 Homo sapiens hypothetical protein FLJ33651 (FLJ33651 ), mRNA
NMJ 82591 Homo sapiens hypothetical protein FLJ37673 (FLJ37673), mRNA
NMJ 82592 Homo sapiens hypothetical protein FLJ39576 (FLJ39576), mRNA
NMJ 82594 Homo sapiens zinc finger protein 454 (ZNF454), mRNA
NMJ82595 Homo sapiens hypothetical protein DKFZp564N2472 (DKFZp564N2472), mF
NMJ 82596 Homo sapiens hypothetical protein FLJ25037 (FLJ25037), mRNA
NMJ 82597 Homo sapiens hypothetical protein FLJ39575 (FLJ39575), mRNA
NMJ 82598 Homo sapiens hypothetical protein FLJ36980 (FLJ36980), mRNA
NMJ 82600 Homo sapiens hypothetical protein LOC286359 (LOC286359), mRNA
NMJ 82603 Homo sapiens hypothetical protein FLJ37874 (FLJ37874), mRNA
NMJ 82605 Homo sapiens hypothetical protein FLJ40448 (FLJ40448), mRNA
NMJ 82606 Homo sapiens hypothetical protein LOC339967 (LOC339967), mRNA
NMJ 82607 Homo sapiens hypothetical protein MGC44287 (MGC44287), mRNA
NMJ82608 Homo sapiens hypothetical protein DKFZp68601689 (DKFZp68601689), mF
NMJ 82609 Homo sapiens hypothetical protein MGC48625 (MGC48625), mRNA
NM 182610 Homo sapiens sterile alpha motif domain containing 7 (SAMD7), mRNA
NMJ 82611 Homo sapiens G protein-coupled receptor 144 (GPR144), mRNA
NM 182612 Homo sapiens hypothetical protein FLJ34283 (FLJ34283), mRNA
NMJ 82613 Homo sapiens hypothetical protein FLJ33915 (FLJ33915), mRNA
NMJ 82614 Homo sapiens hypothetical protein MGC20579 (MGC20579), mRNA
NM 182615 Homo sapiens hypothetical protein MGC40069 (MGC40069), mRNA
NM 182616 Homo sapiens hypothetical protein MGC61550 (MGC61550), mRNA
NMJ 82617 Homo sapiens xenobiotic/medium-chain fatty acid:CoA ligase (HXMA), nude
NM 182619 Homo sapiens secretory protein LOC348174 (LOC348174), mRNA
NM 182620 Homo sapiens family with sequence similarity 33, member A (FAM33A), mRt
NMJ 82621 Homo sapiens hypothetical protein MGC52498 (MGC52498), mRNA
NMJ 82623 Homo sapiens hypothetical protein FLJ36766 (FLJ36766), mRNA
NMJ 82625 Homo sapiens hypothetical protein FLJ40869 (FLJ40869), mRNA
NM 182626 Homo sapiens hypothetical protein FLJ25102 (FLJ25102), mRNA
NMJ 82627 Homo sapiens hypothetical protein MGC64882 (MGC64882), mRNA
NM 182628 Homo sapiens hypothetical protein FLJ40083 (FLJ40083), mRNA
NM 182631 Homo sapiens hypothetical protein LOC348840 (LOC348840), mRNA
NMJ 82632 Homo sapiens solute carrier family 6 (neurotransmitter transporter), member
NM 182633 Homo sapiens hypothetical protein FLJ39963 (FLJ39963), mRNA
NM 182634 Homo sapiens hypothetical protein FLJ36166 (FLJ36166), mRNA
NMJ 82635 Homo sapiens hypothetical protein LOC349236 (LOC349236), mRNA
NM 182637 Homo sapiens Hermansky-Pudlak syndrome 1 (HPS1), transcript variant 2, r
NM 182638 Homo sapiens Hermansky-Pudlak syndrome 1 (HPS1), transcript variant 4, r
NMJ 82639 Homo sapiens Hermansky-Pudlak syndrome 1 (HPS1), transcript variant 3, r
NM 182640 Homo sapiens mitochondrial ribosomal protein S9 (MRPS9), nuclear gene er
NM 182641 Homo sapiens fetal Alzheimer antigen (FALZ), transcript variant 1 , mRNA
NM 182642 Homo sapiens CTD (carboxy-terminal domain, RNA polymerase II, polypeptii
NM 182643 Homo sapiens deleted in liver cancer 1 (DLC1 ), transcript variant 1 , mRNA
NMJ 82644 Homo sapiens EphA3 (EPHA3), transcript variant 2, mRNA
NM 182645 Homo sapiens vestigial like 2 (Drosophila) (VGLL2), transcript variant 1 , mRt
NMJ 82646 Homo sapiens cytoplasmic polyadenylation element binding protein 2 (CPEB
NMJ 82647 Homo sapiens opiate receptor-like 1 (OPRL1 ), transcript variant 1 , mRNA
NM 182648 Homo sapiens bromodomain adjacent to zinc finger domain, 1 A (BAZ1 A), tra
NM 182649 Homo sapiens proliferating cell nuclear antigen (PCNA), transcript variant 2,
NM 182658 Homo sapiens chromosome 20 open reading frame 185 (C20orf185), mRNA
NMJ 82659 Homo sapiens ubiquitously transcribed tetratricopeptide repeat gene, Y-linke
NM 182660 Homo sapiens ubiquitously transcribed tetratricopeptide repeat gene, Y-linke
NMJ 82661 Homo sapiens ceramide kinase (CERK), transcript variant 2, mRNA
NM 182662 Homo sapiens aminoadipate aminotransferase (AADAT), transcript variant 2.
NMJ 82663 Homo sapiens Ras association (RalGDS/AF-6) domain family 5 (RASSF5), ti
NM 182664 Homo sapiens Ras association (RalGDS/AF-6) domain family 5 (RASSF5), ti
NM 182665 Homo sapiens Ras association (RalGDS/AF-6) domain family 5 (RASSF5), ti
NM 182666 Homo sapiens ubiquitin-conjugating enzyme E2E 1 (UBC4/5 homolog, yeast
NM 182676 Homo sapiens phospholipid transfer protein (PLTP), transcript variant 2, mRI
NMJ 82678 Homo sapiens ubiquitin-conjugating enzyme E2E 3 (UBC4/5 homolog, yeast
NMJ 82679 Homo sapiens hypothetical protein FLJ20249 (FLJ20249), transcript variant :
NM 182680 Homo sapiens amelogenin (amelogenesis imperfecta 1 , X-linked) (AMELX), ■
NMJ 82681 Homo sapiens amelogenin (amelogenesis imperfecta 1 , X-linked) (AMELX), •
NM 182682 Homo sapiens ubiquitin-conjugating enzyme E2G 1 (UBC7 homolog, C. eleg
NMJ 82683 Homo sapiens uroplakin 3B (UPK3B), transcript variant 3, mRNA
NM 182684 Homo sapiens uroplakin 3B (UPK3B), transcript variant 2, mRNA
NMJ 82685 Homo sapiens ephrin-A1 (EFNA1), transcript variant 2, mRNA
NM 182686 Homo sapiens polycystic kidney disease 1-like (PKD1-like), transcript variant
NMJ 82687 Homo sapiens membrane-associated tyrosine- and threonine-specific cdc2-iι
NMJ 82688 Homo sapiens ubiquitin-conjugating enzyme E2G 2 (UBC7 homolog, yeast) |
NMJ 82689 Homo sapiens ephrin-A4 (EFNA4), transcript variant 2, mRNA
NMJ 82690 Homo sapiens ephrin-A4 (EFNA4), transcript variant 3, mRNA
NM J 82691 Homo sapiens SFRS protein kinase 2 (SRPK2), transcript variant 2, mRNA
NM_ J 82692 Homo sapiens SFRS protein kinase 2 (SRPK2), transcript variant 1 , mRNA
NM J 82697 Homo sapiens ubiquitin-conjugating enzyme E2H (UBC8 homolog, yeast) (U
NM J 82699 Homo sapiens DEAD (Asp-Glu-Ala-Asp) box polypeptide 53 (DDX53), mRNA
NM J 82700 Homo sapiens Sp8 transcription factor (SP8), transcript variant 1 , mRNA
NM 182701 Homo sapiens glutathione peroxidase 6 (olfactory) (GPX6), mRNA
NM 82702 Homo sapiens testis serine protease 2 (TESSP2), mRNA
NM J 82703 Homo sapiens hypothetical protein LOC348094 (LOC348094), mRNA
NM 82704 Homo sapiens selenoprotein V (SELV), mRNA
NM_ J 82705 Homo sapiens hypothetical protein MGC45871 (MGC45871), mRNA
NM_ 82706 Homo sapiens scribbled homolog (Drosophila) (SCRIB), transcript variant 1 ,
NM 182709 Homo sapiens HIV-1 Tat interacting protein, 60kDa (HTATIP), transcript vari?
NM_ 82710 Homo sapiens HIV-1 Tat interacting protein, 60kDa (HTATIP), transcript vari?
NM 182712 Homo sapiens eukaryotic translation initiation factor 3, subunit 9 eta, 116kDε
NM_ 82715 Homo sapiens synaptophysin-like protein (SYPL), transcript variant 2, mRNA
NM_ 182717 Homo sapiens cAMP responsive element modulator (CREM), transcript varia
NM 82718 Homo sapiens cAMP responsive element modulator (CREM), transcript varia
NM_ 82719 Homo sapiens cAMP responsive element modulator (CREM), transcript varia
NM 182720 Homo sapiens cAMP responsive element modulator (CREM), transcript varia
NM_ 82721 Homo sapiens cAMP responsive element modulator (CREM), transcript varia
NM 82722 Homo sapiens cAMP responsive element modulator (CREM), transcript varia
NM 82723 Homo sapiens cAMP responsive element modulator (CREM), transcript varia
NM_ 82724 Homo sapiens cAMP responsive element modulator (CREM), transcript varia
NM 82725 Homo sapiens cAMP responsive element modulator (CREM), transcript varia
NM 82728 Homo sapiens solute carrier family 7 (cationic amino acid transporter, y+ sys
NM_ 182729 Homo sapiens thioredoxin reductase 1 (TXNRD1), transcript variant 5, mRN/
NM_ 82734 Homo sapiens phospholipase C, beta 1 (phosphoinositide-specific) (PLCB1).
NM 182739 Homo sapiens NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 6, 17
NM_ 82740 Homo sapiens polycystic kidney disease 1-like 2 (PKD1 L2), transcript varianl
NM .182741 Homo sapiens mucin 1 , transmembrane (MUC1), mRNA
NM 82742 Homo sapiens thioredoxin reductase 1 (TXNRD1), transcript variant 2, mRN/
NM_ .182743 Homo sapiens thioredoxin reductase 1 (TXNRD1), transcript variant 4, mRN/
NM 82744 Homo sapiens neuroblastoma, suppression of tumorigenicity 1 (NBL1), trans
NM .182746 Homo sapiens MCM4 minichromosome maintenance deficient 4 (S. cerevisiε
NM .182749 Homo sapiens chromosome 21 open reading frame 127 (C21orf127), transci
NM 182751 Homo sapiens MCM10 minichromosome maintenance deficient 10 (S. cerevi
NM .182752 Homo sapiens hypothetical protein LOC127262 (LOC127262), mRNA
NM 182755 Homo sapiens hypothetical protein LOC220929 (LOC220929), mRNA
NM .182756 Homo sapiens speedy homolog 1 (Drosophila) (SPDY1), mRNA
NM 182757 Homo sapiens IBR domain containing 2 (IBRDC2), mRNA
NM 182758 Homo sapiens hypothetical protein FLJ38736 (FLJ38736), mRNA
NM .182759 Homo sapiens TAFA3 protein (TAFA3), mRNA
NM .182760 Homo sapiens sulfatase modifying factor 1 (SUMF1), mRNA
NM .182761 Homo sapiens hypothetical protein LOC340069 (LOC340069), mRNA
NM .182762 Homo sapiens putative binding protein 7a5 (7A5), mRNA
NM .182763 Homo sapiens myeloid cell leukemia sequence 1 (BCL2-related) (MCL1), tra
NM 182764 Homo sapiens engulfment and cell motility 2 (ced-12 homolog, C. elegans) (I
NM 182765 Homo sapiens HECT domain containing 2 (HECTD2), transcript variant 1 , ml
NM 182766 Homo sapiens hypothetical protein FLJ32940 (DKFZp686H1423), transcript '
NM_ .182767 Homo sapiens solute carrier family 6 (neurotransmitter transporter), member
NM 182769 Homo sapiens cAMP responsive element modulator (CREM), transcript varia
NM_ 182770 Homo sapiens cAMP responsive element modulator (CREM), transcript varia
NM 182771 Homo sapiens cAMP responsive element modulator (CREM), transcript varia
NM .182772 Homo sapiens cAMP responsive element modulator (CREM), transcript varia
NM .182774 Homo sapiens hypothetical protein LOC259173 (FLJ36525), transcript variar
NM_ .1 82775 Homo sapiens hypothetical protein LOC259173 (FLJ36525), transcript variar
NM_ .182776 Homo sapiens MCM7 minichromosome maintenance deficient 7 (S. cerevisiε
NMJ 82777 Homo sapiens ribosomal protein S3A (RPS3A), mRNA
NMJ 82779 Homo sapiens dishevelled, dsh homolog 1 (Drosophila) (DVL1), mRNA
NMJ 82789 Homo sapiens poly(A) binding protein interacting protein 1 (PAIP1), transcrip
NMJ 82790 Homo sapiens pre-B-cell colony enhancing factor 1 (PBEF1 ), transcript varia
NMJ 82791 Homo sapiens hypothetical protein FLJ32855 (FLJ32855), mRNA
NMJ 82792 Homo sapiens thymosin-like 1 (TMSL1 ), mRNA
NMJ 82793 Homo sapiens thymosin-like 2 (TMSL2), mRNA
NMJ 82794 Homo sapiens thymosin-like 4 (TMSL4), mRNA
NMJ 82795 Homo sapiens nucleophosmin/nucleoplasmin, 2 (NPM2), mRNA
NMJ 82796 Homo sapiens methionine adenosyltransferase II, beta (MAT2B), transcript v
NMJ 82797 Homo sapiens phospholipase C, beta 4 (PLCB4), transcript variant 2, mRNA
NMJ 82798 Homo sapiens hypothetical protein FLJ39155 (FLJ39155), transcript variant ,'
NMJ 82799 Homo sapiens hypothetical protein FLJ39155 (FLJ39155), transcript variant :
NMJ 82800 Homo sapiens arsenate resistance protein ARS2 (ARS2), transcript variant 2
NMJ 82801 Homo sapiens hypothetical protein FLJ39155 (FLJ39155), transcript variant <
NMJ 82802 Homo sapiens MCM8 minichromosome maintenance deficient 8 (S. cerevisiε
NMJ 82804 Homo sapiens apolipoprotein B48 receptor (APOB48R), mRNA
NMJ 82810 Homo sapiens activating transcription factor 4 (tax-responsive enhancer elen
NMJ 82811 Homo sapiens phospholipase C, gamma 1 (PLCG1), transcript variant 2, mR
NMJ 82812 Homo sapiens splicing factor 4 (SF4), transcript variant c, mRNA
NMJ 82826 Homo sapiens scavenger receptor class A, member 3 (SCARA3), transcript *
NMJ82827 Homo sapiens FK506 binding protein 9-like (FKBP9L), mRNA
NMJ 82828 Homo sapiens growth differentiation factor 7 (GDF7), mRNA
NMJ 82829 Homo sapiens hypothetical protein LOC158160 (LOC158160), mRNA
NMJ 82830 Homo sapiens MAM domain containing 1 (MAMDC1), mRNA
NMJ 82831 Homo sapiens TNT protein (TNT), mRNA
NMJ 82832 Homo sapiens placenta-specific 4 (PLAC4), mRNA
NMJ 82833 Homo sapiens GDPD domain containing protein (LOC220032), mRNA
NMJ 82835 Homo sapiens sed family domain containing 1 (SCFD1), transcript variant 2
NMJ 82836 Homo sapiens Rab geranylgeranyltransferase, alpha subunit (RABGGTA), tr
NMJ 82838 Homo sapiens solute carrier family 35, member E2 (SLC35E2), mRNA
NMJ 82847 Homo sapiens amiloride-sensitive cation channel 4, pituitary (ACCN4), transi
NMJ82848 Homo sapiens claudin 10 (CLDN10), transcript variant 1 , mRNA
NMJ 82849 Homo sapiens cyclin B1 interacting protein 1 (CCNB1 IP1), transcript variant
NMJ 82850 Homo sapiens cAMP responsive element modulator (CREM), transcript varia
NMJ 82851 Homo sapiens cyclin B1 interacting protein 1 (CCNB1 IP1), transcript variant
NMJ 82852 Homo sapiens cyclin B1 interacting protein 1 (CCNB1 IP1 ), transcript variant ■
NMJ 82853 Homo sapiens cAMP responsive element modulator (CREM), transcript varia
NMJ 82854 Homo sapiens selectin ligand interactor cytoplasmic-1 (SL1C1 ), mRNA
NMJ 82894 Homo sapiens ceh-10 homeo domain containing homolog (C. elegans) (CHX
NMJ 82895 Homo sapiens scavenger receptor class F, member 2 (SCARF2), transcript \
NMJ 82896 Homo sapiens hypothetical protein DKFZp761 H079 (DKFZp761 H079), trans*
NMJ 82898 Homo sapiens cAMP responsive element binding protein 5 (CREB5), mRNA
NMJ 82899 Homo sapiens cAMP responsive element binding protein 5 (CREB5), mRNA
NMJ 82901 Homo sapiens chromosome 11 open reading frame 17 (C11orf17), transcript
NMJ 82902 Homo sapiens kinesin family member 9 (KIF9), mRNA
NMJ 82903 Homo sapiens kinesin family member 9 (KIF9), mRNA
NMJ 82904 Homo sapiens procollagen-proline, 2-oxoglutarate 4-dioxygenase (proline 4-I
NMJ 82905 Homo sapiens CXYorfl -related protein (DKFZp434K1323), mRNA
NMJ 82906 Homo sapiens C-type (calcium dependent, carbohydrate-recognition domain
NMJ 82907 Homo sapiens PR domain containing 1 , with ZNF domain (PRDM1), transcrii
NMJ 82908 Homo sapiens dehydrogenase/reductase (SDR family) member 2 (DHRS2),
NMJ 82909 Homo sapiens downregulated in ovarian cancer 1 (DOC1), transcript variant
NMJ 82910 Homo sapiens spectrin repeat containing, nuclear envelope 2 (SYNE2), tran;
NMJ 82911 Homo sapiens testis specific, 10 (TSGA10), mRNA
NMJ 82912 Homo sapiens spectrin repeat containing, nuclear envelope 2 (SYNE2), trans
NMJ 82913 Homo sapiens spectrin repeat containing, nuclear envelope 2 (SYNE2), trans
NMJ 82914 Homo sapiens spectrin repeat containing, nuclear envelope 2 (SYNE2), trans
NMJ 82915 Homo sapiens dudulin 2 (TSAP6), mRNA
NMJ 82916 Homo sapiens tRNA nucleotidyl transferase, CCA-adding, 1 (TRNT1 ), mRNA
NMJ 82917 Homo sapiens eukaryotic translation initiation factor 4 gamma, 1 (EIF4G1), ti
NM 82918 Homo sapiens v-ets erythroblastosis virus E26 oncogene like (avian) (ERG),
N 82919 Homo sapiens TIR domain containing adaptor inducing interferon-beta (TRIF
NMJ 82920 Homo sapiens a disintegrin-like and metalloprotease (reprolysin type) with th
NM 82921 Homo sapiens a disintegrin-like and metalloprotease (reprolysin type) with th
NMJ 82922 Homo sapiens hypothetical protein FLJ20718 (FLJ20718), mRNA
N J 82923 Homo sapiens kinesin 2 60/70kDa (KNS2), mRNA
N J 82924 Homo sapiens MICAL-like 2 (FLJ23471 ), transcript variant 1 , mRNA
NMJ 82925 Homo sapiens fms-related tyrosine kinase 4 (FLT4), transcript variant 1 , mRI
NMJ 82926 Homo sapiens kinectin 1 (kinesin receptor) (KTN1), mRNA
NMJ 82931 Homo sapiens myeloid/lymphoid or mixed-lineage leukemia 5 (trithorax home
NMJ 82932 Homo sapiens solute carrier family 8 (sodium-calcium exchanger), member
N 82933 Homo sapiens solute carrier family 8 (sodium-calcium exchanger), member J
N J 82934 Homo sapiens myelin-associated oligodendrocyte basic protein (MOBP), mR
NMJ 82935 Homo sapiens myelin-associated oligodendrocyte basic protein (MOBP), mR
NMJ 82936 Homo sapiens solute carrier family 8 (sodium-calcium exchanger), member J
NMJ 82943 Homo sapiens procollagen-lysine, 2-oxoglutarate 5-dioxygenase (lysine hydr
NM 82944 Homo sapiens ninein (GSK3B interacting protein) (NIN), transcript variant 1 ,
NMJ 82945 Homo sapiens ninein (GSK3B interacting protein) (NIN), transcript variant 3,
NMJ82946 Homo sapiens ninein (GSK3B interacting protein) (NIN), transcript variant 5,
NMJ 82947 Homo sapiens RAC/CDC42 exchange factor (GEFT), transcript variant 1 , mF
NMJ 82948 Homo sapiens protein kinase, cAMP-dependent, catalytic, beta (PRKACB), ti
N 82960 Homo sapiens hypothetical protein MGC21644 (MGC21644), transcript varia
NMJ82961 Homo sapiens spectrin repeat containing, nuclear envelope 1 (SYNE1), trans
NMJ 82962 Homo sapiens baculoviral IAP repeat-containing 3 (BIRC3), transcript varianl
NMJ 82964 Homo sapiens neuron navigator 2 (NAV2), transcript variant 1 , mRNA
NMJ 82965 Homo sapiens sphingosine kinase 1 (SPHK1), mRNA
NMJ 82966 Homo sapiens neural precursor cell expressed, developmentally down-regul*
NMJ82970 Homo sapiens regulating synaptic membrane exocytosis 4 (RIMS4), mRNA
NMJ 82971 Homo sapiens cytochrome c oxidase subunit 8C (COX8C), mRNA
NMJ 82972 Homo sapiens interferon regulatory factor 2 binding protein 2 (IRF2BP2), mR
NMJ 82973 Homo sapiens transmembrane serine protease 9 (TMPRSS9), mRNA
NMJ 82974 Homo sapiens galactosyltransferase family 6 domain containing 1 (GLTDC1)
NMJ 82975 Homo sapiens hypothetical protein FLJ20403 (FLJ20403), transcript variant '■
N J 82976 Homo sapiens hypothetical protein FLJ20403 (FLJ20403), transcript variant •
NMJ 82977 Homo sapiens nicotinamide nucleotide transhydrogenase (NNT), mRNA
NMJ 82978 Homo sapiens guanine nucleotide binding protein (G protein), alpha activatin
NMJ 82980 Homo sapiens pregnancy-induced growth inhibitor (OKL38), mRNA
NMJ82981 Homo sapiens pregnancy-induced growth inhibitor (OKL38), mRNA
NMJ 82982 Homo sapiens G protein-coupled receptor kinase 4 (GRK4), transcript varian
NMJ 82983 Homo sapiens hepsin (transmembrane protease, serine 1) (HPN), transcript
NMJ82984 Homo sapiens Hpall tiny fragments locus 9C (HTF9C), transcript variant 1 , r
NMJ 82985 Homo sapiens ring finger protein 36 (RNF36), transcript variant a, mRNA
NMJ83001 Homo sapiens SHC (Src homology 2 domain containing) transforming proteii
NMJ 83002 Homo sapiens solute carrier family 8 (sodium-calcium exchanger), member ϊ
NMJ 83003 Homo sapiens cytochrome c oxidase subunit Vila polypeptide 3 (liver) (COX!
N J 83004 Homo sapiens eukaryotic translation initiation factor 5 (EIF5), transcript varia
NMJ 83005 Homo sapiens ribonuclease P/MRP 38kDa subunit (RPP38), transcript variai
NMJ83006 Homo sapiens discs, large (Drosophila) homolog-associated protein 4 (DLG/
NMJ 83008 Homo sapiens socius (SOC), mRNA
NMJ83009 Homo sapiens DKFZP434I116 protein (DKFZP434I116), transcript variant 2,
NMJ 83010 Homo sapiens trinucleotide repeat containing 5 (TNRC5), mRNA
NMJ 83011 Homo sapiens cAMP responsive element modulator (CREM), transcript varia
NM J83012 Homo sapiens cAMP responsive element modulator (CREM), transcript varia
NM_183013 Homo sapiens cAMP responsive element modulator (CREM), transcript varia
NM_183040 Homo sapiens dystrobrevin binding protein 1 (DTNBP1), transcript variant 2,
NM__183041 Homo sapiens dystrobrevin binding protein 1 (DTNBP1), transcript variant 3,
NM_183043 Homo sapiens ring finger protein (C3H2C3 type) 6 (RNF6), transcript variant
NM_183044 Homo sapiens ring finger protein (C3H2C3 type) 6 (RNF6), transcript variant
NM_183045 Homo sapiens ring finger protein (C3H2C3 type) 6 (RNF6), transcript variant
NM_183047 Homo sapiens protein kinase C binding protein 1 (PRKCBP1), transcript varii
NM_183048 Homo sapiens protein kinase C binding protein 1 (PRKCBP1), transcript varii
NM_183049 Homo sapiens thymosin-like 3 (TMSL3), mRNA
NM_183050 Homo sapiens branched chain keto acid dehydrogenase E1 , beta polypeptid
NM_183057 Homo sapiens vacuolar protein sorting 28 (yeast) (VPS28), transcript variant
NM_183058 Homo sapiens lysozyme-like 2 (LYZL2), mRNA
NM_183059 Homo sapiens chromosome 1 open reading frame 36 (C1orf36), mRNA
NM__183060 Homo sapiens cAMP responsive element modulator (CREM), transcript varia
NM_183062 Homo sapiens marapsin 2 (MPN2), mRNA
NM_183063 Homo sapiens ring finger protein 7 (RNF7), transcript variant 2, mRNA
NM__183065 Homo sapiens hypothetical protein MGC10744 (MGC10744), transcript varia
NM_183075 Homo sapiens cytochrome P450, family 2, subfamily U, polypeptide 1 (CYP2
NM_183078 Homo sapiens ring finger protein (C3HC4 type) 8 (RNF8), transcript variant 2
NM_183079 Homo sapiens prion protein (p27-30) (Creutzfeld-Jakob disease, Gerstmann-
NM_183227 Homo sapiens RAB23, member RAS oncogene family (RAB23), transcript vε
NM_183228 Homo sapiens zinc finger protein, subfamily 1 A, 3 (Aiolos) (ZNFN1 A3), trans*
NM_183229 Homo sapiens zinc finger protein, subfamily 1 A, 3 (Aiolos) (ZNFN1 A3), trans*
NM_183230 Homo sapiens zinc finger protein, subfamily 1 A, 3 (Aiolos) (ZNFN1 A3), trans*
NM_183231 Homo sapiens zinc finger protein, subfamily 1 A, 3 (Aiolos) (ZNFN1 A3), trans*
NM_183232 Homo sapiens zinc finger protein, subfamily 1A, 3 (Aiolos) (ZNFN1A3), trans*
NM_183233 Homo sapiens solute carrier family 22 (organic cation transporter), member 1
NM_183234 Homo sapiens RAB27A, member RAS oncogene family (RAB27A), transcripl
NM_183235 Homo sapiens RAB27A, member RAS oncogene family (RAB27A), transcripl
NM_183236 Homo sapiens RAB27A, member RAS oncogene family (FRAB27A), transcripl
NM_183237 Homo sapiens ring finger protein 7 (RNF7), transcript variant 3, mRNA
NM_183238 Homo sapiens zinc finger protein 605 (ZNF605), mRNA
NM_183239 Homo sapiens glutathione S-transferase omega 2 (GST02), mRNA
NM_183240 Homo sapiens voltage-dependent calcium channel gamma subunit-like prate
NM_183241 Homo sapiens hypothetical protein LOC286257 (LOC286257), mRNA
NM_183242 Homo sapiens BTB (POZ) domain containing 8 (BTBD8), mRNA
NM_183243 Homo sapiens IMP (inosine monophosphate) dehydrogenase 1 (IMPDH1), tr
NM_183244 Homo sapiens phosphatase and actin regulator 3 (PHACTR3), transcript vari
NM_183245 Homo sapiens inversin (INVS), transcript variant 2, mRNA
NM_183246 Homo sapiens phosphatase and actin regulator 3 (PHACTR3), transcript vari
NM_183247 Homo sapiens transmembrane protease, serine 4 (TMPRSS4), transcript var
NM_183323 Homo sapiens poly(A) binding protein interacting protein 1 (PAIP1), transcrip
NM_183337 Homo sapiens regulator of G-protein signalling 11 (RGS11 ), transcript varian
NM_183352 Homo sapiens SEC13-like 1 (S. cerevisiae) (SEC13L1), transcript variant 2, i
NM_183353 Homo sapiens ring finger protein 12 (RNF12), transcript variant 2, mRNA
NM_183356 Homo sapiens asparagine synthetase (ASNS), transcript variant 3, mRNA
NM_183357 Homo sapiens adenylate cyclase 5 (ADCY5), mRNA
NM_183359 Homo sapiens bromodomain containing 8 (BRD8), transcript variant 3, mRN>
NM_183360 Homo sapiens dystrobrevin, beta (DTNB), transcript variant 4, mRNA
NM_183361 Homo sapiens dystrobrevin, beta (DTNB), transcript variant 5, mRNA
NM_183372 Homo sapiens hypothetical protein LOC200030 (LOC200030), mRNA
NM_183373 Homo sapiens chromosome 6 open reading frame 145 (C6orf145), mRNA
NM_183374 Homo sapiens cytochrome P450, family 26, subfamily C, polypeptide 1 (CYP
NM_183375 Homo sapiens epidermis-specific serine protease-like protein (ESSPL), mRN
NM_183376 Homo sapiens arrestin domain containing 4 (ARRDC4), mRNA
NM_183377 Homo sapiens amiloride-sensitive cation channel 1 , neuronal (degenerin) (A(
NM_183378 Homo sapiens ovochymase 1 (OVCH1), mRNA
NMJ 83379 Homo sapiens testis serine protease 1 (TESSP1), mRNA
NM 83380 Homo sapiens dystonin (DST), transcript variant 1 , mRNA
NM 83381 Homo sapiens ring finger protein 13 (RNF13), transcript variant 4, mRNA
NM 83382 Homo sapiens ring finger protein 13 (RNF13), transcript variant 2, mRNA
NM 83383 Homo sapiens ring finger protein 13 (RNF13), transcript variant 3, mRNA
NMJ 83384 Homo sapiens ring finger protein 13 (RNF13), transcript variant 5, mRNA
NM 83385 Homo sapiens peroxisomal acyl-CoA thioesterase (PTE1 ), transcript variant :
NM 83386 Homo sapiens peroxisomal acyl-CoA thioesterase (PTE1 ), transcript variant :
NM 83387 Homo sapiens echinoderm microtubule associated protein like 5 (EML5), mF
NM 83393 Homo sapiens Ca2+-dependent secretion activator (CADPS), transcript variε
NM 83394 Homo sapiens Ca2+-dependent secretion activator (CADPS), transcript variε
NM 83395 Homo sapiens cold autoinflammatory syndrome 1 (CIAS1 ), transcript variant
NMJ 83397 Homo sapiens peroxisomal membrane protein 4, 24kDa (PXMP4), transcript
NMJ 83398 Homo sapiens ring finger protein 14 (RNF14), transcript variant 2, mRNA
NMJ 83399 Homo sapiens ring finger protein 14 (RNF14), transcript variant 3, mRNA
NMJ 83400 Homo sapiens ring finger protein 14 (RNF14), transcript variant 4, mRNA
NMJ 83401 Homo sapiens ring finger protein 14 (RNF14), transcript variant 5, mRNA
NMJ 83404 Homo sapiens retinoblastoma-like 1 (p107) (RBL1), transcript variant 2, mR
NMJ 83412 Homo sapiens F-box protein 44 (FBX044), transcript variant 2, mRNA
NMJ 83413 Homo sapiens F-box protein 44 (FBX044), transcript variant 3, mRNA
NMJ 83414 Homo sapiens ubiquitin protein ligase E3B (UBE3B), transcript variant 2, mR
NMJ 83415 Homo sapiens ubiquitin protein ligase E3B (UBE3B), transcript variant 3, mR
NMJ 83416 Homo sapiens kinesin family member 1B (KIF1B), transcript variant 2, mRN/
NMJ 83418 Homo sapiens molybdenum cofactor synthesis 2 (MOCS2), transcript variant
NMJ 83419 Homo sapiens ring finger protein 19 (RNF19), transcript variant 1 , mRNA
NMJ 83420 Homo sapiens F-box protein 25 (FBX025), transcript variant 2, mRNA
NMJ 83421 Homo sapiens F-box protein 25 (FBX025), transcript variant 1 , mRNA
NMJ 83422 Homo sapiens transforming growth factor beta 1 induced transcript 4 (TGFB'
NMJ 83425 Homo sapiens RNA-binding region (RNP1, RRM) containing 1 (RNPC1), trar
NMJ 84041 Homo sapiens aldolase A, fructose-bisphosphate (ALDOA), transcript variant
NMJ 84042 Homo sapiens Cohen syndrome 1 (COH1), transcript variant 2, mRNA
NMJ 84043 Homo sapiens aldolase A, fructose-bisphosphate (ALDOA), transcript variant
NMJ 84085 Homo sapiens ring finger protein 29 (RNF29), transcript variant 1 , mRNA
NMJ 84086 Homo sapiens ring finger protein 29 (RNF29), transcript variant 3, mRNA
NMJ 84087 Homo sapiens ring finger protein 29 (RNF29), transcript variant 4, mRNA
NMJ 84231 Homo sapiens NCK interacting protein with SH3 domain (NCKIPSD), transcr
NMJ 84234 Homo sapiens RNA-binding region (RNP1 , RRM) containing 2 (RNPC2), trar
NMJ 84237 Homo sapiens RNA-binding region (RNP1 , RRM) containing 2 (RNPC2), trar
NMJ 84241 Homo sapiens RNA-binding region (RNP1 , RRM) containing 2 (RNPC2), trar
NMJ 84244 Homo sapiens RNA-binding region (RNP1 , RRM) containing 2 (RNPC2), trar
NMJ 87841 Homo sapiens ring finger protein 30 (RNF30), transcript variant 2, mRNA
NMJ 94071 Homo sapiens cAMP responsive element binding protein 3-like 2 (CREB3L2)
NMJ 94072 Homo sapiens spermatid-specific linker histone H1-like protein (HILS1), mRI1
NMJ 94247 Homo sapiens heterogeneous nuclear ribonucleoprotein A3 (HNRPA3), mRt>
NMJ 94248 Homo sapiens otoferlin (OTOF), transcript variant 1 , mRNA
NMJ 94249 Homo sapiens dead end homolog 1 (zebrafish) (DND1), mRNA
NMJ 94250 Homo sapiens similar to C630007C17Rik protein (LOC91752), mRNA
NMJ 94251 Homo sapiens G protein-coupled receptor 151 (GPR151), mRNA
N J 94252 Homo sapiens chromosome 9 open reading frame 20 (C9orf20), mRNA
NMJ 94255 Homo sapiens solute carrier family 19 (folate transporter), member 1 (SLC19
NMJ 94259 Homo sapiens ubiquitin-conjugating enzyme E2I (UBC9 homolog, yeast) (UE
NMJ 94260 Homo sapiens ubiquitin-conjugating enzyme E2I (UBC9 homolog, yeast) (UE
NMJ 94261 Homo sapiens ubiquitin-conjugating enzyme E2I (UBC9 homolog, yeast) (UE
N J 94270 Homo sapiens protein containing single MORN motif in testis (MOPT), mRN/
NMJ 94271 Homo sapiens ring finger protein 34 (RNF34), transcript variant 1 , mRNA
NMJ 94276 Homo sapiens hypothetical protein FLJ20209 (FLJ20209), mRNA
NMJ 94277 Homo sapiens hypothetical protein LOC90167 (LOC90167), mRNA
NMJ 94278 Homo sapiens chromosome 14 open reading frame 43 (C14orf43), mRNA
NMJ 94279 Homo sapiens HESB like domain containing 1 (HBLD1), mRNA
NMJ 94281 Homo sapiens chromosome 18 open reading frame 37 (C18orf37), mRNA
NMJ94282 Homo sapiens hypothetical protein DKFZp686L1814 (DKFZp686L1814), mR
NMJ 94283 Homo sapiens hypothetical protein LOC134218 (LOC134218), mRNA
NMJ 94284 Homo sapiens claudin 23 (CLDN23), mRNA
NMJ 94285 Homo sapiens hypothetical protein FLJ39441 (FLJ39441), mRNA
NMJ 94286 Homo sapiens KIAA1853 protein (KIAA1853), mRNA
NMJ94287 Homo sapiens chromosome 14 open reading frame 166B (C14orf166B), mR
NMJ 94288 Homo sapiens hypothetical protein LOC146712 (LOC146712), mRNA
NMJ 94289 Homo sapiens hypothetical protein LOC152195 (LOC152195), mRNA
NMJ94290 Homo sapiens hypothetical protein LOC153684 (LOC153684), mRNA
NMJ 94291 Homo sapiens hypothetical protein BC017881 (LOC157378), mRNA
NMJ 94292 Homo sapiens hypothetical protein DKFZp761A078 (DKFZp761A078), mRN;
NMJ 94293 Homo sapiens cardiomyopathy associated 1 (CMYA1), mRNA
NMJ 94294 Homo sapiens hypothetical protein LOC169355 (LOC169355), mRNA
NMJ 94295 Homo sapiens hypothetical protein DKFZp434H020 (DKFZp434H020), mRN
NMJ 94298 Homo sapiens solute carrier family 16 (monocarboxylic acid transporters), mi
NMJ 94299 Homo sapiens hypothetical protein LOC221711 (LOC221711), mRNA
NMJ 94300 Homo sapiens hypothetical protein LOC223075 (LOC223075), mRNA
NMJ 94302 Homo sapiens hypothetical protein DKFZp434O0527 (DKFZp434O0527), mF
NMJ94303 Homo sapiens chromosome 10 open reading frame 39 (C10orf39), mRNA
NMJ94309 Homo sapiens chromosome 21 open reading frame 125 (C21orf125), mRNA
NMJ 94310 Homo sapiens hypothetical protein LOC284837 (LOC284837), mRNA
NMJ 94312 Homo sapiens hypothetical protein LOC339768 (LOC339768), mRNA
NMJ 94313 Homo sapiens chromosome 9 open reading frame 48 (C9orf48), mRNA
NMJ94314 Homo sapiens FRBZ1 protein (FRBZ1), mRNA
NMJ 94315 Homo sapiens ubiquitin-conjugating enzyme E2, J2 (UBC6 homolog, yeast) (
NMJ 94316 Homo sapiens ubiquitin-conjugating enzyme E2, J2 (UBC6 homolog, yeast) |
NMJ 94317 Homo sapiens hypothetical protein MGC52057 (MGC52057), mRNA
NMJ 94318 Homo sapiens beta 3-glycosyltransferase-like (B3GTL), mRNA
NMJ 94319 Homo sapiens zinc finger protein 542 (ZNF542), mRNA
NMJ 94320 Homo sapiens zinc finger protein 169 (ZNF169), mRNA
NMJ 94322 Homo sapiens otoferlin (OTOF), transcript variant 3, mRNA
NMJ 94323 Homo sapiens otoferlin (OTOF), transcript variant 4, mRNA
NMJ 94324 Homo sapiens hypothetical protein MGC39900 (MGC39900), mRNA
NMJ 94325 Homo sapiens zinc finger protein 30 (KOX 28) (ZNF30), mRNA
NMJ 94326 Homo sapiens hypothetical protein MGC52010 (MGC52010), mRNA
NMJ 94327 Homo sapiens galectin-3 internal gene (GALIG), mRNA
NMJ 94328 Homo sapiens ring finger protein 38 (RNF38), transcript variant 2, mRNA
NMJ 94329 Homo sapiens ring finger protein 38 (RNF38), transcript variant 3, mRNA
NMJ 94330 Homo sapiens ring finger protein 38 (RNF38), transcript variant 5, mRNA
NMJ 94331 Homo sapiens ring finger protein 38 (RNF38), transcript variant 4, mRNA
NMJ 94332 Homo sapiens ring finger protein 38 (RNF38), transcript variant 6, mRNA
NMJ 94352 Homo sapiens ring finger protein 40 (RNF40), transcript variant 2, mRNA
NMJ 94356 Homo sapiens epimorphin (EPIM), transcript variant 2, mRNA
NMJ 94358 Homo sapiens ring finger protein 41 (RNF41), transcript variant 2, mRNA
NMJ 94359 Homo sapiens ring finger protein 41 (RNF41 ), transcript variant 3, mRNA
NMJ 94428 Homo sapiens DEAH (Asp-Glu-Ala-His) box polypeptide 34 (DHX34), transcr
N J 94429 Homo sapiens FGFR1 oncogene partner (FGFR1 OP), transcript variant 2, m
NMJ 94430 Homo sapiens ribonuclease, RNase A family, 4 (RNASE4), transcript variant
NMJ 94431 Homo sapiens ribonuclease, RNase A family, 4 (RNASE4), transcript variant
NMJ 94434 Homo sapiens VAMP (vesicle-associated membrane protein)-associated pro
NMJ 94435 Homo sapiens vasoactive intestinal peptide (VIP), transcript variant 2, mRNA
NMJ 94436 Homo sapiens lactate dehydrogenase D (LDHD), nuclear gene encoding miti
NMJ 94439 Homo sapiens hypothetical protein LOC285498 (LOC285498), mRNA
NMJ 94441 Homo sapiens butyrophilin, subfamily 3, member A1 (BTN3A1), transcript va
NM 94442 Homo sapiens lamin B receptor (LBR), transcript variant 2, mRNA
NM 94447 Homo sapiens C-type (calcium dependent, carbohydrate-recognition domain
NM 94448 Homo sapiens C-type (calcium dependent, carbohydrate-recognition domain
NM 94449 Homo sapiens pleckstrin homology domain containing, family E (with leucine
NM 94450 Homo sapiens C-type (calcium dependent, carbohydrate-recognition domain
NM 94451 Homo sapiens lipoic acid synthetase (LIAS), nuclear gene encoding mitocho
NMJ 94452 Homo sapiens ring finger protein 121 (RNF121 ), transcript variant 2, mRNA
NMJ 94453 Homo sapiens ring finger protein 121 (RNF121), transcript variant 3, mRNA
NMJ 94454 Homo sapiens cerebral cavernous malformations 1 (CCM1), transcript varian
NMJ 94455 Homo sapiens cerebral cavernous malformations 1 (CCM1), transcript varian
NMJ 94456 Homo sapiens cerebral cavernous malformations 1 (CCM1), transcript varian
NMJ 94457 Homo sapiens ubiquitin-conjugating enzyme E2, J2 (UBC6 homolog, yeast) |
NMJ 94458 Homo sapiens ubiquitin-conjugating enzyme E2, J2 (UBC6 homolog, yeast) |
NMJ 94460 Homo sapiens ring finger protein 126 (RNF126), transcript variant 2, mRNA
NMJ 94463 Homo sapiens ring finger protein 128 (RNF128), transcript variant 1 , mRNA
NMJ 97939 Homo sapiens ring finger protein 135 (RNF135), transcript variant 2, mRNA
NMJ 97941 Homo sapiens similar to ADAMTS-10 precursor (A disintegrin and metalloprc
NMJ 97947 Homo sapiens C-type (calcium dependent, carbohydrate-recognition domain
NMJ 97948 Homo sapiens C-type (calcium dependent, carbohydrate-recognition domain
NMJ 97949 Homo sapiens C-type (calcium dependent, carbohydrate-recognition domain
NM 97950 Homo sapiens C-type (calcium dependent, carbohydrate-recognition domain
NMJ 97951 Homo sapiens C-type (calcium dependent, carbohydrate-recognition domain
NMJ 97952 Homo sapiens C-type (calcium dependent, carbohydrate-recognition domain
NMJ 97953 Homo sapiens C-type (calcium dependent, carbohydrate-recognition domain
NMJ 97954 Homo sapiens C-type (calcium dependent, carbohydrate-recognition domain
NM 97955 Homo sapiens normal mucosa of esophagus specific 1 (NMES1 ), transcript \
NMJ 97956 Homo sapiens chromosome 9 open reading frame 90 (C9orf90), mRNA
NMJ 97957 Homo sapiens MAX protein (MAX), transcript variant 6, mRNA
NMJ 97958 Homo sapiens acheron (FLJ11196), transcript variant 2, mRNA
NMJ 97960 Homo sapiens dipeptidylpeptidase 8 (DPP8), transcript variant 3, mRNA
NMJ 97961 Homo sapiens dipeptidylpeptidase 8 (DPP8), transcript variant 4, mRNA
NMJ 97962 Homo sapiens glutaredoxin 2 (GLRX2), transcript variant 2, mRNA
NMJ 97964 Homo sapiens hypothetical protein HSPC268 (HSPC268), mRNA
NM 97965 Homo sapiens sodium-dependent organic anion transporter (SOAT), mRNA
NM 97966 Homo sapiens BH3 interacting domain death agonist (BID), transcript variant
NMJ 97967 Homo sapiens BH3 interacting domain death agonist (BID), transcript variant
NMJ 97968 Homo sapiens zinc finger protein 198 (ZNF198), mRNA
NM 97970 Homo sapiens bol, boule-like (Drosophila) (BOLL), transcript variant 1 , mRNj
NMJ 97972 Homo sapiens non-metastatic cells 7, protein expressed in (nucleoside-diphc
NMJ 97973 Homo sapiens asparagine-linked glycosylation 2 homolog (yeast, alpha-1 ,3-r
NM 97974 Homo sapiens butyrophilin, subfamily 3, member A3 (BTN3A3), transcript va
NMJ 97975 Homo sapiens butyrophilin-like 3 (BTNL3), transcript variant 1 , mRNA
NMJ 97976 Homo sapiens PBX/knotted 1 homeobox 1 (PKNOX1 ), transcript variant 2, m
NMJ 97977 Homo sapiens zinc finger protein 189 (ZNF189), mRNA
NMJ 97978 Homo sapiens hemogen (HEMGN), transcript variant 2, mRNA
N J 98038 Homo sapiens nudix (nucleoside diphosphate linked moiety X)-type motif 9 (I
NMJ 98039 Homo sapiens nudix (nucleoside diphosphate linked moiety X)-type motif 9 (I
NM 98040 Homo sapiens polyhomeotic-like 2 (Drosophila) (PHC2), transcript variant 1 ,
NMJ 98041 Homo sapiens nudix (nucleoside diphosphate linked moiety X)-type motif 6 (I
NMJ 98042 Homo sapiens PDZ and LIM domain 2 (mystique) (PDLIM2), transcript variar
NMJ 98043 Homo sapiens zinc finger, DHHC domain containing 16 (ZDHHC16), transcri
NMJ 98044 Homo sapiens zinc finger, DHHC domain containing 16 (ZDHHC16), transcri
NMJ 98045 Homo sapiens zinc finger, DHHC domain containing 16 (ZDHHC16), transcri
NMJ 98046 Homo sapiens zinc finger, DHHC domain containing 16 (ZDHHC16), transcri
NMJ 98047 Homo sapiens 3-hydroxyisobutyryl-Coenzyme A hydrolase (HIBCH), transcrii
N J 98053 Homo sapiens CD3Z antigen, zeta polypeptide (TiT3 complex) (CD3Z), trans
N J 98055 Homo sapiens zinc finger protein 42 (myeloid-specific retinoic acid-responsiv
NMJ 98056 Homo sapiens sodium channel, voltage-gated, type V, alpha (long QT syndrc
NMJ 98057 Homo sapiens delta sleep inducing peptide, immunoreactor (DSIPI), transcrii
NMJ 98058 Homo sapiens zinc finger protein 266 (ZNF266), mRNA
NMJ 98060 Homo sapiens nebulin-related anchoring protein (NRAP), transcript variant 2
NMJ 98061 Homo sapiens carboxylesterase 2 (intestine, liver) (CES2), transcript variant
NMJ 98066 Homo sapiens glucosamine-phosphate N-acetyltransferase 1 (GNPNAT1), rr
NMJ 98074 Homo sapiens olfactory receptor, family 2, subfamily C, member 3 (OR2C3),
NMJ 98075 Homo sapiens hypothetical protein DKFZp761L1518 (DKFZp761 L1518), mR
NMJ 98076 Homo sapiens family with sequence similarity 36, member A (FAM36A), mRt
NMJ98077 Homo sapiens gm117 (gm117), mRNA
NMJ 98078 Homo sapiens chromosome 21 open reading frame 121 (C21orf121), mRNA
NMJ 98079 Homo sapiens similar to golgi autoantigen, golgin subfamily a (FLJ40113), m
NMJ 98080 Homo sapiens hypothetical protein LOC253827 (LOC253827), mRNA
NMJ 98081 Homo sapiens sex comb on midleg-like 4 (Drosophila) (SCML4), mRNA
NMJ 98082 Homo sapiens hypothetical protein LOC284001 (LOC284001), mRNA
NMJ 98083 Homo sapiens dehydrogenase/reductase (SDR family) member 4 like 2 (DHF
NMJ 98085 Homo sapiens ring finger protein 148 (RNF148), mRNA
NMJ 98086 Homo sapiens jub, ajuba homolog (Xenopus laevis) (JUB), transcript variant
NMJ 98087 Homo sapiens zinc finger protein 200 (ZNF200), mRNA
NMJ 98088 Homo sapiens zinc finger protein 200 (ZNF200), mRNA
NMJ 98089 Homo sapiens zinc finger protein 155 (pHZ-96) (ZNF155), transcript variant .
NMJ 98097 Homo sapiens chromosome 7 open reading frame 28B (C7orf28B), mRNA
NMJ 98098 Homo sapiens aquaporin 1 (channel-forming integral protein, 28kDa) (AQP1
NMJ 98120 Homo sapiens estrogen receptor binding site associated, antigen, 9 (EBAG9
NMJ 98123 Homo sapiens CUB and Sushi multiple domains 3 (CSMD3), transcript variai
NMJ 98124 Homo sapiens CUB and Sushi multiple domains 3 (CSMD3), transcript variai
NMJ 98125 Homo sapiens TYRO protein tyrosine kinase binding protein (TYROBP), tran
NMJ98128 Homo sapiens ring finger protein 138 (RNF138), transcript variant 2, mRNA
NMJ 98129 Homo sapiens laminin, alpha 3 (LAMA3), transcript variant 1 , mRNA
NMJ 98138 Homo sapiens SEC31-like 2 (S. cerevisiae) (SEC31L2), transcript variant 2, i
NMJ98139 Homo sapiens semenogelin I (SEMG1), transcript variant 2, mRNA
NMJ 98141 Homo sapiens glucosidase, alpha; neutral C (GANC), mRNA
NMJ98147 Homo sapiens hypothetical protein LOC116236 (LOC116236), mRNA
NMJ98148 Homo sapiens carboxypeptidase X (M14 family), member 2 (CPXM2), mRNA
NMJ 98149 Homo sapiens chromosome 1 open reading frame 40 (C1orf40), mRNA
NMJ98150 Homo sapiens hypothetical protein DKFZp313G1735 (DKFZp313G1735), mF
NMJ 98151 Homo sapiens hypothetical protein LOC253012 (LOC253012), mRNA
NMJ 98152 Homo sapiens urotensin ll-related peptide (URP), mRNA
NMJ 98153 Homo sapiens triggering receptor expressed on myeloid cells-like 4 (TREMU
NMJ98154 Homo sapiens hypothetical protein LOC339168 (LOC339168), mRNA
NMJ98155 Homo sapiens chromosome 21 open reading frame 33 (C21orf33), nuclear g
NMJ 98156 Homo sapiens von Hippel-Lindau syndrome (VHL), transcript variant 2, mRN
NMJ 98157 Homo sapiens ubiquitin-conjugating enzyme E2L 3 (UBE2L3), transcript varii
NMJ 98158 Homo sapiens microphthalmia-associated transcription factor (MITF), transci
NMJ 98159 Homo sapiens microphthalmia-associated transcription factor (MITF), transcr
NMJ 98173 Homo sapiens transcription factor CP2-like 4 (TFCP2L4), transcript variant 2
NMJ98174 Homo sapiens transcription factor CP2-like 4 (TFCP2L4), transcript variant 3
NMJ 98175 Homo sapiens non-metastatic cells 1 , protein (NM23A) expressed in (NME1)
NMJ 98177 Homo sapiens microphthalmia-associated transcription factor (MITF), transcr
NMJ 98178 Homo sapiens microphthalmia-associated transcription factor (MITF), transcr
NMJ98179 Homo sapiens G protein-coupled receptor 103 (GPR103), mRNA
NMJ98180 Homo sapiens P518 precursor protein (P518), mRNA
NMJ98181 Homo sapiens similar to golgi autoantigen, golgin subfamily a, 2; SY11 prate
NMJ 98182 Homo sapiens transcription factor CP2-like 2 (TFCP2L2), transcript variant 2
NMJ 98183 Homo sapiens ubiquitin-conjugating enzyme E2L 6 (UBE2L6), transcript varii
NMJ 98184 Homo sapiens osteocrin (OSTN), mRNA
NMJ 98185 Homo sapiens oviductin protease (OVTN), mRNA
NMJ 98186 Homo sapiens astrotactin 2 (ASTN2), transcript variant 2, mRNA
NMJ 98187 Homo sapiens astrotactin 2 (ASTN2), transcript variant 3, mRNA
NMJ 98188 Homo sapiens astrotactin 2 (ASTN2), transcript variant 4, mRNA
NMJ 98189 Homo sapiens COP9 constitutive photomorphogenic homolog subunit 8 (Ara
NMJ 98194 Homo sapiens stomatin (STOM), transcript variant 2, mRNA
NMJ 98195 Homo sapiens ubiquitin-activating enzyme E1C (UBA3 homolog, yeast) (UBE
NMJ 98196 Homo sapiens CD96 antigen (CD96), transcript variant 1, mRNA
NMJ 98197 Homo sapiens ubiquitin-activating enzyme E1C (UBA3 homolog, yeast) (UBE
NMJ 98201 Homo sapiens processing of precursor 5, ribonuclease P/MRP subunit (S. ce
NMJ 98202 Homo sapiens processing of precursor 5, ribonuclease P/MRP subunit (S. ce
NMJ 98204 Homo sapiens transcription factor-like 4 (TCFL4), transcript variant 2, mRNA
NMJ 98205 Homo sapiens transcription factor-like 4 (TCFL4), transcript variant 1 , mRNA
NMJ98207 Homo sapiens LAG1 longevity assurance homolog 1 (S. cerevisiae) (LASS1]
NMJ 98212 Homo sapiens caveolin 2 (CAV2), transcript variant 2, mRNA
NMJ 98213 Homo sapiens 2'-5'-oligoadenylate synthetase-like (OASL), transcript variant
NMJ 98215 Homo sapiens family with sequence similarity 13, member C1 (FAM13C1), tr
NMJ 98216 Homo sapiens small nuclear ribonucleoprotein polypeptides B and B1 (SNRF
NMJ 98217 Homo sapiens inhibitor of growth family, member 1 (ING1 ), transcript variant
NMJ 98218 Homo sapiens inhibitor of growth family, member 1 (ING1), transcript variant
NMJ 98219 Homo sapiens inhibitor of growth family, member 1 (ING1), transcript variant
NMJ 98220 Homo sapiens small nuclear ribonucleoprotein polypeptide B" (SNRPB2), tra
NMJ 98225 Homo sapiens Rho-related BTB domain containing 1 (RHOBTB1), transcript
NMJ 98227 Homo sapiens regulator of G-protein signalling 12 (RGS12), transcript varian
NMJ 98229 Homo sapiens regulator of G-protein signalling 12 (RGS12), transcript varian
NMJ98230 Homo sapiens regulator of G-protein signalling 12 (RGS12), transcript varian
NMJ 98232 Homo sapiens ribonuclease, RNase A family, 1 (pancreatic) (RNASE1), trans
NMJ 98234 Homo sapiens ribonuclease, RNase A family, 1 (pancreatic) (RNASE1), trans
NMJ 98235 Homo sapiens ribonuclease, RNase A family, 1 (pancreatic) (RNASE1), trans
NMJ 98236 Homo sapiens Rho guanine nucleotide exchange factor (GEF) 11 (ARHGEF'
NMJ 98239 Homo sapiens WNT1 inducible signaling pathway protein 3 (WISP3), transcr
NMJ 98240 Homo sapiens restin (Reed-Steinberg cell-expressed intermediate filament-a
NMJ 98241 Homo sapiens eukaryotic translation initiation factor 4 gamma, 1 (EIF4G1), tr
NMJ 98242 Homo sapiens eukaryotic translation initiation factor 4 gamma, 1 (EIF4G1), ti
NMJ 98243 Homo sapiens ankyrin repeat and SOCS box-containing 7 (ASB7), transcript
NMJ 98244 Homo sapiens eukaryotic translation initiation factor 4 gamma, 1 (EIF4G1 ), ti
NMJ 98252 Homo sapiens gelsolin (amyloidosis, Finnish type) (GSN), transcript variant 2
NMJ 98253 Homo sapiens telomerase reverse transcriptase (TERT), transcript variant 3,
NMJ 98254 Homo sapiens telomerase reverse transcriptase (TERT), transcript variant 4,
NMJ 98255 Homo sapiens telomerase reverse transcriptase (TERT), transcript variant 2,
NMJ 98256 Homo sapiens E2F transcription factor 6 (E2F6), transcript variant b, mRNA
NMJ 98257 Homo sapiens E2F transcription factor 6 (E2F6), transcript variant e, mRNA
NMJ 98258 Homo sapiens E2F transcription factor 6 (E2F6), transcript variant 4, mRNA
NMJ 98261 Homo sapiens similar to splicing factor, arginine/serine-rich 4 (FLJ11021), trε
NMJ98262 Homo sapiens similar to splicing factor, arginine/serine-rich 4 (FLJ11021), trε
NMJ98263 Homo sapiens similar to splicing factor, arginine/serine-rich 4 (FLJ11021), trε
NMJ 98264 Homo sapiens chromosome 1 open reading frame 2 (C1orf2), transcript varis
NMJ 98265 Homo sapiens SP011 meiotic protein covalently bound to DSB-like (S. cerev
NMJ 98266 Homo sapiens inhibitor of growth family, member 3 (ING3), transcript variant
NMJ 98267 Homo sapiens inhibitor of growth family, member 3 (ING3), transcript variant
NMJ 98268 Homo sapiens homeodomain interacting protein kinase 1 (HIPK1), transcript
NMJ 98269 Homo sapiens homeodomain interacting protein kinase 1 (HIPK1), transcript
NMJ 98270 Homo sapiens Nance-Horan syndrome (congenital cataracts and dental anoi
NMJ 98271 Homo sapiens leiomodin 3 (fetal) (LMOD3), mRNA
NMJ 98274 Homo sapiens SET and MYND domain containing 1 (SMYD1), mRNA
NMJ 98275 Homo sapiens hypothetical protein LOC196264 (LOC196264), mRNA
NMJ 98276 Homo sapiens transmembrane protein 17 (TMEM17), mRNA
NMJ 98277 Homo sapiens solute carrier family 37 (glycerol-3-phosphate transporter), me
NMJ 98278 Homo sapiens hypothetical protein LOC255743 (LOC255743), mRNA
NMJ 98279 Homo sapiens chromosome X open reading frame 23 (CXorf23), mRNA
NMJ 98281 Homo sapiens hypothetical protein LOC285513 (LOC285513), mRNA
NMJ 98282 Homo sapiens hypothetical protein LOC340061 (LOC340061), mRNA
NMJ 98283 Homo sapiens EGF-like-domain, multiple 11 (EGFL11 ), mRNA
NMJ98284 Homo sapiens hypothetical protein LOC349114 (LOC349114), mRNA
NMJ 98285 Homo sapiens hypothetical protein LOC349136 (LOC349136), mRNA
NMJ 98287 Homo sapiens inhibitor of growth family, member 4 (ING4), transcript variant
NMJ 98289 Homo sapiens cell death-inducing DFFA-like effector a (CIDEA), transcript vs
NMJ 98291 Homo sapiens v-src sarcoma (Schmidt-Ruppin A-2) viral oncogene homolog
NMJ 98309 Homo sapiens tetratricopeptide repeat domain 8 (TTC8), transcript variant 1 ,
NMJ 98310 Homo sapiens tetratricopeptide repeat domain 8 (TTC8), transcript variant 2,
NMJ 98312 Homo sapiens TAK1 -binding protein 3 (TAB3), transcript variant 2, mRNA
NMJ 98315 Homo sapiens loss of heterozygosity, 11 , chromosomal region 2, gene A (LC
NMJ 98316 Homo sapiens tensin like C1 domain containing phosphatase (TENC1), trans
NMJ 98317 Homo sapiens kelch-like 17 (Drosophila) (KLHL17), mRNA
NMJ98318 Homo sapiens HMT1 hnRNP methyltransferase-like 2 (S. cerevisiae) (HRMT
NMJ 98319 Homo sapiens HMT1 hnRNP methyltransferase-like 2 (S. cerevisiae) (HRMT
NMJ 98320 Homo sapiens carboxypeptidase M (CPM), transcript variant 2, mRNA
NMJ 98321 Homo sapiens UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylga
NMJ 98324 Homo sapiens citrate synthase (CS), nuclear gene encoding mitochondrial pi
NMJ 98325 Homo sapiens E2F transcription factor 6 (E2F6), transcript variant c, mRNA
NMJ 98327 Homo sapiens suppression of tumorigenicity 7 like (ST7L), transcript variant
NMJ 98328 Homo sapiens suppression of tumorigenicity 7 like (ST7L), transcript variant
NMJ 98329 Homo sapiens ubiquitin-activating enzyme E1-domain containing 1 (UBE1 DC
NMJ 98330 Homo sapiens inositol polyphosphate-5-phosphatase F (INPP5F), transcript
NMJ98331 Homo sapiens inositol polyphosphate-5-phosphatase F (INPP5F), transcript
NMJ 98333 Homo sapiens purinergic receptor P2Y, G-protein coupled, 10 (P2RY10), trai
NMJ 98334 Homo sapiens glucosidase, alpha; neutral AB (GANAB), mRNA
NMJ 98335 Homo sapiens glucosidase, alpha; neutral AB (GANAB), mRNA
NMJ 98336 Homo sapiens insulin induced gene 1 (INSIG1), transcript variant 2, mRNA
NMJ 98337 Homo sapiens insulin induced gene 1 (INSIG1), transcript variant 3, mRNA
NMJ 98353 Homo sapiens potassium channel tetramerisation domain containing 8 (KCT
NMJ 98376 Homo sapiens calcium channel, voltage-dependent, alpha 1G subunit (CACf
NMJ 98377 Homo sapiens calcium channel, voltage-dependent, alpha 1G subunit (CACf
NMJ 98378 Homo sapiens calcium channel, voltage-dependent, alpha 1G subunit (CAd*
NMJ 98379 Homo sapiens calcium channel, voltage-dependent, alpha 1G subunit (CACf
NMJ 98380 Homo sapiens calcium channel, voltage-dependent, alpha 1G subunit (CACC
NMJ 98381 Homo sapiens E74-like factor 5 (ets domain transcription factor) (ELF5), tran
NMJ98382 Homo sapiens calcium channel, voltage-dependent, alpha 1 G subunit (CACt
NMJ 98383 Homo sapiens calcium channel, voltage-dependent, alpha 1G subunit (CACC
NMJ 98384 Homo sapiens calcium channel, voltage-dependent, alpha 1G subunit (CACC
NMJ 98385 Homo sapiens calcium channel, voltage-dependent, alpha 1G subunit (CACI1
NMJ 98386 Homo sapiens calcium channel, voltage-dependent, alpha 1G subunit (CACf
NMJ 98387 Homo sapiens calcium channel, voltage-dependent, alpha 1 G subunit (CACF
NMJ 98388 Homo sapiens calcium channel, voltage-dependent, alpha 1 G subunit (CACI1
NMJ98389 Homo sapiens lung type-l cell membrane-associated glycoprotein (T1A-2), tr
NMJ 98390 Homo sapiens c-Maf-inducing protein (CMIP), transcript variant C-mip, mRN;
NMJ 98391 Homo sapiens fibronectin leucine rich transmembrane protein 3 (FLRT3), tra
NMJ98392 Homo sapiens transcription factor 21 (TCF21), transcript variant 1 , mRNA
NMJ98393 Homo sapiens testis expressed sequence 14 (TEX14), transcript variant 1, rr
NMJ 98394 Homo sapiens chromosome 9 open reading frame 85 (C9orf85), transcript vs
NMJ 98395 Homo sapiens Ras-GTPase-activating protein SH3-domain-binding protein (<
NMJ 98396 Homo sapiens calcium channel, voltage-dependent, alpha 1G subunit (CACI'
NMJ 98397 Homo sapiens calcium channel, voltage-dependent, alpha 1G subunit (CACI'
NMJ 98398 Homo sapiens serologically defined breast cancer antigen 84 (SDBCAG84),
NMJ 98399 Homo sapiens cyclic AMP-regulated phosphoprotein, 21 kD (ARPP-21), tran;
NMJ98400 Homo sapiens neural precursor cell expressed, developmentally down-reguls
NMJ98401 Homo sapiens hypothetical protein LOC157567 (LOC157567), mRNA
NMJ 98402 Homo sapiens protein tyrosine phosphatase-like (proline instead of catalytic .
NMJ 98403 Homo sapiens monocyte to macrophage differentiation-associated 2 (MMD2
NMJ 98404 Homo sapiens potassium channel tetramerisation domain containing 4 (KCT
NMJ98406 Homo sapiens progestin and adipoQ receptor family member VI (PAQR6), tn
NMJ98407 Homo sapiens growth hormone secretagogue receptor (GHSR), transcript va
NMJ 98426 Homo sapiens putative breast adenocarcinoma marker (32kD) (BC-2), transc
NMJ 98427 Homo sapiens brevican (BCAN), transcript variant 2, mRNA
NMJ 98428 Homo sapiens parathyroid hormone-responsive B1 gene (B1), transcript vari
NMJ98430 Homo sapiens regulator of G-protein signalling 12 (RGS12), transcript varian
NMJ 98431 Homo sapiens heat shock 70kDa protein 4 (HSPA4), transcript variant 2, mR
NMJ 98432 Homo sapiens regulator of G-protein signalling 12 (RGS12), transcript varian
NMJ 98433 Homo sapiens serine/threonine kinase 6 (STK6), transcript variant 1 , mRNA
NMJ 98434 Homo sapiens serine/threonine kinase 6 (STK6), transcript variant 3, mRNA
NMJ 98435 Homo sapiens serine/threonine kinase 6 (STK6), transcript variant 4, mRNA
NMJ 98436 Homo sapiens serine/threonine kinase 6 (STK6), transcript variant 5, mRNA
NMJ 98437 Homo sapiens serine/threonine kinase 6 (STK6), transcript variant 6, mRNA
NMJ 98439 Homo sapiens kelch repeat and BTB (POZ) domain containing 3 (KBTBD3),
NMJ 98440 Homo sapiens chromosome 22 open reading frame 14 (C22orf14), transcript
NMJ 98441 Homo sapiens FLJ40296 protein (FLJ40296), mRNA
NMJ 98442 Homo sapiens FLJ45651 protein (FLJ45651), mRNA
NMJ 98443 Homo sapiens MRCC2446 (UNQ2446), mRNA
NMJ 98444 Homo sapiens GPAD9366 (UNQ9366), mRNA
NMJ 98445 Homo sapiens FLJ45909 protein (FLJ45909), mRNA
NMJ 98446 Homo sapiens FLJ45459 protein (FLJ45459), mRNA
NMJ 98447 Homo sapiens FLJ42654 protein (FLJ42654), mRNA
NMJ98448 Homo sapiens LPPM429 (UNQ429), mRNA
NMJ 98449 Homo sapiens similar to embigin (MGC71 45), mRNA
NMJ 98450 Homo sapiens AAIR8193 (UNQ8193), mRNA
NMJ 98451 Homo sapiens forkhead box R2 (FOXR2), mRNA
NMJ98452 Homo sapiens pregnancy upregulated non-ubiquitously expressed CaM kina
NMJ 98457 Homo sapiens zinc finger protein 600 (ZNF600), mRNA
NMJ 98458 Homo sapiens zinc finger protein 497 (ZNF497), mRNA
NMJ 98459 Homo sapiens FLJ37099 protein (FLJ37099), mRNA
NMJ 98460 Homo sapiens hypothetical protein DKFZp686G0786 (DKFZp686G0786), mF
NMJ 98461 Homo sapiens FLJ45273 protein (FLJ45273), mRNA
N J 98462 Homo sapiens FLJ46154 protein (FLJ46154), mRNA
NMJ 98463 Homo sapiens FLJ42117 protein (FLJ42117), mRNA
NMJ 98464 Homo sapiens tryptophan/serine protease (UNQ9391), mRNA
NMJ 98465 Homo sapiens Nik related kinase (NRK), mRNA
NM J98466 Homo sapiens FLJ37183 protein (FLJ37183), mRNA
NMJ 98467 Homo sapiens FLJ42526 protein (FLJ42526), mRNA
NMJ 98468 Homo sapiens chromosome 6 open reading frame 167 (C6orf167), mRNA
NMJ 98469 Homo sapiens chromosome 9 open reading frame 18 (C9orf18), mRNA
NMJ 98471 Homo sapiens FLJ46061 protein (FLJ46061), mRNA
NMJ 98472 Homo sapiens chromosome 10 open reading frame 125 (C1 Oorfl 25), mRNA
NMJ 98473 Homo sapiens FLJ46111 protein (FLJ46111), mRNA
NMJ 98474 Homo sapiens olfactomedin-like 1 (OLFML1), mRNA
NMJ 98476 Homo sapiens FLJ41131 protein (FLJ41131), mRNA
NMJ 98477 Homo sapiens DMC (UNQ473), mRNA
NMJ98478 Homo sapiens NTPase, KAP family P-loop domain containing 1 (NKPD1), ml
NMJ98479 Homo sapiens FLJ40321 protein (FLJ40321), mRNA
NMJ98480 Homo sapiens zinc finger protein 615 (ZNF615), mRNA
NMJ 98481 Homo sapiens LAIR hlog (UNQ3033), mRNA
NMJ 98482 Homo sapiens similar to B-cell linker; B cell linker protein (LOC284948), tram
NMJ 98483 Homo sapiens FLJ46536 protein (FLJ46536), mRNA
NMJ 98484 Homo sapiens zinc finger protein 621 (ZNF621 ), mRNA
NMJ 98485 Homo sapiens FLJ41238 protein (FLJ41238), mRNA
NMJ 98486 Homo sapiens ribosomal protein L7-like 1 (RPL7L1 ), mRNA
NMJ 98488 Homo sapiens FLJ46072 protein (FLJ46072), mRNA
NMJ98489 Homo sapiens similar to DLNB14 (DLNB14), mRNA
NMJ 98490 Homo sapiens RAB43, member RAS oncogene family (RAB43), mRNA
NMJ 98491 Homo sapiens FLJ44299 protein (FLJ44299), mRNA
NMJ 98492 Homo sapiens liver and lymph node sinusoidal endothelial cell C-type lectin I
NMJ 98493 Homo sapiens FLJ45235 protein (FLJ45235), mRNA
NMJ 98494 Homo sapiens FLJ16030 protein (FLJ16030), mRNA
NMJ 98495 Homo sapiens CTAGE family, member 4 (CTAGE4), mRNA
NMJ 98496 Homo sapiens A-domain containing protein similar to matrilin and collagen (/
NMJ 98498 Homo sapiens Similar to RIKEN cDNA 1810046K07 gene (MGC50104), mRf
NMJ 98499 Homo sapiens FLJ46156 protein (FLJ46156), mRNA
NMJ 98501 Homo sapiens FLJ42461 protein (FLJ42461 ), mRNA
NMJ 98502 Homo sapiens FLJ43826 protein (FLJ43826), mRNA
NMJ 98503 Homo sapiens sodium- and chloride-activated ATP-sensitive potassium char
NMJ 98504 Homo sapiens progestin and adipoQ receptor family member IX (PAQR9), m
NMJ 98506 Homo sapiens FLJ44691 protein (FLJ44691), mRNA
NMJ 98507 Homo sapiens HGS_RE408 (UNQ1912), mRNA
NMJ 98508 Homo sapiens FLJ44186 protein (FLJ44186), mRNA
NMJ 98510 Homo sapiens ITI-like protein (UNQ6369), mRNA
NMJ 98511 Homo sapiens FLJ42925 protein (FLJ42925), mRNA
NMJ 98512 Homo sapiens FLJ25989 protein (FLJ25989), mRNA
NMJ 98513 Homo sapiens CGI-72 protein (CGI-72), transcript variant 3, mRNA
NMJ 98514 Homo sapiens NHL repeat containing 2 (NHLRC2), mRNA
NMJ 98516 Homo sapiens UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylga
NMJ 98517 Homo sapiens FLJ00332 protein (FLJ00332), mRNA
NMJ 98519 Homo sapiens similar to Cytochrome c, somatic (MGC12965), mRNA
NMJ 98520 Homo sapiens FLJ44112 protein (FLJ44112), mRNA
NMJ 98521 Homo sapiens FLJ25323 protein (FLJ25323), mRNA
NMJ 98524 Homo sapiens LOC161577 protein (LOC161577), mRNA
NMJ 98525 Homo sapiens similar to kinesin family member 21 A; N-5 kinesin (LOC3746S
NMJ 98526 Homo sapiens hypothetical protein DKFZp547K1113 (DKFZp547K1113), mF
NMJ98527 Homo sapiens Similar to RIKEN cDNA 1110033009 gene (MGC45386), mRI
NMJ 98529 Homo sapiens FLJ46247 protein (FLJ46247), mRNA
NMJ98531 Homo sapiens ATPase, Class II, type 9B (ATP9B), mRNA
NMJ 98532 Homo sapiens chromosome 19 open reading frame 35 (C19orf35), mRNA
NMJ 98533 Homo sapiens short-chain dehydrogenase/reductase 10 (SCDR10), transcrif
NMJ 98534 Homo sapiens FLJ35784 protein (FLJ35784), mRNA
NMJ 98537 Homo sapiens FLJ44968 protein (FLJ44968), mRNA
NMJ 98538 Homo sapiens HLAR698 (UNQ698), mRNA
NMJ 98539 Homo sapiens zinc finger protein 568 (ZNF568), mRNA
NMJ 98540 Homo sapiens UDP-Gal:betaGal beta 1 ,3-galactosyltransferase polypeptide
NMJ 98541 Homo sapiens insulin growth factor-like family member 1 (IGFL1 ), mRNA
NMJ 98542 Homo sapiens similar to hypothetical protein FLJ23233 (MGC4728), mRNA
NMJ98543 Homo sapiens L0C148872 protein (LOC148872), mRNA
NMJ 98544 Homo sapiens cortistatin (CORT), transcript variant 1 , mRNA
NMJ 98545 Homo sapiens hypothetical gene supported by AK075558; BC021286 (LOC3
NMJ 98546 Homo sapiens hypothetical protein LOC374955 (LOC374955), mRNA
NMJ 98547 Homo sapiens FLJ46354 protein (FLJ46354), mRNA
NMJ 98549 Homo sapiens FLJ35093 protein (FLJ35093), mRNA
NMJ 98550 Homo sapiens FLJ36760 protein (FLJ36760), mRNA
NMJ 98552 Homo sapiens hypothetical gene supported by BC009447 (MGC15887), mRI
NMJ 98553 Homo sapiens FLJ30851 protein (FLJ30851), mRNA
NMJ 98554 Homo sapiens thyroid adenoma associated (THADA), mRNA
NMJ 98557 Homo sapiens FLJ45645 protein (FLJ45645), mRNA
NMJ 98559 Homo sapiens hypothetical gene supported by BC052750 (MGC50811 ), mRI
NMJ 98560 Homo sapiens lipoma HMGIC fusion partner-like protein 4 (LOC375323), mF
NMJ 98562 Homo sapiens FLJ43654 protein (FLJ43654), mRNA
NMJ 98563 Homo sapiens Similar to RIKEN cDNA 1810038N08 gene (MGC52022), mRI
NMJ 98564 Homo sapiens FLJ44290 protein (FLJ44290), mRNA
NMJ 98565 Homo sapiens ELLP3030 (UNQ3030), mRNA
NMJ 98566 Homo sapiens FLJ32363 protein (FLJ32363), mRNA
NMJ98567 Homo sapiens FLJ44216 protein (FLJ44216), mRNA
NMJ 98568 Homo sapiens gap junction protein, beta 7 (GJB7), mRNA
NMJ 98569 Homo sapiens G protein-coupled receptor 126 (GPR126), mRNA
NMJ 98570 Homo sapiens PSST739 (UNQ739), mRNA
NMJ 98571 Homo sapiens FLJ39237 protein (FLJ39237), mRNA
NMJ 98572 Homo sapiens similar to Putative protein C21orf56 (MGC61633), mRNA
NMJ 98573 Homo sapiens GAAI470 (UNQ470), mRNA
NMJ 98577 Homo sapiens FLJ46361 protein (FLJ46361), mRNA
NMJ 98580 Homo sapiens solute carrier family 27 (fatty acid transporter), member 1 (SLl
NMJ 98581 Homo sapiens zinc finger CCCH type domain containing 6 (ZC3HDC6), mRh
NMJ98582 Homo sapiens FLJ43374 protein (FLJ43374), mRNA
NMJ 98584 Homo sapiens carbonic anhydrase XIII (CA13), mRNA
NMJ 98585 Homo sapiens GLSR2492 (UNQ2492), mRNA
NMJ98586 Homo sapiens NHL repeat containing 1 (NHLRC1), mRNA
NMJ 98587 Homo sapiens regulator of G-protein signalling 12 (RGS12), transcript varian
NMJ 98589 Homo sapiens basigin (OK blood group) (BSG), transcript variant 2, mRNA
NMJ 98590 Homo sapiens basigin (OK blood group) (BSG), transcript variant 3, mRNA
NMJ 98591 Homo sapiens basigin (OK blood group) (BSG), transcript variant 4, mRNA
NMJ 98593 Homo sapiens C1q and tumor necrosis factor related protein 1 (C1QTNF1), t
NMJ 98594 Homo sapiens C1q and tumor necrosis factor related protein 1 (C1QTNF1), r
NMJ 98595 Homo sapiens actin filament associated protein (AFAP), transcript variant 2,
NMJ 98596 Homo sapiens sulfatase 2 (SULF2), transcript variant 2, mRNA
NMJ 98597 Homo sapiens SEC24 related gene family, member C (S. cerevisiae) (SEC2<
NMJ 98679 Homo sapiens Rap guanine nucleotide exchange factor (GEF) 1 (RAPGEF1)
NMJ 98681 Homo sapiens putative NFkB activating protein (KIAA0720), transcript varian
NMJ 98682 Homo sapiens glycophorin E (GYPE), transcript variant 2, mRNA
NMJ 98686 Homo sapiens RAB15, member RAS onocogene family (RAB15), mRNA
NMJ 98687 Homo sapiens keratin associated protein 10-4 (KRTAP10-4), mRNA
NMJ 98688 Homo sapiens keratin associated protein 10-6 (KRTAP10-6), mRNA
NMJ 98689 Homo sapiens keratin associated protein 10-7 (KRTAP10-7), mRNA
NMJ 98690 Homo sapiens keratin associated protein 10-9 (KRTAP10-9), mRNA
NMJ 98691 Homo sapiens keratin associated protein 10-1 (KRTAP10-1), mRNA
NMJ 98692 Homo sapiens keratin associated protein 10-11 (KRTAP10-11), mRNA
NMJ 98693 Homo sapiens keratin associated protein 10-2 (KRTAP10-2), mRNA
NMJ 98694 Homo sapiens keratin associated protein 10-5 (KRTAP10-5), mRNA
NMJ 98695 Homo sapiens keratin associated protein 10-8 (KRTAP10-8), mRNA
NMJ 98696 Homo sapiens keratin associated protein 10-3 (KRTAP10-3), mRNA
NMJ 98697 Homo sapiens keratin associated protein 12-3 (KRTAP12-3), mRNA
NMJ 98698 Homo sapiens keratin associated protein 12-4 (KRTAP12-4), mRNA
NMJ 98699 Homo sapiens keratin associated protein 10-12 (KRTAP10-12), mRNA
NMJ 98700 Homo sapiens CUG triplet repeat, RNA binding protein 1 (CUGBP1), transcri
NMJ 98704 Homo sapiens short-chain dehydrogenase/reductase 10 (SCDR10), transcrif
NMJ 98705 Homo sapiens short-chain dehydrogenase/reductase 10 (SCDR10), transcrif
NMJ 98706 Homo sapiens short-chain dehydrogenase/reductase 10 (SCDR10), transcrif
NMJ 98707 Homo sapiens short-chain dehydrogenase/reductase 10 (SCDR10), transcrif
NMJ 98708 Homo sapiens short-chain dehydrogenase/reductase 10 (SCDR10), transcrif
NMJ 98709 Homo sapiens arylsulfatase B (ARSB), transcript variant 2, mRNA
NMJ 98712 Homo sapiens prostaglandin E receptor 3 (subtype EP3) (PTGER3), transcrii
NMJ 98713 Homo sapiens prostaglandin E receptor 3 (subtype EP3) (PTGER3), transcrii
NMJ 98714 Homo sapiens prostaglandin E receptor 3 (subtype EP3) (PTGER3), transcrii
NMJ 98715 Homo sapiens prostaglandin E receptor 3 (subtype EP3) (PTGER3), transcrii
NMJ 98716 Homo sapiens prostaglandin E receptor 3 (subtype EP3) (PTGER3), transcrii
NMJ 98717 Homo sapiens prostaglandin E receptor 3 (subtype EP3) (PTGER3), transcrii
NMJ 98718 Homo sapiens prostaglandin E receptor 3 (subtype EP3) (PTGER3), transcrii
NMJ 98719 Homo sapiens prostaglandin E receptor 3 (subtype EP3) (PTGER3), transcrii
NMJ 98720 Homo sapiens prostaglandin E receptor 3 (subtype EP3) (PTGER3), transcrii
NMJ 98721 Homo sapiens collagen, type XXV, alpha 1 (COL25A1), transcript variant 1 , r
NMJ 98722 Homo sapiens amphoterin-induced gene and ORF 3 (AMIG03), mRNA
NMJ 98723 Homo sapiens transcription elongation factor A (Sll), 2 (TCEA2), transcript vs
NMJ 98793 Homo sapiens CD47 antigen (Rh-related antigen, integrin-associated signal '
NMJ 98794 Homo sapiens mitogen-activated protein kinase kinase kinase kinase 5 (MAF
NMJ 98795 Homo sapiens tudor domain containing 1 (TDRD1), mRNA
NMJ 98797 Homo sapiens prostaglandin E synthase (PTGES), transcript variant 2, mRN.
NMJ 98798 Homo sapiens ankyrin repeat domain 5 (ANKRD5), transcript variant 2, mRN
NMJ 98799 Homo sapiens breast carcinoma amplified sequence 4 (BCAS4), transcript v,
NMJ 98822 Homo sapiens ATP synthase, H+ transporting, mitochondrial F0 complex, su
NMJ 98827 Homo sapiens G protein-coupled receptor 133 (GPR133), mRNA
NMJ 98828 Homo sapiens similar to microtubule associated testis specific serine/threoni
NMJ 98829 Homo sapiens ras-related C3 botulinum toxin substrate 1 (rho family, small G
NMJ 98830 Homo sapiens ATP citrate lyase (ACLY), transcript variant 2, mRNA
NMJ 98833 Homo sapiens serine (or cysteine) proteinase inhibitor, clade B (ovalbumin),
NMJ 98834 Homo sapiens acetyl-Coenzyme A carboxylase alpha (ACACA), transcript va
NMJ 98835 Homo sapiens acetyl-Coenzyme A carboxylase alpha (ACACA), transcript va
NMJ 98836 Homo sapiens acetyl-Coenzyme A carboxylase alpha (ACACA), transcript va
NMJ 98837 Homo sapiens acetyl-Coenzyme A carboxylase alpha (ACACA), transcript va
NMJ 98838 Homo sapiens acetyl-Coenzyme A carboxylase alpha (ACACA), transcript va
NMJ 98839 Homo sapiens acetyl-Coenzyme A carboxylase alpha (ACACA), transcript va
NMJ 98841 Homo sapiens chromosome 9 open reading frame 10 opposite strand (C9orf
NMJ 98843 Homo sapiens surfactant, pulmonary-associated protein B (SFTPB), transcrii
NMJ 98844 Homo sapiens zona pellucida binding protein 2 (ZPBP2), transcript variant 1 ,
NMJ 98845 Homo sapiens sialic acid binding Ig-like iectin 6 (SIGLEC6), transcript varian
NMJ 98846 Homo sapiens sialic acid binding Ig-like Iectin 6 (SIGLEC6), transcript varian
N J 98847 Homo sapiens FLJ22794 protein (FLJ22794), transcript variant 2, mRNA
NMJ98849 Homo sapiens similar to seven in absentia 2 (LOC283514), mRNA
NMJ 98850 Homo sapiens pleckstrin homology-like domain, family B, member 3 (PHLDE
NMJ 98851 Homo sapiens hypothetical protein LOC348645 (LOC348645), mRNA
NMJ 98853 Homo sapiens tripartite motif-containing 50C (TRIM50C), mRNA
NMJ 98855 Homo sapiens zinc finger protein 211 (ZNF211), transcript variant 2, mRNA
NMJ 98856 Homo sapiens CAP-binding protein complex interacting protein 1 (FLJ23588;
NMJ 98857 Homo sapiens similar to sodium- and chloride-dependent creatine transporte
NMJ 98859 Homo sapiens prickle-like 2 (Drosophila) (PRICKLE2), mRNA
NMJ 98867 Homo sapiens hypothetical protein MGC15677 (MGC15677), transcript varia
NMJ 98868 Homo sapiens KIAA0676 protein (KIAA0676), transcript variant 1 , mRNA
NMJ 98880 Homo sapiens FLJ20259 protein (FLJ20259), transcript variant 2, mRNA
N J 98881 Homo sapiens FLJ20298 protein (FLJ20298), transcript variant 2, mRNA
NMJ 98883 Homo sapiens metaxin 1 (MTX1), transcript variant 2, mRNA
NMJ 98887 Homo sapiens nucleoporin 43kDa (NUP43), transcript variant 1 , mRNA
NMJ 98889 Homo sapiens ankyrin repeat domain 17 (ANKRD17), transcript variant 2, ml
NMJ 98890 Homo sapiens APG16 autophagy 16-like (S. cerevisiae) (APG16L), transcrip
N J 98892 Homo sapiens BMP2 inducible kinase (BMP2K), transcript variant 1 , mRNA
NMJ 98893 Homo sapiens zinc finger protein 160 (ZNF160), transcript variant 2, mRNA
NMJ 98896 Homo sapiens RAB6A, member RAS oncogene family (RAB6A), transcript vi
NMJ 98897 Homo sapiens fibroblast growth factor (acidic) intracellular binding protein (F
NMJ 98900 Homo sapiens formin-like 3 (FMNL3), transcript variant 2, mRNA
NMJ 98901 Homo sapiens sorcin (SRI), transcript variant 2, mRNA
NMJ 98902 Homo sapiens transmembrane 4 superfamily member 8 (TM4SF8), transcrip
NMJ 98903 Homo sapiens gamma-aminobutyric acid (GABA) A receptor, gamma 2 (GAE
N J 98904 Homo sapiens gamma-aminobutyric acid (GABA) A receptor, gamma 2 (GAE
NMJ 98907 Homo sapiens hypothetical protein LOC253982 (LOC253982), mRNA
NMJ 98920 Homo sapiens chromosome 6 open reading frame 157 (C6orf157), mRNA
NMJ 98923 Homo sapiens MAS-related GPR, member D (MRGPRD), mRNA
NMJ 98924 Homo sapiens hypothetical protein MGC45477 (MGC45477), mRNA
NMJ 98925 Homo sapiens sema domain, immunoglobulin domain (Ig), transmembrane c
NMJ 98926 Homo sapiens hypothetical protein LOC55924 (LOC55924), transcript varian
NMJ 98928 Homo sapiens interferon-inducible protein X (IFIX), transcript variant a2, mRI
NMJ 98929 Homo sapiens interferon-inducible protein X (IFIX), transcript variant b1 , mRI
NMJ98930 Homo sapiens interferon-inducible protein X (IFIX), transcript variant b2, mRI
NMJ 98935 Homo sapiens synovial sarcoma translocation gene on chromosome 18-like
NMJ 98938 Homo sapiens prostaglandin E synthase 2 (PTGES2), transcript variant 2, m
NMJ 98939 Homo sapiens prostaglandin E synthase 2 (PTGES2), transcript variant 3, m
NMJ 98940 Homo sapiens prostaglandin E synthase 2 (PTGES2), transcript variant 4, m
NM J98941 Homo sapiens tumor differentially expressed 1 (TDE1 ), transcript variant 2, n
NMJ 98943 Homo sapiens CXYorfl -related protein (MGC52000), mRNA
NMJ 98944 Homo sapiens olfactory receptor, family 7, subfamily C, member 1 (OR7C1),
NMJ 98945 Homo sapiens amyotrophic lateral sclerosis 2 (juvenile) chromosome region,
NMJ 98946 Homo sapiens lipocalin 6 (LCN6), mRNA
NMJ 98947 Homo sapiens cancer-associated nucleoprotein (CANP), mRNA
NMJ 98948 Homo sapiens nudix (nucleoside diphosphate linked moiety X)-type motif 1 (I
NMJ 98949 Homo sapiens nudix (nucleoside diphosphate linked moiety X)-type motif 1 (I
NMJ 98950 Homo sapiens nudix (nucleoside diphosphate linked moiety X)-type motif 1 (I
NMJ 98951 Homo sapiens transglutaminase 2 (C polypeptide, protein-glutamine-gamma
NMJ 98952 Homo sapiens nudix (nucleoside diphosphate linked moiety X)-type motif 1 (I
NMJ 98953 Homo sapiens nudix (nucleoside diphosphate linked moiety X)-type motif 1 (I
NMJ 98954 Homo sapiens nudix (nucleoside diphosphate linked moiety X)-type motif 1 (I
NMJ98955 Homo sapiens mannosyl (alpha-1 ,6-)-glycoprotein beta-1 ,6-N-acetyl-glucosa
NMJ 98956 Homo sapiens Sp8 transcription factor (SP8), transcript variant 2, mRNA
NMJ 98963 Homo sapiens DEAH (Asp-Glu-Ala-Asp/His) box polypeptide 57 (DHX57), tra
NMJ 98964 Homo sapiens parathyroid hormone-like hormone (PTHLH), transcript varian
NMJ 98965 Homo sapiens parathyroid hormone-like hormone (PTHLH), transcript varian
NMJ 98966 Homo sapiens parathyroid hormone-like hormone (PTHLH), transcript varian
NMJ98968 Homo sapiens DAZ interacting protein 1 (DZIP1), mRNA
NMJ 98969 Homo sapiens amino-terminal enhancer of split (AES), transcript variant 1 , rr
NMJ 98970 Homo sapiens amino-terminal enhancer of split (AES), transcript variant 3, rr
NMJ 98971 Homo sapiens MBD2 (methyl-CpG-binding protein)-interacting zinc finger pre
NMJ 98973 Homo sapiens MAP kinase interacting serine/threonine kinase 1 (MKNK1), rr
NMJ 98974 Homo sapiens PTK9 protein tyrosine kinase 9 (PTK9), transcript variant 2, m
NMJ98976 Homo sapiens TH 1-like (Drosophila) (TH1L), transcript variant 1, mRNA
NMJ 98977 Homo sapiens Rho guanine nucleotide exchange factor (GEF) 1 (ARHGEF1]
NMJ 98988 Homo sapiens leukocyte receptor cluster (LRC) member 9 (LENG9), mRNA
NMJ 98989 Homo sapiens deleted in lymphocytic leukemia 7 (DLEU7), mRNA
NMJ 98990 Homo sapiens N-acyl-phosphatidylethanolamine-hydrolyzing phospholipase
NMJ 98991 Homo sapiens potassium channel tetramerisation domain containing 1 (KCT
NMJ 98992 Homo sapiens synaptotagmin X (SYT10), mRNA
NMJ98993 Homo sapiens SH3 and cysteine rich domain 2 (STAC2), mRNA
NMJ 98994 Homo sapiens transglutaminase 6 (TGM6), mRNA
NMJ 98995 Homo sapiens chromosome 18 open reading frame 34 (C18orf34), mRNA
NMJ 98996 Homo sapiens membrane-associated phospholipase A1 beta (LOC375108),
NMJ 98998 Homo sapiens aquaporin 12 (AQP12), mRNA
NMJ 98999 Homo sapiens prestin (motor protein) (PRES), transcript variant a, mRNA
NMJ 99000 Homo sapiens lipoma HMGIC fusion partner-like 3 (LHFPL3), mRNA
NMJ 99001 Homo sapiens Similar to RIKEN cDNA 2310002J15 gene (MGC59937), mRI
NMJ 99002 Homo sapiens Rho guanine nucleotide exchange factor (GEF) 1 (ARHGEF1]
NMJ99003 Homo sapiens THAP domain containing, apoptosis associated protein 1 (TH,
NMJ 99004 Homo sapiens arrestin, beta 2 (ARRB2), transcript variant 2, mRNA
NMJ 99005 Homo sapiens zinc finger protein 322B (ZNF322B), mRNA
NMJ 99006 Homo sapiens cortistatin (CORT), transcript variant 3, mRNA
NMJ 99037 Homo sapiens sodium channel, voltage-gated, type I, beta (SCN1B), transcri
NMJ 99039 Homo sapiens kelch-like 5 (Drosophila) (KLHL5), transcript variant b, mRNA
NMJ99040 Homo sapiens nudix (nucleoside diphosphate linked moiety X)-type motif 4 (I
NMJ99043 Homo sapiens chromosome 14 open reading frame 102 (C14orf102), transci
NMJ 99044 Homo sapiens MGC22960 gene (MGC22960), mRNA
NMJ 99046 Homo sapiens testis/prostate/placenta-expressed protein (TEPP), transcript '
NMJ 99047 Homo sapiens TATA box binding protein like 2 (TBPL2), mRNA
NMJ99050 Homo sapiens chromosome 21 open reading frame 25 (C21orf25), mRNA
NMJ 99051 Homo sapiens DBCCR1 -like (DBCCR1L), mRNA
NMJ 99052 Homo sapiens chromosome 20 open reading frame 7 (C20orf7), transcript vε
NMJ99053 Homo sapiens FLJ12716 protein (FLJ12716), transcript variant 2, mRNA
NMJ 99054 Homo sapiens MAP kinase-interacting serine/threonine kinase 2 (MKNK2), π
NMJ99069 Homo sapiens nuclear protein E3-3 (DKFZP564J0123), transcript variant 1 , r
NMJ 99070 Homo sapiens nuclear protein E3-3 (DKFZP564J0123), transcript variant 2, r
NMJ 99071 Homo sapiens chromosome 21 open reading frame 58 (C21orf58), transcripl
NMJ 99072 Homo sapiens l-mfa domain-containing protein (HIC), mRNA
NMJ99073 Homo sapiens nuclear protein E3-3 (DKFZP564J0123), transcript variant 3, r
NMJ99074 Homo sapiens nuclear protein E3-3 (DKFZP564J0123), transcript variant 4, r
NMJ 99075 Homo sapiens CBF1 interacting corepressor (CIR), transcript variant 2, mRN
NMJ 99076 Homo sapiens cyclin M2 (CNNM2), transcript variant 2, mRNA
NMJ 99077 Homo sapiens cyclin M2 (CNNM2), transcript variant 3, mRNA
NMJ 99078 Homo sapiens cyclin M3 (CNNM3), transcript variant 2, mRNA
NMJ99121 Homo sapiens von Willebrand factor A domain-related protein (WARP), trans
NMJ99122 Homo sapiens transforming growth factor beta regulator 4 (TBRG4), transcrii
NMJ99123 Homo sapiens chromosome 14 open reading frame 154 (C14orf154), transci
NMJ 99124 Homo sapiens hypothetical protein FLJ23554 (FLJ23554), transcript variant . '
NMJ 99126 Homo sapiens zinc finger protein 585A (ZNF585A), transcript variant 2, mRN
NMJ 99127 Homo sapiens gamma-glutamyltransferase-like 4 (GGTL4), transcript variant
NMJ 99129 Homo sapiens ubiquitin-conjugating enzyme variant Kua (Kua), mRNA
NMJ 99131 Homo sapiens ventral anterior homeobox 1 (VAX1), mRNA
NMJ99133 Homo sapiens hypothetical protein LOC134145 (LOC134145), mRNA
NMJ 99135 Homo sapiens FOXD4-like 2 (FOXD4L2), mRNA
NMJ 99136 Homo sapiens hypothetical protein MGC72075 (MGC72075), mRNA
NMJ 99138 Homo sapiens hypothetical protein FLJ25477 (FLJ25477), transcript variant ;
NMJ 99139 Homo sapiens XIAP associated factor-1 (HSXIAPAF1), transcript variant 2, n
NMJ99141 Homo sapiens coactivator-associated arginine methyltransferase 1 (CARM1)
NMJ99144 Homo sapiens ubiquitin-conjugating enzyme E2 variant 1 (UBE2V1), transcri
NMJ 99160 Homo sapiens LIM homeobox 6 (LHX6), transcript variant 2, mRNA
NMJ 99161 Homo sapiens serum amyloid A1 (SAA1), transcript variant 2, mRNA
NMJ 99162 Homo sapiens ADP-ribosylhydrolase like 1 (ADPRHL1 ), transcript variant 2, i
NMJ 99163 Homo sapiens CXYorfl- related protein (FLJ25222), mRNA
NMJ 99165 Homo sapiens adenylosuccinate synthase like 1 (ADSSL1), transcript varianl
NMJ 99166 Homo sapiens aminolevulinate, delta-, synthase 1 (ALAS1), transcript varian
NMJ 99167 Homo sapiens clusterin-like 1 (retinal) (CLUL1), transcript variant 2, mRNA
NMJ99168 Homo sapiens chemokine (C-X-C motif) ligand 12 (stromal cell-derived factoi
NMJ99169 Homo sapiens transmembrane, prostate androgen induced RNA (TMEPAI), 1
NMJ 99170 Homo sapiens transmembrane, prostate androgen induced RNA (TMEPAI), 1
NMJ99171 Homo sapiens transmembrane, prostate androgen induced RNA (TMEPAI), 1
NMJ99173 Homo sapiens bone garnma-carboxyglutamate (gla) protein (osteocalcin) (B(
NMJ 99175 Homo sapiens chromosome 21 open reading frame 123 (C21orf123), mRNA
NMJ 99176 Homo sapiens mitochondrial ribosome recycling factor (MRRF), transcript va
NMJ 99177 Homo sapiens mitochondrial ribosome recycling factor (MRRF), transcript va
NMJ 99179 Homo sapiens kin of IRRE like 2 (Drosophila) (KIRREL2), transcript variant 2
NMJ99180 Homo sapiens kin of IRRE like 2 (Drosophila) (KIRREL2), transcript variant 3
NMJ 99181 Homo sapiens FLJ44670 protein (FLJ44670), mRNA
NMJ99182 Homo sapiens hLAT1-3TM (IMAA), mRNA
NMJ 99183 Homo sapiens testis serine protease 5 (TESSP5), mRNA
NMJ 99184 Homo sapiens chromosome 6 open reading frame 108 (C6orf108), transcripl
NMJ 99185 Homo sapiens nucleophosmin (nucleolar phosphoprotein B23, numatrin) (NF
NMJ 99186 Homo sapiens 2,3-bisphosphoglycerate mutase (BPGM), transcript variant 2
NMJ99187 Homo sapiens keratin 18 (KRT18), transcript variant 2, mRNA
NMJ 99188 Homo sapiens c-Mpl binding protein (LOC113251), transcript variant 2, mRN
NMJ 99190 Homo sapiens c-Mpl binding protein (LOC113251), transcript variant 3, mRN
NMJ 99191 Homo sapiens brain and reproductive organ-expressed (TNFRSF1 A modulat
NMJ99192 Homo sapiens brain and reproductive organ-expressed (TNFRSF1A modulal
NMJ 99193 Homo sapiens brain and reproductive organ-expressed (TNFRSF1A modulal
NMJ99194 Homo sapiens brain and reproductive organ-expressed (TNFRSF1A modulat
NMJ 99202 Homo sapiens Theg homolog (mouse) (THEG), transcript variant 2, mRNA
NMJ 99203 Homo sapiens ubiquitin-conjugating enzyme E2 variant 1 (Kua-UEV), transcr
NMJ 99204 Homo sapiens dehydrogenase/reductase (SDR family) member 9 (DHRS9),
NMJ 99205 Homo sapiens p30 DBC protein (DBC-1 ), transcript variant 2, mR A
NMJ 99206 Homo sapiens T-cell leukemia/lymphoma 1 B (TCL1 B), transcript variant 2, m
NMJ 99227 Homo sapiens methionine aminopeptidase 1 D (MAPI D), mRNA
NMJ 99228 Homo sapiens thrombopoietin (myeloproliferative leukemia virus oncogene Ii
NMJ 99229 Homo sapiens ribulose-5-phosphate-3-epimerase (RPE), transcript variant 1
NMJ 99231 Homo sapiens glial cell derived neurotrophic factor (GDNF), transcript varian
NMJ 99232 Homo sapiens allantoicase (ALLC), transcript variant 2, mRNA
NMJ 99234 Homo sapiens glial cell derived neurotrophic factor (GDNF), transcript varian
NMJ 99235 Homo sapiens collectin sub-family member 11 (COLEC11 ), transcript variant
NMJ99242 Homo sapiens unc-13 homolog D (C. elegans) (UNC13D), mRNA
NMJ 99243 Homo sapiens G protein-coupled receptor 150 (GPR150), mRNA
NMJ 99244 Homo sapiens forkhead box protein D4b (FOXD4b), mRNA
NMJ 99245 Homo sapiens vesicle-associated membrane protein 1 (synaptobrevin 1) (VA
NMJ 99246 Homo sapiens cyclin G1 (CCNG1), transcript variant 2, mRNA
NMJ 99247 Homo sapiens calcium channel, voltage-dependent, beta 1 subunit (CACNB'
NMJ 99248 Homo sapiens calcium channel, voltage-dependent, beta 1 subunit (CACNB'
NMJ99249 Homo sapiens multidrug resistance-related protein (MGC13170), mRNA
NMJ99250 Homo sapiens multidrug resistance-related protein (MGC13170), mRNA
NMJ 99254 Homo sapiens transmembrane phosphoinositide 3-phosphatase and tensin \
NMJ 99255 Homo sapiens transmembrane phosphoinositide 3-phosphatase and tensin t
NMJ 99259 Homo sapiens transmembrane phosphatase with tensin homology (TPTE), tr
NMJ 99260 Homo sapiens transmembrane phosphatase with tensin homology (TPTE), tr
NMJ 99261 Homo sapiens transmembrane phosphatase with tensin homology (TPTE), tr
NMJ 99262 Homo sapiens Sp6 transcription factor (SP6), mRNA
NMJ 99263 Homo sapiens thrombospondin, type I, domain containing 1 (THSD1), transc
NMJ99265 Homo sapiens thrombospondin, type I, domain containing 3 (THSD3), transc
NMJ 99280 Homo sapiens similar to RIKEN cDNA 4632412N22 gene (LOC165186), mR
NMJ 99282 Homo sapiens likely ortholog of rat CIN85-associated multi-domain containin
NMJ 99285 Homo sapiens hypothetical LOC284338 (MGC70924), mRNA
NMJ 99286 Homo sapiens STELLA mRNA (LOC338759), mRNA
NMJ 99290 Homo sapiens alpha-NAC protein (MGC71999), mRNA
NMJ 99292 Homo sapiens tyrosine hydroxylase (TH), transcript variant 1 , mRNA
NMJ 99293 Homo sapiens tyrosine hydroxylase (TH), transcript variant 3, mRNA
NMJ 99294 Homo sapiens cortistatin (CORT), transcript variant 4, mRNA
NMJ 99295 Homo sapiens cortistatin (CORT), transcript variant 5, mRNA
NMJ99296 Homo sapiens thrombospondin, type I, domain containing 3 (THSD3), transc
NMJ 99297 Homo sapiens thymocyte protein thy28 (THY28), transcript variant 2, mRNA
NMJ 99298 Homo sapiens thymocyte protein thy28 (THY28), transcript variant 3, mRNA
NMJ 99320 Homo sapiens PHD protein Jade-1 (JADE1), transcript variant L, mRNA
NMJ 99321 Homo sapiens zona pellucida binding protein 2 (ZPBP2), transcript variant 2,
NMJ99324 Homo sapiens HIV-1 induced protein HIN-1 (HSH1N1), transcript variant 1, m
NMJ 99326 Homo sapiens protein phosphatase 2A 48 kDa regulatory subunit (PR48), tra
NMJ 99327 Homo sapiens sprouty homolog 1 , antagonist of FGF signaling (Drosophila) I
NMJ 99328 Homo sapiens claudin 8 (CLD N8), mRNA
NMJ 99329 Homo sapiens solute carrier family 43, member 3 (SLC43A3), mRNA
NMJ 99330 Homo sapiens homer homolog 2 (Drosophila) (HOMER2), transcript variant .
NMJ 99331 Homo sapiens homer homolog 2 (Drosophila) (HOMER2), transcript variant '.
NMJ 99332 Homo sapiens homer homolog 2 (Drosophila) (HOMER2), transcript variant <■
NMJ99334 Homo sapiens thyroid hormone receptor, alpha (erythroblastic leukemia viral
NMJ 99335 Homo sapiens FYN binding protein (FYB-120/130) (FYB), mRNA
NMJ 99336 Homo sapiens hypothetical protein DKFZp434N062 (DKFZp434N062), mRN.
NMJ99337 Homo sapiens similar to RIKEN cDNA 1810059G22 (LOC374395), mRNA
NMJ 99338 Homo sapiens FLJ35171 protein (FLJ35171), mRNA
NMJ 99339 Homo sapiens hypothetical protein LOC374768 (LOC374768), mRNA
NMJ 99341 Homo sapiens hypothetical protein LOC374920 (LOC374920), mRNA
NMJ 99342 Homo sapiens hypothetical protein LOC374969 (LOC374969), mRNA
NMJ 99343 Homo sapiens FLJ90637 protein (FLJ90637), mRNA
NMJ 99344 Homo sapiens hypothetical protein LOC375035 (LOC375035), mRNA
NMJ 99345 Homo sapiens similar to phosphatidylinositol 4-kinase alpha (LOC375133), n
NMJ 99346 Homo sapiens profilin family, member 4 (PFN4), mRNA
NMJ 99348 Homo sapiens protocadherin protein CDHJ (CDHJ), transcript variant 2, mRt
NMJ 99349 Homo sapiens kielin-like (LOC375616), mRNA
NMJ 99350 Homo sapiens hypothetical protein LOC375759 (LOC375759), mRNA
NMJ 99351 Homo sapiens chromosome 1 open reading frame 32 (C1orf32), mRNA
NMJ99352 Homo sapiens putative UST1-like organic anion transporter (LOC387601), m
NMJ99353 Homo sapiens proline-rich protein BstNI subfamily 1 (PRB1), transcript variai
NMJ 99354 Homo sapiens proline-rich protein BstNI subfamily 1 (PRB1), transcript variai
NMJ 99355 Homo sapiens a disintegrin-like and metalloprotease (reprolysin type) with th
NMJ 99356 Homo sapiens thrombopoietin (myeloproliferative leukemia virus oncogene Ii
NMJ99357 Homo sapiens similar to human GTPase-activating protein (ARHGAP11 A), π
NMJ 99358 Homo sapiens forkhead box D4 like 3 (FOXD4L3), mRNA
NMJ 99359 Homo sapiens tumor protein D52-like 2 (TPD52L2), transcript variant 6, mRr>
NMJ 99360 Homo sapiens tumor protein D52-like 2 (TPD52L2), transcript variant 1 , mR
NMJ 99361 Homo sapiens tumor protein D52-like 2 (TPD52L2), transcript variant 2, mR
NMJ 99362 Homo sapiens tumor protein D52-like 2 (TPD52L2), transcript variant 3, mRf
NMJ 99363 Homo sapiens tumor protein D52-like 2 (TPD52L2), transcript variant 4, mR
NMJ 99367 Homo sapiens spastic paraplegia 7, paraplegin (pure and complicated autosi
NMJ 99368 Homo sapiens transient receptor potential cation channel, subfamily C, mem
NMJ 99413 Homo sapiens protein regulator of cytokinesis 1 (PRC1), transcript variant 2,
NMJ99414 Homo sapiens protein regulator of cytokinesis 1 (PRC1), transcript variant 3,
NMJ 99415 Homo sapiens likely ortholog of mouse ubiquitin conjugating enzyme 7 intera
NMJ 99416 Homo sapiens papillary renal cell carcinoma (translocation-associated) (PRC
NMJ 99417 Homo sapiens nuclear protein E3-3 (DKFZP564J0123), transcript variant 5, r
NMJ 99418 Homo sapiens prolylcarboxypeptidase (angiotensinase C) (PRCP), transcripl
NMJ 99420 Homo sapiens polymerase (DNA directed), theta (POLQ), transcript variant 2
NMJ 99421 Homo sapiens suppressor of cytokine signaling 4 (SOCS4), transcript varianl
NMJ 99423 Homo sapiens WW domain containing E3 ubiquitin protein ligase 2 (WWP2)
NMJ 99424 Homo sapiens WW domain containing E3 ubiquitin protein ligase 2 (WWP2)
NMJ 99425 Homo sapiens visual system homeobox 1 homolog, CHX10-like (zebrafish) C
NMJ 99426 Homo sapiens zinc finger protein 64 homolog (mouse) (ZFP64), transcript va
NMJ 99427 Homo sapiens zinc finger protein 64 homolog (mouse) (ZFP64), transcript va
NMJ 99436 Homo sapiens spastic paraplegia 4 (autosomal dominant; spastin) (SPG4), ti
NMJ 99437 Homo sapiens PR domain containing 10 (PRDM10), transcript variant 2, mRI
NMJ99438 Homo sapiens PR domain containing 10 (PRDM10), transcript variant 3, mRI
NMJ99439 Homo sapiens PR domain containing 10 (PRDM10), transcript variant 4, mRI
NMJ 99440 Homo sapiens heat shock 60kDa protein 1 (chaperonin) (HSPD1), nuclear gι
NMJ 99441 Homo sapiens zinc finger protein 334 (ZNF334), transcript variant 2, mRNA
NMJ 99442 Homo sapiens coatomer protein complex, subunit epsilon (COPE), transcript
NMJ 99443 Homo sapiens ubiquitin specific protease 4 (proto-oncogene) (USP4), transc
NMJ 99444 Homo sapiens coatomer protein complex, subunit epsilon (COPE), transcript
NMJ 99450 Homo sapiens zinc finger protein 365 (ZNF365), transcript variant B, mRNA
NMJ 99451 Homo sapiens zinc finger protein 365 (ZNF365), transcript variant C, mRNA
NMJ 99452 Homo sapiens zinc finger protein 365 (ZNF365), transcript variant D, mRNA
NMJ 99453 Homo sapiens 5-hydroxytryptamine (serotonin) receptor 4 (HTR4), transcript
NMJ 99454 Homo sapiens PR domain containing 16 (PRDM16), transcript variant 2, mRI
NMJ 99456 Homo sapiens testis/prostate/placenta-expressed protein (TEPP), transcript >
NMJ 99459 Homo sapiens chromosome 10 open reading frame 71 (C10orf71), mRNA
NMJ 99460 Homo sapiens hypothetical protein LOC283439 (LOC283439), mRNA
NMJ 99461 Homo sapiens nanos homolog 1 (Drosophila) (NANOS1), mRNA
NMJ 99462 Homo sapiens receptor interacting protein kinase 5 (RIPK5), transcript variar
NMJ 99464 Homo sapiens potassium channel regulator (KCNRG), mRNA
NMJ 99478 Homo sapiens proteolipid protein 1 (Pelizaeus-Merzbacher disease, spastic |
NMJ 99482 Homo sapiens preimpiantation protein 3 (PREI3), transcript variant 2, mRNA
NMJ 99483 Homo sapiens chromosome 20 open reading frame 24 (C20orf24), transcripl
NMJ 99484 Homo sapiens chromosome 20 open reading frame 24 (C20orf24), transcript
NMJ 99485 Homo sapiens chromosome 20 open reading frame 24 (C20orf24), transcript
NMJ 99487 Homo sapiens chromosome 20 open reading frame 44 (C20orf44), transcript
NMJ 99511 Homo sapiens steroid sensitive gene 1 (URB), transcript variant 1 , mRNA
NMJ99512 Homo sapiens steroid sensitive gene 1 (URB), transcript variant 2, mRNA
NMJ 99513 Homo sapiens chromosome 20 open reading frame 44 (C20orf44), transcript
NM_201222 Homo sapiens melanoma antigen, family D, 2 (MAGED2), transcript variant
NM_201224 Homo sapiens DEAD (Asp-Glu-Ala-Asp) box polypeptide 47 (DDX47), transc
NM_201252 Homo sapiens aflatoxin B1 aldehyde reductase 3 (AFAR3), mRNA
NM_201253 Homo sapiens crumbs homolog 1 (Drosophila) (CRB1), transcript variant 2, r
NM_201259 Homo sapiens homolog of yeast TIM14 (TIM14), transcript variant 2, mRNA
NM_201260 Homo sapiens homolog of yeast TIM14 (TIM14), transcript variant 3, mRNA
NM_201261 Homo sapiens homolog of yeast TIM14 (TIM14), transcript variant 4, mRNA
NM_201262 Homo sapiens DnaJ (Hsp40) homolog, subfamily C, member 12 (DNAJC12).
NM_201263 Homo sapiens tryptophanyl tRNA synthetase 2 (mitochondrial) (WARS2), nu
NM_201264 Homo sapiens neuropilin 2 (NRP2), transcript variant 6, mRNA
NM_201265 Homo sapiens bone marrow stromal cell-derived ubiquitin-like (BMSC-UbP),
NM_201266 Homo sapiens neuropilin 2 (NRP2), transcript variant 1 , mRNA
NM_201267 Homo sapiens neuropilin 2 (NRP2), transcript variant 5, mRNA
NM_201268 Homo sapiens membrane-bound transcription factor protease, site 1 (MBTPi
NM_201269 Homo sapiens zinc finger motif enhancer binding protein 2 (Zep-2), transcrip'
NM_201274 Homo sapiens myosin phosphatase-Rho interacting protein (M-RIP), mRNA
NM_201277 Homo sapiens calponin 2 (CNN2), transcript variant 2, mRNA
NM_201278 Homo sapiens myotubularin related protein 2 (MTMR2), transcript variant 2, i
NM_201279 Homo sapiens neuropilin 2 (NRP2), transcript variant 3, mRNA
NM_201280 Homo sapiens muted homolog (mouse) (MUTED), mRNA
NM_201281 Homo sapiens myotubularin related protein 2 (MTMR2), transcript variant 3, i
NM_201282 Homo sapiens epidermal growth factor receptor (erythroblastic leukemia vira
NM_201283 Homo sapiens epidermal growth factor receptor (erythroblastic leukemia vira
NM_201284 Homo sapiens epidermal growth factor receptor (erythroblastic leukemia vira
NM_201286 Homo sapiens ubiquitin specific protease 51 (USP51), mRNA
NM_201348 Homo sapiens proline arginine-rich end leucine-rich repeat protein (PRELP),
NM_201349 Homo sapiens docking protein 2, 56kDa (DOK2), transcript variant 2, mRNA
NM_201377 Homo sapiens cancer susceptibility candidate 2 (CASC2), mRNA
NM_201378 Homo sapiens plectin 1 , intermediate filament binding protein 500kDa (PLEC
NM_201379 Homo sapiens plectin 1 , intermediate filament binding protein 500kDa (PLEC
NM_201380 Homo sapiens plectin 1 , intermediate filament binding protein 500kDa (PLEC
NM_201381 Homo sapiens plectin 1 , intermediate filament binding protein 500kDa (PLEC
NM_201382 Homo sapiens plectin 1 , intermediate filament binding protein 500kDa (PLEC
NM_201383 Homo sapiens plectin 1 , intermediate filament binding protein 500kDa (PLEC
NM_201384 Homo sapiens plectin , intermediate filament binding protein 500kDa (PLEC
NM_201397 Homo sapiens glutathione peroxidase 1 (GPX1), transcript variant 2, mRNA
NM_201398 Homo sapiens FAD-synthetase (PP591), transcript variant 2, mRNA
NM_201399 Homo sapiens DPH2-like 2 (S. cerevisiae) (DPH2L2), transcript variant 2, mF
NM_201400 Homo sapiens hypothetical protein SB153 (SB153), transcript variant 1 , mRI*
NM_201402 Homo sapiens deubiquitinating enzyme 3 (DUB3), mRNA
NM_201403 Homo sapiens MOB1 , Mps One Binder kinase activator-like 2C (yeast) (MOE
NM_201412 Homo sapiens LUC7-like (S. cerevisiae) (LUC7L), transcript variant 2, mRNA
NM_201413 Homo sapiens amyloid beta (A4) precursor protein (protease nexin-ll, Alzheii
NM_201414 Homo sapiens amyloid beta (A4) precursor protein (protease nexin-ll, Alzheii
NM_201428 Homo sapiens reticulon 3 (RTN3), transcript variant 2, mRNA
NM_201429 Homo sapiens reticulon 3 (RTN3), transcript variant 3, mRNA
NM_201430 Homo sapiens reticulon 3 (RTN3), transcript variant 4, mRNA
NM_201431 Homo sapiens Ras association (RalGDS/AF-6) domain family 6 (RASSF6), ti
NM_201432 Homo sapiens growth arrest-specific 7 (GAS7), transcript variant b, mRNA
NM_201433 Homo sapiens growth arrest-specific 7 (GAS7), transcript variant c, mRNA
NM_201434 Homo sapiens RAB5C, member RAS oncogene family (RAB5C), transcript v
NM_201435 Homo sapiens testis-specific protein TSP-NY (TSP-NY), transcript variant 2,
NM_201436 Homo sapiens H2A histone family, member V (H2AFV), transcript variant 3, i
NM_201437 Homo sapiens transcription elongation factor A (Sll), 1 (TCEA1 ), transcript vs
NM_201438 Homo sapiens periphilin 1 (PPHLN1), transcript variant 5, mRNA
NM_201439 Homo sapiens periphilin 1 (PPHLN1 ), transcript variant 3, mRNA
NM_201440 Homo sapiens periphilin 1 (PPHLN1), transcript variant 4, mRNA
NM_201441 Homo sapiens TEA domain family member 4 (TEAD4), transcript variant 2, rr
NM_201442 Homo sapiens complement component 1 , s subcomponent (C1S), transcript
NM_201443 Homo sapiens TEA domain family member 4 (TEAD4), transcript variant 3, π
NM_201444 Homo sapiens diacylglycerol kinase, alpha 80kDa (DGKA), transcript variant
NM_201445 Homo sapiens diacylglycerol kinase, alpha 80kDa (DGKA), transcript variant
NM_201446 Homo sapiens EGF-like-domain, multiple 7 (EGFL7), transcript variant 2, mR
NM_201453 Homo sapiens dopamine responsive protein (LOC220869), mRNA
NM_201515 Homo sapiens periphilin 1 (PPHLN1), transcript variant 2, mRNA
NM_201516 Homo sapiens H2A histone family, member V (H2AFV), transcript variant 4, i
NM_201517 Homo sapiens H2A histone family, member V (H2AFV), transcript variant 5, i
NM_201520 Homo sapiens similar to 1810012H11 Rik (FLJ40217), mRNA
NM_201521 Homo sapiens kinesin-like 8 (KNSL8), transcript variant 1 , mRNA
NM_201522 Homo sapiens kinesin-like 8 (KNSL8), transcript variant 2, mRNA
NM_201523 Homo sapiens kinesin-like 8 (KNSL8), transcript variant 3, mRNA
NM_201524 Homo sapiens G protein-coupled receptor 56 (GPR56), transcript variant 2, r
NM_201525 Homo sapiens G protein-coupled receptor 56 (GPR56), transcript variant 3, r
NM_201526 Homo sapiens immunoglobulin superfamily containing leucine-rich repeat (IS
NM_201532 Homo sapiens diacylglycerol kinase, zeta 104kDa (DGKZ), transcript variant
NM_201533 Homo sapiens diacylglycerol kinase, zeta 104kDa (DGKZ), transcript variant
NM_201535 Homo sapiens NDRG family member 2 (NDRG2), transcript variant 1 , mRNA
NM_201536 Homo sapiens NDRG family member 2 (NDRG2), transcript variant 3, mRNA
NM_201537 Homo sapiens NDRG family member 2 (NDRG2), transcript variant 4, mRNA
NM_201538 Homo sapiens NDRG family member 2 (NDRG2), transcript variant 5, mRNA
NM_201539 Homo sapiens NDRG family member 2 (NDRG2), transcript variant 6, mRNA
NM_201540 Homo sapiens NDRG family member 2 (NDRG2), transcript variant 7, mRNA
NM_201541 Homo sapiens NDRG family member 2 (NDRG2), transcript variant 8, mRNA
NM_201542 Homo sapiens mediator of RNA polymerase II transcription, subunit 8 homoli
NM_201543 Homo sapiens Iectin, galactoside-binding, soluble, 8 (galectin 8) (LGALS8), t
NM_201544 Homo sapiens Iectin, galactoside-binding, soluble, 8 (galectin 8) (LGALS8), t
NM_201545 Homo sapiens Iectin, galactoside-binding, soluble, 8 (galectin 8) (LGALS8), t
NM_201546 Homo sapiens hypothetical protein LOC200008 (LOC200008), mRNA
NM_201548 Homo sapiens retinitis pigmentosa 26 (autosomal recessive) (RP26), mRNA
NM_201550 Homo sapiens leucine rich repeat containing 10 (LRRC10), mRNA
NM_201552 Homo sapiens fibrinogen-like 1 (FGL1 ), transcript variant 3, mRNA
NM_201553 Homo sapiens fibrinogen-like 1 (FGL1 ), transcript variant 4, mRNA
NM_201554 Homo sapiens diacylglycerol kinase, alpha 80kDa (DGKA), transcript variant
NM_201555 Homo sapiens four and a half LIM domains 2 (FHL2), transcript variant 2, mF
NM_201556 Homo sapiens four and a half LIM domains 2 (FHL2), transcript variant 3, mF
NM_201557 Homo sapiens four and a half LIM domains 2 (FHL2), transcript variant 4, mF
NM_201559 Homo sapiens forkhead box 03A (FOX03A), transcript variant 2, mRNA
NM_201563 Homo sapiens Fc fragment of IgG, low affinity lie, receptor for (CD32) (FCGF
NM_201564 Homo sapiens chromosome 10 open reading frame 94 (C10orf94), mRNA
NM_201565 Homo sapiens hypothetical gene supported by BC039313 (L0C284861 ), R
NM_201566 Homo sapiens solute carrier family 16 (monocarboxylic acid transporters), mi
NM_201567 Homo sapiens cell division cycle 25A (CDC25A), transcript variant 2, mRNA
NM_201568 Homo sapiens chromosome 1 open reading frame 16 (C1orf16), transcript vs
NM_201569 Homo sapiens chromosome 1 open reading frame 16 (C1orf16), transcript vs
NM_201570 Homo sapiens calcium channel, voltage-dependent, beta 2 subunit (CACNB;
NM_201571 Homo sapiens calcium channel, voltage-dependent, beta 2 subunit (CACNB;
NM_201572 Homo sapiens calcium channel, voltage-dependent, beta 2 subunit (CACNB;
NM_201574 Homo sapiens solute carrier family 4, anion exchanger, member 3 (SLC4A3)
NM_201575 Homo sapiens seizure related 6 homolog (mouse)-like 2 (SEZ6L2), mRNA
NM_201589 Homo sapiens v-maf musculoaponeurotic fibrosarcoma oncogene homolog /
NM_201590 Homo sapiens calcium channel, voltage-dependent, beta 2 subunit (CACNB;
NM_201591 Homo sapiens glycoprotein M6A (GPM6A), transcript variant 2, mRNA
NM_201592 Homo sapiens glycoprotein M6A (GPM6A), transcript variant 3, mRNA
NM_201593 Homo sapiens calcium channel, voltage-dependent, beta 2 subunit (CACNB;
NM_201594 Homo sapiens similar to B-cell linker; B cell linker protein (LOC284948), tran:
NM_201595 Homo sapiens general transcription factor IIA, 1, 19/37kDa (GTF2A1), transc
NM_201596 Homo sapiens calcium channel, voltage-dependent, beta 2 subunit (CACNB;
NM_201597 Homo sapiens calcium channel, voltage-dependent, beta 2 subunit (CACNB;
NM_201598 Homo sapiens hypothetical protein SB153 (SB153), transcript variant 2, mRf*
NM_201599 Homo sapiens zinc finger protein 261 (ZNF261 ), transcript variant 2, mRNA
NM_201612 Homo sapiens IKK interacting protein (IKIP), transcript variant 2, mRNA
NM_201613 Homo sapiens IKK interacting protein (IKIP), transcript variant 3.1 , mRNA
NM_201614 Homo sapiens IKK interacting protein (IKIP), transcript variant 3.2, mRNA
NM__201623 Homo sapiens myeloid inhibitory C-type lectin-like receptor (MICL), transcripl
NM_201624 Homo sapiens ubiquitin specific protease 33 (USP33), transcript variant 2, m
NM_201625 Homo sapiens myeloid inhibitory C-type lectin-like receptor (MICL), transcripl
NM_201626 Homo sapiens ubiquitin specific protease 33 (USP33), transcript variant 3, m
NM_201627 Homo sapiens tripartite motif-containing 41 (TRIM41 ), transcript variant 2, ml
NM_201628 Homo sapiens hypothetical protein FLJ43806 (FLJ43806), mRNA
NM_201629 Homo sapiens tight junction protein 2 (zona occludens 2) (TJP2), transcript v
NM_201630 Homo sapiens leucine rich repeat neuronal 5 (LRRN5), transcript variant 2, n
NM_201631 Homo sapiens transglutaminase 5 (TGM5), transcript variant 1, mRNA
NM_201632 Homo sapiens transcription factor 7 (T-cell specific, HMG-box) (TCF7), transi
NM_201633 Homo sapiens transcription factor 7 (T-cell specific, HMG-box) (TCF7), transi
NM_201634 Homo sapiens transcription factor 7 (T-cell specific, HMG-box) (TCF7), transi
NM_201636 Homo sapiens thromboxane A2 receptor (TBXA2R), transcript variant 1 , mRt
NM_201647 Homo sapiens STAM binding protein (STAMBP), transcript variant 2, mRNA
NM_201648 Homo sapiens glycine-N-acyltransferase (GLYAT), nuclear gene encoding m
NM_201649 Homo sapiens solute carrier family 6 (neurotransmitter transporter, glycine), i
NM_201650 Homo sapiens B7 gene (B7), transcript variant 1 , mRNA
NM_201651 Homo sapiens solute carrier family 28 (sodium-coupled nucleoside transporti
NM_201653 Homo sapiens eosinophil chemotactic cytokine (CHIA), mRNA
NM_201994 Homo sapiens vesicle-associated membrane protein 4 (VAMP4), transcript v
NM_201995 Homo sapiens splicing factor 1 (SF1), transcript variant 2, mRNA
NM_201997 Homo sapiens splicing factor 1 (SF1), transcript variant 4, mRNA
NM_201998 Homo sapiens splicing factor 1 (SF1 ), transcript variant 3, mRNA
NM_201999 Homo sapiens E74-like factor 2 (ets domain transcription factor) (ELF2), tran
NM_202000 Homo sapiens SA hypertension-associated homolog (rat) (SAH), transcript v.
NM_202001 Homo sapiens excision repair cross-complementing rodent repair deficiency,
NM_202002 Homo sapiens forkhead box M1 (FOXM1 ), transcript variant 1 , mRNA
NM_202003 Homo sapiens forkhead box M1 (FOXM1 ), transcript variant 3, mRNA
NM_202004 Homo sapiens hemochromatosis type 2 (juvenile) (HFE2), transcript variant (
NM_202467 Homo sapiens regulator of G-protein signalling 19 interacting protein 1 (RGS
NM >02468 Homo sapiens regulator of G-protein signalling 19 interacting protein 1 (RGS
NM_202469 Homo sapiens regulator of G-protein signalling 19 interacting protein 1 (RGS
NM_202470 Homo sapiens regulator of G-protein signalling 19 interacting protein 1 (RGS
NM_202494 Homo sapiens regulator of G-protein signalling 19 interacting protein 1 (RGS
NM_202758 Homo sapiens rTS beta protein (HSRTSBETA), transcript variant 1 , mRNA
NM_203281 Homo sapiens BMX non-receptor tyrosine kinase (BMX), mRNA
NM_203282 Homo sapiens zinc finger protein 539 (ZNF539), mRNA
NM_203283 Homo sapiens recombining binding protein suppressor of hairless (Drosophil
NM_203284 Homo sapiens recombining binding protein suppressor of hairless (Drosophil
NM_203285 Homo sapiens poliovirus receptor-related 1 (herpesvirus entry mediator C; nc
NM_203286 Homo sapiens poliovirus receptor-related 1 (herpesvirus entry mediator C; n<
NM_203287 Homo sapiens pregnancy specific beta-1 -glycoprotein 11 (PSG11), transcripl
NM_203288 Homo sapiens retinitis pigmentosa 9 (autosomal dominant) (RP9), mRNA
NM_203289 Homo sapiens POU domain, class 5, transcription factor 1 (POU5F1 ), transc
NM_203290 Homo sapiens polymerase (RNA) I polypeptide C, 30kDa (POLR1C), transcr
NM_203291 Homo sapiens retinoblastoma binding protein 8 (RBBP8), transcript variant 2
NM_203292 Homo sapiens retinoblastoma binding protein 8 (RBBP8), transcript variant 3
NM_203293 Homo sapiens tripartite motif-containing 7 (TRIM7), transcript variant 1 , mRN
NM_203294 Homo sapiens tripartite motif-containing 7 (TRIM7), transcript variant 5, mRN
NM_203295 Homo sapiens tripartite motif-containing 7 (TRIM7), transcript variant 4, mRN
NM_203296 Homo sapiens tripartite motif-containing 7 (TRIM7), transcript variant 3, mRN
NM_203297 Homo sapiens tripartite motif-containing 7 (TRIM7), transcript variant 2, mRN
NM_203298 Homo sapiens coiled-coii-helix-coiled-coil-helix domain containing 1 (CHCHC
NM_203299 Homo sapiens hypothetical protein MGC41945 (MGC41945), mRNA
NM_203301 Homo sapiens F-box protein 33 (FBX033), mRNA
NM_203302 Homo sapiens similar to RPL23AP7 protein (MGC70863), transcript variant 2
NM_203303 Homo sapiens cellular nucleic acid binding protein-like (L0C389874), mRNA
NM_203304 Homo sapiens ring finger and KH domain containing 1 (RKHD1), mRNA
NM_203305 Homo sapiens hypothetical protein MGC50853 (MGC50853), mRNA
NM_203306 Homo sapiens hypothetical protein MGC39606 (MGC39606), mRNA
NM_203307 Homo sapiens hypothetical protein MGC35402 (MGC35402), mRNA
NM_203308 Homo sapiens ribosomal protein L13A-like (MGC34774), mRNA
NM_203309 Homo sapiens hypothetical MGC48595 (MGC48595), mRNA
NM_203311 Homo sapiens similar to Taxol resistant associated protein 3 (TRAG-3) (LOC
NM_203314 Homo sapiens 3-hydroxybutyrate dehydrogenase (heart, mitochondrial) (BDF
NM_203315 Homo sapiens 3-hydroxybutyrate dehydrogenase (heart, mitochondrial) (BDF
NM_203316 Homo sapiens dolichyl-phosphate (UDP-N-acetylglucosamine) N-acetylglucc
NM_203318 Homo sapiens myosin XVIIIA (MY018A), transcript variant 2, mRNA
NM_203326 Homo sapiens 5-azacytidine induced 2 (AZI2), transcript variant 2, mRNA
NM_203327 Homo sapiens solute carrier family 23 (nucleobase transporters), member 2 *
NM_203329 Homo sapiens CD59 antigen p18-20 (antigen identified by monoclonal antibc
NM_203330 Homo sapiens CD59 antigen p18-20 (antigen identified by monoclonal antibc
NM_203331 Homo sapiens CD59 antigen p18-20 (antigen identified by monoclonal antibc
NM_203339 Homo sapiens clusterin (complement lysis inhibitor, SP-40,40, sulfated glyco
NM_203341 Homo sapiens 15 kDa selenoprotein (SEP15), transcript variant 2, mRNA
NM_203342 Homo sapiens erythrocyte membrane protein band 4.1 (elliptocytosis 1 , RH-I
NM_203343 Homo sapiens erythrocyte membrane protein band 4.1 (elliptocytosis 1 , RH-I
NM_203344 Homo sapiens SERTA domain containing 3 (SERTAD3), transcript variant 2,
NM_203346 Homo sapiens high density lipoprotein binding protein (vigilin) (HDLBP), mRf
NM_203347 Homo sapiens MSFL2541 (UNQ2541 ), mRNA
NM_203348 Homo sapiens hypothetical MGC50722 (MGC50722), mRNA
NM_203349 Homo sapiens rai-like protein (RaLP), mRNA
NM_203350 Homo sapiens zinc finger protein 265 (ZNF265), transcript variant 1 , mRNA
NM_203351 Homo sapiens mitogen-activated protein kinase kinase kinase 3 (MAP3K3), 1
NM_203352 Homo sapiens PDZ and LIM domain 7 (enigma) (PDLIM7), transcript variant
NM_203353 Homo sapiens PDZ and LIM domain 7 (enigma) (PDLIM7), transcript variant
NM_203354 Homo sapiens meningioma expressed antigen 6 (coiled-coil proline-rich) (MC
NM_203355 Homo sapiens meningioma expressed antigen 6 (coiled-coil proline-rich) (MC
NM_203356 Homo sapiens meningioma expressed antigen 6 (coiled-coil proline-rich) (MC
NM_203357 Homo sapiens meningioma expressed antigen 6 (coiled-coil proline-rich) (MC
NM_203364 Homo sapiens membrane component, chromosome 11 , surface marker 1 (M
NM_203365 Homo sapiens Ras association (RalGDS/AF-6) and pleckstrin homology dorr
NM_203370 Homo sapiens similar to RIKEN cDNA 6530418L21 (LOC389119), mRNA
NM_203371 Homo sapiens similar to RIKEN cDNA 111O018M03 (LOC387758), mRNA
NM_203372 Homo sapiens acyl-CoA synthetase long-chain family member 3 (ACSL3), tn
NM_203373 Homo sapiens hypothetical protein LOC283807 (LOC283807), mRNA
NM_203374 Homo sapiens similar to zinc finger protein (LOC147808), mRNA
NM_203375 Homo sapiens hypothetical gene supported by BC001801 (LOC284912), mR
NM_203376 Homo sapiens similar to RIKEN cDNA 4930429020 (LOC388730), mRNA
NM_203377 Homo sapiens myoglobin (MB), transcript variant 2, mRNA
NM_203378 Homo sapiens myoglobin (MB), transcript variant 3, mRNA
NM_203379 Homo sapiens acyl-CoA synthetase long-chain family member 5 (ACSL5), tπ
NM_203380 Homo sapiens acyl-CoA synthetase long-chain family member 5 (ACSL5), tπ
NM_203381 Homo sapiens protein for MGC71805 (MGC71805), mRNA
NM_203382 Homo sapiens alpha-methylacyl-CoA racernase (AMACR), transcript variant
NM_203383 Homo sapiens ribonuclease/angiogenin inhibitor (RNH), transcript variant 2, i
NM_203384 Homo sapiens ribonuclease/angiogenin inhibitor (RNH), transcript variant 3, 1
NM_203385 Homo sapiens ribonuclease/angiogenin inhibitor (RNH), transcript variant 4, i
NM_203386 Homo sapiens ribonuclease/angiogenin inhibitor (RNH), transcript variant 5, 1
NM_203387 Homo sapiens ribonuclease/angiogenin inhibitor (RNH), transcript variant 6, 1
NM_203388 Homo sapiens ribonuclease/angiogenin inhibitor (RNH), transcript variant 7, 1
NM_203389 Homo sapiens ribonuclease/angiogenin inhibitor (RNH), transcript variant 8, 1
NM_203390 Homo sapiens similar to RIKEN cDNA 300OO04N20 (LOC389677), mRNA
NM_203391 Homo sapiens glycerol kinase (GK), transcript variant 1 , mRNA
NM_203392 Homo sapiens hypothetical gene supported by BC031617 (LOC284123), mR
NM_203393 Homo sapiens hypothetical gene supported by BC031661 (LOC389458), mR
NM_203394 Homo sapiens E2F transcription factor 7 (E2F7), mRNA
NM_203395 Homo sapiens iodotyrosine dehalogenase 1 (DEHAL1), mRNA
NM_203399 Homo sapiens stathmin 1/oncoprotein 18 (STMN1), transcript variant 2, mRI\
NM_203400 Homo sapiens similar to candidate mediator of the p53-dependent G2 arrest
NM_203401 Homo sapiens stathmin 1/oncoprotein 18 (STMN1), transcript variant 1 , mRh
NM_203402 Homo sapiens similar to CG10671-like (LOC161247), mRNA
NM_203403 Homo sapiens chromosome 9 open reading frame 150 (C9orf150), mRNA
NM_203405 Homo sapiens similar to RIKEN cDNA 231OO02B14 (LOC388818), mRNA
NM_203406 Homo sapiens similar to metallo-beta-lactamase superfamily protein (LOC15
NM_203407 Homo sapiens similar to CG32656-PA (LOC340602), mRNA
NM_203408 Homo sapiens similar to hypothetical protein FLJ35782 (LOC158724), mRN/
NM_203411 Homo sapiens similar to RIKEN cDNA 2600017H02 (LOC92162), mRNA
NM_203412 Homo sapiens similar to RIKEN cDNA 493O522D07 (LOC164153), mRNA
NM_203413 Homo sapiens S-phase 2 protein (DERP6), mRNA
NM_203414 Homo sapiens S-phase 2 protein (DERP6), mRNA
NM_203415 Homo sapiens S-phase 2 protein (DERP6), mRNA
NM_203416 Homo sapiens CD163 antigen (CD163), transcript variant 2, mRNA
NM_203417 Homo sapiens Down syndrome critical region gene 1 (DSCR1 ), transcript vai
NM_203418 Homo sapiens Down syndrome critical region gene 1 (DSCR1), transcript vai
NM_203419 Homo sapiens hypothetical protein LOC28S016 (LOC286016), mRNA
NM_203422 Homo sapiens similar to hypothetical protein (LOC221091), mRNA
NM_203423 Homo sapiens hypothetical gene supported by BC031673 (LOC389199), mR
NM_203424 Homo sapiens hypothetical protein MGC50809 (LOC389123), mRNA
NM_203425 Homo sapiens hypothetical gene supported by BC046200 (LOC388407), mR
NM_203426 Homo sapiens hypothetical protein FLJ90297 (LOC388152), mRNA
NM_203428 Homo sapiens Down syndrome critical region gene 8 (DSCR8), transcript vai
NM_203429 Homo sapiens Down syndrome critical region gene 8 (DSCR8), transcript vai
NM_203430 Homo sapiens peptidylprolyl isomerase A (cyclophilin A) (PPIA), transcript vε
NM_203431 Homo sapiens peptidylprolyl isomerase A (cyclophilin A) (PPIA), transcript vε
NM_203433 Homo sapiens Down syndrome critical region gene 2 (DSCR2), transcript vai
NM_203434 Homo sapiens similar to RIKEN cDNA 2610524G09 (LOC389792), mRNA
NM_203436 Homo sapiens achaete-scute complex-like 4 (Drosophila) (ASCL4), mRNA
NM_203437 Homo sapiens aftiphilin protein (AFTIPHILIN), transcript variant 1 , mRNA
NM__203438 Homo sapiens chromosome 10 open reading frame 4 (C10orf4), transcript vs
NM_203439 Homo sapiens chromosome 10 open reading frame 4 (C10orf4), transcript vs
NM_203440 Homo sapiens chromosome 10 open reading frame 4 (C10orf4), transcript vs
NM__203441 Homo sapiens chromosome 10 open reading frame 4 (C10orf4), transcript vs
NM_203444 Homo sapiens ATP-binding cassette, sub-family B (MDFVTAP), member 9 (A
NM_203445 Homo sapiens ATP-binding cassette, sub-family B (MDR/TAP), member 9 (A
NM__203446 Homo sapiens synaptojanin 1 (SYNJ1), transcript variant 2, mRNA
NM_203447 Homo sapiens dedicator of cytokinesis 8 (DOCK8), mRNA
NM__203448 Homo sapiens hypothetical protein MGC21881 (MGC21881), mRNA
NM_203451 Homo sapiens hypothetical LOC40O120 (LOC400120), mRNA
NM__203452 Homo sapiens hypothetical protein LOC403312 (MGC39545), mRNA
NM_203453 Homo sapiens hypothetical LOC403313 (LOC403313), mRNA
NM_203454 Homo sapiens hypothetical protein MGC26594 (MGC26594), mRNA
NM_203456 Homo sapiens peptidylprolyl isomerase E (cyclophilin E) (PPIE), transcript vε
NM_203457 Homo sapiens peptidylprolyl isomerase E (cyclophilin E) (PPIE), transcript vε
NM_203458 Homo sapiens similar to NOTCH2 protein (N2N), mRNA
NM_203459 Homo sapiens KIAA1078 protein (KIAA1078), mRNA
NM_203462 Homo sapiens PP784 protein (PP784), transcript variant 2, mRNA
NM_203463 Homo sapiens LAG1 longevity assurance homolog 6 (S. cerevisiae) (LASS6)
NM_203466 Homo sapiens peptidylprolyl isomerase (cyclophilin) like 5 (PPIL5), transcripl
NM_203467 Homo sapiens peptidylprolyl isomerase (cyclophilin) like 5 (PPIL5), transcripl
NM_203468 Homo sapiens ectonucleoside triphosphate diphosphohydrolase 2 (ENTPD2]
NM_203471 Homo sapiens Iectin, galactoside-binding, soluble, 14 (LGALS14), transcript
NM_203472 Homo sapiens selenoprotein S (SELS), transcript variant 1 , mRNA
NM_203473 Homo sapiens porcupine homolog (Drosophila) (PORCN), transcript variant I
NM_203474 Homo sapiens porcupine homolog (Drosophila) (PORCN), transcript variant (
NM_203475 Homo sapiens porcupine homolog (Drosophila) (PORCN), transcript variant I
NM_203476 Homo sapiens porcupine homolog (Drosophila) (PORCN), transcript variant I
NM_203477 Homo sapiens similar to RPL23AP7 protein (MGC70863), transcript variant 1
NM_203481 Homo sapiens hypothetical LOC403340 (MGC70870), mRNA
NM_203486 Homo sapiens delta-like 3 (Drosophila) (DLL3), transcript variant 2, mRNA
NM_203487 Homo sapiens protocadherin 9 (PCDH9), transcript variant 1 , mRNA
NM_203488 Homo sapiens acylphosphatase 1 , erythrocyte (common) type (ACYP1 ), tran
NM_203494 Homo sapiens ubiquitin specific protease 50 (USP50), mRNA
NM_203495 Homo sapiens COMM domain containing 6 (COMMD6), transcript variant 2, i
NM_203497 Homo sapiens COMM domain containing 6 (COMMD6), transcript variant 1 , i
NM_203499 Homo sapiens DEAD (Asp-Glu-Ala-Asp) box polypeptide 42 (DDX42), transc
NM_203500 Homo sapiens kelch-like ECH-associated protein 1 (KEAP1 ), transcript varia
NM_203503 Homo sapiens C-type (calcium dependent, carbohydrate-recognition domain
NM_203504 Homo sapiens Ras-GTPase activating protein SH3 domain-binding protein 2
NM_203505 Homo sapiens Ras-GTPase activating protein SH3 domain-binding protein 2
NM_203506 Homo sapiens growth factor receptor-bound protein 2 (GRB2), transcript vari
NM_203510 Homo sapiens transmembrane 6 superfamily member 2 (TM6SF2), transcrip
NM_205543 Homo sapiens amyotrophic lateral sclerosis 2 (juvenile) chromosome region,
NM_205545 Homo sapiens RGTR430 (UNQ430), mRNA
NM_205548 Homo sapiens AASA9217 (UNQ9217), mRNA
NM_205767 Homo sapiens QIL1 protein (QIL1), mRNA
NM_205768 Homo sapiens zinc finger protein 238 (ZNF238), transcript variant 1 , mRNA
NM_205833 Homo sapiens immunoglobulin superfamily, member 1 (1GSF1), transcript va
NM_205834 Homo sapiens liver-specific bHLH-Zip transcription factor (LISCH7), transcrif
NM_205835 Homo sapiens liver-specific bHLH-Zip transcription factor (LISCH7), transcrif
NM_205836 Homo sapiens F-box protein 38 (FBX038), transcript variant 2, mRNA
NM_205837 Homo sapiens leukocyte specific transcript 1 (LST1 ), transcript variant 2, mR
NM_205838 Homo sapiens leukocyte specific transcript 1 (LST1 ), transcript variant 3, mR
NM_205839 Homo sapiens leukocyte specific transcript 1 (LST1 ), transcript variant 4, mR
NM_205840 Homo sapiens leukocyte specific transcript 1 (LST1 ), transcript variant 5, mR
NM_205841 Homo sapiens protease inhibitor H (MGC21394), mRNA
NM_205842 Homo sapiens NCK-associated protein 1 (NCKAP1), transcript variant 2, mR
NM_205843 Homo sapiens nuclear factor l/C (CCAAT-binding transcription factor) (NFIC)
NM_205845 Homo sapiens aldo-keto reductase family 1 , member C2 (dihydrodiol dehydn
NM_205846 Homo sapiens hypothetical protein MGC21644 (MGC21644), transcript varia
NM_205847 Homo sapiens GDP-mannose pyrophosphorylase A (GMPPA), transcript vari
NM_205848 Homo sapiens synaptotagmin VI (SYT6), mRNA
NM_205849 Homo sapiens family with sequence similarity 9, member B (FAM9B), mRNA
NM_205850 Homo sapiens solute carrier family 24, member 5 (SLC24A5), mRNA
NM_205852 Homo sapiens macrophage antigen h (UNQ5782), mRNA
NM_205853 Homo sapiens musculoskeletal, embryonic nuclear protein 1 (MUSTN1), mR
NM_205854 Homo sapiens GSGL541 (UNQ541), mRNA
NM_205855 Homo sapiens HWKM1940 (UNQ1940), mRNA
NM_205856 Homo sapiens PNPK6288 (LOC389852), mRNA
NM_205857 Homo sapiens FBI4 protein (FBI4), mRNA
NM_205858 Homo sapiens neuromedin B (NMB), transcript variant 2, mRNA
NM_205859 Homo sapiens olfactory receptor, family 2, subfamily K, member 2 (OR2K2),
NM_205860 Homo sapiens nuclear receptor subfamily 5, group A, member 2 (NR5A2), tn
NM_205861 Homo sapiens dehydrodolichyl diphosphate synthase (DHDDS), transcript vε
NM_205862 Homo sapiens UDP glycosyltransferase 1 family, polypeptide A6 (UGT1 A6),
NM_205863 Homo sapiens amyotrophic lateral sclerosis 2 (juvenile) chromosome region,
NM_205864 Homo sapiens cancer/testis antigen 3 (CTAG3), transcript variant 2, mRNA
NM_206538 Homo sapiens hypothetical protein LOC284361 (LOC284361), transcript vari
NM_206539 Homo sapiens EGF-like-domain, multiple 9 (EGFL9), transcript variant 2, mR
NM_206594 Homo sapiens estrogen-related receptor gamma (ESRRG), transcript varianl
NM_206595 Homo sapiens estrogen-related receptor gamma (ESRRG), transcript varianl
NM_206808 Homo sapiens citrate lyase beta like (CLYBL), transcript variant 2, mRNA
NM_206809 Homo sapiens myelin oligodendrocyte glycoprotein (MOG), transcript variant
NM_206810 Homo sapiens myelin oligodendrocyte glycoprotein (MOG), transcript variant
NM_206811 Homo sapiens myelin oligodendrocyte glycoprotein (MOG), transcript variant
NM_206812 Homo sapiens myelin oligodendrocyte glycoprotein (MOG), transcript variant
NM_206813 Homo sapiens myelin oligodendrocyte glycoprotein (MOG), transcript variant
NM_206814 Homo sapiens myelin oligodendrocyte glycoprotein (MOG), transcript variant
NM_206817 Homo sapiens osteoclast-associated receptor (OSCAR), transcript variant 2,
NM_206818 Homo sapiens osteoclast-associated receptor (OSCAR), transcript variant 1 ,
NM_206819 Homo sapiens myosin binding protein C, slow type (MYBPC1 ), transcript vari
NM_206820 Homo sapiens myosin binding protein C, slow type (MYBPC1), transcript vari
NM_206821 Homo sapiens myosin binding protein C, slow type (MYBPC1), transcript vari
NM_206824 Homo sapiens vitamin K epoxide reductase complex, subunit 1 (VKORC1), ti
NM_206825 Homo sapiens nucleostemin (NS), transcript variant 2, mRNA
NM_206826 Homo sapiens nucleostemin (NS), transcript variant 3, mRNA
NM_206827 Homo sapiens RAS-like, family 11, member A (RASL11A), mRNA
NM_206828 Homo sapiens NACHT, leucine rich repeat and PYD containing 7 (NALP7), ti
NM_206831 Homo sapiens zinc finger, CSL domain containing 2 (ZCSL2), mRNA
NM_206832 Homo sapiens AWKS9372 (UNQ9372), mRNA
NM_206833 Homo sapiens cortexin 1 (CTXN1), mRNA
NM_206834 Homo sapiens chromosome 6 open reading frame 201 (C6orf201), mRNA
NM_206835 Homo sapiens TNF receptor-associated factor 7 (TRAF7), transcript variant ;
NM_206836 Homo sapiens peroxisomal D3,D2-enoyl-CoA isomerase (PECI), transcript vi
NM_206837 Homo sapiens oxidored-nitro domain-containing protein (NOR1), transcript vi
NM_206838 Homo sapiens similar to Hypothetical protein CBG21647 (LOC390511), mRh
NM_206839 Homo sapiens mortality factor 4 like 1 (MORF4L1 ), transcript variant 2, mRN.
NM_206840 Homo sapiens nuclear VCP-Iike (NVL), transcript variant 2, mRNA
NM_206841 Homo sapiens Fraser syndrome 1 (FRAS1 ), transcript variant 2, mRNA
NM_206852 Homo sapiens reticulon 1 (RTN1), transcript variant 3, mRNA
NM_206853 Homo sapiens quaking homolog, KH domain RNA binding (mouse) (QKI), tre
NM_206854 Homo sapiens quaking homolog, KH domain RNA binding (mouse) (QKI), trε
NM_206855 Homo sapiens quaking homolog, KH domain RNA binding (mouse) (QKI), trε
NM_206857 Homo sapiens reticulon 1 (RTN1), transcript variant 2, mRNA
NM_206858 Homo sapiens similar to protein phosphatase 1 , regulatory (inhibitor) subunit
NM_206860 Homo sapiens transforming, acidic coiled-coil containing protein 2 (TACC2),
NM_206861 Homo sapiens transforming, acidic coiled-coil containing protein 2 (TACC2),
NM_206862 Homo sapiens transforming, acidic coiled-coil containing protein 2 (TACC2),
NM_206866 Homo sapiens BTB and CNC homology 1 , basic leucine zipper transcription '
NM_206873 Homo sapiens protein phosphatase 1 , catalytic subunit, alpha isoform (PPP1
NM_206876 Homo sapiens protein phosphatase 1 , catalytic subunit, beta isoform (PPP1C
NM_206877 Homo sapiens protein phosphatase 1 , catalytic subunit, beta isoform (PPP1C
NM_206880 Homo sapiens olfactory receptor, family 2, subfamily V, member 2 (OR2V2),
NM_206883 Homo sapiens prestin (motor protein) (PRES), transcript variant b, mRNA
NM_206884 Homo sapiens prestin (motor protein) (PRES), transcript variant c, mRNA
NM_206885 Homo sapiens prestin (motor protein) (PRES), transcript variant d, mRNA
NM_206886 Homo sapiens sarcoma antigen NY-SAR-41 (NY-SAR-41 ), mRNA
NM_206887 Homo sapiens Down syndrome cell adhesion molecule (DSCAM), transcript ■
NM_206889 Homo sapiens chromosome 21 open reading frame 106 (C21orf106), transci
NM_206890 Homo sapiens chromosome 21 open reading frame 106 (C21orf106), transci
NM_206891 Homo sapiens chromosome 21 open reading frame 106 (C21orf106), transci
NM_206892 Homo sapiens malate dehydrogenase 1B, NAD (soluble) (MDH1 B), mRNA
NM_206893 Homo sapiens membrane-spanning 4-domains, subfamily A, member 10 (Mi
NM_206894 Homo sapiens hypothetical protein LOC388536 (MGC62100), mRNA
NM_206895 Homo sapiens ASCL830 (UNQ830), mRNA
NM_206898 Homo sapiens chromosome 21 open reading frame 61 (C21orf61), transcript
NM_206900 Homo sapiens reticulon 2 (RTN2), transcript variant 2, mRNA
NM_206901 Homo sapiens reticulon 2 (RTN2), transcript variant 3, mRNA
NM_206902 Homo sapiens reticulon 2 (RTN2), transcript variant 4, mRNA
NM_206907 Homo sapiens protein kinase, AMP-activated, alpha 1 catalytic subunit (PRK
NM_206908 Homo sapiens chromosome 6 open reading frame 216 (C6orf216), transcript
NM_206909 Homo sapiens pleckstrin and Sec7 domain containing 3 (PSD3), transcript vs
NM_206910 Homo sapiens chromosome 6 open reading frame 216 (C6orf216), transcript
NM_206911 Homo sapiens chromosome 6 open reading frame 216 (C6orf216), transcripl
NM_206912 Homo sapiens chromosome 6 open reading frame 216 (C6orf216), transcripl
NM_206914 Homo sapiens hepatocellularcarcinoma-associated antigen HCA557a (DKF2
NM_206915 Homo sapiens nerve growth factor receptor (TNFRSF16) associated protein
NM_206917 Homo sapiens nerve growth factor receptor (TNFRSF16) associated protein
NM_206918 Homo sapiens chromosome 14 open reading frame 66 (C14orf66), mRNA
NM_206919 Homo sapiens ADP-ribosylation factor-like 9 (ARL9), mRNA
NM_206920 Homo sapiens apical early endosomal glycoprotein precursor (AEGP), mRN/
NM_206921 Homo sapiens chromosome 6 open reading frame 204 (C6orf204), mRNA
NM_206922 Homo sapiens thymus LIM protein TLP-A (TLP), mRNA
NM_206923 Homo sapiens YY2 transcription factor (YY2), mRNA
NM_206925 Homo sapiens carbonic anhydrase XII (CA12), transcript variant 2, mRNA
NM_206926 Homo sapiens selenoprotein N, 1 (SEPN1 ), transcript variant 2, mRNA
NM_206927 Homo sapiens synaptotagmin-like 2 (SYTL2), transcript variant c, mRNA
NM_206928 Homo sapiens synaptotagmin-like 2 (SYTL2), transcript variant d, mRNA
NM_206929 Homo sapiens synaptotagmin-like 2 (SYTL2), transcript variant e, mRNA
NM_206930 Homo sapiens synaptotagmin-like 2 (SYTL2), transcript variant f , mRNA
NM_206933 Homo sapiens Usher syndrome 2A (autosomal recessive, mild) (USH2A), tra
NM_206937 Homo sapiens ligase IV, DNA, ATP-dependent (LIG4), mRNA
NM_206938 Homo sapiens membrane-spanning 4-domains, subfamily A, member 7 (MS'
NM_206939 Homo sapiens membrane-spanning 4-domains, subfamily A, member 7 (MS'
NM_206940 Homo sapiens membrane-spanning 4-domains, subfamily A, member 7 (MS'
NM_206943 Homo sapiens latent transforming growth factor beta binding protein 1 (LTBF
NM_206944 Homo sapiens transient receptor potential cation channel, subfamily M, mem
NM_206945 Homo sapiens transient receptor potential cation channel, subfamily M, mem
NM_206946 Homo sapiens transient receptor potential cation channel, subfamily M, mem
NM_206947 Homo sapiens transient receptor potential cation channel, subfamily M, mem
NM_206948 Homo sapiens transient receptor potential cation channel, subfamily M, mem
NM_206949 Homo sapiens family with sequence similarity 14, member B (FAM14B), mRt
NM_206953 Homo sapiens preferentially expressed antigen in melanoma (PRAME), trans
NM_206954 Homo sapiens preferentially expressed antigen in melanoma (PRAME), trans
NM_206955 Homo sapiens preferentially expressed antigen in melanoma (PRAME), trans
NM_206956 Homo sapiens preferentially expressed antigen in melanoma (PRAME), trans
NM_206961 Homo sapiens leukocyte tyrosine kinase (LTK), transcript variant 2, mRNA
NM_206962 Homo sapiens HMT1 hnRNP methyltransferase-like 1 (S. cerevisiae) (HRMT
NM_206963 Homo sapiens retinoic acid receptor responder (tazarotene induced) 1 (RARI
NM_206964 Homo sapiens family with sequence similarity 3, member B (FAM3B), transci
NM_206965 Homo sapiens formiminotransferase cyclodeaminase (FTCD), transcript varii
NM_206966 Homo sapiens similar to AVLV472 (MGC23985), mRNA
NM_206967 Homo sapiens MGC17624 protein (MGC17624), mRNA
NM_206994 Homo sapiens gonadotropin-releasing hormone (type 2) receptor 2 (GNRHR
NM_206996 Homo sapiens projection protein PF6 (PF6), mRNA
NM_206997 Homo sapiens G protein-coupled receptor 152 (GPR152), mRNA
NM_206998 Homo sapiens secretoglobin family 1 D member 4 (SCGB1 D4), mRNA
NM_206999 Homo sapiens KIAA1007 protein (KIAA1007), transcript variant 2, mRNA
NM_207002 Homo sapiens BCL2-like 11 (apoptosis facilitator) (BCL2L11 ), transcript variε
NM_207003 Homo sapiens BCL2-like 11 (apoptosis facilitator) (BCL2L11), transcript varis
NM_207005 Homo sapiens upstream transcription factor 1 (USF1), transcript variant 2, m
NM_207006 Homo sapiens hypothetical protein MGC14128 (BJ-TSA-9), transcript variant
NM_207007 Homo sapiens chemokine (C-C motif) ligand 4-like 1 , telomeric (CCL4L1), ml
NM_207009 Homo sapiens family with sequence similarity 45, member A (FAM45A), mRf
NM_207012 Homo sapiens adaptor-related protein complex 3, mu 1 subunit (AP3M1), tra
NM_207013 Homo sapiens transcription elongation factor B (SIN), polypeptide 2 (18kDa, i
NM_207014 Homo sapiens hypothetical protein FLJ23129 (FLJ23129), transcript variant . '
NM_207015 Homo sapiens N-acetylated alpha-linked acidic dipeptidase 2 (NAALADL2), i
NM_207032 Homo sapiens endothelin 3 (EDN3), transcript variant 2, mRNA
NM_207033 Homo sapiens endothelin 3 (EDN3), transcript variant 3, mRNA
NM_207034 Homo sapiens endothelin 3 (EDN3), transcript variant 4, mRNA
NM_207035 Homo sapiens NPD014 protein (NPD014), transcript variant 1 , mRNA
NM_207036 Homo sapiens transcription factor 12 (HTF4, helix-loop-helix transcription fac
NM_207037 Homo sapiens transcription factor 12 (HTF4, helix-loop-helix transcription fac
NM_207038 Homo sapiens transcription factor 12 (HTF4, helix-loop-helix transcription fac
NM_207040 Homo sapiens transcription factor 12 (HTF4, helix-loop-helix transcription fac
NM_207042 Homo sapiens endosulfine alpha (ENSA), transcript variant 1 , mRNA
NM_207043 Homo sapiens endosulfine alpha (ENSA), transcript variant 2, mRNA
NM_207044 Homo sapiens endosulfine alpha (ENSA), transcript variant 4, mRNA
NM_207045 Homo sapiens endosulfine alpha (ENSA), transcript variant 5, mRNA
NM_207046 Homo sapiens endosulfine alpha (ENSA), transcript variant 6, mRNA
NM_207047 Homo sapiens endosulfine alpha (ENSA), transcript variant 7, mRNA
NM_207102 Homo sapiens F-box and WD-40 domain protein 12 (FBXW12), mRNA
NM_207103 Homo sapiens DTFT5783 (UNQ5783), mRNA
NM_207106 Homo sapiens three prime repair exonuclease 2 (TREX2), transcript variant '
NM_207107 Homo sapiens three prime repair exonuclease 2 (TREX2), transcript variant f
NM_207108 Homo sapiens astrotactin (ASTN), transcript variant 2, mRNA
NM_207111 Homo sapiens TRIAD3 protein (TRIAD3), transcript variant 1 , mRNA
NM_207112 Homo sapiens hydroxyacylglutathione hydrolase-like (HAGHL), transcript var
NM_207113 Homo sapiens solute carrier family 37 (glycerol-3-phosphate transporter), me
NM_207115 Homo sapiens zinc finger protein 580 (ZNF580), transcript variant 2, mRNA
NM_207116 Homo sapiens TRIAD3 protein (TRIAD3), transcript variant 2, mRNA
NM_207117 Homo sapiens chromosome 14 open reading frame 68 (C14orf68), mRNA
NM_207118 Homo sapiens general transcription factor IIH, polypeptide 5 (GTF2H5), mRh
NM_207119 Homo sapiens leucine rich repeat containing 20 (LRRC20), transcript variant
NM_207121 Homo sapiens chromosome 20 open reading frame 55 (C20orf55), transcript
NM_207122 Homo sapiens exostoses (multiple) 2 (EXT2), transcript variant 2, mRNA
NM_207123 Homo sapiens GRB2-associated binding protein 1 (GAB1), transcript variant
NM_207125 Homo sapiens polyamine oxidase (exo-N4-amino) (PAOX), transcript variant
NM_207126 Homo sapiens polyamine oxidase (exo-N4-amino) (PAOX), transcript variant
NM_207127 Homo sapiens polyamine oxidase (exo-N4-amino) (PAOX), transcript variant
NM_207128 Homo sapiens polyamine oxidase (exo-N4-amino) (PAOX), transcript variant
NM_207129 Homo sapiens polyamine oxidase (exo-N4-amino) (PAOX), transcript variant
NM_207168 Homo sapiens endosulfine alpha (ENSA), transcript variant 8, mRNA
NM_207170 Homo sapiens GCIP-interacting protein p29 (P29), transcript variant 2, mRN>
NM_207171 Homo sapiens pogo transposable element with ZNF domain (POGZ), transcr
NM_207172 Homo sapiens G protein-coupled receptor 154 (GPR154), transcript variant 1
NM_207173 Homo sapiens G protein-coupled receptor 154 (GPR154), transcript variant 2
NM_207174 Homo sapiens ATP-binding cassette, sub-family G (WHITE), member 1 (ABC
NM_207181 Homo sapiens nephronophthisis 1 (juvenile) (NPHP1 ), transcript variant 2, m
NM_207186 Homo sapiens olfactory receptor, family 10, subfamily A, member 4 (ORIOA"'
NM_207189 Homo sapiens bromodomain, testis-specific (BRDT), transcript variant 1 , mR
NM_207191 Homo sapiens a disintegrin and metalloproteinase domain 15 (metargidin) (A
NM_207194 Homo sapiens a disintegrin and metalloproteinase domain 15 (metargidin) (A
NM_207195 Homo sapiens a disintegrin and metalloproteinase domain 15 (metargidin) (A
NM_207196 Homo sapiens a disintegrin and metalloproteinase domain 15 (metargidin) (A
NM_207197 Homo sapiens a disintegrin and metalloproteinase domain 15 (metargidin) (A
NM_207283 Homo sapiens AAA1 protein (AAA1 ), transcript variant IX, mRNA
NM_207284 Homo sapiens AAA1 protein (AAA1), transcript variant II, mRNA
NM_207285 Homo sapiens AAA1 protein (AAA1), transcript variant III, mRNA
NM_207286 Homo sapiens AAA1 protein (AAA1 ), transcript variant IV, mRNA
NM_207287 Homo sapiens AAA1 protein (AAA1), transcript variant V, mRNA
NM_207288 Homo sapiens AAA1 protein (AAA1 ), transcript variant VI, mRNA
NM_207289 Homo sapiens AAA1 protein (AAA1 ), transcript variant VII, mRNA
NM_207290 Homo sapiens AAA1 protein (AAA1 ), transcript variant VIII, mRNA
NM_207291 Homo sapiens upstream transcription factor 2, c-fos interacting (USF2), trans
NM_207292 Homo sapiens muscleblind-like (Drosophila) (MBNL1), transcript variant 2, m
NM_207293 Homo sapiens muscleblind-like (Drosophila) (MBNL1), transcript variant 3, m
NM_207294 Homo sapiens muscleblind-like (Drosophila) (MBNL1), transcript variant 4, m
NM_207295 Homo sapiens muscleblind-like (Drosophila) (MBNL1), transcript variant 5, m
NM_207296 Homo sapiens muscleblind-like (Drosophila) (MBNL1), transcript variant 6, m
NM_207297 Homo sapiens muscleblind-like (Drosophila) (MBNL1), transcript variant 7, m
NM_207299 Homo sapiens plasticity related gene 3 (PRG-3), transcript variant 1 , mRNA
NM_207303 Homo sapiens attractin-like 1 (ATRNL1 ), mRNA
NM_207304 Homo sapiens muscleblind-like 2 (Drosophila) (MBNL2), transcript variant 3,
NM_207305 Homo sapiens forkhead box D4 (FOXD4), mRNA
NM_207306 Homo sapiens KIAA0495 (KIAA0495), mRNA
NM_207307 Homo sapiens hypothetical protein LOC90288 (LOC90288), mRNA
NM_207308 Homo sapiens nuclear pore membrane glycoprotein 210-like (LOC91181), m
NM_207309 Homo sapiens UDP-N-acteylglucosamine pyrophosphorylase 1-like 1 (UAP1
NM_207310 Homo sapiens hypothetical protein DKFZp434E2321 (DKFZp434E2321), mF
NM_207311 Homo sapiens hypothetical protein LOC92558 (LOC92558), mRNA
NM_207312 Homo sapiens similar to alpha tubulin (LOC112714), mRNA
NM_207313 Homo sapiens hypothetical protein LOC124842 (LOC124842), mRNA
NM_207314 Homo sapiens VNFT9373 (UNQ9373), mRNA
NM_207315 Homo sapiens hypothetical protein LOC129607 (LOC129607), mRNA
NM_207316 Homo sapiens SRSR846 (UNQ846), mRNA
NM_207317 Homo sapiens FLJ32921 protein (FLJ32921), mRNA
NM_207319 Homo sapiens FLJ32867 protein (FLJ32867), mRNA
NM_207320 Homo sapiens FLJ25831 protein (FLJ25831), mRNA
NM_207321 Homo sapiens chromosome 10 open reading frame 129 (C1 Oorfl 29), mRNA
NM_207322 Homo sapiens hypothetical LOC145741 (LOC145741 ), mRNA
NM_207323 Homo sapiens hypothetical protein DKFZp667M2411 (DKFZp667M2411 ), ml
NM_207324 Homo sapiens hypothetical protein LOC147650 (LOC147650), mRNA
NM_207325 Homo sapiens hypothetical protein LOC147991 (LOC147991), mRNA
NM_207326 Homo sapiens hypothetical protein LOC149134 (LOC149134), mRNA
NM_207327 Homo sapiens similar to RIKEN cDNA 2210021 J22 (LOC150383), mRNA
NM_207328 Homo sapiens hypothetical protein LOC150763 (LOC150763), mRNA
NM_207329 Homo sapiens myeloid-associated differentiation marker-like (MYADML), mR
NM_207330 Homo sapiens hypothetical protein LOC152519 (LOC152519), mRNA
NM_207331 Homo sapiens hypothetical protein LOC153561 (LOC153561), mRNA
NM_207332 Homo sapiens hypothetical protein LOC157697 (LOC157697), mRNA
NM_207333 Homo sapiens hypothetical protein LOC162967 (LOC162967), mRNA
NM_207334 Homo sapiens family with sequence similarity 43, member B (FAM43B), mRr
NM_207335 Homo sapiens FLJ46299 protein (FLJ46299), mRNA
NM_207336 Homo sapiens likely ortholog of mouse zinc finger protein EZI (EZI), mRNA
NM_207337 Homo sapiens hypothetical protein LOC196394 (LOC196394), mRNA
NM_207338 Homo sapiens likely ortholog of mouse klotho lactase-phlorizin hydrolase rel;
NM_207339 Homo sapiens similar to PAGE-5 protein (MGC62094), mRNA
NM_207340 Homo sapiens hypothetical protein LOC254359 (LOC254359), mRNA
NM_207341 Homo sapiens similar to ZP1 precursor (MGC87693), mRNA
NM_207343 Homo sapiens hypothetical protein DKFZp547C195 (DKFZp547C195), mRN.
NM_207344 Homo sapiens hypothetical protein LOC283377 (LOC283377), mRNA
NM_207345 Homo sapiens HEEE9341 (UNQ9341), mRNA
NM_207346 Homo sapiens likely homolog of yeast SEN54 (SEN54L), mRNA
NM_207347 Homo sapiens chromosome 18 open reading frame 30 (C18orf30), mRNA
NM_207348 Homo sapiens hypothetical protein LOC284723 (LOC284723), mRNA
NM_207349 Homo sapiens hypothetical protein LOC284739 (LOC284739), mRNA
NM_207350 Homo sapiens similar to FRG1 protein (FSHD region gene 1 protein) (MGC7.
NM_207351 Homo sapiens hypothetical protein FLJ33674 (FLJ33674), mRNA
NM_207352 Homo sapiens cytochrome P450, family 4, subfamily V, polypeptide 2 (CYP4
NM_207353 Homo sapiens ubiquitin-conjugating enzyme UbcM2 (LOC286480), mRNA
NM_207354 Homo sapiens hypothetical protein LOC338692 (LOC338692), mRNA
NM_207355 Homo sapiens protein expressed in prostate, ovary, testis, and placenta (PO
NM_207356 Homo sapiens hypothetical protein LOC339448 (LOC339448), mRNA
NM_207357 Homo sapiens hypothetical protein LOC339524 (LOC339524), mRNA
NM_207358 Homo sapiens hypothetical protein LOC339789 (LOC339789), mRNA
NM_207359 Homo sapiens glutamate decarboxylase-like 1 (GADL1), mRNA
NM_207362 Homo sapiens similar to 2010300C02Rik protein (MGC42367), mRNA
NM_207363 Homo sapiens Nck-associated protein 5 (NAP5), mRNA
NM_207364 Homo sapiens G protein-coupled receptor 148 (GPR148), mRNA
NM_207365 Homo sapiens similar to Arylacetamide deacetylase (AADAC) (MGC72001 ),
NM_207366 Homo sapiens FLJ44060 protein (FLJ44060), mRNA
NM_207367 Homo sapiens FLJ42291 protein (FLJ42291), mRNA
NM_207368 Homo sapiens hypothetical protein LOC348262 (LOC348262), mRNA
NM_207370 Homo sapiens G protein-coupled receptor 153 (GPR1 53), mRNA
NM_207371 Homo sapiens FLJ45187 protein (FLJ45187), mRNA
NM_207372 Homo sapiens SH2 domain containing 4B (SH2D4B), mRNA
NM_207373 Homo sapiens chromosome 10 open reading frame 99 (C10orf99), mRNA
NM_207374 Homo sapiens olfactory receptor (UNQ6469), mRNA
NM_207375 Homo sapiens INPE5792 (UNQ5792), mRNA
NM_207376 Homo sapiens hypothetical protein (LOC387882), mRNA
NM_207377 Homo sapiens TIMM9 (UNQ9438), mRNA
NM_207378 Homo sapiens serine (or cysteine) proteinase inhibitor, clade A (alpha-1 anti|
NM_207379 Homo sapiens FLJ42486 protein (FLJ42486), mRNA
NM_207380 Homo sapiens FLJ43339 protein (FLJ43339), mRNA
NM_207381 Homo sapiens FLJ41287 protein (FLJ41287), mRNA
NM_207382 Homo sapiens FLJ43276 protein (FLJ43276), mRNA
NM_207383 Homo sapiens FLJ42289 protein (FLJ42289), mRNA
NM_207384 Homo sapiens QRWT5810 (UNQ5810), mRNA
NM_207385 Homo sapiens FLJ26184 protein (FLJ26184), mRNA
NM_207386 Homo sapiens FLJ45455 protein (FLJ45455), mRNA
NM_207387 Homo sapiens FLJ35696 protein (FLJ35696), mRNA
NM_207388 Homo sapiens FLJ31222 protein (FLJ31222), mRNA
NM_207389 Homo sapiens FLJ44861 protein (FLJ44861), mRNA
NM_207390 Homo sapiens FLJ45910 protein (FLJ45910), mRNA
NM_207391 Homo sapiens FLJ45744 protein (FLJ45744), mRNA
NM_207392 Homo sapiens KIPV467 (UNQ467), mRNA
NM_207393 Homo sapiens insulin growth factor-like family member 3 (IGFL3), mRNA
NM_207394 Homo sapiens FLJ45949 protein (FLJ45949), mRNA
NM_207395 Homo sapiens FLJ45850 protein (FLJ45850), mRNA
NM_207396 Homo sapiens FLJ46380 protein (FLJ46380), mRNA
NM_207397 Homo sapiens EAPG6122 (UNQ6122), mRNA
NM_207398 Homo sapiens FLJ38822 protein (FLJ38822), mRNA
NM_207399 Homo sapiens FLJ36116 protein (FLJ36116), mRNA
NM_207400 Homo sapiens FLJ39739 protein (FLJ39739), mRNA
NM_207401 Homo sapiens FLJ45717 protein (FLJ45717), mRNA
NM_207403 Homo sapiens FLJ42986 protein (FLJ42986), mRNA
NM_207404 Homo sapiens FLJ45880 protein (FLJ45880), mRNA
NM_207405 Homo sapiens FLJ46481 protein (FLJ46481), mRNA
NM_207406 Homo sapiens FLJ43965 protein (FLJ43965), mRNA
NM_207407 Homo sapiens FLJ16046 protein (FLJ16046), mRNA
NM_207408 Homo sapiens FLJ27505 protein (FLJ27505), mRNA
NM_207409 Homo sapiens AAAL3045 (UNQ3045), mRNA
NM_207410 Homo sapiens IVFI9356 (UNQ9356), mRNA
NM_207411 Homo sapiens HARL2754 (UNQ2754), mRNA
NM_207412 Homo sapiens FLJ43582 protein (FLJ43582), mRNA
NM_207413 Homo sapiens RPLK9433 (UNQ9433), mRNA
NM_207414 Homo sapiens FLJ43860 protein (FLJ43860), mRNA
NM_207416 Homo sapiens FLJ44082 protein (FLJ44082), mRNA
NM_207417 Homo sapiens FLJ46082 protein (FLJ46082), mRNA
NM_207418 Homo sapiens Similar to RIKEN cDNA 2700049P18 gene (MGC57827), mRf
NM_207419 Homo sapiens C1q and tumor necrosis factor related protein 8 (C1QTNF8), r
NM_207420 Homo sapiens intestinal facultative glucose transporter 7 (SLC2A7), mRNA
NM_207421 Homo sapiens peptidylarginine deimiπase type 6 (PADI6), mRNA
NM_207422 Homo sapiens FLJ44635 protein (FLJ44635), mRNA
NM_207423 Homo sapiens FLJ45983 protein (FLJ45983), mRNA
NM_207424 Homo sapiens FLJ40536 protein (FLJ40536), mRNA
NM_207426 Homo sapiens FLJ46831 protein (FLJ46831), mRNA
NM_207427 Homo sapiens hypothetical gene supported by AY129010 (LOC399851), mR
NM_207428 Homo sapiens FLJ45212 protein (FLJ45212), mRNA
NM_207429 Homo sapiens FLJ45803 protein (FLJ45803), mRNA
NM_207430 Homo sapiens FLJ46266 protein (FLJ46266), mRNA
NM_207432 Homo sapiens FLJ45436 protein (FLJ45436), mRNA
NM_207433 Homo sapiens FLJ44874 protein (FLJ44874), mRNA
NM_207434 Homo sapiens FLJ46363 protein (FLJ46363), mRNA
NM_207435 Homo sapiens FLJ40142 protein (FLJ40142), mRNA
NM_207436 Homo sapiens FLJ42957 protein (FLJ42957), mRNA
NM_207437 Homo sapiens FLJ43486 protein (FLJ43486), mRNA
NM_207438 Homo sapiens FLJ43808 protein (FLJ43808), mRNA
NM_207439 Homo sapiens FLJ46358 protein (FLJ46358), mRNA NM_207440 Homo sapiens FLJ26443 protein (FLJ26443), mRNA NM_207441 Homo sapiens FLJ42220 protein (FLJ42220), mRNA NM_207442 Homo sapiens FLJ39779 protein (FLJ39779), mRNA NM_207443 Homo sapiens FLJ45244 protein (FLJ45244), mRNA NM_207444 Homo sapiens FLJ35695 protein (FLJ35695), mRNA NM_207445 Homo sapiens FLJ39531 protein (FLJ39531), mRNA NM_207446 Homo sapiens hypothetical gene supported by AK075564; BC0SO873 (LOC4 NM_207447 Homo sapiens IFMQ9370 (UNQ9370), mRNA NM_207448 Homo sapiens FLJ45256 protein (FLJ45256), mRNA NM_207449 Homo sapiens FLJ44674 protein (FLJ44674), mRNA NM_207450 Homo sapiens FLJ27243 protein (FLJ27243), mRNA NM_207451 Homo sapiens FLJ45121 protein (FLJ45121), mRNA NM_207452 Homo sapiens FLJ45200 protein (FLJ45200), mRNA NM_207453 Homo sapiens FLJ35934 protein (FLJ35934), mRNA NM_207454 Homo sapiens FLJ44815 protein (FLJ44815), mRNA NM_207458 Homo sapiens FLJ46026 protein (FLJ46026), mRNA NM_207459 Homo sapiens FLJ35767 protein (FLJ35767), mRNA NM_207460 Homo sapiens FLJ44313 protein (FLJ44313), mRNA NM_207461 Homo sapiens FLJ44881 protein (FLJ44881), mRNA NM_207462 Homo sapiens FLJ45684 protein (FLJ45684), mRNA NM_207463 Homo sapiens FLJ46230 protein (FLJ46230), mRNA NM_207464 Homo sapiens FLJ40008 protein (FLJ40008), mRNA NM_207465 Homo sapiens FLJ45337 protein (FLJ45337), mRNA NM_207466 Homo sapiens FLJ46489 protein (FLJ46489), mRNA NM_207467 Homo sapiens FLJ35530 protein (FLJ35530), mRNA NM_207468 Homo sapiens FLJ43505 protein (FLJ43505), mRNA NM_207469 Homo sapiens KFLL827 (UNQ827), mRNA NM_207470 Homo sapiens FLJ45832 protein (FLJ45832), mRNA NM_207471 Homo sapiens FLJ42200 protein (FLJ42200), mRNA NM_207472 Homo sapiens FLJ46020 protein (FLJ46020), mRNA NM_207473 Homo sapiens FLJ41733 protein (FLJ41733), mRNA NM_207474 Homo sapiens FLJ42953 protein (FLJ42953), mRNA NM_207475 Homo sapiens FLJ90680 protein (FLJ90680), mRNA NM_207477 Homo sapiens FLJ27365 protein (FLJ27365), mRNA NM_207478 Homo sapiens FLJ44385 protein (FLJ44385), mRNA NM_207479 Homo sapiens FLJ41046 protein (FLJ41046), mRNA NM_207480 Homo sapiens AILT5830 (UNQ5830), mRNA NM_207481 Homo sapiens FLJ34870 protein (FLJ34870), mRNA NM_207482 Homo sapiens FLJ44048 protein (FLJ44048), mRNA NM_207483 Homo sapiens FLJ45964 protein (FLJ45964), mRNA NM_207484 Homo sapiens FLJ40712 protein (FLJ40712), mRNA NM_207485 Homo sapiens FLJ41327 protein (FLJ41327), mRNA NM 207486 Homo sapiens FLJ44076 protein (FLJ44076), mRNA NMI207487 Homo sapiens FLJ46211 protein (FLJ46211), mRNA NM 207488 Homo sapiens FLJ42393 protein (FLJ42393), mRNA NM_207489 Homo sapiens FLJ35816 protein (FLJ35816), mRNA NM 207490 Homo sapiens FLJ45721 protein (FLJ45721), mRNA NMI207491 Homo sapiens similar to KIAA1680 protein (MGC48628), mRNA NM_207492 Homo sapiens FLJ44477 protein (FLJ44477) mRNA NM~207493 Homo sapiens FLJ44896 protein (FLJ44896) mRNA NM 207494 Homo sapiens FLJ40092 protein (FLJ40092) mRNA NM_207495 Homo sapiens hypothetical protein DKFZp686l15217 (DKFZp686l15217), ml NM_207496 Homo sapiens chromosome 6 open reading frame 214 (C6orf214), mRNA NM~207497 Homo sapiens FLJ43752 protein (FLJ43752), mRNA NM θ7498 Homo sapiens FLJ43093 protein (FLJ43093), mRNA NM 207499 Homo sapiens FLJ41841 protein (FLJ41841), mRNA
NM_207500 Homo sapiens FLJ44955 protein (FLJ44955), mRNA
NM_207501 Homo sapiens FLJ27255 protein (FLJ27255), mRNA
NM_207502 Homo sapiens chromosome 6 open reading frame 122 (C6orf122), mRNA
NM_207503 Homo sapiens FLJ42280 protein (FLJ42280), mRNA
NM_207504 Homo sapiens FLJ46365 protein (FLJ46365), mRNA
NM_207505 Homo sapiens FLJ45248 protein (FLJ45248), mRNA
NM_207506 Homo sapiens sterile alpha motif domain containing 12 (SAMD12), mRNA
NM_207507 Homo sapiens FLJ45202 protein (FLJ45202), mRNA
NM_207508 Homo sapiens FLJ45478 protein (FLJ45478), mRNA
NM_207509 Homo sapiens FLJ46836 protein (FLJ46836), mRNA
NM_207510 Homo sapiens FLJ45224 protein (FLJ45224), mRNA
NM_207511 Homo sapiens FLJ36268 protein (FLJ36268), mRNA
NM_207512 Homo sapiens nuclear RNA export factor-like (LOC401610), mRNA
NM_207513 Homo sapiens POTE14 (LOC404785), mRNA
NM_207514 Homo sapiens hypothetical protein FLJ20186 (FLJ20186), transcript variant •
NM_207517 Homo sapiens ADAMTS-like 3 (ADAMTSL3), mRNA
NM_207518 Homo sapiens protein kinase, cAMP-dependent, catalytic, alpha (PRKACA),
NM_207519 Homo sapiens zeta-chain (TCR) associated protein kinase 70kDa (ZAP70), t
NM_207520 Homo sapiens reticulon 4 (RTN4), transcript variant 4, mRNA
NM_207521 Homo sapiens reticulon 4 (RTN4), transcript variant 5, mRNA
NM_207577 Homo sapiens microtubule-associated protein 6 (MAP6), transcript variant 2,
NM_207578 Homo sapiens protein kinase, cAMP-dependent, catalytic, beta (PRKACB), ti
NM_207581 Homo sapiens similar to Numb-interacting homolog gene (LOC405753), mRI
NM_207582 Homo sapiens HERV-FRD provirus ancestral Env polyprotein (HERV-FRD), I
NM_207584 Homo sapiens interferon (alpha, beta and omega) receptor 2 (IFNAR2), trans
NM_207585 Homo sapiens interferon (alpha, beta and omega) receptor 2 (IFNAR2), trans
NM_207627 Homo sapiens ATP-binding cassette, sub-family G (WHITE), member 1 (ABC
NM_207628 Homo sapiens ATP-binding cassette, sub-family G (WHITE), member 1 (ABC
NM_207629 Homo sapiens ATP-binding cassette, sub-family G (WHITE), member 1 (ABC
NM_207630 Homo sapiens ATP-binding cassette, sub-family G (WHITE), member 1 (ABC
NM_207644 Homo sapiens similar to hypothetical protein LOC192734 (LOC388886), mRI
NM_207645 Homo sapiens similar to expressed sequence AI593442 (LOC399947), mRN
NM_207646 Homo sapiens eosinophil lysophospholipase-like (LOC400696), mRNA
NM_207647 Homo sapiens similar to fibronectin type 3 and SPRY domain-containing prol
NM_207660 Homo sapiens nuclear protein UKp68 (FLJ11806), transcript variant 2, mRN/
NM_207661 Homo sapiens nuclear protein UKp68 (FLJ11806), transcript variant 3, mRN/
NM_207662 Homo sapiens nuclear protein UKp68 (FLJ11806), transcript variant 4, mRN
NM_207672 Homo sapiens GRIP1 associated protein 1 (GRIPAP1), transcript variant 2, r
NM_212460 Homo sapiens ADP-ribosylation factor-like 4A (ARL4A), transcript variant 2, i
NM_212461 Homo sapiens protein kinase, AMP-activated, gamma 1 non-catalytic subuni
NM_212464 Homo sapiens calpain 3, (p94) (CAPN3), transcript variant 8, mRNA
NM_212465 Homo sapiens calpain 3, (p94) (CAPN3), transcript variant 7, mRNA
NM_212467 Homo sapiens calpain 3, (p94) (CAPN3), transcript variant 9, mRNA
NM_212469 Homo sapiens choline kinase alpha (CHKA), transcript variant 2, mRNA
NM_212471 Homo sapiens protein kinase, cAMP-dependent, regulatory, type I, alpha (tis:
NM_212472 Homo sapiens protein kinase, cAMP-dependent, regulatory, type I, alpha (tis.
NM_212474 Homo sapiens fibronectin 1 (FN1), transcript variant 6, mRNA
NM_212475 Homo sapiens fibronectin 1 (FN1), transcript variant 2, mRNA
NM_212476 Homo sapiens fibronectin 1 (FN1), transcript variant 5, mRNA
NM_212478 Homo sapiens fibronectin 1 (FN1 ), transcript variant 4, mRNA
NM_212479 Homo sapiens zinc finger, MYND domain containing 11 (ZMYND11), transcri
NM_212481 Homo sapiens AT rich interactive domain 5A (MRF1-like) (ARID5A), transcrif
NM_212482 Homo sapiens fibronectin 1 (FN1), transcript variant 1 , mRNA
NM_212492 Homo sapiens G protein pathway suppressor 1 (GPS1), transcript variant 1 , i
NM_212502 Homo sapiens PCTAIRE protein kinase 3 (PCTK3), transcript variant 2, mR
NM_212503 Homo sapiens PCTAIRE protein kinase 3 (PCTK3), transcript variant 1 , mRN
NM_212530 Homo sapiens cell division cycle 25B (CDC25B), transcript variant 5, mRNA
NM_212533 Homo sapiens ATP-binding cassette, sub-family A (ABC1 ), member 2 (ABC/
NM_212535 Homo sapiens protein kinase C, beta 1 (PRKCB1), transcript variant 1 , mRN,
NM_212539 Homo sapiens protein kinase C, delta (PRKCD), transcript variant 2, mRNA
NM_212540 Homo sapiens E2F transcription factor 6 (E2F6), transcript variant f, mRNA
NM_212543 Homo sapiens UDP-Gal:betaGlcNAc beta 1 ,4- galactosyltransferase, polypei
NM_212550 Homo sapiens biogenesis of lysosome-related organelles complex-1 , subunr
NM_212551 Homo sapiens hypothetical protein SB145 (SB145), mRNA
NM_212552 Homo sapiens similar to RIKEN 1810056020 (LOC388962), mRNA
NM_212553 Homo sapiens deubiquitinating enzyme DUB4 (DUB4), mRNA
NM_212554 Homo sapiens similar to CG9643-PA (LOC399818), mRNA
NM_212555 Homo sapiens LVLF3112 (UNQ3112), mRNA
NM_212556 Homo sapiens ankyrin repeat and SOCS box-containing 18 (ASB18), mRNA
NM_212557 Homo sapiens RSTI689 (UNQ689), mRNA
NM_212558 Homo sapiens similar to RIKEN A930001M12 (LOC401498), mRNA
NM_212559 Homo sapiens X Kell blood group precursor-related, X-linked (XKRX), mRNA
NM_213560 Homo sapiens protein kinase N1 (PKN1), transcript variant 1, mRNA
NM_213566 Homo sapiens DNA fragmentation factor, 45kDa, alpha polypeptide (DFFA),
NM_213568 Homo sapiens solute carrier family 39 (zinc transporter), member 3 (SLC39A
NM_213569 Homo sapiens nebulette (NEBL), transcript variant 2, mRNA
NM_213589 Homo sapiens Ras association (RalGDS/AF-6) and pleckstrin homology dorr
NM_213590 Homo sapiens ret finger protein 2 (RFP2), transcript variant 3, mRNA
NM_213593 Homo sapiens deiodinase, iodothyronine, type I (DI01), transcript variant 2, i
NM_213594 Homo sapiens regulatory factor X, 4 (influences HLA class II expression) (RF
NM_213596 Homo sapiens forkhead box N4 (F0XN4), mRNA
NM_213597 Homo sapiens hypothetical protein LOC124751 (LOC124751), mRNA
NM_213598 Homo sapiens zinc finger protein 543 (ZNF543), mRNA
NM_213599 Homo sapiens transmembrane protein 16E (TMEM16E), mRNA
NM_213600 Homo sapiens hypothetical protein LOC255189 (LOC255189), mRNA
NM_213601 Homo sapiens hypothetical protein LOC283578 (LOC283578), mRNA
NM_213602 Homo sapiens CD33 antigen-like 3 (CD33L3), mRNA
NM_213603 Homo sapiens hypothetical protein LOC285989 (LOC285989), mRNA
NM_213604 Homo sapiens thrombospondin, type I, domain containing 6 (THSD6), mRNA
NM_213605 Homo sapiens zinc finger protein 517 (ZNF517), mRNA
NM_213606 Homo sapiens similar monocarboxylate transporter (LOC387700), mRNA
NM_213607 Homo sapiens similar to RIKEN 4933439F11 (LOC388389), mRNA
NM_213608 Homo sapiens IIDS6411 (UNQ6411), mRNA
NM_213609 Homo sapiens TAFA1 protein (TAFA1), mRNA
NM_213611 Homo sapiens solute carrier family 25 (mitochondrial carrier; phosphate carri
NM_213612 Homo sapiens solute carrier family 25 (mitochondrial carrier; phosphate carri
NM_213613 Homo sapiens solute carrier family 26 (sulfate transporter), member 1 (SLC2
NM_213618 Homo sapiens suppression of tumorigenicity 5 (ST5), transcript variant 3, mF
NM_213619 Homo sapiens ATPase, H+ transporting, lysosomal 50/57kDa, V1 subunit H >
NM_213620 Homo sapiens ATPase, H+ transporting, lysosomal 50/57kDa, V1 subunit H *
NM_213621 Homo sapiens 5-hydroxytryptamine (serotonin) receptor 3A (HTR3A), transci
NM_213622 Homo sapiens STAM binding protein (STAMBP), transcript variant 3, mRNA
NM_213631 Homo sapiens chromosome 20 open reading frame 132 (C20orf132), transci
NM_213632 Homo sapiens chromosome 20 open reading frame 132 (C20orf132), transcr
NM_213633 Homo sapiens pregnancy specific beta-1 -glycoprotein 4 (PSG4), transcript vi
NM_213636 Homo sapiens PDZ and LIM domain 7 (enigma) (PDLIM7), transcript variant
NM_213645 Homo sapiens tryptophanyl-tRNA synthetase (WARS), transcript variant 3, rr
NM_213646 Homo sapiens tryptophanyl-tRNA synthetase (WARS), transcript variant 4, rr
NM_213647 Homo sapiens fibroblast growth factor receptor 4 (FGFR4), transcript variant
NM_213648 Homo sapiens transcription factor 7 (T-cell specific, HMG-box) (TCF7), transi
NM_213649 Homo sapiens sideroflexin 4 (SFXN4), transcript variant 1, mRNA
NM_213650 Homo sapiens sideroflexin 4 (SFXN4), transcript variant 3, mRNA
NM_213651 Homo sapiens solute carrier family 25 (mitochondrial carrier; phosphate carri
NM_213652 Homo sapiens hemochromatosis type 2 (juvenile) (HFE2), transcript variant <
NM_213653 Homo sapiens hemochromatosis type 2 (juvenile) (HFE2), transcript variant s
NM_213654 Homo sapiens armadillo repeat containing 8 (ARMC8), mRNA
NM_213655 Homo sapiens hereditary sensory neuropathy, type II (HSN2), mRNA
NM_213656 Homo sapiens type I hair keratin KA35 (KA35), mRNA
NM_213657 Homo sapiens killer cell lectin-like receptor subfamily C, member 1 (KLRC1 ),
NM_213658 Homo sapiens killer cell lectin-like receptor subfamily C, member 1 (KLRC1),
NM_213662 Homo sapiens signal transducer and activator of transcription 3 (acute-phase
NM_213674 Homo sapiens tropomyosin 2 (beta) (TPM2), transcript variant 2, mRNA
NM_213720 Homo sapiens chromosome 22 open reading frame 16 (C22orf16), mRNA
NM_213723 Homo sapiens chromosome 13 open reading frame 25 (C13orf25), transcript
NM_213724 Homo sapiens chromosome 13 open reading frame 25 (C13orf25), transcript
NM_213725 Homo sapiens ribosomal protein, large, P1 (RPLP1), transcript variant 2, R
NM_213726 Homo sapiens inhibitor of CDK interacting with cyclin A1 (INCA1 ), mRNA
NM_214461 Homo sapiens MGC50273 protein (MGC50273), mRNA
NM_214462 Homo sapiens dapper homolog 2, antagonist of beta-catenin (xenopus) (DAC
NM_214675 Homo sapiens CD209 antigen-like (CD209L), transcript variant 2, mRNA
NM_214676 Homo sapiens CD209 antigen-like (CD209L), transcript variant 3, mRNA
NM_214677 Homo sapiens CD209 antigen-like (CD209L), transcript variant 4, mRNA
NM_214678 Homo sapiens CD209 antigen-like (CD209L), transcript variant 5, mRNA
NM_214679 Homo sapiens CD209 antigen-like (CD209L), transcript variant 6, mRNA
NM_214710 Homo sapiens protease, serine-like 1 (PRSSL1), mRNA
NM_214711 Homo sapiens hypothetical LOC401137 (LOC401137), mRNA
XM_001279 Homo sapiens phosphoprotein enriched in astrocytes 15 (PEA15), mRNA
XM_001290 Homo sapiens ATP-binding cassette, sub-family A (ABC1), member 4 (ABC/
XM_001296 Homo sapiens cytosolic acyl coenzyme A thioester hydrolase (HBACH), mRt
XM_001322 Homo sapiens coagulation factor III (thromboplastin, tissue factor) (F3), mRr>
XM_001393 Homo sapiens peroxiredoxin 1 (PRDX1), mRNA
XM_001442 Homo sapiens phosphoglucomutase 1 (PGM1), mRNA
XM_001463 Homo sapiens ATPase, H+ transporting, lysosomal (vacuolar proton pump) 2
XM_001527 Homo sapiens polymyositis/scleroderma autoantigen 2 (100kD) (PMSCL2), r
XM_001541 Homo sapiens heterogeneous nuclear ribonucleoprotein R (HNRPR), mRNA
XM_001607 Homo sapiens growth arrest and DNA-damage-inducible, alpha (GADD45A),
XM_001644 Homo sapiens protein phosphatase 1, regulatory (inhibitor) subunit 8 (PPP1F
XM_001654 Homo sapiens chromosome 1 open reading frame 8 (C1orf8), mRNA
XM_001655 Homo sapiens HSPC034 protein (LOC51668), mRNA
XM_001677 Homo sapiens involucrin (IVL), mRNA
XM_001690 Homo sapiens cytochrome b5 reductase 1 (B5R.1) (LOC51706), mRNA
XM_007651 Homo sapiens similar to Sorbitol dehydrogenase (L-iditol 2-dehydrogenase) i
XM_010658 Homo sapiens similar to protein phosphatase 1 , regulatory (inhibitor) subunit
XM_012219 Homo sapiens similar to Phosphoglycerate mutase 1 (Phosphoglycerate mut
XM_015334 Homo sapiens family with sequence similarity 10, member A3 (FAM1 OA3), m
XM_015717 Homo sapiens similar to 40S ribosomal protein S7 (S8) (LOC149224), mRN/
XM_016093 Homo sapiens similar to eukaryotic initiation factor 5A isoform I variant A (LC
XM_016113 Homo sapiens similar to 40S ribosomal protein S10 (LOC158104), mRNA
XM_016532 Homo sapiens similar to hepatitis C virus core-binding protein 6; cervical can
XM_016548 Homo sapiens chromodomain protein, Y chromosome, 2 related (LOC20361
XM_016713 Homo sapiens similar to ribosomal protein S3a; 40S ribosomal protein S3a; \
XM_01 374 Homo sapiens similar to Nonhistone chromosomal protein HMG-17 (High-mc
XM_017661 Homo sapiens similar to 40S ribosomal protein S26 (LOC158200), mRNA
XM_017966 Homo sapiens similar to Reticulon protein 3 (Neuroendocrine-specific proteir
XM_018399 Homo sapiens hypothetical protein LOC144983 (LOC144983), mRNA
XM_018432 Homo sapiens similar to 60S ribosomal protein L7 (LOC146110), mRNA
XM_018487 Homo sapiens similar to omega protein (LOC91353), mRNA
XM_027045 Homo sapiens cut-like 2 (Drosophila) (CUTL2), mRNA
XM~027074 Homo sapiens l(3)mbt-like 3 (Drosophila) (L3MBTL3), mRNA
XM_027105 Homo sapiens KIAA0767 protein (KIAA0767), mRNA
XM_027162 Homo sapiens DMRT-like family A2 (DMRTA2), mRNA
XMJ327236 Homo sapiens tetratricopeptide repeat domain 9 (TTC9), mRNA
XM_027237 Homo sapiens mitogen-activated protein kinase kinase kinase 9 (MAP3K9), i
XM_027307 Homo sapiens pleckstrin homology domain containing, family G (with RhoGe
XM_027330 Homo sapiens RNA-binding region (RNP1 , RRM) containing 7 (RNPC7), mR
XM_027658 Homo sapiens fibronectin type III domain containing 1 (FNDC1), mRNA
XM_028067 Homo sapiens midnolin (MIDN), mRNA
XM_028217 Homo sapiens hypothetical LOC90024 (LOC90024), mRNA
XM_028253 Homo sapiens chromosome 19 open reading frame 7 (C19orf7), mRNA
XM_028413 Homo sapiens KIAA1374 protein (KIAA1374), mRNA
XM_028522 Homo sapiens myosin heavy chain Myr 8 (MYR8), mRNA
XM_028810 Homo sapiens KIAA1755 protein (KIAA1755), mRNA
XM_029084 Homo sapiens hypothetical protein FLJ21438 (FLJ21438), mRNA
XM_029101 Homo sapiens KIAA0947 protein (KIAA0947), mRNA
XM_029323 Homo sapiens hypothetical protein LOC90133 (LOC90133), mRNA
XM_029353 Homo sapiens KIAA1509 (KIAA1509), mRNA
XM_029429 Homo sapiens KIAA1328 protein (KIAA1328), mRNA
XM_029438 Homo sapiens KIAA0397 gene product (KIAA0397), mRNA
XM_029805 Homo sapiens similar to ribosomal protein L7 (LOC90193), mRNA
XM_029962 Homo sapiens potassium channel, subfamily T, member 1 (KCNT1), mRNA
XM_030300 Homo sapiens netrin receptor Unc5h1 (KIAA1976), mRNA
XM_030378 Homo sapiens zinc finger protein 527 (ZNF527), mRNA
XM_030445 Homo sapiens chromosome 10 open reading frame 75 (C10orf75), mRNA
XM_030559 Homo sapiens par-6 partitioning defective 6 homolog beta (C. elegans) (PAR
XM_030577 Homo sapiens ATPase, Class II, type 9A (ATP9A), mRNA
XM_030665 Homo sapiens KIAA1229 protein (KIAA1229), mRNA
XM_030669 Homo sapiens hypothetical protein LOC90288 (LOC90288), mRNA
XM_030729 Homo sapiens hypothetical protein DKFZp434M 117 (DKFZp434H 117), mR
XM_030892 Homo sapiens hypothetical protein LOC90317 (LOC90317), mRNA
XM_030893 Homo sapiens similar to ribosomal protein L37 (LOC147655), mRNA
XM_030896 Homo sapiens hypothetical protein LOC90321 (LOC90321), mRNA
XM_030958 Homo sapiens hypothetical protein LOC90333 (LOC90333), mRNA
XM_031009 Homo sapiens similar to fer-1 like protein 3 (LOC90342), mRNA
XM_031102 Homo sapiens WD repeat domain 22 (WDR22), mRNA
XM_031104 Homo sapiens UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylga
XM_031246 Homo sapiens roundabout, axon guidance receptor, homolog 2 (Drosophila)
XM_031342 Homo sapiens zinc finger, SWIM domain containing 4 (ZSWIM4), mRNA
XM_031357 Homo sapiens KIAA0802 protein (KIAA0802), mRNA
XM_031401 Homo sapiens EGF-like-domain, multiple 3 (EGFL3), mRNA
XM_031553 Homo sapiens U2-associated SR140 protein (SR140), mRNA
XM_031561 Homo sapiens TRAF4 associated factor 1 (FLJ14502), mRNA
XM_031689 Homo sapiens MAX gene associated (MGA), mRNA
XM_031706 Homo sapiens likely ortholog of mouse mitogen activated protein kinase bine
XM_031744 Homo sapiens START domain containing 9 (STARD9), mRNA
XM_031975 Homo sapiens similar to Ribulose-phosphate 3-epimerase (Ribulose-5-phosf
XM_032059 Homo sapiens similar to BC37295_3 (LOC90485), mRNA
XM_032181 Homo sapiens KIAA1233 protein (KIAA1233), mRNA
XM_032278 Homo sapiens signal-induced proliferation-associated 1 like 3 (SIPA1 L3), mF
XM_032397 Homo sapiens DKFZP564I122 protein (DKFZP564I122), mRNA
XM_032542 Homo sapiens FLJ41352 protein (FLJ41352), mRNA
XMJD32571 Homo sapiens KIAA0888 protein (KIAA0888), mRNA
XM_032678 Homo sapiens hypothetical protein LOC90576 (LOC90576), mRNA
XM_032693 Homo sapiens KIAA0420 gene product (KIAA0420), mRNA
XM_032812 Homo sapiens similar to hypothetical protein (LOC388506), mRNA
XM_032901 Homo sapiens KIAA0226 gene product (KIAA0226), mRNA
XM_032945 Homo sapiens chromosome 21 open reading frame 25 (C21orf25), mRNA
XM_032996 Homo sapiens KIAA0819 protein (KIAA0819), mRNA
XM_032997 Homo sapiens flavoprotein oxidoreductase MICAL3 (MICAL3), mRNA
XM_033113 Homo sapiens KIAA0789 gene product (KIAA0789), mRNA
XM_033173 Homo sapiens protocadherin 19 (PCDH19), mRNA
XM_033370 Homo sapiens zinc finger homeobox 2 (ZFHX2), mRNA
XM_033371 Homo sapiens chromosome 14 open reading frame 120 (C14orf120), mRNA
XM_ 033391 Homo sapiens protein phosphatase 1 , regulatory (inhibitor) subunit 3E (PPP-
XM_033704 Homo sapiens cDN A DKFZp434C184 gene (DKFZp434C184), mRNA
XM_033853 Homo sapiens hypothetical zinc finger protein FLJ20573 (FLJ20573), mRNA
XM_034086 Homo sapiens KIAA1107 protein (KIAA1107), mRNA
XM_034262 Homo sapiens KIAA1727 protein (KIAA1727), mRNA
XMJD34274 Homo sapiens v-myb myeloblastosis viral oncogene homolog (avian)-like 1 (I
XM_034594 Homo sapiens KIAA1604 protein (KIAA1604), mRNA
XM_034623 Homo sapiens similar to small nuclear ribonucleoprotein E (LOC158352), mF
XM_034640 Homo sapiens similar to ribosomal protein L4; 60S ribosomal protein L4; hon
XM_034717 Homo sapiens KIAA0493 protein (KIAA0493), mRNA
XM_034819 Homo sapiens KIAA0326 protein (KIAA0326), mRNA
XM_034872 Homo sapiens septin 8 (SEPT8), mRNA
XM_034904 Homo sapiens KIAA0912 protein (KIAA0912), mRNA
XM_035037 Homo sapiens low density lipoprotein receptor-related protein 4 (LRP4), mRf
XM_035299 Homo sapiens zinc finger, SWIM domain containing 6 (ZSWIM6), mRNA
XM_035371 Homo sapiens KIAA1643 protein (KI AA1643), mRNA
XM_035405 Homo sapiens KIAA1384 protein (KIAA1384), mRNA
XM_035497 Homo sapiens KIAA1602 protein (KIAA1602), mRNA
XM_035527 Homo sapiens hypothetical protein FLJ10980 (FLJ10980), mRNA
XM_035572 Homo sapiens chromosome 4 open reading frame 9 (C4orf9), mRNA
XM_035601 Homo sapiens SAP90/PSD-95-associated protein 3 (SAPAP3), mRNA
XM_035825 Homo sapiens KIAA0143 protein (KIAA0143), mRNA
XMJD35863 Homo sapiens zinc finger protein 37a (KOX 21 ) (ZNF37A), mRNA
XM_035946 Homo sapiens KIAA1613 protein (KIAA1613), mRNA
XM_035953 Homo sapiens chromosome 9 open reading frame 11 (C9orf11 ), mRNA
XM_036115 Homo sapiens KIAA1753 protein (KIAA1753), mRNA
XM_036218 Homo sapiens zinc finger protein 506 (ZNF506), mRNA
XM_036299 Homo sapiens KI AA1522 protein (KIAA1522), mRNA
XM_036612 Homo sapiens hypothetical LOC91170 (LOC91170), mRNA
XM_036708 Homo sapiens KIAA0368 (KIAA0368), mRNA
XM_036729 Homo sapiens ubiquitin specific protease 41 (USP41), mRNA
XM_036740 Homo sapiens nuclear pore membrane glycoprotein 210-like (LOC91181), m
XM_036936 Homo sapiens KIAA1666 protein (KIAA1666), mRNA
XM_036942 Homo sapiens similar to hypothetical protein (LOC150221), mRNA
XM_036988 Homo sapiens KIAA1000 protein (KIAA1000), mRNA
XM_037493 Homo sapiens SH3 and multiple ankyrin repeat domains 3 (SHANK3), mRN/
XM_037523 Homo sapiens KIAA1076 protein (KIAA1076), mRNA
XM_037557 Homo sapiens KIAA0984 protein (KIAA0984), mRNA
XM_037759 Homo sapiens KIAA0376 protein (KIAA0376), mRNA
XM_037817 Homo sapiens hypothetical protein FLJ31033 (FLJ31033), mRNA
XM_038063 Homo sapiens UDP-N-acteylglucosamine pyrophosphorylase 1-like 1 (UAP1
XM_038150 Homo sapiens microtubule associated serine/threonine kinase 3 (MAST3), rr
XM_038288 Homo sapiens KIAA0191 protein (KIAA0191), mRNA
XM_038291 Homo sapiens hypothetical protein FLJ13456 (FLJ13456), mRNA
XM_038298 Homo sapiens hypothetical protein DKFZp434E2321 (DKFZp434E2321), mF
XM_038436 Homo sapiens KIAA1786 protein (KIAA1786), mRNA
XM_038520 Homo sapiens KIAA0542 gene product (KIAA0542), mRNA
XM_038567 Homo sapiens metastasis associated family, member 3 (MTA3), mRNA
XM_038576 Homo sapiens hypothetical protein BC007901 (LOC91461), mRNA
XM_038604 Homo sapiens unc-13 homolog A (C. elegans) (UNC13A), mRNA
XM_038664 Homo sapiens KIAA0564 protein (KIAA0564), mRNA
XM_038920 Homo sapiens Nedd4 binding protein 3 (N4BP3), mRNA
XM_038999 Homo sapiens NEDD4-related E3 ubiquitin ligase NEDL2 (NEDL2), mRNA
XM_039169 Homo sapiens KI AA1276 protein (KIAA1276), mRNA
XMJD39218 Homo sapiens similar to ribosomal protein S2; 40S ribosomal protein S2 (LO
XM_039385 Homo sapiens KI AA1093 protein (KIAA1093), mRNA
XM_039393 Homo sapiens plexin A4 (PLXNA4), mRNA
XM_039495 Homo sapiens DNA segment, Chr 15, Wayne State University 75, expressed
XM_039515 Homo sapiens G2 protein (G2), mRNA
XM_039548 Homo sapiens SMYD family member 5 (SMYD5), mRNA
XM_039570 Homo sapiens SEC15-like 2 (S. cerevisiae) (SEC15L2), mRNA
XMJD39627 Homo sapiens contactin 3 (plasmacytoma associated) (CNTN3), mRNA
XM_039676 Homo sapiens KIAA1240 protein (KIAA1240), mRNA
XM_039698 Homo sapiens KI AA1432 (KIAA1432), mRNA
XM_039702 Homo sapiens similar to ribosomal protein S3a; 40S ribosomal protein S3a; \
XM_039721 Homo sapiens similar to MGC5244 protein (LOC91632), mRNA
XM_039733 Homo sapiens KIAA0953 (KIAA0953), mRNA
XM_039762 Homo sapiens myelin transcription factor 1-like (MYT1 L), mRNA
XM_039796 Homo sapiens Traf2 and NCK interacting kinase (K1AA0551 ), mRNA
XM_039877 Homo sapiens mucin 5, subtype B, tracheobronchial (MUC5B), mRNA
XM_039908 Homo sapiens hypothetical protein BC007307 (LOC91664), mRNA
XM_039922 Homo sapiens similar to FLJ00050 protein (LOC401873), mRNA
XM_040149 Homo sapiens similar to E74-like factor 2 (ets domain transcription factor); n<
XM_040265 Homo sapiens KIAA0217 protein (KIAA0217), mRNA
XM_040383 Homo sapiens KI AA1677 (KI AA1677), mRNA
XM_040486 Homo sapiens KIAA1789 protein (KIAA1789), mRNA
XM_040527 Homo sapiens tenascin N (TNN), mRNA
XM_040592 Homo sapiens zinc finger protein 469 (ZNF469), mRNA
XM_040910 Homo sapiens chromosome 14 open reading frame 73 (C14orf73), mRNA
XM_041018 Homo sapiens KI AA0367 (KIAA0367), mRNA
XM_041020 Homo sapiens similar to protein 40kD (LOC158473), mRNA
XM_041116 Homo sapiens chromosome 14 open reading frame 171 (C14orf171), mRNA
XM_041126 Homo sapiens KIAA1486 protein (KIAA1486), mRNA
XM_041162 Homo sapiens Nedd4 family interacting protein 2 (NDFIP2), mRNA
XM_041191 Homo sapiens KI AA0931 protein (KIAA0931 ), mRNA
XM_041221 Homo sapiens similar to RNA-binding protein S1 , serine-rich domain; SR pro
XM_041363 Homo sapiens likely ortholog of mouse semaF cytoplasmic domain associate
XM_041964 Homo sapiens KIAA0523 protein (KIAA0523), mRNA
XM_042066 Homo sapiens mitogen-activated protein kinase kinase kinase 1 (MAP3K1 ), i
XM_0421 8 Homo sapiens similar to AKAP-binding sperm protein ropporin (LOC152015)
XMJD42234 Homo sapiens similar to RIKEN cDNA 4933437K13 (LOC92017), mRNA
XM_042301 Homo sapiens KIAA1546 protein (K1AA1546), mRNA
XM_042323 Homo sapiens calmodulin binding transcription activator 1 (CAMTA1), mRNA
XM_042500 Homo sapiens similar to ribosomal protein S2; 40S ribosomal protein S2 (LO
XM_042635 Homo sapiens KIAA1069 protein (KIAA1069), mRNA
XM_042661 Homo sapiens KIAA1530 protein (KIAA1530), mRNA
XM_042685 Homo sapiens KIAA1414 protein (KIAA1414), mRNA
XMJ342698 Homo sapiens ubiquitin specific protease 22 (USP22), mRNA
XM_042833 Homo sapiens KIAA0295 protein (KIAA0295), mRNA
XM_042936 Homo sapiens glutamate receptor interacting protein 2 (GRIP2), mRNA
XM_042978 Homo sapiens KIAA1817 protein (KIAA1817), mRNA
XM_043118 Homo sapiens KIAA0286 protein (KIAA0286), mRNA
XM_043272 Homo sapiens KIAA0346 protein (KIAA0346), mRNA
XM_043492 Homo sapiens KIAA1728 protein (KIAA1728), mRNA
XM_043493 Homo sapiens synaptic vesicle protein 2C (SV2C), mRNA
XM_043500 Homo sapiens similar to death-associated protein (LOC92196), mRNA
XM_043613 Homo sapiens glutamate receptor, ionotropic, delta 1 (GRID1), mRNA
XM_043624 Homo sapiens hypothetical protein DKFZp434E1822 (DKFZp434E1822), mF
XM_043653 Homo sapiens hypothetical protein FLJ10097 (FLJ10097), mRNA
XMJ343739 Homo sapiens hypothetical cardiac/skeletal muscle-expressed ORF (LOC92!
XM_043863 Homo sapiens hypothetical protein DKFZp434H2226 (DKFZp434H2226), mF
XM_043979 Homo sapiens similar to FLJ 12363 protein (LOC92267), mRNA
XM_043989 Homo sapiens hypothetical protein LOC92270 (LOC92270), mRNA
XM_044062 Homo sapiens hypothetical protein DKFZp76102018 (DKFZp76102018), mf
XM_044166 Homo sapiens hypothetical protein LOC92312 (L0C92312), mRNA
XM_044178 Homo sapiens KIAA1211 protein (KIAA1211), mRNA
XM_044196 Homo sapiens DKFZP434C212 protein (DKFZP434C212), mRNA
XM_044212 Homo sapiens KIAA1862 protein (KIAA1862), mRNA
XM_044334 Homo sapiens RIM binding protein 2 (KIAA0318), mRNA
XM_044434 Homo sapiens KIAA1458 protein (KIAA1458), mRNA
X _044461 Homo sapiens KIAA1102 protein (KIAA1102), mRNA
XM_044580 Homo sapiens KIAA1024 protein (KIAA1024), mRNA
XM_044622 Homo sapiens collagen, type XIV, alpha 1 (undulin) (COL14A1), mRNA
XM_044632 Homo sapiens KIAA0556 protein (KIAA0556), mRNA
XM_044727 Homo sapiens myotubularin related protein 7 (MTMR7), mRNA
XM_044836 Homo sapiens KIAA1340 protein (KI AA1340), mRNA
XM_044921 Homo sapiens KIAA1442 protein (KIAA1442), mRNA
XM_045086 Homo sapiens KIAA1764 protein (KIAA1764), mRNA
XM_045113 Homo sapiens astrotactin (ASTN), mRNA
XM_045271 Homo sapiens KIAA1580 protein (KIAA1580), mRNA
XM_045283 Homo sapiens similar to IK cytokine; arginine/glutamic acid/aspartic acid repi
XM_045290 Homo sapiens similar to basic leucine zipper and W2 domains 1 (LOC15157
XM_045308 Homo sapiens PHD finger protein 19 (PHF19), mRNA
XM_045421 Homo sapiens chromosome 20 open reading frame 194 (C20orf194), mRNA
XM_045423 Homo sapiens KIAA0701 protein (KIAA0701), mRNA
XM_045581 Homo sapiens likely ortholog of mouse 5-azacytidine induced gene 1 (AZI1),
XM_045705 Homo sapiens similar to homologue of MJD, high homology to a genomic se*
XM_045712 Homo sapiens KIAA0316 gene product (KIAA0316), mRNA
XM_045787 Homo sapiens hypothetical protein LOC92558 (LOC92558), mRNA
XM_045792 Homo sapiens GCN1 general control of amino-acid synthesis 1-like 1 (yeast)
XM_045907 Homo sapiens KIAA1170 protein (KIAA1170), mRNA
XM_045911 Homo sapiens tomosyn-like (KIAA1006), mRNA
XM_046099 Homo sapiens similar to small nuclear ribonucleoprotein E (LOC148064), mF
XM_046264 Homo sapiens DKFZP434B172 protein (DKFZP434B172), mRNA
XM_046305 Homo sapiens KIAA1205 (KIAA1205), mRNA
XM_046390 Homo sapiens zinc finger protein 473 (ZNF473), mRNA
XM_046437 Homo sapiens chromosome 20 open reading frame 50 (C20orf50), mRNA
XM_046531 Homo sapiens KIAA1614 protein (KIAA1614), mRNA
XM_046570 Homo sapiens KIAA1679 protein (KIAA1679), mRNA
XM_046581 Homo sapiens zinc finger, SWIM domain containing 5 (ZSWIM5), mRNA
XM_046600 Homo sapiens KIAA1272 protein (KI AA1272), mRNA
XM_046677 Homo sapiens solute carrier family 39 (zinc transporter), member 14 (SLC39
XM_046685 Homo sapiens KIAA1399 protein (KIAA1399), mRNA
XM_046751 Homo sapiens protein tyrosine phosphatase, receptor type, f polypeptide (PT
XM_046808 Homo sapiens neurofascin (NFASC), mRNA
XM_046861 Homo sapiens KRAB box containing C2H2 type zinc finger bA526D8.4 (BA5!
XM_047025 Homo sapiens ornithine aminotransferase-like 1 (OATL1 ), mRNA
XM_047083 Homo sapiens similar to tubulin, beta 5 (LOC92755), mRNA
XM_047214 Homo sapiens KIAA0930 protein (KIAA0930), mRNA
XM_047325 Homo sapiens THO complex 2 (THOC2), mRNA
XM_047355 Homo sapiens KIAA1765 protein (KIAA1765), mRNA
XM_047357 Homo sapiens KIAA0342 gene product (KIAA0342), mRNA
XM_047462 Homo sapiens Spir-2 protein (Spir-2), mRNA
XM_047499 Homo sapiens hypothetical protein LOC149603 (LOC149603), mRNA
XMJD47550 Homo sapiens zinc finger protein 492 (ZNF492), mRNA
XM_047554 Homo sapiens similar to Zinc finger protein 492 (LOC148198), mRNA
XM_047610 Homo sapiens KIAA1086 (KIAA1086), mRNA
XM_047617 Homo sapiens KIAA1349 protein (KIAA1349), mRNA
XMJD47707 Homo sapiens solute carrier family 39 (zinc transporter), member 10 (SLC39
XM_047734 Homo sapiens similar to hect domain and RLD 2 (LOC146489), mRNA
XM_047770 Homo sapiens similar to alpha2-glucosyltransferase (LOC144245), mRNA
XM_047995 Homo sapiens odd Oz/ten-m homolog 2 (ODZ2), mRNA
XM_048070 Homo sapiens zinc finger protein 292 (ZNF292), mRNA
XM_048104 Homo sapiens filaggrin (FLG), mRNA
XM_048128 Homo sapiens KIAA1596 (KIAA1596), mRNA
XM_048235 Homo sapiens Huntingtin interacting protein M (HYPM), mRNA
XM_048362 Homo sapiens KIAA1543 (KIAA1543), mRNA
XM_048462 Homo sapiens RUN and SH3 domain containing 2 (RUSC2), mRNA
XM_048592 Homo sapiens KI AA1045 (KI AA1045), mRNA
XM_048675 Homo sapiens KI AA1238 protein (KI AA1238), mRNA
XM_048721 Homo sapiens hypothetical protein DKFZp762K222 (DKFZp762K222), mRN;
XM_048747 Homo sapiens KIAA1223 protein (KI AA1223), mRNA
XM_048774 Homo sapiens KIAA1332 protein (KIAA1332), mRNA
XM_048786 Homo sapiens K1AA1061 protein (KIAA1061), mRNA
XM_048825 Homo sapiens KI AA1026 protein (KI AA1026), mRNA
XM_048898 Homo sapiens heat shock 70kDa protein 12A (HSPA12A), mRNA
XM_049037 Homo sapiens trinucleotide repeat containing 9 (TNRC9), mRNA
XM_049078 Homo sapiens KI AA1239 protein (KI AA1239), mRNA
XM_049237 Homo sapiens KIAA0841 (KIAA0841), mRNA
XM_049349 Homo sapiens KIAA0534 protein (KIAA0534), mRNA
XM_049351 Homo sapiens KIAA1600 protein (KIAA1600), mRNA
XM_049380 Homo sapiens KIAA0339 gene product (KIAA0339), mRNA
XM_049384 Homo sapiens chromosome 7 open reading frame 3 (C7orf3), mRNA
XM D49575 Homo sapiens similar to succinate dehydrogenase flavoprotein subunit (LOG
XM_049619 Homo sapiens PR domain containing 6 (PRDM6), mRNA
XM_049695 Homo sapiens vang-like 2 (van gogh, Drosophila) (VANGL2), mRNA
XM_049952 Homo sapiens hypothetical protein FLJ23529 (FLJ23529), mRNA
XM_050041 Homo sapiens myosin ID (MY01 D), mRNA
XM_050219 Homo sapiens synaptopodin 2 (SYNP02), mRNA
XM_050278 Homo sapiens kinesin family member 26A (KIF26A), mRNA
XM_050325 Homo sapiens KI AA1126 protein (KI AA1126), mRNA
XM_050478 Homo sapiens KIAA1202 protein (KIAA1202), mRNA
XM_050561 Homo sapiens SIN3 homolog B, transcriptional regulator (yeast) (SIN3B), mF
XM_050564 Homo sapiens similar to RIKEN cDNA 2410004L22 gene (M. musculus) (MG
XM_050625 Homo sapiens secreted frizzled-related protein 2 (SFRP2), mRNA
XM_050644 Homo sapiens Ki AA1623 (KIAA1623), mRNA
XM_050846 Homo sapiens Indian hedgehog homolog (Drosophila) (IHH), mRNA
XM_051017 Homo sapiens KIAA0657 protein (KIAA0657), mRNA
XM_051081 Homo sapiens KIAA0608 protein (KIAA0608), mRNA
XM_051091 Homo sapiens KIAA1040 protein (KIAA1040), mRNA
XM_051197 Homo sapiens KIAA1005 protein (KIAA1005), mRNA
XM_051200 Homo sapiens fatso (FTO), mRNA
XM_051221 Homo sapiens SPHK1 (sphingosine kinase type 1) interacting protein (SKIP)
XM_051264 Homo sapiens thioredoxin reductase 3 (TXNRD3), mRNA
XM_051271 Homo sapiens family with sequence similarity 10, member A6 (FAM10A6),
XM_051699 Homo sapiens KIAA1344 (KIAA1344), mRNA
XM_051862 Homo sapiens hypothetical protein from EUROIMAGE 588495 (LOC58489),
XM_051956 Homo sapiens similar to KIAA0592 protein (LOC387680), mRNA
XM_052561 Homo sapiens KIAA1337 protein (KIAA1337), mRNA
XM_052597 Homo sapiens ubiquitin specific protease 53 (USP53), mRNA
XM_052620 Homo sapiens mannosidase alpha class 2B member 2 (KIAA0935), mRNA
XM_053074 Homo sapiens translocase of inner mitochondrial membrane 50 homolog (ye
XM_053177 Homo sapiens similar to alpha tubulin (LOC112714), mRNA
XM_053966 Homo sapiens hypothetical protein LOC113230 (LOC113230), mRNA
XM_054284 Homo sapiens alpha-tubulin isotype H2-alpha (H2-ALPHA), mRNA
XM_054313 Homo sapiens similar to T-complex protein 1 (LOC155100), mRNA
XM_054983 Homo sapiens KIAA1952 protein (KIAA1952), mRNA
XM_055095 Homo sapiens KIAA1906 protein (KIAA1906), mRNA
XM_055481 Homo sapiens KIAA1915 protein (KIAA1915), mRNA
XM J55636 Homo sapiens KIAA1912 protein (KIAA1912), mRNA
XM_055725 Homo sapiens similar to unc-93 homolog B1 ; unc93 (C.elegans) homolog B;
XM_055866 Homo sapiens lemur tyrosine kinase 3 (LMTK3), mRNA
XM_056254 Homo sapiens heparan sulfate (glucosamine) 3-O-sulfotransferase 4 (HS3S1
XM_056282 Homo sapiens KI AA1904 protein (KI AA1904), mRNA
XM_056298 Homo sapiens KI AA1889 protein (KI AA1889), mRNA
XM_056434 Homo sapiens tetratricopeptide repeat domain 6 (TTC6), mRNA
XM_056455 Homo sapiens Melanoma associated gene (D2S448), mRNA
XM_056680 Homo sapiens hypothetical protein LOC115749 (LOC115749), mRNA
XM_056681 Homo sapiens similar to ribosomal protein L14; 60S ribosomal protein L14 (L
XM_057040 Homo sapiens KIAA1922 protein (KIAA1922), mRNA
XM_057107 Homo sapiens KI AA1937 protein (KIAA1937), mRNA
XM_057296 Homo sapiens hypothetical protein LOC116064 (LOC116064), mRNA
XM_058332 Homo sapiens similar to hypothetical protein MGC45962 (LOC118670), mRf>
XM_058335 Homo sapiens similar to ARF GTPase-activating protein (LOC118704), mRN
XM_058404 Homo sapiens hypothetical protein LOC119548 (LOC119548), mRNA
XM_058426 Homo sapiens hypothetical protein FLJ00012 (FLJ00012), mRNA
XM_058513 Homo sapiens hypothetical protein DKFZp434H2111 (DKFZp434H2111 ), mF
XM_058581 Homo sapiens similar to hypothetical protein 9530023G02 (LOC121642), mF
XMJ358611 Homo sapiens hypothetical LOC150928 (LOC150928), mRNA
XM_058628 Homo sapiens chromosome 14 open reading frame 109 (C14orf109), mRNA
XM_058661 Homo sapiens chromosome 14 open reading frame 35 (C14orf35), mRNA
XM_058677 Homo sapiens similar to 60S ribosomal protein L21 (LOC123031), mRNA
XM_058719 Homo sapiens similar to RIKEN cDNA C630028N24 gene (LOC123688), mR
XM_058720 Homo sapiens similar to junction-mediating and regulatory protein p300 JMY
XM_058721 Homo sapiens hypothetical protein LOC123722 (LOC123722), mRNA
XM_058743 Homo sapiens hypothetical protein LOC123876 (LOC123876), mRNA
XM_058857 Homo sapiens hypothetical LOC124871 (LOC124871 ), mRNA
XM_058879 Homo sapiens hypothetical protein LOC124976 (LOC124976), mRNA
XM_058931 Homo sapiens similar to hypothetical protein B230399E16 (LOC125704), mF
XM_058956 Homo sapiens Purkinje cell protein 2 (PCP2), mRNA
XM_058961 Homo sapiens trafficking protein particle complex 5 (TRAPPC5), mRNA
XM_058964 Homo sapiens hypothetical protein LOC89887 (LOC89887), mRNA
XM_058967 Homo sapiens similar to Elongation factor 1-delta (EF-1-delta) (Antigen NY-C
XM_058997 Homo sapiens hypothetical protein LOC126167 (LOC126167), mRNA
XM_058999 Homo sapiens hypothetical protein LOC126208 (LOC126208), mRNA
XM_059037 Homo sapiens similar to LIM domains containing 1 (LOC126374), mRNA
XM_059047 Homo sapiens hypothetical LOC126435 (LOC126435), mRNA
XM_059051 Homo sapiens hypothetical protein LOC126520 (LOC126520), mRNA
XM_059061 Homo sapiens hypothetical protein LOC126661 (LOC126661), mRNA
XM_059074 Homo sapiens hypothetical protein LOC126755 (LOC126755), mRNA
XM_059095 Homo sapiens formin binding protein 2 (FNBP2), mRNA
XM_059104 Homo sapiens similar to CG5435-PA (LOC127003), mRNA
XM_059132 Homo sapiens similar to RIKEN cDNA 4930549C01 (LOC127309), mRNA
XM_059140 Homo sapiens similar to dJ39G22.2 (novel protein) (LOC127391), mRNA
XM_059166 Homo sapiens similar to KIAA1697 protein (LOC127602), mRNA
XM_059256 Homo sapiens hypothetical LOC128499 (LOC128499), mRNA
XM_059267 Homo sapiens similar to RIKEN cDNA 2210009G21 (LOC128710), mRNA
XM_059318 Homo sapiens KIAA1941 protein (KIAA1941), mRNA
XM_059341 Homo sapiens hypothetical protein LOC129293 (LOC129293), mRNA
XM_059368 Homo sapiens hypothetical protein LOC129607 (LOC129607), mRNA
XM_059384 Homo sapiens hypothetical LOC129881 (LOC129881), mRNA
XMJD59396 Homo sapiens hypothetical LOC130063 (LOC130063), mRNA
XM_059399 Homo sapiens similar to Calcium and integrin-binding protein 1 (Calmyrin) (C
XM_059438 Homo sapiens similar to CG14894-PA (LOC130502), mRNA
XM_059461 Homo sapiens similar to RIKEN cDNA A230078I05 gene (LOC130612), mRh
XM_059462 Homo sapiens hypothetical LOC130643 (LOC130643), mRNA
XM_059473 Homo sapiens hypothetical LOC130839 (LOC130839), mRNA
XM_059482 Homo sapiens FLJ00133 protein (FLJ00133), mRNA
XM_059492 Homo sapiens hypothetical LOC131076 (LOCI 31076), mRNA
XM_059548 Homo sapiens similar to SRSR846 (LOC131920), mRNA
XM_059578 Homo sapiens similar to hypothetical protein A430083B19 (LOC132203), mF
XM_059598 Homo sapiens similar to unc-93 homolog B1 ; unc93 (C.elegans) homolog B;
XM_059608 Homo sapiens hypothetical LOC132870 (LOC132870), mRNA
XM_059617 Homo sapiens similar to MGC69138 protein (LOC132946), mRNA
XM_059669 Homo sapiens similar to ribosomal protein S3a; 40S ribosomal protein S3a; \
XM_059672 Homo sapiens hypothetical LOC133874 (LOC133874), mRNA
XM_059689 Homo sapiens similar to 3110006E14Rik protein (LOC134111), mRNA
XM_059702 Homo sapiens hypothetical protein FLJ36748 (FLJ36748), mRNA
XM_059729 Homo sapiens interleukin-1 receptor-associated kinase 1 binding protein 1 (I
XM_059730 Homo sapiens chromosome 6 open reading frame 159 (C6orf159), mRNA
XM_059776 Homo sapiens FK506 binding protein 1 C (FKBP1 C), mRNA
XM_059830 Homo sapiens similar to RIKEN cDNA 1700016G05 (LOC136242), mRNA
XM_059832 Homo sapiens hypothetical protein LOC136288 (LOC136288), mRNA
XM_059909 Homo sapiens hypothetical LOC137485 (LOC137485), mRNA
XM_059923 Homo sapiens similar to Nuclear receptor binding factor-2 (LOC137829), mR
XM_059929 Homo sapiens hypothetical protein LOC137886 (LOC137886), mRNA
XM_059954 Homo sapiens chromosome 9 open reading frame 57 (C9orf57), mRNA
XM_059956 Homo sapiens similar to RIKEN cDNA 1700028P14 (LOC138255), mRNA
XM_059972 Homo sapiens hypothetical protein LOC138428 (LOC138428), mRNA
XM_059987 Homo sapiens ankyrin repeat domain 19 (ANKRD19), mRNA
XM_060020 Homo sapiens hypothetical protein BC016683 (LOC139231 ), mRNA
XM_060054 Homo sapiens similar to XAGE-5 protein (LOC139793), mRNA
XM_060087 Homo sapiens similar to template acyivating factor-l alpha (LOC126598), mF
XM_060104 Homo sapiens similar to RIKEN cDNA 5430400H23 (LOC126637), mRNA
XM_060171 Homo sapiens similar to erythrocyte membrane-associated giant protein anti*
XM_060278 Homo sapiens similar to tight junction protein 3 (zona occludens 3) (LOC126
XM_060301 Homo sapiens similar to Olfactory receptor 2M6 (LOC127059), mRNA
XM_060303 Homo sapiens similar to Olfactory receptor 2M6 (LOC127062), mRNA
XM_060305 Homo sapiens similar to seven transmembrane helix receptor (LOC127064),
XM_060307 Homo sapiens similar to Olfactory receptor 5BF1 (LOC127066), mRNA
XM_060309 Homo sapiens similar to seven transmembrane helix receptor (LOC127068),
XMJ360310 Homo sapiens similar to seven transmembrane helix receptor (LOC127069),
XM_060315 Homo sapiens similar to Olfactory receptor 2T4 (LOC127074), mRNA
XM_060316 Homo sapiens olfactory receptor, family 2, subfamily T, member 1 (OR2T1 ),
XM_060318 Homo sapiens similar to Olfactory receptor 2T11 (LOC127077), mRNA
XM_060328 Homo sapiens similar to 60S ACIDIC RIBOSOMAL PROTEIN P1 (LOC1270Ϊ
XM_060417 Homo sapiens similar to 60S ribosomal protein L36 (LOC127295), mRNA
XM_060458 Homo sapiens similar to Olfactory receptor 10J5 (LOC127385), mRNA
XM_060509 Homo sapiens S100 calcium binding protein A7-like 2 (S100A7L2), mRNA
XM_060535 Homo sapiens similar to ribosomal protein L18a; 60S ribosomal protein L18a
XM_060537 Homo sapiens similar to iGb3 synthase (LOC127550), mRNA
XM_060563 Homo sapiens similar to seven transmembrane helix receptor (LOC127608),
XM_060569 Homo sapiens similar to seven transmembrane helix receptor (LOC127614),
XMJ360572 Homo sapiens similar to Olfactory receptor 5AV1 (LOC127617), mRNA
XM_060580 Homo sapiens similar to olfactory receptor GA_x6K02SYYHDF-1415-2371 (L
XM_060597 Homo sapiens similar to zinc finger protein 135 (clone pHZ-17); zinc finger pi
XM_060880 Homo sapiens similar to dJ675G8.1 (novel zinc finger protein) (LOC128208),
XM_060887 Homo sapiens similar to peptidyl-Pro cis trans isomerase (LOC128192), mRI
XM_060943 Homo sapiens similar to Nuclear transport factor 2 (NTF-2) (Placental proteir
XM_060945 Homo sapiens similar to Olfactory receptor 10T2 (LOC128360), mRNA
XM_060951 Homo sapiens similar to Olfactory receptor 6P1 (LOC128366), mRNA
XM_060952 Homo sapiens similar to seven transmembrane helix receptor (LOC128367),
XM_060953 Homo sapiens similar to Olfactory receptor 10Z1 (LOC128368), mRNA
XM_060955 Homo sapiens similar to seven transmembrane helix receptor (LOC128370),
XM_060956 Homo sapiens similar to Olfactory receptor 6K6 (LOC128371 ), mRNA
XM_060957 Homo sapiens similar to Olfactory receptor 6N1 (LOC128372), mRNA
XM_060970 Homo sapiens paired related homeobox-like 1 (PRRXL1), mRNA
XM_061055 Homo sapiens hypothetical protein FLJ32938 (FLJ32938), mRNA
XM_061222 Homo sapiens similar to hypothetical protein 9930115F20 (LOC118934), mR
XM_061427 Homo sapiens similar to Small nuclear ribonucleoprotein Sm D2 (snRNP con
XM_061542 Homo sapiens similar to 40S ribosomal protein S8 (LOC119563), mRNA
XM_061562 Homo sapiens similar to RIKEN cDNA 4632411J06 (LOC119593), mRNA
XM_061610 Homo sapiens similar to Olfactory receptor 52E2 (LOC119678), mRNA
XM_061611 Homo sapiens similar to Olfactory receptor 52J3 (LOC119679), mRNA
XM_061614 Homo sapiens similar to Olfactory receptor 51L1 (LOC119682), mRNA
XM_061619 Homo sapiens similar to Olfactory receptor 51A7 (LOC119687), mRNA
XM_061624 Homo sapiens similar to Olfactory receptor 51 S1 (LOC119692), mRNA
XM_061626 Homo sapiens similar to Olfactory receptor 51 F2 (LOC119694), mRNA
XM_061627 Homo sapiens similar to Olfactory receptor 52R1 (LOC119695), mRNA
XM_061628 Homo sapiens similar to seven transmembrane helix receptor (LOC119696),
XM_061656 Homo sapiens similar to olfactory receptor MOR232-3 (LOC119749), mRNA
XM_061666 Homo sapiens similar to Olfactory receptor 4X2 (LOC119764), mRNA
XM_061674 Homo sapiens similar to seven transmembrane helix receptor (LOC119772),
XM_061676 Homo sapiens similar to Olfactory receptor 52K2 (LOC119774), mRNA
XM_061677 Homo sapiens similar to seven transmembrane helix receptor (LOC119775),
XM_061849 Homo sapiens similar to FLJ10251 protein (LOC120082), mRNA
XM_061864 Homo sapiens similar to fat3; fat3 protein (LOC120105), mRNA
XM_061871 Homo sapiens FAT tumor suppressor homolog 3 (Drosophila) (FAT3), mRN/
XM_061880 Homo sapiens similar to autoantigen NOR-90 (LOC120126), mRNA
XM_061888 Homo sapiens similar to autoantigen NOR-90 (LOC120144), mRNA
XM_061890 Homo sapiens similar to tripartite motif-containing 43 (LOC120146), mRNA
XM_061930 Homo sapiens similar to Homeobox protein DBX1 (LOC120237), mRNA
XM_062025 Homo sapiens similar to Hnrpal protein (LOC120364), mRNA
XM_062162 Homo sapiens similar to Olfactory receptor 8I2 (LOC120586), mRNA
XM_062263 Homo sapiens similar to seven transmembrane helix receptor (LOC120787),
XM_062269 Homo sapiens similar to Olfactory receptor 56A4 (LOC120793), mRNA
XM_062272 Homo sapiens similar to Olfactory receptor 56A1 (LOC120796), mRNA
XM_062285 Homo sapiens similar to Olfactory receptor 2D3 (LOC120775), mRNA
XM_062286 Homo sapiens similar to seven transmembrane helix receptor (LOC120776),
XM_062300 Homo sapiens similar to RING finger protein 18 (Testis-specific ring-finger pr
XM_062437 Homo sapiens similar to Keratin, type I cytoskeletal 18 (Cytokeratin 18) (K18
XM_062467 Homo sapiens similar to seven transmembrane helix receptor (LOC121129),
XM_062468 Homo sapiens similar to seven transmembrane helix receptor (LOC121130),
XM_062520 Homo sapiens similar to Sucrase-isomaltase, intestinal (LOC121216), mRNA
XM_062553 Homo sapiens similar to Olfactory receptor 10AD1 (LOC121275), mRNA
XM_062594 Homo sapiens similar to seven transmembrane helix receptor (LOC121360),
XM_062598 Homo sapiens similar to Olfactory receptor 10A7 (LOC121364), mRNA
XM_062645 Homo sapiens similar to solute carrier family 9, member 7; nonselective sodi
XM_062735 Homo sapiens forkhead box N4 (FOXN4), mRNA
XM_062788 Homo sapiens similar to histidine-rich protein (LOC121792), mRNA
XM_062871 Homo sapiens hypothetical protein FLJ40176 (FLJ40176), mRNA
XMJ362872 Homo sapiens hypothetical protein LOC121952 (LOC121952), mRNA
XM_062890 Homo sapiens similar to peptidyl-Pro cis trans isomerase (LOC121981 ), mRI
XM_062912 Homo sapiens similar to Mitochondrial import receptor subunit TOM22 homol
XM_062966 Homo sapiens similar to MAP/microtubule affinity-regulating kinase 3 (LOCH
XM_063084 Homo sapiens similar to peptidylprolyl isomerase A (cyclophilin A) (LOC1222
XM_063123 Homo sapiens similar to hypothetical protein ZC477.8 - Caenorhabditis elegj
XM_063138 Homo sapiens similar to RIKEN cDNA 1110013H04 (LOC122438), mRNA
XM_063202 Homo sapiens similar to 60S ribosomal protein L23a (LOC122585), mRNA
XM_063287 Homo sapiens similar to RIKEN cDNA 5830406J20 (LOC122706), mRNA
XM_063308 Homo sapiens similar to Olfactory receptor 4K14 (LOC122740), mRNA
XM_063310 Homo sapiens similar to Olfactory receptor 4L1 (LOC122742), mRNA
XM_063315 Homo sapiens similar to Olfactory receptor 11H6 (LOC122748), mRNA
XM_063336 Homo sapiens similar to cytochrome c oxidase subunit IV (COXIV) pseudoge
XM_063481 Homo sapiens similar to hypothetical gene supported by AK044523 (LOC12J
XM_063630 Homo sapiens similar to 60S ribosomal protein L29 (Cell surface heparin bin*
XM J63871 Homo sapiens similar to ENSANGP00000013733 (LOC123855), mRNA
XM_063919 Homo sapiens similar to neuronal nonacetlycholine binding subunit (LOC123
XM J64003 Homo sapiens similar to KIAA0565 protein (LOC124149), mRNA
XM_064062 Homo sapiens similar to putative G-protein coupled receptor (LOC124274), r
XM_064152 Homo sapiens sarcalumenin (SRL), mRNA
XM_0641 7 Homo sapiens similar to Olfactory receptor 4D2 (LOC124538), mRNA
XM_064190 Homo sapiens hypothetical protein FLJ40311 (FLJ40311), mRNA
XM_064257 Homo sapiens similar to HESB like domain containing 2 (LOC124667), mRN
XM D64265 Homo sapiens similar to smooth muscle and non-muscle myosin alkali light c
XM_064298 Homo sapiens hypothetical protein LOC124751 (LOC124751), mRNA
XM_064333 Homo sapiens hypothetical protein LOC124842 (LOC124842), mRNA
XM_064689 Homo sapiens hypothetical LOC125595 (LOC125595), mRNA
XM_064856 Homo sapiens hypothetical protein LOC125893 (LOC125893), mRNA
XM_064859 Homo sapiens similar to 40S ribosomal protein S15a (LOC125910), mRNA
XM_064865 Homo sapiens zinc finger protein 543 (ZNF543), mRNA
XM_064879 Homo sapiens similar to seven transmembrane helix receptor (LOC125958),
XM_064883 Homo sapiens similar to Olfactory receptor 7G1 (Olfactory receptor 19-15) (C
XM__064884 Homo sapiens similar to Olfactory receptor 1 M1 (Olfactory receptor 19-6) (01
XM_064903 Homo sapiens similar to KIAA2033 protein (LOC126017), mRNA
XM_065006 Homo sapiens similar to ribosomal protein S4, X-linked (LOC126235), mRN/
XM_065026 Homo sapiens similar to FKSG27 (LOC126298), mRNA
XM_065050 Homo sapiens similar to Olfactory receptor 111 (Olfactory receptor 19-20) (OI
XM_065124 Homo sapiens similar to zinc finger protein 91 (HPF7, HTF10) (LOC126502),
XM__065153 Homo sapiens similar to Olfactory receptor 10H4 (LOC126541), mRNA
XM_065166 Homo sapiens KIAA1957 (KIAA1957), mRNA
XM_065237 Homo sapiens similar to FKSG30 (LOC129439), mRNA
XM_ 065278 Homo sapiens similar to hypothetical protein (LOC129521), mRNA
XM_065316 Homo sapiens similar to acidic integral membrane protein (LOC129614), mR
XM__065332 Homo sapiens similar to MECT1 protein (LOC129656), mRNA
XM_065348 Homo sapiens casprδ protein (casprδ), mRNA
XM_065416 Homo sapiens similar to fibulin 1 isoform C precursor (LOC129804), mRNA
XM__065445 Homo sapiens similar to autoantigen NOR-90 (LOC129870), mRNA
XM_065555 Homo sapiens similar to Olfactory receptor 9A4 (LOC130075), mRNA
XM_065722 Homo sapiens similar to 40S ribosomal protein SA (P40) (34/67 kDa laminin
XM_065743 Homo sapiens similar to SWI/SNF-related matrix-associated actin-dependen
XM_065750 Homo sapiens similar to Uncharacterized hematopoietic stem/progenitor cell.
XM_065828 Homo sapiens similar to 60S acidic ribosomal protein P1 (LOC130678), mRf
XM_065899 Homo sapiens similar to 60S ribosomal protein L23a (LOC130773), mRNA
XM_065998 Homo sapiens chromosome 20 open reading frame 148 (C20orf148), mRNA
XM_066003 Homo sapiens chromosome 20 open reading frame 122 (C20orf122), mRNA
XM_066040 Homo sapiens ribosomal protein S4-like (RPS4L), mRNA
XM_066058 Homo sapiens chromosome 20 open reading frame 174 (C20orf174), mRNA
XM_066069 Homo sapiens similar to hypothetical protein (LOC128629), mRNA
XM_066102 Homo sapiens ribosomal protein L7a like 2 (RPL7AL2), mRNA
XM_066139 Homo sapiens ribosomal protein L7a-like 3 (RPL7AL3), mRNA
XM_066162 Homo sapiens solute carrier family 25 (mitochondrial carrier; adenine nucleo'
XM 366176 Homo sapiens similar to bA218C14.1 (novel protein similar to mouse cystatir
XM_066177 Homo sapiens similar to bA218C14.1 (novel protein similar to mouse cystatir
XM_066189 Homo sapiens gamma-glutamyltransferase-like activity 3 (GGTLA3), mRNA
XM_066243 Homo sapiens hypothetical LOC128939 (LOC128939), mRNA
XM_066339 Homo sapiens hypothetical protein similar to topoisomerase (DNA) III beta (F
XM_066350 Homo sapiens similar to Ovis aries Y chromosome repeat region OY11.1 (3'(
XM_066351 Homo sapiens hypothetical gene similar to gamma-glutamyltransferase-like s
XM_066443 Homo sapiens similar to hypothetical protein MGC15827 (LOC139046), mRf
XMJD66452 Homo sapiens similar to plasmolipin (LOC139061 ), mRNA
XM_066457 Homo sapiens similar to SPANX N member 2 (LOC139067), mRNA
XM_066469 Homo sapiens similar to MAGE family testis and tumor-specific protein (LOC
XM_066484 Homo sapiens similar to testis expressed sequence 13A (LOC139116), mRN
XM_066534 Homo sapiens similar to Diacylglycerol kinase, delta (Diglyceride kinase) (DC
XM_066585 Homo sapiens similar to testis expressed sequence 13A (LOC139263), mRN
XM_066621 Homo sapiens similar to envelope protein (LOC139302), mRNA
XM_066685 Homo sapiens similar to KIAA1387 protein (LOC139420), mRNA
XM_066690 Homo sapiens similar to H326 (LOC139425), mRNA
XM_066695 Homo sapiens similar to ferritin heavy chain - chicken (LOC139431), mRNA
XM_066701 Homo sapiens similar to melanoma antigen, family B, 4; melanoma-associati
XM_066752 Homo sapiens similar to E2F transcription factor 6 isoform a (LOC139542), n
XM_066765 Homo sapiens similar to bA351 K23.4 (novel protein) (LOC139562), mRNA
XM_066859 Homo sapiens similar to zinc finger protein 92 (LOC139735), mRNA
XM_066946 Homo sapiens hypothetical protein LOC139886 (LOC139886), mRNA
XM_067076 Homo sapiens similar to testis specific protein, Y-linked (LOC140103), mRN/
XM_067176 Homo sapiens similar to peptidyl-Pro cis trans isomerase (LOC131055), mRI
XM_067193 Homo sapiens similar to microtubule-associated protein 6 (LOC131086), mR
XM_067228 Homo sapiens similar to otolin-1 (LOC131149), mRNA
XM_067369 Homo sapiens similar to abnormal cell LINeage LIN-41 , heterochronic gene;
XM_067448 Homo sapiens similar to MEST (LOC131572), mRNA
XM_067503 Homo sapiens similar to peptidyl-Pro cis trans isomerase (LOC131691), mRI
XM_067585 Homo sapiens hypothetical protein LOC131873 (LOC131873), mRNA
XM_067605 Homo sapiens similar to hypothetical protein (LOC131909), mRNA
XM_067904 Homo sapiens similar to Transcription factor BTF3 homolog 3 (LOC132556),
XM_067994 Homo sapiens similar to heat shock factor binding protein 1 (LOC132706), m
XM_068121 Homo sapiens similar to hornerin (LOC132969), mRNA
XM_068229 Homo sapiens similar to 9530003A05 protein (LOC133185), mRNA
XM_068430 Homo sapiens similar to 60S acidic ribosomal protein P1 (LOC133609), mRf
XM_068602 Homo sapiens hypothetical LOC133923 (LOC133923), mRNA
XM_068632 Homo sapiens similar to hypothetical protein MGC52498 (LOC133993), mRf
XM_068681 Homo sapiens similar to seven transmembrane helix receptor (LOC134082),
XM_068682 Homo sapiens similar to Olfactory receptor 2Y1 (LOC134083), mRNA
XM_068889 Homo sapiens similar to eukaryotic translation initiation factor 3 subunit k; mi
XM_068903 Homo sapiens similar to Ten-m2 (LOC134541), mRNA
XM_069035 Homo sapiens chromosome 6 open reading frame 213 (C6orf213), mRNA
XM_069595 Homo sapiens similar to Olfactory receptor 4F3 (LOC135896), mRNA
XM_069609 Homo sapiens similar to Olfactory receptor 9A2 (LOC135924), mRNA
XM_069612 Homo sapiens similar to OG-2 homeodomain protein-like; similar to U65067
XM_069616 Homo sapiens similar to seven transmembrane helix receptor (LOC135941),
XM_069619 Homo sapiens similar to olfactory receptor MOR261-13 (LOC135944), mRN/
XM_069621 Homo sapiens similar to Olfactory receptor 6B1 (Olfactory receptor 7-3) (OR;
XM_069623 Homo sapiens similar to Olfactory receptor 2F2 (Olfactory receptor 7-1 ) (OR]
XM_069728 Homo sapiens similar to beta-glucuronidase (LOC136132), mRNA
XM_069734 Homo sapiens similar to ribosomal protein L18; 60S ribosomal protein L18 (L
XM_069743 Homo sapiens similar to RIKEN cDNA 1500011 L16 (LOC136157), mRNA
XM_069842 Homo sapiens similar to 60S ribosomal protein L15 (LOC136321), mRNA
XM_070233 Homo sapiens similar to ribosomal protein L10a (LOC137107), mRNA
XM_070277 Homo sapiens otoconin 90 (OC90), mRNA
XMJ370619 Homo sapiens similar to homeobox protein NFOC2-6 (LOC137814), mRNA
XM_071013 Homo sapiens similar to bA62C3.1 (similar to testicular serine protease) (LO*
XM_071061 Homo sapiens AT rich interactive domain 3C (BRIGHT- like) (ARID3C), mRN
XM_071093 Homo sapiens similar to Olfactory receptor 13C5 (LOC138799), mRNA
XM_071096 Homo sapiens similar to Olfactory receptor 13C8 (LOC138802), mRNA
XM_071097 Homo sapiens similar to Olfactory receptor 13C3 (LOC138803), mRNA
XM_071098 Homo sapiens similar to Olfactory receptor 13C4 (LOC138804), mRNA
XM_071099 Homo sapiens similar to Olfactory receptor 13F1 (LOC138805), mRNA
XM_071150 Homo sapiens similar to Olfactory receptor 1 L8 (LOC138881), mRNA
XM_071151 Homo sapiens similar to Olfactory receptor 1 N2 (LOC138882), mRNA
XM_071173 Homo sapiens similar to Hkrlp (LOC138932), mRNA
XM_071201 Homo sapiens similar to Centaurin gamma 2 (LOC138972), mRNA
XM_071712 Homo sapiens hypothetical protein LOC120376 (LOC120376), mRNA
XM_071793 Homo sapiens chromosome 14 open reading frame 28 (C14orf28), mRNA
XM_071866 Homo sapiens cerebellar degeneration-related protein 2, 62kDa (CDR2), mR
XM_072402 Homo sapiens aminoacylase 1 -like 2 (ACY1 L2), mRNA
XM_072554 Homo sapiens similar to RIKEN cDNA 4833436C18 gene (LOC138729), mR
XM_084000 Homo sapiens mitochondrial carrier triple repeat 2 (MCART2), mRNA
XMJ384357 Homo sapiens similar to Hypothetical protein MGC56918 (LOC142827), mRf
XM_084377 Homo sapiens similar to Triacylglycerol lipase, gastric precursor (Gastric lipa
XM_084445 Homo sapiens similar to ARF GTPase-activating protein (LOC143158), mRN
XM_084467 Homo sapiens similar to eukaryotic initiation factor 5A isoform I variant A (LC
XM_084482 Homo sapiens AT rich interactive domain 5B (MRF1-like) (ARID5B), mRNA
XM_084514 Homo sapiens heat shock 90kDa protein 1 , alpha-like 3 (HSPCAL3), mRNA
XM_084529 Homo sapiens KIAA0298 gene product (KIAA0298), mRNA
XM_084530 Homo sapiens KIAA0033 protein (KIAA0033), mRNA
XM_084578 Homo sapiens PTPRF interacting protein, binding protein 2 (liprin beta 2) (PF
XM_084672 Homo sapiens similar to CDNA sequence BC021608 (LOC143941 ), mRNA
XM_084845 Homo sapiens similar to Interferon-induced transmembrane protein 3 (Interfe
XM_084852 Homo sapiens hypothetical LOC144404 (LOC144404), mRNA
XM_084868 Homo sapiens similar to MGC76214 protein (LOC144448), mRNA
XM_084990 Homo sapiens hypothetical LOC144962 (LOC144962), mRNA
XM_085028 Homo sapiens ATPase, Class VI, type 11 A (ATP11 A), mRNA
XM_085127 Homo sapiens KIAA0599 (KIAA0599), mRNA
XM_085138 Homo sapiens similar to ribosomal protein L3; 60S ribosomal protein L3; HIV
XM_085175 Homo sapiens tetratricopeptide repeat domain 7 like 1 (TTC7L1), mRNA
XM_085200 Homo sapiens hypothetical LOC145660 (LOC145660), mRNA
XM_085231 Homo sapiens hypothetical protein LOC145783 (LOC145783), mRNA
XM_085234 Homo sapiens unc-13 homolog C (C. elegans) (UNC13C), mRNA
XM_085236 Homo sapiens hypothetical LOC145788 (LOC145788), mRNA
XM_085261 Homo sapiens mesoderm posterior 2 (MESP2), mRNA
XM_085290 Homo sapiens similar to golgin-67 isoform c (LOC145988), mRNA
XM_085316 Homo sapiens similar to RIKEN cDNA 1810007E14; EST AA238765 (LOC14
XM_085347 Homo sapiens similar to hypothetical protein FLJ10815 (LOC146167), mRN/
XM_085367 Homo sapiens FLJ40162 protein (FLJ40162), mRNA
XM_085375 Homo sapiens zinc finger protein 90 homolog (mouse) (ZFP90), mRNA
XM_085383 Homo sapiens hypothetical protein LOC146206 (LOC146206), mRNA
XM_085463 Homo sapiens similar to CDNA sequence BC038613 (LOC146439), mRNA
XM_085507 Homo sapiens zinc finger protein 500 (ZNF500), mRNA
XM_085517 Homo sapiens hypothetical LOC146599 (LOC146599), mRNA
XM_085578 Homo sapiens FLJ46675 protein (FLJ46675), mRNA
XM_085596 Homo sapiens zinc finger protein 18 (KOX 11) (ZNF18), mRNA
XM_085606 Homo sapiens similar to CDRT15 protein (LOC146822), mRNA
XM_085634 Homo sapiens hypothetical protein LOC146909 (LOC146909), mRNA
XM_085689 Homo sapiens potassium channel tetramerisation domain containing 11 (KC
XM_085722 Homo sapiens similar to Tripartite motif protein 16 (Estrogen-responsive B b(
XM_085724 Homo sapiens hypothetical LOC147151 (LOC147151), mRNA
XM_085775 Homo sapiens similar to 60S ribosomal protein L7a (Surfeit locus protein 3) (
XM_085777 Homo sapiens similar to additional sex combs like 2; polycomb group protein
XM_085824 Homo sapiens hypothetical protein LOC147650 (LOC147650), mRNA
XM_085830 Homo sapiens hypothetical LOC147649 (LOC147649), mRNA
XM_085831 Homo sapiens hypothetical protein LOC147645 (LOC147645), mRNA
XM_085833 Homo sapiens hypothetical protein LOC147646 (LOC147646), mRNA
XM_085836 Homo sapiens KIAA1956 protein (KI AA1956), mRNA
XMJ385851 Homo sapiens similar to zinc finger protein 285 (LOC147711), mRNA
XM_085870 Homo sapiens similar to complement C3 protein (GPC3) precursor (LOC147
XM_085929 Homo sapiens Meisl , myeloid ecotropic viral integration site 1 homolog 3 (m
XMJ385932 Homo sapiens similar to VPRI645 (LOC147920), mRNA
XM_085967 Homo sapiens hypothetical LOC147942 (LOC147942), mRNA
XM_086001 Homo sapiens similar to Placental tissue protein 13 (Placenta protein 13) (G;
XM_086046 Homo sapiens hypothetical protein FLJ30663 (FLJ30663), mRNA
XM_086095 Homo sapiens hypothetical protein LOC148203 (LOC148203), mRNA
XM_086186 Homo sapiens hypothetical protein FLJ13815 (FLJ13815), mRNA
XM_086188 Homo sapiens dnaj-like protein (LOC148418), mRNA
XM_086257 Homo sapiens similar to ribosomal protein S15; rat insulinoma gene (LOC14;
XM_086287 Homo sapiens similar to Osteotesticular phosphatase; protein tyrosine phosp
XM_086308 Homo sapiens hypothetical LOC148766 (LOC148766), mRNA
XMJD86343 Homo sapiens similar to 60S ribosomal protein L17 (L23) (LOC148854), mRI
XM_086344 Homo sapiens similar to LIM homeo domain transcription factor (LOC148864
XM_086360 Homo sapiens hypothetical LOC148915 (LOC148915), mRNA
XM_086402 Homo sapiens hypothetical LOC149018 (LOC149018), mRNA
XM_086409 Homo sapiens KIAA2025 protein (KIAA2025), mRNA
XM_086494 Homo sapiens similar to similarity to monoubiquitin/carboxy-extension proteir
XM_086604 Homo sapiens similar to CHIA protein (LOC149620), mRNA
XM_086616 Homo sapiens hypothetical LOC149643 (LOC149643), mRNA
XM_086622 Homo sapiens hypothetical LOC149659 (LOC149659), mRNA
XM_086637 Homo sapiens similar to RIKEN cDNA 1700049M11 (LOC149709), mRNA
XM_086648 Homo sapiens similar to dJ579F20.1 (high-mobility group (nonhistone chrom
XM_086650 Homo sapiens protein phosphatase 4, regulatory subunit 1-like (PPP4R1 L), r
XM_086725 Homo sapiens similar to bB329D4.2.1 (novel protein similar to a truncated ni
XM_086732 Homo sapiens hypothetical LOC149950 (LOC149950), mRNA
XM_086761 Homo sapiens hypothetical protein LOC150084 (LOC150084), mRNA
XM_086826 Homo sapiens hypothetical protein LOC150368 (LOC150368), mRNA
XM_086876 Homo sapiens similar to MGC5244 protein (LOC150207), mRNA
XM_086879 Homo sapiens hypothetical LOC150371 (LOC150371 ), mRNA
XM_086894 Homo sapiens hypothetical protein LOC150297 (LOC150297), mRNA
XM_086905 Homo sapiens similar to RIKEN cDNA 2210021 J22 (LOC150383), mRNA
XM_086931 Homo sapiens similar to epsilon isoform of 14-3-3 protein (LOC150498), mRI
XM_086937 Homo sapiens similar to hypothetical protein A230046P18 (LOC150519), mF
XM_086996 Homo sapiens hypothetical protein LOC150763 (LOC150763), mRNA
XM_087056 Homo sapiens KIAA1841 protein (KIAA1841 ), mRNA
XM_087062 Homo sapiens similar to 60S acidic ribosomal protein P1 (LOC150978), mRf
XM_087089 Homo sapiens KIAA0007 protein (KIAA0007), mRNA
XM_087097 Homo sapiens hypothetical LOC151111 (LOC151111), mRNA
XM_087137 Homo sapiens protein phosphatase 1 regulatory subunit 1A (LOC151242), rr
XM_087141 Homo sapiens hypothetical LOC151256 (LOC151256), mRNA
XM_087167 Homo sapiens similar to KIAA1641 protein (LOC389008), mRNA
XM_087171 Homo sapiens myeloid-associated differentiation marker-like (MYADML), mR
XM_087182 Homo sapiens hypothetical LOC151363 (LOC151363), mRNA
XM_087200 Homo sapiens hypothetical LOC151443 (LOC151443), mRNA
XM_087208 Homo sapiens hypothetical LOC151451 (LOC151451), mRNA
XM_087225 Homo sapiens similar to male-specific lethal 3-like 1 isoform a; drosophila M!
XM_087254 Homo sapiens ATPase, Class VI, type 11B (ATP11 B), mRNA
XM_087353 Homo sapiens KIAA0794 protein (KIAA0794), mRNA
XM_087384 Homo sapiens hypothetical protein LOC152098 (LOC152098), mRNA
XM_087386 Homo sapiens HEG homolog (HEG), mRNA
XM_087483 Homo sapiens hypothetical protein LOC152519 (LOC152519), mRNA
XM_087490 Homo sapiens similar to RIKEN cDNA 4933434I20 (LOC152586), mRNA
XM_087499 Homo sapiens similar to 60S ribosomal protein L7a (Surfeit locus protein 3) (
XM_087500 Homo sapiens similar to NEFA-interacting nuclear protein NIP30 (LOC15266
XM_087553 Homo sapiens similar to WW45 protein (LOC152891 ), mRNA
XM_087593 Homo sapiens K1AA1430 protein (KIAA1430), mRNA
XM_087671 Homo sapiens hypothetical LOC153441 (LOC153441), mRNA
XM_087672 Homo sapiens KIAA1935 protein (KI AA1935), mRNA
XM_087761 Homo sapiens similar to protein related with psoriasis (LOC153770), mRNA
XM_087762 Homo sapiens hypothetical LOC153778 (LOC153778), mRNA
XM_087800 Homo sapiens similar to CGI-62 protein (LOC153918), mRNA
XM_087804 Homo sapiens synaptotagmin-like 3 (SYTL3), mRNA
XM_087859 Homo sapiens similar to 60S ribosomal protein L21 (LOC154165), mRNA
XM_087901 Homo sapiens similar to RIKEN cDNA 2410004A20 (LOC154288), mRNA
XM_087928 Homo sapiens hypothetical protein LOC154449 (LOC154449), mRNA
XM_088066 Homo sapiens similar to 60S ribosomal protein L35 (LOC154880), mRNA
XM_088072 Homo sapiens hypothetical LOC154907 (LOC154907), mRNA
XM_088118 Homo sapiens family with sequence similarity 10, member A7 (FAM10A7), m
XM_088140 Homo sapiens hypothetical protein LOC155054 (LOC155054), mRNA
XM_088142 Homo sapiens chromosome 7 open reading frame 32 (C7orf32), mRNA
XM_088143 Homo sapiens similar to hypothetical protein 4931409K22 (LOC155046), mR
XM_088315 Homo sapiens KIAA0870 protein (KIAA0870), mRNA
XM_088331 Homo sapiens hypothetical protein LOC157570 (LOC157570), mRNA
XM_088367 Homo sapiens similar to SPC18 protein (LOC157708), mRNA
XM_088376 Homo sapiens chromosome 8 open reading frame 7 (C8orf7), mRNA
XM_088459 Homo sapiens KIAA0310 (KIAA0310), mRNA
XM_088491 Homo sapiens similar to Olfactory receptor 1Q1 (Olfactory receptor TPCR101
XM_088516 Homo sapiens hypothetical LOC158226 (LOC158226), mRNA
XM_088525 Homo sapiens chromosome 9 open reading frame 28 (C9orf28), mRNA
XM_088551 Homo sapiens KIAA2026 (KIAA2026), mRNA
XM_088566 Homo sapiens K1AA1958 (KIAA1958), mRNA
XM_088567 Homo sapiens zinc finger protein 483 (ZNF483), mRNA
XM_088578 Homo sapiens RAD26L hypothetical protein, alternatively spliced product; sir
XM_088636 Homo sapiens cylicin, basic protein of sperm head cytoskeleton 1 (CYLC1), i
XM_088677 Homo sapiens similar to UPF3 regulator of nonsense transcripts homolog B I
XM_088679 Homo sapiens hypothetical LOC158812 (LOC158812), mRNA
XM_088680 Homo sapiens hypothetical LOC158813 (LOC158813), mRNA
XM_088683 Homo sapiens similar to bA351 K23.5 (novel protein) (LOC158835), mRNA
XM_088684 Homo sapiens similar to Ab2-183 (LOC158830), mRNA
XM_088686 Homo sapiens hypothetical LOC158825 (LOC158825), mRNA
XM_088691 Homo sapiens hypothetical protein LOC158833 (LOC158833), mRNA
XM_088726 Homo sapiens hypothetical LOC158957 (LOC158957), mRNA
XM_088735 Homo sapiens hypothetical protein LOC158983 (LOC158983), mRNA
XM_088768 Homo sapiens similar to F-box only protein 25 (LOC159176), mRNA
XM_088797 Homo sapiens similar to BC035954 protein (LOC163301 ), mRNA
XM_088817 Homo sapiens similar to Hypothetical protein DJ845024.1 (LOC374949), mF
XM_088951 Homo sapiens olfactomedin 3 (OLFM3), mRNA
XM_089081 Homo sapiens deleted in neuroblastoma 5 (DNB5), mRNA
XM_089243 Homo sapiens similar to cDNA sequence BC022623 (LOC163933), mRNA
XM_089281 Homo sapiens similar to AD-003 protein (LOC149281), mRNA
XM_089307 Homo sapiens similar to implantation-related RGS2-like protein (LOC164036
XM_089384 Homo sapiens similar to RIKEN cDNA A430025D11 (LOC164118), mRNA
XM_089747 Homo sapiens hypothetical protein FLJ35908 (FLJ35908), mRNA
XM_089858 Homo sapiens similar to Olfactory receptor 52B4 (LOC143496), mRNA
XM_089863 Homo sapiens similar to Olfactory receptor 52I2 (LOC143502), mRNA
XM_089866 Homo sapiens similar to RIKEN cDNA 1500011L16 (LOC143506), mRNA
XM_090203 Homo sapiens similar to Olfactory receptor 2AG1 (HT3) (LOC144125), mRN;
XM_090294 Homo sapiens hypothetical protein FLJ38508 (FLJ38508), mRNA
XM )90844 Homo sapiens hypothetical protein LOC161291 (LOC161291), mRNA
XM_090885 Homo sapiens chromosome 14 open reading frame 42 (C14orf42), mRNA
XM_091156 Homo sapiens similar to Adenosine deaminase CG11994-PA (LOC161823),
XM_091331 Homo sapiens hypothetical protein LOC162073 (LOC162073), mRNA
XM_091809 Homo sapiens similar to WW domain binding protein 2 (LOC147468), mRNA
XMJD91830 Homo sapiens similar to G protein-coupled receptor 124 (LOC162835), mRN
XM_091886 Homo sapiens similar to Zinc finger protein KM 8 (HKr18) (LOC162962), mRt
XMJ391914 Homo sapiens hypothetical protein LOC162993 (LOC162993), mRNA
XM_092019 Homo sapiens similar to BC331191 (LOC163131), mRNA
XM_092241 Homo sapiens similar to Olfactory receptor 6B3 (LOC150681), mRNA
XM_092267 Homo sapiens similar to keratin 8; cytokeratin 8; keratin, type II cytoskeletal I
XM_092342 Homo sapiens hypothetical protein FLJ39061 (FLJ39061), mRNA
XM_092553 Homo sapiens similar to autoantigen NOR-90 (LOC151320), mRNA
XM_092681 Homo sapiens similar to MLRQ subunit of the NADH ubiquinone oxidoreduct
XM_092778 Homo sapiens hypothetical protein LOC164395 (LOC164395), mRNA
XM_092995 Homo sapiens zinc finger protein 21 (KOX 14) (ZNF21), mRNA
XMJD93024 Homo sapiens hypothetical protein LOC169981 (LOC169981), mRNA
XM_093087 Homo sapiens similar to transcription factor (p38 interacting protein) (LOC17
XM_093644 Homo sapiens similar to NACHT-, LRR- and PYD-containing protein 2 (PYRI
XM_093813 Homo sapiens similar to hypothetical protein (LOC166348), mRNA
XM_093839 Homo sapiens KIAA0826 protein (KIAA0826), mRNA
XM_093895 Homo sapiens KIAA0882 protein (KIAA0882), mRNA
XM_094066 Homo sapiens similar to RIKEN cDNA 5430419M09 (LOC152877), mRNA
XM_094074 Homo sapiens similar to embryonic blastocoelar extracellular matrix protein r.
XMJ394581 Homo sapiens SEC24 related gene family, member A (S. cerevisiae) (SEC2^
XM_094794 Homo sapiens dapper homolog 2, antagonist of beta-catenin (xenopus) (DAC
XM_095568 Homo sapiens hypothetical protein DKFZp762C1112 (DKFZp762C1112), mF
XM_095746 Homo sapiens forkhead box D4 (FOXD4), mRNA
XM_095965 Homo sapiens hypothetical protein LOC169834 (LOC169834), mRNA
XM_095991 Homo sapiens chromosome 9 open reading frame 81 (C9orf81), mRNA
XM_096317 Homo sapiens chromosome 10 open reading frame 73 (C10orf73), mRNA
XM_096376 Homo sapiens hypothetical LOC143034 (LOC143034), mRNA
XMJ396472 Homo sapiens similar to RIKEN cDNA 1700021 K07 (LOC143678), mRNA
XM_096516 Homo sapiens hypothetical LOC143970 (LOC143970), mRNA
XM_096642 Homo sapiens hypothetical LOC144631 (LOC144631), mRNA
XM_096676 Homo sapiens hypothetical LOC144762 (LOC144762), mRNA
XM_096688 Homo sapiens hypothetical protein LOC144920 (LOC144920), mRNA
XM_096733 Homo sapiens chromosome 14 open reading frame 72 (C14orf72), mRNA
XM_096734 Homo sapiens hypothetical LOC145197 (LOC145197), mRNA
XM_096852 Homo sapiens hypothetical LOC145741 (LOC145741), mRNA
XM_096864 Homo sapiens hypothetical LOC145780 (LOC145780), mRNA
XM_096883 Homo sapiens hypothetical LOC145846 (LOC145846), mRNA
XM_096885 Homo sapiens similar to ENSANGP00000021391 (LOC145853), mRNA
XM_096919 Homo sapiens similar to SH2 domain protein 2A (T cell-specific adapter prot<
XM_097065 Homo sapiens hypothetical LOC146701 (LOC146701), mRNA
XM_097265 Homo sapiens hypothetical protein LOC147670 (LOC147670), mRNA
XM_097278 Homo sapiens hypothetical LOC147710 (LOC147710), mRNA
XMJ397347 Homo sapiens hypothetical LOC147941 (LOC147941), mRNA
XM_097351 Homo sapiens hypothetical LOC147975 (LOC147975), mRNA
XM_097580 Homo sapiens hypothetical protein LOC149086 (LOC149086), mRNA
XM_097622 Homo sapiens similar to RIKEN cDNA C030014K22 gene (LOC149297), mR
XM_097725 Homo sapiens hypothetical LOC149738 (LOC149738), mRNA
XM_097729 Homo sapiens hypothetical LOC149704 (LOC149704), mRNA
XM_097736 Homo sapiens chromosome 20 open reading frame 82 (C20orf82), mRNA
XM_097753 Homo sapiens hypothetical LOC149913 (LOC149913), mRNA
XM_097792 Homo sapiens hypothetical LOC150051 (LOC150051 ), mRNA
XM_097886 Homo sapiens hypothetical protein LOC150223 (LOC150223), mRNA
XM_097977 Homo sapiens hypothetical protein LOC150946 (LOC150946), mRNA
XM_098008 Homo sapiens hypothetical LOC151154 (LOC151154), mRNA
XM_098030 Homo sapiens hypothetical LOC151261 (LOC151261), mRNA
XM_098117 Homo sapiens hypothetical LOC151760 (LOC151760), mRNA
XM_098163 Homo sapiens hypothetical LOC152118 (LOC152118), mRNA
XM_098164 Homo sapiens hypothetical LOC152122 (LOC152122), mRNA
XM_098238 Homo sapiens SH3 domain protein D19 (DKFZp434D0215), mRNA
XM_098317 Homo sapiens hypothetical LOC153134 (LOC153134), mRNA
XM_098350 Homo sapiens hypothetical LOC153297 (LOC153297), mRNA
XM_098368 Homo sapiens KIAA1317 protein (KIAA1317), mRNA
XM_098406 Homo sapiens hypothetical LOC153630 (LOC153630), mRNA
XM_098450 Homo sapiens hypothetical LOC153959 (LOC153959), mRNA
XM_098512 Homo sapiens hypothetical LOC154323 (LOC154323), mRNA
XM_098625 Homo sapiens hypothetical LOC154872 (LOC154872), mRNA
XM_098762 Homo sapiens KIAA1416 protein (KIAA1416), mRNA
XM_098828 Homo sapiens hypothetical LOC157813 (LOC157813), mRNA
XM_098847 Homo sapiens hypothetical LOC157943 (LOC157943), mRNA
XM_098940 Homo sapiens similar to zinc finger protein 11 b (KOX 2) (LOC158431 ), mRN.
XM_098980 Homo sapiens hypothetical LOC158730 (LOC158730), mRNA
XM_099034 Homo sapiens hypothetical LOC159170 (LOC159170), mRNA
XM 04657 Homo sapiens similar to RIKEN cDNA 1700019P01 (LOC164714), mRNA
XM_106386 Homo sapiens KIAA1345 protein (KIAA1345), mRNA
XM_113228 Homo sapiens similar to Proprotein convertase subtilisin/kexin type 7 precurs
XM_113596 Homo sapiens similar to CG32542-PA (LOC196752), mRNA
XM_113625 Homo sapiens hypothetical protein LOC195977 (LOC195977), mRNA
XM_113641 Homo sapiens hypothetical protein LOC196051 (LOC196051), mRNA
XM_113678 Homo sapiens nucleoporin 160kDa (NUP160), mRNA
XM_113696 Homo sapiens hypothetical protein LOC196337 (LOC196337), mRNA
XM_113706 Homo sapiens dynein, axonemal, heavy polypeptide 10 (DNAH10), mRNA
XM_113708 Homo sapiens hypothetical protein LOC196394 (LOC196394), mRNA
XM_113743 Homo sapiens hypothetical protein DKFZp313M0720 (DKFZp313M0720), ml
XM_113763 Homo sapiens chromosome 14 open reading frame 125 (C14orf125), mRNA
XM_113776 Homo sapiens hypothetical protein LOC196913 (LOC196913), mRNA
XM_113796 Homo sapiens hypothetical protein LOC196996 (LOC196996), mRNA
XM_113825 Homo sapiens similar to RIKEN cDNA 4930424G05 (LOC197135), mRNA
XM_113871 Homo sapiens hypothetical protein LOC197350 (LOC197350), mRNA
XM_113912 Homo sapiens similar to DKFZP566O084 protein (LOC201140), mRNA
XM_113916 Homo sapiens similar to hypothetical protein A930006D11 (LOC201181), mF
XM_113947 Homo sapiens KIAA0565 gene product (KIAA0565), mRNA
XM_113967 Homo sapiens similar to Rab12 protein (LOC201475), mRNA
XM_113971 Homo sapiens similar to B230208J24Rik protein (LOC201501), mRNA
XM_113978 Homo sapiens hypothetical protein LOC284352 (LOC284352), mRNA
XM_114000 Homo sapiens ankyrin repeat domain 24 (ANKRD24), mRNA
XM_114047 Homo sapiens similar to KI AA0454 protein (LOC199882), mRNA
XM_114067 Homo sapiens similar to expressed sequence AV028368 (LOC199953), mRf
XM_114087 Homo sapiens KIAA1836 protein (KIAA1836), mRNA
XM_114090 Homo sapiens similar to KIAA0454 protein (LOC200019), mRNA
XM_114129 Homo sapiens hypothetical LOC200159 (LOC200159), mRNA
XM_114152 Homo sapiens hypothetical protein LOC200205 (LOC200205), mRNA
XM_114156 Homo sapiens hypothetical protein LOC200213 (LOC200213), mRNA
XM_114158 Homo sapiens similar to Polyadenylate-binding protein 2 (Poly(A)-binding pre
XM_114166 Homo sapiens similar to KIAA0386 (LOC200230), mRNA
XM_114222 Homo sapiens similar to hypothetical protein (LOC200373), mRNA
XM_114272 Homo sapiens selective LIM binding factor, rat homolog (SLB), mRNA
XM_114297 Homo sapiens hypothetical protein FLJ10599 (FLJ10599), mRNA
XM_114301 Homo sapiens similar to beta-1 ,4-mannosyltransferase; beta-1 ,4 mannosyltrs
XM_114303 Homo sapiens GRP1 -binding protein GRSP1 (GRSP1), mRNA
XM_114317 Homo sapiens similarto RIKEN CDNA 3110001N18 (LOC200916), mRNA
XM_114355 Homo sapiens similar to esterase/N-deacetylase (EC 3.5.1.-), 50K hepatic - r
XM_114415 Homo sapiens similarto Glycerol kinase, testis specific 1 (ATP:glycerol 3-phι
XM_114418 Homo sapiens KIAA1729 protein (KIAA1729), mRNA
XM_114430 Homo sapiens hypothetical protein LOC202051 (LOC202051 ), mRNA
XM_114432 Homo sapiens KIAA1281 protein (KIAA1281), mRNA
XM_114447 Homo sapiens KIAA1999 protein (KIAA1999), mRNA
XM_114456 Homo sapiens hypothetical protein LOC202181 (LOC202181 ), mRNA
XM_114481 Homo sapiens hypothetical LOC202404 (LOC202404), mRNA
XM_114560 Homo sapiens similar to hypothetical protein MGC35361 (LOC202802), mRf
XM_114611 Homo sapiens hypothetical protein KIAA1833 (KIAA1833), mRNA
XM_114618 Homo sapiens hypothetical protein LOC203069 (LOC203069), mRNA
XM_114621 Homo sapiens similar to RIKEN cDNA 4930578I06 (LOC203076), mRNA
XM_114685 Homo sapiens chromosome 9 open reading frame 21 (C9orf21), mRNA
XM_114723 Homo sapiens similar to PAGE-5 protein (LOC203569), mRNA
XM_114735 Homo sapiens complement component (3b/4b) receptor 1-like (CR1 L), mRN-
XM_114973 Homo sapiens hypothetical protein LOC203806 (LOC203806), mRNA
XMJ14987 Homo sapiens similarto RIKEN cDNA 1500011L16 (LOC196120), mRNA
XM_115009 Homo sapiens hypothetical protein LOC203859 (LOC203859), mRNA
XM_115092 Homo sapiens similar to Olfactory receptor 56B4 (LOC196335), mRNA
XM_115100 Homo sapiens similar to autoantigen NOR-90 (LOC196346), mRNA
XM_115715 Homo sapiens similar to ENSANGP00000000189 (LOC200493), mRNA
XM_115760 Homo sapiens similar to h2-calponin (LOC205272), mRNA
XM_115769 Homo sapiens similar to chromosome 20 open reading frame 81 (LOC20059
XMJ 15897 Homo sapiens similar to high mobility group protein homolog HMG4 (LOC20:
XM_115925 Homo sapiens similar to trophinin; melanoma antigen, family D, 3; trophinin-.
XM_115974 Homo sapiens similar to hypothetical protein H41 (LOC200842), mRNA
XM_116036 Homo sapiens similar to Gamma-aminobutyric-acid receptor rho-3 subunit pr
XM_116384 Homo sapiens similar to Transcription initiation factor TFIID 28 kDa subunit (
XMJ 16396 Homo sapiens similar to peptidylprolyl isomerase A (cyclophilin A) (LOC2022
XM_116497 Homo sapiens chromosome 6 open reading frame 163 (C6orf163), mRNA
XM_116623 Homo sapiens similar to Ubiquinol-cytochrome C reductase iron-sulfur subur
XMJ16936 Homo sapiens similarto RIKEN CDNA2310038H17 (LOC196541), mRNA
XMJ 16970 Homo sapiens similar to hypothetical protein (LOC196994), mRNA
XM_116971 Homo sapiens hypothetical protein LOC196993 (LOC196993), mRNA
XM_116980 Homo sapiens hypothetical LOC197049 (LOC197049), mRNA
XMJ 17014 Homo sapiens hypothetical LOC197317 (LOC197317), mRNA
XM_117030 Homo sapiens hypothetical LOC197387 (LOC197387), mRNA
XMJ 17044 Homo sapiens hypothetical LOC201109 (LOC201109), mRNA
XMJ 17056 Homo sapiens hypothetical LOC201201 (LOC201201 ), mRNA
XMJ 17100 Homo sapiens hypothetical LOC201484 (LOC201484), mRNA
XMJ 17112 Homo sapiens hypothetical LOC199680 (LOC199680), mRNA
XMJ 17117 Homo sapiens hypothetical gene FLJ13072 (FLJ13072), mRNA
XMJ 17152 Homo sapiens hypothetical LOC199897 (LOC199897), mRNA
XMJ 17174 Homo sapiens hypothetical protein LOC200010 (LOC200010), mRNA
XMJ 17213 Homo sapiens hypothetical LOC200292 (LOC200292), mRNA
XMJ 17224 Homo sapiens similar to RIKEN cDNA 0610009J22 (LOC200312), mRNA
XMJ 17236 Homo sapiens hypothetical LOC200475 (LOC200475), mRNA
XMJ 17239 Homo sapiens hypothetical LOC200491 (LOC200491 ), mRNA
XMJ 17257 Homo sapiens hypothetical LOC200624 (LOC200624), mRNA
XMJ 17266 Homo sapiens hypothetical LOC200726 (LOC200726), mRNA
XMJ 17268 Homo sapiens hypothetical LOC200731 (LOC200731), mRNA
XMJ 17294 Homo sapiens hypothetical protein LOC200933 (LOC200933), mRNA
XMJ 17408 Homo sapiens hypothetical LOC202546 (LOC202546), mRNA
XMJ 17451 Homo sapiens hypothetical LOC202775 (LOC202775), mRNA
XMJ 17514 Homo sapiens hypothetical LOC203235 (LOC203235), mRNA
XMJ 17548 Homo sapiens hypothetical LOC203413 (LOC203413), mRNA
XMJ 65401 Homo sapiens similar to heat shock 70kD protein binding protein; progestero
XMJ 65448 Homo sapiens similar to BLOCK 23 (LOC220717), mRNA
XMJ 65511 Homo sapiens similar to Fatty acid-binding protein, epidermal (E-FABP) (Psc
XMJ 65534 Homo sapiens similarto methyltransferase-like protein 1 isoform c; D1075-lil*
XMJ 65921 Homo sapiens similar to HP95 (LOC219392), mRNA
XMJ 65973 Homo sapiens ubiquitin specific protease 24 (USP24), mRNA
XMJ 66086 Homo sapiens neuregulin 3 (NRG3), mRNA
XMJ 66090 Homo sapiens placenta-specific 9 (PLAC9), mRNA
XMJ 66103 Homo sapiens DNA2 DNA replication helicase 2-like (yeast) (DNA2L), mRN/
XMJ 66110 Homo sapiens G protein-coupled receptor 158 (GPR158), mRNA
XMJ 66125 Homo sapiens KIAA protein (similar to mouse paladin) (KIAA1274), mRNA
XMJ 66132 Homo sapiens KIAA1462 (KIAA1462), mRNA
XMJ 66138 Homo sapiens ankyrin repeat domain 16 (ANKRD16), mRNA
XMJ 66140 Homo sapiens Scm-like with four mbt domains 2 (SFMBT2), mRNA
XMJ 66160 Homo sapiens similar to chromosome 6 open reading frame 182 (LOC22101
XMJ 66164 Homo sapiens hypothetical protein LOC219854 (LOC219854), mRNA
XMJ 66203 Homo sapiens similar to RIKEN cDNA 1110030K22 (LOC219537), mRNA
XMJ 66213 Homo sapiens KIAA0937 protein (KIAA0937), mRNA
XMJ 66227 Homo sapiens macrophage expressed gene 1 (MPEG1), mRNA
XMJ 66254 Homo sapiens odd Oz/ten-m homolog 4 (ODZ4), mRNA
XMJ 66256 Homo sapiens microtubule-associated protein 6 (MAP6), mRNA
XMJ 66270 Homo sapiens KIAA0774 (KIAA0774), mRNA
XMJ 66300 Homo sapiens absent in melanoma 1 (AIM1), mRNA
XMJ 66320 Homo sapiens KIAA1553 (KIAA1553), mRNA
XMJ 66346 Homo sapiens chromosome 6 open reading frame 129 (C6orf129), mRNA
XMJ 66372 Homo sapiens leucine rich repeat and fibronectin type III domain containing I
XMJ 66376 Homo sapiens KIAA1949 protein (KIAA1949), mRNA
XMJ 66420 Homo sapiens RPEL repeat containing 1 (RPEL1), mRNA
XMJ 66432 Homo sapiens hypothetical protein LOC221442 (LOC221442), mRNA
XMJ 66443 Homo sapiens tudor domain containing 6 (TDRD6), mRNA
XMJ 66450 Homo sapiens bromodomain and PHD finger containing, 3 (BRPF3), mRNA
XMJ 66451 Homo sapiens KI AA1586 (KIAA1586), mRNA
XMJ 66453 Homo sapiens tau tubulin kinase 1 (TTBK1 ), mRNA
XMJ 66479 Homo sapiens KIAA0240 (KIAA0240), mRNA
XMJ 66508 Homo sapiens TWIST neighbor (TWISTNB), mRNA
XMJ 66523 Homo sapiens tweety homolog 3 (Drosophila) (TTYH3), mRNA
XMJ 66527 Homo sapiens KIAA0415 gene product (KIAA0415), mRNA
XMJ66529 Homo sapiens glucocorticoid induced transcript 1 (GLCCI1), mRNA
XMJ 66532 Homo sapiens KIAA1950 protein (KIAA1950), mRNA
XMJ 66571 Homo sapiens KIAA0363 protein (KIAA0363), mRNA
XMJ 66573 Homo sapiens KIAA0895 protein (KIAA0895), mRNA
XMJ 66630 Homo sapiens similar to KIAA2020 protein (LOC387692), mRNA
XMJ 66659 Homo sapiens hypothetical protein LOC220213 (LOC220213), mRNA
XMJ 66707 Homo sapiens olfactory receptor, family 13, subfamily A, member 1 (OR13A'
XMJ 66720 Homo sapiens similar to Protein C21orf59 (LOC220998), mRNA
XMJ 66747 Homo sapiens similar to KIAA1838 (LOC219797), mRNA
XMJ 66757 Homo sapiens similar to Olfactory receptor 8B12 (LOC219858), mRNA
XMJ 66767 Homo sapiens similar to seven transmembrane helix receptor (LOC219865),
XMJ 66776 Homo sapiens similarto Olfactory receptor 10G8 (LOC219869), mRNA
XMJ 66777 Homo sapiens similar to Olfactory receptor 10G9 (LOC219870), mRNA
XMJ 66780 Homo sapiens similar to Olfactory receptor 10S1 (LOC219873), mRNA
XMJ 66781 Homo sapiens similar to Olfactory receptor 6T1 (LOC219874), mRNA
XMJ 66782 Homo sapiens similar to Olfactory receptor 4D5 (LOC219875), mRNA
XMJ 66805 Homo sapiens similar to seven transmembrane helix receptor (LOC21941 ),
XMJ 66808 Homo sapiens simi ar to Olfactory receptor 5AS1 (LOC219447), mRNA XMJ 66813 Homo sapiens simi ar to Olfactory receptor 8K5 (LOC219453), mRNA XMJ 66818 Homo sapiens simil lar to hypothetical protein FLJ13194 (LOC219462), mRN/ XMJ 66820 Homo sapiens simi! ar to Olfactory receptor 5T2 (LOC219464), mRNA XMJ 66823 Homo sapiens simi ar to Olfactory receptor 8H1 (LOC219469), mRNA XMJ 66825 Homo sapiens simi ar to Olfactory receptor 8K3 (LOC219473), mRNA XMJ 66829 Homo sapiens simi ar to Olfactory receptor 8J1 (LOC219477), mRNA XMJ 66831 Homo sapiens simi ar to seven transmembrane helix receptor (LOC219479), XMJ 66834 Homo sapiens simi ar to Olfactory receptor 5M3 (LOC219482), mRNA XMJ 66835 Homo sapiens simi ar to Olfactory receptor 5M8 (LOC219484), mRNA XMJ 66845 Homo sapiens simi ar to Olfactory receptor 5AR1 (LOC219493), mRNA XMJ 66856 Homo sapiens simi ar to expressed sequence AI841794 (LOC219527), mRN XMJ 66868 Homo sapiens simi ar to Olfactory receptor 4C16 (LOC219428), mRNA XMJ 66869 Homo sapiens simi ar to seven transmembrane helix receptor (LOC219429), XMJ 66871 Homo sapiens simi ar to Olfactory receptor 4S2 (LOC219431 ), mRNA XMJ 66872 Homo sapiens simi ar to Olfactory receptor 4C6 (LOC219432), mRNA XMJ 66877 Homo sapiens simi ar to Olfactory receptor 5D14 (LOC219436), mRNA XMJ 66878 Homo sapiens simi ar to Olfactory receptor 5L1 (OST262) (LOC219437), mR XMJ 66879 Homo sapiens simi ar to Olfactory receptor 5D18 (LOC219438), mRNA XMJ 66898 Homo sapiens simi ar to Olfactory receptor 5A2 (LOC219981), mRNA XMJ 66899 Homo sapiens simi ar to Olfactory receptor 5A1 (OST181) (LOC219982), mR XMJ 66900 Homo sapiens simi ar to Olfactory receptor 4D6 (LOC219983), mRNA XMJ 66903 Homo sapiens simi ar to seven transmembrane helix receptor (L0C219986), XMJ 66910 Homo sapiens simi ar to Olfactory receptor 6Q1 (LOC219952), mRNA XMJ 66912 Homo sapiens simi ar to Olfactory receptor 911 (LOC219954), mRNA XMJ 66914 Homo sapiens simi ar to Olfactory receptor 9Q1 (LOC219956), mRNA XMJ 66915 Homo sapiens simi ar to seven transmembrane helix receptor (LOC219957), XMJ 66916 Homo sapiens simi ar to Olfactory receptor 1S2 (LOC219958), mRNA XMJ 66917 Homo sapiens simi ar to Olfactory receptor 1S1 (LOC219959), mRNA XMJ 66918 Homo sapiens simi ar to Olfactory receptor 10Q1 (LOC219960), mRNA XMJ 66923 Homo sapiens simi ar to Olfactory receptor 5B17 (LOC219965), mRNA XMJ 66926 Homo sapiens simi ar to olfactory receptor MOR202-4 (LOC219968), mRNA XMJ 66966 Homo sapiens simi lar to Meningioma-expressed antigen 6/11 (MEA6) (MEA1 XMJ 66971 Homo sapiens simi ar to Leucine-rich repeat protein SHOC-2 (Ras-binding pi XMJ 67001 Homo sapiens simi ar to 40S ribosomal protein S26 (LOC219542), mRNA XMJ 67044 Homo sapiens solute carrier family 35, member F1 (SLC35F1), mRNA XMJ 67072 Homo sapiens benzodiazapine receptor (peripheral)-like 1 (BZRPL1), mRNA XMJ 67147 Homo sapiens zinc finger protein 390 (ZNF390), mRNA XMJ 67149 Homo sapiens chromosome 6 open reading frame 194 (C6orf194), mRNA XMJ 67152 Homo sapiens similar to Zinc finger protein 192 (LD5-1) (LOC222701 ), mRNi XMJ 67254 Homo sapiens similar to tropomyosin 3 (LOC221875), mRNA XMJ 67275 Homo sapiens similar to ribosomal protein L23 (LOC222901), mRNA XMJ 67709 Homo sapiens hypothetical protein LOC221061 (LOC221061), mRNA XMJ 67711 Homo sapiens integrin, alpha 8 (ITGA8), mRNA XMJ 67908 Homo sapiens hypothetical LOC221140 (LOC221140), mRNA XMJ 68030 Homo sapiens zinc finger protein 319 (ZNF319), mRNA XMJ 68053 Homo sapiens chromosome 6 open reading frame 184 (C6orf184), mRNA XMJ 68055 Homo sapiens chromosome 6 open reading frame 185 (C6orf185), mRNA XMJ 68060 Homo sapiens chromosome 6 open reading frame 154 (C6orf154), mRNA XMJ 68073 Homo sapiens hypothetical LOC221344 (LOC221344), mRNA XMJ 68101 Homo sapiens hypothetical LOC221766 (LOC221766), mRNA XMJ 68223 Homo sapiens similar to RIKEN cDNA A530016O06 gene (LOC221813), mR XMJ 68302 Homo sapiens zinc finger protein 36 (KOX 18) (ZNF36), mRNA XMJ 68354 Homo sapiens similar to SMT3 suppressor of mif two 3 homolog 2 (LOC222C XMJ 68521 Homo sapiens hypothetical LOC222190 (LOC222190), mRNA XMJ 68578 Homo sapiens mucin 3B (MUC3B), mRNA XM 168583 Homo sapiens mucin 17 (MUC17), mRNA
XMJ 68585 Homo sapiens similar to mucin 11 (LOC219612), mRNA
XMJ 68590 Homo sapiens zuotin related factor 1 (ZRF1), mRNA
XMJ 69057 Homo sapiens hypothetical LOC219908 (LOC219908), mRNA
XMJ 69227 Homo sapiens similar to proline rich antigen 2 (LOC220061 ), mRNA
XMJ 69258 Homo sapiens similar to KIAA0543 protein (LOC219638), mRNA
XMJ 69434 Homo sapiens similar to TAR DNA binding protein (LOC219414), mRNA
XMJ 70525 Homo sapiens similar to ARF GTPase-activating protein (LOC255319), mRN
XMJ 70536 Homo sapiens similar to eukaryotic translation initiation factor 2, subunit 3 gε
XMJ 70597 Homo sapiens similar to peptidylprolyl isomerase A (LOC256374), mRNA
XMJ 70620 Homo sapiens similar to 60S ribosomal protein L21 (LOC256457), mRNA
XMJ 70637 Homo sapiens similar to beta-tubulin 4Q (LOC253936), mRNA
XMJ 70658 Homo sapiens tangerin (DKFZp762C186), mRNA
XMJ 70659 Homo sapiens similar to 2010003K11 Rik protein (LOC254439), mRNA
XMJ 70667 Homo sapiens hypothetical protein LOC254359 (LOC254359), mRNA
XMJ 70708 Homo sapiens hypothetical LOC255411 (LOC255411), mRNA
XMJ 70736 Homo sapiens hypothetical protein LOC253512 (LOC253512), mRNA
XMJ 70749 Homo sapiens similar to GARNL1 protein (LOC387984), mRNA
XMJ 70754 Homo sapiens similar to serine (or cysteine) proteinase inhibitor, clade A (alp
XMJ 70777 Homo sapiens similar to Microsomal signal peptidase 25 kDa subunit (SPase
XMJ 70840 Homo sapiens similar to CDRT15 protein (LOC256223), mRNA
XMJ 70842 Homo sapiens hypothetical protein FLJ40244 (FLJ40244), mRNA
XMJ 70909 Homo sapiens similar to hypothetical protein MGC20470 (LOC257177), mRf
XMJ 70950 Homo sapiens similar to OSJNBa0043A12.32 (LOC254897), mRNA
XMJ 70994 Homo sapiens hypothetical LOC255349 (LOC255349), mRNA
XMJ 71008 Homo sapiens similar to high mobility group AT-hook 1 (LOC257200), mRNA
XMJ 71013 Homo sapiens similar to Gamma-2-syntrophin (G2SYN) (Syntrophin 5) (SYN
XMJ 71032 Homo sapiens hypothetical protein LOC255812 (LOC255812), mRNA
XMJ 71040 Homo sapiens similar to MEGF6 (LOC253820), mRNA
XMJ 71054 Homo sapiens KIAA0527 protein (KIAA0527), mRNA
XMJ 71060 Homo sapiens hypothetical protein MGC50836 (MGC50836), mRNA
XMJ 71068 Homo sapiens hypothetical protein LOC253017 (LOC253017), mRNA
XMJ 71078 Homo sapiens similar to ALGV3072 (LOC255324), mRNA
XMJ 71081 Homo sapiens similar to ras-related C3 botulinum toxin substrate 1 isoform F
XMJ 71094 Homo sapiens similar to eukaryotic translation initiation factor elF4E-1 (LOCi
XMJ 71105 Homo sapiens hypothetical LOC255338 (LOC255338), mRNA
XM 71150 Homo sapiens similar to bA145E8.1 (KIAA1074) (LOC254027), mRNA
XMJ 71154 Homo sapiens similar to myelin protein zero-like 1 ; protein zero related (LOC
XMJ71156 Homo sapiens similar to bA145E8.1 (KIAA1074) (LOC254750), mRNA
XMJ 71158 Homo sapiens similar to ribosomal protein S2; 40S ribosomal protein S2 (MG
XMJ 71163 Homo sapiens similar to Six transmembrane epithelial antigen of prostate (L(
XMJ 71165 Homo sapiens similar to cAMP-dependent protein kinase type l-beta regulate
XMJ 71171 Homo sapiens similar to hypothetical protein MGC49416 (LOC255374), mRf
XMJ71207 Homo sapiens similar to RIKEN cDNA 4930429J24 (LOC255220), mRNA
XMJ 71224 Homo sapiens coactivator associated arginine methyltransferase 1-like (CAF
XMJ 71410 Homo sapiens hypothetical protein DKFZp667B0210 (DKFZp667B0210), mF
XMJ 71424 Homo sapiens similar to olfactory receptor (LOC256892), mRNA
XMJ 71447 Homo sapiens similar to Transcription elongation factor B polypeptide 2 (RN;
XMJ 71489 Homo sapiens similar to seven transmembrane helix receptor (LOC256144),
XMJ71490 Homo sapiens similar to Olfactory receptor 4S1 (LOC256148), mRNA
XMJ 71495 Homo sapiens similar to Claudin-22 (LOC255244), mRNA
XMJ 71503 Homo sapiens similar to large subunit ribosomal protein L36a (LOC255701),
XMJ 71528 Homo sapiens similar to seven transmembrane helix receptor (LOC255725),
XMJ 71536 Homo sapiens mas-related G protein-coupled MRGE (MRGE), mRNA
XMJ 71578 Homo sapiens similar to olfactory receptor MOR109-1 (LOC254783), mRNA
XMJ 71581 Homo sapiens similar to olfactory receptor MOR111-4 (LOC254786), mRNA
XMJ 71590 Homo sapiens similar to ribosomal protein L22 (LOC256441), mRNA
XMJ 71766 Homo sapiens similar to high mobility group protein homolog HMG4 (LOC25I
XMJ 71855 Homo sapiens similar to myeloid-associated differentiation marker (LOC255.
XMJ71892 Homo sapiens similar to ribosomal protein L31 (LOC253013), mRNA
XMJ 71973 Homo sapiens similar to zinc finger protein 91 (HPF7, HTF10) (LOC253342),
XMJ 72230 Homo sapiens similar to ribosomal protein S17 (LOC257039), mRNA
XMJ 72341 Homo sapiens hypothetical protein FLJ35036 (FLJ35036), mRNA
XMJ 72389 Homo sapiens similar to Tryptophan-rich protein (Congenital heart disease 5
XMJ 72751 Homo sapiens similar to Olfactory receptor 1 L4 (Olfactory receptor 9-E) (ORi
XMJ 72801 Homo sapiens KIAA1210 protein (KIAA1210), mRNA
XMJ 72852 Homo sapiens hypothetical LOC256676 (LOC256676), mRNA
XMJ 72855 Homo sapiens hypothetical LOC255849 (LOC255849), mRNA
XMJ 72860 Homo sapiens hypothetical LOC255649 (LOC255649), mRNA
XMJ 72861 Homo sapiens similar to ZP1 precursor (LOC255714), mRNA
XMJ 72874 Homo sapiens hypothetical LOC253724 (LOC253724), mRNA
XMJ 72889 Homo sapiens hypothetical LOC256176 (LOC256176), mRNA
XMJ 72917 Homo sapiens hypothetical LOC256453 (LOC256453), mRNA
XMJ 72929 Homo sapiens hypothetical protein LOC255189 (LOC255189), mRNA
XMJ 72931 Homo sapiens hypothetical protein LOC254559 (LOC254559), mRNA
XMJ 72968 Homo sapiens hypothetical protein LOC253962 (LOC253962), mRNA
XMJ 72995 Homo sapiens hypothetical LOC255809 (LOC255809), mRNA
XMJ 73012 Homo sapiens hypothetical LOC256686 (LOC256686), mRNA
XMJ 73015 Homo sapiens hypothetical LOC256483 (LOC256483), mRNA
XMJ 73036 Homo sapiens hypothetical protein LOC255654 (LOC255654), mRNA
XMJ 73063 Homo sapiens hypothetical LOC253662 (LOC253662), mRNA
XMJ 73068 Homo sapiens hypothetical LOC253584 (LOC253584), mRNA
XMJ 73083 Homo sapiens hypothetical protein LOC255025 (LOC255025), mRNA
XMJ 73084 Homo sapiens hypothetical protein LOC254827 (LOC254827), mRNA
XMJ 73087 Homo sapiens hypothetical protein LOC255798 (LOC255798), mRNA
XMJ 73105 Homo sapiens hypothetical LOC256283 (LOC256283), mRNA
XMJ 73119 Homo sapiens hypothetical LOC255130 (LOC255130), mRNA
XMJ 73120 Homo sapiens hypothetical LOC254938 (LOC254938), mRNA
XMJ 73132 Homo sapiens similar to unc-93 homolog B1 ; unc93 (C.elegans) homolog B;
XMJ 73135 Homo sapiens hypothetical LOC256880 (LOC256880), mRNA
XMJ 73140 Homo sapiens hypothetical LOC253254 (LOC253254), mRNA
XMJ 73160 Homo sapiens hypothetical LOC255187 (LOC255187), mRNA
XMJ 73164 Homo sapiens hypothetical LOC256096 (LOC256096), mRNA
XMJ 73166 Homo sapiens chromosome 6 open reading frame 191 (C6orf191), mRNA
XMJ 73173 Homo sapiens amine oxidase, flavin containing 1 (AOF1), mRNA
XMJ 75125 Homo sapiens hemicentin-2 (DKFZp434P0216), mRNA
XM_208028 Homo sapiens similar to double homeobox protein (LOC283039), mRNA
XM_208035 Homo sapiens similar to hypothetical protein FLJ10817 (LOC347806), mRN/
XMJ.08042 Homo sapiens similar to 40S ribosomal protein S25 (LOC283114), mRNA
XM_208043 Homo sapiens similar to RING finger protein 18 (Testis-specific ring-finger pr
XM_208058 Homo sapiens similar to NADPH oxidase 4 variant (LOC283247), mRNA
XM_208060 Homo sapiens similar to tripartite motif-containing 51 (LOC283257), mRNA
XM_208061 Homo sapiens similar to tripartite motif protein 48; TRIM48 (LOC283259), mF
XM_208072 Homo sapiens similar to ribosomal protein L13a; 60S ribosomal protein L13a
XM_208080 Homo sapiens similar to Polyhomeotic-like protein 1 (Early development regi
XM_208097 Homo sapiens similar to telomeric repeat binding factor 1 isoform 2; Telomer
XM_208108 Homo sapiens similar to synaptogyrin 2; cellugyrin (LOC283698), mRNA
XM_208116 Homo sapiens similar to elongation factor Sill p15 subunit (LOC283747), mF
XM_208142 Homo sapiens similar to hypothetical protein (LOC283957), mRNA
XMJ-08145 Homo sapiens similar to rhophilin-like protein; RhoB effector; rhophilin-2; rho
XM_208151 Homo sapiens similar to solute carrier family 16, member 6; monocarboxylati
XM_208185 Homo sapiens similar to large subunit ribosomal protein L36a (LOC284230),
XM_208200 Homo sapiens similar to Heterogeneous nuclear ribonucleoprotein A1 (Helix-
XM_208203 Homo sapiens similar to methyl-CpG binding domain protein 3-like 2 (LOC28
XM_208204 Homo sapiens similar to actin-related protein 2 (LOC284441 ), mRNA
XM_208213 Homo sapiens similar to fatty acid omega-hydroxylase CYP4A11 (LOC28454
XM_208234 Homo sapiens similar to unactive progesterone receptor, 23 kD; likely ortholc
XM_208250 Homo sapiens similar to FRG1 protein (FSHD region gene 1 protein) (LOC2£
XM_208261 Homo sapiens similar to lymphocyte activation-associated protein; kelch (Drc
XM_208270 Homo sapiens acrosin-like protease (bA395L14.13), mRNA
XM_208281 Homo sapiens similar to ribosomal protein L18a; 60S ribosomal protein L18a
XM_208300 Homo sapiens similar to 60S ribosomal protein L23a (LOC285214), mRNA
XM_208312 Homo sapiens similar to unc-93 homolog B1 ; unc93 (C.elegans) homolog B;
XM_208313 Homo sapiens similar to tropomyosin 4 (LOC285321), mRNA
XM_208316 Homo sapiens similar to vascular endothelial zinc finger 1 (LOC285388), mR
XM_208317 Homo sapiens similar to 40S ribosomal protein S20 (LOC285384), mRNA
XM_208319 Homo sapiens similar to Epidermal Langerhans cell protein LCP1 (LOC2854
XM_208320 Homo sapiens similar to mitochondrial translational release factor 1-like (LOC
XM_208333 Homo sapiens hypothetical protein MGC48637 (MGC48637), mRNA
XM_208352 Homo sapiens alcohol dehydrogenase 5B (ADH5B), mRNA
XM_208356 Homo sapiens similar to cytochrome c oxidase subunit Via polypeptide 1 pre
XM_208361 Homo sapiens similar to RPL6 protein (LOC285900), mRNA
XMJ.08373 Homo sapiens similar to Heterogeneous nuclear ribonucleoprotein A1 (Helix-
XM_208403 Homo sapiens similar to Von Ebners gland protein precursor (VEG protein) ('
XM_208423 Homo sapiens similar to ribosomal protein S2; 40S ribosomal protein S2 (LO
XM_208431 Homo sapiens similar to ubiquitin-conjugating enzyme UbcM2 (LOC286480),
XM J_08438 Homo sapiens similar to Tetratricopeptide repeat protein 3 (TPR repeat prate
XM_208443 Homo sapiens Ras-like GTPase-like (LOC286526), mRNA
XM_208502 Homo sapiens similar to hypothetical protein 9330140G23 (LOC283071), mF
XM_208522 Homo sapiens KIAA1394 protein (KIAA1394), mRNA
XM_208524 Homo sapiens hypothetical protein LOC283129 (LOC283129), mRNA
XM_208529 Homo sapiens hypothetical protein DKFZp547C195 (DKFZp547C195), mRN
XM_208541 Homo sapiens similar to Olfactory receptor 8D2 (Olfactory receptor-like prote
XM_208543 Homo sapiens similar to Olfactory receptor 8D1 (Olfactory receptor-like prote
XM_208545 Homo sapiens similar to bone morphogenetic protein receptor, type IA precu
XM_208554 Homo sapiens similar to Protein farnesyltransferase/geranylgeranyltransferai
XM_208563 Homo sapiens hypothetical LOC283202 (LOC283202), mRNA
XM_208604 Homo sapiens similar to Olfactory receptor 10A4 (HP2) (Olfactory receptor-lil
XM_208612 Homo sapiens hypothetical LOC283321 (LOC283321), mRNA
XM_208613 Homo sapiens similar to 60 kDa heat shock protein, mitochondrial precursor
XM_208635 Homo sapiens similar to cDNA sequence BC030307 (LOC283350), mRNA
XM_208647 Homo sapiens hypothetical protein LOC283377 (LOC283377), mRNA
XM_208658 Homo sapiens similar to Succinyl-CoA ligase [GDP-forming] beta-chain, mito
XM_208667 Homo sapiens similar to HEEE9341 (LOC283420), mRNA
XM_208690 Homo sapiens similar to 40S ribosomal protein S26 (LOC283479), mRNA
XM_208731 Homo sapiens chromosome 14 open reading frame 68 (C14orf68), mRNA
XM_208746 Homo sapiens hypothetical protein LOC283578 (LOC283578), mRNA
XM_208766 Homo sapiens KIAA0284 (KIAA0284), mRNA
XM_208778 Homo sapiens hypothetical LOC283677 (LOC283677), mRNA
XM_208798 Homo sapiens similar to hypothetical protein FLJ35785 (LOC283717), mRN/
XM_208809 Homo sapiens similar to hypothetical protein D030069K18 (LOC283726), mF
XM_208835 Homo sapiens similar to hypothetical protein FLJ36144 (LOC283767), mRN/
XM_208847 Homo sapiens similar to testicular Metalloprotease-like, Disintegrin-like, Cyst
XM_208859 Homo sapiens similar to ARHGAP21 protein (LOC283816), mRNA
XM_208863 Homo sapiens similar to nuclear pore complex interacting protein (LOC3481J
XM_208887 Homo sapiens hypothetical protein LOC283871 (LOC283871), mRNA
XM_208889 Homo sapiens similar to MGC9515 protein (LOC388240), mRNA
XM_208891 Homo sapiens similar to nuclear pore complex interacting protein (LOC2838
XM_208908 Homo sapiens hypothetical protein LOC283922 (LOC283922), mRNA
XM_208927 Homo sapiens hypothetical protein FLJ36208 (FLJ36208), mRNA
XM_208930 Homo sapiens similar to RIKEN cDNA 4930511J11 (LOC283953), mRNA
XM_208944 Homo sapiens hypothetical protein LOC283989 (LOC283989), mRNA
XM_208990 Homo sapiens hypothetical LOC284067 (LOC284067), mRNA
XM_208993 Homo sapiens Similar to hypothetical gene supported by AL050367; AK0229
XM_209012 Homo sapiens similar to keratin 17 (LOC284089), mRNA
XM_209041 Homo sapiens similar to KIAA1503 protein (LOC284158), mRNA
XM_209073 Homo sapiens hypothetical protein LOC284207 (LOC284207), mRNA
XM_209076 Homo sapiens similar to Ankyrin repeat domain protein 18A (LOC284232), rr
XM_209083 Homo sapiens similar to telomeric repeat binding factor 1 isoform 2; Telomer
XM_209097 Homo sapiens similar to FLJ10101 protein (LOC284269), mRNA
XM_209104 Homo sapiens similar to Placental thrombin inhibitor (Cytoplasmic antiproteir
XM_209111 Homo sapiens hypothetical protein LOC284307 (LOC284307), mRNA
XM_209138 Homo sapiens similar to LL5 beta (LOC388548), mRNA
XM_209140 Homo sapiens hypothetical protein LOC284323 (LOC284323), mRNA
XM_209145 Homo sapiens similar to RIKEN cDNA 0610012D14 (LOC284363), mRNA
XM_209149 Homo sapiens similar to hypothetical protein FLJ31030 (LOC284318), mRN/
XM_209155 Homo sapiens hypothetical protein LOC284371 (LOC284371 ), mRNA
XM_209163 Homo sapiens similar to solute carrier family 7, member 3; amino acid transp
XM_209178 Homo sapiens similar to 60S ribosomal protein L10 (QM protein homolog) (L
XM_209180 Homo sapiens similar to FKSG60 (LOC284397), mRNA
XM_209187 Homo sapiens similar to BC022651 protein (LOC284417), mRNA
XM_209196 Homo sapiens similar to HSPC323 (LOC284422), mRNA
XM_209204 Homo sapiens hypothetical protein MGC26694 (MGC26694), mRNA
XM_209227 Homo sapiens similar to KIAA0456 protein (LOC391087), mRNA
XM_209234 Homo sapiens hypothetical protein DKFZp434E1410 (DKFZp434E1410), mF
XM_209252 Homo sapiens similar to Rl KEN cDN A 4930522H 14 (LOC284546), mRNA
XM_209337 Homo sapiens similar to coiled-coil-helix-coiled-coil-helix domain containing I
XM_209363 Homo sapiens protein tyrosine phosphatase, non-receptor type substrate 1-li
XM_209384 Homo sapiens hypothetical LOC284859 (LOC284859), mRNA
XM_209394 Homo sapiens hypothetical LOC284874 (LOC284874), mRNA
XM_209408 Homo sapiens similar to bA436C9.2 (PUTATIVE novel protein similar to part
XM_209423 Homo sapiens similar to bA395L14.5 (novel phosphoglucomutase like proteii
XM_209429 Homo sapiens similar to ARHQ protein (LOC284988), mRNA
XM_209489 Homo sapiens similar to CG14853-PB (LOC285141), mRNA
XM_209490 Homo sapiens hypothetical protein LOC285148 (LOC285148), mRNA
XM_209500 Homo sapiens similar to 60S ribosomal protein L10 (QM protein homolog) (L
XM_209505 Homo sapiens similar to K1AA0445 protein (LOC285188), mRNA
XM_209509 Homo sapiens similar to RIKEN cDNA 0710001 B24 (LOC285193), mRNA
XM_209550 Homo sapiens similar to Carboxypeptidase N 83 kDa chain (Carboxypeptidai
XM_209554 Homo sapiens similar to FSHD Region Gene 2 protein (LOC285299), mRNA
XM_209559 Homo sapiens similar to MGC27169 protein (LOC285303), mRNA
XM_209563 Homo sapiens hypothetical LOC285311 (LOC285311 ), mRNA
XM_209569 Homo sapiens similar to ataxin-1 ubiquitin-like interacting protein (LOC28532
XM_209579 Homo sapiens hypothetical protein LOC285346 (LOC285346), mRNA
XM_209597 Homo sapiens similar to beta-1 ,4-mannosyltransferase; beta-1 ,4 mannosyltπ
XM_209604 Homo sapiens hypothetical LOC285423 (LOC285423), mRNA
XM_209607 Homo sapiens hypothetical protein LOC285429 (LOC285429), mRNA
XM_209612 Homo sapiens hypothetical protein LOC285440 (LOC285440), mRNA
XM_209616 Homo sapiens similar to APACD protein (LOC285453), mRNA
XM_209640 Homo sapiens hypothetical protein LOC285501 (LOC285501), mRNA
XM_209643 Homo sapiens similar to hypothetical protein FLJ20035 (LOC285510), mRN/
XM_209655 Homo sapiens similar to beta-1 ,4-mannosyltransferase; beta-1 ,4 mannosyltr;
XM_209656 Homo sapiens hypothetical protein LOC285550 (LOC285550), mRNA
XM_209668 Homo sapiens similar to RIKEN cDNA 1700007I06 (LOC285588), mRNA
XM_209695 Homo sapiens similar to Chromosome-associated kinesin KIF4A (Chromokin
XM_209700 Homo sapiens similar to FLJ00052 protein (LOC285647), mRNA
XM_209704 Homo sapiens similar to hypothetical protein (LOC285658), mRNA
XM_209719 Homo sapiens hypothetical protein LOC285679 (LOC285679), mRNA
XM_209728 Homo sapiens similar to C-terminal binding protein 2 isoform 2; ribeye (LOG.
XM_209741 Homo sapiens similar to Translationally controlled tumor protein (TCTP) (p23
XM_209753 Homo sapiens similar to FLJ00254 protein (LOC285770), mRNA
XM_209824 Homo sapiens similar to matrilin 2 precursor (LOC285929), mRNA
XM_209827 Homo sapiens similar to 40S ribosomal protein S26 (LOC285938), mRNA
XM_209870 Homo sapiens similar to FLJ 10408 protein (LOC286040), mRNA
XM_209889 Homo sapiens similar to hypothetical protein (LOC286076), mRNA
XM_209893 Homo sapiens similar to TBP-associated factor 15 isoform 1 ; TAF15 RNA po
XM_209902 Homo sapiens zinc finger protein 252 (ZNF252), mRNA
XM_209910 Homo sapiens similarto chromosome 15 open reading frame 2 (LOC286129
XM_209913 Homo sapiens similar to ring finger protein 5 (LOC286140), mRNA
XM_209918 Homo sapiens similarto RIKEN cDNA 4930533G20 (L0C286151), mRNA
XM_209920 Homo sapiens similar to mCBP (LOC286157), mRNA
XM_209936 Homo sapiens similar to RIKEN cDNA 1700011J18 (LOC286187), mRNA
XM_209941 Homo sapiens hypothetical protein LOC286207 (LOC286207), mRNA
XMJ209955 Homo sapiens similar to beta-tubulin 4Q (LOC286222), mRNA
XM_209967 Homo sapiens similar to NAD-dependent malic enzyme, mitochondrial precu
XMJ210022 Homo sapiens GTPase activating RANGAP domain-like 1 (GARNL1), mRNA
XM_210035 Homo sapiens similar to putative UST1-like organic anion transporter (LOC2I
XM_210042 Homo sapiens similar to mitochondrial ribosome recycling factor isoform 1 (L
XM_210048 Homo sapiens hypothetical protein LOC286436 (LOC286436), mRNA
XM_210054 Homo sapiens similar to dJ341D10.1 (novel protein) (LOC286453), mRNA
XM_210062 Homo sapiens ras-related C3 botulinum toxin substrate family member 4 (RA
XM_210082 Homo sapiens similar to 40S ribosomal protein S26 (LOC286513), mRNA
XM_210094 Homo sapiens similar to 40S ribosomal protein S26 (LOC286539), mRNA
XM_210119 Homo sapiens similar to Olfactory receptor 5AT1 (LOC284532), mRNA
XM_210168 Homo sapiens similar to Olfactory receptor 4C13 (LOC283092), mRNA
XM_210169 Homo sapiens similar to Olfactory receptor 4C12 (LOC283093), mRNA
XM_210180 Homo sapiens similar to odorant receptor HOR3beta2 (LOC283110), mRNA
XM_210181 Homo sapiens similar to odorant receptor HOR3beta1 (LOC283111), mRNA
XM_210184 Homo sapiens similar to tripartite motif-containing 43 (LOC283117), mRNA
XM_210186 Homo sapiens similar to Olfactory receptor 8B4 (LOC283162), mRNA
XM_210193 Homo sapiens similar to Olfactory receptor 9G4 (LOC283189), mRNA
XM_210196 Homo sapiens similar to seven transmembrane helix receptor (LOC283193),
XM_210242 Homo sapiens similarto olfactory receptor GA_x6K02T2PULF-11031172-11(
XM_210282 Homo sapiens similar to Olfactory receptor 4K5 (LOC283621 ), mRNA
XM_210324 Homo sapiens similarto Vacuolar ATP synthase 16 kDa proteolipid subunit (
XM_210334 Homo sapiens similar to 60S ribosomal protein L29 (Cell surface heparin bin*
XM _210365 Homo sapiens similar to Ribosomal protein L24-like (L0C284288), mRNA
XM_210382 Homo sapiens similar to Olfactory receptor 2Z1 (LOC284383), mRNA
XM_210394 Homo sapiens similar to Contains similarity to extensin (atExtl ) from Arabido
XM_210400 Homo sapiens similar to autoantigen NOR-90 (LOC285031 ), mRNA
XM_210411 Homo sapiens similar to Oligophrenin 1 (LOC285101), mRNA
XM_210501 Homo sapiens similarto HSPC182 protein (LOC286528), mRNA
XM_210515 Homo sapiens similar to testis specific protein, Y-linked (LOC286561 ), mRN/
XM_210543 Homo sapiens similar to SPBPJ4664.02 (LOC285253), mRNA
XM_210557 Homo sapiens similar to heterochromatin-specific nonhistone protein (LOC2J
XM_210559 Homo sapiens similar to Retinoic acid receptor beta (RAR-beta) (RAR-epsilo
XM_210562 Homo sapiens hypothetical protein LOC285335 (LOC285335), mRNA
XM_210576 Homo sapiens similar to RIKEN cDNA 4921517D21 (LOC285405), mRNA
XM_210581 Homo sapiens claudin 22 (CLDN22), mRNA
XM_210613 Homo sapiens similar to Transcription initiation factor TFIID 28 kDa subunit (
XM_210642 Homo sapiens similar to Transcription initiation factor TFIID 28 kDa subunit (
XM_210752 Homo sapiens similar to Olfactory receptor 13C9 (LOC286362), mRNA
XM_210755 Homo sapiens similar to Olfactory receptor 13D1 (LOC286365), mRNA
XM_210787 Homo sapiens similar to Olfactory receptor 10H5 (LOC284433), mRNA
XM_210826 Homo sapiens similar to Wiskott-AIdrich syndrome protein family member 4 (
XM_210856 Homo sapiens similar to Ras suppressor protein 1 (LOC283029), mRNA
XM_210860 Homo sapiens hypothetical LOC283034 (LOC283034), mRNA
XM_210876 Homo sapiens hypothetical LOC283065 (LOC283065), mRNA
XM_210906 Homo sapiens hypothetical LOC283166 (LOC283166), mRNA
XM_210908 Homo sapiens similar to RIKEN cDNA A930008A22 (LOC283157), mRNA
XM_211009 Homo sapiens hypothetical LOC283389 (LOC283389), mRNA
XM_211028 Homo sapiens hypothetical protein LOC283403 (LOC283403), mRNA
XM_211040 Homo sapiens hypothetical LOC283440 (LOC283440), mRNA
XM_211079 Homo sapiens hypothetical LOC283530 (LOC283530), mRNA
XM_211086 Homo sapiens hypothetical LOC283553 (LOC283553), mRNA
XM_211088 Homo sapiens hypothetical LOC283604 (LOC283604), mRNA
XM_211089 Homo sapiens hypothetical LOC283586 (LOC283586), mRNA
XM_211090 Homo sapiens hypothetical protein LOC283587 (LOC283587), mRNA
XM_211092 Homo sapiens hypothetical LOC283583 (LOC283583), mRNA
XM_211108 Homo sapiens hypothetical LOC283584 (LOC283584), mRNA
XM_211174 Homo sapiens hypothetical LOC283710 (LOC283710), mRNA
XM_211197 Homo sapiens hypothetical LOC283780 (LOC283780), mRNA
XM_211244 Homo sapiens hypothetical LOC283895 (LOC283895), mRNA
XM_211246 Homo sapiens similar to BTG3 associated nuclear protein isoform a; BANP r
XM_211251 Homo sapiens hypothetical LOC283902 (LOC283902), mRNA
XM_211287 Homo sapiens hypothetical protein LOC283999 (LOC283999), mRNA
XM_211291 Homo sapiens hypothetical protein LOC283994 (LOC283994), mRNA
XM_211305 Homo sapiens hypothetical protein LOC284021 (LOC284021 ), mRNA
XM_211339 Homo sapiens hypothetical LOC284120 (LOC284120), mRNA
XM_211345 Homo sapiens hypothetical LOC284134 (LOC284134), mRNA
XM_211367 Homo sapiens hypothetical LOC284184 (LOC284184), mRNA
XM_211403 Homo sapiens CD33 antigen-like 3 (CD33L3), mRNA
XM_211408 Homo sapiens hypothetical LOC284260 (LOC284260), mRNA
XM_211413 Homo sapiens hypothetical LOC284275 (LOC284275), mRNA
XM_211422 Homo sapiens hypothetical LOC284303 (LOC284303), mRNA
XM_211432 Homo sapiens hypothetical LOC284321 (LOC284321), mRNA
XM_211447 Homo sapiens hypothetical LOC284409 (LOC284409), mRNA
XM_211460 Homo sapiens hypothetical protein LOC284434 (LOC284434), mRNA
XM_211509 Homo sapiens hypothetical LOC284527 (LOC284527), mRNA
XM_211518 Homo sapiens hypothetical LOC284555 (LOC284555), mRNA
XM_211529 Homo sapiens hypothetical protein LOC284591 (LOC284591 ), mRNA
XM_211557 Homo sapiens hypothetical LOC284623 (LOC284623), mRNA
XM_211573 Homo sapiens hypothetical LOC284646 (LOC284646), mRNA
XM_211627 Homo sapiens hypothetical LOC284754 (LOC284754), mRNA
XM_211694 Homo sapiens hypothetical LOC284931 (LOC284931 ), mRNA
XM_211707 Homo sapiens hypothetical LOC284968 (LOC284968), mRNA
XM_211736 Homo sapiens hypothetical protein LOC285016 (LOC285016), mRNA
XM_211749 Homo sapiens hypothetical LOC285047 (LOC285047), mRNA
XM_211764 Homo sapiens hypothetical LOC285095 (LOC285095), mRNA
XM_211768 Homo sapiens hypothetical LOC285110 (LOC285110), mRNA
XM_211805 Homo sapiens hypothetical protein LOC285205 (LOC285205), mRNA
XM_211816 Homo sapiens hypothetical LOC285248 (LOC285248), mRNA
XM_211837 Homo sapiens hypothetical LOC285307 (LOC285307), mRNA
XM_211843 Homo sapiens hypothetical protein LOC285326 (LOC285326), mRNA
XM_211853 Homo sapiens similar to hypothetical protein MG11009.4 (LOC285344), mRf
XM_211858 Homo sapiens hypothetical protein FLJ12205 (LOC285359), mRNA
XM_211871 Homo sapiens hypothetical LOC285382 (LOC285382), mRNA
XM_211896 Homo sapiens hypothetical LOC285435 (LOC285435), mRNA
XM_211908 Homo sapiens similar to unc-93 homolog B1 ; unc93 (C.elegans) homolog B;
XM_211923 Homo sapiens hypothetical LOC285509 (LOC285509), mRNA
XM_211983 Homo sapiens hypothetical LOC285694 (LOC285694), mRNA
XM_211988 Homo sapiens hypothetical LOC285711 (LOC285711), mRNA
XM_211995 Homo sapiens hypothetical LOC285721 (LOC285721 ), mRNA
XM_212013 Homo sapiens hypothetical LOC285777 (LOC285777), mRNA
XM_212022 Homo sapiens hypothetical LOC285793 (LOC285793), mRNA
XM_212061 Homo sapiens similar to THAP domain containing 5 (LOC285872), mRNA
XM_212067 Homo sapiens hypothetical LOC285890 (LOC285890), mRNA
XM_212094 Homo sapiens hypothetical LOC285919 (LOC285919), mRNA
XM_212123 Homo sapiens hypothetical LOC285995 (LOC285995), mRNA
XM_212162 Homo sapiens similar to CG3104-PA (LOC286080), mRNA
XM_212170 Homo sapiens hypothetical protein LOC286094 (LOC286094), mRNA
XM_212238 Homo sapiens hypothetical protein LOC286235 (LOC286235), mRNA
XM_212241 Homo sapiens DKFZP434M131 protein (DKFZP434M131), mRNA
XM_212319 Homo sapiens hypothetical LOC286441 (LOC286441), mRNA
XM_212326 Homo sapiens hypothetical LOC286478 (LOC286478), mRNA
XM_212581 Homo sapiens zinc finger protein 311 (ZNF311), mRNA
XM_290185 Homo sapiens similar to hypothetical protein MGC5560 (LOC338598), mRN/
XM_290225 Homo sapiens similar to pote protein; Expressed in prostate, ovary, testis, ar
XM_290331 Homo sapiens gamma-glutamyltransferase 2 (GGT2), mRNA
XM_290342 Homo sapiens similar to immunoglobulin superfamily, member 3; immunoglo
XM_290345 Homo sapiens similar to eukaryotic translation initiation factor 3, subunit 5 ep
XM_290351 Homo sapiens similar to Nedd-4-like ubiquitin-protein ligase WWP1 (WW do
XM_290385 Homo sapiens similar to solute carrier family 29 (nucleoside transporters), m*
XM_290389 Homo sapiens similar to solute carrier family 29 (nucleoside transporters), m*
XM_290401 Homo sapiens hypothetical protein LOC340318 (LOC340318), mRNA
XM_290462 Homo sapiens KIAA1674 (KI AA1674), mRNA
XM_290463 Homo sapiens family with sequence similarity 22, member A (FAM22A), mRt
XM_290482 Homo sapiens hypothetical protein FLJ 10824 (FLJ 10824), mRNA
XM_290501 Homo sapiens hypothetical LOC338661 (LOC338661), mRNA
XM_290502 Homo sapiens KIAA1030 protein (KIAA1030), mRNA
XM_290506 Homo sapiens splicing factor 3b, subunit 2, 145kDa (SF3B2), mRNA
XM_290509 Homo sapiens hypothetical protein LOC338692 (LOC338692), mRNA
XM_290516 Homo sapiens myotonic dystrophy protein kinase like protein (HSMDPKIN), i
XM_290517 Homo sapiens KIAA0404 protein (KIAA0404), mRNA
XM_290527 Homo sapiens ubiquitin specific protease 35 (USP35), mRNA
XM_290536 Homo sapiens CTD-binding SR-like protein rA9 (KIAA1542), mRNA
XM_290546 Homo sapiens KIAA0830 protein (KIAA0830), mRNA
XM_290547 Homo sapiens similar to Oligophrenin 1 (LOC338734), mRNA
XM_290552 Homo sapiens cyclic nucleotide gated channel alpha 4 (CNGA4), mRNA
XM_290558 Homo sapiens similar to C1 q-related factor precursor (LOC338761 ), mRNA
XM_290559 Homo sapiens glutamate receptor interacting protein 1 (GRIP1), mRNA
XM_290579 Homo sapiens hypothetical protein LOC338797 (LOC338797), mRNA
XM_290592 Homo sapiens huntingtin interacting protein-1 -related (HIP1R), mRNA
XM_290597 Homo sapiens hypothetical protein LOC283464 (LOC283464), mRNA
XM_290598 Homo sapiens dendrin (DDN), mRNA
XM_290600 Homo sapiens similar to Reticulocalbin 1 precursor (LOC338851), mRNA
XM_290609 Homo sapiens hypothetical LOC338914 (LOC338914), mRNA
XM_290615 Homo sapiens hypothetical protein DKFZp762F0713 (DKFZp762F0713), mR
XM_290626 Homo sapiens similar to chromosome 1 open reading frame 36 (LOC338934
XM_290629 Homo sapiens chromosome 14 open reading frame 78 (C14orf78), mRNA
XM_290631 Homo sapiens glucuronyl C5-epimerase (GLCE), mRNA
XM_290645 Homo sapiens hypothetical protein DKFZp434J0617 (DKFZp434J0617), mR
XM_290660 Homo sapiens similar to HP95 (LOC339003), mRNA
XM_290667 Homo sapiens KIAA0350 protein (KIAA0350), mRNA
XM_290670 Homo sapiens KIAA0220 protein (KIAA0220), mRNA
XM_290671 Homo sapiens hypothetical protein LOC339047 (LOC339047), mRNA
XM_290684 Homo sapiens DKFZP434I216 protein (DKFZP434I216), mRNA
XM_290702 Homo sapiens similar to My016 protein (LOC339088), mRNA
XM_290704 Homo sapiens hypothetical protein FLJ12270 (FLJ12270), mRNA
XM_290712 Homo sapiens hypothetical protein MGC46336 (MGC46336), mRNA
XM_290714 Homo sapiens RAB43, member RAS oncogene family (RAB43), mRNA
XM_290722 Homo sapiens similar to RIKEN cDNA 2610003J06 (LOC339123), mRNA
XM_290732 Homo sapiens KIAA1917 protein (KIAA1917), mRNA
XMJ.90734 Homo sapiens similar to ataxin 2 binding protein 1 isoform gamma; hexariboi
XM_290737 Homo sapiens KIAA1871 protein (KIAA1871), mRNA
XM_290743 Homo sapiens similar to hypothetical protein FLJ36492 (LOC339184), mRN/
XM_290755 Homo sapiens hypothetical protein FLJ35848 (FLJ35848), mRNA
XM_290758 Homo sapiens KIAA0553 protein (KIAA0553), mRNA
XM_290768 Homo sapiens KIAA1554 protein (KIAA1554), mRNA
XM_290777 Homo sapiens hypothetical protein LOC339231 (LOC339231), mRNA
XM_290780 Homo sapiens similar to Envoplakin (210 kDa paraneoplastic pemphigus ant
XM_290781 Homo sapiens similar to Keratin, type I cytoskeletal 14 (Cytokeratin 14) (K14
XM_290782 Homo sapiens similar to Keratin, type I cytoskeletal 16 (Cytokeratin 16) (K16
XMJ-90786 Homo sapiens similar to endozepine-like protein (LOC339253), mRNA
XM_290793 Homo sapiens kinase suppressor of ras (KSR), mRNA
XM_290795 Homo sapiens hypothetical protein DKFZp667M2411 (DKFZp667M2411 ), ml
XM_290799 Homo sapiens KIAA1501 protein (K1AA1501), mRNA
XM_290809 Homo sapiens TAF4b RNA polymerase II, TATA box binding protein (TBP)-a
XM_290811 Homo sapiens KIAA1713 protein (KIAA1713), mRNA
XM_290817 Homo sapiens hypothetical protein FLJ34907 (FLJ34907), mRNA
XM_290818 Homo sapiens Spir-1 protein (Spir-1), mRNA
XM_290820 Homo sapiens hypothetical protein FLJ10211 (FLJ10211 ), mRNA
XM_290822 Homo sapiens hypothetical protein LOC284367 (LOC284367), mRNA
XM_290829 Homo sapiens homolog of Drosophila Intersex (Intersex), mRNA
XM_290831 Homo sapiens hypothetical protein LOC339321 (LOC339321), mRNA
XM_290835 Homo sapiens zinc finger protein 181 (HHZ181) (ZNF181), mRNA
XM_290838 Homo sapiens hypothetical protein LOC339324 (LOC339324), mRNA
XM_290842 Homo sapiens leucine rich repeat and fibronectin type III domain containing '
XM_290848 Homo sapiens hypothetical protein LOC339344 (LOC339344), mRNA
XM_290850 Homo sapiens glutamate receptor, ionotropic, N-methyl-D-aspartate 3B (GRI
XM_290854 Homo sapiens G protein-coupled receptor 108 (GPR108), mRNA
XM_290865 Homo sapiens similar to Zinc finger protein 93 (Zinc finger protein HTF34) (L
XM_290866 Homo sapiens similar to 4930572L20Rik protein (LOC339377), mRNA
XM_290867 Homo sapiens RalGEF-like protein 3, mouse homolog (RGL3), mRNA
XM_290872 Homo sapiens similar to Pyruvate kinase, M2 isozyme (LOC339395), mRNA
XM_290902 Homo sapiens hypothetical protein LOC339448 (LOC339448), mRNA
XM_290922 Homo sapiens hypothetical protein MGC27277 (MGC27277), mRNA
XM_290923 Homo sapiens KI AA1639 protein (KIAA1639), mRNA
XM_290925 Homo sapiens hypothetical LOC339494 (LOC339494), mRNA
XM_290927 Homo sapiens similar to bA476H5.3 (novel protein similar to septin) (LOC33!
XM_290936 Homo sapiens hypothetical protein MGC35030 (MGC35030), mRNA
XM_290941 Homo sapiens prion protein interacting protein (PRNPIP), mRNA
XM_290944 Homo sapiens KIAA0842 protein (KIAA0842), mRNA
XM_290948 Homo sapiens hypothetical protein LOC343071 (LOC343071), mRNA
XM_290949 Homo sapiens similar to hypothetical protein (LOC339553), mRNA
XM_290972 Homo sapiens novel C3HC4 type Zinc finger (ring finger) (FLJ12747), mRNA
XM _290973 Homo sapiens solute carrier family 35, member E4 (SLC35E4), mRNA
XM_290985 Homo sapiens hypothetical protein LOC339692 (LOC339692), mRNA
XM_290994 Homo sapiens similar to speckle-type POZ protein (LOC339744), mRNA
XM_291001 Homo sapiens myosin VIIB (MY07B), mRNA
XM_291005 Homo sapiens similar to 25-hydroxyvitamin D-1 alpha hydroxylase, mitochon
XMJ291007 Homo sapiens hypothetical protein LOC339766 (LOC339766), mRNA
XM_291015 Homo sapiens likely homolog of rat kinase D-interacting substance of 220 kC
XMJ291016 Homo sapiens similarto RIKEN cDNA 1700093K21 (LOC339804), mRNA
XM_291017 Homo sapiens similar to PRO1094 (LOC339793), mRNA
XM_291019 Homo sapiens hypothetical protein FLJ13305 (FLJ13305), mRNA
XM_291020 Homo sapiens KIAA2012 protein (LOC339809), mRNA
XM_291028 Homo sapiens hypothetical protein DKFZp434A128 (DKFZp434A128), mRN;
XM_291054 Homo sapiens similar to beta-1 ,4-mannosyltransferase; beta-1 ,4 mannosyltπ
XM_291055 Homo sapiens KIAA1268 protein (KIAA1268), mRNA
XM_291057 Homo sapiens cytoplasmic linker associated protein 2 (CLASP2), mRNA
XM_291059 Homo sapiens hypothetical protein LOC339896 (LOC339896), mRNA
XM_291062 Homo sapiens similar to K06A9.1b.p (KIAA2018), mRNA
XM_291063 Homo sapiens hypothetical protein LOC339903 (LOC339903), mRNA
XM_291064 Homo sapiens KIAA0540 protein (KIAA0540), mRNA
XM_291074 Homo sapiens hypothetical protein FLJ33674 (FLJ33674), mRNA
XM_291075 Homo sapiens hypothetical LOC339926 (LOC339926), mRNA
XM_291077 Homo sapiens hypothetical protein BC009862 (LOC90113), mRNA
XM_291080 Homo sapiens KIAA0804 protein (KIAA0804), mRNA
XM_291085 Homo sapiens KIAA1204 protein (CDGAP), mRNA
XM_291088 Homo sapiens similar to hypothetical protein SB153 isoform 1 (LOC339944),
XM_291095 Homo sapiens similar to glutamate receptor, ionotropic, N-methyl D-aspartati
XM_291099 Homo sapiens similar to Hypothetical protein MGC38937 (LOC339977), mRf
XM_291105 Homo sapiens transcriptional adaptor 2 (ADA2 homolog, yeast)-beta (MGC2
XM_291106 Homo sapiens KIAA0232 gene product (KIAA0232), mRNA
XM_291111 Homo sapiens G protein-coupled receptor 125 (GPR125), mRNA
XM_291117 Homo sapiens similar to succinate dehydrogenase flavoprotein subunit (LOC
XM_291120 Homo sapiens hypothetical protein LOC340024 (LOC340024), mRNA
XM_291128 Homo sapiens KIAA1311 protein (KIAA1311), mRNA
XM_291137 Homo sapiens similar to hypothetical protein D630003M21 (LOC389256), mF
XM_291139 Homo sapiens similar to RIKEN cDNA 9330196J05 (LOC340075), mRNA
XM_291141 Homo sapiens KIAA0303 protein (KIAA0303), mRNA
XM_291142 Homo sapiens hypothetical protein BC014311 (LOC115548), mRNA
XM_291144 Homo sapiens similar to bA110H4.2 (similar to membrane protein) (LOC340C
XM_291154 Homo sapiens similar to hypothetical protein D730019B10 (LOC340152), mF
XM_291159 Homo sapiens ion transporter protein (DKFZP434F011), mRNA
XM_291161 Homo sapiens similar to developmental pluripotency associated 5; embryonε
XM_291169 Homo sapiens similar to precursor peptide (LOC340204), mRNA
XM_291170 Homo sapiens similar to Hypothetical protein KIAA0036 (LOC340192), mRN;
XM_291181 Homo sapiens similar to LINE-1 REVERSE TRANSCRIPTASE HOMOLOG (I
XM_291200 Homo sapiens similar to CAGL79 (LOC340221), mRNA
XM_291202 Homo sapiens zinc finger protein 479 (ZNF479), mRNA
XM_291204 Homo sapiens hypothetical LOC340228 (LOC340228), mRNA
XM_291208 Homo sapiens similar to hypothetical ZNF-like protein (LOC340246), mRNA
XM_291222 Homo sapiens DKFZP586J0619 protein (DKFZP586J0619), mRNA
XM_291223 Homo sapiens myosin IG (MY01G), mRNA
XM_291241 Homo sapiens intracellular membrane-associated calcium-independent phos
XM_291247 Homo sapiens similar to Piccolo protein (Aczonin) (LOC389530), mRNA
XM_291253 Homo sapiens KIAA0146 protein (KIAA0146), mRNA
XM_291261 Homo sapiens zinc finger protein 517 (ZNF517), mRNA
XM_291262 Homo sapiens zinc finger protein 251 (ZNF251), mRNA
XM_291266 Homo sapiens 5-oxoprolinase (ATP-hydrolysing) (OPLAH), mRNA
XM_291268 Homo sapiens glutamate receptor, ionotropic, N-methyl D-asparate-associati
XM_291269 Homo sapiens hypothetical protein KIAA1875 (KIAA1875), mRNA
XM_291270 Homo sapiens similar to unnmaed protein product (LOC340393), mRNA
XM_291277 Homo sapiens hypothetical protein DKFZp761 P0423 (DKFZp761 P0423), mF
XM_291282 Homo sapiens hypothetical protein LOC157697 (LOC157697), mRNA
XM_291291 Homo sapiens KIAA0725 protein (KIAA0725), mRNA
XM_291314 Homo sapiens F-box only protein 10 (FBXO10), mRNA
XM_291315 Homo sapiens KIAA1815 (KIAA1815), mRNA
XM J291321 Homo sapiens similar to Nance-Horan syndrome protein (LOC340527), mR
XM_291322 Homo sapiens KIAA2001 protein (KIAA2001), mRNA
XM_291326 Homo sapiens KIAA2022 protein (KIAA2022), mRNA
XM_291334 Homo sapiens similar to hypothetical protein MGC15737 (LOC340543), mRf
XM_291335 Homo sapiens hypothetical protein LOC340542 (LOC340542), mRNA
XM_291339 Homo sapiens hypothetical protein LOC340562 (LOC340562), mRNA
XM_291344 Homo sapiens hypothetical protein FLJ 12649 (FLJ 12649), mRNA
XM_291345 Homo sapiens KIAA0522 protein (KIAA0522), mRNA
XM ?91346 Homo sapiens similar to carbonic anhydrase VB, mitochondrial precursor; ca
XM_291378 Homo sapiens similar to C4b-binding protein alpha chain precursor (C4bp) (F
XM_291387 Homo sapiens similar to DAAT9248 (LOC339398), mRNA
XM_291392 Homo sapiens similar to Arylacetamide deacetylase (AADAC) (LOC343066),
XM_291394 Homo sapiens hypothetical protein LOC343068 (LOC343068), mRNA
XM_291395 Homo sapiens similar to Heterogeneous nuclear ribonucleoprotein C-like dJ£
XM_291396 Homo sapiens similar to Hypothetical protein DJ845024.2 (LOC400735), mF
XMJ291400 Homo sapiens similar to chromosome 14 open reading frame 24 (LOC34308
XM_291419 Homo sapiens G protein-coupled receptor 153 (GPR153), mRNA
XM_291428 Homo sapiens similar to ribosomal protein L26 (LOC343153), mRNA
XM_291435 Homo sapiens similar to Olfactory receptor 6F1 (LOC343169), mRNA
XM_291436 Homo sapiens similar to Olfactory receptor 5AY1 (LOC343170), mRNA
XM_291437 Homo sapiens olfactory receptor, family 1 , subfamily C, member 1 (OR1 C1 ),
XM_291438 Homo sapiens similar to seven transmembrane helix receptor (LOC343171 ),
XM__291439 Homo sapiens similar to seven transmembrane helix receptor (LOC343172),
XM _291441 Homo sapiens similar to Olfactory receptor 2T3 (LOC343173), mRNA
XM_291464 Homo sapiens similar to KIAA0433 protein (LOC343221), mRNA
XM_291485 Homo sapiens similar to Myosin-binding protein H (MyBP-H) (H-protein) (LO(
XM_291508 Homo sapiens similar to Farnesyl pyrophosphate synthetase (FPP synthetas
XM_291533 Homo sapiens similar to basic transcription factor 3 (LOC343363), mRNA
XM_291543 Homo sapiens similar to KIAA0454 protein (LOC343381 ), mRNA
XM_291544 Homo sapiens similar to peptidyl-Pro cis trans isomerase (LOC343384), mRI
XM_291548 Homo sapiens similar to Olfactory receptor 10R2 (LOC343406), mRNA
XM_291569 Homo sapiens similar to IFGP6 (LOC343413), mRNA
XM_291577 Homo sapiens similar to RIKEN cDNA 1810009010 (LOC339521), mRNA
XM_291584 Homo sapiens similar to Phosphorylase B kinase alpha regulatory chain, ske
XM_291607 Homo sapiens similar to dJ1182A14.5.1 (novel gene (isoform 1)) (L0C3435C
XM_291623 Homo sapiens similar to hypothetical protein 4833401 D15 (L0C343521 ), mF
XM_291625 Homo sapiens similar to Hypothetical protein DJ845024.2 (L0C374947), mF
XM_291627 Homo sapiens similar to oral cancer overexpressed 2; transmembrane protei
XM_291638 Homo sapiens hypothetical protein LOC343070 (LOC343070), mRNA
XM_291643 Homo sapiens similar to Ig kappa chain (L0C339562), mRNA
XM_291645 Homo sapiens similar to Olfactory receptor 2T5 (LOC343563), mRNA
XM_291663 Homo sapiens similar to bA304l5.1 (novel lipase) (LOC340654), mRNA
XM_291671 Homo sapiens hypothetical protein LOC282996 (LOC282996), mRNA
XM_291697 Homo sapiens similar to RIKEN cDNA A930010E21 (LOC340745), mRNA
XM_291698 Homo sapiens similar to Rpl7a protein (LOC340749), mRNA
XM_291704 Homo sapiens similar to double homeobox, 4; double homeobox protein 4 (L
XM_291716 Homo sapiens similar to NK-type homeobox (LOC340784), mRNA
XM_291723 Homo sapiens protein RAKc (LOC340811 ), mRNA
XM_291725 Homo sapiens alpha 2,8-sialyltransferase (ST8SIA-VI), mRNA
XM_291726 Homo sapiens similarto protein of unknown function (LOC340843), mRNA
XM_291729 Homo sapiens TAF3 RNA polymerase II, TATA box binding protein (TBP)-as
XM_291741 Homo sapiens dual specificity phosphatase and pro isomerase domain contε
XM_291745 Homo sapiens similarto 40S ribosomal protein S26 (LOC338611), mRNA
XM_291757 Homo sapiens similar to protein of unknown function (LOC340893), mRNA
XM_291763 Homo sapiens similar to bA508N22.1 (HSPC025) (LOC340947), mRNA
XM_291767 Homo sapiens similar to hypothetical protein FLJ10213 (LOC340900), mRN/
XM_291770 Homo sapiens similar to breast cancer antigen NY-BR-1 (LOC340913), mRN
XM_291771 Homo sapiens similar to HSPC132 (LOC338617), mRNA
XM_291786 Homo sapiens similar to Phosphorylase B kinase gamma catalytic chain, ske
XM_291793 Homo sapiens similar to Chain A, Crystal Structure Of The Radixin Ferm Dor
XM_291806 Homo sapiens similar to seven transmembrane helix receptor (LOC340980),
XM_291816 Homo sapiens otogelin (OTOG), mRNA
XM_291838 Homo sapiens similar to Olfactory receptor 8D4 (LOC338662), mRNA
XM_291857 Homo sapiens similar to ENSANGP00000020885 (LOC341098), mRNA
XM_291859 Homo sapiens olfactory receptor, family 5, subfamily F, member 1 (OR5F1),
XM_291862 Homo sapiens similar to Olfactory receptor 5AP2 (LOC338675), mRNA
XM_291885 Homo sapiens similar to Tryptophanyl-tRNA synthetase (Tryptophan-tRNA Ii
XM_291892 Homo sapiens similar to olfactory receptor MOR101-1 (LOC341152), mRNA
XM_291924 Homo sapiens similar to Olfactory receptor 4F3 (LOC338718), mRNA
XM_291943 Homo sapiens similar to Elongation factor 1 -alpha 2 (EF-1 -alpha-2) (Elongati
XM_291947 Homo sapiens hephaestin-like (LOC341208), mRNA
XM_291974 Homo sapiens similar to hypothetical protein FLJ33167 (LOC338750), mRN/
XM_291977 Homo sapiens similar to Olfactory receptor 52L1 (LOC338751 ), mRNA
XM_291980 Homo sapiens similar to Olfactory receptor 2AG1 (HT3) (LOC338755), mRN;
XM_291981 Homo sapiens similar to hP4 olfactory receptor (LOC341276), mRNA
XM_291986 Homo sapiens olfactory receptor, family 10, subfamily A, member 3 (OR10AC
XM_291989 Homo sapiens similar to hypothetical protein (LOC338756), mRNA
XM_291991 Homo sapiens similar to protein tyrosine phosphatase, receptor type, Q isofo
XM_292012 Homo sapiens similar to Polyadenylate-binding protein 1 (Poly(A)-binding pre
XM_292021 Homo sapiens similar to hypothetical protein (LOC341346), mRNA
XM_292023 Homo sapiens similar to ribosomal protein L31 (LOC341356), mRNA
XM_292027 Homo sapiens similar to Dag1-prov protein (LOC341370), mRNA
XM_292029 Homo sapiens hypothetical LOC341371 (LOC341371), mRNA
XM_292035 Homo sapiens similar to olfactory specific medium-chain acyl CoA synthetas*
XM_292046 Homo sapiens similar to ribosomal protein L31 (LOC341412), mRNA
XM_292048 Homo sapiens similar to Heat shock factor protein 2 (HSF 2) (Heat shock trai
XM_292049 Homo sapiens similar to olfactory receptor MOR114-1 (LOC341416), mRNA
XM_292051 Homo sapiens similar to Olfactory receptor 6C4 (LOC341418), mRNA
XM_292064 Homo sapiens similar to fertilin alpha subunit (LOC338792), mRNA
XM_292085 Homo sapiens similar to peptidyl-Pro cis trans isomerase (LOC341457), mRI
XM_292093 Homo sapiens liver-specific organic anion transporter 3 (LST-3), mRNA
XM_292098 Homo sapiens similar to hypothetical protein (LOC341461), mRNA
XM _292109 Homo sapiens similar to 60S ribosomal protein L23a (LOC341511 ), mRNA
XM_292122 Homo sapiens similar to neurofilament-like protein (LOC338829), mRNA
XM_292136 Homo sapiens similar to seven transmembrane helix receptor (LOC341568),
XM_292160 Homo sapiens similar to Serine/threonine-protein kinase Nek1 (NimA-related
XM_292184 Homo sapiens similar to immune-responsive gene 1 (LOC341720), mRNA
XM_292193 Homo sapiens hypothetical protein DKFZp686J0811 (DKFZp686J0811 ), mR
XM_292197 Homo sapiens similar to bA215B13.2 (fumarate hydratase (FH) pseudogene]
XM_292210 Homo sapiens similar to ribosomal protein S12 (LOC338870), mRNA
XM_292225 Homo sapiens similar to putative pancreatic ribonuclease (LOC338879), mRI
XM_292227 Homo sapiens similar to Olfactory receptor 6S1 (LOC341799), mRNA
XM_292260 Homo sapiens solute carrier family 35, member F4 (SLC35F4), mRNA
XM_292301 Homo sapiens similar to developmental pluripotency associated 5; embryonε
XM_292357 Homo sapiens similar to Golgi autoantigen, golgin subfamily A member 6 (G<
XM_292384 Homo sapiens similar to zinc finger protein 29 (LOC342132), mRNA
XM_292389 Homo sapiens similar to ATP-binding cassette, sub-family B, member 10, mil
XM_292394 Homo sapiens similar to hypothetical protein FLJ35785 (LOC342167), mRN/
XM_292468 Homo sapiens similar to hypothetical protein (LOC342293), mRNA
XM_292503 Homo sapiens similar to hypothetical protein (LOC342355), mRNA
XM_292504 Homo sapiens similar to hypothetical protein FLJ35867 (LOC342357), mRN/
XM_292512 Homo sapiens similar to Ataxin-1 (Spinocerebellar ataxia type 1 protein hom<
XM_292527 Homo sapiens similar to Hypothetical protein MGC67567 (LOC342409), mRt
XM_292562 Homo sapiens similar to c439A6.1 (novel protein similar to heparan sulfate (c
XM_292573 Homo sapiens similar to testicular Metalloprotease-like, Disintegrin-like, Cyst
XMJ.92596 Homo sapiens similar to peptidyl-Pro cis trans isomerase (LOC342541), mRI
XM_292624 Homo sapiens similar to hypothetical protein 4932411 E22 (LOC342600), mR
XM_292627 Homo sapiens olfactory receptor, family 4, subfamily D, member 1 (OR4D1),
XM_292664 Homo sapiens similar to NADH-ubiquinone oxidoreductase 15 kDa subunit ((
XM_292674 Homo sapiens similar to leucine-rich repeat domain-containing protein (LOG!
XM_292678 Homo sapiens similar to RIKEN CDNA 2610040E16 (LOC339291), mRNA
XM _292700 Homo sapiens similar to 40S ribosomal protein S2 (LOC342808), mRNA
XM_292707 Homo sapiens similar to TFIIH basal transcription factor complex p62 subuni
XM_292717 Homo sapiens similar to KIAA1074 protein (LOC342850), mRNA
XM_292723 Homo sapiens similar to zinc finger protein 495 (LOC342933), mRNA
XM_292724 Homo sapiens similar to zinc finger protein 495 (LOC342934), mRNA
XM_292729 Homo sapiens similar to double homeobox protein (LOC342939), mRNA
XM_292740 Homo sapiens hypothetical protein LOC342892 (LOC342892), mRNA
XM_292745 Homo sapiens similar to F-box only protein 2 (LOC342897), mRNA
XM_292765 Homo sapiens zinc finger protein 404 (ZNF404), mRNA
XM_292778 Homo sapiens leucine-rich repeats and immunoglobulin-like domains 4 (LRK
XM_292779 Homo sapiens hypothetical LOC342918 (LOC342918), mRNA
XM_292784 Homo sapiens similar to hypothetical protein (LOC339351 ), mRNA
XM_292785 Homo sapiens similar to contains transmembrane (TM) region (LOC342865).
XM_292796 Homo sapiens similar to ret finger protein-like 1 (LOC342931), mRNA
XM_292803 Homo sapiens similar to Hypothetical protein CBG18249 (LOC342959), mRf
XM_292810 Homo sapiens similar to Zinc finger protein 20 (Zinc finger protein KOX13) (C
XM_292813 Homo sapiens similar to hypothetical protein FLJ38281 (LOC342972), mRN/
XM_292817 Homo sapiens similar to actin 3 - fruit fly (Drosophila melanogaster) (fragmer
XM_292819 Homo sapiens nanos homolog 3 (Drosophila) (NANOS3), mRNA
XM_292820 Homo sapiens hypothetical LOC342979 (LOC342979), mRNA
XM_292824 Homo sapiens similar to hypothetical protein MGC45408 (LOC342969), mRf
XM_292832 Homo sapiens similar to hypothetical protein FLJ12488 (LOC342991), mRN/
XM_292836 Homo sapiens similar to ribosomal protein L34; 60S ribosomal protein L34 (L
XM_292850 Homo sapiens similar to pMesogeninl (LOC343930), mRNA
XM_292873 Homo sapiens similar to 2010300C02Rik protein (LOC343990), mRNA
XM_292889 Homo sapiens similar to Gnotl homeodomain protein (LOC344022), mRNA
XM_292895 Homo sapiens similar to zinc finger protein 135 (clone pHZ-17); zinc finger pi
XM_292943 Homo sapiens Nck-associated protein 5 (NAP5), mRNA
XM_292957 Homo sapiens similar to anaphase promoting complex subunit 1 ; anaphase-|
XM_292958 Homo sapiens similar to Foxl 1c protein (LOC344167), mRNA
XM_292963 Homo sapiens similar to peptidylprolyl isomerase A (cyclophilin A) (LOC3441
XM_292968 Homo sapiens similar to Homeobox even-skipped homolog protein 2 (EVX-2
XM_292982 Homo sapiens similar to pote protein; Expressed in prostate, ovary, testis, ar
XM_293018 Homo sapiens similar to Fatty acid-binding protein, epidermal (E-FABP) (Psc
XM_293026 Homo sapiens similar to UNR-interacting protein (WD-40 repeat protein PT-\ι
XM_293029 Homo sapiens similar to cyclin-dependent kinase-like 1 (CDC2-related kinasi
XM_293034 Homo sapiens similarto RIKEN cDNA 2010316F05 (LOC344405), mRNA
XM_293042 Homo sapiens similar to ribosomal protein S12 (LOC344423), mRNA
XM_293090 Homo sapiens FLJ00204 protein (FLJ00204), mRNA
XM_293092 Homo sapiens G protein-coupled receptor 148 (GPR148), mRNA
XM_293097 Homo sapiens similar to VsaA -like protein (LOC343565), mRNA
XM_293104 Homo sapiens similar to dJ132F21.2 (Contains a novel protein similar to the
XM_293106 Homo sapiens similar to hypothetical protein FLJ32191 (LOC343593), mRN/
XM_293121 Homo sapiens similar to bA379F14.2 (novel protein) (LOC343629), mRNA
XM_293123 Homo sapiens similar to dJ1100H13.4 (putative RhoGAP domain containing
XM_293157 Homo sapiens similar to beta-galactoside alpha-2,3-sialyltransferase (LOC3^
XM_293160 Homo sapiens similar to dJ310O13.4 (novel protein similar to predicted C. el*
XM_293177 Homo sapiens similar to Zinc finger protein 85 (Zinc finger protein HPF4) (HI
XM_293225 Homo sapiens similar to RIKEN cDNA 4930583C14 (LOC343854), mRNA
XM_293226 Homo sapiens similar to POM121 membrane glycoprotein-like 1 (LOC34385!
XM_293276 Homo sapiens similar to Heat shock 27 kDa protein (HSP 27) (Stress-respon
XM_293284 Homo sapiens similar to Zinc finger protein 81 (HFZ20) (LOC347344), mRN/
XM_293293 Homo sapiens similar to BMP-2 inducible protein kinase (BIKe) (HRIHFB201
XM_293312 Homo sapiens similar to H3 histone, family 3B (LOC347376), mRNA
XM_293320 Homo sapiens similar to bA370B6.1 (similar to histone H2B) (LOC347393), n
XM_293325 Homo sapiens similar to ENSANGP00000015797 (LOC347411 ), mRNA
XM_293332 Homo sapiens similar to RIKEN cDNA 1700113017 (LOC340549), mRNA
XM_293334 Homo sapiens similar to KIAA1726 protein (LOC340554), mRNA
XM_293342 Homo sapiens similar to TGF beta-inducible nuclear protein 1 (Hairy cell leul<
XM_293352 Homo sapiens similar to P38IP protein (LOC347438), mRNA
XM_293354 Homo sapiens similar to H326 (LOC347442), mRNA
XM_293360 Homo sapiens similar to hSIAHI (LOC340571), mRNA
XM_293366 Homo sapiens similar to Olfactory receptor 13H1 (LOC347468), mRNA
XM_293380 Homo sapiens similar to hypothetical protein A630014H24 (LOC347454), mF
XM_293387 Homo sapiens similar to KIAA1892-like (LOC340578), mRNA
XM_293396 Homo sapiens similar to Heat shock transcription factor, Y-linked (Heat sho
XM_293398 Homo sapiens RAB41 , member RAS homolog family (RAB41 ), mRNA
XM_293401 Homo sapiens similar to arylsulfatase (LOC347527), mRNA
XM_293405 Homo sapiens similar to hypothetical protein D430021108 (LOC340595), mR
XM_293407 Homo sapiens similar to melanoma antigen 2 (LOC347541 ), mRNA
XM_293412 Homo sapiens similar to ribosomal protein L18a; 60S ribosomal protein L18a
XM_293416 Homo sapiens similar to nuclear protein p30 (LOC347549), mRNA
XM_293449 Homo sapiens similar to CYorf16 protein (LOC389915), mRNA
XM_293460 Homo sapiens similar to testis specific protein, Y-linked (LOC347596), mRN/
XM_293514 Homo sapiens similar to CG17293-PA (LOC344620), mRNA
XM_293529 Homo sapiens similar to RIKEN cDNA 4930558021 (LOC344657), mRNA
XM_293542 Homo sapiens similar to Elongation factor 1 -gamma (EF-1 -gamma) (eEF-1 B
XM_293565 Homo sapiens similar to seven transmembrane helix receptor (LOC344729),
XM_293570 Homo sapiens similar to Heterogeneous nuclear ribonucleoprotein A1 (Helix-
XM_293577 Homo sapiens similar to Arylacetamide deacetylase (AADAC) (LOC344752),
XM_293580 Homo sapiens G protein-coupled receptor 149 (GPR149), mRNA
XM_293581 Homo sapiens similar to seven transmembrane helix receptor (LOC344760),
XM_293596 Homo sapiens similar to peptidylprolyl isomerase A (LOC344797), mRNA
XM_293599 Homo sapiens type II transmembrane serine protease 7 (TMPRSS7), mRNA
XM_293633 Homo sapiens similar to Protein MGC35450/QtsA-16602 (LOC344892), mRf
XM_293656 Homo sapiens similar to ENSANGP00000007226 (LOC339951 ), mRNA
XM_293669 Homo sapiens similar to actinin, alpha 4 (LOC344978), mRNA
XM_293671 Homo sapiens similar to GTP-binding protein SAR1a (COPII-associated sma
XM_293680 Homo sapiens similar to MGC53273 protein (LOC345051 ), mRNA
XM_293687 Homo sapiens similar to RIKEN cDNA 5730467H21 (LOC345079), mRNA
XM_293715 Homo sapiens similar to CG13722-PA (LOC345156), mRNA
XM_293745 Homo sapiens hypothetical LOC345222 (LOC345222), mRNA
XM_293801 Homo sapiens similar to CG32656-PA (LOC345375), mRNA
XM_293802 Homo sapiens similar to UDP-glucuronosyltransferase 2B15 precursor, micrc
XM_293821 Homo sapiens similar to profilin 3 (LOC345456), mRNA
XM_293828 Homo sapiens similar to hypothetical protein 9630041 N07 (LOC345462), mF
XM_293829 Homo sapiens similar to UNR protein (LOC345463), mRNA
XM_293868 Homo sapiens similar to hypothetical protein (LOC345537), mRNA
XM_293875 Homo sapiens similar to RIKEN cDNA B130016O10 gene (LOC345557), mR
XM_293886 Homo sapiens similar to Ubiquitin carboxyl-terminal hydrolase 7 (Ubiquitin th
XM_293893 Homo sapiens LRG-47-like protein (LRG47), mRNA
XM_293903 Homo sapiens similar to fibrillarin (LOC345630), mRNA
XM_293911 Homo sapiens hypothetical protein FLJ40191 (FLJ40191), mRNA
XM_293918 Homo sapiens similar to geminin (LOC345643), mRNA
XM_293923 Homo sapiens similar to protease (prosome, macropain) 26S subunit, ATPas
XM_293924 Homo sapiens similar to RIKEN cDNA 4732495G21 gene (LOC345651), mR
XM_293927 Homo sapiens similar to template acyivating factor-l alpha (LOC345659), mF
XM_293937 Homo sapiens similar to RIKEN cDNA 0610012A05 (LOC345711), mRNA
XM_293943 Homo sapiens similar to histone (15.4 kD) (his-72) (LOC340096), mRNA
XM_293971 Homo sapiens similar to hypothetical protein (LOC345778), mRNA
XM_293976 Homo sapiens similar to RIKEN cDNA 6430502M16 gene (LOC340120), mR
XM_293984 Homo sapiens similar to Transcription factor BTF3 (RNA polymerase B trans
XM_294017 Homo sapiens solute carrier family 35, member D3 (SLC35D3), mRNA
XM_294019 Homo sapiens similar to Ect2 protein (LOC345930), mRNA
XM_294070 Homo sapiens similar to Glyceraldehyde 3-phosphate dehydrogenase, liver (
XM_294077 Homo sapiens similar to dJ153G14.3 (novel C2H2 type Zinc Finger protein) (
XM_294093 Homo sapiens similar to bA145L22.2 (novel KRAB box containing C2H2 type
XM_294139 Homo sapiens chromosome 6 open reading frame 143 (C6orf143), mRNA
XM_294165 Homo sapiens similar to septin 10 isoform 1 (LOC346288), mRNA
XM_294209 Homo sapiens similar to Unc4.1 homeobox (LOC340260), mRNA
XM_294219 Homo sapiens similar to RIKEN cDNA A930017N06 gene (LOC346355), mR
XM_294247 Homo sapiens similar to Splicing factor, arginine/serine-rich, 46kD (LOC346^
XM_294249 Homo sapiens similar to GluR-delta2 philic-protein (LOC340265), mRNA
XM_294261 Homo sapiens similar to hypothetical protein FLJ22527 (LOC346545), mRN/
XM_294265 Homo sapiens similar to envelope protein (LOC346547), mRNA
XM_294310 Homo sapiens similar to Olfactory receptor 6V1 (LOC346517), mRNA
XM_294311 Homo sapiens similar to Histidine triad nucleotide-binding protein 1 (Adenosi
XM_294316 Homo sapiens similar to Olfactory receptor 2A12 (LOC346525), mRNA
XM_294318 Homo sapiens similar to Olfactory receptor 2A1 (LOC346528), mRNA
XM_294319 Homo sapiens similar to Importin alpha-2 subunit (Karyopherin alpha-2 subui
XM_294328 Homo sapiens similar to Protein C6orf66 (HSPC125) (My013 protein) (LOC3.
XM_294353 Homo sapiens similar to RIKEN cDNA 6332401019 gene (LOC340344), mR
XM_294354 Homo sapiens similar to 40S ribosomal protein S2 (LOC346643), mRNA
XM_294357 Homo sapiens hypothetical protein LOC346653 (LOC346653), mRNA
XM _294365 Homo sapiens similar to Na+/L-ascorbic acid transporter 1 ; SVCT1 (LOC346
XM_294370 Homo sapiens guanine nucleotide binding protein, alpha transducing 3 (GNA
XM_294383 Homo sapiens similar to seven transmembrane helix receptor (LOC346708),
XM_294387 Homo sapiens similar to hypothetical protein 8230402K04 (LOC340359), mR
XM_294438 Homo sapiens similar to 60S RIBOSOMAL PROTEIN L5 (LOC346848), mRf
XM_294450 Homo sapiens similar to solute carrier family 16 (monocarboxylic acid transpi
XM_294456 Homo sapiens similar to putative nuclear protein (4B256) (LOC346910), mRI
XM_294468 Homo sapiens similar to H2A histone family, member Z (LOC346990), mRN/
XM_294473 Homo sapiens similar to ribosomal protein L37 (LOC346950), mRNA
XM_294476 Homo sapiens similar to Interferon-induced transmembrane protein 3 (Interfe
XM_294480 Homo sapiens similar to pote protein; Expressed in prostate, ovary, testis, ar
XM_294521 Homo sapiens FLJ43950 protein (FLJ43950), mRNA
XM_294531 Homo sapiens similar to Olfactory receptor 1 J1 (LOC347168), mRNA
XM_294533 Homo sapiens olfactory receptor, family 1 , subfamily J, member 4 (OR1 J4), r
XM_294534 Homo sapiens similar to Olfactory receptor 1 B1 (Olfactory receptor 9-B) (OR!
XM_294540 Homo sapiens similar to cancer related gene-liver 1 (LOC340485), mRNA
XM_294567 Homo sapiens similar to bA113024.1 (similar to insulin-like growth factor bin
XM_294568 Homo sapiens similar to transcription elongation factor IIS - mouse (LOC340
XM_294574 Homo sapiens similar to A-kinase anchor protein 8; A-kinase anchor protein,
XM_294581 Homo sapiens similar to ribosomal protein L36; 60S ribosomal protein L36 (L
XM_294584 Homo sapiens similar to hypothetical protein FLJ40432 (LOC347262), mRN/
XM_294590 Homo sapiens similar to bA13B9.3 (novel protein similar to KRT8) (LOC3472
XM_294592 Homo sapiens similar to RIKEN cDNA 2310039E09 (LOC347273), mRNA
XM_294634 Homo sapiens similar to alpha-2 macroglobulin family protein VIP (LOC3402
XM_294666 Homo sapiens hypothetical LOC338616 (LOC338616), mRNA
XM_294675 Homo sapiens hypothetical protein LOC338667 (LOC338667), mRNA
XM_294680 Homo sapiens hypothetical protein LOC338694 (LOC338694), mRNA
XM_294688 Homo sapiens hypothetical LOC338731 (LOC338731 ), mRNA
XM_294692 Homo sapiens hypothetical LOC338749 (LOC338749), mRNA
XM_294723 Homo sapiens hypothetical LOC338825 (LOC338825), mRNA
XM_294743 Homo sapiens hypothetical L0C338918 (LOC338918), mRNA
XM_294750 Homo sapiens hypothetical L0C338951 (LOC338951), mRNA
XM_294751 Homo sapiens similar to RIKEN cDNA 4930425N13 (LOC338949), mRNA
XM_294765 Homo sapiens similar to FLJ40113 protein (LOC388154), mRNA
XM_294775 Homo sapiens hypothetical LOC339022 (LOC339022), mRNA
XM_294778 Homo sapiens hypothetical LOC339025 (LOC339025), mRNA
XM_294794 Homo sapiens similar to putative membrane-bound dipeptidase-2 (LOC3390
XM_294802 Homo sapiens hypothetical LOC339077 (LOC339077), mRNA
XM_294854 Homo sapiens hypothetical LOC339209 (LOC339209), mRNA
XM_294867 Homo sapiens hypothetical LOC339226 (LOC339226), mRNA
XM_294894 Homo sapiens hypothetical LOC339281 (LOC339281), mRNA
XM_294906 Homo sapiens hypothetical LOC339306 (LOC339306), mRNA
XM_294910 Homo sapiens hypothetical LOC339352 (LOC339352), mRNA
XM_294914 Homo sapiens hypothetical LOC339358 (LOC339358), mRNA
XM_294919 Homo sapiens hypothetical protein LOC339366 (LOC339366), mRNA
XM_294922 Homo sapiens hypothetical LOC339375 (LOC339375), mRNA
XM_294960 Homo sapiens hypothetical LOC339453 (LOC339453), mRNA
XM_294963 Homo sapiens LOC388583 (LOC388583), mRNA
XM_294993 Homo sapiens hypothetical protein LOC339529 (LOC339529), mRNA
XM_294997 Homo sapiens hypothetical LOC339541 (LOC339541), mRNA
XM_295007 Homo sapiens hypothetical LOC339583 (LOC339583), mRNA
XM_295017 Homo sapiens chromosome 21 open reading frame 54 (C21orf54), mRNA
XM_295034 Homo sapiens hypothetical LOC339693 (LOC339693), mRNA
XM_295058 Homo sapiens hypothetical LOC339760 (LOC339760), mRNA
XM_295059 Homo sapiens hypothetical LOC339771 (LOC339771 ), mRNA
XM_295062 Homo sapiens hypothetical LOC339782 (LOC339782), mRNA
XM_295091 Homo sapiens hypothetical LOC339875 (LOC339875), mRNA
XM_295096 Homo sapiens hypothetical LOC339899 (LOC339899), mRNA
XM_295097 Homo sapiens hypothetical LOC339902 (LOC339902), mRNA
XM_295120 Homo sapiens hypothetical LOC339985 (LOC339985), mRNA
XM_295126 Homo sapiens hypothetical LOC339997 (LOC339997), mRNA
XM_295146 Homo sapiens hypothetical LOC340065 (LOC340065), mRNA
XM_295155 Homo sapiens hypothetical protein LOC340094 (LOC340094), mRNA
XM_295166 Homo sapiens hypothetical LOC340148 (LOC340148), mRNA
XM_295169 Homo sapiens hypothetical LOC340149 (LOC340149), mRNA
XM_295178 Homo sapiens hypothetical LOC340171 (LOC340171), mRNA
XM_295195 Homo sapiens similar to Matn2-prov protein (LOC340267), mRNA
XM_295200 Homo sapiens hypothetical LOC340286 (LOC340286), mRNA
XM_295205 Homo sapiens hypothetical LOC340349 (LOC340349), mRNA
XM_295213 Homo sapiens hypothetical LOC340346 (LOC340346), mRNA
XM_295252 Homo sapiens hypothetical LOC340450 (LOC340450), mRNA
XM_295257 Homo sapiens hypothetical LOC340477 (LOC340477), mRNA
XM_295261 Homo sapiens hypothetical LOC340511 (LOC340511 ), mRNA
XM_295263 Homo sapiens hypothetical protein LOC340508 (LOC340508), mRNA
XM_295270 Homo sapiens hypothetical protein LOC340581 (LOC340581), mRNA
XM_295309 Homo sapiens similar to putative neuronal cell adhesion molecule (LOC3430
XM_295598 Homo sapiens hypothetical LOC343484 (LOC343484), mRNA
XMJ.95865 Homo sapiens similar to apical early endosomal glycoprotein precursor (LOC
XM -96117 Homo sapiens similar to Heterogeneous nuclear ribonucleoprotein A1 (Helix-
XM_296315 Homo sapiens similar to 60S ribosomal protein L35 (LOC341604), mRNA
XM_296817 Homo sapiens similar to hypothetical protein DKFZp434P0316 (L0C342346)
XM_297205 Homo sapiens hypothetical LOC342900 (LOC342900), mRNA
XM_297687 Homo sapiens similar to transmembrane protein 16C; chromosome 11 open
XM_297816 Homo sapiens similar to dJ824F16.3 (novel protein similar to mouse thrombc
XM_298045 Homo sapiens hypothetical LOC347475 (LOC347475), mRNA
XM_298053 Homo sapiens hypothetical LOC347487 (LOC347487), mRNA
XM_298151 Homo sapiens hypothetical LOC344595 (LOC344595), mRNA
XM_298233 Homo sapiens similar to hypothetical protein FLJ38348 (LOC344709), mRN/
XM_301210 Homo sapiens similar to testis-specific protein NYD-TSP1 (LOC388775), mR
XM_350780 Homo sapiens KIAA0592 protein (KIAA0592), mRNA
XM_350880 Homo sapiens ras homolog gene family, member C like 1 (ARHCL1), mRNA
XM_351335 Homo sapiens similar to hypothetical protein KIAA0454 - human (fragment) (
XM_351366 Homo sapiens similar to dJ309K20.1.1 (novel protein similar to dysferiin, isof
XM_351473 Homo sapiens similar to GLYCEROL-3-PHOSPHATE ACYLTRANSFERASE
XM_351574 Homo sapiens similar to Succinate dehydrogenase [ubiquinone] flavoprotein
XM_351617 Homo sapiens similarto dJ28l24.1.2 (Spinal Muscular Atrophy region (SMA3
XM_351723 Homo sapiens similar to tripartite motif protein TRIM50B (LOC375593), mRN
XM_351854 Homo sapiens similar to cDNA sequence BC034076 (LOC389797), mRNA
XM_351948 Homo sapiens similar to Transcript Y 7 protein (LOC389916), mRNA
XM_352159 Homo sapiens similar to hypothetical protein LOC339047 (LOC388215), mRI
XM_352463 Homo sapiens similar to myoferlin isoform b; fer-1-like 3 (LOC391408), mRN
XM_352847 Homo sapiens hypothetical protein LOC340529 (LOC340529), mRNA
XM_353628 Homo sapiens similar to tripartite motif protein TRIM50B (LOC378125), mRN
XM 370532 Homo sapiens similar to ubiquitin-conjugating enzyme E2 variant 1 isoform a
XM_370533 Homo sapiens similar to eukaryotic initiation factor 4B (LOC387637), mRNA
XM_370534 Homo sapiens similarto RIKEN cDNA E130012A19 (LOC387640), mRNA
XM 370536 Homo sapiens similar to RIKEN cDNA 4921522E24 (LOC387642), mRNA
XM_370537 Homo sapiens similar to amyloid beta (A4) precursor protein-binding, family I
XM__370538 Homo sapiens similar to amyloid beta (A4) precursor protein-binding, family I
XMJ370541 Homo sapiens FLJ44037 protein (FLJ44037), mRNA
XM_370542 Homo sapiens similar to supervillin isoform 2; membrane-associated F-actin
XM_370543 Homo sapiens similar to Pre-B cell enhancing factor precursor (LOC387651)
XMJ370545 Homo sapiens similar to bA291 L22.2 (similar to CDC10 (cell division cycle 1(
XM_370554 Homo sapiens similar to ARF GTPase-activating protein (LOC387671 ), mRN
XM 370555 Homo sapiens similar to Glutamate dehydrogenase, mitochondrial precursor
XM_370557 Homo sapiens similar to KIAA0592 protein (LOC387676), mRNA
XM 370560 Homo sapiens similar to ADP-ribosylation factor-like protein 4 (LOC387684),
XM_370562 Homo sapiens LOC399782 (LOC387688), mRNA
XM_370564 Homo sapiens similar to BMS1-like, ribosome assembly protein; ribosome bii
XM_370565 Homo sapiens similar to hypothetical protein FLJ20967 (LOC387694), mRN/
XM_370566 Homo sapiens similar to RLLV1833 (LOC387695), mRNA
XM_370567 Homo sapiens KIAA1975 protein similarto MRIP2 (KIAA1975), mRNA
XM_370569 Homo sapiens similar to expressed sequence AW210596 (LOC387700), mR
XM_370570 Homo sapiens similar to hypothetical protein FLJ25224 (LOC387702), mRN/
XM_370571 Homo sapiens similar to ATP-dependent DNA helicase II, 70 kDa subunit (Li
XM_370573 Homo sapiens similar to KIAA1345 protein (LOC387707), mRNA
XMJ370575 Homo sapiens hypothetical protein MGC11279 (MGC11279), mRNA
XM_370577 Homo sapiens similar to RIKEN cDNA 6430537H07 gene (LOC387712), mR
XM_370580 Homo sapiens similar to Hmx2 protein (LOC387716), mRNA
XM_370582 Homo sapiens hypothetical gene supported by BC062717 (LOC387718), mR
XM_370584 Homo sapiens hypothetical gene supported by AK127642 (LOC387720), mR
XM_370585 Homo sapiens similar to bA442018.2 (novel protein) (LOC387721 ), mRNA
XM_370586 Homo sapiens LOC399826 (LOC387723), mRNA
XM_370589 Homo sapiens similar to double homeobox protein (LOC387727), mRNA
XM_370591 Homo sapiens similar to hypothetical protein MGC5560 (LOC387728), mRN/
XM_370593 Homo sapiens similar to double homeobox protein (LOC387729), mRNA
XM_370597 Homo sapiens similar to putative haemopoietic membrane protein (LOC3877
XM 370603 Homo sapiens similar to SB153 protein (LOC387745), mRNA
XM_370605 Homo sapiens similar to seven transmembrane helix receptor (LOC387748),
XM_370606 Homo sapiens hypothetical gene supported by BX647806 (LOC387749), mR
XM_370607 Homo sapiens similar to SI:zC220F6.1 (novel protein similar to human dyneii
XM_370610 Homo sapiens similar to 60S ribosomal protein L23a (LOC387752), mRNA
XM_370611 Homo sapiens similar to 60S ribosomal protein L21 (LOC387753), mRNA
XM_370612 Homo sapiens calcitonin-related polypeptide, beta (CALCB), mRNA
XM_370613 Homo sapiens similar to RIKEN cDNA 3830422K02 (LOC387755), mRNA
XM_370615 Homo sapiens hypothetical protein DKFZp686024166 (DKFZp686024166),
XM_370616 Homo sapiens hypothetical protein L0C338645 (L0C338645), mRNA
XM 370618 Homo sapiens hypothetical protein FLJ20294 (FLJ20294), mRNA
XM_370619 Homo sapiens similar to mitochondrial carrier protein MGC4399 (LOC387761
XM_370621 Homo sapiens similar to tripartite motif protein 48; TRIM48 (LOC387770), mF
XM_370622 Homo sapiens similar to seven transmembrane helix receptor (LOC387772),
XM_370623 Homo sapiens similar to MGC15937 protein (LOC387773), mRNA
XM_370629 Homo sapiens similar to organic anion transporter 6 (LOC387775), mRNA
XM_370630 Homo sapiens protein phosphatase 1 , regulatory (inhibitor) subunit 14B (PPF
XM_370631 Homo sapiens similar to p33 ringo (LOC387778), mRNA
XM_370632 Homo sapiens similar to phosphoglycerate mutase 1 (brain) (LOC387779), rr
XM_370633 Homo sapiens similar to solute carrier family 22 member 6; organic cationic t
XM_370634 Homo sapiens similar to UBIQUITIN-LIKE PROTEIN FUBI (LOC387781), mF
XM_370635 Homo sapiens KIAA0280 protein (KIAA0280), mRNA
XM_370636 Homo sapiens similar to RIKEN cDNA 2610209A20 (LOC387787), mRNA
XM_370638 Homo sapiens LOC399929 (LOC387790), mRNA
XM_370639 Homo sapiens similar to Mitochondrial import receptor subunit TOM20 homol
XM_370642 Homo sapiens LOC399932 (LOC387795), mRNA
XM_370644 Homo sapiens similar to tripartite motif-containing 43 (LOC387800), mRNA
XM_370648 Homo sapiens LOC399943 (LOC387804), mRNA
XM_370649 Homo sapiens similar to ARP2/3 complex 21 kDa subunit (p21-ARC) (Actin-r
XM_370651 Homo sapiens hypothetical protein FLJ32810 (FLJ32810), mRNA
XM_370652 Homo sapiens dynein, cytoplasmic, heavy polypeptide 2 (DNCH2), mRNA
XM_370653 Homo sapiens KIAA1826 protein (KIAA1826), mRNA
XM_370654 Homo sapiens KI AA1726 protein (KIAA1726), mRNA
XM_370657 Homo sapiens placenta expressed transcript 1 (PLET1), mRNA
XM_370658 Homo sapiens similar to hypothetical protein FLJ25224 (LOC387811), mRN/
XM_370660 Homo sapiens KIAA1201 protein (KIAA1201), mRNA
XM_370661 Homo sapiens similar to seven transmembrane helix receptor (LOC387815),
XM_370662 Homo sapiens olfactory receptor, family 8, subfamily G, member 2 (0R8G2),
XM_370663 Homo sapiens retinoblastoma inhibiting gene 1 (RBIG1), mRNA
XM_370664 Homo sapiens similar to hypothetical protein (LOC387816), mRNA
XM_370665 Homo sapiens similar to DnaJ (Hsp40) homolog, subfamily B, member 6 isof
XM_370666 Homo sapiens similar to hypothetical protein (LOC387822), mRNA
XM_370667 Homo sapiens KI AA1110 protein (KI AA1110), mRNA
XM_370668 Homo sapiens similar to ribosomal protein L13a; 60S ribosomal protein L13a
XM_370669 Homo sapiens homeobox C14 (LOC360030), mRNA
XM_370672 Homo sapiens similar to hypothetical protein SB153 isoform 1 (LOC387830),
XM_370674 Homo sapiens similar to helicase (LOC387832), mRNA
XM_370675 Homo sapiens similar to DKFZp434C0631 protein (LOC387834), mRNA
XM_370678 Homo sapiens similar to INPE5792 (LOC387836), mRNA
XM_370681 Homo sapiens similar to ribosomal protein L13a; 60S ribosomal protein L13a
XM_370682 Homo sapiens KIAA1467 protein (KIAA1467), mRNA
XM_370684 Homo sapiens similar to Elongation factor 1 -alpha 1 (EF-1 -alpha-1 ) (Elongati
XM_370685 Homo sapiens hypothetical protein LOC144363 (LOC144363), mRNA
XM_370686 Homo sapiens similar to Rl KEN cDNA 2210417D09 (LOC387849), mRNA
XM_370687 Homo sapiens similar to ribosomal protein L13a; 60S ribosomal protein L13a
XM_370688 Homo sapiens similar to Adenylate kinase isoenzyme 4, mitochondrial (ATP-
XM_370690 Homo sapiens AT rich interactive domain 2 (ARID, RFX-like) (ARID2), mRN/
XM_370691 Homo sapiens similar to expressed sequence AI836003 (LOC387856), mRN
XM_370692 Homo sapiens hypothetical protein LOC121006 (LOC121006), mRNA
XM_370693 Homo sapiens POU domain, class 6, transcription factor 1 (POU6F1), mRNA
XM_370695 Homo sapiens similar to K+ channel tetramerization protein (LOC387861), m
XM_370696 Homo sapiens hypothetical protein FLJ34236 (FLJ34236), mRNA
XM _370697 Homo sapiens similar to 40S ribosomal protein SA (P40) (34/67 kDa laminin
XM_370699 Homo sapiens protein tyrosine phosphatase, receptor type, Q (PTPRQ), mRI
XM_370702 Homo sapiens FYVE, RhoGEF and PH domain containing 6 (FGD6), mRNA
XM_370704 Homo sapiens similar to 10 KD HEAT SHOCK PROTEIN, MITOCHONDRIAL
XM_370705 Homo sapiens LOC400067 (LOC387881 ), mRNA
XM_370707 Homo sapiens similar to hypothetical protein C130069F04 (LOC387890), mF
XM_370709 Homo sapiens LOC400085 (LOC387894), mRNA
XM_370710 Homo sapiens similar to protein 40kD (LOC387902), mRNA
XM_370711 Homo sapiens similar to hypothetical protein (LOC387904), mRNA
XM_370713 Homo sapiens similar to bA271 B5.1 (similar to ribosomal protein S7) (LOC3J
XM_370714 Homo sapiens similarto Ferritin heavy chain (Ferritin H subunit) (LOC38790!
XM_370715 Homo sapiens similar to hypothetical protein MGC48915 (LOC387911), mRf
XM_370716 Homo sapiens similar to TPTE and PTEN homologous inositol lipid phosphat
XM_370718 Homo sapiens similar to WGAR9166 (LOC387914), mRNA
XM_370721 Homo sapiens similar to bA251J8.3.1 (novel protein, isoform 1) (LOC387920
XM_370722 Homo sapiens similar to RIKEN cDNA 8030451 K01 (LOC387921 ), mRNA
XM_370723 Homo sapiens similar to tubulin, beta 5 (LOC387922), mRNA
XM_370724 Homo sapiens similar to ribosome associated membrane protein 4 (LOC387!
XM_370725 Homo sapiens similar to KIAA1612 protein (LOC387924), mRNA
XM_370726 Homo sapiens similar to BB049667 protein (LOC387927), mRNA
XM_370727 Homo sapiens similar to ribosomal protein L13a; 60S ribosomal protein L13a
XM_370728 Homo sapiens similar to heterogeneous nuclear ribonucleoprotein A3 (LOC3
XM_370729 Homo sapiens similar to Fatty acid-binding protein, epidermal (E-FABP) (Psc
XM_370731 Homo sapiens similar to expressed sequence AW121567 (LOC387944), mR
XM_370732 Homo sapiens similar to expressed sequence AW121567 (LOC387945), mR
XM_370733 Homo sapiens LOC400166 (LOC387949), mRNA
XM_370734 Homo sapiens hypothetical gene supported by BC034570 (LOC387952), mR
XM_370737 Homo sapiens hypothetical protein FLJ 10357 (FLJ 10357), mRNA
XM_370738 Homo sapiens helicase with SNF2 domain 1 (HELSNF1 ), mRNA
XM_370754 Homo sapiens thiamine triphosphatase (THTPA), mRNA
XM_370756 Homo sapiens KIAA1305 (KIAA1305), mRNA
XM_370758 Homo sapiens hypothetical gene supported by BX248251 (LOC387978), mR
XM_370759 Homo sapiens similar to RIKEN cDNA D930036F22 gene (LOC387979), R
XM_370760 Homo sapiens similar to 60S RIBOSOMAL PROTEIN L12 (LOC387982), mR
XM_370762 Homo sapiens similar to TIMM9 (LOC387990), mRNA
XM_370763 Homo sapiens similarto ribosomal protein L31 (LOC387991), mRNA
XM_370765 Homo sapiens papilin, proteoglycan-like sulfated glycoprotein (PAPLN), mRf
XM_370767 Homo sapiens chromosome 14 open reading frame 46 (C14orf46), mRNA
XM_370768 Homo sapiens similar to Acylphosphatase, organ-common type isozyme (Ac;
XM_370769 Homo sapiens hypothetical protein LOC161394 (LOC161394), mRNA
XM_370772 Homo sapiens similar to protease inhibitor (LOC388007), mRNA
XM_370776 Homo sapiens similar to RTI1 (LOC388015), mRNA
XM_370777 Homo sapiens Similarto Lysophospholipase (LOC374569), mRNA
XM_370778 Homo sapiens similar to expressed sequence AI839735 (LOC388021 ), mRN
XM_370779 Homo sapiens hypothetical gene supported by AK131040 (LOC388022), mR
XM_370781 Homo sapiens similar to Ig alpha-2 chain C region (LOC388025), mRNA
XM_370782 Homo sapiens similar to Ig epsilon chain C region (LOC388026), mRNA
XM_370785 Homo sapiens chromosome 14 open reading frame 81 (C14orf81), mRNA
XM_370826 Homo sapiens similar to breast cancer antigen NY-BR-1 (LOC388065), mRN
XM_370829 Homo sapiens similar to carnitine deficiency-associated gene expressed in v
XM_370830 Homo sapiens similar to breast cancer anti-estrogen resistance 1 ; Crk-assoc
XM_370831 Homo sapiens similarto Ribosome biogenesis protein BMS1 homolog (LOC
XM_370832 Homo sapiens similar to Ribosome biogenesis protein BMS1 homolog (LOC
XM_370833 Homo sapiens similar to 40S ribosomal protein S8 (LOC388076), mRNA
XM_370834 Homo sapiens similar to immunoglobulin heavy chain variable region (LOC3!
XM_370835 Homo sapiens similar to FLJ27099 protein (LOC388078), mRNA
XM_370836 Homo sapiens similar to ZCCHC2 protein (LOC388079), mRNA
XM_370837 Homo sapiens similar to hypothetical protein (LOC388080), mRNA
XM_370838 Homo sapiens hypothetical protein LOC339005 (LOC339005), mRNA
XM_370839 Homo sapiens similar to golgin-67 isoform b (LOC388084), mRNA
XM_370840 Homo sapiens similar to hypothetical protein (LOC388085), mRNA
XM_370843 Homo sapiens similar to hypothetical protein (LOC388092), mRNA
XM_370844 Homo sapiens similar to hypothetical protein (LOC388094), mRNA
XM_370845 Homo sapiens similar to hyperpolarization activated cyclic nucleotide-gated p
XM_370846 Homo sapiens similar to hypothetical protein FLJ35785 (LOC388098), mRN/
XM_370848 Homo sapiens similar to chromosome 1 open reading frame 37 (LOC388104
XM_370849 Homo sapiens similar to hyperpolarization activated cyclic nucleotide-gated p
XM_370851 Homo sapiens similar to hypothetical protein FLJ35785 (LOC388109), mRN/
XM_370852 Homo sapiens similar to 4930563P21 Rik protein (LOC388110), mRNA
XM_370853 Homo sapiens similar to Nanog homeobox; homeobox transcription factor Ns
XM_370855 Homo sapiens similar to RIKEN cDNA 2600011 L02 (LOC388115), mRNA
XM_370856 Homo sapiens similar to RIKEN cDNA 6720467C03 (LOC388116), mRNA
XM_370858 Homo sapiens similar to kinesin-like protein (LOC388118), mRNA
XM_370863 Homo sapiens ATPase, Class I, type 8B, member 4 (ATP8B4), mRNA
XM_370864 Homo sapiens similar to TNF-induced protein (LOC388121 ), mRNA
XM_370865 Homo sapiens similar to Laminin receptor 1 (LOC388122), mRNA
XM_370866 Homo sapiens hypothetical protein FLJ25756 (FLJ25756), mRNA
XM_370867 Homo sapiens hypothetical protein FLJ20086 (FLJ20086), mRNA
XM_370868 Homo sapiens similar to hypothetical LOC237397 (LOC388125), mRNA
XM_370871 Homo sapiens hypothetical protein LOC145837 (LOC145837), mRNA
XM_370872 Homo sapiens similar to 60S ribosomal protein L17 (L23) (LOC388132), mRI
XM_370873 Homo sapiens similar to RIKEN cDNA 6030419C18 gene (LOC388135), mR
XM_370876 Homo sapiens similar to Golgi autoantigen, golgin subfamily A member 6 (Gc
XM_370878 Homo sapiens KIAA2002 protein (KIAA2002), mRNA
XM_370879 Homo sapiens similar to 60S ribosomal protein L21 (LOC388143), mRNA
XM_370880 Homo sapiens mesoderm development candidate 2 (MESDC2), mRNA
XM_370881 Homo sapiens similar to FLJ40113 protein (LOC388146), mRNA
XM_370882 Homo sapiens similar to ribosomal protein L9; 60S ribosomal protein L9 (LO<
XM_370883 Homo sapiens LOC400417 (LOC388148), mRNA
XM_370886 Homo sapiens similar to FLJ40113 protein (LOC388151), mRNA
XM_370887 Homo sapiens LOC400422 (LOC388153), mRNA
XM_370893 Homo sapiens similar to hypothetical protein FLJ13119 (LOC388159), mRN/
XM_370894 Homo sapiens LOC400427 (LOC388160), mRNA
XM_370895 Homo sapiens LOC400432 (LOC388163), mRNA
XM_370897 Homo sapiens similarto RIKEN cDNA 3010021 M21 (LOC388165), mRNA
XM_370898 Homo sapiens similarto FLJ40113 protein (LOC388166), mRNA
XM_370899 Homo sapiens similar to FLJ40113 protein (LOC388167), mRNA
XM_370904 Homo sapiens similar to hypothetical protein 4921538011 (LOC388173), mF
XM_370905 Homo sapiens similar to Golgi autoantigen, golgin subfamily a, 2; golgin-95; ι
XM_370906 Homo sapiens similarto hypothetical protein FLJ20147 (LOC388175), mRN/
XM_370908 Homo sapiens similar to cyclin-E binding protein 1 (H. sapiens) (MGC14386)
XM_370909 Homo sapiens similar to H2A histone family, member V isoform 2; histone H!
XM_370910 Homo sapiens similar to KIAA0974 protein (LOC388181 ), mRNA
XM_370911 Homo sapiens hypothetical gene supported by AK124283 (LOC388182), mR
XM_370917 Homo sapiens similar to hypothetical protein (LOC388189), mRNA
XM_370918 Homo sapiens hypothetical protein DKFZp434P162 (DKFZp434P162), mRN;
XM_370924 Homo sapiens LOC400486 (LOC388199), mRNA
XM_370925 Homo sapiens hypothetical protein LOC283951 (LOC283951), mRNA
XM_370926 Homo sapiens similar to KIAA0445 protein (LOC388202), mRNA
XM_370927 Homo sapiens ring finger protein 151 (RNF151), mRNA
XM_370928 Homo sapiens KIAA1171 protein (KIAA1171), mRNA
XM_370930 Homo sapiens similarto RIKEN cDNA 1520401A03 gene (LOC388205), mRI
XM_370931 Homo sapiens olfactory receptor, family 1 , subfamily F, member 2 (OR1 F2),
XM_370932 Homo sapiens hypothetical protein FLJ39639 (FLJ39639), mRNA
XM_370934 Homo sapiens similar to CG15828-PA (LOC388210), mRNA
XM_370935 Homo sapiens LOC400498 (LOC388211 ), mRNA
XM_370938 Homo sapiens similar to QRWT5810 (LOC388218), mRNA
XM_370939 Homo sapiens similar to KIAA0220 (LOC388221 ), mRNA
XM_370942 Homo sapiens similar to carbonic anhydrase VA, mitochondrial precursor; ca
XM_370943 Homo sapiens similar to MGC9515 protein (LOC400510), mRNA
XM_370944 Homo sapiens hypothetical protein LOC146177 (LOC146177), mRNA
XM_370946 Homo sapiens similar to protein kinase/ribonuclease IRE1 beta (LOC388226
XM_370947 Homo sapiens ER to nucleus signalling 2 (ERN2), mRNA
XM_370948 Homo sapiens similar to SH3-binding kinase (LOC388228), mRNA
XM_370949 Homo sapiens similar to Group X secretory phospholipase A2 precursor (Phe
XM_370952 Homo sapiens similar to MGC9515 protein (LOC388233), mRNA
XM_370958 Homo sapiens similar to nuclear pore complex interacting protein (LOC3882;
XM_370959 Homo sapiens similar to RRN3 (LOC388238), mRNA
XM_370965 Homo sapiens similar to hypothetical protein BC011981 (LOC388242), mRN.
XM_370966 Homo sapiens similar to carbonic anhydrase VA, mitochondrial precursor; ca
XM_370967 Homo sapiens hypothetical protein LOC124411 (LOC124411), mRNA
XM_370968 Homo sapiens similar to KIAA1501 protein (LOC388248), mRNA
XM_370972 Homo sapiens similar to Adrenoleukodystrophy protein (ALDP) (LOC388253;
XM_370973 Homo sapiens similar to KIAA1501 protein (LOC388255), mRNA
XM_370974 Homo sapiens similar to NY-REN-7 antigen (LOC388258), mRNA
XM_370975 Homo sapiens similar to protein kinase related to Raf protein kinases; Metho
XM_370977 Homo sapiens similar to immunoglobulin heavy chain VH3 (LOC388264), mF
XM_370980 Homo sapiens similar to RAB41 (LOC388271 ), mRNA
XM_370981 Homo sapiens similar to RIKEN cDNA 4921524J17 (LOC388272), mRNA
XM_370982 Homo sapiens similar to Heterogeneous nuclear ribonucleoprotein A1 (Helix-
XM_370984 Homo sapiens LOC388284 (LOC388284), mRNA
XM_370986 Homo sapiens similar to hypothetical protein (L1 H 3 region) - human (LOC3E
XM_370987 Homo sapiens LOC388289 (LOC388289), mRNA
XM_370988 Homo sapiens similar to protein 40kD (LOC388290), mRNA
XM_370991 Homo sapiens similar to hypothetical protein - fruit fly (Drosophila melanogas
XM_370992 Homo sapiens LOC388298 (LOC388298), mRNA
XM_370993 Homo sapiens similar to coenzyme A diphosphatase (LOC388299), mRNA
XM_370995 Homo sapiens snail homolog 3 (Drosophila) (SNAI3), mRNA
XM_370997 Homo sapiens similar to Brain-type organic cation transporter (Solute carrier
XM_371001 Homo sapiens LOC388317 (LOC388317), mRNA
XM_371006 Homo sapiens similar to RIKEN cDNA C730027E14 (LOC388323), mRNA
XM_371008 Homo sapiens similar to Death effector filament-forming Ced-4-like apoptosis
XM_371009 Homo sapiens LOC388327 (LOC388327), mRNA
XM_371010 Homo sapiens hypothetical protein MGC49942 (MGC49942), mRNA
XM_371012 Homo sapiens similar to ENSANGP00000015193 (LOC388329), mRNA
XM_371013 Homo sapiens similar to Gag-Pro-Pol protein (LOC388332), mRNA
XM_371014 Homo sapiens similar to Williams Beuren syndrome chromosome region 19 I
XM_371015 Homo sapiens ubiquitin specific protease 43 (USP43), mRNA
XM_371016 Homo sapiens similar to RIKEN cDNA A730055C05 gene (LOC388335), mR
XM_371017 Homo sapiens similar to hypothetical protein D430041 B17 (LOC388336), mF
XM_371018 Homo sapiens similar to CDRT15 protein (LOC388337), mRNA
XM_371019 Homo sapiens similar to ribosomal protein (LOC388339), mRNA
XM_371020 Homo sapiens LOC388341 (LOC388341 ), mRNA
XM_371023 Homo sapiens similar to ribosomal protein L13; 60S ribosomal protein L13; b
XM_371024 Homo sapiens similar to poly(A) binding protein interacting protein 1 isoform
XM_371026 Homo sapiens similar to hypothetical protein FLJ10847 (LOC388351), mRN/
XM_371028 Homo sapiens similar to AF038169 protein (LOC388353), mRNA
XM_371032 Homo sapiens similar to bB329D4.2.1 (novel protein similar to a truncated ni
XM_371034 Homo sapiens similar to PDZ and LIM domain 1 (elfin); carboxy terminal LIM
XM_371035 Homo sapiens similar to 40S ribosomal protein S7 (S8) (LOC388363), mRN/
XM_371036 Homo sapiens KIAA0100 gene product (K1AA0100), mRNA
XM_371039 Homo sapiens hypothetical protein MGC19764 (MGC19764), mRNA
XM_371043 Homo sapiens similar to TBC1 domain family member 3 (Rab GTPase-activε
XM_371046 Homo sapiens similar to TBC1 domain family member 3 (Rab GTPase-activε
XM_371052 Homo sapiens Nek, Ash and phospholipase C binding protein (NAP4), mRN/
XM_371053 Homo sapiens LOC388381 (LOC388381 ), mRNA
XM_371054 Homo sapiens LOC388382 (LOC388382), mRNA
XM_371056 Homo sapiens similar to RIKEN cDNA 4933439F11 (LOC388389), mRNA
XM_371057 Homo sapiens hypothetical protein LOC201175 (LOC201175), mRNA
XM_371058 Homo sapiens similar to KIAA0563 gene product (LOC388391 ), mRNA
XM_371059 Homo sapiens similar to FALZ protein (LOC388392), mRNA
XM_371062 Homo sapiens similar to Hypothetical protein KIAA0563 (LOC388395), mRN;
XM_371063 Homo sapiens similar to KIAA0563 gene product (LOC388396), mRNA
XM_371066 Homo sapiens similar to SMT3 suppressor of mif two 3 homolog 2 (LOC388S
XM_371067 Homo sapiens similar to Eukaryotic translation initiation factor 4E (elF4E) (el
XM_371068 Homo sapiens similar to ribosomal protein L7 (LOC388401), mRNA
XM_371069 Homo sapiens similar to 40S ribosomal protein S2 (LOC388402), mRNA
XM_371070 Homo sapiens similar to Yippee-like protein 2 (DiGeorge syndrome-related p
XM_371074 Homo sapiens putative ankyrin-repeat containing protein (DKFZP564D166),
XM_371077 Homo sapiens similar to PP905 (LOC388413), mRNA
XM_371078 Homo sapiens similar to galectin 3 binding protein; L3 antigen; Mac-2-bindinc
XM_371079 Homo sapiens Fas binding protein 1 (FBF-1 ), mRNA
XM_371081 Homo sapiens LOC388424 (LOC388424), mRNA
XM_371082 Homo sapiens hypothetical protein FLJ20753 (FLJ20753), mRNA
XM_371083 Homo sapiens LOC388428 (LOC388428), mRNA
XM_371084 Homo sapiens KIAA1447 protein (KIAA1447), mRNA
XM_371085 Homo sapiens hypothetical protein LOC339229 (LOC339229), mRNA
XM_371086 Homo sapiens similar to dysferlin-interacting protein 1 (LOC388432), mRNA
XM_371088 Homo sapiens similar to hypothetical protein DKFZp434F142 (LOC388437),
XM_371089 Homo sapiens similar to MEGF11 protein (LOC388438), mRNA
XM_371092 Homo sapiens similar to cytokine (LOC388440), mRNA
XM_371097 Homo sapiens LOC388444 (LOC388444), mRNA
XM_371098 Homo sapiens hypothetical protein LOC348262 (LOC348262), mRNA
XM_371105 Homo sapiens LOC388457 (LOC388457), mRNA
XM_371106 Homo sapiens LOC388458 (LOC388458), mRNA
XM_371107 Homo sapiens similar to 60S ribosomal protein L6 (TAX-responsive enhance
XM_371108 Homo sapiens similar to KIAA1314 protein (LOC388462), mRNA
XM_371109 Homo sapiens hypothetical protein LOC284221 (LOC284221 ), mRNA
XM_371110 Homo sapiens similar to acyl-malonyl condensing enzyme (LOC388463), mF
XM_371111 Homo sapiens similar to breast cancer antigen NY-BR-1.1 (LOC388469), mF
XM_371113 Homo sapiens similar to 60S RIBOSOMAL PROTEIN L21 (LOC388471), mR
XM_371114 Homo sapiens formin homology 2 domain containing 3 (FHOD3), mRNA
XM_371115 Homo sapiens similar to 60S ribosomal protein L7a (Surfeit locus protein 3) (
XM_371116 Homo sapiens myosin VB (MY05B), mRNA
XM_371117 Homo sapiens similar to Nonhistone chromosomal protein HMG-14 (High-mc
XM_371118 Homo sapiens similar to serologically defined colon cancer antigen 3 (LOC35
XM_371120 Homo sapiens thioredoxin-like 4 (TXNL4), mRNA
XM_371121 Homo sapiens similar to bA476H5.3 (novel protein similar to septin) (LOC38!
XM_371122 Homo sapiens similar to Dip1 -associated protein C (LOC388488), mRNA
XM_371125 Homo sapiens similar to KIAA1683 protein (LOC388491), mRNA
XM_371130 Homo sapiens similar to methyl-CpG binding domain protein 3-like 2 (LOC38
XM_371132 Homo sapiens FLJ38144 protein (FLJ38144), mRNA
XM_371134 Homo sapiens similar to complement C3-Q2 (LOC388503), mRNA
XM_371138 Homo sapiens hypothetical protein LOC284390 (LOC284390), mRNA
XM_371139 Homo sapiens hypothetical protein FLJ14959 (FLJ14959), mRNA
XM_371140 Homo sapiens similar to zinc finger protein 433 (LOC388507), mRNA
XM_371141 Homo sapiens similar to ribosomal protein L17 (LOC388508), mRNA
XM_371142 Homo sapiens similar to hypothetical protein FLJ38281 (LOC388509), mRN/
XM_371143 Homo sapiens similar to Asialoglycoprotein receptor 2 (IHepatic Iectin H2) (AJ
XM_371145 Homo sapiens similar to Cytochrome P450 4F12 (CYPIVF12) (LOC388514),
XM_371146 Homo sapiens KIAA1683 (KIAA1683), mRNA
XM_371147 Homo sapiens similar to MOST-1 protein (LOC388517), mRNA
XM_371150 Homo sapiens zinc finger protein 90 (HTF9) (ZNF90), mRNA
XM_371151 Homo sapiens similar to 40S ribosomal protein S16 (LOC388519), mRNA
XM__371152 Homo sapiens zinc finger protein 486 (ZNF486), mRNA
XM_371153 Homo sapiens similarto hypothetical protein (LOC388521), mRNA
XM 371154 Homo sapiens similar to Zinc finger protein 429 (LOC388522), mRNA
XM 371155 Homo sapiens similar to 40S ribosomal protein SA (P40) (34/67 kDa laminin
XM_371157 Homo sapiens similarto CG14939-PA (LOC388526), mRNA
XM_371158 Homo sapiens hypothetical protein LOC147991 (LOC147991), mRNA
XM 371159 Homo sapiens similar to regulator of G protein signaling 9-binding protein; R<
XM_371160 Homo sapiens similar to 60S ribosomal protein L21 (LOC388532), mRNA
XM 371161 Homo sapiens similar to KIPV467 (LOC388533), mRNA
XM_371164 Homo sapiens NYD-SP11 protein (NYD-SP11 ), mRNA
XM_371165 Homo sapiens similar to SPRED-3 (LOC388538), mRNA
XM_371167 Homo sapiens syncollin (SYCN), mRNA
XM_371170 Homo sapiens similar to Zinc finger protein 216 (LOC388545), mRNA
XM_371174 Homo sapiens zinc finger protein 283 (ZNF283), mRNA
XM_371175 Homo sapiens zinc finger protein 229 (ZNF229), mRNA
XM_371176 Homo sapiens similar to CEACAM5 protein (LOC388550), mRNA
XM_371177 Homo sapiens similar to carcinoembryonic antigen-related cell adhesion mot
XM_371178 Homo sapiens similar to BC043666 protein (LOC388552), mRNA
XM_371179 Homo sapiens F-box only protein 34-like (FBX034L), mRNA
XM_371181 Homo sapiens nanos homolog 2 (Drosophila) (NANOS2), mRNA
XM_371182 Homo sapiens similar to BC282485 J (LOC388554), mRNA
XM_371183 Homo sapiens similar to RPRC483 (LOC388555), mRNA
XM_371184 Homo sapiens KIAA1183 protein (KIAA1183), mRNA
XM_371187 Homo sapiens hypothetical gene MGC45922 (MGC45922), mRNA
XM_371190 Homo sapiens hypothetical protein LOC162967 (LOC162967), mRNA
XM_371191 Homo sapiens similar to RIKEN cDNA 1300007C21 (LOC388560), mRNA
XM_371192 Homo sapiens similar to KIAA2033 protein (LOC388561 ), mRNA
XM_371195 Homo sapiens hypothetical protein MGC35045 (MGC35045), mRNA
XM_371196 Homo sapiens LOC388564 (LOC388564), mRNA
XM_371197 Homo sapiens similar to zinc finger protein 111 (LOC388565), mRNA
XM_371198 Homo sapiens similar to Zinc finger protein 471 (EZFIT-related protein 1 ) (LC
XM_371200 Homo sapiens similar to R30217J (LOC388567), mRNA
XM_371201 Homo sapiens similar to Zinc finger protein 324 (Zinc finger protein ZF5128)
XM_371202 Homo sapiens CXYorfl -related protein (FLJ00038), mRNA
XM_371204 Homo sapiens similar to 60S ribosomal protein L23a (LOC388574), mRNA
XM_371205 Homo sapiens similar to hypothetical protein DKFZp434F142 (LOC388576),
XM_371206 Homo sapiens similar to F-box only protein 25 (LOC388578), mRNA
XM_371207 Homo sapiens similar to beta-tubulin 4Q (LOC388579), mRNA
XM_371208 Homo sapiens similar to RIKEN cDNA 1110035L05 (LOC388581), mRNA
XM_371210 Homo sapiens taste receptor, type 1, member 3 (TAS1R3), mRNA
XM_371214 Homo sapiens KIAA0450 gene product (KIAA0450), mRNA
XM_371215 Homo sapiens similar to hairy and enhancer of split 5 (LOC388585), mRNA
XM_371216 Homo sapiens LOC388591 (LOC388591), mRNA
XM_371221 Homo sapiens similar to WD repeat domain 9 isoform A; WD repeat domain
XM_371222 Homo sapiens similar to Hypothetical protein MGC37938 (LOC388595), mRf
XM_371223 Homo sapiens similar to 40S ribosomal protein S16 (LOC388596), mRNA
XM_371225 Homo sapiens hypothetical protein LOC284729 (LOC284729), mRNA
XM_371227 Homo sapiens ciliary rootlet coiled-coil, rootletin (CROCC), mRNA
XM_371230 Homo sapiens similar to heparan-sulfate 6-sulfotransferase (LOC388605), m
XM_371232 Homo sapiens similar to Succinate dehydrogenase [ubiquinone] cytochrome
XM_371234 Homo sapiens similar to RhCE protein (LOC388607), mRNA
XM_371235 Homo sapiens similar to mutant membrane protein RhCe (LOC388608), mRI
XM_371236 Homo sapiens hypothetical protein FLJ10747 (FLJ10747), mRNA
XM_371237 Homo sapiens cation channel, sperm associated 4 (CATSPER4), mRNA
XM_371238 Homo sapiens LOC388610 (LOC388610), mRNA
XM_371239 Homo sapiens similar to EAPG6122 (LOC388611), mRNA
XM_371241 Homo sapiens similar to RIKEN cDNA 9330177P20 (LOC388618), mRNA
XM_371243 Homo sapiens similar to 60S ribosomal protein L21 (LOC388621 ), mRNA
XM_371244 Homo sapiens LOC388624 (LOC388624), mRNA
XM_371245 Homo sapiens similar to FLJ00408 protein (LOC388625), mRNA
XM_371246 Homo sapiens hypothetical protein FLJ21156 (FLJ21156), mRNA
XM_371248 Homo sapiens LOC374973 (LOC374973), mRNA
XM_371249 Homo sapiens similar to cytochrome P450 4Z1 (LOC388629), mRNA
XM_371250 Homo sapiens LOC388630 (LOC388630), mRNA
XM_371252 Homo sapiens similar to heterogeneous nuclear ribonucleoprotein A3 (LOC3
XM_371253 Homo sapiens similar to Low-density lipoprotein receptor-related protein 2 pr
XM_371254 Homo sapiens similar to Ubiquitin carboxyl-terminal hydrolase 24 (Ubiquitin t
XM_371257 Homo sapiens KI AA1799 protein (KI AA1799), mRNA
XM_371258 Homo sapiens hypothetical protein FLJ10770 (KIAA1579), mRNA
XM_371259 Homo sapiens hypothetical protein DKFZp547l048 (DKFZp547l048), mRNA
XM_371260 Homo sapiens similar to crystallin, zeta; NADPH:quinone reductase (LOC38J
XM_371261 Homo sapiens similar to Triosephosphate isomerase (TIM) (LOC388642), ml
XM_371262 Homo sapiens EGF, latrophilin and seven transmembrane domain containing
XM_371263 Homo sapiens mucolipin 2 (MCOLN2), mRNA
XM_371265 Homo sapiens similar to Guanylate binding protein 4 (LOC388646), mRNA
XM_371267 Homo sapiens hypothetical protein LOC164045 (LOC164045), mRNA
XM_371268 Homo sapiens similar to 1700028K03 protein (LOC388649), mRNA
XM_371269 Homo sapiens similar to RIKEN cDNA 2900024C23 (LOC388650), mRNA
XM_371273 Homo sapiens similar to 40S ribosomal protein SA (P40) (34/67 kDa laminin
XM_371276 Homo sapiens similar to hypothetical protein MGC8902 (LOC388658), mRN/
XM_371277 Homo sapiens similar to RIKEN cDNA C230093N12 (LOC388659), mRNA
XM_371278 Homo sapiens similar to mitsugumin29 (LOC388660), mRNA
XM_371279 Homo sapiens amphoterin-induced gene (KIAA1163), mRNA
XM_371280 Homo sapiens similar to Orphan sodium- and chloride-dependent neurotrans
XM_371281 Homo sapiens similar to DKFZP564K247 protein (LOC388665), mRNA
XM_371283 Homo sapiens LOC388666 (LOC388666), mRNA
XM_371284 Homo sapiens T-box 15 (TBX15), mRNA
XM_371285 Homo sapiens 3-beta-hydroxysteroid dehydrogenase, tissue-type heart (LOC
XM_371286 Homo sapiens hypothetical protein MGC45731 (MGC45731), mRNA
XM_371288 Homo sapiens similar to cyclophilin-LC; cyclophilin homolog overexpressed i
XM_371291 Homo sapiens similar to hypothetical protein FLJ21308 (LOC388673), mRN/
XM_371292 Homo sapiens LOC388674 (LOC388674), mRNA
XM_371299 Homo sapiens similar to KIAA0454 protein (LOC388681), mRNA
XM_371301 Homo sapiens similar to hypothetical protein FLJ21308 (LOC388685), mRN/
XM_371302 Homo sapiens similar to cyclophilin-LC; cyclophilin homolog overexpressed i
XM_371304 Homo sapiens similar to cyclophilin-LC; cyclophilin homolog overexpressed i
XM_371305 Homo sapiens similar to hypothetical protein KIAA0454 - human (fragment) (
XM_371306 Homo sapiens similar to KIAA0454 protein (LOC388689), mRNA
XM_371310 Homo sapiens similar to hypothetical protein SB145 (LOC388695), mRNA
XM_371311 Homo sapiens hypothetical protein FLJ36032 (FLJ36032), mRNA
XM_371312 Homo sapiens hypothetical protein FLJ39117 (FLJ39117), mRNA
XM_371313 Homo sapiens similar to dJ14N1.2 (novel S-100/ICaBP type calcium binding
XM_371314 Homo sapiens similar to skin-specific protein (LOC388699), mRNA
XM_371315 Homo sapiens LOC388701 (LOC388701), mRNA
XM_371316 Homo sapiens similar to 40S ribosomal protein SA (P40) (34/67 kDa laminin
XM_371320 Homo sapiens FLJ00193 protein (FLJ00193), mRNA
XM_371326 Homo sapiens similar to Putative dimethylaniline monooxygenase [N-oxide fc
XM_371328 Homo sapiens hypothetical gene supported by BC007071 (dJ383J4.3), mRN
XM_371329 Homo sapiens similar to Protein translation factor SUM homolog (Suhisol) (
XM_371330 Homo sapiens similar to bA92K2.2 (similar to ubiquitin) (LOC388720), mRN/
XM_371331 Homo sapiens similar to Hypothetical protein CBG13135 (LOC388722), mRf
XM_371332 Homo sapiens kinesin family member 21 B (KIF21 B), mRNA
XM_371333 Homo sapiens similar to hypothetical protein FLJ37794 (LOC388724), mRN/
XM_371334 Homo sapiens similar to mKIAA1151 protein (LOC388725), mRNA
XM_371335 Homo sapiens similar to osteotesticular protein tyrosine phosphatase (LOC3i
XM 371336 Homo sapiens similar to alpha tubulin (LOC388728), mRNA
XM_371338 Homo sapiens hypothetical protein LOC93273 (LOC93273), mRNA
XM_371339 Homo sapiens similar to 40S ribosomal protein S25 (LOC388733), mRNA
XM 371340 Homo sapiens similar to Px19-like protein (25 kDa protein of relevant evolutic
XM_371343 Homo sapiens similar to KIAA0663 protein (LOC388739), mRNA
XM_371344 Homo sapiens similar to calpain 8 (LOC388743), mRNA
XM 371346 Homo sapiens hypothetical protein LOC347813 (LOC347813), mRNA
XM 371350 Homo sapiens similar to RIKEN cDNA 1810063B05 (LOC388753), mRNA
XM_371351 Homo sapiens similar to 60S ribosomal protein L35 (LOC388754), mRNA
XM_371352 Homo sapiens formin 2 (FMN2), mRNA
XM 371353 Homo sapiens LOC388756 (LOC388756), mRNA
XM_371354 Homo sapiens hypothetical protein FLJ10157 (FLJ10157), mRNA
XM 371355 Homo sapiens LOC388759 (LOC388759), mRNA
XM_371356 Homo sapiens similar to olfactory receptor GA_x6K02T2NHDJ-9721756-972.
XM 371357 Homo sapiens similar to Olfactory receptor 2M6 (LOC388762), mRNA
XM 371358 Homo sapiens olfactory receptor, family 2, subfamily M, member 4 (OR2M4),
XM_371359 Homo sapiens similar to Ankrd3-prov protein (LOC388763), mRNA
XM 371369 Homo sapiens C219-reactive peptide (FLJ39207), mRNA
XM_371374 Homo sapiens similar to hypothetical protein (LOC388774), mRNA
XM 371380 Homo sapiens S100 calcium binding protein A13 (S100A13), mRNA
XM_371384 Homo sapiens similar to AG02 (LOC388776), mRNA
XM_371385 Homo sapiens similar to hypothetical protein DJ328E19.C1.1 (LOC388777),
XM 371388 Homo sapiens DKFZp434D177-like (DKFZp434D177-like), mRNA
XM_371390 Homo sapiens similar to hypothetical protein (LOC388783), mRNA
XM_371391 Homo sapiens similarto dJ680N4.2 (ubiquitin-conjugating enzyme E2D 3 (he
XM_371395 Homo sapiens similar to dJ1093G12.6 (A novel protein) (LOC388794), mRN;
XM 371397 Homo sapiens similar to hypothetical protein FLJ33620 (LOC388795), mRN/
XM_371398 Homo sapiens myosin, heavy polypeptide 7B, cardiac muscle, beta (MYH7B]
XM 371399 Homo sapiens chromosome 20 open reading frame 142 (C20orf142), mRNA
XM 371401 Homo sapiens chromosome 20 open reading frame 106 (C20orf106), mRNA
XM 371402 Homo sapiens similar to dJ1153D9.4 (novel protein) (LOC388799), mRNA
XM_371403 Homo sapiens LOC388802 (LOC388802), mRNA
XM 371405 Homo sapiens similar to bA476H5.3 (novel protein similar to septin) (LOC38:
XM_371407 Homo sapiens similar to Ankyrin repeat domain protein 18A (LOC388812), rr
XM 371409 Homo sapiens similar to peptidylprolyl isomerase A (cyclophilin A) (LOC388£
XM 371411 Homo sapiens similar to GNGT1 protein (LOC388819), mRNA
XM 371413 Homo sapiens similar to Protein CGI-27 (C21orf19-like protein) (LOC388822
XM_371416 Homo sapiens similar to C21orf258 (LOC388828), mRNA
XM 371417 Homo sapiens KIAA0179 (KIAA0179), mRNA
XM 371418 Homo sapiens similar to PRED59 (LOC388830), mRNA
XM_371421 Homo sapiens similar to hypothetical protein (LOC388840), mRNA
XM 371423 Homo sapiens similar to hypothetical protein DKFZp434P211.1 - human (fra<
XM_371424 Homo sapiens similar to breakpoint cluster region isoform 1 (LOC388847), rr
XM_371429 Homo sapiens similar to hypothetical protein (LOC388852), mRNA
XM 371430 Homo sapiens similar to sushi domain containing 2; Sushi domain (SCR repj
XM 371431 Homo sapiens similar to Gamma-glutamyltranspeptidase 1 precursor (Gamrr
XM_371436 Homo sapiens similar to E2F transcription factor 6 isoform a (LOC388861 ), n
XM_371437 Homo sapiens similar to dJ831C21.3 (novel protein similar to DKFZP434P21
XM_371455 Homo sapiens LOC388882 (LOC388882), mRNA
XM 371459 Homo sapiens hypothetical protein MGC1842 (MGC1842), mRNA
XM 371460 Homo sapiens LOC388886 (LOC388886), mRNA
XM_371461 Homo sapiens KIAA1671 protein (KIAA1671), mRNA
XM 371463 Homo sapiens similar to hypothetical protein 4930562D19 (LOC388891), mF
XM 371466 Homo sapiens LOC388900 (LOC388900), mRNA
XM 371468 Homo sapiens hypothetical protein MGC40042 (MGC40042), mRNA
XM 371469 Homo sapiens LOC388906 (LOC388906), mRNA
XM_371470 Homo sapiens similar to ribosomal protein L5; 60S ribosomal protein L5 (LO(
XM_371471 Homo sapiens chromosome 22 open reading frame 1 (C22orf1), mRNA
XM_371474 Homo sapiens plexin B2 (PLXNB2), mRNA
XM_371476 Homo sapiens similar to Doublecortin domain-containing protein 2 (RU2S prc
XM_371477 Homo sapiens similar to RIKEN cDNA 5830483C08 gene (LOC388926), mR
XM_371478 Homo sapiens LOC388927 (LOC388927), mRNA
XM_371479 Homo sapiens LOC388928 (LOC388928), mRNA
XM_371480 Homo sapiens similar to tudor domain containing 6 protein (LOC388929), mF
XM_371481 Homo sapiens LOC388931 (LOC388931), mRNA
XM_371484 Homo sapiens selenoprotein 1, 1 (K1AA1724), mRNA
XM_371485 Homo sapiens similarto RIKEN cDNA 1700001 C02 (LOC388936), mRNA
XM 371486 Homo sapiens similar to phospholipase B (LOC388937), mRNA
XM_371487 Homo sapiens phospholipase B1 (PLB1), mRNA
XM 371488 Homo sapiens similar to CDNA sequence BC027072 (LOC388939), mRNA
XM_371489 Homo sapiens similarto hypothetical protein (L1H 3 region) - human (LOC3E
XM_371490 Homo sapiens similar to cysteine and histidine-rich domain (CHORD)-contaiι
XM_371491 Homo sapiens LOC388946 (LOC388946), mRNA
XM_371492 Homo sapiens similar to signal-transducing adaptor protein-2; brk kinase sut
XM__371493 Homo sapiens similar to hypothetical protein (LOC388950), mRNA
XM__371494 Homo sapiens similar to Testis-specific Y-encoded-like protein 1 (TSPY-like
XM__371495 Homo sapiens similar to 40S ribosomal protein SA (P40) (34/67 kDa laminin
XM_371496 Homo sapiens similar to Px19-like protein (25 kDa protein of relevant evolutic
XM 371497 Homo sapiens similar to expressed sequence C79663 (LOC388957), mRNA
XM_371500 Homo sapiens similarto hypothetical protein 4921507A12 (LOC388960), mR
XM_371501 Homo sapiens hypothetical protein MGC22014 (MGC22014), mRNA
XM_371502 Homo sapiens similar to RIKEN cDNA 1810056020 (LOC388962), mRNA
XM_371503 Homo sapiens similar to Retinol dehydrogenase 12 (LOC388963), mRNA
XM_371504 Homo sapiens similar to hepatitis C virus core-binding protein 6; cervical can
XMJ371505 Homo sapiens similar to Phosphatidylethanolamine-binding protein (PEBP) (
XM_371506 Homo sapiens LOC388969 (LOC388969), mRNA
XM_371511 Homo sapiens similar to anaphase promoting complex subunit 1 ; anaphase-|
XM_371513 Homo sapiens similar to RAN-binding protein 2-like 1 isoform 1; sperm meml
XM_371514 Homo sapiens similar to WW domain binding protein 1 (LOC388975), mRNA
XM_371515 Homo sapiens similar to protein that is immuno-reactive with anti-PTH polycli
XM_371517 Homo sapiens similar to immunoglobulin kappa light chain VC region (LOC31
XM_371534 Homo sapiens similar to hypothetical protein (LOC389000), mRNA
XM_371535 Homo sapiens similar to hypothetical protein DKFZp434A1 1 (LOC389002),
XM_371536 Homo sapiens similar to tripartite motif-containing 43 (LOC389004), mRNA
XM_371537 Homo sapiens similar to RING finger protein 18 (Testis-specific ring-finger pr
XM_371539 Homo sapiens similar to hypothetical protein (LOC389007), mRNA
XM_371540 Homo sapiens ankyrin-related (UNQ2430), mRNA
XM_371542 Homo sapiens RW1 protein (RW1), mRNA
XMJ371543 Homo sapiens LOC389012 (LOC389012), mRNA
XM_371544 Homo sapiens similar to Sodium/hydrogen exchanger 4 (Na(+)/H(+) exchang
XM_3 1546 Homo sapiens similarto Elongation factor 1 -alpha 1 (EF-1 -alpha-1) (Elongati
XM_371547 Homo sapiens similar to Elongation factor 1-alpha 1 (EF-1 -alpha-1) (Elongati
XM_37 552 Homo sapiens similar to RAN-binding protein 2-like 1 isoform 1; sperm meml
XM_371555 Homo sapiens similar to myosin-Vllb (LOC389031), mRNA
XM_371558 Homo sapiens similar to FKSG30 (LOC389036), mRNA
XM_371561 Homo sapiens similar to CDNA sequence BC043098 (LOC389039), mRNA
XM_371564 Homo sapiens similar to sequence-specific single-stranded-DNA-binding pro
XM_371567 Homo sapiens similar to pote protein; Expressed in prostate, ovary, testis, ar
XM_371568 Homo sapiens similar to pote protein; Expressed in prostate, ovary, testis, ar
XM_371569 Homo sapiens similar to Probable mitochondrial import receptor subunit TOI\,
XM_371571 Homo sapiens similar to breast cancer antigen NY-BR-1 (LOC389052), mRN
XM_37 572 Homo sapiens similar to heterogeneous nuclear ribonucleoprotein K (LOC38
XM_371573 Homo sapiens neurexophilin 2 (NXPH2), mRNA
XM_371575 Homo sapiens formin binding protein 3 (FNBP3), mRNA
XM_371576 Homo sapiens KIAA1189 protein (KIAA1189), mRNA
XM_371577 Homo sapiens similar to RNA polymerase B transcription factor 3 (MGC2390
XM_371581 Homo sapiens similar to zinc finger protein Sp5 (LOC389058), mRNA
XM_371583 Homo sapiens LOC389064 (LOC389064), mRNA
XM_371584 Homo sapiens similar to RIKEN cDNA B830010L13 gene (LOC389065), mRI
XM_371585 Homo sapiens LOC389066 (LOC389066), mRNA
XM_371586 Homo sapiens hypothetical protein FLJ25415 (FLJ25415), mRNA
XM_371588 Homo sapiens similar to Seienide.water dikinase 1 (Selenophosphate synthe
XM_371590 Homo sapiens KIAA1571 protein (KIAA1571), mRNA
XM_371591 Homo sapiens similar to RIKEN cDNA 9430067K14 gene; Ras GTPase-activ
XM_371592 Homo sapiens similar to RIKEN cDNA D630023F18 (LOC389073), mRNA
XM_371593 Homo sapiens similar to REGULATED ENDOCRINE SPECIFIC PROTEIN 1t
XM_371594 Homo sapiens similar to CAVP-target protein (CAVPT) (LOC389076), mRNA
XM_371595 Homo sapiens dedicator of cytokinesis 10 (DOCK10), mRNA
XM_371600 Homo sapiens similar to enterokinase (LOC389083), mRNA
XM_371603 Homo sapiens similar to hypothetical protein FLJ40243 (LOC389085), mRN/
XM_371604 Homo sapiens hypothetical protein FLJ37034 (FLJ37034), mRNA
XM_371605 Homo sapiens hypothetical protein LOC151174 (LOC151174), mRNA
XM_371606 Homo sapiens similar to seven transmembrane helix receptor (LOC389090),
XM_371614 Homo sapiens hypothetical protein FLJ10707 (FLJ10707), mRNA
XM_371617 Homo sapiens TBP-interacting protein (TIP120B), mRNA
XM_371618 Homo sapiens similar to nucleoporin 210; nuclear pore membrane glycoprote
XM_371619 Homo sapiens FYVE, RhoGEF and PH domain containing 5 (FGD5), mRNA
XM_371621 Homo sapiens similar to RIKEN cDNA B830010L13 gene (LOC389099), mRI
XM_371622 Homo sapiens similar to 60S ribosomal protein L23a (LOC389101), mRNA
XM_371623 Homo sapiens similar to YPLR6490 (LOC389102), mRNA
XM_371625 Homo sapiens LOC389106 (LOC389106), mRNA
XM_371626 Homo sapiens similar to Hsp70/Hsp90 organizing protein homolog CG2720-I
XM_371629 Homo sapiens similar to RIKEN cDNA 1110038F21 (LOC389111 ), mRNA
XM_371630 Homo sapiens similar to ribosomal protein S27 (LOC389112), mRNA
XM_371631 Homo sapiens similarto hypothetical protein FLJ35863 (LOC389114), mRN/
XM_371632 Homo sapiens FLJ36157 protein (FLJ36157), mRNA
XM_371638 Homo sapiens similar to mouse fat 1 cadherin (LOC389117), mRNA
XM_371639 Homo sapiens similar to VLLR9392 (LOC389118), mRNA
XM_371641 Homo sapiens similar to RIKEN cDNA 4921517D21 (LOC389120), mRNA
XM_371643 Homo sapiens similar to hypothetical protein MGC39725 (LOC389124), mRf
XM_371645 Homo sapiens similar to 40S ribosomal protein S10 (LOC389127), mRNA
XM_371647 Homo sapiens similar to CG9996-PA (LOC389129), mRNA
XM_371649 Homo sapiens similar to hypothetical protein SB153 isoform 1 (LOC389130),
XM_371653 Homo sapiens similar to hypothetical protein FLJ11292 (LOC389135), mRN/
XM_371655 Homo sapiens similar to SVH-B (LOC389137), mRNA
XM_371656 Homo sapiens similar to olfactory receptor GA_x54KRFPKG5P-55348161-5£
XM_371658 Homo sapiens similar to laminin receptor 1 (ribosomal protein SA); P40-3, fui
XM_371660 Homo sapiens similar to seven transmembrane helix receptor (LOC389144),
XM_371662 Homo sapiens hypothetical protein LOC255330 (LOC255330), mRNA
XM_371663 Homo sapiens similar to WD repeat domain 10 isoform 3 (LOC389147), mRf
XM_371664 Homo sapiens KIAA1257 protein (KIAA1257), mRNA
XM_371665 Homo sapiens LOC389151 (LOC389151), mRNA
XM_371666 Homo sapiens LOC389152 (LOC389152), mRNA
XM_371668 Homo sapiens similar to 60S ribosomal protein L21 (LOC389156), mRNA
XM 371670 Homo sapiens similar to Plscrl protein (LOC389158), mRNA
XM_371671 Homo sapiens similar to Chromosome 1 open reading frame 37 (LOC38916
XM_371672 Homo sapiens similar to template acyivating factor-l alpha (LOC389168), mF
XM_371674 Homo sapiens similar to hypothetical protein FLJ14957 (LOC389170), mRN/
XM_371677 Homo sapiens similar to phosphoserine aminotransferase isoform 1 (LOC38!
XM 371678 Homo sapiens LOC389174 (LOC389174), mRNA
XM_371679 Homo sapiens similar to ribosomal protein L22 (LOC389175), mRNA
XM_371680 Homo sapiens LOC389177 (LOC389177), mRNA
XM_371681 Homo sapiens similarto RING finger protein 13 (LOC389178), mRNA
XM_371682 Homo sapiens similar to 5-hydroxytryptamine receptor 3 subunit C (LOC389'
XM_371683 Homo sapiens LOC389187 (LOC389187), mRNA
XM_371684 Homo sapiens similar to FSHD Region Gene 2 protein (LOC389192), mRNA
XM_371687 Homo sapiens LOC389197 (LOC389197), mRNA
XM_371690 Homo sapiens LOC389202 (LOC389202), mRNA
XM_371691 Homo sapiens LOC389203 (LOC389203), mRNA
XM_371692 Homo sapiens similar to LINE-1 REVERSE TRANSCRIPTASE HOMOLOG (I
XM_371693 Homo sapiens LOC389206 (LOC389206), mRNA
XM_371694 Homo sapiens similar to ENSANGP00000012385 (LOC389207), mRNA
XM_371695 Homo sapiens similar to RIKEN cDNA 4732406D01 gene (LOC389208), mR
XM_371697 Homo sapiens similar to expressed sequence AW060714 (LOC389211), mR
XM_371698 Homo sapiens similar to KIAA1680 protein (LOC401145), mRNA
XM_371701 Homo sapiens similar to template acyivating factor-l alpha (LOC389217), mF
XM_371702 Homo sapiens similar to Ribosomal protein L7A CG3314-PD (LOC389218), i
XM 371705 Homo sapiens LOC389221 (LOC389221), mRNA
XM_371706 Homo sapiens hypothetical protein KIAA1109 (KIAA1109), mRNA
XM 371709 Homo sapiens ring finger protein 150 (RNF150), mRNA
XM 371710 Homo sapiens LOC389227 (LOC389227), mRNA
XM_371711 Homo sapiens similar to GRIK2 protein (LOC389228), mRNA
XM_371714 Homo sapiens similar to ring finger protein 129 (LOC389239), mRNA
XM_371715 Homo sapiens similar to alpha NAC/1.9.2. protein (LOC389240), mRNA
XM_371717 Homo sapiens odd Oz Ten-m homolog 3 (ODZ3), mRNA
XM__371718 Homo sapiens similar to vesicle-associated soluble NSF attachment protein I
XM_371719 Homo sapiens similar to RSTI689 (LOC389255), mRNA
XM 371722 Homo sapiens similar to Leucine-rich repeat-containing protein 14 (LOC3892
XM_371725 Homo sapiens similar to hypothetical protein (LOC389261 ), mRNA
XM_371726 Homo sapiens similar to antifreeze glycoprotein precursor - black rockcod (Li
XM_371728 Homo sapiens similar to CDNA sequence BC012016 (LOC389276), mRNA
XM_371729 Homo sapiens similar to RPE-spondin (LOC389279), mRNA
XM 371731 Homo sapiens similar to bA110H4.2 (similar to membrane protein) (LOC389I
XM_371732 Homo sapiens similar to RIKEN cDNA C230086A09 gene (LOC389282), mR
XM_371733 Homo sapiens hypothetical protein FLJ23577 (FLJ23577), mRNA
XM_371736 Homo sapiens similar to FKSG62 (LOC389286), mRNA
XM_371738 Homo sapiens similar to annexin II receptor (LOC389289), mRNA
XM_371740 Homo sapiens hypothetical protein FLJ23563 (FLJ23563), mRNA
XM_371741 Homo sapiens similar to hypothetical protein (LOC389293), mRNA
XM_371743 Homo sapiens similar to bA110H4.2 (similar to membrane protein) (LOC3892
XM_371749 Homo sapiens similar to bA110H4.2 (similar to membrane protein) (LOC389;
XM_371754 Homo sapiens similar to nonhistone chromosomal protein HMG-17 - rat (LOC
XM_371755 Homo sapiens similar to Rho-guanine nucleotide exchange factor (Rho-inter;
XM_371757 Homo sapiens similar to 60S ribosomal protein L7 (LOC389305), mRNA
XM_371758 Homo sapiens similar to ribosomal protein L10a (LOC389308), mRNA
XM_371759 Homo sapiens hypothetical protein FLJ 11292 (FLJ 11292), mRNA
XM_371760 Homo sapiens hypothetical protein LOC116068 (LOC116068), mRNA
XM_371761 Homo sapiens KIAA0825 protein (KIAA0825), mRNA
XM__371762 Homo sapiens similar to Proteasome activator complex subunit 2 (Proteason
XM_371763 Homo sapiens LOC389313 (LOC389313), mRNA
XM_371764 Homo sapiens similar to NADH dehydrogenase subunit 5 (LOC389314), mRI
XM_371765 Homo sapiens LOC389315 (LOC389315), mRNA
XM_371768 Homo sapiens LOC389320 (LOC389320), mRNA
XM_371769 Homo sapiens LOC389321 (LOC389321), mRNA
XM_371770 Homo sapiens similar to heterogeneous nuclear ribonucleoprotein K (LOC38
XM_371771 Homo sapiens similar to hypothetical protein (LOC389323), mRNA
XM_371772 Homo sapiens LOC389326 (LOC389326), mRNA
XM_371776 Homo sapiens similar to hypothetical protein 4933429F08 (LOC389337), mR
XM_371777 Homo sapiens hypothetical protein LOC348938 (LOC348938), mRNA
XM_371778 Homo sapiens similar to RIKEN CDNA 4921536K21 (LOC389341), mRNA
XM_371781 Homo sapiens similar to 60S ribosomal protein L10 (QM protein homolog) (L
XM_371782 Homo sapiens similar to hypothetical protein (LOC389346), mRNA
XM_371783 Homo sapiens similar to NY-REN-7 antigen (LOC389347), mRNA
XM_371790 Homo sapiens similar to 6-pyruvoyl-tetrahydropterin synthase (LOC389351),
XM_371791 Homo sapiens similar to High mobility group protein 4 (HMG-4) (High mobilitj
XM_371793 Homo sapiens similar to bA476l 15.3 (novel protein similar to septin) (LOC38!
XM_371796 Homo sapiens similar to Protein phosphatase 1 , regulatory subunit 3D (Prote
XM_371797 Homo sapiens LOC389365 (LOC389365), mRNA
XM_371798 Homo sapiens similar to C6orf52 protein (LOC389366), mRNA
XM_371801 Homo sapiens O-acyltransferase (membrane bound) domain containing 1 (0
XM_371809 Homo sapiens chromosome 6 open reading frame 205 (C6orf205), mRNA
XM_371812 Homo sapiens major histocompatibility complex, class II, DQ alpha 1 (HLA-D
XM_371813 Homo sapiens kinesin family member C1 (KIFC1), mRNA
XM_371814 Homo sapiens similar to Rps15a protein (LOC389382), mRNA
XM_371815 Homo sapiens similar to AAAL3045 (LOC389383), mRNA
XM_371816 Homo sapiens similar to RIKEN cDNA 4930539E08 (LOC389384), mRNA
XM_37181 Homo sapiens similar to Cytosol aminopeptidase (Leucine aminopeptidase) i
XM_371818 Homo sapiens similar to Cytosol aminopeptidase (Leucine aminopeptidase) i
XM_371819 Homo sapiens similar to 60S ribosomal protein L12 (LOC389387), mRNA
XM_371820 Homo sapiens similar to hypothetical protein BC006605 (LOC389389), mRN,
XM_371822 Homo sapiens chromosome 6 open reading frame 110 (C6orf110), mRNA
XM_371823 Homo sapiens similar to hypothetical protein DKFZp434D2328 (LOC389394]
XM_371824 Homo sapiens similar to heterogeneous nuclear ribonucleoprotein A3 (LOC3
XM_371825 Homo sapiens similar to BXMAS2-10 protein (LOC389396), mRNA
XM_371826 Homo sapiens similar to I VFI9356 (LOC389400), mRNA
XM_371829 Homo sapiens similar to ENSANGP00000009924 (LOC389405), mRNA
XM_371832 Homo sapiens KIAA1411 (KIAA1411), mRNA
XM_371835 Homo sapiens inhibitor of Bruton agammaglobulinemia tyrosine kinase (IBTH
XM_371837 Homo sapiens similar to oxidoreductase UCPA (LOC389416), mRNA
XM_371838 Homo sapiens ubiquitin specific protease 45 (USP45), mRNA
XM_371841 Homo sapiens similar to hypothetical protein (L1 H 3 region) - human (LOC3S
XM_371842 Homo sapiens LOC389421 (LOC389421), mRNA
XM_371843 Homo sapiens similar to ribosomal protein S27a (LOC389425), mRNA
XM_371844 Homo sapiens TSPY-like (TSPYL), mRNA
XM_371845 Homo sapiens similar to MGC32805 protein (LOC389427), mRNA
XM_371846 Homo sapiens similar to ribosomal protein L5; 60S ribosomal protein L5 (LO<
XM_371847 Homo sapiens similar to RIKEN cDNA 2310057J18 (LOC389429), mRNA
XM_371848 Homo sapiens chromosome 6 open reading frame 115 (C6orf115), mRNA
XM_371849 Homo sapiens chromosome 6 open reading frame 198 (C6orf198), mRNA
XM_371850 Homo sapiens similar to hypothetical protein 9130014G24 (LOC389431), mF
XM_371851 Homo sapiens similar to RIKEN cDNA E130306M17 gene (LOC389432), mR
XM_371853 Homo sapiens similar to 60S ribosomal protein L27a (LOC389435), mRNA
XM_371856 Homo sapiens similar to frazzled CG8581-PA (LOC389444), mRNA
XM_371857 Homo sapiens similar to 60S ribosomal protein L21 (LOC389445), mRNA
XM_371858 Homo sapiens similar to apolipoprotein(a) (EC 3.4.21.-) - rhesus macaque (fi
XM_371863 Homo sapiens family with sequence similarity 20, member C (FAM20C), mRI
XM_371873 Homo sapiens similar to zinc finger protein 316; kruppel-related zinc finger pi
XM_371874 Homo sapiens similar to Matn2-prov protein (LOC389462), mRNA
XM_371877 Homo sapiens KIAA0960 protein (KIAA0960), mRNA
XM_371878 Homo sapiens hypothetical protein FLJ14712 (FLJ14712), mRNA
XM_371879 Homo sapiens LOC389466 (LOC389466), mRNA
XM_371884 Homo sapiens similar to 40S ribosomal protein S26 (LOC389472), mRNA
XM_371885 Homo sapiens similar to Neuronal protein 3.1 (p311 protein) (LOC389473), n
XM_371889 Homo sapiens similar to RP9 protein (LOC389477), mRNA
XM_371891 Homo sapiens KIAA0877 protein (KIAA0877), mRNA
XM_371897 Homo sapiens hypothetical protein DKFZp76112123 (DKFZp76112123), mRN
XM_371901 Homo sapiens similar to PRO0758 (LOC389490), mRNA
XM_371904 Homo sapiens similar to Protein p8 (Candidate of metastasis 1 ) (LOC389492
XM_371917 Homo sapiens similar to Williams Beuren syndrome chromosome region 19 I
XM_371923 Homo sapiens similar to opposite strand transcription unit to Stag3; Gats pro
XM_371925 Homo sapiens similar to transcription factor GTF2IRD2 (LOC389524), mRNA
XM_371930 Homo sapiens similar to PMS4 homolog mismatch repair protein - human (L(
XM_371933 Homo sapiens protein phosphatase 1 , regulatory (inhibitor) subunit 9A (PPP'
XM_371936 Homo sapiens similar to hypothetical protein MGC56855 (LOC389538), mRf
XM_371939 Homo sapiens similar to CG14977-PA (LOC389541), mRNA
XM_371943 Homo sapiens similar to Williams-Beuren syndrome critical region protein 19
XM_371948 Homo sapiens similar to zinc finger protein 312; forebrain embryonic zinc finς
XM_371949 Homo sapiens similar to cardiac leiomodin (LOC389550), mRNA
XM_371953 Homo sapiens KIAA1466 protein (KIAA1466), mRNA
XM_371954 Homo sapiens nucleoporin 205kDa (NUP205), mRNA
XM_371956 Homo sapiens KIAA1549 protein (KIAA1549), mRNA
XM_371960 Homo sapiens KIAA1277 (KIAA1277), mRNA
XM_371995 Homo sapiens similar to hypothetical protein MGC41943 (LOC389592), mRf
XM_372002 Homo sapiens similar to amyotrophic lateral sclerosis 2 (juvenile) chromoson
XM_372004 Homo sapiens LOC389602 (LOC389602), mRNA
XM_372005 Homo sapiens LOC389603 (LOC389603), mRNA
XM_372006 Homo sapiens similar to vasoactive intestinal peptide receptor 2 (LOC38960'
XM_372009 Homo sapiens similar to bA476H5.3 (novel protein similar to septin) (LOC38!
XM_372010 Homo sapiens LOC389607 (LOC389607), mRNA
XM_372011 Homo sapiens similar to HARL2754 (LOC389610), mRNA
XM_372013 Homo sapiens similar to FLJ10408 protein (LOC389611), mRNA
XM_372017 Homo sapiens similar to seven transmembrane helix receptor (LOC389616),
XM_372018 Homo sapiens similar to hypothetical protein FLJ10408 (LOC389617), mRN/
XM_372019 Homo sapiens similar to FLJ10408 protein (LOC389618), mRNA
XM_372024 Homo sapiens LOC389622 (LOC389622), mRNA
XM_372025 Homo sapiens similar to Activated RNA polymerase II transcriptional coactiv;
XM_372026 Homo sapiens similar to seven transmembrane helix receptor (LOC389626),
XM_372027 Homo sapiens LOC389627 (LOC389627), mRNA
XM_372028 Homo sapiens similar to FLJ10408 protein (LOC389630), mRNA
XM_372030 Homo sapiens similar to FLJ10408 protein (LOC389633), mRNA
XM_372031 Homo sapiens mitochondrial tumor suppressor gene 1 (MTSG1), mRNA
XM_372035 Homo sapiens LOC389643 (LOC389643), mRNA
XM_372036 Homo sapiens similar to 60S ribosomal protein L5 (LOC389644), mRNA
XM_372037 Homo sapiens LOC389649 (LOC389649), mRNA
XM_372038 Homo sapiens hypothetical protein FLJ32731 (FLJ32731), mRNA
XM_372039 Homo sapiens similar to hypothetical protein (L1 H 3 region) - human (LOC3S
XM_372040 Homo sapiens similar to asparagine synthetase; glutamine-dependent aspar
XM_372041 Homo sapiens similar to RPLK9433 (LOC389658), mRNA
XM_372042 Homo sapiens similar to polycystin 1-like 3 (LOC389660), mRNA
XM_372045 Homo sapiens similar to hypothetical protein (LOC389663), mRNA
XM_372046 Homo sapiens similar to tropomyosin 4 (LOC389667), mRNA
XM_372047 Homo sapiens similar to hypothetical protein FLJ10307 (LOC389668), mRN/
XM_372048 Homo sapiens similar to 40S ribosomal protein SA (P40) (34/67 kDa laminin
XM_372050 Homo sapiens similar to Heterogeneous nuclear ribonucleoprotein A1 (Helix-
XM_372054 Homo sapiens LOC389678 (LOC389678), mRNA
XM_372055 Homo sapiens similar to hypothetical protein (LOC389679), mRNA
XM_372058 Homo sapiens Pvt1 oncogene homolog, MYC activator (mouse) (PVT1), mRI
XM_372060 Homo sapiens similar to FLJ46354 protein (LOC389690), mRNA
XM_372062 Homo sapiens similar to FLJ46354 protein (LOC389694), mRNA
XM_372063 Homo sapiens similar to epiplakin (LOC389697), mRNA
XM_372069 Homo sapiens similar to prat GOR (LOC389699), mRNA
XM_372074 Homo sapiens similar to Selenoprotein T (LOC389704), mRNA
XM_372075 Homo sapiens similar to RIKEN cDNA 3110001 D03 gene (M. musculus) (MG
XM_372076 Homo sapiens similar to 2700029M09Rik protein (LOC389705), mRNA
XM_372077 Homo sapiens LOC389708 (LOC389708), mRNA
XM_372078 Homo sapiens similar to hypothetical protein (L1 H 3 region) - human (LOC36
XM_372083 Homo sapiens KIAA1161 (KIAA1161), mRNA
XM_372086 Homo sapiens similar to RAB1 B, member RAS oncogene family; small GTP-
XM_372088 Homo sapiens similar to cell recognition molecule CASPR3 isoform 1 (LOC3
XM_372089 Homo sapiens similar to chromosome 9 open reading frame 36 (LOC389723
XM_372090 Homo sapiens similar to cell recognition molecule CASPR3 isoform 1 (LOC3
XM_372092 Homo sapiens similar to FK506-binding protein 4 (Peptidyl-prolyl cis-trans isc
XM_372094 Homo sapiens similar to chromosome 9 open reading frame 36 (LOC389730
XM_372097 Homo sapiens similar to cell recognition molecule CASPR3 isoform 1 (LOC3
XM_372099 Homo sapiens similar to bA251017.4 (similar to methylenetetrahydrofolate d
XM_372100 Homo sapiens LOC389739 (LOC389739), mRNA
XM_372102 Homo sapiens LOC389742 (LOC389742), mRNA
XM_372103 Homo sapiens LOC389744 (LOC389744), mRNA
XM_372104 Homo sapiens similar to breast cancer antigen NY-BR-1 (LOC389745), mRN
XM_372108 Homo sapiens similar to Ribosome biogenesis protein BMS1 homolog (LOC^
XM_372109 Homo sapiens similar to mitochondrial C1-tetrahydrofolate synthase (LOC38'
XM_372110 Homo sapiens aquaporin 7-like (LOC375719), mRNA
XM_372111 Homo sapiens similar to mitochondrial C1-tetrahydrofolate synthase (LOC38'
XM_372112 Homo sapiens similar to phosphoglucomutase 5 (LOC389753), mRNA
XM_372114 Homo sapiens similar to mitochondrial C1-tetrahydrofolate synthase (LOC38
XM_372116 Homo sapiens similar to COBW domain containing protein 3 (LOC389760), r
XM_372117 Homo sapiens similar to CG3073-PA (LOC375743), mRNA
XM_372118 Homo sapiens similar to RIKEN cDNA 1700013B16 (LOC389761), mRNA
XM_372119 Homo sapiens LOC389762 (LOC389762), mRNA
XM_372120 Homo sapiens similar to RIKEN cDNA 1700013B16 (LOC389763), mRNA
XM_372121 Homo sapiens similar to kinesin family member 27 (LOC389764), mRNA
XM_372122 Homo sapiens similar to kinesin family member 27 (LOC389765), mRNA
XM_372123 Homo sapiens similar to RIKEN cDNA 4921517D22 (LOC389766), mRNA
XM_372124 Homo sapiens zinc finger, CCHC domain containing 6 (ZCCHC6), mRNA
XM_372125 Homo sapiens similar to potassium channel tetramerisation domain containir
XM_372128 Homo sapiens similar to Osteotesticular phosphatase; protein tyrosine phosp
XM_372132 Homo sapiens similar to RIKEN cDNA 2810453106 (LOC389776), mRNA
XM_372133 Homo sapiens KIAA1529 (KIAA1529), mRNA
XM_372137 Homo sapiens similar to RIKEN cDNA 4732481 H14 (LOC389785), mRNA
XM_372138 Homo sapiens LOC389786 (LOC389786), mRNA
XM_372140 Homo sapiens LOC389789 (LOC389789), mRNA
XM_372141 Homo sapiens similar to phosphatidylinositol phosphate kinase-like protein (I
XM_372142 Homo sapiens LOC389791 (LOC389791), mRNA
XM_372143 Homo sapiens hypothetical protein LOC375757 (LOC375757), mRNA
XM_372148 Homo sapiens similar to CDNA sequence BC034076 (LOC389796), mRNA
XM_372150 Homo sapiens LOC389799 (LOC389799), mRNA
XM_372154 Homo sapiens similar to bA74P14.2 (novel protein) (LOC389803), mRNA
XM_372157 Homo sapiens similar to HSPC324 (LOC389811 ), mRNA
XM_372159 Homo sapiens similar to CG15216-PA (LOC389813), mRNA
XM_372160 Homo sapiens similar to LPAL6438 (LOC389814), mRNA
XM_372161 Homo sapiens similar to CDNA sequence BC004853 (LOC389816), mRNA
XM_372162 Homo sapiens LOC389817 (LOC389817), mRNA
XM_372163 Homo sapiens similar to cell recognition molecule CASPR3 isoform 1 (LOC3
XM_372168 Homo sapiens LOC389821 (LOC389821), mRNA
XM_372169 Homo sapiens similar to hypothetical protein (LOC389822), mRNA
XM_372175 Homo sapiens LOC389823 (LOC389823), mRNA
XM_372177 Homo sapiens similar to Surfeit locus protein 1 (LOC389825), mRNA
XM_372180 Homo sapiens similar to RIKEN cDNA 1110002H13 (LOC389827), mRNA
XM_372181 Homo sapiens similar to MSTP058 (LOC389828), mRNA
XM_372182 Homo sapiens hypothetical protein FLJ39378 (FLJ39378), mRNA
XM_372186 Homo sapiens similar to hypothetical protein MGC27019 (LOC389830), mRf
XM_372190 Homo sapiens similar to AF038169 protein (LOC389832), mRNA
XM_372191 Homo sapiens similarto hypothetical protein MGC27019 (LOC389833), mRf
XM_372192 Homo sapiens similar to RIKEN cDNA 2700049P18 (LOC389835), mRNA
XM_372193 Homo sapiens KIAA1751 protein (KIAA1751), mRNA
XM_372194 Homo sapiens hypothetical protein MGC13275 (MGC13275), mRNA
XM_372195 Homo sapiens agrin (AGRN), mRNA
XM_372197 Homo sapiens similar to Glutathione peroxidase 1 (GSHPx-1) (Cellular glutat
XM_372198 Homo sapiens hypothetical protein MGC17403 (MGC17403), mRNA
XM_372199 Homo sapiens similar to MAP/ERK kinase kinase 5; apoptosis signal regulati
XM_372200 Homo sapiens similar to Ran-specific GTPase-activating protein (Ran bindini
XM_372201 Homo sapiens similar to hypothetical protein FLJ35782 (LOC389843), mRN/
XM_372202 Homo sapiens similar to ferritin, heavy polypeptide-like 17 (LOC389844), mR
XM_372203 Homo sapiens similar to Amine oxidase [flavin-containing] A (Monoamine oxi
XM_372204 Homo sapiens similar to 40S ribosomal protein SA (P40) (34/67 kDa laminin
XM_372205 Homo sapiens similar to hypothetical protein, MGC:7199 (LOC389850), mRf
XM_372208 Homo sapiens similar to dJ54B20.3 (novel protein similar to lysozyme C (1
XM_372209 Homo sapiens similar to Acetyl-coenzyme A acyltransferase 2 (LOC389854),
XM_372210 Homo sapiens protein phosphatase 1 , regulatory (inhibitor) subunit 3F (PPP'
XM_372212 Homo sapiens similar to G antigen 7 (LOC389855), mRNA
XM_372213 Homo sapiens similar to ubiquitin specific protease 27, X chromosome (LOC
XM_372214 Homo sapiens similar to nuclear protein p30 (LOC389857), mRNA
XM_372223 Homo sapiens similar to 28S ribosomal protein S18c, mitochondrial precurso
XM_372224 Homo sapiens similar to PAGE-5 protein (LOC389860), mRNA
XM_372226 Homo sapiens similar to Zinc finger X-linked protein ZXDB (LOC389862), mF
XM_372227 Homo sapiens similar to PAI-1 mRNA-binding protein; chromodomain helica;
XM_372231 Homo sapiens hypothetical protein FLJ20105 (FLJ20105), mRNA
XM_372233 Homo sapiens similar to Seienide.water dikinase 1 (Selenophosphate synthe
XM_372239 Homo sapiens hypothetical protein BC007652 (LOC92129), mRNA
XM_372245 Homo sapiens similar to R28830J (LOC389885), mRNA
XM_372247 Homo sapiens similar to RIKEN cDNA 1110012005 (LOC389887), mRNA
XM_372248 Homo sapiens similar to FLJ20527 protein (LOC389888), mRNA
XM_372253 Homo sapiens similar to ENSANGP00000013187 (LOC389891), mRNA
XM_372254 Homo sapiens similar to Mothers against decapentaplegic homolog interactir
XM_372255 Homo sapiens similar to MGC68553 protein (LOC389895), mRNA
XM_372257 Homo sapiens similar to ubiquitin-conjugating enzyme E2N (LOC389898), m
XM_372258 Homo sapiens similar to HS1 binding protein (LOC389899), mRNA
XM_372261 Homo sapiens similar to DKFZP586L0724 protein (LOC389900), mRNA
XM_372262 Homo sapiens similar to ATP-dependent DNA helicase II, 70 kDa subunit (LL
XM_372267 Homo sapiens zinc finger protein 275 (ZNF275), mRNA
XM_372268 Homo sapiens similar to Extracellular matrix protein 2 precursor (Matrix glycc
XM_372272 Homo sapiens LOC389905 (LOC389905), mRNA
XM_372273 Homo sapiens similar to Serine/threonine protein kinase PRKX (Protein kinai
XM_372274 Homo sapiens similar to Serine/threonine protein kinase PRKX (Protein kinai
XM_372275 Homo sapiens LOC389911 (LOC389911 ), mRNA
XM_372282 Homo sapiens cytokine receptor-like factor 2 (CRLF2), mRNA
XM_372286 Homo sapiens neuroligin 4, Y linked (NLGN4Y), mRNA
XM_372289 Homo sapiens chromosome Y open reading frame 15A (CYorf15A), mRNA
XM_372292 Homo sapiens similar to RNA binding motif protein, Y chromosome, family 1
XM_372295 Homo sapiens similar to bA476H5.3 (novel protein similar to septin) (LOC38!
XM_372296 Homo sapiens similar to deleted in azoospermia (LOC389926), mRNA
XM_372302 Homo sapiens similar to 3-hydroxyhexobarbital dehydrogenase 1/3-alpha, 17
XM_372303 Homo sapiens similar to proline-rich proteoglycan 2 (LOC389936), mRNA
XM_372304 Homo sapiens similar to SI:dZ211013.2 (novel protein) (LOC389938), mRN/
XM_372305 Homo sapiens similar to Gliacolin (LOC389941), mRNA
XM_372306 Homo sapiens similar to polyketide synthase (LOC389944), mRNA
XM_372307 Homo sapiens LOC389950 (LOC389950), mRNA
XM_372308 Homo sapiens similar to Zinc finger protein 37A (Zinc finger protein KOX21 ) i
XM_372309 Homo sapiens similar to KIAA1975 protein (LOC389952), mRNA
XM_372310 Homo sapiens similar to Ig kappa chain V region (Z4) - human (LOC389953)
XM_372311 Homo sapiens similar to cubilin; intrinsic factor-cobalamin receptor; intrinsic f
XM_372312 Homo sapiens similar to KIAA0592 protein (LOC389959), mRNA
XM_372314 Homo sapiens similar to RIKEN cDNA 2610524G07 (LOC389972), mRNA
XM_372315 Homo sapiens similar to 60S ribosomal protein L12 (LOC389974), mRNA
XM_372316 Homo sapiens similar to Ribosome biogenesis protein BMS1 homolog (LOC;
XM_372317 Homo sapiens similar to hypothetical protein DKFZp586O0120.1 - human (fn
XM_372319 Homo sapiens similar to Gene with similarity to rat kidney-specific (KS) gene
XM_372320 Homo sapiens similar to bA182L21.1 (novel protein similarto hypothetical pr*
XM_372321 Homo sapiens similar to bA182L21.1 (novel protein similar to hypothetical pr*
XM_372322 Homo sapiens similar to nuclear DNA-binding protein; small unique nuclear r
XM_372323 Homo sapiens similar to nuclear DNA-binding protein; small unique nuclear r
XM_372324 Homo sapiens similar to lipase A precursor; Lipase A, lysosomal acid, choles
XM_372325 Homo sapiens similar to Tebp-pending-prov protein (LOC390000), mRNA
XM_372328 Homo sapiens similar to peptidylprolyl isomerase A (cyclophilin A) (LOC390C
XM_372329 Homo sapiens similar to Mitochondrial import receptor subunit TOM22 homol
XM_372330 Homo sapiens similar to 40S ribosomal protein S26 (LOC390009), mRNA
XM 372331 Homo sapiens similar to Sax-1 (LOC390010), mRNA
XM_372334 Homo sapiens similar to double homeobox protein (LOC390016), mRNA
XM 372335 Homo sapiens similar to double homeobox protein (LOC390017), mRNA
XM_372337 Homo sapiens similar to sperm associated AWN protein (LOC390020), mRN
XM_372340 Homo sapiens similar to double homeobox protein (LOC390024), mRNA
XM_372341 Homo sapiens similar to Potential carboxypeptidase-like protein X2 precurso
XM_372343 Homo sapiens similar to RIKEN cDNA 1500011L16 (LOC390031), mRNA
XM_372345 Homo sapiens similar to RIKEN cDNA 1500011 L16 (LOC390033), mRNA
XM_372346 Homo sapiens similar to olfactory receptor MOR16-1 (LOC390034), mRNA
XM_372347 Homo sapiens similar to Olfactory receptor 52K1 (LOC390036), mRNA
XM_372348 Homo sapiens similar to Olfactory receptor 5211 (LOC390037), mRNA
XM_372349 Homo sapiens sim ilar to seven transmembrane helix receptor (LOC390038),
XM_372350 Homo sapiens similar to olfactory receptor MOR11-2 (LOC390039), mRNA
XM_372351 Homo sapiens similar to olfactory receptor MOR8-1 (LOC390046), mRNA
XM_372352 Homo sapiens similar to odorant receptor HOR3beta5 (LOC390054), mRNA
XM 372353 Homo sapiens sim ilar to HOR5Beta6 (LOC390058), mRNA
XM_372354 Homo sapiens si ilar to HOR5Beta7 (LOC390059), mRNA
XM_372355 Homo sapiens similar to Olfactory receptor 51Q1 (LOC390061), mRNA
XM_372356 Homo sapiens similar to Olfactory receptor 5111 (HOR5beta11 ) (LOC390063
XM_372357 Homo sapiens similar to Olfactory receptor 5112 (HOR5beta12) (LOC390064
XM_372358 Homo sapiens similar to HOR5Beta13 (LOC390065), mRNA
XM_372359 Homo sapiens similar to Olfactory receptor 52D1 (HOR5beta14) (LOC39006I
XM_372360 Homo sapiens similar to seven transmembrane helix receptor (LOC390067),
XM 372361 Homo sapiens similar to Olfactory receptor 52N4 (LOC390072), mRNA
XM_372362 Homo sapiens similar to Olfactory receptor 56B2 (LOC390073), mRNA
XM_372364 Homo sapiens similar to Olfactory receptor 52N1 (LOC390075), mRNA
XM_372365 Homo sapiens similar to Olfactory receptor 52N2 (LOC390077), mRNA
XM_372366 Homo sapiens similar to Olfactory receptor 52E6 (LOC390078), mRNA
XMJ372367 Homo sapiens similar to Olfactory receptor 52E6 (LOC390079), mRNA
XM_372368 Homo sapiens similar to Olfactory receptor 52E4 (LOC390081 ), mRNA
XM_372369 Homo sapiens similar to Olfactory receptor 52E5 (LOC390082), mRNA
XM_372370 Homo sapiens similar to Olfactory receptor 56A6 (LOC390083), mRNA
XM_372371 Homo sapiens similar to Olfactory receptor 56A4 (LOC390084), mRNA
XM_372372 Homo sapiens si ilar to seven transmembrane helix receptor (LOC390091 ),
XM_372373 Homo sapiens similar to Olfactory receptor 10A6 (LOC390093), mRNA
XM__372374 Homo sapiens similar to large subunit ribosomal protein L36a (LOC390096),
XM_372375 Homo sapiens simi to 1-aminocyclopropane-1-carboxylate synthase (LOC XM_372376 Homo sapiens simi to Olfactory receptor 4X1 (LOC390113), mRNA XM_372377 Homo sapiens simi to RIKEN CDNA 0610012A05 (LOC390120), mRNA XM_372378 Homo sapiens simi to RIKEN cDNA 0610012A05 (LOC390121), mRNA XM_372379 Homo sapiens simi to hypothetical protein FLJ14082 (LOC390124), mRN/ XM_372380 Homo sapiens simi to RIKEN cDNA 0610012A05 (LOC390125), mRNA XM_372381 Homo sapiens simi to olfactory receptor MOR234-3 (LOC390128), mRNA XM_372382 Homo sapiens simi to olfactory receptor MOR231-3 (LOC390133), mRNA XM_372384 Homo sapiens simi to Olfactory receptor 4A5 (LOC390137), mRNA XM_372385 Homo sapiens simi to olfactory receptor (LOC390138), mRNA XM_372386 Homo sapiens simi to Olfactory receptor 5D13 (LOC390142), mRNA XM_372387 Homo sapiens olfactory receptor, family 5, subfamily L, member 2 (OR5L2), i XM_372388 Homo sapiens simi to Olfactory receptor 5D16 (LOC390144), mRNA XM_372389 Homo sapiens simi to seven transmembrane helix receptor (LOC390148), XM_372390 Homo sapiens simil to Olfactory receptor 8H2 (LOC390151), mRNA XM_372391 Homo sapiens simi to Olfactory receptor 8H3 (LOC390152), mRNA XM_372393 Homo sapiens simi to Olfactory receptor 5T3 (LOC390154), mRNA XM_372394 Homo sapiens simi to seven transmembrane helix receptor (LOC390155), XM_372395 Homo sapiens simi to Olfactory receptor 8K1 (LOC390157), mRNA XM_372396 Homo sapiens simi to ribosomal protein L5; 60S ribosomal protein L5 (LO< XM_372397 Homo sapiens simi to Olfactory receptor 5M9 (LOC390162), mRNA XM_372399 Homo sapiens simi to seven transmembrane helix receptor (LOC390166), XM_372400 Homo sapiens simi to Olfactory receptor 5M1 (OST050) (LOC390167), mF XM_372401 Homo sapiens simi to Olfactory receptor 5M1 (OST050) (LOC390168), mF XM_372402 Homo sapiens simi to olfactory receptor GA_x6K02T2Q125-47402610-47^ XM_372403 Homo sapiens simi to Olfactory receptor 9G1 (LOC390174), mRNA XM_372404 Homo sapiens simi to GTP-binding protein alpha-s subunit (LOC390175), XM_372405 Homo sapiens simi to Olfactory receptor 5AK2 (LOC390181), mRNA XM_372406 Homo sapiens simi to Olfactory receptor 5B2 (OST073) (LOC390186), mR XM_372409 Homo sapiens simi to Olfactory receptor 5B16 (LOC390191), mRNA XM_372410 Homo sapiens LOC3901 92 (LOC390192), mRNA XM_372411 Homo sapiens simi to Olfactory receptor 5AN1 (LOC390195), mRNA XM_372412 Homo sapiens simi to seven transmembrane helix receptor (LOC390197), XM_372413 Homo sapiens simi to Olfactory receptor 4D9 (LOC390199), mRNA XM_372414 Homo sapiens simi to seven transmembrane helix receptor (LOC390201), XM_372415 Homo sapiens simi to membrane-spanning 4-domains, subfamily A, memt XM_372416 Homo sapiens simi to leucine-rich repeat-containing 10; leucine-rich repeε XM_372418 Homo sapiens simi to Double C2, gamma (LOC390213), mRNA XM_372420 Homo sapiens simi to chromosome 11 open reading frame2; chromosome XM_372423 Homo sapiens simi to tripartite motif-containing 43 (LOC390231), mRNA XM_372424 Homo sapiens simi to tripartite motif-containing 43 (LOC390233), mRNA XM_372425 Homo sapiens simi to RIKEN cDNA 0610012A05 (LOC390234), mRNA XM_372426 Homo sapiens simi to RIKEN cDNA 0610012A05 (LOC390237), mRNA XM_372427 Homo sapiens simi to tripartite motif-containing 43 (LOC390238), mRNA XM_372428 Homo sapiens simi to folate receptor 3 (LOC390243), mRNA XM_372429 Homo sapiens simi to FLJ10251 protein (LOC390245), mRNA XM_372430 Homo sapiens simi to RIKEN cDNA 2210418010 (LOC390246), mRNA XM_372431 Homo sapiens simi to putative protein family member (XC187) (LOC3902E XM_372432 Homo sapiens simi to programmed cell death-2/Rp8 homolog (LOC39025I XM_372433 Homo sapiens simi to brain-specific homeodomain protein (LOC390259), i XM_372434 Homo sapiens simi to Olfactory receptor 6X1 (LOC390260), mRNA XM_372435 Homo sapiens simi to Olfactory receptor 6M1 (LOC390261), mRNA XM_372436 Homo sapiens simi to Olfactory receptor 10G4 (LOC390264), mRNA XM_372437 Homo sapiens simi to Olfactory receptor 10G7 (LOC390265), mRNA XM_372438 Homo sapiens simi to Olfactory receptor 10D4 (LOC390266), mRNA XM_372441 Homo sapiens simi to seven transmembrane helix receptor (LOC390271), XM 372442 Homo sapiens simi to seven transmembrane helix receptor (LOC390272),
XM_372443 Homo sapiens lar to Olfactory receptor 8A1 (OST025) (LOC390275), mR XM_372444 Homo sapiens lar to Bcl-2 homologous antagonist/killer (Apoptosis regul; XM_372445 Homo sapiens lar to retinitis pigmentosa GTPase regulator (LOC390278; XM_372447 Homo sapiens lar to eukaryotic translation initiation factor 3, subunit 5 (e XM_372448 Homo sapiens lar to signal recognition particle 14kDa (homologous Alu F XM_372449 Homo sapiens lar to olfactory receptor GA_x6K02T2PVTD-14054886-14' XM_372450 Homo sapiens lar to Heterogeneous nuclear ribonucleoprotein A1 (Helix- XM_372452 Homo sapiens lar to Peptidylprolyl isomerase A (cyclophilin A) (LOC390. XM_372453 Homo sapiens lar to RIKEN cDNA 1110014F12 (LOC390300), mRNA XM_372456 Homo sapiens lar to transient receptor protein 6 (LOC390310), mRNA XM_372457 Homo sapiens lar to olfactory receptor like protein (LOC390313), mRNA XM_372459 Homo sapiens lar to olfactory receptor MOR111-1 (LOC390321), mRNA XM_372460 Homo sapiens lar to olfactory receptor MOR112-1 (LOC390323), mRNA XM_372461 Homo sapiens lar to olfactory receptor MOR109-1 (LOC390324), mRNA XM_372462 Homo sapiens lar to olfactory receptor GA_x6K02T2PULF-11304679-11! XM_372463 Homo sapiens lar to olfactory receptor MOR108-4 (LOC390326), mRNA XM_372464 Homo sapiens lar to olfactory receptor GA_x6K02T2PULF-11553313-11! XM_372465 Homo sapiens lar to olfactory receptor MOR114-12 (LOC390328), mRN/ XM_372466 Homo sapiens lar to poly(A) binding protein, cytoplasmic 4 isoform 2 (LO XM_372468 Homo sapiens lar to T-box transcription factor TBX20 (LOC390338), mRI XM_372469 Homo sapiens lar to Gag-Pro-Pol-Env protein (LOC390342), mRNA XM_372470 Homo sapiens lar to rriicrotubule-associated proteins 1A/1 B light chain 3 XM_372471 Homo sapiens lar to 60S ribosomal protein L10 (QM protein) (Tumor sup XM_372472 Homo sapiens lar to Small nuclear ribonucleoprotein Sm D2 (snRNP con XM_372473 Homo sapiens lar to eukaryotic translation elongation factor 1 alpha 1 ; C XM_372474 Homo sapiens lar to ribosomal protein L9 (LOC390353), mRNA XM_372476 Homo sapiens lar to dynein, axonemal, heavy chain 11 ; situs inversus vii XM_372480 Homo sapiens lar to KIAA1786 protein (LOC390365), mRNA XM_372482 Homo sapiens lar to 0S ribosomal protein S6 (Phosphoprotein NP33) (L XM_372483 Homo sapiens lar to proline rich protein 2 (LOC390371), mRNA XM_372485 Homo sapiens lar to N litochondrial ornithine transporter 1 (Solute carrier' XM_372486 Homo sapiens lar to Avlpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1 ,3-r XM_372487 Homo sapiens lar to chromosome 9 open reading frame 12; 1 ,3,4,5,6-pe XM_372488 Homo sapiens lar to istone 1 , H2bc; H2B histone family, member S (LO XM_372490 Homo sapiens lar to Striatin (LOC390387), mRNA XM_372491 Homo sapiens lar to g lyceraldehyde-3-phosphate dehydrogenase (phosp XM_372493 Homo sapiens lar to A.DP.ATP carrier protein T2 - human (LOC390405), XM_372494 Homo sapiens lar to chromosome 15 open reading frame 2 (LOC390414 XM_372496 Homo sapiens lar to ADP-ribosylation factor 4 (LOC390423), mRNA XM_372497 Homo sapiens lar to rnKIAA0324 protein (LOC390426), mRNA XM_372498 Homo sapiens ^390427 (LOC390427), mRNA XM_372499 Homo sapiens lar to Olfactory receptor 4M1 (LOC390428), mRNA XM_372500 Homo sapiens lar to Olfactory receptor 4N2 (LOC390429), mRNA XM_372501 Homo sapiens lar to Olfactory receptor 4K2 (LOC390431), mRNA XM_372502 Homo sapiens lar to olfactory receptor MOR239-6 (LOC390432), mRNA XM_372503 Homo sapiens lar to Olfactory receptor 4K13 (LOC390433), mRNA XM_372504 Homo sapiens lar to Olfactory receptor 4K17 (LOC390436), mRNA XM_372505 Homo sapiens lar to Olfactory receptor 4N5 (LOC390437), mRNA XM_372506 Homo sapiens lar to Olfactory receptor 11G2 (LOC390439), mRNA XM_372507 Homo sapiens lar to Olfactory receptor 11 H4 (LOC390442), mRNA XM_372508 Homo sapiens lar to Ribonuclease-like protein 9 precursor (LOC390443) XM_372509 Homo sapiens lar to Olfactory receptor 5AU1 (LOC390445), mRNA XM_372521 Homo sapiens lar to SMT3 suppressor of mif two 3 homolog 2 (LOC3904 XM_372522 Homo sapiens lar to Plasmodium falciparum trophozoite antigen r45-like XM_372524 Homo sapiens ^390481 (LOC390481), mRNA XM_372525 Homo sapiens lar to ribosomal protein L31 (LOC390485), mRNA XM 372527 Homo sapiens lar to 60S ribosomal protein L21 (LOC390488), mRNA
XM_372528 Homo sapiens hypothetical protein FLJ36749 (FLJ36749), mRNA
XM_372532 Homo sapiens similar to Alpha-1 -antitrypsin-related protein precursor (LOC3!
XM_372534 Homo sapiens similar to proline-rich glycoprotein (sgp158) (LOC390507), mF
XM_372535 Homo sapiens similar to MGC53446 protein (LOC390508), mRNA
XM_372536 Homo sapiens similar to chloride intracellular channel 6; chloride channel for
XM_372542 Homo sapiens similar to protein kinase related to Raf protein kinases; Metho
XM_372543 Homo sapiens similar to immunoglobulin heavy-chain-2 light-chain-2 VH seg
XM_372544 Homo sapiens similar to hect domain and RLD 2 (LOC390533), mRNA
XM_372547 Homo sapiens similar to CDC42-binding protein kinase alpha isoform A; ser-
XM_372548 Homo sapiens similar to Olfactory receptor 4M2 (LOC390538), mRNA
XM_372549 Homo sapiens similar to seven transmembrane helix receptor (LOC390539),
XM_372550 Homo sapiens similar to salivary proline-rich protein (LOC390540), mRNA
XM_372553 Homo sapiens similar to neuronal nonacetlycholine binding subunit (LOC390
XM_372555 Homo sapiens similar to hect domain and RLD 2 (LOC390549), mRNA
XM_372556 Homo sapiens similar to hect domain and RLD 2 (LOC390551 ), mRNA
XM_372559 Homo sapiens similar to Dynamin-1 (D100) (Dynamin, brain) (B-dynamin) (L<
XM_372560 Homo sapiens similar to hect (homologous to the E6-AP (UBE3A) carboxyl tc
XM_372562 Homo sapiens similar to neuronal nicotinic acetylcholine receptor beta4 subi
XM_372563 Homo sapiens similar to amyloid beta A4 precursor protein-binding, family A,
XM_372565 Homo sapiens similar to amyloid beta A4 precursor protein-binding, family A,
XM_372566 Homo sapiens similar to GLP_457J3116J 1626 (LOC390570), mRNA
XM_372568 Homo sapiens similar to casein kinase 1 , alpha 1 (LOC390575), mRNA
XM_372569 Homo sapiens similar to KIAA1728 protein (LOC390577), mRNA
XM_372570 Homo sapiens similar to zinc finger protein 444; endothelial zinc finger protei
XM_372573 Homo sapiens similar to hypothetical protein BC009980 (LOC390594), mRN,
XM_372574 Homo sapiens similar to ubiquitin associated protein (LOC390595), mRNA
XM_372575 Homo sapiens similar to cell division cycle 2-like 1 (PITSLRE proteins) isofor
XM_372576 Homo sapiens similar to RIKEN cDNA C230094B15 (LOC390598), mRNA
XM_372577 Homo sapiens similar to High mobility group protein 1 (HMG-1) (Amphoterin)
XM_372578 Homo sapiens hypothetical protein LOC123346 (LOC123346), mRNA
XM_372579 Homo sapiens similar to Dynamin-1 (D100) (Dynamin, brain) (B-dynamin) (L(
XM_372580 Homo sapiens similar to Dynamin-1 (D100) (Dynamin, brain) (B-dynamin) (L(
XM_372581 Homo sapiens similar to hypothetical protein FLJ20452 (LOC390611 ), mRN/
XM_372583 Homo sapiens similar to Dynamin-1 (D100) (Dynamin, brain) (B-dynamin) (L(
XM_372584 Homo sapiens similar to RIKEN cDNA 6430502M16 gene (LOC390616), mR
XM_372585 Homo sapiens similar to hypothetical protein DKFZp434H 020 (LOC390623),
XM_372586 Homo sapiens similar to golgi autoantigen golgin subfamily a2-like (LOC390*
XM_372588 Homo sapiens similar to hypothetical protein (LOC390633), mRNA
XM_372589 Homo sapiens similar to RIKEN cDNA D330012F22 gene (LOC390637), mR
XM_372591 Homo sapiens similar to Golgi autoantigen, golgin subfamily a, 2; golgln-95; ι
XM_372592 Homo sapiens hypothetical protein LOC145814 (LOC145814), mRNA
XM_372593 Homo sapiens similar to Serine/threonine protein kinase PRKX (Protein kinai
XM_372596 Homo sapiens similar to Olfactory receptor 4F6 (LOC390648), mRNA
XM_372597 Homo sapiens similar to Olfactory receptor 4F15 (LOC390649), mRNA
XM_372598 Homo sapiens similar to Olfactory receptor 4F14 (LOC390650), mRNA
XM_372599 Homo sapiens similar to olfactory receptor MOR245-8 (LOC390651 ), mRNA
XM_372601 Homo sapiens similar to hect domain and RLD 2 (LOC390654), mRNA
XM_372606 Homo sapiens similar to C1q Related (LOC390664), mRNA
XM_372607 Homo sapiens similar to Neuronal pentraxin II precursor (NP-II) (NP2) (LOC2
XM_372608 Homo sapiens similar to hypothetical protein 6820428D13 (LOC390668), mF
XM_372609 Homo sapiens similar to ATP-binding cassette transporter ABCA3 (LOC3906
XM_372611 Homo sapiens similar to Hypothetical protein MGC56918 (LOC390680), mRf
XM_372614 Homo sapiens similar to NAD(P) dependent steroid dehydrogenase-like (LO(
XM_372615 Homo sapiens LOC390684 (LOC390684), mRNA
XM_372616 Homo sapiens similar to CDC37-like gene (LOC390688), mRNA
XM_372617 Homo sapiens similar to Protein C20orf27 (LOC390690), mRNA
XM_372618 Homo sapiens similar to Filamin B (FLN-B) (Beta-filamin) (Actin-binding like |
XM_372622 Homo sapiens similar to hypothetical protein MGC5244 (LOC390697), mRN/ XM_372625 Homo sapiens similarto protein kinase related to Raf protein kinases; Metho XM_372626 Homo sapiens similar to protein phosphatase 2A 48 kDa regulatory subunit ii XM_372628 Homo sapiens similar to Ig heavy chain - human (fragment) (LOC390709), m XM_372629 Homo sapiens similarto Ig H-chain V-region (DP-40) (LOC390710), mRNA XM_372630 Homo sapiens similar to immunoglobulin M chain (LOC390712), mRNA XM_372631 Homo sapiens similar to immunoglobulin M chain (LOC390713), mRNA XM_372632 Homo sapiens similar to Ig H-chain V-region (DP-39) (LOC390714), mRNA XM_372633 Homo sapiens similar to Ig heavy chain - human (fragment) (LOC390715), m XM_372634 Homo sapiens similar to double homeobox protein (LOC390718), mRNA XM_372637 Homo sapiens similar to Rab31-like (LOC390728), mRNA XM_372638 Homo sapiens similar to 60S ribosomal protein L10 (QM protein homolog) (L XM_372639 Homo sapiens similar to carboxylesterase 3 (LOC390732), mRNA XM_372640 Homo sapiens similar to chromosome 15 open reading frame 2 (LOC390734 XM_372641 Homo sapiens similar to Spermatogenesis associated protein 2 (Spermatoge XM_372642 Homo sapiens similar to MGC9515 protein (LOC390737), mRNA XM_372643 Homo sapiens similar to mKIAA1261 protein (LOC390738), mRNA XM_372644 Homo sapiens similar to Serine/Threonine protein kinase (LOC390743), mRf XM_372646 Homo sapiens similar to proline rich protein (LOC390747), mRNA XM_372647 Homo sapiens hypothetical protein LOC348180 (LOC348180), mRNA XM_372648 Homo sapiens similar to embryonic poly(A) binding protein 2 (LOC390748), r XM_372650 Homo sapiens similar to putative protein (LOC390753), mRNA XM_372651 Homo sapiens similar to hypothetical class II basic helix-loop-helix protein (L> XM_372654 Homo sapiens similar to RIKEN CDNA 2810408A11 (LOC390760), mRNA XM_372655 Homo sapiens similar to RIKEN cDNA 9930039A11 gene (LOC390761), mRI XM_372657 Homo sapiens similar to 60S ribosomal protein L23a (LOC390765), mRNA XM_372658 Homo sapiens similar to Ribonuclease 1-11 (RNase H1) (Ribonuclease H type XM_372660 Homo sapiens similar to Ubiquitin-conjugating enzyme E2-24 kDa (Ubiquitin- XM_372662 Homo sapiens similar to ubiquitin specific protease 6; tre-2 oncogene; hyper| XM_372663 Homo sapiens similar to Ribosomal protein S6 kinase I (S6K) (P70-S6K) (LO XM_372666 Homo sapiens similar to Chemokine (C-C motif) ligand 3-like 1 (LOC390788) XM_372668 Homo sapiens similar to ADP-ribosylation factor-like 8; ADP-ribosylation-like XM_372669 Homo sapiens similar to type I hair keratin 6 (LOC390792), mRNA XM_372670 Homo sapiens similar to keratin associated protein 2-4; keratin associated pr XM_372673 Homo sapiens similar to hypothetical protein A930006D11 (LOC390796), mF XM_372675 Homo sapiens similar to wingless-type MMTV integration site family, membei XM_372676 Homo sapiens similar to S100 calcium-binding protein A10; S100 calcium-bir XM_372677 Homo sapiens similar to DEAH (Asp-Glu-Ala-His) box polypeptide 40 (LOC3! XM_372678 Homo sapiens similar to Monocarboxylate transporter 7 (MCT 7) (MCT 6) (LC XM_372679 Homo sapiens similar to cleavage and polyadenylation specific factor 4; mus XM_372680 Homo sapiens similar to Hypothetical protein KIAA0176 (LOC390814), mRNj XM_372682 Homo sapiens similar to L-threonine aldolase (LOC390816), mRNA XM_372685 Homo sapiens similar to aminopeptidase puromycin sensitive; puromycin-sei XM_372687 Homo sapiens similar to hypothetical protein BC007436 (LOC390824), mRN, XM_372689 Homo sapiens similar to mitochondrial carrier triple repeat 1 (LOC390828), π XM_372692 Homo sapiens similar to KIAA0563-related gene (LOC390845), mRNA XM_372693 Homo sapiens similar to 27 kDa Golgi SNARE protein (Golgi SNAP receptor XM_372695 Homo sapiens similar to Chain A, Crystal Structure Of The R463a Mutant Of XM_372696 Homo sapiens similar to FKSG30 (LOC390861), mRNA XM_372697 Homo sapiens similar to minus agglutinin (LOC390864), mRNA XM_372698 Homo sapiens similar to hypothetical protein (LOC390865), mRNA XM_372700 Homo sapiens similar to immediate early protein homolog (LOC390869), mR XM_372701 Homo sapiens similar to CXYorfl -related protein (LOC390871), mRNA XM_372702 Homo sapiens similar to onecut 3 (LOC390874), mRNA XM_372703 Homo sapiens similar to fos39347_1 (LOC390875), mRNA XM_372704 Homo sapiens similar to ribosomal protein L35 (LOC390876), mRNA XM 372705 Homo sapiens similar to adenylate kinase (EC 2.7.4.3), cytosolic - common c
XM_372707 Homo sapiens similar to nuclear factor, interleukin 3, regulated (LOC390880; XM_372708 Homo sapiens similar to Olfactory receptor 7G2 (Olfactory receptor 19-13) (C XM_372709 Homo sapiens similar to Olfactory receptor 7G3 (OST085) (LOC390883), mF XM_372711 Homo sapiens similarto 60S RIBOSOMAL PROTEIN L12 (LOC390891), mR XM_372712 Homo sapiens similarto Olfactory receptor 7A10 (OST027) (LOC390892), m XM_372714 Homo sapiens similar to Olfactory receptor 7A2 (LOC390894), mRNA XM_372715 Homo sapiens similar to olfactory receptor, family 7, subfamily A, member 17 XM_372716 Homo sapiens widely-interspaced zinc finger motifs (WIZ), mRNA XM_372719 Homo sapiens similar to Zinc finger protein 492 (LOC390906), mRNA XM_372720 Homo sapiens similar to Zinc finger protein 93 (Zinc finger protein HTF34) (L XM_372721 Homo sapiens similar to cytochrome P450 monooxygenase (LOC390914), rr XM_372723 Homo sapiens similar to RP2 protein, testosterone-regulated - ricefield mous XM_372724 Homo sapiens similar to Zinc finger protein 132 (LOC390920), mRNA XM_372726 Homo sapiens similar to hypothetical protein (LOC390927), mRNA XM_372727 Homo sapiens similar to RIKEN cDNA C330005M16 (LOC390928), mRNA XM_372728 Homo sapiens similar to HSPC270 (LOC390929), mRNA XM_372730 Homo sapiens similar to Erf (LOC390937), mRNA XM_372731 Homo sapiens similar to pregnancy specific beta-1 -glycoprotein 1 (LOC3909 XM_372732 Homo sapiens similar to R28379_1 (LOC390940), mRNA XM_372733 Homo sapiens LOC390941 (LOC390941), mRNA XM_372735 Homo sapiens similar to paraneoplastic neuronal antigen MM2 (LOC390945] XM_372737 Homo sapiens similar to heterogeneous nuclear ribonucleoprotein M isoform XM_372740 Homo sapiens similar to ENSANGP00000004655 (LOC390955), mRNA XM_372741 Homo sapiens similar to peptidyl-Pro cis trans isomerase (LOC390956), mRI XM_372745 Homo sapiens similar to Zinc finger protein KM 8 (HKr18) (LOC390963), mRf XM_372746 Homo sapiens similar to YME1-like 1 ; ATP-dependent metalloprotease FtsH' XM_372748 Homo sapiens similar to 1060P1 1.3 (killer inhibitory receptor 2-2-2 (KIR222) XM_372749 Homo sapiens similarto protein kinase Bsk146 (LOC390975), mRNA XM_372750 Homo sapiens similar to ret finger protein-like 1 (LOC390976), mRNA XM_372751 Homo sapiens similar to zinc finger protein 495 (LOC390977), mRNA XM_372753 Homo sapiens similar to Zinc finger protein 264 (LOC390980), mRNA XM_372754 Homo sapiens similar to 5830417C01 Rik protein (LOC390981), mRNA XM_372755 Homo sapiens similar to RIKEN cDNA B230396O12 (LOC390988), mRNA XM_372757 Homo sapiens similar to helix-loop-helix transcription factor (LOC390992), m XM_372758 Homo sapiens similar to Solute carrier family 2, facilitated glucose transporte XM_372759 Homo sapiens similar to 60S ribosomal protein L10 (QM protein) (Tumor sup XM_372760 Homo sapiens similar to Hypothetical protein DJ1198H6.2 (LOC390999), mF XM_372761 Homo sapiens similar to Hypothetical protein DJ845024.2 (LOC391001), mF XM_372762 Homo sapiens similar to Hypothetical protein DJ1198H6.2 (LOC391002), mF XM_372763 Homo sapiens similarto hypothetical protein (LOC391003), mRNA XM_372764 Homo sapiens similar to Hypothetical protein DJ845024.2 (LOC391004), mF XM_372765 Homo sapiens similar to KIAA1026 protein (LOC391005), mRNA XM_372767 Homo sapiens similar to Peptidyl arginine deiminase, egg and embryo abunc XM_372768 Homo sapiens similar to Putative dynein light chain protein DJ8B22.1 (LOC3 XM_372769 Homo sapiens similar to Group I IC secretory phospholipase A2 precursor (PI XM_372770 Homo sapiens similar to hypothetical protein AE2 (LOC391015), mRNA XM_372771 Homo sapiens similar to secreted acid phosphatase 2 (SAP2) (LOC391021), XM_372773 Homo sapiens similar to ribosomal protein L18a; 60S ribosomal protein L18a XM_372774 Homo sapiens hypothetical protein DJ159A19.3 (DJ159A19.3), mRNA XM_372775 Homo sapiens similar to protein tyrosine phosphatase, receptor type, U isofo XM_372776 Homo sapiens similar to chromosome 14 open reading frame 138 (LOC3910 XM_372777 Homo sapiens similar to forkhead box protein 06 (LOC391030), mRNA XM_372778 Homo sapiens similar to 40S ribosomal protein S15a (LOC391035), mRNA XM_372779 Homo sapiens similar to RIKEN cDNA 5730434I03 gene (LOC391040), mRf* XM_372780 Homo sapiens similar to Solute carrier family 2, facilitated glucose transporte XM_372781 Homo sapiens similar to NAD-dependent deacetylase sirtuin 5 (SIR2-like pro XM 372784 Homo sapiens similar to Stromal cell derived factor receptor 2 (LOC391059),
XM_372785 Homo sapiens simi ar to peptidylprolyl isomerase A (cyclophilin A) (LOC391C XM_372788 Homo sapiens simi ar to SNAGI (LOC391086), mRNA XM_372789 Homo sapiens simi ar to hypothetical protein MGC8902 (LOC391088), mRN/ XM_372792 Homo sapiens simi ar to Hypothetical protein DJ845024.1 (LOC391092), mF XM_372795 Homo sapiens simil ar to histone protein Hist2h3c1 (LOC391097), mRNA XM_372796 Homo sapiens simi arto Olfactory receptor 10K1 (LOC391107), mRNA XM_372797 Homo sapiens simi ar to Olfactory receptor 10K1 (LOC391109), mRNA XM_372798 Homo sapiens simi ar to seven transmembrane helix receptor (LOC391110), XM_372799 Homo sapiens simi ar to Olfactory receptor 6Y1 (LOC391112), mRNA XM_372800 Homo sapiens simil ar to Olfactory receptor 6K3 (LOC391114), mRNA XM_372801 Homo sapiens simi ar to porcine serum amyloid P component (SAP) (LOC39 XM_372802 Homo sapiens simi ar to hypothetical protein A030011 M19 (LOC391123), mF XM_372803 Homo sapiens simi ar to ribosomal protein S17 (LOC391130), mRNA XM_372804 Homo sapiens simi ar to hypothetical protein BC015183 (LOC391133), mRN, XM_372805 Homo sapiens simi ar to dJ612B18.1 (similar to 40S ribosomal protein) (LOC XM_372806 Homo sapiens simi ar to Multisynthetase complex auxiliary component p43 (I XM_372807 Homo sapiens simi ar to Rho-GTPase-activating protein 8 (LOC391144), mR XM_372809 Homo sapiens LOC391156 (LOC391156), mRNA XM_372811 Homo sapiens similar to ribosomal protein L31 (LOC391161), mRNA XM_372812 Homo sapiens similar to Glyceraldehyde 3-phosphate dehydrogenase, liver ( XM_372813 Homo sapiens similar to Voltage-dependent anion-selective channel protein XM_372815 Homo sapiens similar to 40S ribosomal protein S26 (LOC391165), mRNA XM_372816 Homo sapiens similar to hypothetical protein D11 Ertd497e (LOC391169), mF XM_372817 Homo sapiens similar to hypothetical protein (LOC391170), mRNA XM_372818 Homo sapiens similar to kinesin-like protein (103.5 kD) (klp-6) (LOC391185), XM_372820 Homo sapiens similar to Olfactory receptor 11 L1 (LOC391189), mRNA XM_372821 Homo sapiens similar to Olfactory receptor 2L8 (LOC391190), mRNA XM_372822 Homo sapiens similar to Olfactory receptor 2AK2 (LOC391191), mRNA XM_372823 Homo sapiens olfactory receptor, family 2, subfamily L, member 2 (OR2L2), i XM_372824 Homo sapiens simi. lar to seven transmembrane helix receptor (LOC391192), XM_372825 Homo sapiens simil ar to Olfactory receptor 2M6 (LOC391194), mRNA XM_372826 Homo sapiens simi ar to seven transmembrane helix receptor (LOC391195), XM_372827 Homo sapiens simi ar to Olfactory receptor 2M7 (LOC391196), mRNA XM_372833 Homo sapiens simi ar to Hypothetical protein FLJ10709 (LOC391201), mRN; XM_372834 Homo sapiens simi ar to Tissue alpha-L-fucosidase precursor (Alpha-L-fucos XM_372838 Homo sapiens simil ar to RIKEN cDNA B230396O12 (LOC391205), mRNA XM_372840 Homo sapiens simil ar to ribosomal protein L36; 60S ribosomal protein L36 (L XM_372841 Homo sapiens simi ar to Olfactory receptor 2T3 (LOC391210), mRNA XM_372842 Homo sapiens simil ar to Olfactory receptor 2G3 (LOC391211 ), mRNA XM_372843 Homo sapiens simi ar to Olfactory receptor 2T5 (LOC391212), mRNA XM_372845 Homo sapiens simil ar to seven transmembrane helix receptor (LOC391214), XM_372858 Homo sapiens simi ar to scratch; scratch 1 (LOC391226), mRNA XM_372859 Homo sapiens simi ar to Homeobox protein Nkx-2.4 (Homeobox protein NKX XM_372863 Homo sapiens simi ar to nodulin (LOC391240), mRNA XM_372864 Homo sapiens simil ar to Soggy-1 protein precursor (SGY-1) (LOC391241), π XM_372866 Homo sapiens simi ar to LPIN3 (LOC391248), mRNA XM_372869 Homo sapiens simi ar to dJ601O1.1 (novel protein with Kunitz/Bovine pancre XM_372870 Homo sapiens simi ar to Ubiquitin-like protein SMT3C precursor (Ubiquitin-hc XM_372871 Homo sapiens simi ar to bA164D18.1 (novel protein similar to KIAA0233) (LC XM_372873 Homo sapiens simi ar to putative glycine-rich protein (LOC391261), mRNA XM_372874 Homo sapiens simi ar to salivary proline-rich protein (LOC391262), mRNA XM_372875 Homo sapiens simi ar to KIAA0685 protein (LOC391268), mRNA XM_372876 Homo sapiens simi ar to hypothetical protein DKFZp434A171 (LOC391269), XM_372878 Homo sapiens simi lar to 60S ribosomal protein L23a (LOC391282), mRNA XM_372879 Homo sapiens chromosome 21 open reading frame 57 (C21orf57), mRNA XM_372880 Homo sapiens LOC391284 (LOC391284), mRNA XM 372882 Homo sapiens hypothetical protein LOC128954 (LOC128954), mRNA
XM_372884 Homo sapiens cat eye syndrome chromosome region, candidate 2 (CECR2).
XM_372887 Homo sapiens similar to minus agglutinin ( OC391296), mRNA
XM_372889 Homo sapiens similar to KIAA0649 protein (LOC391298), mRNA
XM_372890 Homo sapiens LOC391299 (LOC391299), mRNA
XM_372891 Homo sapiens LOC391303 (LOC391303), mRNA
XM_372892 Homo sapiens similar to KIAA0649 protein (LOC391304), mRNA
XM_372893 Homo sapiens similar to sushi domain containing 2; Sushi domain (SCR rep«
XM_372894 Homo sapiens similar to Hypothetical protein DJ1198H6.2 (LOC391306), mF
XM_372900 Homo sapiens similar to D-dopachrome tautomerase (Phenylpyruvate tauton
XM_372901 Homo sapiens similar to turn- transplantation antigen P198 (LOC391328), ml
XM_372905 Homo sapiens similar to KIAA0563 gene product (LOC391335), mRNA
XM_372907 Homo sapiens similar to erythrocyte membrane-associated giant protein antii
XM_372908 Homo sapiens similar to vitelline envelope sperm lysin receptor (LOC391340
XM_372910 Homo sapiens similar to Hypothetical protei n CBG24508 (LOC391342), mR
XM_372911 Homo sapiens similar to CG2839-PA (L0C391343), mRNA
XM_372913 Homo sapiens similar to Mucin 2 precursor (Intestinal mucin 2) (LOC391347
XM_372914 Homo sapiens similar to MGC52970 protein (LOC391348), mRNA
XM_372916 Homo sapiens similar to peptidyl-Pro cis trans isomerase (LOC391352), mRI
XM_372917 Homo sapiens similar to Cerebellar-degene ration-related antigen 1 (CDR34)
XM_372918 Homo sapiens similar to Mothers against decapentaplegic homolog interactir
XM_372919 Homo sapiens similar to ENSANGP0000O018456 (LOC391356), mRNA
XM_372920 Homo sapiens similar to Ubiquinol-cytochrome C reductase complex 11 kDa
XM_372921 Homo sapiens similar to hypothetical protein HSPC152 (LOC391358), mRN/
XM_372922 Homo sapiens similarto Epithelial membrane protein-2 (EMP-2) (XMP protei
XM_372923 Homo sapiens similar to class II basic helix-loop-helix protein TCF23 (LOC3S
XM_372924 Homo sapiens similar to human alpha-catenin (LOC391362), mRNA
XM_372926 Homo sapiens similar to ribosomal protein S12 (LOC391370), mRNA
XM_372927 Homo sapiens tetratricopeptide repeat domain 7 (TTC7), mRNA
XM_372928 Homo sapiens similar to C-terminal binding protein 2 isoform 2; ribeye (LOCc
XM_372933 Homo sapiens similar to immunoglobulin kappa light chain variable region O'
XM_372940 Homo sapiens similar to Adrenoleukodystrophy protein (ALDP) (LOC391403
XM_372941 Homo sapiens similar to Ig kappa chain V region (Z4) - human (LOC391405)
XM_372942 Homo sapiens similar to immunoglobulin anti-gp96/grp94 variable region of t
XM_372945 Homo sapiens similar to phenol sulfotransferase (LOC391418), mRNA
XM_372946 Homo sapiens similar to sulfotransferase 1 C (LOC391419), mRNA
XM_372947 Homo sapiens similar to Ribosome biogenesis protein BMS1 homolog (LOC,
XM_372950 Homo sapiens similar to ENSANGP00000OO4103 (LOC391426), mRNA
XM_372952 Homo sapiens similar to Ig kappa chain precursor V region (orphon V108) - \
XM_372953 Homo sapiens similarto Seienide.water dikinase 1 (Selenophosphate synthe
XM_372954 Homo sapiens similar to hypothetical protein (LOC391429), mRNA
XM_372957 Homo sapiens similar to FKSG30 (L0C391 42), mRNA
XM_372958 Homo sapiens similar to PRED65 (L0C391 445), mRNA
XM_372959 Homo sapiens similar to rRNA intron-encoded homing endonuclease (LOC3J
XM_372963 Homo sapiens similar to Kinesin heavy chain isoform 5C (Kinesin heavy chai
XM_372964 Homo sapiens similar to ATP synthase alpha chain, mitochondrial precursor
XM_372965 Homo sapiens similar to KIAA0181 (LOC391456), mRNA
XM_372966 Homo sapiens similar to protein 40kD (LOC391462), mRNA
XM_372967 Homo sapiens similar to diml ; diml (S. pombe) (LOC391466), mRNA
XM_372969 Homo sapiens similar to DYStrophin related (417.4 kD) (dys-1 ) (LOC391475]
XM_372970 Homo sapiens similar to Alpha-endosulfine (LOC391481 ), mRNA
XM_372973 Homo sapiens similar to guanidinoacetate methyltransferase; GAMT (LOC39
XM_372974 Homo sapiens similar to UDP-glucuronosyltransferase 1-5 precursor, micros*
XM_372975 Homo sapiens similar to TAR DNA-binding protein-43 (TDP-43) (LOC391494
XM_372977 Homo sapiens similar to olfactory receptor MOR208-2 (LOC391496), mRNA
XM_372978 Homo sapiens similar to ENSANGP00000OO7346 (LOC391497), mRNA
XM_372979 Homo sapiens similar to Involucrin (LOC391498), mRNA
XM_372980 Homo sapiens similar to EST gb|ATTS113G comes from this gene. (LOC391-
XM_372981 Homo sapiens similar to Hypothetical protein CBG20540 (LOC391501 ), mR
XM_372982 Homo sapiens similar to Gamma-2-syntrophin (G2SYN) (Syntrophin 5) (SYN
XM_372983 Homo sapiens syntrophin, gamma 2 (SNTG2), mRNA
XM_372984 Homo sapiens similar to Drosophila melanogaster CG8797 gene product-rel;
XM_372985 Homo sapiens similar to CHCHD4 protein (LOC391510), mRNA
XM_372986 Homo sapiens similar to WD repeat domain 10 isoform 3 (LOC391512), mRf
XM_372987 Homo sapiens similar to ENSANGP00000014786 (LOC391513), mRNA
XM_372988 Homo sapiens similar to peptidyl-Pro cis trans isomerase (LOC391532), mRI
XM_372991 Homo sapiens similar to coated vesicle membrane protein (LOC391540), mF
XM_372992 Homo sapiens similar to hypothetical protein FLJ23834 (LOC391542), mRN/
XM_372993 Homo sapiens similar to MGC39820 protein (LOC391544), mRNA
XM_372995 Homo sapiens similar to Msx2-interacting protein (SMART/HDAC1 associate
XM_372996 Homo sapiens similar to hypothetical protein DKFZp434L0850 (LOC391555)
XM_372997 Homo sapiens olfactory receptor, family 5, subfamily K, member 1 (OR5K1 ),
XM_372998 Homo sapiens similar to MLRQ subunit of the NADH ubiquinone oxidoreduct
XM_373001 Homo sapiens similar to Histone H2B.n (H2B/n) (H2B.2) (LOC391566), mRN
XM_373002 Homo sapiens similar to Succinate dehydrogenase [ubiquinone] iron-sulfur p
XM_373003 Homo sapiens similar to peptidyl-Pro cis trans isomerase (LOC391587), mRI
XM_373004 Homo sapiens similar to KIAA1552 protein (LOC391594), mRNA
XM_373005 Homo sapiens similar to alpha-1 platein precursor (LOC391601), mRNA
XM_373006 Homo sapiens similar to Probable ATP-dependent helicase DHX34 (DEAH-b
XM_373007 Homo sapiens LOC391610 (LOC391610), mRNA
XM_373009 Homo sapiens HPX-153 homeobox (HSPX153), mRNA
XM_373016 Homo sapiens similar to deubiquitinating enzyme 3 (LOC391622), mRNA
XM_373021 Homo sapiens similar to deubiquitinating enzyme 3 (LOC391628), mRNA
XM_373022 Homo sapiens similar to seven transmembrane helix receptor (LOC391630),
XM_373025 Homo sapiens similar to SNAG1 (LOC391652), mRNA
XM_373026 Homo sapiens similar to Hypothetical protein MGC66426 (LOC391654), mRf
XM_373027 Homo sapiens similar to 40S ribosomal protein S15a (LOC391656), mRNA
XM_373028 Homo sapiens similar to UDP-glucuronosyltransferase (LOC391661 ), mRNA
XM_373029 Homo sapiens similar to Synaptic glycoprotein SC2 (LOC391676), mRNA
XM_373030 Homo sapiens hypothetical protein LOC285556 (LOC285556), mRNA
XM_373031 Homo sapiens similar to TUBULIN BETA CHAIN (LOC391692), mRNA
XM_373033 Homo sapiens similar to ribosomal protein S23 (LOC391701 ), mRNA
XM_373035 Homo sapiens similar to RIKEN cDNA 9930021 J17 (LOC391705), mRNA
XM_373036 Homo sapiens similar to Chromatin accessibility complex protein 1 (CHRAC-
XM_373037 Homo sapiens similar to ring finger protein 129 (LOC391711), mRNA
XM_373038 Homo sapiens similar to ring finger protein 129 (LOC391712), mRNA
XM_373039 Homo sapiens similar to ring finger protein 129 (LOC391714), mRNA
XM_373040 Homo sapiens similar to RING finger protein 15 (Zinc finger protein RoRet) ("
XM_373041 Homo sapiens similar to Claudin-22 (LOC391721 ), mRNA
XM_373042 Homo sapiens similar to myosin:SUBUNIT=regulatory light chain (LOC39172
XM_373043 Homo sapiens similar to Heslike (LOC391723), mRNA
XM_373053 Homo sapiens similar to Microneme antigen (LOC391733), mRNA
XM_373056 Homo sapiens similar to Transcription initiation factor TFIID 28 kDa subunit (
XM_373057 Homo sapiens similar to Transcription initiation factor TFIID 28 kDa subunit (
XM_373058 Homo sapiens similar to Transcription initiation factor TFIID 28 kDa subunit (
XM_373059 Homo sapiens similar to Transcription initiation factor TFIID 28 kDa subunit (
XM_373061 Homo sapiens similar to Transcription initiation factor TFIID 28 kDa subunit (
XM_373073 Homo sapiens similar to Transcription initiation factor TFIID 28 kDa subunit (
XM_373075 Homo sapiens similar to Transcription initiation factor TFIID 28 kDa subunit (
XM_373076 Homo sapiens similar to TAF11 RNA polymerase II, TATA box binding protei
XM_373077 Homo sapiens similar to Transcription initiation factor TFIID 28 kDa subunit (
XM_373078 Homo sapiens similar to Transcription initiation factor TFIID 28 kDa subunit (
XM_373079 Homo sapiens similar to histone (15.4 kD) (his-72) (LOC391769), mRNA
XM_373080 Homo sapiens similar to ENSANGP00000014197 (LOC391770), mRNA
XM_373082 Homo sapiens similar to CG12279-PA (LOC391784), mRNA
XM_373084 Homo sapiens similar to ribosomal protein L31 (LOC391789), mRNA
XM_373085 Homo sapiens similar to cadherin 12, type 2 preproprotein; Br-cadherin; cadt
XM_373087 Homo sapiens similar to cadherin 12, type 2 preproprotein; Br-cadherin; cadt
XM_373088 Homo sapiens similar to cadherin 12, type 2 preproprotein; Br-cadherin; cadt
XM_373089 Homo sapiens similar to hypothetical protein FLJ10891 (LOC391796), mRN/
XM_373090 Homo sapiens LOC391797 (LOC391797), mRNA
XM_373091 Homo sapiens similar to 60S ribosomal protein L10 (QM protein homolog) (L
XM_373092 Homo sapiens similar to peptidyl-Pro cis trans isomerase (LOC391807), mRI
XM_373093 Homo sapiens similar to 28 kDa heat- and acid-stable phosphoprotein (PDGI
XM_373094 Homo sapiens similar to protease (prosome, macropain) 26S subunit, ATPas
XM_373096 Homo sapiens similar to 60S RIBOSOMAL PROTEIN L21 (LOC391823), mR
XM_373097 Homo sapiens similar to Thyroid hormone receptor-associated protein compl
XM_373098 Homo sapiens similar to beta-tropomyosin (LOC391844), mRNA
XM_373099 Homo sapiens similar to hypothetical protein (LOC391847), mRNA
XM_373100 Homo sapiens similar to 60S ribosomal protein L10 (QM protein homolog) (L
XM_373101 Homo sapiens similar to neuralized homolog (LOC391849), mRNA
XM_373103 Homo sapiens similar to Beta-1 ,4-galactosyltransferase 7 (Beta-1 ,4-GalTase
XM_373106 Homo sapiens similar to Tb-291 membrane associated protein (LOC391859)
XM_373107 Homo sapiens similar to cAMP-regulated phosphoprotein 19 (ARPP-19) (LOi
XM_373109 Homo sapiens hypothetical protein L0C34O156 (LOC340156), mRNA
XM_373156 Homo sapiens hypothetical protein MGC26484 (MGC26484), mRNA
XM_373157 Homo sapiens similar to Phosphorylase B kinase gamma catalytic chain, ske
XM_373170 Homo sapiens hypothetical protein LOC54103 (LOC54103), mRNA
XM_373171 Homo sapiens similar to dynein, cytoplasmic, light peptide; 8kD LC; dynein L
XM_373178 Homo sapiens similar to Zn-alpha-2-glycoprotein precursor (LOC392082), ml
XM_373180 Homo sapiens similar to Cohesin subunit SA-3 (Stromal antigen 3) (SCC3 he
XM_373190 Homo sapiens similar to dJ753D5.2 (novel protein similar to RPS17 (40S ribc
XM_373209 Homo sapiens similar to seven transmembrane helix receptor (LOC392133),
XM_373210 Homo sapiens similar to olfactory receptor OR261-1 (LOC392138), mRNA
XM_373214 Homo sapiens similar to Mtr3 (mRNA transport regulator 3)-homolog; Mtr3 (n
XM_373219 Homo sapiens similar to gastrulation brain homeobox 1 (LOC392152), mRN/
XM_373223 Homo sapiens similar to polycystic kidney disease 1-like 3 (LOC392159), mF
XM_373224 Homo sapiens similar to cAMP-dependent protein kinase type l-beta regulat
XM_373227 Homo sapiens similar to Olfactory receptor 4F3 (LOC392167), mRNA
XM_373233 Homo sapiens similar to 60S ribosomal protein L10 (QM protein) (Tumor sup
XM_373234 Homo sapiens similar to demidefensin 3 (LOC392182), mRNA
XM_373236 Homo sapiens similar to chromosome 11 open reading frame2; chromosome
XM_373238 Homo sapiens similar to deubiquitinating enzyme 3 (LOC392188), mRNA
XM_373239 Homo sapiens similar to ubiquitin-specific protease 17-like protein (LOC3921
XM_373241 Homo sapiens similar to beta-1 ,4-mannosyltransferase; beta-1 ,4 mannosyltr;
XM_373242 Homo sapiens similar to 60S ribosomal protein L10 (QM protein homolog) (L
XM_373243 Homo sapiens similar to deubiquitinating enzyme 3 (LOC392197), mRNA
XM_373245 Homo sapiens similar to 60S ribosomal protein L32 (LOC392202), mRNA
XM_373246 Homo sapiens similar to Ac2-210 (LOC392208), mRNA
XM_373248 Homo sapiens similar to VENT-like homeobox 2; hemopoietic progenitor hon
XM_373249 Homo sapiens similar to Ig lambda light chain leader and V-region (LOC392.
XM_373250 Homo sapiens similar to Band 4.1-like protein 5 (LOC392218), mRNA
XM_373252 Homo sapiens similar to coiled-coil-helix-coiled-coil-helix domain containing !
XM_373253 Homo sapiens similar to RIKEN cDNA 0610038D11 (LOC392222), mRNA
XM_373255 Homo sapiens similar to hypothetical class II basic helix-loop-helix protein (U
XM_373257 Homo sapiens similar to large subunit ribosomal protein L36a (LOC392248),
XM_373259 Homo sapiens similar to glyceraldehyde-3-phosphate dehydrogenase (LOC3
XM_373260 Homo sapiens similar to growth differentiation factor 16 (LOC392255), mRN/
XM_373263 Homo sapiens similar to laminin-binding protein (LOC392262), mRNA
XM_373265 Homo sapiens similar to cDNA sequence BC024139 (LOC392274), mRNA
XM_373266 Homo sapiens similar to sphingomyelin phosphodiesterase 3, neutral membr
XM_373270 Homo sapiens similar to Rab coupling protein; Rab-interacting recycling proti
XM_373275 Homo sapiens milar to peptidyl-Pro cis trans isomerase (LOC392285), mRI XM_373276 Homo sapiens milar to chromosome 15 open reading frame 12; mitochondi XM_373277 Homo sapiens milar to microtubule-associated proteins 1A/1B light chain 3 XM_373281 Homo sapiens milar to Chloride intracellular channel protein 4 (Intracellular XM_373283 Homo sapiens milar to Ferritin light chain (Ferritin L subunit) (LOC392299), XM_373284 Homo sapiens milar to High mobility group protein 4-like (HMG-4L) (LOC39 XM_373287 Homo sapiens milar to RIKEN CDNA 4930412F15 gene (LOC392307), mRI XM_373288 Homo sapiens milar to Olfactory receptor 70 (LOC392308), mRNA XM_373289 Homo sapiens milar to Olfactory receptor 13J1 (LOC392309), mRNA XM_373290 Homo sapiens milarto high mobility group protein homolog HMG4 (LOC39; XM_373295 Homo sapiens milar to hypothetical protein MGC17986 (LOC392332), mRf XM_373297 Homo sapiens LOC392339 (LOC392339), mRNA XM_373298 Homo sapiens milar to adenylate kinase 3 alpha like (LOC392347), mRNA XM_373300 Homo sapiens milarto tMDC I (LOC392351), mRNA XM_373301 Homo sapiens milar to peptidyl-Pro cis trans isomerase (LOC392352), mRI XM_373302 Homo sapiens milar to ORF2 (LOC392355), mRNA XM_373303 Homo sapiens milar to Eukaryotic translation initiation factor 3 subunit 1 (el XM_373304 Homo sapiens milar to Cathepsin L precursor (Major excreted protein) (MEI XM_373305 Homo sapiens milar to chromosome 15 open reading frame 2 (LOC392364 XM_373308 Homo sapiens milar to Olfactory receptor 1 3C2 (LOC392376), mRNA XM_373309 Homo sapiens milarto constitutive photomorphogenic protein (LOC392379 XM_373310 Homo sapiens milar to large subunit ribosomal protein L36a (LOC392381), XM_373311 Homo sapiens milar to ribosomal protein L31 (LOC392382), mRNA XM_373312 Homo sapiens milar to 60S ribosomal protein L32 (LOC392384), mRNA XM_373313 Homo sapiens olfactory receptor, family 1 , subfamily J, member 5 (OR1 J5), r XM_373314 Homo sapiens similar to G protein-coupled receptor homolog clone H8 (LOC XM_373315 Homo sapiens similar to Olfactory receptor 1 L6 (LOC392390), mRNA XM_373316 Homo sapiens similar to Olfactory receptor 5C1 (Olfactory receptor 9-F) (OR! XM_373317 Homo sapiens similar to Olfactory receptor 1 K1 (LOC392392), mRNA XM_373319 Homo sapiens similar to hemicentin (LOC392395), mRNA XM_373320 Homo sapiens similar to Von Ebners gland protein precursor (VEG protein) (' XM_373322 Homo sapiens similarto Putative MUP-like lipocalin precursor (LOC392399), XM_373324 Homo sapiens similar to Ankyrin repeat domain protein 5 (LOC392404), mRr XM_373325 Homo sapiens similar to Pim1 (LOC392405), mRNA XM_373326 Homo sapiens similar to hypothetical protein (LOC392406), mRNA XM_373327 Homo sapiens similar to Glutamate [NMDA] receptor subunit zeta 1 precurso XM_373328 Homo sapiens similar to hypothetical protein FLJ20433 (LOC392409), mRN/ XM_373329 Homo sapiens similar to NADPH-dependent FMN and FAD containing oxidor XM_373331 Homo sapiens similarto paranemin (LOC392411), mRNA XM_373334 Homo sapiens similar to MGC43306 protein (LOC392413), mRNA XM_373335 Homo sapiens similar to Phenylalanine-4-hydroxylase (PAH) (Phe-4-monoox XM_373336 Homo sapiens similar to 1060P11.3 (killer inhibitory receptor 2-2-2 (KIR222) XM_373337 Homo sapiens similar to protein kinase C, zeta (LOC392420), mRNA XM_373338 Homo sapiens similar to bA92K2.2 (similar to ubiquitin) (LOC392425), mRN/ XM_373339 Homo sapiens similar to Nucleolar phosphoprotein p130 (Nucleolar 130 kDa XM_373340 Homo sapiens similarto melanoma antigen, family B, 6 (LOC392433), mRN/ XM_373341 Homo sapiens similar to PR264/SC35 (LOC392439), mRNA XM_373343 Homo sapiens similar to 60S ribosomal protein L32 (LOC392447), mRNA XM_373344 Homo sapiens similar to CpG binding protein (Protein containing PHD finger XM_373345 Homo sapiens similar to synovial sarcoma, X breakpoint 6 (LOC392462), mF XM_373347 Homo sapiens similar to RIKEN cDNA 2010001 H14 (LOC392465), mRNA XM_373348 Homo sapiens similar to melanoma antigen (LOC392466), mRNA XM_373349 Homo sapiens similar to melanoma antigen, family B, 4; melanoma-associati XM_373350 Homo sapiens similar to nuclear protein p30 (LOC392468), mRNA XM_373351 Homo sapiens similar to dJ834A16.1 (similar to PGAM) (LOC392473), mRN/ XM_373352 Homo sapiens similar to XAGE-5 protein (LOC392475), mRNA XM 373353 Homo sapiens similar to Probable G protein-coupled receptor GPR83 precur
XM_373354 Homo sapiens similar to ribosomal protein L31 (LOC392487), mRNA
XM_373355 Homo sapiens similar to Ras homolog gene family, member G (LOC392489)
XM_373356 Homo sapiens similar to Translationally controlled tumor protein (TCTP) (p23
XM_373357 Homo sapiens LOC392491 (LOC392491 ), mRNA
XM_373358 Homo sapiens similar to hypothetical protein DKFZp761 H079 isoform 1 (LOC
XM_373359 Homo sapiens similar to histone H2B-related protein (LOC392512), mRNA
XM_373362 Homo sapiens similarto dJ820B18.1 (similar to nuclear cap binding protein)
XM_373365 Homo sapiens similar to Serine/threonine-protein kinase PRP4 homolog (PR
XM_373366 Homo sapiens similar to beta-tubulin 1 (LOC392528), mRNA
XM_373367 Homo sapiens similar to RIKEN cDNA 2900070E19 (LOC392531 ), mRNA
XM_373368 Homo sapiens similar to Histone H3.3 (LOC392533), mRNA
XM_373369 Homo sapiens similar to Heat shock cognate 71 kDa protein (LOC392535), r
XM_373370 Homo sapiens similar to TJP4 protein (LOC392539), mRNA
XM_373372 Homo sapiens similar to hypothetical protein FLJ20527 (LOC392546), mRN/
XM_373373 Homo sapiens similar to Glyceraldehyde 3-phosphate dehydrogenase, liver (
XM_373374 Homo sapiens similar to Ras-related protein Rab-28 (Rab-26) (LOC392551 ),
XM_373378 Homo sapiens similar to hypothetical protein MGC15827 (LOC392559), mRf>
XM_373381 Homo sapiens H2A histone family, member B (H2AFB), mRNA
XM_373382 Homo sapiens similar to neurofilament-like protein (LOC392563), mRNA
XM_373384 Homo sapiens similar to testis expressed sequence 13A (LOC392566), mRN
XM_373388 Homo sapiens similar to Menkes Disease (ATP7A) (LOC392570), mRNA
XM_373391 Homo sapiens similar to Zinc finger protein ZFD25 (LOC392576), mRNA
XM_373395 Homo sapiens similar to testis specific protein, Y-linked (LOC392582), mRN/
XM_373397 Homo sapiens similar to testis specific protein, Y-linked (LOC392584), mRN/
XM_373398 Homo sapiens similar to Testis-Specific Protein Y (TSPY) (LOC392586), mR
XM_373399 Homo sapiens similar to Sedlin (Trafficking protein particle complex protein 2
XM_373407 Homo sapiens nuclear receptor subfamily 2, group F, member 6 (NR2F6), m
XM_373413 Homo sapiens hypothetical protein FLJ20847 (FLJ20847), mRNA
XM_373419 Homo sapiens valyl-tRNA synthetase 2-like (VARS2L), mRNA
XM_373431 Homo sapiens hypothetical protein BCO04360 (LOC87769), mRNA
XM_373433 Homo sapiens hypothetical protein BCO02926 (LOC90379), mRNA
XM_373440 Homo sapiens chondroitin sulfate synthase 3 (CSS3), mRNA
XM_373444 Homo sapiens LOC399709 (LOC387630), mRNA
XM_373445 Homo sapiens LOC399710 (LOC387631 ), mRNA
XM_373446 Homo sapiens LOC399711 (LOC387632), mRNA
XM_373447 Homo sapiens LOC399714 (LOC387633), mRNA
XM_373448 Homo sapiens LOC399722 (LOC387636), mRNA
XM_373449 Homo sapiens LOC399724 (LOC387638), mRNA
XM_373450 Homo sapiens LOC399725 (LOC387639), mRNA
XM_373451 Homo sapiens hypothetical gene supported by AK056080; AL832706; BC01 <
XM_373452 Homo sapiens LOC399735 (LOC387649), mRNA
XM_373453 Homo sapiens LOC399741 (LOC387654), mRNA
XM_373454 Homo sapiens LOC399743 (LOC387656), mRNA
XM_373456 Homo sapiens hypothetical gene supported by BX538120 (LOC387662), mR
XM_373460 Homo sapiens LOC399764 (LOC387675), mRNA
XM_373461 Homo sapiens LOC399775 (LOC387683), mRNA
XM_373463 Homo sapiens LOC399779 (LOC387686), mRNA
XM_373464 Homo sapiens LOC399781 (LOC387687), mRNA
XM_373465 Homo sapiens LOC399784 (LOC387689), mRNA
XM_373466 Homo sapiens LOC399787 (LOC387690), mRNA
XM_373468 Homo sapiens LOC399794 (LOC387697), mRNA
XM_373469 Homo sapiens LOC399797 (LOC387701 ), mRNA
XM_373470 Homo sapiens LOC399801 (LOC387705), mRNA
XM_373471 Homo sapiens LOC399802 (LOC387706), mRNA
XM_373472 Homo sapiens LOC399805 (LOC387709), mRNA
XM_373474 Homo sapiens LOC399808 (LOC387710), mRNA
XM_373475 Homo sapiens LOC399809 (LOC38771 1 ), mRNA
XM_373477 Homo sapiens LOC399813 (LOC387715), mRNA
XM_373478 Homo sapiens LOC399817 (LOC387717), mRNA
XM_373479 Homo sapiens LOC399825 (LOC387722), mRNA
XM_373480 Homo sapiens LOC399828 (LOC387724), mRNA
XM_373481 Homo sapiens hypothetical gene supported by BC027847; BC047942 (LOCό
XM_373483 Homo sapiens LOC399847 (LOC387736), mRNA
XM_373484 Homo sapiens LOC399848 (LOC387737), mRNA
XM_373485 Homo sapiens LOC399849 (LOC387739), mRNA
XM_373486 Homo sapiens hypothetical gene supported by AK124823 (LOC387741 ), mR
XM_373487 Homo sapiens hypothetical gene supported by AK124823 (LOC387742), mR
XM_373488 Homo sapiens LOC399854 (LOC387743), mRNA
XM_373489 Homo sapiens LOC399856 (LOC387746), mRNA
XM_373490 Homo sapiens LOC399868 (LOC387754), mRNA
XM_373491 Homo sapiens LOC399871 (LOC387756), mRNA
XM_373492 Homo sapiens LOC399874 (LOC387757), mRNA
XM_373494 Homo sapiens LOC399878 (LOC387760), mRNA
XM_373495 Homo sapiens LOC399880 (LOC387761 ), mRNA
XM_373496 Homo sapiens LOC399882 (LOC387762), mRNA
XM_373497 Homo sapiens hypothetical gene supported by BC05256O (LOC387763), mR
XM_373498 Homo sapiens LOC399885 (LOC387764), mRNA
XM_373499 Homo sapiens LOC399887 (LOC387765), mRNA
XM_373500 Homo sapiens LOC399896 (LOC387771 ), mRNA
XM_373502 Homo sapiens LOC399902 (LOC387776), mRNA
XM_373503 Homo sapiens LOC399903 (LOC387777), mRNA
XM_373505 Homo sapiens LOC399910 (LOC387783), mRNA
XM_373506 Homo sapiens LOC399911 (LOC387784), mRNA
XM_373507 Homo sapiens LOC399916 (LOC387785), mRNA
XM_373509 Homo sapiens hypothetical gene supported by BC065746 (LOC387789), mR
XM_373511 Homo sapiens LOC399934 (LOC387796), mRNA
XM_373512 Homo sapiens LOC399935 (LOC387797), mRNA
XM_373513 Homo sapiens LOC399952 (LOC387810), mRNA
XM_373515 Homo sapiens LOC399958 (LOC387814), mRNA
XM_373516 Homo sapiens LOC399966 (LOC387817), mRNA
XM_373517 Homo sapiens LOC399970 (LOC387819), mRNA
XM_373518 Homo sapiens hypothetical gene supported by BC039676 (LOC387821), mR
XM_373519 Homo sapiens LOC399976 (LOC387823), mRNA
XM_373520 Homo sapiens LOC399977 (LOC387824), mRNA
XM_373521 Homo sapiens LOC399989 (LOC387826), mRNA
XM_373522 Homo sapiens hypothetical protein LOC283314 (LOC283314), mRNA
XM_373524 Homo sapiens LOC399997 (LOC387833), mRNA
XM_373525 Homo sapiens LOC400005 (LOC387838), mRNA
XM_373527 Homo sapiens LOC400016 (LOC387846), mRNA
XM_373529 Homo sapiens LOC400017 (LOC387848), mRNA
XM_373530 Homo sapiens LOC400028 (LOC387853), mRNA
XM_373531 Homo sapiens LOC400029 (LOC387854), mRNA
XM_373533 Homo sapiens LOC400034 (LOC387859), mRNA
XM_373534 Homo sapiens LOC400042 (LOC387863), mRNA
XM_373535 Homo sapiens LOC400044 (LOC387864), mRNA
XM_373537 Homo sapiens LOC400049 (LOC387866), mRNA
XM_373538 Homo sapiens LOC400054 (LOC387869), mRNA
XM_373539 Homo sapiens LOC400057 (LOC387871 ), mRNA
XM_373540 Homo sapiens hypothetical gene supported by BC044741 (LOC387872), mR
XM_373541 Homo sapiens LOC400060 (LOC387873), mRNA
XM_373542 Homo sapiens LOC400062 (LOC387875), mRNA
XM_373543 Homo sapiens LOC400063 (LOC387876), mRNA
XM_373544 Homo sapiens hypothetical gene supported by AK056999; BC013920 (LOC3
XM_373545 Homo sapiens LOC400071 (LOC387883), mRNA
XM_373546 Homo sapiens LOC400072 (LOC387884), mRNA
XM_373547 Homo sapiens LOC400074 (LOC387885), mRNA
XM_373548 Homo sapiens LOC400075 (LOC387886), mRNA
XM_373549 Homo sapiens LOC400076 (LOC387887), mRNA
XM_373550 Homo sapiens LOC400078 (LOC387888), mRNA
XM_373553 Homo sapiens hypothetical gene supported by BC040060 (LOC387895), mR
XM_373554 Homo sapiens LOC400089 (LOC387896), mRNA
XM_373555 Homo sapiens LOC400090 (LOC387897), mRNA
XM_373556 Homo sapiens LOC400091 (LOC387898), mRNA
XM_373557 Homo sapiens LOC400100 (LOC387901), mRNA
XM_373558 Homo sapiens LOC400102 (LOC387903), mR A
XM_373559 Homo sapiens LOC400104 (LOC387905), mRNA
XM_373560 Homo sapiens LOC400107 (LOC387910), mRNA
XM_373561 Homo sapiens hypothetical gene supported by AK000246 (LOC387915), mR
XM_373562 Homo sapiens LOC400116 (LOC387917), mRNA
XM_373563 Homo sapiens LOC400117 (LOC387918), mRNA
XM_373564 Homo sapiens LOC400133 (LOC387925), mRNA
XM_373566 Homo sapiens LOC400138 (LOC387931 ), mRNA
XM_373567 Homo sapiens LOC400139 (LOC387932), mRNA
XM_373568 Homo sapiens LOC400143 (LOC387935), mRNA
XM_373569 Homo sapiens LOC400147 (LOC387937), mRNA
XM_373570 Homo sapiens LOC400149 (LOC387939), mRNA
XM_373571 Homo sapiens LOC400150 (LOC387940), mRNA
XM_373572 Homo sapiens LOC400152 (LOC387941), mRNA
XM_373573 Homo sapiens LOC400153 (LOC387942), mRNA
XM_373574 Homo sapiens LOC400155 (LOC387943), mRNA
XM_373575 Homo sapiens LOC400159 (LOC387946), mRNA
XM_373576 Homo sapiens LOC400160 (LOC387947), mRNA
XM_373577 Homo sapiens LOC400162 (LOC387948), mRNA
XM_373578 Homo sapiens LOC400168 (LOC387950), mRNA
XM_373580 Homo sapiens hypothetical protein LOC254028 (LOC254028), mRNA
XM_373581 Homo sapiens LOC400176 (LOC387957), mRNA
XM_373583 Homo sapiens LOC400196 (LOC387974), mRNA
XM_373585 Homo sapiens LOC400198 (LOC387977), mRNA
XM_373587 Homo sapiens LOC400202 (LOC387981), mRNA
XM_373588 Homo sapiens LOC400204 (LOC387983), mRNA
XM_373593 Homo sapiens LOC400215 (LOC387989), mRNA
XM_373594 Homo sapiens LOC400220 (LOC387992), mRNA
XM_373595 Homo sapiens hypothetical gene supported by BC033546 (LOC387993), mR
XM_373596 Homo sapiens LOC400225 (LOC387994), mRNA
XM_373599 Homo sapiens LOC400232 (LOC388001 ), mRNA
XM_373600 Homo sapiens LOC400233 (LOC388002), mRNA
XM_373601 Homo sapiens LOC400235 (LOC388003), mRNA
XM_373602 Homo sapiens LOC400240 (LOC388006), mRNA
XM_373603 Homo sapiens chromosome 14 open reading frame 86 (C14orf86), mRNA
XM_373604 Homo sapiens hypothetical gene supported by BC019017 (LOC388009), mR
XM_373605 Homo sapiens LOC400245 (LOC388010), mRN A
XM_373606 Homo sapiens hypothetical gene supported by AK091668 (LOC388011), mR
XM_373607 Homo sapiens LOC400248 (LOC388014), mRNA
XM_373608 Homo sapiens LOC400251 (LOC388016), mRNA
XM_373609 Homo sapiens LOC400252 (LOC388017), mRNA
XM_373610 Homo sapiens LOC400253 (LOC388018), mRNA
XM_373611 Homo sapiens LOC400254 (LOC388019), mRNA
XM_373615 Homo sapiens similar to HSPC053 (LOC388066), mRNA
XM_373616 Homo sapiens hypothetical gene supported by AK056084; NM_152518 (LOC
XM_373617 Homo sapiens LOC400321 (LOC388081), mRNA
XM_373622 Homo sapiens LOC400330 (LOC388091 ), mRNA
XM_373623 Homo sapiens LOC400337 (LOC388096), mRNA XM_373624 Homo sapiens hypothetical protein LOC283697 (LOC283697), mRNA XM_373625 Homo sapiens hypothetical gene supported by BC035481 (LOC388113), mR XM_373626 Homo sapiens LOC400362 (LOC388114), mRNA XM_373627 Homo sapiens LOC400365 (LOC388117), mRNA XM_373628 Homo sapiens LOC400375 (LOC388123), mRNA XM_373630 Homo sapiens LOC400379 (LOC388126), mRNA XM_373631 Homo sapiens hypothetical gene supported by BC033162 (LOC388129), mR XM_373632 Homo sapiens LOC400384 (LOC388131), mRNA XM_373633 Homo sapiens LOC400390 (LOC388134), mRNA XM_373634 Homo sapiens LOC400394 (LOC388136), mRNA XM_373635 Homo sapiens LOC400401 (LOC388139), mRNA XM_373636 Homo sapiens LOC400402 (LOC388140), mRNA XM_373637 Homo sapiens LOC400404 (LOC388141), mRNA XM_373638 Homo sapiens LOC400407 (LOC388142), mRNA XM_373639 Homo sapiens LOC400412 (LOC388144), mRNA XM_373641 Homo sapiens LOC400425 (LOC388158), mRNA XM_373645 Homo sapiens LOC400440 (LOC388169), mRNA XM_373646 Homo sapiens LOC400441 (LOC388170), mRNA XM_373647 Homo sapiens LOC400443 (LOC388171), mRNA XM_373648 Homo sapiens LOC400449 (LOC388176), mRNA XM_373649 Homo sapiens LOC400457 (LOC388179), mRNA XM_373650 Homo sapiens LOC400458 (LOC388180), mRNA XM_373653 Homo sapiens hypothetical gene supported by BC054509 (LOC388193), mR XM_373654 Homo sapiens hypothetical gene supported by BC028568 (LOC388197), mR XM_373655 Homo sapiens LOC400485 (LOC388198), mRNA XM_373657 Homo sapiens LOC400489 (LOC388201), mRNA XM_373659 Homo sapiens LOC400491 (LOC388204), mRNA XM_373660 Homo sapiens LOC400494 (LOC388206), mRNA XM_373662 Homo sapiens LOC400495 (LOC388208), mRNA XM_373666 Homo sapiens LOC400501 (LOC388214), mRNA XM_373667 Homo sapiens LOC400504 (LOC388216), mRNA XM_373668 Homo sapiens LOC388219 (LOC388219), mRNA XM_373669 Homo sapiens LOC388220 (LOC388220), mRNA XM_373670 Homo sapiens LOC388227 (LOC388227), mRNA XM_373671 Homo sapiens LOC388231 (LOC388231), mRNA XM_373672 Homo sapiens LOC388235 (LOC388235), mRNA XM_373673 Homo sapiens LOC388244 (LOC388244), mRNA XM_373674 Homo sapiens LOC388245 (LOC388245), mRNA XM_373675 Homo sapiens hypothetical protein LOC90835 (LOC90835), mRNA XM_373676 Homo sapiens LOC388251 (LOC388251), mRNA XM_373677 Homo sapiens LOC388260 (LOC388260), mRNA XM_373679 Homo sapiens LOC388267 (LOC388267), mRNA XM_373681 Homo sapiens LOC388269 (LOC388269), mRNA XM_373682 Homo sapiens LOC388270 (LOC388270), mRNA XM_373683 Homo sapiens LOC388273 (LOC388273), mRNA XM_373684 Homo sapiens LOC388274 (LOC388274), mRNA XM_373685 Homo sapiens LOC388276 (LOC388276), mRNA XM_373686 Homo sapiens LOC388277 (LOC388277), mRNA XM_373687 Homo sapiens LOC388278 (LOC388278), mRNA XM_373688 Homo sapiens LOC388279 (LOC388279), mRNA XM_373689 Homo sapiens LOC388281 (LOC388281), mRNA XM_373690 Homo sapiens LOC388282 (LOC388282), mRNA XM_373691 Homo sapiens LOC388283 (LOC388283), mRNA XM_373692 Homo sapiens LOC388291 (LOC388291), mRNA XM_373693 Homo sapiens LOC388295 (LOC388295), mRNA XM 373694 Homo sapiens LOC388296 (LOC388296), mRNA
XM_373695 Homo sapiens LOC388300 (LOC388300), mRNA
XM_373696 Homo sapiens LOC388301 (LOC388301), mRNA
XM_373697 Homo sapiens LOC388302 (LOC388302), mRNA
XM_373698 Homo sapiens LOC388303 (LOC388303), mRNA
XM_373699 Homo sapiens LOC388304 (LOC388304), mRNA
XM_373700 Homo sapiens LOC388305 (LOC388305), mRNA
XM_373701 Homo sapiens LOC388306 (LOC388306), mRNA
XM_373702 Homo sapiens LOC388309 (LOC388309), mRNA
XM_373703 Homo sapiens LOC388311 (LOC388311 ), mRNA
XM_373704 Homo sapiens LOC388312 (LOC388312), mRNA
XM_373705 Homo sapiens LOC388314 (LOC388314), mRNA
XM_373707 Homo sapiens LOC388318 (LOC388318), mRNA
XM_373708 Homo sapiens LOC388319 (LOC388319), mRNA
XM_373709 Homo sapiens LOC388324 (LOC388324), mRNA
XM_373711 Homo sapiens LOC388330 (LOC388330), mRNA
XM_373713 Homo sapiens LOC388334 (LOC388334), mRNA
XM_373714 Homo sapiens LOC388338 (LOC388338), mRNA
XM_373715 Homo sapiens LOC388340 (LOC388340), mRNA
XM_373717 Homo sapiens LOC388347 (LOC388347), mRNA
XM_373718 Homo sapiens LOC388348 (LOC388348), mRNA
XM_373719 Homo sapiens LOC388350 (LOC388350), mRNA
XM_373721 Homo sapiens LOC388358 (LOC388358), mRNA
XM_373722 Homo sapiens LOC388362 (LOC388362), mRNA
XM_373723 Homo sapiens LOC388365 (LOC388365), mRNA
XM_373724 Homo sapiens LOC388366 (LOC388366), mRNA
XM_373726 Homo sapiens LOC388375 (LOC388375), mRNA
XM_373727 Homo sapiens LOC388376 (LOC388376), mRNA
XM_373728 Homo sapiens LOC388380 (LOC388380), mRNA
XM_373730 Homo sapiens LOC388384 (LOC388384), mRNA
XM_373731 Homo sapiens keratin associated protein 4-9 (KRTAP4-9), mRNA
XM_373733 Homo sapiens LOC388387 (LOC388387), mRNA
XM_373734 Homo sapiens LOC388388 (LOC388388), mRNA
XM_373735 Homo sapiens LOC388390 (LOC388390), mRNA
XM_373737 Homo sapiens LOC388397 (LOC388397), mRNA
XM_373740 Homo sapiens LOC388406 (LOC388406), mRNA
XM_373741 Homo sapiens LOC388410 (LOC388410), mRNA
XM_373742 Homo sapiens hypothetical protein MGC40489 (MGC40489), mRNA
XM_373744 Homo sapiens LOC388414 (LOC388414), mRNA
XM_373745 Homo sapiens LOC388415 (LOC388415), mRNA
XM_373746 Homo sapiens LOC388416 (LOC388416), mRNA
XM_373747 Homo sapiens LOC388417 (LOC388417), mRNA
XM_373748 Homo sapiens LOC388418 (LOC388418), mRNA
XM_373749 Homo sapiens LOC388420 (LOC388420), mRNA
XM_373750 Homo sapiens LOC388421 (LOC388421), mRNA
XM_373751 Homo sapiens LOC388422 (LOC388422), mRNA
XM_373753 Homo sapiens LOC388425 (LOC388425), mRNA
XM_373754 Homo sapiens LOC388426 (LOC388426), mRNA
XM_373755 Homo sapiens LOC388427 (LOC388427), mRNA
XM_373756 Homo sapiens LOC388429 (LOC388429), mRNA
XM_373757 Homo sapiens LOC388430 (LOC388430), mRNA
XM_373758 Homo sapiens LOC388431 (LOC388431), mRNA
XM_373760 Homo sapiens hypothetical protein BC009233 (LOC92659), mRNA
XM_373761 Homo sapiens LOC388434 (LOC388434), mRNA
XM_373763 Homo sapiens LOC388436 (LOC388436), mRNA
XM_373765 Homo sapiens LOC388439 (LOC388439), mRNA
XM_373766 Homo sapiens LOC388443 (LOC388443), mRNA
XM_373767 Homo sapiens LOC388445 (LOC388445), mRNA
XM 373769 Homo sapiens LOC388453 (LOC388453) mRNA
XM 373770 Homo sapiens LOC388454 (LOC388454) mRNA
XM 373771 Homo sapiens LOC388456 (LOC388456) mRNA
XM_373772 Homo sapiens LOC388459 (LOC388459) mRNA
XM 373773 Homo sapiens LOC388461 (LOC388461) mRNA
XM_373774 Homo sapiens LOC388465 (LOC388465) mRNA
XM 373775 Homo sapiens LOC388467 (LOC388467) mRNA
XM 373776 Homo sapiens LOC388473 (LOC388473) mRNA
XM_373778 Homo sapiens LOC388477 (LOC388477) mRNA
XM 373779 Homo sapiens LOC388480 (LOC388480) mRNA
XM_373780 Homo sapiens LOC388481 (LOC388481) mRNA
XM 373782 Homo sapiens LOC388483 (LOC388483) mRNA
XM 373783 Homo sapiens LOC388484 (LOC388484) mRNA
XM_373784 Homo sapiens LOC388485 (LOC388485) mRNA
XM 373785 Homo sapiens LOC388487 (LOC388487) mRNA
XM 373786 Homo sapiens LOC388489 (LOC388489) mRNA
XM_373787 Homo sapiens LOC388490 (LOC388490) mRNA
XM 373788 Homo sapiens LOC388494 (LOC388494) mRNA
XM 373789 Homo sapiens LOC388496 (LOC388496) mRNA
XM_373790 Homo sapiens LOC388497 (LOC388497) mRNA
XM_373792 Homo sapiens LOC388500 (LOC388500) mRNA
XM 373793 Homo sapiens LOC388504 (LOC388504) mRNA
XM 373795 Homo sapiens LOC388515 (LOC388515) mRNA
XM_373796 Homo sapiens LOC388516 (LOC388516) mRNA
XM 373797 Homo sapiens LOC388518 (LOC388518) mRNA
XM_373798 Homo sapiens hypothetical gene supported by BC030765 (LOC374890), mR
XM 373799 Homo sapiens LOC388527 (LOC388527) mRNA
XM_373800 Homo sapiens LOC388528 (LOC388528) mRNA
XM_373801 Homo sapiens LOC388529 (LOC388529) mRNA
XM 373802 Homo sapiens LOC388530 (LOC388530) mRNA
XM_373803 Homo sapiens LOC388535 (LOC388535) mRNA
XM_373804 Homo sapiens LOC388539 (LOC388539) mRNA
XM 373805 Homo sapiens LOC388540 (LOC388540) mRNA
XM_373808 Homo sapiens LOC388549 (LOC388549) mRNA
XM 373809 Homo sapiens LOC388553 (LOC388553) mRNA
XM 373810 Homo sapiens hypothetical protein LOC199800 (LOC199800), mRNA
XM_373811 Homo sapiens LOC388563 (LOC388563) mRNA
XM 373812 Homo sapiens LOC388568 (LOC388568) mRNA
XM_373814 Homo sapiens LOC388572 (LOC388572) mRNA
XM 373815 Homo sapiens LOC388573 (LOC388573) mRNA
XM_373817 Homo sapiens LOC388577 (LOC388577) mRNA
XM_373818 Homo sapiens LOC388580 (LOC388580) mRNA
XM_373820 Homo sapiens LOC388584 (LOC388584) mRNA
XM_373821 Homo sapiens LOC388586 (LOC388586) mRNA
XM_373822 Homo sapiens LOC388587 (LOC388587) mRNA
XM 373823 Homo sapiens LOC388588 (LOC388588) mRNA
XM_373824 Homo sapiens LOC388589 (LOC388589) mRNA
XM 373825 Homo sapiens LOC388590 (LOC388590) mRNA
XM_373826 Homo sapiens LOC388593 (LOC388593) mRNA
XM 373827 Homo sapiens LOC388597 (LOC388597) mRNA
XM 373828 Homo sapiens LOC388601 (LOC388601) mRNA
XM_373831 Homo sapiens LOC388604 (LOC388604) mRNA
XM_373832 Homo sapiens LOC388609 (LOC388609) mRNA
XM_373833 Homo sapiens LOC388612 (LOC388612) mRNA
XM 373835 Homo sapiens LOC388614 (LOC388614) mRNA
XM 373836 Homo sapiens LOC388615 (LOC388615) , mRNA
XM 373837 Homo sapiens LOC388616 (LOC388616) , mRNA
XM_373838 Homo sapiens LOC388617 LOC388617 mRNA XM_373839 Homo sapiens LOC388622 LOC388622 mRNA XM_373840 Homo sapiens LOC388623 LOC388623: mRNA XM_373843 Homo sapiens LOC388628 LOC388628 mRNA XM_373844 Homo sapiens LOC388635 LOC388635: mRNA XM_373845 Homo sapiens LOC388638 LOC388638 mRNA XM_373846 Homo sapiens LOC388639 LOC388639 mRNA XM_373847 Homo sapiens hypothetical protein LOC339468 (LOC339468), mRNA XM_373848 Homo sapiens LOC388641 LOC388641 mRNA XM_373849 Homo sapiens LOC388643 LOC388643 mRNA XM_373850 Homo sapiens LOC388644 LOC388644 mRNA XM_373852 Homo sapiens LOC388648 LOC388648: mRNA XM_373853 Homo sapiens LOC388651 LOC388651 mRNA XM_373854 Homo sapiens LOC388652 LOC388652 mRNA XM_373855 Homo sapiens LOC388655 LOC388655 mRNA XM_373856 Homo sapiens LOC388661 LOC388661 mRNA XM_373857 Homo sapiens LOC388663 LOC388663 mRNA XM_373858 Homo sapiens LOC388667 LOC388667 mRNA XM_373859 Homo sapiens LOC388668 LOC388668 mRNA XM_373862 Homo sapiens LOC388679 LOC388679 mRNA XM_373864 Homo sapiens LOC388684 LOC388684 mRNA XM_373865 Homo sapiens LOC388690 LOC388690 mRNA XM_373866 Homo sapiens LOC388692 LOC388692 mRNA XM_373867 Homo sapiens LOC388696 LOC388696; mRNA XM_373868 Homo sapiens similar to homerin (LOC388697) , mRNA XM_373870 Homo sapiens LOC388706 LOC388706 mRNA XM_373871 Homo sapiens LOC388709 LOC388709 mRNA XM_373872 Homo sapiens LOC388710 LOC388710 mRNA XM_373873 Homo sapiens LOC388713 LOC388713 mRNA XM_373875 Homo sapiens LOC388715 LOC388715 mRNA XM_373876 Homo sapiens LOC388716 LOC388716 mRNA XM_373877 Homo sapiens LOC388717 LOC388717 mRNA XM_373878 Homo sapiens LOC388719 LOC388719 mRNA XM_373879 Homo sapiens LOC388721 LOC388721 mRNA XM_373880 Homo sapiens LOC388723 LOC388723 mRNA XM_373881 Homo sapiens LOC388727 LOC388727 mRNA XM_373882 Homo sapiens LOC388729 LOC388729 mRNA XM_373883 Homo sapiens LOC388731 LOC388731 mRNA XM_373884 Homo sapiens LOC388732 LOC388732 mRNA XM_373885 Homo sapiens LOC388735 LOC388735 mRNA XM_373886 Homo sapiens LOC388738 LOC388738 mRNA XM_373889 Homo sapiens LOC388745 LOC388745 mRNA XM_373890 Homo sapiens LOC388746 LOC388746 mRNA XM_373893 Homo sapiens LOC388751 LOC388751 mRNA XM_373894 Homo sapiens LOC388752 LOC388752 mRNA XM_373896 Homo sapiens LOC388755 LOC388755 mRNA XM_373898 Homo sapiens LOC388758 LOC388758 mRNA XM_373903 Homo sapiens LOC388779 LOC388779; mRNA XM_373904 Homo sapiens LOC388780 LOC388780; mRNA XM_373905 Homo sapiens LOC388785 LOC388785 mRNA XM_373906 Homo sapiens LOC388786 LOC388786 mRNA XM_373907 Homo sapiens LOC388787 LOC388787 mRNA XM_373908 Homo sapiens LOC388789 LOC388789; mRNA XM_373909 Homo sapiens LOC388790 LOC388790 mRNA XM_373910 Homo sapiens LOC388791 LOC388791 mRNA XM_373911 Homo sapiens LOC388792 LOC388792 mRNA XM 373913 Homo sapiens LOC388793 LOC388793 mRNA
XM 373914 Homo sapiens LOC388796 [LOC388796) mRNA
XM_373915 Homo sapiens LOC388798 [LOC388798) mRNA
XM_373916 Homo sapiens LOC388800 LOC388800) mRNA
XM_373917 Homo sapiens LOC388801 LOC388801) mRNA
XM_373920 Homo sapiens LOC388805 LOC388805) mRNA
XM_373921 Homo sapiens LOC388806 LOC388806) mRNA
XM 373922 Homo sapiens LOC388807 [LOC388807) mRNA
XM_373924 Homo sapiens hypothetical Drotein LOC2J 34739 (LOC284739), mRNA
XM_373925 Homo sapiens LOC388813 10C388813) mRNA
XM 373926 Homo sapiens LOC388814 10C388814) mRNA
XM_373927 Homo sapiens LOC388815 LOC388815) mRNA
XM 373928 Homo sapiens LOC388816 LOC388816) mRNA
XM 373930 Homo sapiens LOC388820 LOC388820) mRNA
XM_373931 Homo sapiens LOC388823 LOC388823) mRNA
XM 373932 Homo sapiens LOC388824 LOC388824) mRNA
XM 373933 Homo sapiens LOC388825 LOC388825) mRNA
XM 373936 Homo sapiens LOC388829 LOC388829) mRNA
XM_373938 Homo sapiens LOC388831 LOC388831) mRNA
XM_373939 Homo sapiens LOC388832 LOC388832) mRNA
XM_373940 Homo sapiens LOC388833 LOC388833) mRNA
XM_373941 Homo sapiens LOC388834 LOC388834) mRNA
XM 373942 Homo sapiens LOC388839 LOC388839) mRNA
XM_373943 Homo sapiens LOC388843 LOC388843) mRNA
XM 373944 Homo sapiens LOC388845 LOC388845) mRNA
XM 373945 Homo sapiens LOC388849 LOC388849) mRNA
XM_373947 Homo sapiens LOC388862 LOC388862) mRNA
XM_373949 Homo sapiens LOC388885 LOC388885) mRNA
XM 373950 Homo sapiens LOC388887 LOC388887) mRNA
XM_373951 Homo sapiens LOC388888 LOC388888) mRNA
XM_373952 Homo sapiens LOC388889 LOC388889) mRNA
XM_373953 Homo sapiens LOC388890 LOC388890) mRNA
XM_373954 Homo sapiens LOC388893 LOC388893) mRNA
XM_373955 Homo sapiens LOC388895 LOC388895) mRNA
XM_373957 Homo sapiens LOC388897 LOC388897) mRNA
XM_373958 Homo sapiens LOC388898 LOC388898) mRNA
XM_373959 Homo sapiens LOC388899 LOC388899) mRNA
XM_373962 Homo sapiens LOC388903 LOC388903) mRNA
XM_373964 Homo sapiens LOC388910 LOC388910) mRNA
XM_373965 Homo sapiens LOC388911 LOC388911) mRNA
XM_373966 Homo sapiens LOC388912 LOC388912) mRNA
XM_373967 Homo sapiens LOC388914 LOC388914) mRNA
XM_373968 Homo sapiens LOC388915 LOC388915) mRNA
XM_373969 Homo sapiens LOC388916 LOC388916) mRNA
XM_373970 Homo sapiens LOC388917 LOC388917) mRNA
XM_373971 Homo sapiens LOC388918 LOC388918) mRNA
XM_373972 Homo sapiens LOC388920 LOC388920) mRNA
XM 373973 Homo sapiens LOC388923 LOC388923) mRNA
XM_373974 Homo sapiens LOC388924 LOC388924) mRNA
XM_373975 Homo sapiens LOC388930 LOC388930) mRNA
XM_373977 Homo sapiens LOC388934 LOC388934) mRNA
XM_373978 Homo sapiens LOC388935 LOC388935) mRNA
XM 373979 Homo sapiens LOC388938 LOC388938) mRNA
XM 373980 Homo sapiens LOC388940 LOC388940) mRNA
XM_373981 Homo sapiens LOC388942 LOC388942) mRNA
XM_373982 Homo sapiens LOC388944 LOC388944) mRNA
XM_373983 Homo sapiens LOC388945 [LOC388945) mRNA
XM_373984 Homo sapiens LOC388947 [LOC388947) mRNA
XM_373985 Homo sapiens LOC388948 LOC388948) mRNA
XM 373986 Homo sapiens LOC388952 rLOC388952) mRNA
XM_373989 Homo sapiens LOC388959 LOC388959) mRNA
XM 373990 Homo sapiens LOC388961 LOC388961) mRNA
XM_373991 Homo sapiens LOC388964 LOC388964) mRNA
XM_373992 Homo sapiens LOC388967 LOC388967) mRNA
XM_373995 Homo sapiens LOC389003 LOC389003) mRNA
XM_373997 Homo sapiens LOC389010 LOC389010) mRNA
XM_373998 Homo sapiens LOC389011 LOC389011) mRNA
XM 374001 Homo sapiens LOC389019 LOC389019) mRNA
XM_374002 Homo sapiens LOC389023 LOC389023) mRNA
XM_374003 Homo sapiens LOC389024 LOC389024) mRNA
XM_374004 Homo sapiens LOC389025 LOC389025) mRNA
XM_374005 Homo sapiens LOC389027 LOC389027) mRNA
XM_374006 Homo sapiens LOC389028 LOC389028) mRNA
XM 374007 Homo sapiens LOC389029 LOC389029) mRNA
XM 374008 Homo sapiens LOC389030 LOC389030) mRNA
XM 374009 Homo sapiens LOC389032 LOC389032) mRNA
XM_374010 Homo sapiens LOC389033 LOC389033) mRNA
XM 374011 Homo sapiens LOC389038 LOC389038) mRNA
XM_374012 Homo sapiens LOC389043 LOC389043) mRNA
XM_374013 Homo sapiens LOC389048 LOC389048) mRNA
XM_374014 Homo sapiens LOC389050 LOC389050) mRNA
XM 374015 Homo sapiens LOC389051 LOC389051) mRNA
XM_374016 Homo sapiens LOC389055 LOC389055) mRNA
XM_374017 Homo sapiens LOC389059 LOC389059) mRNA
XM 374019 Homo sapiens LOC389062 LOC389062) mRNA
XM_374020 Homo sapiens hypothetical ( jene supports >d by BC013438 (LOC375295), mR
XM_374021 Homo sapiens LOC389067 LOC389067) mRNA
XM 374022 Homo sapiens LOC389070 LOC389070) mRNA
XM_374023 Homo sapiens LOC389071 LOC389071) mRNA
XM 374025 Homo sapiens LOC389077 LOC389077) mRNA
XM_374026 Homo sapiens LOC389081 LOC389081) mRNA
XM_374028 Homo sapiens LOC389087 LOC389087) mRNA
XM 374029 Homo sapiens LOC389089 LOC389089) mRNA
XM_374033 Homo sapiens LOC389095 LOC389095) mRNA
XM 374035 Homo sapiens LOC389100 LOC389100) mRNA
XM 374036 Homo sapiens LOC389103 LOC389103) mRNA
XM_374037 Homo sapiens LOC389105 LOC389105) mRNA
XM 374038 Homo sapiens LOC389108 LOC389108) mRNA
XM_374042 Homo sapiens LOC389131 LOC389131) mRNA
XM_374045 Homo sapiens LOC389139 LOC389139) mRNA
XM 374046 Homo sapiens LOC389142 LOC389142) mRNA
XM_374047 Homo sapiens LOC389143 LOC389143) mRNA
XM_374048 Homo sapiens LOC389145 LOC389145) mRNA
XM 374049 Homo sapiens LOC389146 LOC389146) mRNA
XM 374050 Homo sapiens LOC389148 LOC389148) mRNA
XM_374051 Homo sapiens LOC389149 LOC389149) mRNA
XM 374052 Homo sapiens LOC389150 LOC389150) mRNA
XM 374053 Homo sapiens LOC389153 LOC389153) mRNA
XM_374054 Homo sapiens LOC389154 LOC389154) mRNA
XM 374055 Homo sapiens LOC389159 T.OC389159) mRNA
XM_374056 Homo sapiens LOC389160 LOC389160) mRNA
XM_374057 Homo sapiens LOC389161 LOC389161) mRNA
XM_374058 Homo sapiens LOC389162 LOC389162) mRNA
XMJ374059 Homo sapiens LOC389163 (LOC389163) mRNA
XM_374060 Homo sapiens LOC389166 10C389166) mRNA
XM_374061 Homo sapiens LOC389167 (LOC389167), mRNA
XM_374062 Homo sapiens LOC389172 (LOC389172), mRNA
XM_374064 Homo sapiens LOC389182 (LOC389182), mRNA
XM_374065 Homo sapiens LOC389183 (LOC389183), mRNA
XM_374066 Homo sapiens LOC389184 (LOC389184), mRNA
XM_374067 Homo sapiens LOC389185 (LOC389185), mRNA
XM_374068 Homo sapiens LOC389186 (LOC389186), mRNA
XM_374069 Homo sapiens hypothetical protein LOC254808 (LOC254808), mRNA
XM_374070 Homo sapiens LOC389188 (LOC389188), mRNA
XM_374071 Homo sapiens LOC389189 (LOC389189), mRNA
XM_374075 Homo sapiens LOC389196 (LOC389196), mRNA
XM_374076 Homo sapiens LOC389198 (LOC389198), mRNA
XM_374077 Homo sapiens LOC389201 (LOC389201 ), mRNA
XM_374078 Homo sapiens hypothetical protein LOC285548 (LOC285548), mRNA
XM_374079 Homo sapiens LOC389204 (LOC389204), mRNA
XM_374080 Homo sapiens KIAA0114 gene product (KIAA0114), mRNA
XM_374081 Homo sapiens LOC389209 (LOC389209), mRNA
XM_374082 Homo sapiens LOC389212 (LOC389212), mRNA
XM_374086 Homo sapiens LOC389224 (LOC389224), mRNA
XM_374088 Homo sapiens LOC389230 (LOC389230), mRNA
XM_374089 Homo sapiens LOC389231 (LOC389231 ), mRNA
XM_374090 Homo sapiens LOC389232 (LOC389232), mRNA
XM_374091 Homo sapiens LOC389233 (LOC389233), mRNA
XM_374092 Homo sapiens LOC389234 (LOC389234), mRNA
XM_374093 Homo sapiens LOC389236 (LOC389236), mRNA
XM_374094 Homo sapiens LOC389237 (LOC389237), mRNA
XM_374095 Homo sapiens LOC389242 (LOC389242), mRNA
XM_374096 Homo sapiens LOC389243 (LOC389243), mRNA
XM_374097 Homo sapiens LOC389244 (LOC389244), mRNA
XM_374098 Homo sapiens LOC389245 (LOC389245), mRNA
XM_374099 Homo sapiens LOC389247 (LOC389247), mRNA
XM_374100 Homo sapiens LOC389248 (LOC389248), mRNA
XM_374101 Homo sapiens LOC389250 (LOC389250), mRNA
XM_374103 Homo sapiens LOC389252 (LOC389252), mRNA
XM_374104 Homo sapiens LOC389253 (LOC389253), mRNA
XM_374105 Homo sapiens LOC389259 (LOC389259), mRNA
XM_374106 Homo sapiens LOC389262 (LOC389262), mRNA
XM_374107 Homo sapiens LOC389263 (LOC389263), mRNA
XM_374108 Homo sapiens LOC389264 (LOC389264), mRNA
XM_374109 Homo sapiens LOC389266 (LOC389266), mRNA
XM_374110 Homo sapiens LOC389268 (LOC389268), mRNA
XM_374111 Homo sapiens LOC389269 (LOC389269), mRNA
XM_374112 Homo sapiens LOC389270 (LOC389270), mRNA
XM_374113 Homo sapiens LOC389271 (LOC389271), mRNA
XM_374114 Homo sapiens LOC389272 (LOC389272), mRNA
XM_374115 Homo sapiens LOC389273 (LOC389273), mRNA
XM_374117 Homo sapiens LOC389275 (LOC389275), mRNA
XM_374118 Homo sapiens LOC389285 (LOC389285), mRNA
XM_374120 Homo sapiens LOC389288 (LOC389288), mRNA
XM_374121 Homo sapiens LOC389290 (LOC389290), mRNA
XM_374123 Homo sapiens LOC389295 (LOC389295), mRNA
XM_374124 Homo sapiens LOC389300 (LOC389300), mRNA
XM_374125 Homo sapiens LOC389302 (LOC389302), mRNA
XM_374127 Homo sapiens LOC389306 (LOC389306), mRNA
XM_374128 Homo sapiens LOC389307 (LOC389307), mRNA
XM_374129 Homo sapiens LOC389309 (LOC389309), mRNA
XM_374130 Homo sapiens LOC389310 (LOC389310), mRNA
XM_374131 Homo sapiens LOC389311 (LOC389311 mRNA XM_374132 Homo sapiens LOC389316 (LOC389316 mRNA XM_374133 Homo sapiens LOC389318 (LOC389318 mRNA XM_374134 Homo sapiens LOC389319 (LOC389319 mRNA XM_374135 Homo sapiens LOC389324 (LOC389324; mRNA XM_374136 Homo sapiens LOC389325 (LOC389325 mRNA XM_374137 Homo sapiens LOC389328 (LOC389328 mRNA XM_374138 Homo sapiens LOC389329 (LOC389329 mRNA XM_374139 Homo sapiens LOC389330 (LOC389330 mRNA XM_374140 Homo sapiens LOC389331 (LOC389331 mRNA XM_374141 Homo sapiens LOC389332 (LOC389332 mRNA XM_374142 Homo sapiens LOC389333 (LOC389333 mRNA XM_374143 Homo sapiens LOC389334 (LOC389334 mRNA XM_374144 Homo sapiens LOC389335 (LOC389335 mRNA XM_374145 Homo sapiens LOC389338 (LOC389338 mRNA XM_374146 Homo sapiens LOC389339 (LOC389339 mRNA XM_374147 Homo sapiens LOC389340 (LOC389340 mRNA XM_374148 Homo sapiens LOC389343 (LOC389343 mRNA XM_374149 Homo sapiens LOC389344 (LOC389344 mRNA XM_374150 Homo sapiens LOC389345 (LOC389345 mRNA XM_374151 Homo sapiens LOC389348 (LOC389348; mRNA XM_374152 Homo sapiens LOC389353 (LOC389353; mRNA XM_374153 Homo sapiens LOC389354 (LOC389354 mRNA XM_374154 Homo sapiens LOC389357 (LOC389357; mRNA XM_374155 Homo sapiens LOC389358 (LOC389358; mRNA XM_374156 Homo sapiens LOC389359 (LOC389359 mRNA XM_374157 Homo sapiens LOC389360 (LOC389360 mRNA XM_374158 Homo sapiens LOC389361 (LOC389361 mRNA XM_374159 Homo sapiens LOC389362 (LOC389362 mRNA XM_374161 Homo sapiens LOC389369 (LOC389369 mRNA XM_374162 Homo sapiens LOC389370 (LOC389370; mRNA XM_374163 Homo sapiens LOC389372 (LOC389372 mRNA XM_374164 Homo sapiens LOC389380 (LOC389380 mRNA XM_374165 Homo sapiens LOC389381 (LOC389381 mRNA XM_374167 Homo sapiens LOC389390 (LOC389390; mRNA XM_374168 Homo sapiens LOC389392 (LOC38939 mRNA XM_374169 Homo sapiens LOC389393 (LOC389393 mRNA XM_374170 Homo sapiens LOC389398 (LOC389398 mRNA XM_374171 Homo sapiens LOC389399 (LOC389399 mRNA XM_374172 Homo sapiens LOC389403 (LOC389403; mRNA XM_374173 Homo sapiens LOC389409 (LOC389409, mRNA XM_374174 Homo sapiens LOC389411 (LOC389411 mRNA XM_374175 Homo sapiens LOC389413 (LOC389413; mRNA XM_374176 Homo sapiens LOC389414 (LOC389414, mRNA XM_374177 Homo sapiens LOC389415 (LOC389415; mRNA XM_374178 Homo sapiens LOC389420 (LOC389420; mRNA XM_374179 Homo sapiens LOC389422 (LOC389422 mRNA XM_374180 Homo sapiens LOC389423 (LOC389423 mRNA XM_374181 Homo sapiens LOC389426 (LOC389426 mRNA XM_374183 Homo sapiens LOC389433 (LOC389433 mRNA XM_374184 Homo sapiens LOC389436 (LOC389436 mRNA XM_374185 Homo sapiens LOC389437 (LOC389437 mRNA XM_374186 Homo sapiens LOC389438 (LOC389438 mRNA XM_374187 Homo sapiens LOC389439 (LOC389439 mRNA XM_374188 Homo sapiens LOC389440 (LOC389440 mRNA XM_374189 Homo sapiens LOC389441 (LOC389441 mRNA XM 374190 Homo sapiens LOC389442 (LOC389442 mRNA
XM_374191 Homo sapiens LOC389443 (LOC389443 ), mRNA XM_374192 Homo sapiens LOC389447 (LOC389447 ), mRNA XM_374193 Homo sapiens LOC389449 (LOC389449 ), mRNA XM_374194 Homo sapiens LOC389450 (LOC389450 ), mRNA XM_374196 Homo sapiens LOC389452 (LOC389452 ), mRNA XM_374198 Homo sapiens LOC389455 (LOC389455 ), mRNA XM_374199 Homo sapiens LOC389456 (LOC389456 ), mRNA XM_374200 Homo sapiens LOC389457 (LOC389457 ), mRNA XM_374202 Homo sapiens similar to Zinc finger 268 (Zinc finger protein HZF3) (L XM_374203 Homo sapiens LOC389469 (LOC389469 i), mRNA XM_374204 Homo sapiens LOC389470 (LOC389470 ), mRNA XM_374218 Homo sapiens LOC389506 (LOC389506 ), mRNA XM_374222 Homo sapiens LOC389526 (LOC389526 ), mRNA XM_374226 Homo sapiens LOC389533 (LOC389533 ), mRNA XM_374227 Homo sapiens LOC389534 (LOC389534 ), mRNA XM_374232 Homo sapiens LOC389556 (LOC389556 ), mRNA XM_374236 Homo sapiens LOC389562 (LOC389562 ), mRNA XM_374242 Homo sapiens LOC389605 (LOC389605 ), mRNA XM_374243 Homo sapiens LOC389608 (LOC389608 ), mRNA XM_374244 Homo sapiens LOC389609 (LOC389609 ), mRNA XM_374245 Homo sapiens LOC389612 (LOC389612 ), mRNA XM_374247 Homo sapiens LOC389623 (LOC389623 ), mRNA XM_374248 Homo sapiens LOC389624 (LOC389624 ), mRNA XM_374249 Homo sapiens similar to ι ZNF169 protein (LOC389628), mRNA XM_374250 Homo sapiens LOC389629 (LOC389629 ), mRNA XM_374251 Homo sapiens LOC389632 (LOC389632 ), mRNA XM_374252 Homo sapiens LOC389634 (LOC389634 ), mRNA XM_374253 Homo sapiens LOC389635 (LOC389635 ), mRNA XM_374254 Homo sapiens LOC389636 (LOC389636 ), mRNA XM_374255 Homo sapiens LOC389637 (LOC389637 '), mRNA XM_374256 Homo sapiens LOC389638 (LOC389638 ), mRNA XM_374257 Homo sapiens hypothetical protein LOC2861 14 (LOC286114), mRNA XM_374258 Homo sapiens LOC389639 (LOC389639 ), mRNA XM_374259 Homo sapiens LOC389640 (LOC389640 ), mRNA XM_374260 Homo sapiens LOC389641 (LOC389641 ), mRNA XM_374261 Homo sapiens LOC389642 (LOC389642 ), mRNA XM_374262 Homo sapiens LOC389645 (LOC389645 ), mRNA XM_374263 Homo sapiens LOC389647 (LOC389647 ), mRNA XM_374264 Homo sapiens LOC389648 (LOC389648; ), mRNA XM_374266 Homo sapiens LOC389650 (LOC389650; ), mRNA XM_374267 Homo sapiens LOC389654 (LOC389654 ), mRNA XM_374268 Homo sapiens LOC389656 (LOC389656 ), mRNA XM_374269 Homo sapiens LOC389657 (LOC389657 ■;), mRNA XM_374270 Homo sapiens LOC389659 (LOC389659 ), mRNA XM_374271 Homo sapiens LOC389665 (LOC389665; ), mRNA XM_374272 Homo sapiens LOC389666 (LOC389666 ■;), mRNA XM_374273 Homo sapiens LOC389669 (LOC389669 ), mRNA XM_374274 Homo sapiens LOC389670 (LOC389670 ), mRNA XM_374275 Homo sapiens LOC389671 (LOC389671 ), mRNA XM_374276 Homo sapiens LOC389676 (LOC389676 ), mRNA XM_374277 Homo sapiens LOC389683 (LOC389683 ), mRNA XM_374278 Homo sapiens LOC389684 (LOC389684 ), mRNA XM_374279 Homo sapiens LOC389685 (L0C389685; ), mRNA XM_374280 Homo sapiens LOC389686 (LOC389686 ), mRNA XM_374281 Homo sapiens LOC389687 (LOC389687 ), mRNA XM_374282 Homo sapiens LOC389688 (LOC389688 ), mRNA XM 374283 Homo sapiens LOC389689 (LOC389689 ), mRNA
XM_374284 Homo sapiens LOC389691 LOC389691) mRNA
XM_374285 Homo sapiens LOC389693 LOC389693) mRNA
XM 374286 Homo sapiens LOC389695 LOC389695) mRNA
XM 374287 Homo sapiens LOC389696 LOC389696) mRNA
XM_374289 Homo sapiens LOC389702 LOC389702) mRNA
XM 374290 Homo sapiens LOC389703 LOC389703) mRNA
XM_374291 Homo sapiens LOC389706 LOC389706) mRNA
XM 374292 Homo sapiens LOC389707 LOC389707) mRNA
XM_374293 Homo sapiens LOC389718 LOC389718) mRNA
XM 374294 Homo sapiens LOC389720 LLOC389720) mRNA
XM_374295 Homo sapiens LOC389767 LOC389767) mRNA
XM 374296 Homo sapiens LOC389771 LOC389771) mRNA
XM 374297 Homo sapiens LOC389773 LOC389773) mRNA
XM_374298 Homo sapiens LOC389774 LOC389774) mRNA
XM_374299 Homo sapiens LOC389775 .LOC389775) mRNA
XM 374300 Homo sapiens LOC389777 LOC389777) mRNA
XM 374301 Homo sapiens LOC389778 LOC389778) mRNA
XM_374302 Homo sapiens LOC389782 LOC389782) mRNA
XM_374303 Homo sapiens LOC389783 LOC389783) mRNA
XM_374304 Homo sapiens LOC389784 LOC389784) mRNA
XM_374305 Homo sapiens LOC389793 LOC389793) mRNA
XM_374306 Homo sapiens LOC389794 LOC389794) mRNA
XM_374307 Homo sapiens LOC389801 LOC389801) mRNA
XM 374308 Homo sapiens LOC389804 LOC389804) mRNA
XM 374309 Homo sapiens LOC389806 LOC389806) mRNA
XM_374313 Homo sapiens LOC389810 LOC389810) mRNA
XM 374315 Homo sapiens LOC389815 LOC389815) mRNA
XM_374317 Homo sapiens LOC389831 LOC389831) mRNA
XM_374318 Homo sapiens LOC389834 LOC389834) mRNA
XM 374319 Homo sapiens hypothetical | Drotein LOC3C 59457 (LOC339457), mRNA
XM 374323 Homo sapiens LOC389841 I LOC389841) mRNA
XM_374325 Homo sapiens LOC389846 { LOC389846) mRNA
XM 374326 Homo sapiens LOC389858 LOC389858) mRNA
XM_374327 Homo sapiens LOC389861 LOC389861) mRNA
XM_374328 Homo sapiens LOC389863 LOC389863) mRNA
XM 374329 Homo sapiens LOC389865 LOC389865) mRNA
XM_374330 Homo sapiens LOC389867 < LOC389867) mRNA
XM_374331 Homo sapiens LOC389868 LOC389868) mRNA
XM_374333 Homo sapiens LOC389878 LOC389878) mRNA
XM 374336 Homo sapiens LOC389889 I LOC389889) mRNA
XM_374337 Homo sapiens LOC389890 LOC389890) mRNA
XM 374338 Homo sapiens LOC389893 LOC389893) mRNA
XM 374339 Homo sapiens LOC389896 J.OC389896) mRNA
XM_374340 Homo sapiens LOC389897 LOC389897) mRNA
XM_374341 Homo sapiens LOC389908 LOC389908) mRNA
XM 374342 Homo sapiens LOC389909 LOC389909) mRNA
XM 374343 Homo sapiens LOC389910 J_OC389910) mRNA
XM_374349 Homo sapiens LOC389931 LOC389931) mRNA
XM 374352 Homo sapiens LOC390367 LOC390367) mRNA
XM 374354 Homo sapiens LOC390550 J.OC390550) mRNA
XM_374355 Homo sapiens LOC390565 (LOC390565) mRNA
XM_374356 Homo sapiens LOC390568 LOC390568) mRNA
XM_374357 Homo sapiens LOC390749 (LOC390749) mRNA
XM_374358 Homo sapiens LOC390751 LLOC390751) mRNA
XM_374359 Homo sapiens LOC390821 (LOC390821) mRNA
XM_374361 Homo sapiens LOC390870 (LOC390870) mRNA
XM_374362 Homo sapiens LOC390989 (LOC390989) mRNA
XM_374364 Homo sapiens LOC391067 (LOC391067), mRNA
XM_374365 Homo sapiens LOC391238 (LOC391238), mRNA
XM_374367 Homo sapiens similar to hypothetical protein AN2833.2 (LOC391727), mRN/
XM_374368 Homo sapiens LOC391728 (LOC391728), mRNA
XM_374369 Homo sapiens LOC391729 (LOC391729), mRNA
XM_374370 Homo sapiens LOC392160 (LOC392160), mRNA
XM_374371 Homo sapiens LOC392177 (LOC392177), mRNA
XM_374372 Homo sapiens LOC392238 (LOC392238), mRNA
XM_374377 Homo sapiens LOC392580 (LOC392580), mRNA
XM_374378 Homo sapiens LOC392581 (LOC392581 ), mRNA
XM_374379 Homo sapiens LOC392583 (LOC392583), mRNA
XM_374382 Homo sapiens similar to cAMP-dependent protein kinase type l-beta regulate
XM_374386 Homo sapiens similar to RIKEN cDNA A930017N06 gene (LOC392617), mR
XM_374388 Homo sapiens similar to KIAA1691 protein (LOC392619), mRNA
XM_374389 Homo sapiens KIAA1950 protein (KI AA1950), mRNA
XM_374396 Homo sapiens ubiquitin specific protease 42 (USP42), mRNA
XM_374399 Homo sapiens similar to matrilin 2 precursor (LOC392630), mRNA
XM_374401 Homo sapiens similar to alpha-2 macroglobulin family protein VIP (LOC3926
XM_374404 Homo sapiens KIAA0960 protein (KIAA0960), mRNA
XM_374405 Homo sapiens similar to Neurogenic locus notch homolog protein 1 precurso
XM_374406 Homo sapiens similar to RIKEN cDNA A530016O06 gene (LOC392636), R
XM_374413 Homo sapiens similar to Neuronal protein 3.1 (p311 protein) (LOC392642), n
XM_374414 Homo sapiens similar to mKIAA0038 protein (LOC392647), mRNA
XM_374419 Homo sapiens similar to RP9 protein (LOC392652), mRNA
XM_374422 Homo sapiens KIAA0877 protein (KIAA0877), mRNA
XM_374425 Homo sapiens KIAA0895 protein (KIAA0895), mRNA
XM_374427 Homo sapiens similar to T-cell receptor gamma chain V region PT-gamma-1)
XM_374428 Homo sapiens LOC392661 (LOC392661), mRNA
XM_374430 Homo sapiens hypothetical protein DKFZp761 l2123 (DKFZp76112123), mRN
XM_374431 Homo sapiens similar to myosin IG (LOC392665), mRNA
XM_374432 Homo sapiens similar to KIAA0363 (LOC392666), mRNA
XM_374434 Homo sapiens LOC392668 (LOC392668), mRNA
XM_374435 Homo sapiens hypothetical protein LOC136288 (LOC136288), mRNA
XM_374437 Homo sapiens LOC392670 (LOC392670), mRNA
XM_374439 Homo sapiens similar to LanC lantibiotic synthetase component C-like 2; tesl
XM_374460 Homo sapiens similar to Williams-Beuren syndrome critical region protein 19
XM_374461 Homo sapiens similar to PMS5 homolog mismatch repair protein - human (L(
XM_374473 Homo sapiens similar to general transcription factor II, i isoform 1 ; BTK-asso*
XM_374475 Homo sapiens similar to PMS2L13 (LOC392729), mRNA
XM_374479 Homo sapiens similar to Williams-Beuren syndrome critical region protein 19
XM_374481 Homo sapiens similar to deltex 2 (LOC392736), mRNA
XM_374483 Homo sapiens similar to 60S ribosomal protein L7a (Surfeit locus protein 3) (
XM_374484 Homo sapiens similar to Piccolo protein (Aczonin) (LOC392742), mRNA
XM_374487 Homo sapiens similar to Six transmembrane epithelial antigen of prostate (L(
XM_374489 Homo sapiens hypothetical protein FLJ39885 (FLJ39885), mRNA
XM_374490 Homo sapiens similar to ribosomal protein S27 (LOC392748), mRNA
XM_374491 Homo sapiens protein phosphatase 1 , regulatory (inhibitor) subunit 9A (PPP-
XM_374496 Homo sapiens similar to ZNF36 protein (LOC392756), mRNA
XM_374498 Homo sapiens similar to CG14977-PA (LOC392758), mRNA
XM_374501 Homo sapiens similar to intestinal mucin 3 (LOC392762), mRNA
XM_374502 Homo sapiens similar to intestinal mucin (LOC392763), mRNA
XM_374506 Homo sapiens similar to Williams-Beuren syndrome critical region protein 19
XM_374514 Homo sapiens similar to laminin beta-4 chain precursor (LOC392775), mRN/
XM_374517 Homo sapiens similar to zinc finger protein 312; forebrain embryonic zinc fin
XM_374518 Homo sapiens similar to cardiac leiomodin (LOC392780), mRNA
XM_374519 Homo sapiens similar to 40S ribosomal protein S2 (LOC392781 ), mRNA
XM_374522 Homo sapiens similar to Filamin C (Gamma-filamin) (Filamin 2) (Protein FLN
XM_374526 Homo sapiens similar to eukaryotic translation elongation factor 1 beta 2; eul
XM_374529 Homo sapiens KIAA1466 protein (KIAA1466), mRNA
XM_374530 Homo sapiens nucleoporin 205kDa (NUP205), mRNA
XM_374538 Homo sapiens KIAA1277 (KI AA1277), mRNA
XM_374579 Homo sapiens hypothetical protein LOC155054 (LOC155054), mRNA
XM_374581 Homo sapiens similar to KIAA1862 protein (LOC392836), mRNA
XM_374583 Homo sapiens similar to actin-related protein Arp11 (LOC392838), mRNA
XM_374584 Homo sapiens similar to amyotrophic lateral sclerosis 2 (juvenile) chromoson
XM_374585 Homo sapiens similar to hypothetical protein 4931409K22 (LOC392842), mR
XM_374586 Homo sapiens similarto hypothetical protein FLJ22527 (LOC392843), mRN/
XM_374589 Homo sapiens LOC392848 (LOC392848), mRNA
XM_374590 Homo sapiens similarto C7orf3 protein (LOC392850), mRNA
XM_374591 Homo sapiens LOC392851 (LOC392851), mRNA
XM_374592 Homo sapiens similar to THAP domain containing 5 (LOC392852), mRNA
XM_374600 Homo sapiens similar to GluR-delta2 phllic-protein (LOC392862), mRNA
XM_374603 Homo sapiens similar to cell division cycle 10 homolog (LOC392884), mRNA
XM_374604 Homo sapiens similar to T-cell receptor (V-J-C) precursor (LOC392887), mR
XM_374605 Homo sapiens similar to T-cell receptor gamma chain V region PT-gamma-11
XM_374607 Homo sapiens similar to Splicing factor, arginine/serine-rich, 46kD (LOC392Ϊ
XM_374608 Homo sapiens hypothetical protein MGC26484 (MGC26484), mRNA
XM_374625 Homo sapiens similar to hypothetical protein (LOC392943), mRNA
XM_374627 Homo sapiens guanine nucleotide binding protein, alpha transducing 3 (GNA
XM_374628 Homo sapiens similar to dynein, cytoplasmic, light peptide; 8kD LC; dynein L
XM_374636 Homo sapiens similar to acyl CoA:monoacylglycerol acyltransferase 2 (LOCJ
XM_374637 Homo sapiens similar to alpha-2-glycoprotein 1 , zinc; Alpha-2-glycoprotein, z
XM_374638 Homo sapiens similar to Cohesin subunit SA-3 (Stromal antigen 3) (SCC3 he
XM_374646 Homo sapiens similar to ribosomal protein L18; 60S ribosomal protein L18 (L
XM_374648 Homo sapiens similar to RIKEN cDNA 1500011 L16 (LOC392982), mRNA
XM_374650 Homo sapiens similar to ribosomal protein S14 (LOC392988), mRNA
XM_374653 Homo sapiens similar to CG6293-PA (LOC392993), mRNA
XM 374655 Homo sapiens similar to dJ753D5.2 (novel protein similar to RPS17 (40S ribc
XM_374657 Homo sapiens similar to dGTPase (EC 3.1.5.1 ) - mouse (fragment) (LOC393
XM 374680 Homo sapiens similar to seven transmembrane helix receptor (LOC393037),
XM_374682 Homo sapiens similar to Olfactory receptor 6B1 (Olfactory receptor 7-3) (OR;
XM_374683 Homo sapiens similar to olfactory receptor MOR261-13 (LOC393046), mRN/
XM_374684 Homo sapiens similar to olfactory receptor MOR261-1 (LOC393047), mRNA
XM_374687 Homo sapiens similar to seven transmembrane helix receptor (LOC393051 ),
XM_374689 Homo sapiens similar to seven transmembrane helix receptor (LOC393053),
XM_374694 Homo sapiens similar to Mtr3 (mRNA transport regulator 3)-homolog; Mtr3 (n
XM_374699 Homo sapiens similar to gastrulation brain homeobox 1 (LOC393070), mRN/
XM_374702 Homo sapiens similar to envelope protein (LOC393073), mRNA
XM_374705 Homo sapiens similarto polycystic kidney disease 1-like 3 (LOC393078), mF
XM_374706 Homo sapiens similar to 60S RIBOSOMAL PROTEIN L21 (LOC393079), mR
XM_374710 Homo sapiens LOC392620 (LOC392620), mRNA
XM_374711 Homo sapiens LOC392621 (LOC392621), mRNA
XM__374713 Homo sapiens similar to Zinc finger protein 268 (Zinc finger protein HZF3) (L
XM_374715 Homo sapiens LOC392641 (LOC392641), mRNA
XM_374721 Homo sapiens LOC392657 (LOC392657), mRNA
XM_374722 Homo sapiens LOC392659 (LOC392659), mRNA
XM_374730 Homo sapiens LOC392702 (LOC392702), mRNA
XM_374734 Homo sapiens LOC392726 (LOC392726), mRNA
XMJ374735 Homo sapiens LOC392730 (LOC392730), mRNA
XM_374741 Homo sapiens LOC392749 (LOC392749), mRNA
XM_374751 Homo sapiens LOC392790 (LOC392790), mRNA
XM_374752 Homo sapiens LOC392791 (LOC392791), mRNA
XM_374761 Homo sapiens LOC392849 (LOC392849), mRNA
XM_374763 Homo sapiens LOC393076 (LOC393076), mRNA
XM_374764 Homo sapiens LOC393080 (LOC393080), mRNA
XM_374765 Homo sapiens similar to cDNA sequence BC016423 (LOC399712), mRNA
XM_374766 Homo sapiens hypothetical gene supported by AK128185 (LOC399715), mR
XM_374767 Homo sapiens similar to hypothetical protein (LOC399716), mRNA
XM_374768 Homo sapiens USP6 N-terminal like (USP6NL), mRNA
XM_374769 Homo sapiens similar to USP6NL protein (LOC399718), mRNA
XM_374770 Homo sapiens similar to seven transmembrane helix receptor (LOC399719),
XM_374779 Homo sapiens ankyrin repeat domain 26 (ANKRD26), mRNA
XM_374781 Homo sapiens hypothetical protein LOC220906 (LOC220906), mRNA
XM_374782 Homo sapiens similar to supervillin isoform 2; membrane-associated F-actin
XM_374786 Homo sapiens similar to hypothetical protein LOC349114 (LOC399744), mRI
XM_374787 Homo sapiens similar to hypothetical protein FLJ40432 (LOC399747), mRN/
XM_374792 Homo sapiens similar to Hypothetical protein KIAA0514 (LOC399755), ITIRNJ
XM_374799 Homo sapiens similar to ARF GTPase-activating protein (LOC399758), mRN
XM_374801 Homo sapiens similar to ARF GTPase-activating protein (LOC399761 ), mRN
XM_374802 Homo sapiens similar to DKFZP566K0524 protein (LOC399762), mRNA
XM_374803 Homo sapiens similar to LINE-1 REVERSE TRANSCRIPTASE HOMOLOG (I
XM_374807 Homo sapiens similar to ARF GTPase-activating protein (LOC399769), mRN
XM_374809 Homo sapiens similar to activator of S phase kinase (LOC399777), mRNA
XM_374810 Homo sapiens similar to activator of S phase kinase (LOC399778), mRNA
XM_374813 Homo sapiens similar to peptidylprolyl isomerase A (cyclophilin A) (LOC3997
XM_374817 Homo sapiens similar to bA182L21.1 (novel protein similar to hypothetical pr*
XM_374829 Homo sapiens programmed cell death 11 (PDCD11 ), mRNA
XM_374830 Homo sapiens similar to hypothetical protein LOC119395 (LOC399807), mRI
XM_374831 Homo sapiens SH3 multiple domains 1 (SH3MD1), mRNA
XM_374832 Homo sapiens similar to ribosomal protein L13a; 60S ribosomal protein L13a
XM_374835 Homo sapiens similar to RIKEN cDNA 1700022C21 (LOC399814), mRNA
XM_374836 Homo sapiens similar to hypothetical protein FLJ13490 (LOC399815), mRN/
XM_374839 Homo sapiens similar to CG9643-PA (LOC399818), mRNA
XM_374840 Homo sapiens similar to CG15021-PA (LOC399819), mRNA
XM_374842 Homo sapiens hypothetical gene supported by AK094354 (LOC399821 ), mR
XM_374844 Homo sapiens similar to hypothetical protein B130055A05 (LOC399823), mF
XM_374847 Homo sapiens similar to shadow of prion protein; Shadoo (LOC399831 ), mR
XM_374851 Homo sapiens similar to Synaptotagmin XV (SytXV) (ChMOSyt) (LOC399837
XM_374852 Homo sapiens similar to double homeobox protein (LOC399839), mRNA
XM_374854 Homo sapiens similar to nuclear receptor coactivator 4; RET-activating gene
XM__374855 Homo sapiens similar to Glutamate dehydrogenase 1 , mitochondrial precursi
XM_374857 Homo sapiens similar to bA476H5.3 (novel protein similar to septin) (LOC391
XM_374858 Homo sapiens hypothetical gene supported by AK093729; AK128780; AL111
XM_374860 Homo sapiens tumor protein p53 inducible protein 5 (TP53I5), mRNA
XM_374864 Homo sapiens hypothetical gene supported by AK091259 (LOC399857), mR
XM_374873 Homo sapiens similar to RIKEN cDNA 3830422K02 (LOC399870), mRNA
XM_374877 Homo sapiens hypothetical protein DKFZp779M0652 (DKFZp779M0652), ml
XM_374879 Homo sapiens hypothetical protein LOC114971 (LOC114971), mRNA
XM_374880 Homo sapiens hypothetical gene supported by BC065704 (LOC399888), mR
XM_374882 Homo sapiens similar to Metabotropic glutamate receptor 5 precursor (mGlul
XMJ374885 Homo sapiens hypothetical gene supported by AK128188 (LOC399898), mR
XM_374890 Homo sapiens hypothetical gene supported by AK093779 (LOC399900), mR
XM_374893 Homo sapiens LOC399904 (LOC399904), mRNA
XM_374898 Homo sapiens similar to pecanex-like 3 (LOC399909), mRNA
XM_374899 Homo sapiens similar to Gag-Pro-Pol protein (LOC399913), mRNA
XM_374900 Homo sapiens similar to beta-1 ,4-mannosyltransferase; beta-1 ,4 mannosyltrc
XM_374902 Homo sapiens similar to polymerase (LOC399917), mRNA
XM_374904 Homo sapiens similar to SH3 and multiple ankyrin repeat domains protein 2 i
XM_374905 Homo sapiens similar to proline rich synapse associated protein 1 isoform E;
XM_374906 Homo sapiens hypothetical gene supported by AK124096 (LOC399923), mR
XM_374907 Homo sapiens SH3 multiple domains 3 (SH3MD3), mRNA
XM_374909 Homo sapiens similar to hypothetical protein (LOC399925), mRNA
XM_374911 Homo sapiens similar to ribosomal protein S12 (LOC399927), mRNA
XM_374912 Homo sapiens X-ray radiation resistance associated 1 (XRRA1), mRNA
XM_374915 Homo sapiens hypothetical protein LOC283219 (LOC283219), mRNA
XM_374917 Homo sapiens similar to tripartite motif-containing 51 (LOC399937), mRNA
XM_374919 Homo sapiens similar to tripartite motif protein 48; TRIM48 (LOC399939), mF
XM_374920 Homo sapiens similar to tripartite motif-containing 51 (LOC399940), mRNA
XM_374922 Homo sapiens KIAA1731 protein (KIAA1731), mRNA
XM_374927 Homo sapiens KIAA1377 protein (KI AA1377), mRNA
XM_374930 Homo sapiens similar to expressed sequence AI593442 (LOC399947), mRN
XM_374932 Homo sapiens hypothetical gene supported by AB096245 (LOC399948), mR
XM_374933 Homo sapiens similar to RIKEN cDNA 4833427G06 (LOC399949), mRNA
XM_374936 Homo sapiens KIAA1052 protein (KIAA1052), mRNA
XM_374937 Homo sapiens hypothetical gene supported by AK127233 (LOC399957), mR
XM_374944 Homo sapiens similar to LVLF3112 (LOC399967), mRNA
XM_374945 Homo sapiens similar to Seminal vesicle protein 7 precursor (SVS VII) (Caltri
XM_374948 Homo sapiens similar to LINE-1 REVERSE TRANSCRIPTASE HOMOLOG (I
XM_374949 Homo sapiens similar to Sorting nexin 19 (LOC399979), mRNA
XM_374952 Homo sapiens hypothetical gene supported by AK126822 (LOC399990), mR
XM_374959 Homo sapiens similar to DDX11 protein (LOC399999), mRNA
XM_374965 Homo sapiens similar to BC004636 protein (LOC400010), mRNA
XM_374967 Homo sapiens similar to carboxyl-terminal modulator protein isoform a (LOG'
XM_374972 Homo sapiens similar to MUC19 (LOC400023), mRNA
XM_374973 Homo sapiens similar to hypothetical protein (L1 H 3 region) - human (LOC4C
XM_374976 Homo sapiens hypothetical protein LOC283331 (LOC283331), mRNA
XM_374981 Homo sapiens similar to DKFZP564K247 protein (LOC400038), mRNA
XM_374982 Homo sapiens similar to oriLyt TD-element binding protein 7 (LOC400040), n
XM_374983 Homo sapiens KIAA0748 gene product (KIAA0748), mRNA
XM_374985 Homo sapiens KIAA0352 gene product (KIAA0352), mRNA
XM_374987 Homo sapiens similar to 60S ribosomal protein L26 (LOC400055), mRNA
XM_374989 Homo sapiens KIAA0373 gene product (KIAA0373), mRNA
XM_374995 Homo sapiens similar to ribosomal protein L18a; 60S ribosomal protein L18a
XM_374996 Homo sapiens AMP-activated protein kinase family member 5 (ARK5), mRN*
XM_374997 Homo sapiens hypothetical gene supported by AK097461 ; BC046185 (LOC4
XM_374999 Homo sapiens hypothetical gene supported by AK124947 (LOC400077), mR
XM_375000 Homo sapiens KI AA1853 protein (KI AA1853), mRNA
XM_375004 Homo sapiens similar to Dynein heavy chain at 89D CG1842-PA (LOC40008
XM_375005 Homo sapiens similar to Dynein heavy chain at 89D CG1842-PA (LOC40008
XM_375007 Homo sapiens hypothetical gene supported by AK123815 (LOC400093), mR
XM_375013 Homo sapiens hypothetical gene supported by AK127292; AK128225 (LOC4
XM_375018 Homo sapiens hypothetical gene supported by AK092066 (LOC400121), mR
XM_375023 Homo sapiens hypothetical gene supported by AJ412041 (LOC400132), mR
XM_375027 Homo sapiens hypothetical protein LOC283491 (LOC283491), mRNA
XM_375029 Homo sapiens hypothetical protein LOC338862 (LOC338862), mRNA
XM_375031 Homo sapiens hypothetical gene supported by AK093158 (LOC400145), mR
XM_375032 Homo sapiens TBC1 domain family, member 4 (TBC1 D4), mRNA
XM_375033 Homo sapiens hypothetical protein LOC144776 (LOC144776), mRNA
XM_375035 Homo sapiens similar to 40S ribosomal protein S26 (LOC400156), mRNA
XM_375038 Homo sapiens hypothetical gene supported by AK129953 (LOC400165), mR
XM_375039 Homo sapiens similar to LRRGT00052 (LOC400169), mRNA
XM_375041 Homo sapiens similar to CLL-associated antigen KW-1 splice variant 1 (LOC
XM_375042 Homo sapiens DKFZP434B061 protein (DKFZP434B061), mRNA
XM_375045 Homo sapiens chromosome 14 open reading frame 92 (C14orf92), mRNA
XM_375065 Homo sapiens zinc finger protein 409 (ZNF409), mRNA
XM_375067 Homo sapiens similar to peroxisomal short-chain alcohol dehydrogenase; N/
XM_375074 Homo sapiens KIAA0391 (KIAA0391), mRNA
XM_375075 Homo sapiens similar to STELLA (LOC400206), mRNA
XM_375076 Homo sapiens hypothetical gene supported by AK124214 (LOC400207), mR
XM_375077 Homo sapiens chromosome 14 open reading frame 25 (C14orf25), mRNA
XM_375078 Homo sapiens similar to small acidic protein; small acidic protein sid2057p (L
XM_375080 Homo sapiens KIAA0831 (KIAA0831), mRNA
XM_375081 Homo sapiens hypothetical gene supported by BX248296 (LOC400214), mR
XM_375084 Homo sapiens similar to hypothetical protein (LOC400219), mRNA
XM_375085 Homo sapiens KIAA1393 (KI AA1393), mRNA
XM_375086 Homo sapiens zinc finger and BTB domain containing 1 (ZBTB1), mRNA
XM_375087 Homo sapiens pleckstrin homology domain containing, family H (with MyTH4
XM_375088 Homo sapiens hypothetical gene supported by AK097098 (LOC400223), mR
XM_375090 Homo sapiens similar to pleckstrin homology domain protein (5V327) (LOC4i
XM_375099 Homo sapiens hypothetical protein LOC283585 (LOC283585), mRNA
XM_375101 Homo sapiens similar to RIKEN cDNA 0610010D24 (LOC400239), mRNA
XM_375105 Homo sapiens KIAA0329 (KIAA0329), mRNA
XM_375108 Homo sapiens similar to RIKEN cDNA A530016L24 gene (LOC400258), mRI
XM_375145 Homo sapiens similar to breast cancer antigen NY-BR-1 (LOC400296), mRN
XM_375147 Homo sapiens similar to breast cancer anti-estrogen resistance 1 ; Crk-assoc
XM_375148 Homo sapiens similar to Ribosome biogenesis protein BMS1 homolog (LOΦ
XM_375150 Homo sapiens similar to protein kinase CHK2 isoform b; checkpoint-like proti
XM_375152 Homo sapiens similar to hypothetical protein (LOC400304), mRNA
XM_375153 Homo sapiens hypothetical protein DKFZp547L112 (DKFZP547L112), mRN/
XM_375154 Homo sapiens similar to KIAA0125 (LOC400309), mRNA
XM_375157 Homo sapiens similar to Ribosome biogenesis protein BMS1 homolog (LOΦ
XM_375163 Homo sapiens similar to hypothetical protein FLJ36144 (LOC400320), mRN/
XM_375165 Homo sapiens hypothetical protein DKFZp434P162 (LOC390535), mRNA
XM_375170 Homo sapiens similar to KIAA1971 protein (LOC400336), mRNA
XM_375171 Homo sapiens DKFZP434L187 protein (DKFZP434L187), mRNA
XM_375174 Homo sapiens hypothetical gene supported by BC037839 (LOC400340), mR
XM_375176 Homo sapiens similar to RIKEN cDNA 6530401 L14 gene (LOC400342), mRI
XM_375178 Homo sapiens similar to hect domain and RLD 2 (LOC400344), mRNA
XM_375179 Homo sapiens similar to hect domain and RLD 2 (LOC400345), mRNA
XM_375181 Homo sapiens KIAA1018 protein (KIAA1018), mRNA
XM_375183 Homo sapiens hypothetical gene supported by BC037839 (LOC400353), mR
XM_375185 Homo sapiens formin (limb deformity) (FMN), mRNA
XM_375187 Homo sapiens nuclear protein in testis (NUT), mRNA
XM_375190 Homo sapiens hypothetical gene supported by AK093014 (LOC400359), mR
XM_375191 Homo sapiens hypothetical gene supported by BX647708 (LOC400360), mR
XM_375196 Homo sapiens similar to KIAA0377 gene product (LOC400367), mRNA
XM_375200 Homo sapiens similar to ubiquitin specific proteinase 50 (LOC400372), mRN
XM_375203 Homo sapiens hypothetical gene supported by AK126787 (LOC400376), mR
XM_375207 Homo sapiens similar to RIKEN cDNA 1110004B15 (LOC400380), mRNA
XM_375209 Homo sapiens likely ortholog of mouse klotho lactase-phlorizin hydrolase reli
XM_375210 Homo sapiens similar to zinc finger and BTB domain containing 8; BTB/POZ
XM_375224 Homo sapiens similar to cervical cancer suppressor-1 (LOC400410), mRNA
XM_375226 Homo sapiens similar to FLJ40113 protein (LOC400414), mRNA
XM_375228 Homo sapiens similar to RIKEN cDNA 3010021 M21 (LOC400416), mRNA
XM_375230 Homo sapiens similar to KIAA1920 protein (LOC400418), mRNA
XM_375235 Homo sapiens similar to hypothetical protein FLJ22795 (LOC400423), mRN/
XM_375239 Homo sapiens similar to KIAA1920 protein (LOC400428), mRNA
XM_375240 Homo sapiens similar to hypothetical protein FLJ22795 (LOC400429), mRN/
XM_375242 Homo sapiens similar to golgi autoantigen golgin subfamily a2-like (LOC400
XM_375243 Homo sapiens KIAA1920 protein (KI AA1920), mRNA
XM_375246 Homo sapiens similar to hypothetical protein DKFZp434H 020 (LOC400435),
XM_375247 Homo sapiens KIAA0211 gene product (KIAA0211 ), mRNA
XM_375248 Homo sapiens similar to KIAA1920 protein (LOC400436), mRNA
XM_375249 Homo sapiens similar to hypothetical protein FLJ22795 (LOC400437), mRN/
XM_375252 Homo sapiens LOC400442 (LOC400442), mRNA
XM_375260 Homo sapiens hypothetical gene supported by AK075564; BC060873 (LOC4
XM_375261 Homo sapiens similar to Nonhistone chromosomal protein HMG-14 (High-mc
XM_375263 Homo sapiens similar to IFMQ9370 (LOC400454), mRNA
XM_375266 Homo sapiens LOC400461 (LOC400461), mRNA
XM_375268 Homo sapiens similar to RIKEN cDNA 3010021 M21 (LOC400464), mRNA
XM_375269 Homo sapiens similar to Dynamin-1 (D100) (Dynamin, brain) (B-dynamin) (L(
XM_375272 Homo sapiens similar to hypothetical protein FLJ36144 (LOC400468), mRN/
XM_375274 Homo sapiens similar to solute carrier family 22 member 4; organic cation tr.
XM_375275 Homo sapiens similar to hypothetical protein FLJ22795 (LOC400472), mRN/
XM_375276 Homo sapiens similar to KIAA1920 protein (LOC400473), mRNA
XM_375277 Homo sapiens similar to FLJ40113 protein (LOC400477), mRNA
XM_375280 Homo sapiens similar to KIAA1920 protein (LOC400478), mRNA
XM_375282 Homo sapiens similar to DDX11 protein (LOC400479), mRNA
XM_375284 Homo sapiens similar to interleukin 9 receptor (LOC400481), mRNA
XM_375288 Homo sapiens hypothetical protein MGC24381 (MGC24381), mRNA
XM_375292 Homo sapiens similar to hypothetical protein (LOC400492), mRNA
XM_375298 Homo sapiens KIAA1987 protein (KIAA1987), mRNA
XM_375302 Homo sapiens hypothetical gene supported by AK126539 (LOC400499), mR
XM_375305 Homo sapiens similar to TSG118.1 protein (LOC400506), mRNA
XM_375306 Homo sapiens similar to hypothetical protein FLJ20581 (LOC400507), mRN/
XM_375307 Homo sapiens similar to hypothetical protein (LOC400508), mRNA
XM_375308 Homo sapiens similar to FLJ12363 protein (LOC400509), mRNA
XM_375313 Homo sapiens hypothetical protein MGC9515 (MGC9515), mRNA
XM_375316 Homo sapiens hypothetical protein LOC283887 (LOC283887), mRNA
XM_375319 Homo sapiens similar to nuclear pore complex interacting protein (LOC4005'
XM_375320 Homo sapiens similar to apolipoprotein B48 receptor (LOC400514), mRNA
XM_375325 Homo sapiens similar to nuclear pore complex interacting protein (LOC4005'
XM_375330 Homo sapiens similar to MGC9515 protein (LOC400520), mRNA
XM_375331 Homo sapiens similar to PI-3-kinase-related kinase SMG-1 isoform 1 ; lambdi
XM_375333 Homo sapiens hypothetical protein FLJ25404 (FLJ25404), mRNA
XM_375334 Homo sapiens hypothetical protein LOC283901 (LOC283901), mRNA
XM_375341 Homo sapiens similar to Ig heavy chain - human (fragment) (LOC400524), m
XM_375344 Homo sapiens similar to hypothetical protein (LOC400526), mRNA
XM_375349 Homo sapiens similar to protein phosphatase 2A 48 kDa regulatory subunit ii
XM_375351 Homo sapiens similar to KIAA1501 protein (LOC400529), mRNA
XM_375352 Homo sapiens similar to KIAA1501 protein (LOC400530), mRNA
XM_375353 Homo sapiens hypothetical protein LOC146481 (LOC146481), mRNA
XM_375355 Homo sapiens Nedd4 binding protein 1 (N4BP1), mRNA
XM_375357 Homo sapiens similar to hypothetical protein (LOC400537), mRNA
XM_375358 Homo sapiens hypothetical protein FLJ25339 (FLJ25339), mRNA
XM_375359 Homo sapiens brain expressed, associated with Nedd4 (BEAN), mRNA
XM_375360 Homo sapiens hypothetical gene supported by AK130753 (LOC400539), mR
XM_375362 Homo sapiens solute carrier family 7 (cationic amino acid transporter, y+ sys
XM_375363 Homo sapiens similar to PI-3-kinase-related kinase SMG-1 isoform 2; lambdi
XM_375364 Homo sapiens similar to PI-3-kinase-related kinase SMG-1 isoform 2; lambdi
XM_375369 Homo sapiens similar to LHPE306 (LOC400546), mRNA
XM_375373 Homo sapiens similar to LOC93426 protein (LOC400547), mRNA
XM_375375 Homo sapiens KIAA0431 protein (KIAA0431), mRNA
XM_375376 Homo sapiens KIAA0703 gene product (KIAA0703), mRNA
XM_375377 Homo sapiens KIAA0513 gene product (KIAA0513), mRNA
XM_375378 Homo sapiens similar to hypothetical protein (LOC400549), mRNA
XM_375379 Homo sapiens hypothetical gene supported by AK127438 (LOC400555), mR
XM_375383 Homo sapiens hypothetical gene supported by AK126695 (LOC400559), mR
XM_375384 Homo sapiens similar to AFG3(ATPase family gene 3)-like 1 (LOC400563), r
XM_375386 Homo sapiens similar to bA476H5.3 (novel protein similar to septin) (LOC40ι
XM_375387 Homo sapiens hypothetical gene supported by AK128660 (LOC400566), mR
XM_375392 Homo sapiens similar to HSPC296 (LOC400569), mRNA
XM_375397 Homo sapiens KIAA0753 gene product (KIAA0753), mRNA
XM_375399 Homo sapiens similar to DNA segment, Chr 11 , Brigham & Womens Genetic
XM_375404 Homo sapiens hypothetical protein LOC146850 (LOC146850), mRNA
XM_375408 Homo sapiens KIAA0672 gene product (KIAA0672), mRNA
XM_375410 Homo sapiens hypothetical gene supported by AK123100 (LOC400574), mR
XM_375412 Homo sapiens hypothetical gene supported by AK127731 (LOC400576), mR
XM_375418 Homo sapiens similar to GRB2-related adaptor protein (LOC400581 ), mRNA
XM_375423 Homo sapiens similarto RIKEN cDNA 0610013E23 (LOC400585), mRNA
XM_375424 Homo sapiens similar to stearoyl-CoA desaturase; acyl-CoA desaturase; fatt
XM_375426 Homo sapiens similar to 60S ribosomal protein L21 (LOC400587), mRNA
XM_375430 Homo sapiens hypothetical protein LOC201229 (LOC201229), mRNA
XM_375434 Homo sapiens similar to Very hypothetical protein (LOC400590), mRNA
XM_375436 Homo sapiens hypothetical gene supported by AK126768 (LOC400591 ), mR
XM_375438 Homo sapiens similar to Hypothetical protein KIAA0563 (LOC400594), mRNi
XM_375439 Homo sapiens similar to TBC1 domain family member 3 (Rab GTPase-activa
XM_375443 Homo sapiens hypothetical protein LOC284100 (LOC284100), mRNA
XM_375446 Homo sapiens hypothetical protein FLJ11822 (FLJ11822), mRNA
XM_375449 Homo sapiens hypothetical protein LOC284106 (LOC284106), mRNA
XM_375452 Homo sapiens similar to keratin associated protein 9.2 (LOC400596), mRNA
XM_375453 Homo sapiens similar to RIKEN cDNA B830010L13 gene (LOC400597), mRI
XM_375456 Homo sapiens hypothetical protein DKFZp761G2113 (DKFZp761G2113), mF
XM_375469 Homo sapiens ProSAPiP2 protein (ProSAPiP2), mRNA
XM_375471 Homo sapiens KIAA0924 protein (KIAA0924), mRNA
XM_375475 Homo sapiens hypothetical gene supported by AK126318 (LOC400608), mR
XMJ375478 Homo sapiens similar to RIKEN cDNA 1100001 G20 (LOC400610), mRNA
XM_375482 Homo sapiens similar to U5 snRNP-specific protein, 200 kDa; U5 snRNP-sp«
XM_375484 Homo sapiens similar to adapter protein 162 (LOC400615), mRNA
XM_375485 Homo sapiens helicase with zinc finger domain (HELZ), mRNA
XM_375491 Homo sapiens similar to FTO protein (LOC400622), mRNA
XM_375492 Homo sapiens similar to Ammd protein (LOC400625), mRNA
XM_375494 Homo sapiens hypothetical gene supported by AK127919 (LOC400627), mR
XM_375495 Homo sapiens apoptosis-associated tyrosine kinase (AATK), mRNA
XM_375496 Homo sapiens similar to FLJ00403 protein (LOC400628), mRNA
XM_375500 Homo sapiens similar to RIKEN CDNA 3110023B02 (MGC16597), mRNA
XM_375502 Homo sapiens similar to RIKEN cDNA 4921530G04 (LOC400629), mRNA
XM_375511 Homo sapiens similar to TBC1 domain family member 3 (Rab GTPase-activε
XM 375514 Homo sapiens similar to pyrroline-5-carboxylate reductase 1 isoform 2; P5C
XM_375516 Homo sapiens hypothetical protein LOC284121 (LOC284121), mRNA
XM_375517 Homo sapiens similar to prolyl 4-hydroxylase, beta subunit; v-erb-a avian ery
XM_375520 Homo sapiens similar to HSPC214 (LOC400637), mRNA
XM_375527 Homo sapiens hypothetical protein LOC339290 (LOC339290), mRNA
XM_375537 Homo sapiens similar to thiopurine methyltransferase (LOC400650), mRNA
XMJ375543 Homo sapiens similar to 40S ribosomal protein S3a (LOC400652), mRNA
XM_375544 Homo sapiens hypothetical gene supported by AK126293 (LOC400658), mR
XM_375545 Homo sapiens hypothetical gene supported by AK126829 (LOC400661), mR
XM_375548 Homo sapiens similar to bA476l 15.3 (novel protein similar to septin) (LOC40I
XM_375549 Homo sapiens similar to hypothetical protein L0C349114 (LOC400664), mRI
XM_375550 Homo sapiens similar to Protein Sck (LOC400665), mRNA
XM_375551 Homo sapiens hypothetical gene supported by AK127589 (LOC400666), mR
XM_375552 Homo sapiens similar to GLGL782 (LOC400668), mRNA
XM_375553 Homo sapiens KIAA0963 (KIAA0963), mRNA
XM_375557 Homo sapiens NY-REN-24 antigen (NY-REN-24), mRNA
XM_375558 Homo sapiens KIAA1881 (KIAA1881), mRNA
XM_375559 Homo sapiens scaffold attachment factor B2 (SAFB2), mRNA
XM_375560 Homo sapiens similar to expressed sequence AI662250 (LOC400673), mRN
XM_375563 Homo sapiens hypothetical protein FLJ38149 (FLJ38149), mRNA
XM_375568 Homo sapiens ZFP-36 for a zinc finger protein (HSZFP36), mRNA
XM_375569 Homo sapiens hypothetical protein DKFZp434H610 (DKFZp434l1610), mRN
XM_375575 Homo sapiens similar to hypothetical protein (LOC400677), mRNA
XM_375583 Homo sapiens hypothetical gene supported by AK128109 (LOC400679), mR
XM_375589 Homo sapiens hypothetical gene supported by BC041864 (LOC400681), mR
XM_375590 Homo sapiens similar to zinc finger protein 100; zinc finger protein 100 (Y1 ) (
XM_375593 Homo sapiens zinc finger protein 536 (ZNF536), mRNA
XM_375594 Homo sapiens zinc finger protein 507 (ZNF507), mRNA
XM_375599 Homo sapiens hypothetical protein FLJ21369 (FLJ21369), mRNA
XM_375602 Homo sapiens hypothetical gene supported by AK055260 (LOC400687), mR
XM_375603 Homo sapiens similar to comment for location 3447-3655 BLASTX gi|10329C
XM_375604 Homo sapiens similar to Hypothetical zinc finger protein KIAA1559 (LOC400I
XM_375606 Homo sapiens hypothetical protein DKFZp7790175 (DKFZp7790175), mRN
XM_375608 Homo sapiens similar to hypothetical protein (LOC400692), mRNA
XM_375609 Homo sapiens similar to Hypothetical zinc finger protein KIAA0961 (LOC400I
XM_375614 Homo sapiens similar to Placental protein 13-like (Charcot-Leyden crystal pπ
XM_375618 Homo sapiens similar to pregnancy-specific beta-1 glycoprotein C1 - human
XM_375619 Homo sapiens similar to polycythemia rubra vera 1 ; cell surface receptor (LO
XM_375629 Homo sapiens hypothetical gene DKFZp434J0226 (DKFZp434J0226), mRN/
XM_375631 Homo sapiens hypothetical gene supported by AK124070 (LOC400707), mR
XM_375632 Homo sapiens similar to Serine/threonine protein phosphatase 5 (PP5) (Prot
XM_375633 Homo sapiens solute carrier family 8 (sodium-calcium exchanger), member .
XM_375634 Homo sapiens similar to sialic acid binding Ig-like Iectin 11 ; sialic acid-bindinj
XM_375638 Homo sapiens similar to zinc finger protein 534; KFRAB domain only 3 (LOC4ι
XM_375639 Homo sapiens similar to zinc finger protein KR-ZNF1 (LOC400714), mRNA
XM_375640 Homo sapiens similar to hypothetical protein MGC48625 (LOC400715), mRI^
XM_375646 Homo sapiens zinc finger protein 525 (ZNF525), mRNA
XM_375651 Homo sapiens KIAA1115 (KIAA1115), mRNA
XM_375654 Homo sapiens hypothetical protein FLJ35258 (FLJ35258), mRNA
XM_375655 Homo sapiens similar to hypothetical protein A430110N23 (LOC400717), mF
XM_375656 Homo sapiens similar to hypothetical protein A430110N23 (LOC400718), mF
XM_375658 Homo sapiens hypothetical gene supported by AK123294 (LOC400719), mR
XM_375660 Homo sapiens zinc finger protein 264 (ZNF264), mRNA
XM_375663 Homo sapiens similar to hypothetical protein FLJ23506 (LOC400720), mRN/
XM_375664 Homo sapiens similar to KIAA2003 protein (LOC400721 ), mRNA
XM_375665 Homo sapiens hypothetical protein BC012365 (LOC116412), mRNA
XM_375667 Homo sapiens similar to bA476H5.3 (novel protein similar to septin) (LOC40ι
XM_375668 Homo sapiens hypothetical gene supported by AK093729; AK128780; BX64'
XM_375669 Homo sapiens similar to 60S ribosomal protein L23a (LOC400725), mRNA
XM_375670 Homo sapiens hypothetical gene supported by AK093729; BC062355; BX64'
XM_375671 Homo sapiens similar to bA476H5.3 (novel protein similar to septin) (LOC40ι
XM_375678 Homo sapiens similar to KIAA1751 protein (LOC400731 ), mRNA
XM_375681 Homo sapiens KIAA0495 (KIAA0495), mRNA
XM_375682 Homo sapiens glycine-, glutamate-, thienylcyclohexylpiperidine-binding prote
XM_375684 Homo sapiens hairy and enhancer of split (Drosophila) homolog 2 (HES2), rr
XM_375685 Homo sapiens KIAA0469 gene product (KIAA0469), mRNA
XM_375687 Homo sapiens similar to RIKEN cDNA F730108M23 gene (LOC400734), mR
XM_375688 Homo sapiens similar to hypothetical protein (LOC400736), mRNA
XM_375690 Homo sapiens similar to RIKEN cDNA 9030409G11 (LOC400737), mRNA
XM_375695 Homo sapiens hypothetical protein LOC126917 (LOC126917), mRNA
XM_375696 Homo sapiens similar to ribosomal protein S14 (LOC400744), mRNA
XM_375697 Homo sapiens KIAA0459 protein (KIAA0459), mRNA
XM_375698 Homo sapiens hypothetical gene supported by AK124869 (LOC400745), mR
XM_375700 Homo sapiens similar to Hypothetical protein BC005730 (LOC400746), mRN
XM_375707 Homo sapiens hypothetical gene supported by AK054768 (LOC400747), mR
XM_375712 Homo sapiens syndecan 3 (N-syndecan) (SDC3), mRNA
XM_375713 Homo sapiens hypothetical protein LOC284551 (LOC284551), mRNA
XM_375714 Homo sapiens similar to RIKEN cDNA 1700025K23 (LOC400749), mRNA
XM_375718 Homo sapiens inositol polyphosphate-5-phosphatase, 75kDa (INPP5B), mRf
XM_375720 Homo sapiens regulating synaptic membrane exocytosis 3 (RIMS3), mRNA
XM_375726 Homo sapiens KIAA0494 gene product (KIAA0494), mRNA
XM_375729 Homo sapiens chromosome 1 open reading frame 34 (C1orf34), mRNA
XM_375732 Homo sapiens similar to Retrovirus-related Pol polyprotein from transposon .
XM_375737 Homo sapiens DnaJ (Hsp40) homolog, subfamily C, member 6 (DNAJC6), m
XM_375738 Homo sapiens hypothetical gene supported by BC047053 (LOC400757), mR
XM_375744 Homo sapiens hypothetical protein LOC339524 (LOC339524), mRNA
XM_375746 Homo sapiens similar to Interferon-induced guanylate-binding protein 1 (GTF
XM_375747 Homo sapiens similar to Interferon-induced guanylate-binding protein 1 (GTF
XM_375753 Homo sapiens hypothetical gene supported by AK092728 (LOC400765), mR
XM_375754 Homo sapiens hypothetical protein LOC163404 (LOC163404), mRNA
XM_375756 Homo sapiens similar to Stromal cell derived factor receptor 2 (LOC400766),
XM_375761 Homo sapiens similar to Matrin 3 (LOC400767), mRNA
XM_375762 Homo sapiens netrin G1 (NTNG1), mRNA
XM_375770 Homo sapiens leucine-rich repeats and immunoglobulin-like domains 2 (LRK
XM_375774 Homo sapiens similar to hypothetical protein DKFZp434A171 (LOC400773),
XM_375775 Homo sapiens similar to embigin (LOC400774), mRNA
XM_375779 Homo sapiens similar to hypothetical protein AE2 (LOC400776), mRNA
XM_375783 Homo sapiens hypothetical protein LOC149013 (LOC149013), mRNA
XM_375785 Homo sapiens similar to autoantigen La (LOC400779), mRNA
XM_375802 Homo sapiens hypothetical gene supported by BC014333 (LOC400784), mR
XM_375803 Homo sapiens similar to putative UST1-like organic anion transporter (LOC4I
XM_375806 Homo sapiens KIAA0476 gene product (KIAA0476), mRNA
XM_375809 Homo sapiens hypothetical protein LOC126669 (LOC126669), mRNA
XM_375810 Homo sapiens similar to glucocerebrosidase (LOC400787), mRNA
XM_375811 Homo sapiens similar to misato (LOC400788), mRNA
XM_375812 Homo sapiens KIAA0907 protein (KIAA0907), mRNA
XM_375814 Homo sapiens similar to LINE-1 REVERSE TRANSCRIPTASE HOMOLOG (I
XM_375816 Homo sapiens similar to Slamf7 protein (LOC400792), mRNA
XM_375821 Homo sapiens hypothetical gene supported by AK128015 (LOC400799), mR
XM_375825 Homo sapiens kinesin family member 14 (KIF14), mRNA
XM_375833 Homo sapiens hypothetical protein LOC284581 (LOC284581), mRNA
XM_375834 Homo sapiens inhibitor of kappa light polypeptide gene enhancer in B-cells, I
XM_375837 Homo sapiens KIAA0205 gene product (KIAA0205), mRNA
XM_375838 Homo sapiens hypothetical protein LOC128387 (LOC128387), mRNA
XM_375841 Homo sapiens hypothetical gene supported by AK128488 (LOC400804), mR
XM_375842 Homo sapiens similar to hypothetical protein LOC349114 (LOC400805), mRI
XM_375843 Homo sapiens similar to hypothetical protein FLJ25976 (LOC400806), mRN/
XM_375845 Homo sapiens similar to bA476H5.3 (novel protein similar to septin) (LOC40ι
XM_375846 Homo sapiens similar to hypothetical protein FLJ25976 (LOC400808), mRN/
XM_375848 Homo sapiens KIAA0792 gene product (KIAA0792), mRNA
XM_375849 Homo sapiens similar to Hypothetical protein CBG08611 (LOC400809), mRf'
XM_375850 Homo sapiens similar to Ferritin heavy chain (Ferritin H subunit) (LOC4008K
XM_375851 Homo sapiens KIAA0133 gene product (KIAA0133), mRNA
XM_375853 Homo sapiens protein BAP28 (FLJ 10359), mRNA
XM_375856 Homo sapiens similar to bA476H5.3 (novel protein similar to septin) (LOC40ι
XM_375863 Homo sapiens similar to 60S ribosomal protein L23a (LOC400814), mRNA
XM_375865 Homo sapiens similar to BC002216 protein (LOC400815), mRNA
XM_375869 Homo sapiens similar to LOC375080 protein (LOC400818), mRNA
XM_375873 Homo sapiens similar to KIAA0447 protein (LOC400820), mRNA
XM_375875 Homo sapiens similar to hypothetical protein LOC349114 (LOC400821), mRI
XM_375876 Homo sapiens similar to hypothetical protein FLJ25976 (LOC400822), mRN/
XM_375882 Homo sapiens similar to chromosome 14 open reading frame 24 (LOC40082
XM_375885 Homo sapiens similar to C219-reactive peptide (LOC400824), mRNA
XM_375887 Homo sapiens similar to Calcyclin (Prolactin receptor associated protein) (PF
XM_375897 Homo sapiens similar to AG1 (LOC400826), mRNA
XM_375899 Homo sapiens similar to Ba1-651 (LOC400827), mRNA
XM_375902 Homo sapiens similar to beta-defensin 32 precursor (LOC400830), mRNA
XM_375904 Homo sapiens similar to RIKEN cDNA 4933425020 (LOC400833), mRNA
XM_375911 Homo sapiens KIAA0186 gene product (KIAA0186), mRNA
XM_375912 Homo sapiens similar to hypothetical protein FLJ38374 (LOC400840), mRN/
XM_375914 Homo sapiens similar to dJ1184F4.4 (novel protein similar to nucleolar prote
XM_375917 Homo sapiens hypothetical protein LOC149692 (LOC149692), mRNA '
XM_375922 Homo sapiens similar to Protein C20orf85 (LOC400848), mRNA
XM_375925 Homo sapiens similar to cyclin-like F-box (3A784) (LOC400854), mRNA
XM_375928 Homo sapiens similar to hypothetical protein MGC30156 (LOC400855), mRf*
XM_375929 Homo sapiens hypothetical gene supported by AK123815 (LOC400856), mR
XM_375930 Homo sapiens similar to PRED3 (LOC400857), mRNA
XM_375931 Homo sapiens similar to PRED4 (LOC400858), mRNA
XM_375934 Homo sapiens similar to LINE-1 REVERSE TRANSCRIPTASE HOMOLOG (I
XM_375935 Homo sapiens hypothetical protein LOC284825 (LOC284825), mRNA
XM_375936 Homo sapiens chaperonin containing TCP1 , subunit 8 (theta) (CCT8), mRN/5
XM_375941 Homo sapiens hypothetical gene supported by AK127082 (LOC400867), mR
XM_375946 Homo sapiens hypothetical gene supported by AK124122 (LOC400878), mR
XM_375948 Homo sapiens similar to Serine/threonine-protein kinase Nek2 (NimA-related
XM_375951 Homo sapiens similar to Gene with similarity to rat kidney-specific (KS) gene
XM_375953 Homo sapiens hypothetical gene supported by AK129567; NM_2014O1 (LOC
XM_375954 Homo sapiens similar to LOC284861 protein (LOC400887), mRNA
XM_375955 Homo sapiens similar to immunoglobulin superfamily, member 3; immunoglo
XM_375958 Homo sapiens similar to proline dehydrogenase (oxidase) 1 ; proline oxidase
XM_375963 Homo sapiens similar to hypothetical protein LOC145497 (LOC400891 ), mRI
XM_375964 Homo sapiens similar to breakpoint cluster region isoform 1 (LOC400892), rr
XM_375965 Homo sapiens similar to Gamma-glutamyltranspeptidase 1 precursor (Gamrr
XM_375966 Homo sapiens similar to immunoglobulin superfamily, member 3; immunoglo
XM_375993 Homo sapiens similar to a putative protein with homology to a sequence betv
XM_375996 Homo sapiens similar to Gamma-glutamyltransferase-like protein 4 (LOC400
XM_375997 Homo sapiens similar to breakpoint cluster region isoform 1 (LOC400918), rr
XM_376001 Homo sapiens similar to low density lipoprotein receptor-related protein 5; lo\
XM_376003 Homo sapiens hypothetical gene supported by AK056895 (LOC400924), mR
XM_376007 Homo sapiens KIAA0645 gene product (KIAA0645), mRNA
XM_376008 Homo sapiens hypothetical protein LOC91464 (LOC91464), mRNA
XM_376010 Homo sapiens similar to TPTE and PTEN homologous inositol lipid phosphat
XM_376013 Homo sapiens hypothetical protein LOC200321 (LOC200321), mRNA
XM_376018 Homo sapiens KIAA1644 protein (KIAA1644), mRNA
XM_376019 Homo sapiens similar to hypothetical protein (LOC400930), mRNA
XM_376020 Homo sapiens hypothetical gene supported by AK130875 (LOC400931 ), mR
XM_376021 Homo sapiens hypothetical gene supported by AK128136 (LOC400932), mR
XM_376022 Homo sapiens hypothetical gene supported by AK126356 (LOC400934), mR
XM_376023 Homo sapiens zinc finger, BED domain containing 4 (ZBED4), mRNA
XM_376024 Homo sapiens similar to interleukin 17 receptor E; EST AA589509 (LOC400ξ
XM_376031 Homo sapiens hypothetical gene supported by AK123041 (LOC400940), mR
XM_376032 Homo sapiens hypothetical gene supported by AK124409 (LOC400941), mR
XM_376033 Homo sapiens hypothetical protein LOC339789 (LOC339789), mRNA
XM_376034 Homo sapiens similar to AILT5830 (LOC400943), mRNA
XM_376043 Homo sapiens similar to RIKEN cDNA 2310016E02 (LOC400948), mRNA
XM_376044 Homo sapiens SPTF-associated factor 65 gamma (STAF65(gamma)), mRN/
XM_376048 Homo sapiens similar to FKSG60 (LOC400949), mRNA
XM_376049 Homo sapiens hypothetical gene supported by AK124893 (LOC400950), mR
XM_376051 Homo sapiens similar to NGNL6975 (LOC400952), mRNA
XM_376056 Homo sapiens similar to echinoderm microtubule associated protein like 5 (L
XM_376059 Homo sapiens SERTA domain containing 2 (SERTAD2), mRNA
XM_376060 Homo sapiens KIAA0053 gene product (KIAA0053), mRNA
XM_376062 Homo sapiens similar to KIAA1155 protein (LOC400961), mRNA
XM_376068 Homo sapiens similar to LP3727 (LOC400962), mRNA
XM_376072 Homo sapiens hypothetical gene supported by AK098018 (LOC400965), mR
XM_376073 Homo sapiens similar to Ig kappa variable region (LOC400967), mRNA
XM_376074 Homo sapiens LOC400968 (LOC400968), mRNA
XM_376094 Homo sapiens hypothetical protein LOC90499 (LOC90499), mRNA
XM_376097 Homo sapiens similar to seven transmembrane helix receptor (LOC400984),
XM_376099 Homo sapiens similar to Glycerol-3-phosphate acyltransferase, mitochondria
XM_376100 Homo sapiens similar to KIAA1641 protein (LOC400986), mRNA
XM_376101 Homo sapiens similar to LOC375251 protein (LOC400987), mRNA
XM_376106 Homo sapiens similar to RIKEN cDNA 6330578E17 (LOC400989), mRNA
XM_376108 Homo sapiens similar to Sodium/hydrogen exchanger 4 (Na(+)/H(+) exchang
XM_376111 Homo sapiens plasminogen-related protein A (LOC285189), mRNA
XM_376112 Homo sapiens similar to RAN-binding protein 2-like 1 isoform 1 ; sperm meml
XM_376117 Homo sapiens hypothetical gene supported by AK095987 (LOC400994), mR
XM_376118 Homo sapiens hypothetical gene supported by AK095987 (LOC400996), mR
XM_376121 Homo sapiens hypothetical gene supported by AK095987 (LOC400998), mR
XM_376125 Homo sapiens similar to Single-stranded DNA-binding protein, isoform b (LO
XM_376126 Homo sapiens similar to hypothetical protein A230046P18 (LOC401003), mF
XM_376127 Homo sapiens similar to pote protein; Expressed in prostate, ovary, testis, ar
XM_376130 Homo sapiens similar to pote protein; Expressed in prostate, ovary, testis, ar
XM_376139 Homo sapiens hypothetical gene supported by AK123815 (LOC401011), mR
XM_376141 Homo sapiens similar to zinc finger protein 285 (LOC401012), mRNA
XM_376142 Homo sapiens hypothetical gene supported by AK057980; AK092189 (LOC4
XM_376144 Homo sapiens hypothetical protein LOC339745 (LOC339745), mRNA
XM_376148 Homo sapiens similar to RIKEN cDNA 5830415L20 (LOC401015), mRNA
XM_376150 Homo sapiens similar to ribosomal protein S3a; 40S ribosomal protein S3a; \
XM_376154 Homo sapiens similar to ribosomal protein S15; rat insulinoma gene (LOC40
XM_376158 Homo sapiens hypothetical gene supported by AK126104; BX648733 (LOC4
XM_376160 Homo sapiens similar to CDNA sequence BC030440 (LOC401026), mRNA
XM_376161 Homo sapiens similar to IIDS6411 (LOC401027), mRNA
XM_376162 Homo sapiens similar to Nucleophosmin 1 (LOC401028), mRNA
XM_376165 Homo sapiens similar to DAZ associated protein 2; deleted in azoospermia a
XM_376171 Homo sapiens KIAA1843 protein (KIAA1843), mRNA
XM_376172 Homo sapiens zinc finger protein 142 (clone pHZ-49) (ZNF142), mRNA
XM_376178 Homo sapiens thyroid hormone receptor interactor 12 (TRIP12), mRNA
XM_376179 Homo sapiens LOC401034 (LOC401034), mRNA
XM_376180 Homo sapiens similar to archease (LOC401035), mRNA
XM_376185 Homo sapiens similar to ankyrin repeat and SOCS box-containing 18; SOCS
XM_376186 Homo sapiens hypothetical protein LOC93463 (LOC93463), mRNA
XM_376189 Homo sapiens DKFZP586K1520 protein (DKFZP586K1520), mRNA
XM_376190 Homo sapiens hypothetical gene supported by AK125867 (LOC401039), mR
XM_376191 Homo sapiens hypothetical gene supported by AK127861 (LOC401040), mR
XM_376193 Homo sapiens FERM, RhoGEF and pleckstrin domain protein 2 (FARP2), ml
XM_376195 Homo sapiens hypothetical gene supported by AK123321 (LOC401045), mR
XM_376200 Homo sapiens similar to inhibitor of growth family, member 5 (LOC401047), i
XM_376201 Homo sapiens ER degradation enhancer, mannosidase alpha-like 1 (EDEM1
XM_376203 Homo sapiens KIAA0218 gene product (KIAA0218), mRNA
XM_376206 Homo sapiens hypothetical protein LOC285375 (LOC285375), mRNA
XM_376207 Homo sapiens similar to FLJ00274 protein (LOC401054), mRNA
XM_376209 Homo sapiens similar to Nonhistone chromosomal protein HMG-17 (High-mc
XM_376212 Homo sapiens hypothetical protein LOC339862 (LOC339862), mRNA
XM_376225 Homo sapiens similar to serine protease-like 1 (LOC401063), mRNA
XM_376227 Homo sapiens hypothetical gene supported by AK097724 (LOC401064), mR
XM_376232 Homo sapiens Vpr-binding protein (VprBP), mRNA
XM_376233 Homo sapiens similar to hypothetical protein MGC39725 (LOC401067), mRI
XM_376238 Homo sapiens hypothetical protein LOC285331 (LOC285331), mRNA
XM_376239 Homo sapiens similar to FtsJ homolog 2 isoform b; cell division protein FtsJ;
XM_376241 Homo sapiens hypothetical gene supported by AK125779 (LOC401070), mR
XM_376243 Homo sapiens hypothetical gene supported by AK125942 (LOC401072), mR
XM_376247 Homo sapiens similar to double homeobox protein (LOC401074), mRNA
XM_376248 Homo sapiens similar to zinc finger protein 39 (LOC401075), mRNA
XM_376249 Homo sapiens similar to CAP, adenylate cyclase-associated protein 1 ; aden}
XM_376254 Homo sapiens hypothetical protein DKFZp667G2110 (DKFZp667G2110), mF
XM_376256 Homo sapiens hypothetical gene supported by AK126064 (LOC401080), mR
XM_376257 Homo sapiens similarto hypothetical protein FLJ25976 (LOC401082), mRN/
XM_376258 Homo sapiens similar to Nuclear transcription factor Y subunit beta (NF-Y pr<
XM_376267 Homo sapiens similar to hypothetical protein, MNCb-4779 (LOC401087), mR
XM_376268 Homo sapiens hypothetical gene supported by AK127796 (LOC401088), mR
XM_376269 Homo sapiens hypothetical gene supported by AK125319 (LOC401089), mR
XM_376278 Homo sapiens similar to RIKEN cDNA 0610027B03 (LOC401095), mRNA
XM_376280 Homo sapiens hypothetical protein BC010062 (LOC152078), mRNA
XM_376281 Homo sapiens hypothetical gene supported by BC031660 (LOC401097), mR
XM_376284 Homo sapiens hypothetical protein BC011266 (LOC93556), mRNA
XM_376287 Homo sapiens hypothetical gene supported by AK128090 (LOC401100), mR
XM_376290 Homo sapiens hypothetical gene supported by AK124384 (LOC401105), mR
XM_376292 Homo sapiens hypothetical gene supported by AK129507 (LOC401109), mR
XM_376299 Homo sapiens hypothetical gene supported by AK093135 (LOC401114), mR
XM_376300 Homo sapiens hypothetical gene supported by AK124538 (LOC401116), mR
XM_376301 Homo sapiens similar to chromosome 11 open reading frame2; chromosome
XM_376303 Homo sapiens hypothetical protein LOC285484 (LOC285484), mRNA
XM_376305 Homo sapiens similar to deubiquitinating enzyme 3 (LOC401121), mRNA
XM_376306 Homo sapiens similar to hypothetical protein LOC169270 (LOC401122), mRI
XM_376307 Homo sapiens hypothetical protein DKFZp667E0512 (DKFZp667E0512), mF
XM_376309 Homo sapiens hypothetical protein LOC285540 (LOC285540), mRNA
XM_376310 Homo sapiens zinc finger, CCHC domain containing 4 (ZCCHC4), mRNA
XM_376312 Homo sapiens hypothetical gene supported by AK127623 (LOC401123), mR
XM_376314 Homo sapiens similar to TBC1 domain family member 1 (LOC401125), mRN
XM_376317 Homo sapiens similar to SNAG1 (LOC401130), mRNA
XM_376318 Homo sapiens similar to hypothetical protein FLJ30672 (LOC401132), mRN/
XM_376320 Homo sapiens similar to RIKEN cDNA 9930019B18 gene (LOC401136), mRI
XM_376322 Homo sapiens similar to hypothetical protein (LOC401137), mRNA
XM_376323 Homo sapiens similar to RSTI689 (LOC401138), mRNA
XM_376324 Homo sapiens similarto Ameloblastin precursor (LOC401139), mRNA
XM_376325 Homo sapiens hypothetical protein FLJ13105 (FLJ13105), mRNA
XM_376327 Homo sapiens similar to hypothetical protein LOC231503 (LOC401141), mRI
XM_376328 Homo sapiens family with sequence similarity 13, member A1 (FAM13A1), m
XM_376331 Homo sapiens KIAA1680 protein (KIAA1680), mRNA
XM_376333 Homo sapiens similar to elongation factor 1 alpha (LOC401146), mRNA
XM_376334 Homo sapiens similar to hypothetical protein (LOC401147), mRNA
XM_376338 Homo sapiens hypothetical gene supported by AK127273 (LOC401150), mR
XM_376339 Homo sapiens similar to RIKEN cDNA 1810037117 (LOC401152), mRNA
XM_376342 Homo sapiens similarto bA291L22.2 (similarto CDC10 (cell division cycle 1(
XM_376347 Homo sapiens hypothetical gene supported by AK126441 (LOC401157), mR
XM_376348 Homo sapiens similar to hypothetical protein (L1H 3 region) - human (LOC4C
XM_376349 Homo sapiens hypothetical protein LOC201725 (LOC201725), mRNA
XM_376350 Homo sapiens PDZ domain containing guanine nucleotide exchange factor ('
XM_376353 Homo sapiens similar to hypothetical protein FLJ20035 (LOC401160), mRN/
XM_376354 Homo sapiens similar to unactive progesterone receptor, 23 kD; likely ortholc
XM_376355 Homo sapiens KIAA0626 gene product (KIAA0626), mRNA
XM_376364 Homo sapiens hypothetical gene supported by AK126844 (LOC401166), mR
XM_376365 Homo sapiens hypothetical protein BC014011 (LOC116349), mRNA
XM_376366 Homo sapiens DKFZP564I1171 protein (DKFZP564I1171 ), mRNA
XM_376368 Homo sapiens similar to Programmed cell death protein 6 (Probable calcium'
XM_376370 Homo sapiens hypothetical gene supported by AK090679 (LOC401172), mR
XM_376371 Homo sapiens similar to hypothetical protein FLJ36144 (LOC401174), mRN/
XM_376372 Homo sapiens hypothetical protein LOC134121 (LOC134121), mRNA
XM_376376 Homo sapiens similar to SMA3 protein (LOC401179), mRNA
XM_376379 Homo sapiens similar to RIKEN CDNA 4921505C17 (LOC401183), mRNA
XM_376383 Homo sapiens similar to cAMP-specific phosphodiesterase PDE4D7 (LOC40
XM_376386 Homo sapiens similar to DNA segment, Chr 13, Brigham & Womens Genetic
XM_376387 Homo sapiens hypothetical gene supported by AK127903 (LOC401191), mR
XM_376389 Homo sapiens similar to small EDRK-rich factor 1A, telomeric; spinal muscul
XM_376391 Homo sapiens similar to psi neuronal apoptosis inhibitory protein (LOC4011£
XM_376394 Homo sapiens hypothetical gene supported by AK130705 (LOC401195), mR
XM_376395 Homo sapiens similar to POM121 membrane glycoprotein-like 1 (LOC40119I
XM_376397 Homo sapiens hypothetical protein LOC153561 (LOC1 53561), mRNA
XM_376403 Homo sapiens similar to ribosomal protein L7-like 1 (LOC401197), mRNA
XM_376405 Homo sapiens Rho-guanine nucleotide exchange factor (RGNEF), mRNA
XM_376412 Homo sapiens similar to KIAA0825 protein (LOC401202), mRNA
XM_376413 Homo sapiens hypothetical protein DKFZp564C0469 (DKFZp564C0469), mF
XM_376416 Homo sapiens similar to Beta-glucuronidase precursor (Beta-G1) (LOC4012C
XM_376419 Homo sapiens hypothetical protein LOC285638 (LOC285638), mRNA
XM_376420 Homo sapiens similar to 40S ribosomal protein S25 (LOC401206), mRNA
XM_376423 Homo sapiens hypothetical gene supported by AK126569 (LOC401207), mR
XM_376427 Homo sapiens LOC401208 (LOC401208), mRNA
XM_376428 Homo sapiens similar to HYPOTHETICAL PROTEIN ORF-1137 (LOC40120J
XM_376430 Homo sapiens similar to nuclear receptor coactivator 4; RET-activating gene
XM_376433 Homo sapiens hypothetical protein LOC153218 (LOC1 53218), mRNA
XM_376436 Homo sapiens hypothetical protein LOC134466 (LOC1 34466), mRNA
XM_376440 Homo sapiens hypothetical protein LOC285629 (LOC285629), mRNA
XM_376443 Homo sapiens hypothetical gene supported by AK097772 (LOC401217), mR
XM_376444 Homo sapiens hypothetical protein LOC133491 (LOC1 33491), mRNA
XM_376447 Homo sapiens similar to hypothetical protein (LOC401221 ), mRNA
XM_376453 Homo sapiens similar to KIAA0752 protein (LOC401223), mRNA
XM_376454 Homo sapiens similar to acetoacetyl-CoA synthetase; acetoacetate-CoA liga
XM_376458 Homo sapiens hypothetical gene supported by AK093729; AK128780; BX64'
XM_376461 Homo sapiens similar to bA476H5.3 (novel protein similar to septin) (LOC40
XM_376463 Homo sapiens hypothetical protein MGC39372 (MGC39372), mRNA
XM_376464 Homo sapiens similar to HIV TAT specific factor 1 ; cofactor required for Tat ε
XM_376469 Homo sapiens hypothetical gene supported by AK026805 (LOC401236), mR
XM_376471 Homo sapiens similar to chromosome 15 open reading frame 2 (LOC401238
XM_376472 Homo sapiens similar to K1AA0319 (LOC401239), mRNA
XM_376473 Homo sapiens similar to SMA3-like protein bA239L20.1 (LOC401240), mRN/
XM_376474 Homo sapiens integral membrane glycoprotein-like (LOC166994), mRNA
XM_376479 Homo sapiens mediator of DNA damage checkpoint 1 (MDC1), mRNA
XM_376480 Homo sapiens hypothetical gene supported by AK098O12 (LOC401247), mR
XM_376486 Homo sapiens similar to coiled-coil domain 1 protein precursor (LOC401250)
XM_376487 Homo sapiens similar to NG23 (LOC401251), mRNA
XM_376488 Homo sapiens hypothetical gene supported by AK123889 (LOC401252), mR
XM_376491 Homo sapiens hypothetical gene supported by AK125740 (LOC401253), mR
XM_376498 Homo sapiens similar to kinesin-related protein 3A (LOC401259), mRNA
XM_376499 Homo sapiens hypothetical gene supported by AK123643 (LOC401260), mR
XM_376503 Homo sapiens ectonucleotide pyrophosphatase/phosphodiesterase 4 (putatr
XM_376505 Homo sapiens similar to hypothetical protein FLJ30296 (LOC401263), mRN/
XM_376508 Homo sapiens hypothetical gene supported by AK091177 (LOC401265), mR
XM_376516 Homo sapiens myosin VI (MY06), mRNA
XM_376518 Homo sapiens chromosome 6 open reading frame 84 (C6orf84), mRNA
XM_376519 Homo sapiens ankyrin repeat domain 6 (ANKRD6), mRNA
XM_376522 Homo sapiens similar to Heat shock protein 67B2 (LOC401270), mRNA
XM_376525 Homo sapiens zinc finger protein 450 (ZNF450), mRNA
XM_376527 Homo sapiens hypothetical gene supported by AK124171 (LOC401271), mR
XM_376532 Homo sapiens similar to KIAA0408 protein (LOC401272), mRNA
XM_376533 Homo sapiens similar to FLJ44670 protein (LOC401273), mRNA
XM_376535 Homo sapiens chromosome 6 open reading frame 207 (C6orf207), mRNA
XM_376536 Homo sapiens similar to hypothetical protein (LOC401274), mRNA
XM_376537 Homo sapiens BCL2-associated transcription factor 1 (BCLAF1), mRNA
XM_376540 Homo sapiens chromosome 6 open reading frame 56 (C6orf56), mRNA
XM_376541 Homo sapiens hypothetical gene supported by AK126903 (LOC401278), mR
XM_376547 Homo sapiens RNA binding motif protein 16 (RBM16), mRNA
XM_376549 Homo sapiens hypothetical gene supported by AK125637 (LOC401280), mR
XM_376550 Homo sapiens KIAA1423 (KIAA1423), mRNA
XM_376554 Homo sapiens similar to T-complex protein 10A homolog (LOC401285), mRf
XM_376555 Homo sapiens hypothetical gene supported by AK127120 (LOC401286), mR
XM_376556 Homo sapiens chromosome 6 open reading frame 70 (C6orf70), mRNA
XM_376557 Homo sapiens hypothetical gene supported by AK056013 (LOC401288), mR
XM_376558 Homo sapiens hypothetical gene supported by AK127120 (LOC401293), mR
XM_376560 Homo sapiens hypothetical gene supported by AK125637 (LOC401295), mR
XM_376564 Homo sapiens unc-84 homolog A (C. elegans) (UNC84A), mRNA
XM_376565 Homo sapiens hypothetical gene supported by BC031661 (LOC401298), mR
XM_376566 Homo sapiens hypothetical protein LOC285924 (LOC285924), mRNA
XM_376567 Homo sapiens KIAA1856 protein (KIAA1 856), mRNA
XM_376568 Homo sapiens hypothetical gene supported by AK125308 (LOC401300), mR
XM_376569 Homo sapiens hypothetical gene supported by AK123535 (LOC401302), mR
XM_376571 Homo sapiens ubiquitin specific protease 42 (USP42), mRNA
XM_376573 Homo sapiens similar to beta-1 ,4-mannosyltransferase; beta-1 ,4 mannosyltr;
XM_376575 Homo sapiens hypothetical gene supported by AK027125 (LOC401307), mR
XM_376576 Homo sapiens similar to Chain , Heat-Shock Cognate 70kd Protein (44kd Atf
XM_376577 Homo sapiens similar to heat shock 70kDa protein 8 isoform 2; heat shock o
XM_376578 Homo sapiens PHD finger protein 14 (PHF14), mRNA
XM_376585 Homo sapiens similar to Dual specificity protein kinase CLK2 (CDC like kinas
XM_376586 Homo sapiens similarto Hypothetical protein KIAA0087 (HA1002) (LOC4013
XM_376587 Homo sapiens similar to mKIAAO038 protein (LOC401316), mRNA
XM_376588 Homo sapiens KIAA0644 gene product (KIAA0644), mRNA
XM_376589 Homo sapiens KIAA0241 protein (KIAA0241), mRNA
XM_376590 Homo sapiens LOC89231 (LOC89231 ), mRNA
XM_376591 Homo sapiens similar to KIAA0877 protein (LOC401322), mRNA
XM_376593 Homo sapiens similar to RIKEN cDNA 9330128H10 gene (LOC401323), mR
XM_376595 Homo sapiens hypothetical gene supported by AF447883 (LOC401325), mR
XM_376597 Homo sapiens similar to sequence-specific single-stranded-DNA-binding pro
XM_376598 Homo sapiens similar to t-complex 1 ; T-complex locus TCP-1 ; t-complex 1 (a
XM_376600 Homo sapiens similar to RAS p21 protein activator 4; GTPase activating prot
XM_376602 Homo sapiens similar to Ras GTPase-activating protein 4 (RasGAP-activatin
XM_376604 Homo sapiens similar to cell division cycle 10 homolog (LOC401332), mRNA
XM_376605 Homo sapiens similar to hypothetical protein FLJ25976 (LOC401333), mRN/
XM_376607 Homo sapiens hypothetical gene supported by AK126096 (LOC401335), mR
XM_376609 Homo sapiens growth factor receptor-bound protein 10 (GRB10), mRNA
XM_376610 Homo sapiens hypothetical gene supported by AK097404; NM_198284 (LOC
XM_376611 Homo sapiens hypothetical gene supported by AK127870 (LOC401337), mR
XM_376612 Homo sapiens similar to SMT3 suppressor of mif two 3 homolog 2 (LOC4012
XM_376613 Homo sapiens similarto hypothetical protein DKFZp434F142 (LOC401341),
XM_376614 Homo sapiens LOC401350 (LOC40135O), mRNA
XM_376615 Homo sapiens hypothetical gene supported by BC040831 (LOC401351), mR
XM_376616 Homo sapiens hypothetical gene supported by BC040831 (LOC401354), mR
XM_376617 Homo sapiens similar to BC060615 protein (LOC401355), mRNA
XM_376618 Homo sapiens similar to CAGL79 (LOC401356), mRNA
XM_376619 Homo sapiens similar to hypothetical protein LOC285908 (LOC401357), mRI
XM_376621 Homo sapiens similar to hypothetical protein FLJ25037 (LOC401360), mRN/
XM_376622 Homo sapiens similar to hypothetical protein MGC16733 similar to CG1 113
XM_376623 Homo sapiens similar to MGC16733 protein (LOC401362), mRNA
XM_376625 Homo sapiens similar to hypothetical protein FLJ10900 (LOC401369), mRN/
XM_376626 Homo sapiens similar to Williams Beuren syndrome chromosome region 19 I
XM_376628 Homo sapiens similar to Neutrophil cytosol factor 1 (NCF-1) (Neutrophil NAD
XM_376629 Homo sapiens similar to transcription factor GTF2IRD2 (LOC401375), mRNA
XM_376630 Homo sapiens similar to Nuclear envelope pore membrane protein POM 121
XM_376631 Homo sapiens Williams Beuren syndrome chromosome region 24 (WBSCRΣ
XM_376636 Homo sapiens similar to PMS4 (LOC401378), mRNA
XM_376638 Homo sapiens similar to PMS4 (LOC401379), mRNA
XM_376639 Homo sapiens similar to Williams Beuren syndrome chromosome region 19 I
XM_376640 Homo sapiens similar to PMS4 homolog mismatch repair protein - human (L(
XM_376642 Homo sapiens tripartite motif-containing 50B (TRIM50B), mRNA
XM_376643 Homo sapiens similar to hypothetical protein LOC285908 (LOC401383), mRI
XM_376647 Homo sapiens sema domain, immunoglobulin domain (Ig), short basic doma
XM_376648 Homo sapiens similar to hypothetical protein 4932412H11 (LOC401387), mF
XM_376649 Homo sapiens hypothetical protein FLJ39885 (FLJ39885), mRNA
XM_376651 Homo sapiens hypothetical gene supported by AK124274 (LOC401388), mR
XM_376652 Homo sapiens distal-less homeo box 6 (DLX6), mRNA
XM_376653 Homo sapiens similar to chromosome 11 open reading frame2; chromosome
XM_376655 Homo sapiens similar to importin alpha 1 b (LOC401391), mRNA
XM_376656 Homo sapiens hypothetical protein FLJ22037 (FLJ22037), mRNA
XM_376657 Homo sapiens hypothetical protein LOC285989 (LOC285989), mRNA
XM_376658 Homo sapiens similar to Zinc-alpha-2-glycoprotein precursor (Zn-alpha-2-gly
XM_376663 Homo sapiens similar to reverse transcriptase related protein (LOC401395),
XM_376664 Homo sapiens KIAA1218 protein (KIAA1218), mRNA
XM_376665 Homo sapiens hypothetical protein LOC286009 (LOC286009), mRNA
XM_376668 Homo sapiens similar to Serine/threonine-protein kinase tousled-like 2 (Tous
XM_376670 Homo sapiens similar to hypothetical protein FLJ25976 (LOC401402), mRN/
XM_376671 Homo sapiens coatomer protein complex, subunit gamma 2 (COPG2), mRN/
XM_376672 Homo sapiens similar to ribosomal protein S14 (LOC401404), mRNA
XM_376677 Homo sapiens hypothetical protein LOC155006 (LOC155006), mRNA
XM_376679 Homo sapiens hypothetical protein FLJ25778 (FLJ25778), mRNA
XM_376680 Homo sapiens KIAA1718 protein (KIAA1718), mRNA
XM_376681 Homo sapiens similar to RAB19, member RAS oncogene family (LOC40140!
XM_376683 Homo sapiens LCHN protein (LCHN), mRNA
XM_376684 Homo sapiens hypothetical protein LOC93432 (LOC93432), mRNA
XM_376707 Homo sapiens similar to KIAA0738 protein (LOC401426), mRNA
XM_376712 Homo sapiens FLJ43692 protein (FLJ43692), mRNA
XM_376713 Homo sapiens similar to Olfactory receptor 2A7 (LOC401427), mRNA
XM_376715 Homo sapiens similar to seven transmembrane helix receptor (LOC401428),
XM_376716 Homo sapiens similar to KIAA1285 protein (LOC401429), mRNA
XM_376717 Homo sapiens likely ortholog of mouse zinc finger protein EZI (EZI), mRNA
XM_376718 Homo sapiens FLJ45737 protein (FLJ45737), mRNA
XM_376719 Homo sapiens similar to KIAA2036 protein (LOC401430), mRNA
XM_376720 Homo sapiens KIAA0543 protein (KIAA0543), mRNA
XM_376722 Homo sapiens hypothetical protein LOC155036 (LOC155036), mRNA
XM_376724 Homo sapiens KIAA1402 protein (CSGIcA-T), mRNA
XM_376725 Homo sapiens hypothetical gene supported by AK127717 (LOC401433), mR
XM_376727 Homo sapiens hypothetical protein LOC285888 (LOC285888), mRNA
XM_376728 Homo sapiens hypothetical protein LOC155435 (LOC155435), mRNA
XM_376730 Homo sapiens ubiquitin-protein isopeptide ligase (E3) (KIAA0010), mRNA
XM_376736 Homo sapiens similar to Hypothetical protein KIAA0711 (LOC401444), mRN;
XM_376741 Homo sapiens similar to ubiquitin-specific protease 17-like protein (LOC4014
XM_376746 Homo sapiens similar to seven transmembrane helix receptor (LOC401450),
XM_376750 Homo sapiens similar to chromosome 11 open reading frame2; chromosome
XM_376754 Homo sapiens similar to deubiquitinating enzyme 3 (LOC401453), mRNA
XM_376756 Homo sapiens similar to hypothetical protein SB153 isoform 1 (LOC401454),
XM_376757 Homo sapiens similar to KIAA1456 protein (LOC401455), mRNA
XM_376761 Homo sapiens hypothetical gene supported by BC062364; BX647289 (LOC4
XM_376763 Homo sapiens similar to TRANSCRIPTION FACTOR COE2 (EARLY B-CELL
XM_376764 Homo sapiens paraneoplastic antigen MA2 (PNMA2), mRNA
XM_376771 Homo sapiens hypothetical gene supported by AK128232 (LOC401459), mR
XM_376774 Homo sapiens similar to hypothetical protein 4932417K07 (LOC401460), mR
XM_376776 Homo sapiens thymus high mobility group box protein TOX (TOX), mRNA
XM_376780 Homo sapiens similar to Myelin P2 protein (LOC401465), mRNA
XM_376781 Homo sapiens solute carrier family 10 (sodium/bile acid cotransporter family)
XM_376783 Homo sapiens hypothetical gene supported by BC055092 (LOC401466), mR
XM_376784 Homo sapiens similar to prot GOR (LOC401467), mRNA
XM_376785 Homo sapiens similar to NFS1 nitrogen fixation 1 isoform b precursor; cysteii
XM_376786 Homo sapiens similar to Ceruloplasmin precursor (Ferroxidase) (LOC40146ξ
XM_376787 Homo sapiens similar to 40S ribosomal protein S26 (LOC401470), mRNA
XM_376791 Homo sapiens hypothetical gene supported by AK127183 (LOC401472), mR
XM_376793 Homo sapiens similar to RIKEN cDNA A830094I09 gene (LOC401474), mRh
XM_376795 Homo sapiens hypothetical gene supported by AK127771 (LOC401478), mR
XM_376797 Homo sapiens similar to KIAA0870 protein (LOC401479), mRNA
XM_376800 Homo sapiens similar to Hypothetical zinc finger protein KIAA0628 (LOC401-
XM_376801 Homo sapiens hypothetical gene supported by AK091211 ; AK125852 (LOC4
XM_376809 Homo sapiens similar to bA110H4.2 (similar to membrane protein) (LOC401'<
XM_376810 Homo sapiens similar to dJ28l24.1.2 (Spinal Muscular Atrophy region (SMA3
XM_376814 Homo sapiens similar to DDX11 protein (LOC401487), mRNA
XM_376819 Homo sapiens similar to RIKEN cDNA 4933428I03 (LOC401494), mRNA
XM_376821 Homo sapiens chromosome 9 open reading frame 14 (C9orf14), mRNA
XM_376822 Homo sapiens similar to PR02738 (LOC401497), mRNA
XM_376824 Homo sapiens similarto RIKEN cDNA A930001M12 gene (LOC401498), mR
XM_376829 Homo sapiens hypothetical protein LOC158381 (LOC158381), mRNA
XM_376830 Homo sapiens KIAA0258 (KIAA0258), mRNA
XM_376833 Homo sapiens LOC401505 (LOC401505), mRNA
XM__376838 Homo sapiens hypothetical gene supported by AK127145 (LOC401507), mR
XM_376840 Homo sapiens hypothetical gene supported by AK127145 (LOC401508), mR
XM_376841 Homo sapiens similar to DKFZP572C163 protein (LOC401509), mRNA
XM_376843 Homo sapiens similar to LOC286286 protein (LOC401510), mRNA
XM_376846 Homo sapiens similar to DKFZP572C163 protein (LOC401511 ), mRNA
XM_376847 Homo sapiens hypothetical gene supported by AK124538 (LOC401512), mR
XM_376848 Homo sapiens hypothetical gene supported by AK124538 (LOC401514), mR
XM_376850 Homo sapiens hypothetical gene supported by AK124122 (LOC401515), mR
XM_376852 Homo sapiens similarto bA251017.3 (similar to aquaporin 7) (LOC401516),
XM_376855 Homo sapiens similar to keratinocyte growth factor-like protein, group III - hu
XM_376858 Homo sapiens similar to breast cancer antigen NY-BR-1 (LOC401519), mRN
XM_376861 Homo sapiens LOC401520 (LOC401520), mRNA
XM__376863 Homo sapiens similar to Keratinocyte growth factor precursor (KGF) (Fibrobl;
XMJ376866 Homo sapiens similar to bA251017.3 (similar to aquaporin 7) (LOC401524),
XM_376869 Homo sapiens similar to tumor suppressor deleted in oral cancer-related 1 (L
XMJ376872 Homo sapiens LOC401529 (LOC401529), mRNA
XM_376874 Homo sapiens chromosome 9 open reading frame 71 (C9orf71), mRNA
XM_376876 Homo sapiens similar to coiled-coil-helix-coiled-coil-helix domain containing . '
XMJ376880 Homo sapiens hypothetical gene supported by AK127390 (LOC401534), mR
XM_376885 Homo sapiens hypothetical gene supported by AK127445 (LOC401535), mR
XM_376888 Homo sapiens similar to Laminin receptor 1 (LOC401537), mRNA
XM_376890 Homo sapiens similar to CG14980-PB (LOC375601), mRNA
XM_376892 Homo sapiens similar to hypothetical protein (LOC401541), mRNA
XM_376895 Homo sapiens zinc finger protein 510 (ZNF510), mRNA
XM_376898 Homo sapiens thioredoxin domain containing 4 (endoplasmic reticulum) (TXf
XM_376899 Homo sapiens similar to RIKEN cDNA D630039A03 gene (LOC401546), mR
XM__376900 Homo sapiens similar to KIAA0563 gene product (LOC401547), mRNA
XM_376902 Homo sapiens similar to RIKEN cDNA 4732481 H14 (LOC401548), mRNA
XM_376903 Homo sapiens KIAA0674 protein (KIAA0674), mRNA
XM_376905 Homo sapiens EGF-like-domain, multiple 5 (EGFL5), mRNA
XM_376909 Homo sapiens similar to hypothetical protein FLJ25955 (LOC401551), mRN/
XM_376917 Homo sapiens far upstream element (FUSE) binding protein 3 (FUBP3), mRI
XM_376921 Homo sapiens hypothetical protein MGC43306 (MGC43306), mRNA
XM_376924 Homo sapiens chromosome 9 open reading frame 62 (C9oιϊ62), mRNA
XM_376925 Homo sapiens similar to DNL zinc finger (3D41) (LOC401560), mRNA
XM_376930 Homo sapiens hypothetical gene supported by AK127160 (LOC401562), mR
XM_376931 Homo sapiens hypothetical gene supported by AK093587; AK124899 (LOC4
XM_376939 Homo sapiens similar to 4931415M17 protein (LOC401565), mRNA
XM_376947 Homo sapiens similar to surfeit 5 isoform b; surfeit locus protein 5 (LOC4015
XM_376949 Homo sapiens similar to Surfeit locus protein 4 (LOC401567), mRNA
XM_376950 Homo sapiens similar to MGC43306 protein (LOC401568), mRNA
XM_376960 Homo sapiens similar to RIKEN cDNA 2900002H16 (LOC401569), mRNA
XM_376965 Homo sapiens LOC401570 (LOC401570), mRNA
XM_376968 Homo sapiens hypothetical gene supported by AK124122 (LOC401572), mR
XM_376978 Homo sapiens similar to hypothetical protein FLJ33610 (LOC401576), mRN/
XM_376981 Homo sapiens similar to hypothetical protein (L1 H 3 region) - human (LOC4C
XM_376986 Homo sapiens similar to Syntenin 1 (Syndecan binding protein 1 ) (Melanoms
XM_376989 Homo sapiens similar to dJ54B20.4 (novel KRAB box containing C2H2 type ;
XM_377000 Homo sapiens hypothetical gene supported by AK096379 (LOC401589), mR
XM_377002 Homo sapiens hypothetical gene supported by AK096379 (LOC401590), mR
XM_377012 Homo sapiens similar to Spindlin-like protein 2 (SPIN-2) (LOC401591), mRN
XM_377014 Homo sapiens Cdc42 guanine nucleotide exchange factor (GEF) 9 (ARHGEF
XM_377018 Homo sapiens LOC401594 (LOC401594), mRNA
XM_377019 Homo sapiens LOC401595 (LOC401595), mRNA
XM_377024 Homo sapiens similar to adaptor-related protein complex 2, beta 1 subunit; a
XM_377025 Homo sapiens similar to adaptor-related protein complex 2, beta 1 subunit; b
XM_377026 Homo sapiens similar to adaptor-related protein complex 2, beta 1 subunit; b
XM_377027 Homo sapiens similar to heat shock 70kD protein binding protein; progestero
XM_377028 Homo sapiens similar to histone acetyltransferase (LOC401606), mRNA
XM_377031 Homo sapiens similar to nuclear RNA export factor 2; TAP like protein 2 (LO(
XM_377032 Homo sapiens G protein-coupled receptor-associated sorting protein (GASP]
XM_377033 Homo sapiens similar to STELLA (LOC401611), mRNA
XM_377034 Homo sapiens similar to mitochondrial carrier triple repeat 1 (LOC401612), π
XM_377041 Homo sapiens similar to hypothetical protein MGC15416 (LOC401618), mRf
XM_377053 Homo sapiens hypothetical gene supported by BC040297 (LOC401619), mR
XM_377060 Homo sapiens hypothetical protein LOC203547 (LOC203547), mRNA
XM_377062 Homo sapiens similar to LINE-1 REVERSE TRANSCRIPTASE HOMOLOG (I
XM_377071 Homo sapiens similar to LINE-1 REVERSE TRANSCRIPTASE HOMOLOG (I
XM_377072 Homo sapiens similar to LINE-1 REVERSE TRANSCRIPTASE HOMOLOG (I
XM_377073 Homo sapiens similar to CXYorfl -related protein (LOC401624), mRNA
XM_377076 Homo sapiens hypothetical gene supported by AK097803; BC017239 (LOC4
XM_377087 Homo sapiens similar to hypothetical protein FLJ33610 (LOC401627), mRN/
XM_377097 Homo sapiens similar to RNA binding motif protein, Y-linked, family 1 (LOC4I
XM_377098 Homo sapiens similar to Transcript Y 6 protein (LOC401633), mRNA
XM_377102 Homo sapiens LOC401634 (LOC401634), mRNA
XM_377104 Homo sapiens LOC401635 (LOC401635), mRNA
XM_377109 Homo sapiens similar to 40S ribosomal protein SA (P40) (34/67 kDa laminin
XM_377110 Homo sapiens similar to peptidylprolyl isomerase A (cyclophilin A) (LOC4016
XM_377115 Homo sapiens similar to PRED65 (LOC401641 ), mRNA
XM_377117 Homo sapiens similar to bA182L21.1 (novel protein similar to hypothetical pn
XM_377122 Homo sapiens similar to Guanine nucleotide-binding protein G(i), alpha-2 sul
XM_377129 Homo sapiens similar to golgi autoantigen, golgin subfamily a, 7 (LOC40164'
XM_377133 Homo sapiens similar to deleted in malignant brain tumors 1 ; crp-ductin; vorr
XM_377136 Homo sapiens similar to double homeobox protein (LOC401650), mRNA
XM_377137 Homo sapiens similar to double homeobox protein (LOC401651), mRNA
XM_377140 Homo sapiens similar to double homeobox protein (LOC401652), mRNA
XM_377142 Homo sapiens similar to double homeobox protein (LOC401653), mRNA
XM_377143 Homo sapiens similar to double homeobox protein (LOC401654), mRNA
XM_377147 Homo sapiens similar to RIKEN cDNA 1500011 L16 (LOC401660), mRNA
XM_377154 Homo sapiens similar to seven transmembrane helix receptor (LOC401661 ),
XM_377155 Homo sapiens similar to Olfactory receptor 51 H1 (LOC401663), mRNA
XM_377156 Homo sapiens similar to Olfactory receptor 51T1 (LOC401665), mRNA
XM_377158 Homo sapiens similar to Olfactory receptor 51A4 (LOC401666), mRNA
XM_377159 Homo sapiens similar to Olfactory receptor 51A2 (LOC401667), mRNA
XM_377179 Homo sapiens similar to seven transmembrane helix receptor (LOC401675),
XM_377185 Homo sapiens similar to filamin-binding LIM protein-1 ; migfilin (LOC401679),
XM_377189 Homo sapiens similar to Metabotropic glutamate receptor 5 precursor (mGlul
XM_377200 Homo sapiens similar to seven transmembrane helix receptor (LOC401687),
XM_377218 Homo sapiens similar to MGC15937 protein (LOC401694), mRNA
XM_377222 Homo sapiens similar to seven transmembrane helix receptor (LOC401696),
XM_377230 Homo sapiens similar to U2af1 protein (LOC401703), mRNA
XM_377231 Homo sapiens similar to heterogeneous nuclear ribonucleoprotein C isoform
XM_377240 Homo sapiens similar to peptidylprolyl isomerase A (cyclophilin A) (LOC4017
XM_377259 Homo sapiens similar to S-phase kinase-associated protein 1A isoform a; or<
XM_377262 Homo sapiens similar to Hypothetical protein CBG01854 (LOC401716), mRI*
XM_377265 Homo sapiens similar to fidgetin (LOC401720), mRNA
XM_377278 Homo sapiens similar to ribosomal protein L6 (LOC401725), mRNA
XM_377283 Homo sapiens similar to Heat shock cognate 71 kDa protein (LOC401726), r
XM_377285 Homo sapiens similar to 60S ribosomal protein L11 (LOC401727), mRNA
XM_377287 Homo sapiens similar to MALI 3P1.296 (LOC401728), mRNA
XM_377296 Homo sapiens similar to Hypothetical protein CBG23155 (LOC401732), mR
XM_377305 Homo sapiens similar to Hypothetical protein CBG01089 (LOC401740), mRf*
XM_377306 Homo sapiens similar to vav-1 interacting Kruppel-like protein isoform b (LOC
XM_377337 Homo sapiens similar to cerebellin (LOC401766), mRNA
XM_377338 Homo sapiens KIAA0323 (KIAA0323), mRNA
XM_377343 Homo sapiens similar to hypothetical protein DJ667H12.2 (LOC401778), mR
XM_377355 Homo sapiens KIAA0602 protein (KIAA0602), mRNA
XM_377369 Homo sapiens similar to hypothetical protein FLJ36144 (LOC401806), mRN/
XM_377374 Homo sapiens similar to hypothetical protein FLJ36144 (LOC401808), mRN/
XM_377376 Homo sapiens similar to hypothetical protein FLJ36144 (LOC401811), mRN/
XM_377377 Homo sapiens similar to LRRGT00052 (LOC401812), mRNA
XM_377383 Homo sapiens similar to stereocilin (LOC401815), mRNA
XM_377388 Homo sapiens similar to antigen Cs44 (LOC401819), mRNA
XM_377390 Homo sapiens similar to LOC375757 protein (LOC401820), mRNA
XM_377394 Homo sapiens similar to melanoma chondroitin sulfate proteoglycan (LOC40
XM_377395 Homo sapiens similar to MKI67 (FHA domain) interacting nucleolar phosphoi
XM_377407 Homo sapiens similar to hypothetical protein 4732467B22 (LOC401827), mF
XM_377408 Homo sapiens similar to mast cell protease-11 (LOC401828), mRNA
XM_377412 Homo sapiens similar to 3-phosphoinositide dependent protein kinase-1 (hPl
XM_377414 Homo sapiens si ilar to Group X secretory phospholipase A2 precursor (Phe
XM_377416 Homo sapiens similar to hypothetical protein MGC33867 (LOC401833), mRh
XM_377420 Homo sapiens similar to hypothetical protein BC011981 (LOC401837), mRN,
XM_377423 Homo sapiens similar to Immunoglobulin heavy chain (LOC401841), mRNA
XM_377424 Homo sapiens similar to Ig H-chain V-region (DP-40) (LOC401842), mRNA
XM_377425 Homo sapiens similar to protein kinase related to Raf protein kinases; Metho
XM_377426 Homo sapiens similar to IGHV gene product (LOC401845), mRNA
XM_377429 Homo sapiens similar to IGHV gene product (LOC401846), mRNA
XM_377439 Homo sapiens similar to hypothetical protein FLJ36144 (LOC401858), mRN/
XM_377444 Homo sapiens similar to peptidylprolyl isomerase A (cyclophilin A) (LOC401 £
XM_377445 Homo sapiens similar to RIKEN cDNA C330003B14 (LOC401860), mRNA
XM_377446 Homo sapiens similar to RIKEN cDNA C330003B14 (LOC401861), mRNA
XM_377447 Homo sapiens similarto ribosomal protein S1 (LOC401862), mRNA
XM_377451 Homo sapiens LOC401867 (LOC401867), mRNA
XM_377455 Homo sapiens LOC401868 (LOC401868), mRNA
XM_377457 Homo sapiens similar to 40S RIBOSOMAL PROTEIN S12 (LOC401870), mF
XM_377464 Homo sapiens suppressor of Ty 6 homolog (S. cerevisiae) (SUPT6H), mRN/1
XM_377475 Homo sapiens similar to CDNA sequence BC004853 (LOC401883), mRNA
XM_377476 Homo sapiens similar to Arf2-prov protein (LOC401884), mRNA
XM_377477 Homo sapiens similar to 60S ribosomal protein L21 (LOC401885), mRNA
XM_377480 Homo sapiens similar to 60S ribosomal protein L17 (L23) (Amino acid starva
XM_377488 Homo sapiens similar to bA526D8.2 (novel protein similar to KIAA1074) (LO(
XM_377496 Homo sapiens similar to hypothetical protein (LOC401894), mRNA
XM_377498 Homo sapiens KIAA0863 protein (KIAA0863), mRNA
XM_377500 Homo sapiens similar to ribosomal protein S15; rat insulinoma gene (LOC40
XM_377506 Homo sapiens egf-like module containing, mucin-like, hormone receptor-like
XM_377511 Homo sapiens similar to 60S ribosomal protein L10 (QM protein homolog) (L
XM_377512 Homo sapiens similar to glyceraldehyde 3-phosphate dehydrogenase (LOC4
XM_377514 Homo sapiens similarto hypothetical protein FLJ38281 (LOC401898), mRN/
XM_377515 Homo sapiens similar to zinc finger protein 433 (LOC401899), mRNA
XM_377516 Homo sapiens similar to ribosomal protein L28; 60S ribosomal protein L28 (L
XM_377521 Homo sapiens similar to 60S ribosomal protein L23a (LOC401904), mRNA
XM_377522 Homo sapiens similar to fibroblast growth factor receptor 3 (LOC401907), mF
XM_377527 Homo sapiens similar to 60S ribosomal protein L29 (Cell surface heparin bin*
XM_377529 Homo sapiens similar to Zinc finger protein 345 (Zinc finger protein HZF10) (
XM_377533 Homo sapiens similar to Pregnancy-specific beta-1 -glycoprotein 4 precursor
XM_377537 Homo sapiens similar to OPA3 protein; Optic atrophy 3 (Iraqi-Jewish optic ati
XMJ377538 Homo sapiens similar to Mucin 4 (Tracheobronchial mucin) (LOC401923), m
XM__377553 Homo sapiens similar to zinc finger protein (LOC401932), mRNA
XM_377554 Homo sapiens similar to hypothetical protein MGC4734 (LOC401933), mRN/
XM_377555 Homo sapiens similar to RIKEN cDNA 5830442J12 (LOC401934), mRNA
XM_377556 Homo sapiens similar to Hypothetical protein CBG06524 (LOC401936), mRf*
XM_377558 Homo sapiens similar to elongation factor 1 delta (LOC401937), mRNA
XM_377563 Homo sapiens similar to Hypothetical protein DJ845024.1 (LOC401938), mF
XM_377565 Homo sapiens similar to Hypothetical protein DJ845024.1 (LOC401939), F
XM_377566 Homo sapiens similar to dJ845024.1 (Melanoma Preferentially Expressed Ai
XM_377568 Homo sapiens similar to hypothetical protein (LOC401941 ), mRNA
XM_377570 Homo sapiens similarto Hypothetical protein DJ845024.1 (LOC401942), mF
XMJ377577 Homo sapiens hypothetical gene supported by AK127275 (LOC401944), mR
XM_377579 Homo sapiens similar to clCK0721 Q.2 (60S Ribosomal Protein L12 LIKE pro
XM_377580 Homo sapiens similar to DC2 protein (LOC401946), mRNA
XM_377583 Homo sapiens similar to ribosomal protein L27 (LOC401947), mRNA
XM_377585 Homo sapiens similar to receptor tyrosine phosphatase (LOC401948), mRN/
XM_377586 Homo sapiens similar to MGC52970 protein (LOC401949), mRNA
XM_377593 Homo sapiens similar to dJ612B15.1 (novel protein similar to 60S ribosomal
XM_377594 Homo sapiens similar to sporulation-induced transcript 4-associated protein !
XM_377595 Homo sapiens similar to beta-actin (LOC401956), mRNA
XM_377597 Homo sapiens similar to glyceraldehyde 3-phosphate dehydrogenase (LOC4
XM_377599 Homo sapiens similar to hypothetical protein MGC8902 (LOC401962), mRN/
XM_377600 Homo sapiens similar to hypothetical protein MGC8902 (LOC401963), mRN/
XM_377601 Homo sapiens similar to AG3 (LOC401964), mRNA
XM_377611 Homo sapiens similar to Olfactory receptor 10J6 (LOC401973), mRNA
XM_377613 Homo sapiens similar to ribosomal protein S2; 40S ribosomal protein S2 (LO
XM_377618 Homo sapiens LOC401976 (LOC401976), mRNA
XM_377620 Homo sapiens similar to 14.5 kDa translational inhibitor protein (p14.5) (UK1
XM_377630 Homo sapiens similar to RIKEN cDNA 2610020C11 (LOC401983), mRNA
XM_377631 Homo sapiens similar to beta actin (LOC401987), mRNA
XM_377633 Homo sapiens similar to Olfactory receptor 2G2 (LOC401990), mRNA
XM_377635 Homo sapiens similar to CDC-like kinase 3; cdc2/CDC28-like protein kinase
XM_377643 Homo sapiens similar to Olfactory receptor 2T2 (LOC401992), mRNA
XM_377644 Homo sapiens similar to Olfactory receptor 2T5 (LOC401993), mRNA
XM_377645 Homo sapiens similar to seven transmembrane helix receptor (LOC401994),
XM_377649 Homo sapiens similar to 14.5 kDa translational inhibitor protein (p14.5) (UK1
XM_377655 Homo sapiens similarto seven transmembrane helix receptor (LOC401997),
XM_377657 Homo sapiens similar to Olfactory receptor 2T5 (LOC401998), mRNA
XM_377658 Homo sapiens similar to seven transmembrane helix receptor (LOC401999),
XM_377659 Homo sapiens similar to Olfactory receptor 2T11 (LOC402000), mRNA
XM_377660 Homo sapiens similar to seven transmembrane helix receptor (LOC402001 ),
XM_377661 Homo sapiens similar to Olfactory receptor 5BF1 (LOC402002), mRNA
XM_377662 Homo sapiens similar to Olfactory receptor 2T4 (LOC402003), mRNA
XM_377663 Homo sapiens similar to Olfactory receptor 2T1 (Olfactory receptor 1-25) (OF
XM_377664 Homo sapiens similar to Olfactory receptor 2T2 (LOC402006), mRNA
XM_377665 Homo sapiens similar to Olfactory receptor 2T3 (LOC402007), mRNA
XM_377666 Homo sapiens similar to Olfactory receptor 2T5 (LOC402008), mRNA
XM_377668 Homo sapiens similar to hypothetical protein F830045P16 (LOC402009), mF
XM_377675 Homo sapiens similar to dJ1187J4.2 (novel protein similar to rat RYF3) (LOC
XM_377687 Homo sapiens similar to hypothetical protein FLJ21347 (LOC402027), mRN/
XM_377690 Homo sapiens similar to extensin-like protein (LOC402030), mRNA
XM_377691 Homo sapiens LOC402O32 (LOC402032), mRNA
XM_377694 Homo sapiens similar to LOC284861 protein (LOC402034), mRNA
XM_377695 Homo sapiens similar to LOC284861 protein (LOC402035), mRNA
XM_377696 Homo sapiens similar to carbonic anhydrase XV (LOC402036), mRNA
XM_377700 Homo sapiens similar to LOC284861 protein (LOC402038), mRNA
XM_377713 Homo sapiens similar to SRR1-like protein (LOC402055), mRNA
XM_377715 Homo sapiens similar to Small nuclear ribonucleoprotein associated protein I
XM_377716 Homo sapiens similar to 40S ribosomal protein S17 (LOC402057), mRNA
XM_377717 Homo sapiens similar to dJ1119A7.3 (PUTATIVE novel protein similar to HP!
XM_377720 Homo sapiens hypothetical protein BC012882 (LOC150356), mRNA
XM_377721 Homo sapiens similar to Hypothetical protein CBG23588 (LOC402064), mRf*
XM_377725 Homo sapiens similar to LWamide neuropeptide precursor protein (LOC402C
XM_377728 Homo sapiens similar to Hypothetical protein CBG08601 (LOC402067), mRt*
XM_377732 Homo sapiens similar to egg envelope component ZPAX (LOC402068), mRI*
XM_377741 Homo sapiens similar to SPCPB16A4.07c (LOC402072), mRNA
XM_377742 Homo sapiens KIAA1940 protein (KIAA1940), mRNA
XM_377751 Homo sapiens similar to Ig kappa chain V region (Z3) - human (LOC402089)
XM_377754 Homo sapiens LOC402090 (LOC402090), mRNA
XM_377755 Homo sapiens similar to hypothetical protein (LOC402094), mRNA
XM_377756 Homo sapiens similar to dJ908M14.1.3 (ribosomal protein S21 , isoform 3) (L
XM_377760 Homo sapiens similar to ribosomal protein L22 (LOC402098), mRNA
XM_377761 Homo sapiens similar to ribosomal protein L22 (LOC402100), mRNA
XM_377766 Homo sapiens similar to hypothetical protein DKFZp434P0316 (LOC402103)
XM_377768 Homo sapiens similar to hypothetical protein FLJ10462 (LOC402104), mRN/
XM_377771 Homo sapiens similar to 60S ribosomal protein L6 (TAX-responsive enhance
XM_377774 Homo sapiens kinesin family member 5C (KIF5C), mRNA
XM_377776 Homo sapiens similar to Acidic ribosomal phosphoprotein PO (LOC402109),
XM_377778 Homo sapiens LOC402110 (LOC402110), mRNA
XM_377783 Homo sapiens similar to RIKEN cDNA A930041G11 gene (LOC402117), mR
XM_377786 Homo sapiens similar to ribosomal protein L23 (LOC402120), mRNA
XM_377797 Homo sapiens similar to laminin receptor-like protein LAMRL5 (LOC402123),
XM_377803 Homo sapiens similar to RIKEN cDNA 1700112C13 (LOC402128), mRNA
XM_377809 Homo sapiens similar to Olfactory receptor 5K2 (LOC402135), mRNA
XM_377811 Homo sapiens similar to transcription factor INI (LOC402136), mRNA
XM_377815 Homo sapiens similar to chromosome 11 open reading frame2; chromosome
XM_377818 Homo sapiens similar to mesenchymal stem cell protein DSC92; neurite outc
XM_377820 Homo sapiens similar to ribosomal protein L7-like 1 (LOC402152), mRNA
XM_377823 Homo sapiens similar to p53 apoptosis effector related to Pmp22; p53 apopti
XM_377824 Homo sapiens simi lar to Kinesin-like protein KIF3A (Microtubule plus end-din XM_377828 Homo sapiens simi lar to GLP_171_8870_6279 (LOC402160), mRNA XM_377829 Homo sapiens simi lar to hypothetical protein MGC45871 (LOC402161), mR XM_377830 Homo sapiens simi lar to deubiquitinating enzyme 3 (LOC402164), mRNA XM_377831 Homo sapiens simi lar to deubiquitinating enzyme 3 (LOC402165), mRNA XM_377832 Homo sapiens simi lar to deubiquitinating enzyme 3 (LOC402166), mRNA XM_377834 Homo sapiens simi lar to deubiquitinating enzyme 3 (LOC402167), mRNA XM_377835 Homo sapiens simi lar to deubiquitinating enzyme 3 (LOC402168), mRNA XM_377836 Homo sapiens simi lar to deubiquitinating enzyme 3 (LOC402169), mRNA XM_377837 Homo sapiens simi lar to deubiquitinating enzyme 3 (LOC402170), mRNA XM_377841 Homo sapiens simi lar to 60S ribosomal protein L21 (LOC402176), mRNA XM_377845 Homo sapiens simi lar to ribosomal protein S21 ; 40S ribosomal protein S21 (I XM_377847 Homo sapiens simi lar to ras-related C3 botulinum toxin substrate 1 isoform F XM_377849 Homo sapiens simi lar to alanyl trna synthetase (LOC402188), mRNA XM_377861 Homo sapiens simi lar to glycine-rich protein (LOC402194), mRNA XM_377875 Homo sapiens simi lar to Transcription initiation factor TFIID 28 kDa subunit XM_377877 Homo sapiens simi lar to Transcription initiation factor TFIID 28 kDa subunit XM_377878 Homo sapiens simi lar to Transcription initiation factor TFIID 28 kDa subunit XM_377879 Homo sapiens simi lar to Transcription initiation factor TFIID 28 kDa subunit XM_377880 Homo sapiens simi lar to Transcription initiation factor TFIID 28 kDa subunit XM_377881 Homo sapiens simi lar to Transcription initiation factor TFIID 28 kDa subunit XM_377882 Homo sapiens simi lar to Transcription initiation factor TFIID 28 kDa subunit XM_377883 Homo sapiens simi lar to Transcription initiation factor TFIID 28 kDa subunit XM_377884 Homo sapiens simi lar to Transcription initiation factor TFIID 28 kDa subunit XM_377885 Homo sapiens simi lar to Transcription initiation factor TFIID 28 kDa subunit XM_377886 Homo sapiens simi lar to Transcription initiation factor TFIID 28 kDa subunit XM_377887 Homo sapiens simi lar to Transcription initiation factor TFIID 28 kDa subunit XM_377889 Homo sapiens simi lar to Transcription initiation factor TFIID 28 kDa subunit XM_377896 Homo sapiens simi lar to ribosomal protein L13a; 60S ribosomal protein L13a XM_377904 Homo sapiens simi lar to cytoplasmic beta-actin (LOC402218), mRNA XM_377907 Homo sapiens simi lar to beta-glucuronidase (LOC402223), mRNA XM_377910 Homo sapiens simi lar to RIKEN cDNA A730017C20 (LOC402228), mRNA XM_377911 Homo sapiens simi lar to hypothetical protein E230025N22 (LOC402231 ), mF XM_377912 Homo sapiens KIAA01 94 protein (KIAA0194), mRNA XM_377918 Homo sapiens simi lar to Ad 147 (LOC402235), mRNA XM_377919 Homo sapiens simi lar to RIKEN CDNA 2310040C09 (LOC402237), mRNA XM_377920 Homo sapiens simi lar to Selenophosphate synthetase 1 (LOC402238), mRN XM_377924 Homo sapiens simi lar to Olfactory receptor 4F3 (LOC402242), mRNA XM_377925 Homo sapiens simi lar to hypothetical protein FLJ37300 (LOC402244), mRN/ XM_377926 Homo sapiens simi lar to mesenchymal stem cell protein DSC92; neurite outc XM_377927 Homo sapiens simi lar to olfactory receptor MOR267-3 (LOC402246), mRNA XM_377928 Homo sapiens simi lar to Calgizzarin (S100C protein) (MLN 70) (LOC402247) XM_377929 Homo sapiens simi lar to ribosomal protein L31 (LOC402248), mRNA XM_377931 Homo sapiens simi lar to olfactory receptor MOR145-2 (LOC402249), mRNA XM_377932 Homo sapiens simi lar to 4930579E17Rik protein (LOC402250), mRNA XM_377933 Homo sapiens simi lar to MGC76216 protein (LOC402251), mRNA XM_377934 Homo sapiens simi lar to peptidylprolyl isomerase A (LOC402252), mRNA XM_377935 Homo sapiens simi lar to FLJ40113 protein (LOC402253), mRNA XM_377938 Homo sapiens simi lar to ELK1 (LOC402257), mRNA XM_377941 Homo sapiens simi lar to equilibrative nucleoside transporter 4; hENT4 (LOC' XM_377942 Homo sapiens simi lar to vomeronasal receptor V1 RC3 (LOC402273), mRNA XM_377943 Homo sapiens simi lar to Opioid binding protein/cell adhesion molecule precu XM_377944 Homo sapiens simi lar to protein kinase related to Raf protein kinases; Metho XM_377945 Homo sapiens simi lar to metabotropic glutamate receptor 8; G protein-couple XM_377946 Homo sapiens simi lar to GTF2I repeat domain containing 1 isoform 2; Williar XM_377947 Homo sapiens simi lar to opposite strand transcription unit to Stag3; Gats pro XM 377949 Homo sapiens simi lar to GrpE protein homolog 1 , mitochondrial precursor (fV
XM_377950 Homo sapiens similar to GTF2I repeat domain containing 1 isoform 2; Williar
XM_377951 Homo sapiens similar to peptidylprolyl isomerase A (LOC402284), mRNA
XM_377955 Homo sapiens hypothetical protein DKFZP434A0225 (DKFZP434A0225), mF
XM_377956 Homo sapiens similar to RIKEN cDNA 4930511M11 (LOC402286), mRNA
XM_377957 Homo sapiens similar to ribosomal protein S3a; 40S ribosomal protein S3a; \
XM_377958 Homo sapiens similar to Ser/Thr protein kinase PAR-1 Balpha (LOC402289),
XM_377959 Homo sapiens similar to Nuclear protein Hcc-1 (HSPC316) (Proliferation ass*
XM_377961 Homo sapiens similar to opposite strand transcription unit to Stag3; Gats pro
XM_377962 Homo sapiens postmeiotic segregation increased 2-like 1 (PMS2L1), mRNA
XM_377964 Homo sapiens similar to 40S RIBOSOMAL PROTEIN SA (P40) (34/67 KD U
XM_377969 Homo sapiens similar to 60S ribosomal protein L23a (LOC402294), mRNA
XM_377970 Homo sapiens similar to Argininosuccinate synthase (Citrulline-aspartate lig.
XM_377972 Homo sapiens similar to Triosephosphate isomerase (TIM) (LOC402298), ml
XM_377973 Homo sapiens similar to aldo-keto reductase family 1 , member B10; aldose r
XM_377974 Homo sapiens similar to beta-tubulin (LOC402300), mRNA
XM_377976 Homo sapiens similar to Nucleoside diphosphate kinase, mitochondrial preci
XM_377995 Homo sapiens similar to Olfactory receptor 2A1 (LOC402317), mRNA
XM_377997 Homo sapiens similar to 60S ribosomal protein L32 (LOC402318), mRNA
XM_377998 Homo sapiens similar to Huntingtin interacting protein K (LOC402319), mRN.
XM_377999 Homo sapiens similar to BET1 homolog (Golgi vesicular membrane traffickin
XM_378001 Homo sapiens similar to TSH receptor suppressor element-binding protein-1
XM_378002 Homo sapiens similar to ppg3 (LOC402322), mRNA
XM_378003 Homo sapiens similar to S-adenosylmethionine decarboxylase 1 ; S-adenosyl
XM_378007 Homo sapiens similar to hypothetical protein PFL1865w (LOC402324), mRN
XM_378008 Homo sapiens similar to ENSANGP00000017949 (LOC402325), mRNA
XM_378009 Homo sapiens similar to hypothetical protein (LOC402326), mRNA
XM_378010 Homo sapiens similar to stage-specific S antigen homolog (LOC402327), mF
XM_378014 Homo sapiens similar to ubiquitin-specific protease 17-like protein (LOC4023
XM_378015 Homo sapiens similar to chromosome 11 open reading frame2; chromosome
XM_378018 Homo sapiens similar to chromosome 11 open reading frame2; chromosome
XM_378028 Homo sapiens similar to L21 ribosomal protein (LOC402336), mRNA
XM_378031 Homo sapiens similar to short chain dehydrogenase reductase 9 (LOC40233
XM_378033 Homo sapiens similar to chromosome 20 open reading frame 6 (LOC402340
XM_378035 Homo sapiens similar to fatty acid binding protein 9, testis; testis lipid binding
XM_378036 Homo sapiens similar to tropomyosin 3, gamma (LOC402344), mRNA
XM_378043 Homo sapiens similar to RIKEN cDNA 1700091 F14 (LOC402353), mRNA
XM_378044 Homo sapiens similar to hypothetical protein (LOC402354), mRNA
XM_378046 Homo sapiens similar to RIKEN cDNA 1700091F14 (LOC402355), mRNA
XM_378052 Homo sapiens similar to Interferon omega-1 precursor (Interferon alpha-ll-1)
XM_378054 Homo sapiens similar to hypothetical protein (LOC402360), mRNA
XM_378062 Homo sapiens similar to 6-pyruvoyl-tetrahydropterin synthase (LOC402365),
XM_378063 Homo sapiens similar to 6-pyruvoyl-tetrahydropterin synthase (LOC402366),
XM_378064 Homo sapiens LOC402367 (LOC402367), mRNA
XM_378067 Homo sapiens similar to phosphoglucomutase 5 (LOC402368), mRNA
XM_378078 Homo sapiens chromosome 9 open reading frame 4 (C9orf4), mRNA
XM_378080 Homo sapiens similar to beta-1 ,3-N-acetylglucosaminyltransferase 5 (LOC4C
XM_378087 Homo sapiens similar to ligand-independent activating molecule for estrogen
XM_378089 Homo sapiens similar to LOC286220 protein (LOC402381 ), mRNA
XM_378090 Homo sapiens similar to F4N2.10 (LOC402382), mRNA
XM_378102 Homo sapiens similar to ENSANGP00000002367 (LOC402386), mRNA
XM_378103 Homo sapiens similar to hydroxymethylpterin pyrophosphokinase-dihydropte
XM_378113 Homo sapiens similar to LOC142827 protein (LOC402402), mRNA
XM_378116 Homo sapiens similar to DKFZP434O047 protein (LOC402403), mRNA
XM_378118 Homo sapiens similar to PAGE-5 protein (LOC402404), mRNA
XM_378123 Homo sapiens similar to Mitochondrial import receptor subunit TOM20 homol
XM_378124 Homo sapiens similar to bA203H6.1 (KIAA0970 protein) (LOC402414), mRN
XM_378125 Homo sapiens similar to hypothetical protein MGC57211 (LOC402415), mRf*
XM_378128 Homo sapiens similarto dJ19N1.1 (novel protein) (LOC402416), mRNA
XM_378137 Homo sapiens similar to olfactory receptor MOR262-9 (LOC402424), mRNA
XM_378143 Homo sapiens similar to tau tubulin kinase 2; tau-tubulin kinase (LOC402429
XM_378152 Homo sapiens similarto bA203H6.1 (KIAA0970 protein) (LOC402432), mRN
XM_378155 Homo sapiens similar to hydroxymethylpterin pyrophosphokinase-dihydropte
XM_378156 Homo sapiens similar to trophinin; melanoma antigen, family D, 3; trophinin-.
XM_378158 Homo sapiens similar to hypothetical protein FLJ90430 (LOC402437), mRN/
XM_378172 Homo sapiens spermatogenesis-related protein 8 (MGC44294), mRNA
XM_378173 Homo sapiens hypothetical protein MGC18216 (MGC18216), mRNA
XM_378175 Homo sapiens hypothetical protein BC017488 (LOC124446), mRNA
XM_378177 Homo sapiens hypothetical gene supported by NM_078471 (LOC399700), m
XM_378178 Homo sapiens hypothetical protein MGC9913 (MGC9913), mRNA
XM_378180 Homo sapiens hypothetical protein GC10812 (MGC10812), mRNA
XM_378181 Homo sapiens KIAA1041 protein (KIAA1041), mRNA
XM_378183 Homo sapiens hypothetical protein MGC5457 (MGC5457), mRNA
XM_378184 Homo sapiens KIAA1383 protein (KI AA1383), mRNA
XM_378185 Homo sapiens hypothetical gene supported by AL833273; NM_014644 (LOC
XM_378186 Homo sapiens hypothetical protein MGC15634 (MGC15634), mRNA
XM_378187 Homo sapiens hypothetical protein MGC4473 (MGC4473), mRNA
XM_378189 Homo sapiens hypothetical protein MGC15705 (MGC15705), mRNA
XM_378190 Homo sapiens hypothetical protein GC10955 (MGC10955), mRNA
XM_378191 Homo sapiens hypothetical protein PR02964 (PR02964), mRNA
XM_378192 Homo sapiens hypothetical gene supported by NM_015583 (LOC399702), m
XM_378193 Homo sapiens hypothetical protein MGC10981 (MGC10981), mRNA
XM_378194 Homo sapiens hypothetical gene supported by NM_020669 (LOC399703), m
XM_378195 Homo sapiens hypothetical gene supported by NM_001517 (LOC399704), m
XM_378196 Homo sapiens hypothetical protein FLJ36112 (FLJ36112), mRNA
XM_378197 Homo sapiens hypothetical protein FLJ14464 (FLJ14464), mRNA
XM_378199 Homo sapiens hypothetical protein FLJ10232 (FLJ10232), mRNA
XM_378200 Homo sapiens hypothetical gene supported by AK097673 (LOC399706), mR
XM_378201 Homo sapiens hypothetical protein LOC282980 (LOC282980), mRNA
XM_378202 Homo sapiens hypothetical gene supported by AK056101 (LOC399707), mR
XM_378203 Homo sapiens hypothetical gene supported by BC055423 (LOC399708), mR
XM_378207 Homo sapiens hypothetical protein OC338588 (LOC338588), mRNA
XM_378208 Homo sapiens hypothetical gene supported by AK125014 (LOC399713), mR
XM_378210 Homo sapiens hypothetical gene supported by AK128810 (LOC399717), mR
XM_378211 Homo sapiens hypothetical protein OC254312 (LOC254312), mRNA
XM_378215 Homo sapiens hypothetical gene supported by BC040880 (LOC399726), mR
XM_378218 Homo sapiens hypothetical gene su pported by BX537934 (LOC399736), mR
XM_378219 Homo sapiens LOC399737 (LOC399737), mRNA
XM_378223 Homo sapiens hypothetical gene supported by AK093334; AL833330; BC02I
XM_378224 Homo sapiens hypothetical gene supported by X06747; BC012158; NM_002
XM_378226 Homo sapiens hypothetical protein OC170371 (LOC170371 ), mRNA
XM_378227 Homo sapiens hypothetical gene supported by AK093334; AL833330; BC02(
XM_378228 Homo sapiens hypothetical gene supported by AK093334; AL833330; BC02I
XM_378230 Homo sapiens hypothetical protein LOC283025 (LOC283025), mRNA
XM_378232 Homo sapiens hypothetical protein LOC219690 (LOC219690), mRNA
XM_378235 Homo sapiens LOC399785 (L0C399785), mRNA
XM_378236 Homo sapiens hypothetical gene supported by AK091031 (LOC399786), mR
XM_378238 Homo sapiens hypothetical protein LOC283050 (LOC283050), mRNA
XM_378239 Homo sapiens hypothetical gene supported by AK056520; NM_153030 (LOC
XM_378240 Homo sapiens hypothetical protein LOC170425 (LOC170425), mRNA
XM_378247 Homo sapiens hypothetical gene supported by AK123344 (LOC399806), mR
XM_378250 Homo sapiens hypothetical protein LOC92482 (LOC92482), mRNA
XM_378251 Homo sapiens hypothetical protein LOC143188 (LOC143188), mRNA
XM_378255 Homo sapiens hypothetical gene supported by AK128177 (LOC399827), mR
XM_378257 Homo sapiens hypothetical gene supported by AK125849 (LOC399829), mR
XM_378259 Homo sapiens hypothetical gene supported by BX647230 (LOC399832), mR
XM_378260 Homo sapiens hypothetical gene supported by AK126615 (LOC399833), mR
XM_378261 Homo sapiens hypothetical gene supported by BC004945; BC020442; BC06
XM_378262 Homo sapiens hypothetical protein LOC284701 (LOC284701), mRNA
XM_378266 Homo sapiens hypothetical gene supported by AY129010 (LOC399851), mR
XM_378271 Homo sapiens hypothetical gene supported by AK002039 (LOC399865), mR
XM_378272 Homo sapiens hypothetical gene supported by BX647519 (LOC399866), mR
XM_378273 Homo sapiens hypothetical gene supported by AK127718 (LOC399867), mR
XM_378276 Homo sapiens LOC399872 (LOC399872), mRNA
XM_378277 Homo sapiens hypothetical gene supported by AK096475 (LOC399873), mR
XM_378279 Homo sapiens hypothetical gene supported by BC040220 (LOC399875), mR
XM_378280 Homo sapiens hypothetical gene supported by AK023501 (LOC399876), mR
XM_378283 Homo sapiens hypothetical gene supported by AK127155 (LOC399879), mR
XM_378286 Homo sapiens hypothetical gene supported by AK093366 (LOC399884), mR
XM_378288 Homo sapiens hypothetical gene supported by AK123417 (LOC399886), mR
XM_378297 Homo sapiens hypothetical gene supported by X15675 (LOC399912), mRNA
XM_378299 Homo sapiens hypothetical gene supported by AK094674 (LOC399919), mR
XM_378300 Homo sapiens hypothetical gene supported by BC039105 (LOC399920), mR
XM_378301 Homo sapiens hypothetical gene supported by AL832797 (LOC399924), mR
XM_378303 Homo sapiens hypothetical protein LOC283214 (LOC283214), mRNA
XM_378304 Homo sapiens hypothetical gene supported by BC026292 (LOC399930), mR
XM_378305 Homo sapiens LOC399933 (LOC399933), mRNA
XM_378308 Homo sapiens LOC399945 (LOC399945), mRNA
XM_378309 Homo sapiens LOC399951 (LOC399951), mRNA
XM_378311 Homo sapiens hypothetical gene supported by AK124988 (LOC399954), mR
XM_378312 Homo sapiens hypothetical protein LOC283143 (LOC283143), mRNA
XM_378313 Homo sapiens LOC399955 (LOC399955), mRNA
XM_378314 Homo sapiens hypothetical protein LOC283152 (LOC283152), mRNA
XM_378316 Homo sapiens hypothetical gene supported by BX647608 (LOC399959), mR
XM_378317 Homo sapiens LOC399961 (LOC399961), mRNA
XM_378320 Homo sapiens hypothetical gene supported by AK125355 (LOC399971 ), mR
XM_378321 Homo sapiens hypothetical gene supported by AK096370 (LOC399972), mR
XM_378325 Homo sapiens hypothetical gene supported by BC031979 (LOC399978), mR
XM_378326 Homo sapiens hypothetical gene supported by AK127362 (LOC399980), mR
XM_378327 Homo sapiens hypothetical protein LOC283177 (LOC283177), mRNA
XM_378328 Homo sapiens hypothetical gene supported by BC039168 (LOC399982), mR
XM_378329 Homo sapiens hypothetical gene supported by AK090616 (LOC399983), mR
XM_378330 Homo sapiens hypothetical gene supported by AK057909 (LOC399984), mR
XM_378331 Homo sapiens hypothetical gene supported by AK056228 (LOC399986), mR
XM_378332 Homo sapiens LOC399987 (LOC399987), mRNA
XM_378336 Homo sapiens LOC399993 (LOC399993), mRNA
XM_378339 Homo sapiens hypothetical gene supported by AK128230 (LOC400002), mR
XM_378340 Homo sapiens LOC400004 (LOC400004), mRNA
XM_378342 Homo sapiens hypothetical gene supported by BC003510; NM_002823 (LOC
XM_378343 Homo sapiens hypothetical gene supported by AK097936 (LOC400007), mR
XM_378344 Homo sapiens LOC400015 (LOC400015), mRNA
XM_378346 Homo sapiens hypothetical protein LOC196415 (LOC196415), mRNA
XM_378349 Homo sapiens LOC400019 (LOC400019), mRNA
XM_378350 Homo sapiens hypothetical gene supported by BC047417 (LOC400027), mR
XM_378353 Homo sapiens LOC40003O (LOC400030), mRNA
XM_378354 Homo sapiens LOC400031 (LOC400031), mRNA
XM_378355 Homo sapiens hypothetical protein LOC283332 (LOC283332), mRNA
XM_378356 Homo sapiens hypothetical protein LOC283400 (LOC283400), mRNA
XM_378358 Homo sapiens hypothetical gene supported by AK123741 (LOC400041), mR
XM_378360 Homo sapiens hypothetical gene supported by BC009385 (LOC400043), mR
XM_378362 Homo sapiens hypothetical gene supported by AK123272 (LOC400046), mR
XM_378363 Homo sapiens LOC400047 (LOC400047), mRNA
XM_378365 Homo sapiens LOC400050 (LOC40005O), mRNA
XM_378366 Homo sapiens hypothetical gene supported by AK124066 (LOC400051 ), mR
XM_378367 Homo sapiens LOC400053 (LOC400053), mRNA
XM_378368 Homo sapiens hypothetical protein LOC283392 (LOC283392), mRNA
XM_378371 Homo sapiens hypothetical protein L0C338758 (LOC338758), mRNA
XM_378372 Homo sapiens hypothetical protein LOC256021 (LOC256021), mRNA
XM_378374 Homo sapiens hypothetical protein LOC338809 (LOC338809), mRNA
XM_378379 Homo sapiens hypothetical protein LOC283432 (LOC283432), mRNA
XM_378381 Homo sapiens hypothetical gene supported by BX648662 (LOC400070), mR
XM_378388 Homo sapiens hypothetical protein L0C144742 (LOC144742), mRNA
XM_378389 Homo sapiens hypothetical gene supported by AK057632; AL137270; BC05"
XM_378390 Homo sapiens hypothetical protein LOC144678 (LOC144678), mRNA
XM_378392 Homo sapiens hypothetical gene supported by AK094824 (LOC400087), mR
XM_378393 Homo sapiens hypothetical gene supported by AK126855 (LOC400088), mR
XM_378394 Homo sapiens hypothetical protein LOC116437 (LOC116437), mRNA
XM_378398 Homo sapiens LOC400092 (LOC400092), mRNA
XM_378399 Homo sapiens hypothetical gene supported by BC024195 (LOC400099), mR
XM_378404 Homo sapiens hypothetical gene supported by AK098387 (LOC400108), mR
XM_378405 Homo sapiens LOC400111 (LOC40011 ), mRNA
XM_378407 Homo sapiens hypothetical gene supported by BX648491 (LOC400115), mR
XM_378411 Homo sapiens hypothetical gene supported by AK124383 (LOC400123), mR
XM_378412 Homo sapiens LOC400125 (LOC400125), mRNA
XM_378413 Homo sapiens hypothetical gene supported by BC025370 (LOC400128), mR
XM_378414 Homo sapiens hypothetical gene supported by AF529010 (LOC400131 ), mR
XM_378416 Homo sapiens LOC400134 (LOC400134), mRNA
XM_378419 Homo sapiens hypothetical protein LOC144766 (LOC144766), mRNA
XM_378421 Homo sapiens hypothetical gene supported by AK056689 (LOC400144), mR
XM_378425 Homo sapiens LOC400151 (LOC400151), mRNA
XM_378428 Homo sapiens hypothetical gene supported by BC035106 (LOC400154), mR
XM_378430 Homo sapiens hypothetical protein LOC283480 (LOC283480), mRNA
XM_378431 Homo sapiens hypothetical protein LOC283483 (LOC283483), mRNA
XM_378434 Homo sapiens hypothetical gene supported by BC038751 (LOC400161), mR
XM_378436 Homo sapiens hypothetical gene supported by BC034786 (LOC400163), mR
XM_378437 Homo sapiens hypothetical gene supported by BX649107 (LOC400164), mR
XM_378439 Homo sapiens hypothetical gene supported by BC041346 (LOC400167), mR
XM_378441 Homo sapiens LOC400171 (LOC400171), mRNA
XM_378449 Homo sapiens LOC400201 (LOC4002O1 ), mRNA
XM_378452 Homo sapiens hypothetical protein LOC253970 (LOC253970), mRNA
XM_378453 Homo sapiens hypothetical gene supported by AK125955 (LOC400208), mR
XM_378454 Homo sapiens hypothetical protein LOC283547 (LOC283547), mRNA
XM_378455 Homo sapiens hypothetical protein LOC283551 (LOC283551), mRNA
XM_378456 Homo sapiens hypothetical gene supported by AK127576 (LOC400212), mR
XM_378457 Homo sapiens hypothetical gene supported by BC055421 (LOC400213), mR
XM_378460 Homo sapiens hypothetical gene supported by BC037850 (LOC400216), mR
XM_378462 Homo sapiens hypothetical gene supported by AK026100 (LOC400221), mR
XM_378465 Homo sapiens LOC400228 (LOC400228), mRNA
XM_378467 Homo sapiens LOC400231 (LOC400231 ), mRNA
XM_378470 Homo sapiens hypothetical gene supported by BC029835 (LOC400234), mR
XM_378472 Homo sapiens LOC400236 (LOC400236), mRNA
XM_378473 Homo sapiens hypothetical gene supported by AK093266 (LOC400238), mR
XM_378476 Homo sapiens hypothetical gene supported by AK127179 (LOC400242), mR
XM_378477 Homo sapiens LOC400243 (LOC400243), mRNA
XM_378482 Homo sapiens LOC400249 (LOC400249), mRNA
XM_378487 Homo sapiens hypothetical protein LOC145216 (LOC145216), mRNA
XM_378488 Homo sapiens LOC400256 (LOC400256), mRNA
XM_378489 Homo sapiens hypothetical gene supported by AK091459 (LOC400257), mR
XM_378490 Homo sapiens hypothetical gene supported by AK127521 (LOC400262), mR
XM_378491 Homo sapiens hypothetical gene supported by BC033241 (LOC400263), mR
XM_378493 Homo sapiens hypothetical gene supported by AK127783 (LOC400302), mR
XM_378494 Homo sapiens LOC400307 (LOC40O307), mRNA
XM_378496 Homo sapiens hypothetical gene supported by BC047459 (LOC400314), mR
XM_378506 Homo sapiens hypothetical gene supported by AK125576; AL117445; BC03I
XM_378507 Homo sapiens hypothetical protein LOC145845 (LOC145845), mRNA
XM_378511 Homo sapiens hypothetical gene supported by BC031266 (LOC400368), mR
XM_378512 Homo sapiens hypothetical gene supported by BX537772 (LOC400369), mR
XM_378514 Homo sapiens hypothetical protein LOC283663 (LOC283663), mRNA
XM_378515 Homo sapiens hypothetical gene supported by AK091917 (LOC400377), mR
XM_378516 Homo sapiens hypothetical protein LOC255177 (LOC255177), mRNA
XM_378517 Homo sapiens hypothetical protein MGC15885 (MGC15885), mRNA
XM_378522 Homo sapiens hypothetical gene supported by BC043587 (LOC400386), mR
XM_378523 Homo sapiens LOC400388 (LOC40O388), mRNA
XM_378525 Homo sapiens hypothetical protein LOC283731 (LOC283731), mRNA
XM_378526 Homo sapiens LOC400393 (LOC40O393), mRNA
XM_378528 Homo sapiens LOC400398 (LOC40O398), mRNA
XM_378529 Homo sapiens hypothetical gene supported by AK022116 (LOC400400), mR
XM_378532 Homo sapiens hypothetical protein LOC253044 (LOC253044), mRNA
XM_378535 Homo sapiens LOC400411 (LOC40O411 ), mRNA
XM_378538 Homo sapiens hypothetical gene supported by AL137524 (LOC400433), mR
XM_378542 Homo sapiens hypothetical protein LOC283761 (LOC283761), mRNA
XM_378544 Homo sapiens hypothetical protein LOC283682 (LOC283682), mRNA
XM_378545 Homo sapiens hypothetical gene supported by BC040875 (LOC400456), mR
XM_378546 Homo sapiens hypothetical protein LOC145820 (LOC145820), mRNA
XM_378549 Homo sapiens hypothetical protein LOC91948 (LOC91948), mRNA
XM_378550 Homo sapiens hypothetical protein LOC145757 (LOC145757), mRNA
XM_378551 Homo sapiens LOC400463 (LOC40O463), mRNA
XM_378553 Homo sapiens hypothetical gene supported by BC041891 (LOC400475), mR
XM_378558 Homo sapiens hypothetical protein LOC146443 (LOC146443), mRNA
XM_378562 Homo sapiens hypothetical gene supported by AL162011 (LOC400496), mR
XM_378564 Homo sapiens LOC400500 (LOC40O500), mRNA
XM_378567 Homo sapiens hypothetical gene supported by BX640722 (LOC400505), mR
XM_378571 Homo sapiens hypothetical gene supported by AK127191 (LOC400511), mR
XM_378573 Homo sapiens hypothetical gene supported by AK025061 (LOC400512), mR
XM_378576 Homo sapiens hypothetical gene su pported by AK126852 (LOC400516), mR
XM_378577 Homo sapiens hypothetical gene supported by AK123554 (LOC400517), mR
XM_378578 Homo sapiens hypothetical gene supported by BC023258 (LOC400518), mR
XM_378579 Homo sapiens hypothetical gene supported by AK097527 (LOC400522), mR
XM_378582 Homo sapiens LOC400523 (LOC40O523), mRNA
XM_378586 Homo sapiens LOC400531 (LOC40O531), mRNA
XM_378588 Homo sapiens hypothetical gene supported by BC047414 (LOC400532), mR
XM_378589 Homo sapiens hypothetical protein LOC283914 (LOC283914), mRNA
XM_378590 Homo sapiens hypothetical gene su pported by AK129756 (LOC400533), mR
XM_378592 Homo sapiens LOC400534 (LOC40O534), mRNA
XM_378594 Homo sapiens hypothetical gene supported by AK128747 (LOC400535), mR
XM_378595 Homo sapiens hypothetical gene supported by AK057373 (LOC400536), mR
XM_378599 Homo sapiens hypothetical protein LOC283854 (LOC283854), mRNA
XM_378601 Homo sapiens hypothetical gene supported by AK057319 (LOC400538), mR
XM_378606 Homo sapiens hypothetical protein LOC283867 (LOC283867), mRNA
XM_378607 Homo sapiens hypothetical gene supported by AL080152 (LOC400540), mR
XM_378608 Homo sapiens hypothetical gene supported by AK096066 (LOC400541), mR
XM_378609 Homo sapiens hypothetical gene supported by BC064480 (LOC400544), mR
XM_378610 Homo sapiens LOC400545 (LOC400545), mRNA
XM_378617 Homo sapiens hypothetical gene supported by BC040918 (LOC400548), mR
XM_378620 Homo sapiens hypothetical gene supported by AK091834 (LOC400550), mR
XM_378621 Homo sapiens hypothetical gene supported by AK125749 (LOC400551), mR
XM_378623 Homo sapiens hypothetical gene supported by AK055320 (LOC400552), mR
XM_378625 Homo sapiens hypothetical gene supported by AK126852 (LOC400553), mR
XM_378626 Homo sapiens hypothetical gene supported by AK123554 (LOC400554), mR
XM_378628 Homo sapiens hypothetical protein MGC23284 (MGC23284), mRNA
XM_378629 Homo sapiens hypothetical gene supported by AK127064 (LOC400556), mR
XM_378630 Homo sapiens hypothetical gene supported by AK055272 (LOC400557), mR
XM_378631 Homo sapiens hypothetical gene supported by AK130578 (LOC400558), mR
XM_378632 Homo sapiens LOC400560 (LOC40O560), mRNA
XM_378633 Homo sapiens LOC400561 (LOC40O561), mRNA
XM_378634 Homo sapiens LOC400562 (LOC40O562), mRNA
XM_378637 Homo sapiens LOC400565 (LOC40O565), mRNA
XM_378639 Homo sapiens hypothetical gene supported by AK093801 (LOC400567), mR
XM_378642 Homo sapiens hypothetical protein LOC284009 (LOC284009), mRNA
XM_378643 Homo sapiens hypothetical gene supported by BC043554 (LOC400568), mR
XM_378646 Homo sapiens LOC400571 (LOC40O571), mRNA
XM_378648 Homo sapiens LOC400572 (LOC40O572), mRNA
XM_378649 Homo sapiens hypothetical gene supported by BC015790; BC041634 (LOCA
XM_378650 Homo sapiens hypothetical gene supported by BC017752 (LOC400575), mR
XM_378653 Homo sapiens hypothetical gene supported by AK098696 (LOC400577), mR
XM_378655 Homo sapiens hypothetical protein LOC96597 (LOC96597), mRNA
XM_378656 Homo sapiens hypothetical gene supported by AK093253 (LOC400579), mR
XM_378660 Homo sapiens hypothetical gene supported by AK093253 (LOC400584), mR
XM_378661 Homo sapiens hypothetical protein LOC339263 (LOC339263), mRNA
XM_378664 Homo sapiens hypothetical gene supported by AK124344 (LOC400588), mR
XM_378667 Homo sapiens hypothetical protein LOC147004 (LOC147004), mRNA
XM_378668 Homo sapiens hypothetical gene supported by AK125932 (LOC400593), mR
XM_378675 Homo sapiens hypothetical protein LOC147093 (LOC147093), mRNA
XM_378676 Homo sapiens LOC400598 (LOC40O598), mRNA
XM_378678 Homo sapiens hypothetical gene supported by AK055254; BC051705 (LOC4
XM_378680 Homo sapiens hypothetical protein LOC147080 (LOC147080), mRNA
XM_378682 Homo sapiens LOC400602 (LOC40O602), mRNA
XM_378683 Homo sapiens hypothetical gene supported by AK000454 (LOC400603), mR
XM_378684 Homo sapiens hypothefical gene supported by BC039664 (LOC400604), mR
XM_378685 Homo sapiens hypothetical gene supported by BC039326 (LOC400605), mR
XM_378686 Homo sapiens hypothetical gene supported by AK126827 (LOC400606), mR
XM_378687 Homo sapiens hypothetical protein LOC339210 (LOC339210), mRNA
XM_378688 Homo sapiens LOC400607 (LOC40O607), mRNA
XM_378689 Homo sapiens LOC400611 (LOC40O611 ), mRNA
XM_378692 Homo sapiens hypothetical gene supported by AK094767 (LOC400612), mR
XM_378693 Homo sapiens LOC400614 (LOC40O614), mRNA
XM_378694 Homo sapiens hypothetical protein LOC146784 (LOC146784), mRNA
XM_378695 Homo sapiens hypothetical gene supported by BC053686 (LOC400616), mR
XM_378698 Homo sapiens hypothetical gene supported by AK093963 (LOC400617), mR
XM_378700 Homo sapiens hypothetical gene supported by AK094963 (LOC400618), mR
XM_378701 Homo sapiens hypothetical protein LOC146795 (LOC146795), mRNA
XM_378703 Homo sapiens hypothetical gene supported by AK129994 (LOC400619), mR
XM_378705 Homo sapiens hypothetical gene supported by BC035399 (LOC400620), mR
XM_378706 Homo sapiens LOC400621 (LOC40O621), mRNA
XM_378708 Homo sapiens hypothetical gene supported by AK130926 (LOC400623), mR
XM_378709 Homo sapiens hypothetical gene supported by AK127023 (LOC400624), mR
XM_378712 Homo sapiens hypothetical protein LOC146713 (LOC146713), mRNA
XM_378713 Homo sapiens hypothetical gene supported by BX648922 (LOC400626), mR
XM_378723 Homo sapiens hypothetical protein FLJ22659 (FLJ22659), mRNA
XM_378724 Homo sapiens hypothetical gene supported by AL832615 (LOC400630), mR
XM_378727 Homo sapiens hypothetical gene supported by BC053686 (LOC400632), mR
XM_378728 Homo sapiens LOC400633 (LOC400633), mRNA
XM_378730 Homo sapiens LOC400638 (LOC40O638), mRNA
XM_378734 Homo sapiens hypotheti ical protein LOC284214 (LOC284214), mRNA XM_378735 Homo sapiens hypotheti ical gene supported by BC041875; BX648984 (LOC4 XM_378738 Homo sapiens hypotheti cal gene supported by AK095347 (LOC400643), mR XM_378741 Homo sapiens hypotheti ical gene supported by AK126243 (LOC400644), mR XM_378742 Homo sapiens hypotheti cal gene supported by AK127888 (LOC400645), mR XM_378743 Homo sapiens hypotheti cal gene supported by BX640930 (LOC400647), mR XM_378745 Homo sapiens hypotheti ical gene supported by BC031271 (LOC400648), mR XM_378746 Homo sapiens hypotheti ical gene supported by AK126075 (LOC400649), mR XM_378747 Homo sapiens hypotheti cal gene supported by AK094936 (LOC400651), mR XM_378750 Homo sapiens hypotheti cal gene supported by BC047606 (LOC400653), mR XM_378751 Homo sapiens hypotheti cal gene supported by BC042493 (LOC400654), mR XM_378753 Homo sapiens hypotheti ical gene supported by BC013370; BC034583 (LOCA XM_378754 Homo sapiens hypotheti cal gene supported by BC039507 (LOC400656), mR XM_378755 Homo sapiens hypotheti cal gene supported by BC036588 (LOC400657), mR XM_378756 Homo sapiens hypotheti cal protein LOC284274 (LOC284274), mRNA XM_378757 Homo sapiens hypotheti cal gene supported by AK093936 (LOC400659), mR XM_378758 Homo sapiens hypotheti cal gene supported by AK094957 (LOC400660), mR XM_378760 Homo sapiens hypotheti cal gene supported by AK055411 (LOC400662), mR XM_378763 Homo sapiens hypotheti cal protein LOC284240 (LOC284240), mRNA XM_378765 Homo sapiens hypotheti cal gene supported by AK096031 (LOC400667), mR XM_378766 Homo sapiens LOC400669 (LOC400669), mRNA XM_378767 Homo sapiens LOC400670 (LOC400670), mRNA XM_378769 Homo sapiens LOC400671 (LOC400671), mRNA XM_378770 Homo sapiens hypothetical gene supported by AK001151 (LOC400672), mR XM_378776 Homo sapiens LOC400675 (LOC400675), mRNA XM_378777 Homo sapiens hypothetical protein LOC284385 (LOC284385), mRNA XM_378780 Homo sapiens hypothetical protein LOC126536 (LOC126536), mRNA XM_378783 Homo sapiens hypothetical gene supported by AK097381 ; BC040866 (LOC4 XM_378784 Homo sapiens hypothetical gene supported by BC030765 (LOC400683), mR XM_378786 Homo sapiens hypothetical protein LOC148145 (LOC148145), mRNA XM_378787 Homo sapiens hypothetical protein LOC284395 (LOC284395), mRNA XM_378791 Homo sapiens hypothetical protein LOC339316 (LOC339316), mRNA XM_378793 Homo sapiens hypothetical gene supported by BC000922 (LOC400684), mR XM_378794 Homo sapiens hypothetical protein LOC284402 (LOC284402), mRNA XM_378795 Homo sapiens hypothetical gene supported by BC045806 (LOC400685), mR XM_378796 Homo sapiens hypothetical gene supported by AK125858 (LOC400686), mR XM_378798 Homo sapiens hypothetical gene supported by AK092138 (LOC400690), mR XM_378799 Homo sapiens LOC400691 (LOC400691), mRNA XM_378800 Homo sapiens hypothetical gene supported by BC042546 (LOC400694), mR XM_378801 Homo sapiens LOC400695 (LOC400695), mRNA XM_378804 Homo sapiens LOC400700 (LOC400700), mRNA XM_378805 Homo sapiens LOC400701 (LOC400701), mRNA XM_378806 Homo sapiens hypothetical gene supported by AK024119 (LOC400702), mR XM_378807 Homo sapiens hypothetical gene supported by AK054869 (LOC400704), mR XM_378810 Homo sapiens hypothetical gene supported by AK096622 (LOC400706), mR XM_378812 Homo sapiens hypothetical gene supported by AK130360 (LOC400710), mR XM_378815 Homo sapiens LOC400722 (LOC400722), mRNA XM_378820 Homo sapiens hypothetical gene supported by AK097327; BC037297 (LOC4 XM_378822 Homo sapiens hypothetical protein LOC254099 (LOC254099), mRNA XM_378823 Homo sapiens hypothetical protein LOC148413 (LOC148413), mRNA XM_378824 Homo sapiens LOC400729 (LOC400729), mRNA XM_378825 Homo sapiens hypothetical gene supported by AK097814 (LOC400730), mR XM_378828 Homo sapiens hypothetical protein LOC115110 (LOC115110), mRNA XM_378831 Homo sapiens hypothetical gene supported by AK124708 (LOC400732), mR XM_378832 Homo sapiens hypothetical protein LOC284661 (LOC284661), mRNA XM_378835 Homo sapiens LOC400733 (LOC400733), mRNA XM 378837 Homo sapiens hypothetical gene supported by AK091499 (LOC400738), mR
XM_378838 Homo sapiens hypothetical gene supported by AK125737 (LOC400739), mR
XM_378840 Homo sapiens hypothetical gene supported by BC036435 (LOC400740), mR
XM_378841 Homo sapiens LOC400741 (LOC400741), mRNA
XM_378842 Homo sapiens hypothetical gene supported by BC033316 (LOC400742), mR
XM_378843 Homo sapiens hypothetical gene supported by AK127830 (LOC400743), mR
XM_378848 Homo sapiens PNAS-123 (LOC85028), mRNA
XM_378852 Homo sapiens hypothetical gene supported by BC040627 (LOC400748), mR
XM_378855 Homo sapiens hypothetical protein LOC339442 (LOC339442), mRNA
XM_378858 Homo sapiens hypothetical protein LOC339539 (LOC339539), mRNA
XM_378859 Homo sapiens hypothetical gene supported by BC031250 (LOC400751), mR
XM_378860 Homo sapiens hypothetical protein LOC149478 (LOC149478), mRNA
XM_378861 Homo sapiens hypothetical gene supported by BC006119 (LOC400752), mR
XM_378862 Homo sapiens LOC400753 (LOC400753), mRNA
XM_378865 Homo sapiens hypothetical gene supported by BC030752 (LOC400756), mR
XM_378866 Homo sapiens hypothetical protein LOC199899 (LOC199899), mRNA
XM_378873 Homo sapiens LOC400758 (LOC400758), mRNA
XM_378874 Homo sapiens hypothetical gene supported by AK130864 (LOC400761 ), mR
XM_378876 Homo sapiens hypothetical protein LOC149351 (LOC149351), mRNA
XM_378877 Homo sapiens hypothetical gene supported by AL832786 (LOC400762), mR
XM_378879 Homo sapiens hypothetical gene supported by AK000394 (LOC400763), mR
XM_378880 Homo sapiens hypothetical gene supported by AK094796 (LOC400764), mR
XM_378883 Homo sapiens hypothetical gene supported by BC051808 (LOC400768), mR
XM_378886 Homo sapiens hypothetical protein LOC284475 (LOC284475), mRNA
XM_378889 Homo sapiens hypothetical gene supported by AK090412 (LOC400770), mR
XM_378890 Homo sapiens hypothetical gene supported by AK098337; BC022881 (LOC4
XM_378891 Homo sapiens hypothetical gene supported by BC012753 (LOC400772), mR
XM_378892 Homo sapiens LOC400775 (LOC400775), mRNA
XM_378893 Homo sapiens hypothetical gene supported by AK125616 (LOC400777), mR
XM_378894 Homo sapiens LOC400778 (LOC400778), mRNA
XM_378897 Homo sapiens LOC400781 (LOC400781 ), mRNA
XM_378898 Homo sapiens LOC400782 (LOC400782), mRNA
XM_378899 Homo sapiens LOC400783 (LOC400783), mRNA
XM_378901 Homo sapiens hypothetical gene supported by M60502 (LOC400786), mRN/
XM_378903 Homo sapiens LOC400789 (LOC400789), mRNA
XM_378905 Homo sapiens hypothetical gene supported by AK094742; AK128347 (LOC4
XM_378908 Homo sapiens hypothetical gene supported by AK125122 (LOC400793), mR
XM_378909 Homo sapiens hypothetical gene supported by BC030596 (LOC400794), mR
XM_378910 Homo sapiens hypothetical gene supported by AK000073 (LOC400795), mR
XM_378912 Homo sapiens hypothetical protein LOC284688 (LOC284688), mRNA
XM_378914 Homo sapiens KIAA0492 protein (KIAA0492), mRNA
XM_378917 Homo sapiens LOC400796 (LOC400796), mRNA
XM_378918 Homo sapiens hypothetical gene supported by AK125993 (LOC400797), mR
XM_378919 Homo sapiens hypothetical gene supported by AK092849 (LOC400798), mR
XM_378921 Homo sapiens hypothetical protein LOC148756 (LOC148756), mRNA
XM_378923 Homo sapiens hypothetical protein LOC339476 (LOC339476), mRNA
XM_378925 Homo sapiens hypothetical gene supported by AK125573 (LOC400800), mR
XM_378930 Homo sapiens hypothetical gene supported by AK097184 (LOC400801 ), mR
XM_378933 Homo sapiens LOC400802 (LOC4008O2), mRNA
XM_378934 Homo sapiens LOC400803 (LOC4008O3), mRNA
XM_378941 Homo sapiens hypothetical protein LOC339535 (LOC339535), mRNA
XM_378945 Homo sapiens hypothetical protein LOC149134 (LOC149134), mRNA
XM_378946 Homo sapiens hypothetical gene supported by AK096414 (LOC400812), mR
XM_378947 Homo sapiens LOC400813 (LOC400813), mRNA
XM_378949 Homo sapiens similar to hypothetical protein LOC148413 (LOC400817), mRI
XM_378950 Homo sapiens hypothetical gene supported by AK097814 (LOC400819), mR
XM_378951 Homo sapiens hypothetical protein LOC284628 (LOC284628), mRNA
XM_378954 Homo sapiens hypothetical gene supported by AK125616 (LOC400828), mR
XM_378955 Homo sapiens LOC400829 (LOC400829), mRNA
XM_378956 Homo sapiens hypothetical gene supported by AK092524 (LOC400831), mR
XM_378957 Homo sapiens LOC400832 (LOC400832), mRNA
XM_378961 Homo sapiens LOC400835 (LOC400835), mRNA
XM_378964 Homo sapiens hypothetical protein LOC339593 (LOC339593), mRNA
XM_378969 Homo sapiens hypothetical protein LOC284788 (LOC284788), mRNA
XM_378970 Homo sapiens hypothetical gene supported by AK090900 (LOC400839), mR
XM_378971 Homo sapiens hypothetical protein LOC284798 (LOC284798), mRNA
XM_378973 Homo sapiens hypothetical protein LOC284801 (LOC284801 ), mRNA
XM_378974 Homo sapiens hypothetical gene supported by AK127732 (LOC400841 ), mR
XM_378976 Homo sapiens LOC400843 (LOC400843), mRNA
XM_378977 Homo sapiens hypothetical gene supported by AK124127 (LOC400844), mR
XM_378978 Homo sapiens hypothetical gene supported by AK090932 (LOC400845), mR
XM_378980 Homo sapiens hypothetical protein LOC339568 (LOC339568), mRNA
XM_378981 Homo sapiens hypothetical gene supported by AK127953 (LOC400846), mR
XM_378982 Homo sapiens hypothetical gene supported by AL832259; BC035164 (LOC4
XM_378983 Homo sapiens hypothetical protein LOC284749 (LOC284749), mRNA
XM_378985 Homo sapiens hypothetical protein LOC284751 (LOC284751 ), mRNA
XM_378988 Homo sapiens LOC400849 (LOC400849), mRNA
XM_378989 Homo sapiens hypothetical gene supported by AK126744 (LOC400850), mR
XM_378992 Homo sapiens hypothetical gene supported by BC015673 (LOC400851), mR
XM_378993 Homo sapiens hypothetical gene supported by AK124784 (LOC400852), mR
XM_378994 Homo sapiens hypothetical gene supported by AK125376 (LOC400853), mR
XM_379002 Homo sapiens LOC400861 (LOC400861 ), mRNA
XM_379003 Homo sapiens LOC400862 (LOC400862), mRNA
XM_379004 Homo sapiens hypothetical gene supported by AK127913 (LOC400863), mR
XM_379006 Homo sapiens hypothetical gene supported by AK094771 (LOC400865), mR
XM_379009 Homo sapiens hypothetical gene supported by AY204750 (LOC400866), mR
XM_379011 Homo sapiens hypothetical protein LOC284835 (LOC284835), mRNA
XM_379012 Homo sapiens hypothetical gene supported by BC03326O (LOC400868), mR
XM_379014 Homo sapiens LOC400869 (LOC400869), mRNA
XM_379015 Homo sapiens hypothetical gene supported by AK123727 (LOC400870), mR
XM_379016 Homo sapiens hypothetical gene supported by AK057962 (LOC400871 ), mR
XM_379017 Homo sapiens LOC400872 (LOC400872), mRNA
XM_379018 Homo sapiens hypothetical gene supported by BX648824 (LOC400873), mR
XM_379020 Homo sapiens hypothetical gene supported by AK096268 (LOC400874), mR
XM_379021 Homo sapiens chromosome 21 open reading frame 30 (C21orf30), mRNA
XM_379022 Homo sapiens hypothetical gene supported by BC040064 (LOC400875), mR
XM_379023 Homo sapiens hypothetical gene supported by BC036902 (LOC400876), mR
XM_379025 Homo sapiens LOC400877 (LOC400877), mRNA
XM_379029 Homo sapiens hypothetical gene supported by AK096951 (LOC400879), mR
XM_379030 Homo sapiens hypothetical gene supported by AY338954 (LOC400880), mR
XM_379032 Homo sapiens hypothetical gene supported by BC021738 (LOC400885), mR
XM_379036 Homo sapiens LOC400890 (LOC400890), mRNA
XM_379040 Homo sapiens hypothetical gene supported by AK097628 (LOC400919), mR
XM__379041 Homo sapiens hypothetical gene supported by BC040576 (LOC400920), mR
XM 379044 Homo sapiens hypothetical protein LOC284898 (LOC284898), mRNA
XM 379046 Homo sapiens LOC400923 (LOC400923), mRNA
XM_379052 Homo sapiens hypothetical gene supported by AK075161 (LOC400926), mR
XM 379054 Homo sapiens hypothetical protein LOC339674 (LOC339674), mRNA
XM 379055 Homo sapiens hypothetical gene supported by AK130208 (LOC400929), mR
XM_379060 Homo sapiens hypothetical protein LOC339685 (LOC339685), mRNA
XM_379064 Homo sapiens hypothetical protein LOC284933 (LOC284933), mRNA
XM_379065 Homo sapiens hypothetical gene supported by BC033837 (LOC400933), mR
XM_379068 Homo sapiens hypothetical gene supported by BC055007 (LOC400937), mR
XM_379069 Homo sapiens hypothetical protein LOC339822 (LOC339822), mRNA
XM_379072 Homo sapiens LOC400939 (LOC400939), mRNA
XM_379073 Homo sapiens hypothetical protein LOC386597 (LOC386597), mRNA
XM_379074 Homo sapiens hypothetical protein LOC339788 (LOC339788), mRNA
XM_379075 Homo sapiens hypothetical gene supported by BX6 1130 (LOC400942), mR
XM_379077 Homo sapiens LOC400944 (LOC400944), mRNA
XM_379078 Homo sapiens hypothetical gene supported by AK123475 (LOC400945), mR
XM 379079 Homo sapiens hypothetical gene supported by AK022396; AK097927 (LOC4
XM_379080 Homo sapiens LOC400947 (LOC400947), mRNA
XM_379085 Homo sapiens hypothetical protein LOC285043 (LOC285043), mRNA
XM_379086 Homo sapiens hypothetical protein LOC285045 (LOC285045), mRNA
XM_379089 Homo sapiens LOC400951 (LOC400951), mRNA
XM_379094 Homo sapiens hypothetical gene supported by BX647332 (LOC400953), mR
XM_379096 Homo sapiens hypothetical gene supported by AL832565 (LOC400955), mR
XM_379097 Homo sapiens hypothetical gene supported by AK122786 (LOC400957), mR
XM_379098 Homo sapiens hypothetical protein LOC339803 (LOC339803), mRNA
XM_379099 Homo sapiens hypothetical protein LOC339807 (LOC339807), mRNA
XM_379100 Homo sapiens hypothetical gene supported by BC037562 (LOC400958), mR
XM_379101 Homo sapiens hypothetical gene supported by BC033059 (LOC400959), mR
XM_379102 Homo sapiens hypothetical gene supported by BC040598 (LOC400960), mR
XM_379106 Homo sapiens hypothetical gene supported by BC044795 (LOC400964), mR
XM_379108 Homo sapiens LOC400969 (LOC400969), mRNA
XM_379109 Homo sapiens hypothetical gene supported by AK127783 (LOC400983), mR
XM_379111 Homo sapiens hypothetical protein LOC285033 (LOC285033), mRNA
XM_379112 Homo sapiens LOC400988 (LOC400988), mRNA
XM_379113 Homo sapiens hypothetical gene supported by BC040181 (LOC400990), mR
XM_379114 Homo sapiens hypothetical protein LOC150577 (LOC150577), mRNA
XM_379117 Homo sapiens hypothetical protein LOC150568 (LOC150568), mRNA
XM_379118 Homo sapiens hypothetical gene supported by AK095498 (LOC400992), mR
XM_379119 Homo sapiens hypothetical protein LOC285000 (LOC285000), mRNA
XM_379121 Homo sapiens hypothetical gene supported by AK056084; AK095678; NM_1
XM_379122 Homo sapiens hypothetical gene supported by AK125994 (LOC400997), mR
XM_379123 Homo sapiens hypothetical gene supported by AK124342 (LOC400999), mR
XM_379131 Homo sapiens LOC401001 (LOC401001), mRNA
XM_379133 Homo sapiens hypothetical protein LOC151121 (LOC151121), mRNA
XM_379135 Homo sapiens hypothetical gene supported by AK093281 (LOC401005), mR
XM_379136 Homo sapiens hypothetical gene supported by AK093281 (LOC401006), mR
XM_379141 Homo sapiens hypothetical gene supported by BC043549; BX648102 (LOC4
XM_379145 Homo sapiens hypothetical gene supported by AK092134 (LOC401020), mR
XM_379146 Homo sapiens hypothetical gene supported by BC047605 (LOC401021), mR
XM_379147 Homo sapiens hypothetical gene supported by BC030713; BC047481 ( OCA
XM_379149 Homo sapiens LOC401025 (LOC401025), mRNA
XM_379154 Homo sapiens hypothetical protein LOC151300 (LOC151300), mRNA
XM_379156 Homo sapiens LOC401032 (LOC401032), mRNA
XM_379158 Homo sapiens hypothetical gene supported by AK055016 (LOC401033), mR
XM_379159 Homo sapiens hypothetical protein LOC151484 (LOC151484), mRNA
XM_379161 Homo sapiens hypothetical gene supported by AK056246 (LOC401037), mR
XM_379163 Homo sapiens hypothetical gene supported by AK057585 (LOC401038), mR
XM_379164 Homo sapiens hypothetical protein LOC151171 (LOC151171), mRNA
XM_379166 Homo sapiens hypothetical gene supported by AK056439 (LOC401041 ), mR
XM_379167 Homo sapiens LOC401042 (LOC401042), mRNA
XM_379168 Homo sapiens LOC401043 (LOC401043), mRNA
XM_379169 Homo sapiens hypothetical gene supported by AK098031 (LOC401044), mR
XM_379170 Homo sapiens LOC401046 (LOC401046), mRNA
XM_379171 Homo sapiens LOC401048 (LOC401048), mRNA
XM_379172 Homo sapiens hypothetical gene supported by AK124857 (LOC401049), mR
XM_379173 Homo sapiens hypothetical gene supported by AK124088 (LOC401050), mR
XM_379174 Homo sapiens LOC401051 (LOC401051 ), mRNA
XM_379175 Homo sapiens hypothetical gene supported by AK022260 (LOC401052), mR
XM_379177 Homo sapiens hypothetical gene supported by BC041425 (LOC401053), mR
XM_379179 Homo sapiens hypothetical protein L0C152274 (LOC152274), mRNA
XM_379180 Homo sapiens LOC401056 (LOC401056), mRNA
XM_379181 Homo sapiens LOC401057 (LOC401057), mRNA
XM_379182 Homo sapiens LOC401058 (LOC401058), mRNA
XM_379183 Homo sapiens hypothetical protein LOC152024 (LOC152024), mRNA
XM_379184 Homo sapiens hypothetical gene supported by AK096885; AK098084 (LOC4
XM_379189 Homo sapiens hypothetical gene supported by AK057338 (LOC401061), mR
XM_379190 Homo sapiens hypothetical gene supported by AK092973 (LOC401062), mR
XM_379191 Homo sapiens LOC401065 (LOC401065), mRNA
XM 379192 Homo sapiens LOC401066 (LOC401066), mRNA
XM_379194 Homo sapiens hypothetical gene supported by BC028186 (LOC401068), mR
XM_379195 Homo sapiens hypothetical protein LOC285401 (LOC285401 ), mRNA
XM_379196 Homo sapiens hypothetical protein LOC151877 (LOC151877), mRNA
XM_379197 Homo sapiens hypothetical gene supported by AL832401 (LOC401073), mR
XM_379198 Homo sapiens hypothetical protein LOC285286 (LOC285286), mRNA
XM_379200 Homo sapiens LOC401078 (LOC401078), mRNA
XM_379201 Homo sapiens LOC401079 (LOC401079), mRNA
XM_379203 Homo sapiens hypothetical protein LOC348801 (LOC348801), mRNA
XM_379204 Homo sapiens hypothetical protein LOC152225 (LOC152225), mRNA
XM_379205 Homo sapiens hypothetical protein LOC151658 (LOC151658), mRNA
XM_379206 Homo sapiens hypothetical gene supported by AK026416 (LOC401081 ), mR
XM_379207 Homo sapiens hypothetical protein LOC285194 (LOC285194), mRNA
XM_379210 Homo sapiens LOC401085 (LOC401085), mRNA
XM_379213 Homo sapiens LOC401086 (LOC401086), mRNA
XM_379214 Homo sapiens hypothetical protein LOC339942 (LOC339942), mRNA
XM_379215 Homo sapiens hypothetical protein LOC132241 (LOC132241), mRNA
XM_379228 Homo sapiens LOC401093 (LOC401093), mRNA
XM_379229 Homo sapiens LOC401094 (LOC401094), mRNA
XM_379230 Homo sapiens hypothetical protein LOC339894 (LOC339894), mRNA
XM_379231 Homo sapiens hypothetical gene supported by BC034803 (LOC401098), mR
XM_379233 Homo sapiens LOC401099 (LOC401099), mRNA
XM_379234 Homo sapiens LOC401101 (LOC401101), mRNA
XM_379235 Homo sapiens hypothetical gene supported by AK127955 (LOC401103), mR
XM_379240 Homo sapiens LOC401104 (LOC401104), mRNA
XM_379243 Homo sapiens hypothetical gene supported by AK091527 (LOC401106), mR
XM_379244 Homo sapiens hypothetical gene supported by AK127609 (LOC401107), mR
XM_379247 Homo sapiens hypothetical gene supported by AK128780; AL137733 (LOC4
XM_379248 Homo sapiens hypothetical gene supported by AK098259 (LOC401112), mR
XM_379249 Homo sapiens hypothetical gene supported by AK123125 (LOC401113), mR
XM_379250 Homo sapiens hypothetical gene supported by BC038466; BC062790 (LOC4
XM_379252 Homo sapiens hypothetical gene supported by BC017173 (LOC401117), mR
XM_379254 Homo sapiens hypothetical protein LOC339988 (LOC339988), mRNA
XM_379255 Homo sapiens hypothetical gene supported by AK094096 (LOC401119), mR
XM_379256 Homo sapiens hypothetical gene supported by AK127863 (LOC401120), mR
XM_379258 Homo sapiens hypothetical protein LOC285547 (LOC285547), mRNA
XM_379260 Homo sapiens hypothetical protein LOC152742 (LOC152742), mRNA
XM_379262 Homo sapiens LOC401124 (LOC401124), mRNA
XM_379263 Homo sapiens LOC401126 (LOC401126), mRNA
XM_379264 Homo sapiens LOC401128 (LOC401128), mRNA
XM_379265 Homo sapiens LOC401129 (LOC401129), mRNA
XM_379267 Homo sapiens hypothetical gene supported by BC040544 (LOC401134), mR
XM_379268 Homo sapiens hypothetical gene supported by AK093682; AK129519 (LOC4
XM_379270 Homo sapiens casein alpha s2-like A (CSN1S2A), mRNA
XM_379271 Homo sapiens LOC401144 (LOC401144), mRNA
XM_379273 Homo sapiens hypothetical gene supported by AK024248; AL137733 (LOC4
XM_379274 Homo sapiens hypothefical gene supported by BC062741 (LOC401151 ), mR
XM_379275 Homo sapiens LOC401153 (LOC401153), mRNA
XM_379276 Homo sapiens hypothetical gene supported by AK123449; BX6 1014 (LOC4
XM_379278 Homo sapiens LOC401156 (LOC401156), mRNA
XMJ379280 Homo sapiens hypothetical protein LOC285422 (LOC285422), mRNA
XM_379287 Homo sapiens LOC401159 (LOC401159), mRNA
XM_379288 Homo sapiens hypothetical protein LOC340017 (LOC340017), mRNA
XM_379294 Homo sapiens similar to hypothetical protein DKFZp762K222 (LOC401163),
XM_379295 Homo sapiens hypothetical protein LOC285441 (LOC285441 ), mRNA
XM_379298 Homo sapiens hypothetical gene supported by AY494056 (LOC401164), mR
XM_379299 Homo sapiens hypothetical gene supported by BC029568 (LOC401165), mR
XM_379303 Homo sapiens hypothetical gene supported by AK126199 (LOC401168), mR
XM_379306 Homo sapiens hypothetical gene supported by BC034612 (LOC401169), R
XM_379309 Homo sapiens hypothetical gene supported by BC035019 (LOC401171), mR
XM_379313 Homo sapiens hypothetical gene supported by AK126802 (LOC401173), mR
XM_379317 Homo sapiens LOC401175 (LOC401175), mRNA
XM_379318 Homo sapiens hypothetical gene supported by BC043001 (LOC401176), mR
XM_379320 Homo sapiens hypothetical gene supported by BC052942 (LOC401177), mR
XM_379321 Homo sapiens hypothetical protein LOC340107 (LOC340107), mRNA
XM_379322 Homo sapiens hypothetical protein LOC340109 (LOC340109), mRNA
XM_379323 Homo sapiens LOC401178 (LOC401178), mRNA
XM__379324 Homo sapiens hypothetical protein LOC340113 (LOC340113), mRNA
XM_379325 Homo sapiens LOC401180 (LOC401180), mRNA
XM_379326 Homo sapiens hypothetical gene supported by BC028978 (LOC401181 ), mR
XM_379327 Homo sapiens LOC401182 (LOC401182), mRNA
XM_379328 Homo sapiens LOC401184 (LOC401184), mRNA
XM_379331 Homo sapiens hypothetical gene supported by AK057759 (LOC401185), mR
XMJ379332 Homo sapiens LOC401186 (LOC401186), mRNA
XM_379333 Homo sapiens LOC401187 (LOC401187), mRNA
XM_379334 Homo sapiens hypothetical protein LOC257396 (LOC257396), mRNA
XM_379336 Homo sapiens hypothetical gene supported by AK091013 (LOC401188), mR
XM_379339 Homo sapiens LOC401199 (LOC401199), mRNA
XM_379340 Homo sapiens hypothefical protein LOC285713 (LOC285713), mRNA
XM_379343 Homo sapiens hypothetical gene supported by AK092258 (LOC401201 ), R
XM_379347 Homo sapiens LOC401203 (LOC401203), mRNA
XM_379355 Homo sapiens hypothetical protein LOC340074 (LOC340074), mRNA
XM_379359 Homo sapiens hypothetical gene supported by AK022326 (LOC401210), mR
XM 379363 Homo sapiens hypothetical gene supported by BX640700 (LOC401212), mR
XM_379364 Homo sapiens hypothetical gene supported by AK127910 (LOC401213), mR
XM_379366 Homo sapiens hypothetical gene supported by AK092848; AK123816 (LOC4
XM_379368 Homo sapiens LOC401215 (LOC401215), mRNA
XMJ379371 Homo sapiens hypothetical protein LOC285626 (LOC285626), mRNA
XM_379372 Homo sapiens hypothetical gene supported by BC039501 (LOC401216), mR
XM_379373 Homo sapiens hypothetical protein LOC257358 (LOC257358), mRNA
XM_379377 Homo sapiens hypothetical gene supported by BX649016 (LOC401219), mR
XM_379378 Homo sapiens hypothetical gene supported by BC036933 (LOC401220), mR
XM_379380 Homo sapiens hypothetical gene supported by BC036933 (LOC401222), mR
XM_379381 Homo sapiens hypothetical gene supported by AK055745 (LOC401225), mR
XM_379382 Homo sapiens hypothetical gene supported by AK093197; BCO40992 (LOC4
XM_379384 Homo sapiens hypothetical protein LOC285766 (LOC285766), mRNA
XM_379386 Homo sapiens hypothetical protein LOC285768 (LOC285768), mRNA
XM_379391 Homo sapiens hypothetical gene supported by AK128409 (LOC401230), mR
XM_379392 Homo sapiens hypothetical gene supported by AK023629 (LOC401231 ), mR
XM_379393 Homo sapiens hypothetical gene supported by BX640709 (LOC401232), mR
XM_379395 Homo sapiens hypothetical gene supported by BC014487 (LOC401234), mR
XM_379396 Homo sapiens hypothetical protein LOC221710 (LOC221710), mRNA
XM_379398 Homo sapiens hypothetical gene supported by AK026189 (LOC401237), mR
XM_379401 Homo sapiens LOC401241 (LOC401241), mRNA
XM_379402 Homo sapiens hypothetical gene supported by AK0555O3 (LOC401242), mR
XM_379403 Homo sapiens chromosome 6 open reading frame 12 (C6orf12), mRNA
XM_379404 Homo sapiens hypothetical gene supported by AK055657 (LOC401245), mR
XM_379406 Homo sapiens hypothetical gene supported by AL832447 (LOC401254), mR
XM_379408 Homo sapiens LOC401255 (LOC401255), mRNA
XM_379409 Homo sapiens hypothetical gene supported by BX6481 12 (LOC401256), mR
XM_379410 Homo sapiens hypothetical gene supported by BC034770 (LOC401257), mR
XM_379411 Homo sapiens hypothetical gene supported by AK125083 (LOC401258), mR
XM_379413 Homo sapiens hypothetical gene supported by AK098234 (LOC401261 ), mR
XM_379417 Homo sapiens hypothetical gene supported by AK0951 17 (LOC401264), mR
XM_379424 Homo sapiens LOC401268 (LOC401268), mRNA
XM_379430 Homo sapiens hypothetical protein LOC285758 (LOC285758), mRNA
XM_379432 Homo sapiens hypothetical protein LOC285733 (LOC285733), mRNA
XM_379433 Homo sapiens hypothetical protein LOC285735 (LOC285735), mRNA
XM_379434 Homo sapiens hypothetical protein LOC154092 (LOC154092), mRNA
XM_379435 Homo sapiens hypothetical gene supported by AK128874 (LOC401275), mR
XM_379436 Homo sapiens hypothetical gene supported by BC038188 (LOC401276), mR
XM_379437 Homo sapiens hypothetical protein LOC153910 (LOC153910), mRNA
XM_379438 Homo sapiens hypothetical protein LOC285740 (LOC285740), mRNA
XM_379439 Homo sapiens LOC401277 (LOC401277), mRNA
XM_379441 Homo sapiens LOC401279 (LOC401279), mRNA
XM_379450 Homo sapiens hypothetical gene supported by AK130765 (LOC401281), mR
XM_379452 Homo sapiens hypothetical gene supported by AL831931 (LOC401282), mR
XM_379453 Homo sapiens hypothetical gene supported by AL832143 (LOC401283), mR
XM_379454 Homo sapiens hypothetical gene supported by AK095077 (LOC401284), mR
XM_379456 Homo sapiens hypothetical protein LOC154222 (LOC154222), mRNA
XM_379458 Homo sapiens hypothetical gene supported by BX648586 (LOC401287), mR
XM_379459 Homo sapiens hypothetical gene supported by AK095441 (LOC401289), mR
XM_379460 Homo sapiens hypothetical gene supported by AL832760 (LOC401290), mR
XM_379461 Homo sapiens LOC401291 (LOC401291), mRNA
XM_379462 Homo sapiens hypothetical gene supported by BC064362; BX537893 (LOC4
XM_379463 Homo sapiens similar to hypothetical protein LOC154222 (LOC401294), mRI
XM_379467 Homo sapiens hypothetical gene supported by AK125766 (LOC401296), mR
XM_379469 Homo sapiens hypothetical gene supported by BC032734 (LOC401297), mR
XM_379471 Homo sapiens LOC401299 (LOC401299), mRNA
XM_379472 Homo sapiens LOC401301 (LOC401301), mRNA
XM_379474 Homo sapiens hypothetical gene supported by AK127500 (LOC401310), mR
XM_379476 Homo sapiens hypothetical gene supported by BC039682 (LOC401312), mR
XM_379477 Homo sapiens hypothetical protein LOC285941 (LOC285941), mRNA
XM_379478 Homo sapiens hypothetical gene supported by AK093987 (LOC401315), mR
XM_379479 Homo sapiens LOC401317 (LOC401317), mRNA
XM_379480 Homo sapiens LOC401318 (LOC401318), mRNA
XM_379481 Homo sapiens hypothetical gene supported by BC023581 ; BC044638 (LOC4
XM_379482 Homo sapiens hypothetical gene supported by BC016976 (LOC401320), mR
XM_379483 Homo sapiens hypothetical gene supported by AK092714 (LOC401321 ), mR
XM_379484 Homo sapiens hypothetical gene supported by AL832092 (LOC401324), mR
XM_379485 Homo sapiens LOC401328 (LOC401328), mRNA
XM_379486 Homo sapiens hypothetical protein LOC285958 (LOC285958), mRNA
XM_379487 Homo sapiens hypothetical gene supported by AK125311 (LOC401334), mR
XM_379488 Homo sapiens hypothetical gene supported by AK024248; AL137733; BC06:
XM_379489 Homo sapiens LOC401345 (LOC401345), mRNA
XM_379490 Homo sapiens LOC401346 (LOC401346), mRNA
XM_379491 Homo sapiens LOC401348 (LOC401348), mRNA
XM_379492 Homo sapiens hypothetical gene supported by BX648489 (LOC401349), mR
XM_379493 Homo sapiens LOC401352 (LOC401352), mRNA
XM_379494 Homo sapiens LOC401353 (LOC401353), mRNA
XM_379495 Homo sapiens LOC401358 (LOC401358), mRNA
XM_379496 Homo sapiens LOC401359 (LOC401359), mRNA
XM_379498 Homo sapiens LOC401363 (LOC401363), mRNA
XM_379499 Homo sapiens hypothetical gene supported by AK024371 ; BC037920 (LOC4
XM_379500 Homo sapiens hypothetical gene supported by AK054923; AK126730; Nlvl_0
XM_379501 Homo sapiens LOC401367 (LOC401367), mRNA
XM_379502 Homo sapiens LOC401368 (LOC401368), mRNA
XM_379503 Homo sapiens LOC401371 (LOC401371), mRNA
XM_379504 Homo sapiens hypothetical gene supported by AK024602 (LOC401380), R
XM_379506 Homo sapiens LOC401384 (LOC401384), mRNA
XM_379507 Homo sapiens LOC401385 (LOC401385), mRNA
XM_379508 Homo sapiens LOC401386 (LOC401386), mRNA
XM_379510 Homo sapiens hypothetical protein FLJ34048 (FLJ34048), mRNA
XM_379511 Homo sapiens LOC401390 (LOC401390), mRNA
XM_379512 Homo sapiens LOC401394 (LOC401394), mRNA
XM_379513 Homo sapiens LOC401396 (LOC401396), mRNA
XM_379514 Homo sapiens hypothetical protein LOC340340 (LOC340340), mRNA
XM_379515 Homo sapiens hypothetical gene supported by BX537645 (LOC401397), mR
XM_379516 Homo sapiens hypothetical gene supported by BX648695 (LOC401398), R
XM_379517 Homo sapiens hypothetical gene supported by BC063892 (LOC401399), R
XM_379518 Homo sapiens LOC401400 (LOC401400), mRNA
XM_379520 Homo sapiens hypothetical protein FLJ43663 (FLJ43663), mRNA
XM_379521 Homo sapiens LOC401405 (LOC401405), mRNA
XM_379522 Homo sapiens LOC401406 (LOC401406), mRNA
XM_379523 Homo sapiens LOC401407 (LOC401407), mRNA
XM_379524 Homo sapiens LOC401408 (LOC401408), mRNA
XM_379526 Homo sapiens hypothetical gene supported by BX648692 (LOC401410), R
XM_379527 Homo sapiens hypothetical gene supported by BC023225 (LOC401431 ), mR
XM_379528 Homo sapiens hypothetical protein LOC90520 (LOC90520), mRNA
XM_379529 Homo sapiens LOC401432 (LOC401432), mRNA
XM_379530 Homo sapiens hypothetical protein LOC285972 (LOC285972), mRNA
XM_379531 Homo sapiens LOC401434 (LOC401434), mRNA
XM_379532 Homo sapiens LOC401435 (LOC401435), mRNA
XM_379533 Homo sapiens LOC401436 (LOC401436), mRNA
XM_379534 Homo sapiens hypothetical gene supported by AK054822 (LOC401437), R
XM_379535 Homo sapiens hypothetical protein LOC285889 (LOC285889), mRNA
XM_379536 Homo sapiens hypothetical gene supported by BC041429 (LOC401438), mR
XM_379537 Homo sapiens hypothetical gene supported by AY166699 (LOC401439), mR
XM_379539 Homo sapiens LOC401440 (LOC401440), mRNA
XM_379540 Homo sapiens hypothetical protein LOC157693 (LOC157693), mRNA
XM_379541 Homo sapiens hypothetical gene supported by AK127852 (LOC401441 ), mR
XM_379543 Homo sapiens hypothetical gene supported by BC028401 (LOC401442), mR
XM_379545 Homo sapiens hypothetical gene supported by BC030648 (LOC401443), mR
XM_379547 Homo sapiens hypothetical gene supported by AK057888 (LOC401445), mR
XM_379548 Homo sapiens hypothetical gene supported by AK124896; BC037255 (LOC4
XM__379550 Homo sapiens hypothetical gene supported by AK091259 (LOC401448), mR
XM_379551 Homo sapiens hypothetical protein LOC349196 (LOC349196), mRNA
XM_379552 Homo sapiens LOC401449 (LOC401449), mRNA
XM_379553 Homo sapiens similar to hypothetical protein LOC157278 (LOC401451 ), mRI
XM_379554 Homo sapiens hypothetical protein LOC157273 (LOC157273), mRNA
XM_379559 Homo sapiens hypothetical protein LOC157278 (LOC157278), mRNA
XM_379562 Homo sapiens LOC401456 (LOC401456), mRNA
XM_379573 Homo sapiens hypothetical protein LOC286135 (LOC286135), mRNA
XM_379582 Homo sapiens hypothetical protein LOC286177 (LOC286177), mRNA
XM_379583 Homo sapiens hypothetical gene supported by AK124256 (LOC401462), mR
XM 379584 Homo sapiens hypothetical gene supported by BC022555; BC050012 (LOC4
XM_379586 Homo sapiens hypothetical protein LOC286186 (LOC286186), mRNA
XM_379587 Homo sapiens LOC401464 (LOC401464), mRNA
XM_379592 Homo sapiens hypothetical protein LOC286144 (LOC286144), mRNA
XM_379594 Homo sapiens hypothetical protein LOC286149 (LOC286149), mRNA
XM_379595 Homo sapiens hypothetical gene supported by AK125891 (LOC401471), mR
XM_379596 Homo sapiens LOC401473 (LOC401473), mRNA
XM_379597 Homo sapiens hypothetical protein FLJ10489 (FLJ10489), mRNA
XM_379601 Homo sapiens hypothetical gene supported by BC036187; NM_0O5839 (LOC
XM_379603 Homo sapiens hypothetical gene supported by BC009730; BC015157 (LOC4
XM_379605 Homo sapiens LOC401477 (LOC401477), mRNA
XM_379608 Homo sapiens hypothetical gene supported by AK056998 (LOC401480), mR
XM_379609 Homo sapiens hypothetical gene supported by BC041936 (LOC401481 ), mR
XM_379610 Homo sapiens LOC401482 (LOC401482), mRNA
XM_379617 Homo sapiens hypothetical gene supported by AK093004 (LOC401488), mR
XM_379618 Homo sapiens hypothetical gene supported by AY343891 ; AY343892; AY34:
XM_379619 Homo sapiens hypothetical gene supported by BC052949 (LOC401490), mR
XM_379622 Homo sapiens hypothetical gene supported by AK092343 (LOC401491), mR
XM_379623 Homo sapiens hypothetical gene supported by AK123194 (LOC401492), mR
XM_379625 Homo sapiens hypothetical gene supported by BC048267; NMJ 78448 (LOC
XM_379627 Homo sapiens LOC401496 (LOC401496), mRNA
XM_379628 Homo sapiens LOC401499 (LOC401499), mRNA
XM_379629 Homo sapiens hypothetical gene supported by AL512690 (LOC401500), mR
XM_379630 Homo sapiens LOC401503 (LOC401503), mRNA
XM_379632 Homo sapiens hypothetical protein LOC158376 (LOC158376), mRNA
XM_379634 Homo sapiens hypothetical gene supported by AK091718 (LOC401504), mR
XM_379635 Homo sapiens LOC401506 (LOC401506), mRNA
XM_379636 Homo sapiens hypothefical protein LOC158228 (LOC158228), mRNA
XM_379637 Homo sapiens hypothefical gene supported by AK127732 (LOC401513), mR
XM_379638 Homo sapiens hypothetical gene supported by AK026419 (LOC401518), mR
XM_379639 Homo sapiens LOC401522 (LOC401522), mRNA
XM_379640 Homo sapiens hypothetical gene supported by BC044751 ; NMJ 75923 (LOC
XM_379641 Homo sapiens hypothetical gene supported by BC062724 (LOC401527), mR
XM_379642 Homo sapiens hypothetical gene supported by BC032955 (LOC401528), mR
XM_379643 Homo sapiens hypothetical gene supported by BC032955 (LOC401530), mR
XM_379644 Homo sapiens hypothetical gene supported by BC000228 (LOC401532), mR
XM_379645 Homo sapiens hypothetical gene supported by AK094988 (LOC401536), mR
XM_379647 Homo sapiens similar to hypothetical protein LOC286238 (LOC401538), mRI
XM_379648 Homo sapiens hypothetical gene supported by AK124333 (LOC401539), mR
XM_379650 Homo sapiens hypothetical protein LOC340515 (LOC340515), mRNA
XM_379651 Homo sapiens chromosome 9 open reading frame 44 (C9orf44), mRNA
XM_379655 Homo sapiens hypothetical gene supported by BC031969 (LOC401 542), mR
XM_379656 Homo sapiens hypothetical gene supported by AK092137 (LOC401 543), mR
XM_379657 Homo sapiens hypothetical gene supported by BC043559 (LOC401 544), mR
XM_379660 Homo sapiens LOC401545 (LOC401545), mRNA
XM_379664 Homo sapiens hypothetical gene supported by BX647840 (LOC401 549), mR
XM_379665 Homo sapiens hypothetical protein LOC286333 (LOC286333), mRNA
XM_379667 Homo sapiens hypothetical gene supported by BC039180 (LOC401 550), mR
XM_379668 Homo sapiens hypothetical gene supported by AK124723; AL833509 (LOC4
XM_379671 Homo sapiens hypothetical gene supported by BC019073; BC036842; BC04
XM_379672 Homo sapiens hypothetical gene supported by AK128673 (LOC401 554), mR
XM_379676 Homo sapiens hypothetical gene supported by AK127261 (LOC401 557), mR
XM_379677 Homo sapiens hypothetical gene supported by AK094119 (LOC401558), mR
XM_379678 Homo sapiens hypothetical gene supported by BC029166 (LOC401559), mR
XM_379680 Homo sapiens hypothetical gene supported by AK023162 (LOC90120), mRI*.
XM_379682 Homo sapiens hypothetical gene supported by AY129027 (LOC401561 ), mR
XM_379684 Homo sapiens hypothetical protein LOC286238 (LOC286238), mRNA
XM_379686 Homo sapiens hypothetical gene supported by AK074437 (LOC401571 ), mR
XM_379688 Homo sapiens hypothetical gene supported by BX648912 (LOC401573), mR
XM_379690 Homo sapiens hypothetical protein LOC284593 (LOC284593), mRNA
XM_379691 Homo sapiens hypothetical protein LOC284600 (LOC284600), mRNA
XM_379692 Homo sapiens LOC401574 (LOC401574), mRNA
XM_379693 Homo sapiens LOC401575 (LOC401575), mRNA
XM_379694 Homo sapiens hypothetical gene supported by AK125149 (LOC401577), mR
XM_379695 Homo sapiens hypothetical gene supported by BC056508; NM_0O4679 (LOC
XM_379696 Homo sapiens hypothefical gene supported by AK057918 (LOC401579), mR
XM_379697 Homo sapiens hypothetical gene supported by AL832542 (LOC401582), mR
XM_379699 Homo sapiens LOC401583 (LOC401583), mRNA
XM_379700 Homo sapiens hypothetical protein LOC286442 (LOC286442), mRNA
XM_379702 Homo sapiens hypothetical gene supported by AK098783 (LOC401585), mR
XM_379703 Homo sapiens hypothetical gene supported by AK130892 (LOC401587), mR
XM_379704 Homo sapiens hypothetical gene supported by AK056314; BC034616 (LOC4
XM_379705 Homo sapiens hypothetical protein LOC158572 (LOC158572), mRNA
XM_379714 Homo sapiens LOC401596 (LOC401596), mRNA
XM_379715 Homo sapiens hypothetical gene supported by AK125301 (LOC401597), mR
XM_379716 Homo sapiens hypothetical gene supported by AK057746 (LOC401599), mR
XM_379717 Homo sapiens hypothetical gene supported by AJ421269; AL359612; NM_0'
XM_379720 Homo sapiens hypothetical gene supported by BX537697 (LOC401613), mR
XM_379721 Homo sapiens hypothetical gene supported by BX640956 (LOC401615), mR
XM_379722 Homo sapiens hypothetical gene supported by AK057519 (LOC401616), mR
XM_379723 Homo sapiens hypothetical gene supported by AK094280 (LOC401617), mR
XM_379728 Homo sapiens hypothetical protein LOC286411 (LOC286411), mRNA
XM_379730 Homo sapiens hypothetical gene supported by BC010531 (LOC401621), mR
XM_379735 Homo sapiens LOC401626 (LOC401626), mRNA
XM_379736 Homo sapiens hypothefical gene supported by AK125149 (LOC401628), mR
XM_379738 Homo sapiens LOC401629 (LOC401629), mRNA
XM_379739 Homo sapiens LOC401630 (LOC401630), mRNA
XM 379741 Homo sapiens LOC401701 (LOC401701), mRNA
XM_379749 Homo sapiens LOC401880 (LOC401880), mRNA
XM_379760 Homo sapiens LOC402387 (LOC402387), mRNA
XM_379761 Homo sapiens LOC402433 (LOC402433), mRNA
XM 379766 Homo sapiens unc-84 homolog A (C. elegans) (UNC84A), mRNA
XM_379767 Homo sapiens DKFZP586J0619 protein (DKFZP586J0619), mRNA
XM_379771 Homo sapiens KIAA0415 gene product (KIAA0415), mRNA
XM_379772 Homo sapiens hypothetical gene supported by BC031661 (LOC4O2450), mR
XM_379773 Homo sapiens hypothetical protein LOC285924 (LOC285924), mRNA
XM__379774 Homo sapiens KIAA1856 protein (KIAA1856), mRNA
XM_379775 Homo sapiens hypothetical gene supported by AK125308 (LOC402452), mR
XM_379776 Homo sapiens hypothetical gene supported by AK123535 (LOC402454), mR
XM_379777 Homo sapiens similar to Oncomodulin (OM) (Parvalbumin beta) (LOC40245e
XM__379780 Homo sapiens similar to unc-93 homolog B1 ; unc93 (C.elegans) homolog B;
XM_379781 Homo sapiens similar to beta-1 ,4-mannosyltransferase; beta-1 ,4 mannosyltn
XM 379783 Homo sapiens hypothetical gene supported by AK027125 (LOC4O2460), mR
XM_379784 Homo sapiens glucocorticoid induced transcript 1 (GLCCI1), mRNA
XM_379786 Homo sapiens similarto Chain , Heat-Shock Cognate 70kd Protein (44kd Atf
XM_379787 Homo sapiens similar to heat shock 70kDa protein 8 isoform 2; heat shock o
XM 379788 Homo sapiens PHD finger protein 14 (PHF14), mRNA
X 37Q792 Homo sapiens similar to TWIST neighbor (LOC402464), mRNA
XM_379793 Homo sapiens ribosomal protein L21 (RPL21), mRNA
XM_379794 Homo sapiens similar to ribosomal protein L23 (LOC402465), mRNA
XM_379796 Homo sapiens similar to Dual specificity protein kinase CLK2 (CDC like kinas
XM_379797 Homo sapiens similar to FKSG54 (LOC402469), mRNA
XM_379798 Homo sapiens KIAA0087 gene product (KIAA0087), mRNA
XM_379800 Homo sapiens KIAA0644 gene product (K1AA0644), mRNA
XM_379801 Homo sapiens KIAA0241 protein (KIAA0241), mRNA
XM_379802 Homo sapiens LOC89231 (LOC89231), mRNA
XM_379803 Homo sapiens similar to KIAA0877 protein (LOC402477), mRNA
XM 379806 Homo sapiens similar to RP9 protein (LOC402478), mRNA
XM_379807 Homo sapiens similar to RIKEN cDNA 9330128H10 gene (LOC402479), R
XM_379809 Homo sapiens hypothetical gene supported by AF447883 (LOC402481), mR
XM 379812 Homo sapiens similar to TRGV9 (LOC402482), mRNA
XM_379815 Homo sapiens similar to sequence-specific single-stranded-DNA-binding pro
XM 379816 Homo sapiens similar to t-complex 1 ; T-complex locus TCP-1 ; t-complex 1 (a
XM_379817 Homo sapiens similar to RAS p21 protein activator 4; GTPase activating prot
XM 379818 Homo sapiens similar to RAS p21 protein activator 4; GTPase activating prot
XM_379819 Homo sapiens similar to DNA directed RNA polymerase II polypeptide J-relai
XM_379820 Homo sapiens similar to Ras GTPase-activating protein 4 (RasGAP-activatin
XM_379824 Homo sapiens similar to cell division cycle 10 homolog (LOC402491), mRNA
XM_379825 Homo sapiens similar to hypothetical protein FLJ25976 (LOC402492), mRN/
XMJ379827 Homo sapiens hypothetical gene supported by AK126096 (LOC402494), mR
XMJ379830 Homo sapiens similar to KIAA0207 (LOC402495), mRNA
XM_379831 Homo sapiens hypothetical gene supported by AK097404; NMJ98284 (LOC
XM_379832 Homo sapiens hypothetical gene supported by AK127870 (LOC402497), mR
XM_379834 Homo sapiens similar to SMT3 suppressor of mif two 3 homolog 2 (LOC402E
XM_379835 Homo sapiens similar to hypothetical protein DKFZp434F142 (LOC402501 ),
XM_379836 Homo sapiens similar to septin 10 isoform 1 (LOC402502), mRNA
XM 379838 Homo sapiens similar to bA145E8.1 (KIAA1074) (LOC402505), mRNA
XM_379839 Homo sapiens similar to CAGL79 (LOC402508), mRNA
XM_379840 Homo sapiens similar to solute carrier family 29 (nucleoside transporters), m*
XMJ379841 Homo sapiens zinc finger protein 479 (ZNF479), mRNA
XM_379842 Homo sapiens LOC402513 (LOC402513), mRNA
XMJ579843 Homo sapiens hypothetical gene supported by BC040831 (LOC402514), mP
XM_379844 Homo sapiens hypothetical gene supported by BC040831 (LOC402517), mR
XM 379845 Homo sapiens similar to solute carrier family 29 (nucleoside transporters), nr
XM_379846 Homo sapiens similar to BC060615 protein (LOC402519), mRNA
XM_379847 Homo sapiens similar to CAGL79 (LOC402520), mRNA
XM_379848 Homo sapiens similar to hypothetical protein LOC285908 (LOC402521), mRI
XM_379849 Homo sapiens similar to GA binding protein transcription factor, alpha subun
XM_379850 Homo sapiens similar to hypothetical ZNF-like protein (LOC402524), mRNA
XM_379851 Homo sapiens similar to RPL6 protein (LOC402525), mRNA
XM_379852 Homo sapiens similar to envelope protein (LOC402526), mRNA
XM_379853 Homo sapiens similar to myelin protein zero-like 1 ; protein zero related (LOC
XM_379854 Homo sapiens similar to hypothetical protein FLJ25037 (LOC402529), mRN/
XM_379855 Homo sapiens similar to hypothetical protein MGC16733 similar to CG12113
XM_379856 Homo sapiens similarto MGC16733 protein (LOC402531), mRNA
XM_379857 Homo sapiens similar to 60S ribosomal protein L35 (LOC402536), mRNA
XM_379858 Homo sapiens similar to Williams-Beuren syndrome critical region protein 19
XM_379859 Homo sapiens similar to hypothetical protein FLJ 10900 (LOC402540), mRN/
XM_379860 Homo sapiens similar to Williams Beuren syndrome chromosome region 19 1
XM_379861 Homo sapiens similar to FKBP6 protein (LOC402543), mRNA
XM_379862 Homo sapiens similar to Brutons tyrosine kinase-associated protein-135; BAI
XM__379863 Homo sapiens similar to Neutrophil cytosol factor 1 (NCF-1) (Neutrophil NAD
XM _379864 Homo sapiens similar to transcription factor GTF2IRD2 (LOC402546), mRNA
XM__379865 Homo sapiens similar to Nuclear envelope pore membrane protein POM 121
XM_379866 Homo sapiens Williams Beuren syndrome chromosome region 24 (WBSCRϊ
XM_379868 Homo sapiens similar to Neutrophil cytosolic factor 1 (LOC402549), mRNA
XM_379869 Homo sapiens similar to transcription factor GTF2IRD2 (LOC402550), mRNA
XM__379871 Homo sapiens similar to RCC1-like G exchanging factor-like isoform 1 ; RCO
XM_379872 Homo sapiens similar to PMS4 (LOC402552), mRNA
XM_379874 Homo sapiens similar to PMS4 homolog mismatch repair protein - human (L(
XM 379875 Homo sapiens similar to FKBP6 protein (LOC402555), mRNA
XMJ379876 Homo sapiens tripartite motif-containing 50B (TRIM50B), mRNA
XM_379877 Homo sapiens similar to Nuclear envelope pore membrane protein POM 121
XM__379879 Homo sapiens similar to hypothetical protein LOC285908 (LOC402558), mRI
XM_379881 Homo sapiens similar to hypothetical protein (LOC402559), mRNA
XM_379883 Homo sapiens similar to Piccolo protein (Aczonin) (LOC402561 ), mRNA
XM_379884 Homo sapiens sema domain, immunoglobulin domain (Ig), short basic doma
XM_379885 Homo sapiens similar to Heterogeneous nuclear ribonucleoprotein A1 (Helix-
XM_379887 Homo sapiens similar to hypothetical protein 4932412H11 (LOC402565), mF
XM_379891 Homo sapiens hypothetical gene supported by AK124274 (LOC402566), mR
XM_379892 Homo sapiens similar to Homeobox protein DLX-6 (LOC402567), mRNA
XM 379893 Homo sapiens similar to CG14980-PB (LOC375601 ), mRNA
XM_379894 Homo sapiens similar to importin alpha 1b (LOC402569), mRNA
XM_379895 Homo sapiens hypothetical protein FLJ22037 (FLJ22037), mRNA
XM_379896 Homo sapiens hypothetical protein LOC285989 (LOC285989), mRNA
XM_379897 Homo sapiens similar to Zinc-alpha-2-glycoprotein precursor (Zn-alpha-2-gly
XM_379899 Homo sapiens similar to hypothetical protein MGC49416 (LOC402572), mRt*
XM_379901 Homo sapiens hypothetical gene supported by BC031966 (LOC402573), mR
XM_379904 Homo sapiens similar to mucin 11 (LOC402575), mRNA
XM_379905 Homo sapiens similarto intestinal membrane mucin MUC17 (LOC402576), n
XM_379906 Homo sapiens similar to PMS5 homolog mismatch repair protein - human (L<
XM_379908 Homo sapiens similar to DNA directed RNA polymerase II polypeptide J-relal
XM 379909 Homo sapiens similar to M-phase phosphoprotein 11 (LOC402580), mRNA
XM_379910 Homo sapiens similar to reverse transcriptase related protein (LOC402581 ),
XM_379911 Homo sapiens similarto KIAA1218 protein (LOC402584), mRNA
XM_379913 Homo sapiens similar to calcium-independent phospholipase A2 (LOC40258
XM_379914 Homo sapiens hypothetical protein LOC286009 (LOC286009), mRNA
XM__379917 Homo sapiens similarto hyaluronoglucosaminidase 1 isoform 1; hyaluronida:
XM_379919 Homo sapiens similar to hypothetical protein FLJ25976 (LOC402594), mRN/
XM__379920 Homo sapiens similar to hypothetical protein LOC285908 (LOC402595), mRI
XM_379921 Homo sapiens hypothetical protein LOC346653 (LOC346653), mRNA
XM_379923 Homo sapiens KIAA1170 protein (KIAA1170), mRNA
XM 379927 Homo sapiens plexin A4 (PLXNA4), mRNA
XM_379931 Homo sapiens hypothetical protein LOC155006 (LOC155006), mRNA
XM _379932 Homo sapiens KIAA1549 protein (KIAA1549), mRNA
XM_379933 Homo sapiens hypothetical protein FLJ25778 (FLJ25778), mRNA
XM_379934 Homo sapiens similar to A630082K20Rik protein (LOC402600), mRNA
XM_379935 Homo sapiens similar to RAB19, member RAS oncogene family (LOC40260'
XM_379936 Homo sapiens similar to hypothetical protein (LOC402602), mRNA
XM_379938 Homo sapiens LCHN protein (LCHN), mRNA
XM_379939 Homo sapiens similar to RIKEN cDNA 1700016G05 (LOC402604), mRNA
XM_379940 Homo sapiens hypothetical protein LOC93432 (LOC93432), mRNA
XM_379954 Homo sapiens similar to KIAA0738 protein (LOC402618), mRNA
XM_379958 Homo sapiens similar to hypothetical protein MGC41943 (LOC402619), mRI*^
XM 379959 Homo sapiens FLJ43692 protein (FLJ43692), mRNA
XM_379962 Homo sapiens similar to KIAA1285 protein (LOC402620), mRNA
XM_379964 Homo sapiens likely ortholog of mouse zinc finger protein EZI (EZI), mRNA
XM~379965 Homo sapiens FLJ45737 protein (FLJ45737), mRNA
XM_379966 Homo sapiens similar to KIAA2036 protein (LOC402621), mRNA
XM 379967 Homo sapiens KIAA0543 protein (KIAA0543), mRNA
XM 379968 Homo sapiens chromosome 7 open reading frame 32 (C7orf32), mRNA
XM 379970 Homo sapiens similar to Zinc finger protein 84 (Zinc finger protein HPF2) (LC
XM 379974 Homo sapiens KIAA1402 protein (CSGIcA-T), mRNA
XM_379975 Homo sapiens hypothetical gene supported by AK12771 (LOC402625), mR
XM_379976 Homo sapiens similar to Fatty acid-binding protein, epidermal (E-FABP) (Psc
XM_379977 Homo sapiens similar to T-complex protein 1 (LOC402629), mRNA
XM_379979 Homo sapiens hypothetical protein LOC285888 (LOC285888), mRNA
XM_379980 Homo sapiens hypothetical protein LOC155435 (LOC155435), mRNA
XM_379983 Homo sapiens ubiquitin-protein isopeptide ligase (E3) (KIAA0010), mRNA
XM_379986 Homo sapiens similar to hypothetical protein FLJ37300 (LOC402633), mRN/
XM_379987 Homo sapiens similar to mesenchymal stem cell protein DSC92; neurite outc
XM_379988 Homo sapiens similar to galectin-related inter-fiber protein (LOC402635), mF
XM_379989 Homo sapiens similar to olfactory receptor MOR267-3 (LOC402636), mRNA
XM_379990 Homo sapiens similar to Calgizzarin (S100C protein) <MLN 70) (LOC402637)
XM_379991 Homo sapiens similar to ribosomal protein L31 (LOC402638), mRNA
XM_379993 Homo sapiens similar to olfactory receptor M0R145-2 (LOC402639), mRNA
XM_379994 Homo sapiens similar to 4930579E17Rik protein (LOC402640), mRNA
XM_379995 Homo sapiens similar to MGC76216 protein (LOC402641), mRNA
XM_379996 Homo sapiens similar to solute carrier family 40 (iron-regulated transporter),
XM_379997 Homo sapiens similar to tropomyosin 3 (LOC402643> , mRNA
XM_379998 Homo sapiens similar to peptidylprolyl isomerase A (LOC402644), mRNA
XM_379999 Homo sapiens similar to FLJ40113 protein (LOC402S45), mRNA
XM_380002 Homo sapiens similar to ELK1 (LOC402648), mRNA
XM_380005 Homo sapiens similar to equilibrative nucleoside transporter 4; hENT4 (LOG
XM_380006 Homo sapiens similar to beta-glucuronidase (LOC402662), mRNA
XM _380007 Homo sapiens similar to vomeronasal receptor V1 RC 3 (LOC402663), mRNA
XM_380008 Homo sapiens similar to Opioid binding protein/cell adhesion molecule precu
XM_380009 Homo sapiens similar to protein kinase related to RaF protein kinases; Metho
XM_380010 Homo sapiens similar to metabotropic glutamate receptor 8; G protein-coup
XM_380011 Homo sapiens similar to GTF2I repeat domain contai ring 1 isoform 2; Williar
XM_380012 Homo sapiens similar to opposite strand transcription unit to Stag3; Gats pro
XM_380013 Homo sapiens similar to GrpE protein homolog 1 , mitochondrial precursor (tV
XM_380014 Homo sapiens similar to GTF2I repeat domain contai ring 1 isoform 2; Williar
XM_380015 Homo sapiens similar to peptidylprolyl isomerase A (LOC402673), mRNA
XM_380018 Homo sapiens hypothetical protein DKFZP434A0225 (DKFZP434A0225), mF
XM_380019 Homo sapiens similarto RIKEN cDNA 4930511M11 CLOC402675), mRNA
XM_380020 Homo sapiens similar to Protein C6orf66 (HSPC125) (My013 protein) (LOC
XM_380021 Homo sapiens similar to ribosomal protein S3a; 40S ribosomal protein S3a; \
XM_380022 Homo sapiens similar to Ser/Thr protein kinase PAR- 1 Balpha (LOC402679),
XM_380024 Homo sapiens similar to opposite strand transcription unit to Stag3; Gats pro
XM_380025 Homo sapiens similar to PMS2L13 (LOC402681), mRNA
XM _380026 Homo sapiens similar to RIKEN cDNA 2700038N03 CLOC402682), mRNA
XM_380028 Homo sapiens similar to 40S RIBOSOMAL PROTEIM SA (P40) (34/67 KD L/
XM_380031 Homo sapiens similar to KIAA0538 protein (LOC402Θ84), mRNA
XM_380032 Homo sapiens similar to Ras GTPase-activating protein 4 (RasGAP-activatin
XM_380033 Homo sapiens similar to 60S ribosomal protein L23a (LOC402686), mRNA
XM_380034 Homo sapiens similar to Argininosuccinate synthase (Citrulline-aspartate lig.
XM_380036 Homo sapiens similar to RIKEN cDNA 6332401019 gene (LOC402689), mR
XM_380038 Homo sapiens similar to Triosephosphate isomerase (TIM) (LOC402691 ), ml
XM_380039 Homo sapiens similar to aldo-keto reductase family 1 , member B10; aldose r
XM _380040 Homo sapiens similar to beta-tubulin (LOC402693), mRNA
XM_380042 Homo sapiens similar to 60S ribosomal protein L15 ( LOC402694), mRNA
XM_380044 Homo sapiens similar to 60S ribosomal protein L17 ( L23) (Amino acid starva
XM_380045 Homo sapiens similar to Nucleoside diphosphate kinase, mitochondrial preci
XM_380047 Homo sapiens similar to Nucleoside diphosphate kinase, mitochondrial preci
XM_380048 Homo sapiens similar to Olfactory receptor 9A4 (LOC402698), mRNA
XM_380055 Homo sapiens similar to Olfactory receptor 9A2 (LOC402707), mRNA
XM _380056 Homo sapiens similar to Olfactory receptor 6V1 (LOC402708), mRNA
XM_380057 Homo sapiens similar to Histidine triad nucleotide-binding protein 1 (Adenosi
XM_380059 Homo sapiens similar to Olfactory receptor 2F2 (Olfactory receptor 7-1 ) (OR'i
XM_380063 Homo sapiens similar to Olfactory receptor 2A12 (LOC402711), mRNA
XM_380067 Homo sapiens similar to Olfactory receptor 2A1 (LOC402712), mRNA
XM_380068 Homo sapiens similar to Olfactory receptor 2A1 (LOC402713), mRNA
XM_380069 Homo sapiens similar to OG-2 homeodomain protein-like; similar to U65067
XM_380070 Homo sapiens similar to Importin alpha-2 subunit (Karyopherin alpha-2 subui
XM _380072 Homo sapiens similar to 60S ribosomal protein L32 CLOC402716), mRNA
XM_380073 Homo sapiens similar to Huntingtin interacting protei n K (LOC402717), mRN,
XM_380074 Homo sapiens similar to BET1 homolog (Golgi vesicular membrane traffickin
Page 403
XM_380076 Homo sapiens similar to TSH receptor suppressor element-binding protein-1
XM_380077 Homo sapiens similar to ppg3 (LOC402720), mRNA
XM_380078 Homo sapiens similar to S-adenosylmethionine decarboxylase 1 ; S-adenosyl
XM_380081 Homo sapiens hypothetical protein FLJ36112 (FLJ36112), mRNA
XM_380082 Homo sapiens hypothetical gene supported by AK125766 (LOC402448), mR
XM_380085 Homo sapiens hypothetical gene supported by BC032734 (LOC402449), mR
XM_380087 Homo sapiens LOC402451 (LOC402451), mRNA
XM_380088 Homo sapiens LOC402453 (LOC402453), mRNA
XM_380089 Homo sapiens hypothetical gene supported by AK127500 (LOC402463), mR
XM_380091 Homo sapiens hypothetical gene supported by BC039682 (LOC402466), mR
XM_380092 Homo sapiens hypothetical protein LOC285941 (LOC285941), mRNA
XM_380093 Homo sapiens hypothetical gene supported by BC025338 (LOC402470), mR
XM_380094 Homo sapiens hypothetical gene supported by AK093987 (LOC402471 ), mR
XM_380095 Homo sapiens LOC402472 (LOC402472), mRNA
XM_380096 Homo sapiens LOC402473 (LOC402473), mRNA
XM_380097 Homo sapiens hypothetical gene supported by BC023581 ; BC044638 (LOC4
XM_380098 Homo sapiens hypothetical gene supported by BC016976 (LOC402475), mR
XM_380099 Homo sapiens hypothetical gene supported by AK092714 (LOC402476), mR
XM_380100 Homo sapiens hypothetical gene supported by AL832092 (LOC402480), mR
XM_380103 Homo sapiens hypothetical gene supported by AK127273 (LOC402483), mR
XM_380104 Homo sapiens LOC402485 (LOC402485), mRNA
XM_380105 Homo sapiens hypothetical protein LOC285958 (LOC285958), mRNA
XM_380106 Homo sapiens hypothetical gene supported by AK125311 (LOC402493), mR
XM_380107 Homo sapiens hypothefical gene supported by AK024248; AL137733; BC06:
XM_380108 Homo sapiens LOC402506 (LOC402506), mRNA
XM_380109 Homo sapiens LOC402507 (LOC402507), mRNA
XM_380110 Homo sapiens LOC402511 (LOC402511 ), mRNA
XM_380111 Homo sapiens hypothetical gene supported by BX648489 (LOC402512), mR
XM_380112 Homo sapiens LOC402515 (LOC402515), mRNA
XM_380113 Homo sapiens LOC402516 (LOC402516), mRNA
XM_380114 Homo sapiens LOC402523 (LOC402523), mRNA
XM_380115 Homo sapiens LOC402528 (LOC402528), mRNA
XM_380117 Homo sapiens LOC402532 (LOC402532), mRNA
XM_380118 Homo sapiens hypothetical gene supported by AK024371 ; BC037920 (LOC4
XM_380119 Homo sapiens hypothetical gene supported by AK054923; AK126730; NM_0
XM_380120 Homo sapiens LOC402537 (LOC402537), mRNA
XM_380121 Homo sapiens LOC402538 (LOC402538), mRNA
XM_380122 Homo sapiens LOC402542 (LOC402542), mRNA
XM_380125 Homo sapiens LOC402553 (LOC402553), mRNA
XM_380126 Homo sapiens hypothetical gene supported by AK091784 (LOC402557), mR
XM_380127 Homo sapiens LOC402560 (LOC402560), mRNA
XM_380128 Homo sapiens LOC402563 (LOC402563), mRNA
XM_380129 Homo sapiens LOC402564 (LOC402564), mRNA
XM_380131 Homo sapiens hypothefical protein FLJ34048 (FLJ34048), mRNA
XM_380133 Homo sapiens LOC402568 (LOC402568), mRNA
XM_380134 Homo sapiens LOC402574 (LOC402574), mRNA
XM_380135 Homo sapiens LOC402578 (LOC402578), mRNA
XM_380136 Homo sapiens LOC402582 (LOC402582), mRNA
XM_380137 Homo sapiens hypothetical protein LOC340340 (LOC340340), mRNA
XM _380138 Homo sapiens LOC402586 (LOC402586), mRNA
XM_380139 Homo sapiens hypothetical gene supported by BX537645 (LOC402587), mR
XM_380140 Homo sapiens LOC402589 (LOC402589), mRNA
XM_380141 Homo sapiens hypothetical gene supported by BX648695 (LOC402590), mR
XM_380142 Homo sapiens hypothetical gene supported by BC063892 (LOC402591), mR
XM_380143 Homo sapiens LOC402592 (LOC402592), mRNA
XM_380146 Homo sapiens hypothetical gene supported by AK125651 ; BC039420 (LOC4
XM_380147 Homo sapiens LOC402597 (LOC402597), mRNA
XM_380148 Homo sapiens LOC402598 (LOC402598), mRNA
XM_380149 Homo sapiens LOC402599 (LOC402599), mRNA
XM_380150 Homo sapiens hypothetical gene supported by BX648692 (LOC402603), mR
XM_380151 Homo sapiens LOC402617 (LOC402617), mRNA
XM_380152 Homo sapiens hypothetical protein LOC90520 (LOC90520), mRNA
XM_380153 Homo sapiens LOC402622 (LOC402622), mRNA
XM_380l54 Homo sapiens hypothetical protein LOC285972 (LOC285972), mRNA
XM_380155 Homo sapiens LOC402626 (LOC402626), mRNA
XM_380156 Homo sapiens LOC402627 (LOC402627), mRNA
XM_380157 Homo sapiens LOC402630 (LOC402630), mRNA
XM_380158 Homo sapiens hypothetical gene supported by AY166699 (LOC402631 ), mR
XM_380159 Homo sapiens hypothetical gene supported by AK054822 (LOC402632), mR
XM_380160 Homo sapiens hypothetical protein LOC285889 (LOC285889), mRNA
XM_380162 Homo sapiens hypothetical protein LOC154822 (LOC154822), mRNA
XM_380170 Homo sapiens phosphodiesterase 4D interacting protein (myomegalin) (PDE
XM_380171 Homo sapiens iroquois homeobox protein 1 (1RX1), mRNA
XM_380173 Homo sapiens hypothetical protein MGC39830 (MGC39830), mRNA
XM_380174 Homo sapiens similar to aquaporin 11 (LOC285192), mRNA
XM_380175 Homo sapiens hypothetical protein MGC22265 (MGC22265), mRNA
XM_380176 Homo sapiens similar to hypothetical protein D030010E02 (LOC401952), mF
NC_000001 Homo sapiens chromosome 1 , complete sequence
NC_000002 Homo sapiens chromosome 2, complete sequence
NC_000003 Homo sapiens chromosome 3, complete sequence
NC_000004 Homo sapiens chromosome 4, complete sequence
NC_000005 Homo sapiens chromosome 5, complete sequence
NC_000006 Homo sapiens chromosome 6, complete sequence
NC_000007 Homo sapiens chromosome 7, complete sequence
NC_000008 Homo sapiens chromosome 8, complete sequence
NCJ300009 Homo sapiens chromosome 9, complete sequence
NC_000010 Homo sapiens chromosome 10, complete sequence
NC_000011 Homo sapiens chromosome 11 , complete sequence
NC_000012 Homo sapiens chromosome 12, complete sequence
NC_000013 Homo sapiens chromosome 13, complete sequence
NC_000014 Homo sapiens chromosome 14, complete sequence
NC_000015 Homo sapiens chromosome 15, complete sequence
NCJ000016 Homo sapiens chromosome 16, complete sequence
NC_00001 Homo sapiens chromosome 17, complete sequence
NC_000018 Homo sapiens chromosome 18, complete sequence ,
NC_000019 Homo sapiens chromosome 19, complete sequence
NC_000020 Homo sapiens chromosome 20, complete sequence
NC_000021 Homo sapiens chromosome 21 , complete sequence
NC_000022 Homo sapiens chromosome 22, complete sequence
NC_000023 Homo sapiens chromosome X, complete sequence
NC_000024 Homo sapiens chromosome Y, complete sequence
NC_001807 Homo sapiens mitochondrion, complete genome
NG_000002 Homo sapiens immunoglobulin lambda locus (IGL@) on chromosome 22
NG_000004 Homo sapiens cytochrome P450, family 3, subfamily A (CYP3A) on chromos
NGJ300OO6 Homo sapiens alpha globin region (HBA@) on chromosome 16
NG_000007 Homo sapiens beta globin region (HBB@) on chromosome 11
NG 00008 Homo sapiens cytochrome P450, family 2, subfamily A (CYP2A) on chromos
NG_000009 Homo sapiens small histone family cluster (HFS@) on chromosome 6
NG_000012 Homo sapiens protocadherin gamma cluster (PCDHG@) on chromosome 5
NG_000013 Homo sapiens MHC class III complement gene cluster, monomodular haplot
NG_000016 Homo sapiens genomic protocadherin alpha cluster (PCDHA@) on chromosi
NG_000017 Homo sapiens protocadherin beta cluster (PCDHB@) on chromosome 5
NG_000018 Homo sapiens type I (acidic) hair keratin gene cluster (KRTHA.1@) on chron
NG_000019 Homo sapiens chorionic gonadotropin beta region (CGB@) on chromosome
NG_000827 Homo sapiens genomic histone family microcluster (HFM@) on chromosome
NG D00833 Homo sapiens immunoglobulin kappa locus, distal duplicated V-cluster (IGK-
NG_000834 Homo sapiens immunoglobulin kappa locus, proximal V-cluster and J-C clusl
NG_000837 Homo sapiens surfeit locus (SURF@) on chromosome 9
NG_000839 Homo sapiens cystatin locus (CST@) on chromosome 20
NGJD00840 Homo sapiens actin, beta pseudogene 8 (ACTBP8) on chromosome 6
NG_000841 Homo sapiens actin, gamma pseudogene 2 (ACTGP2) on chromosome Y
NG_000842 Homo sapiens actin, gamma pseudogene 3 (ACTGP3) on chromosome 20
NGJ300843 Homo sapiens adenosine A2b receptor pseudogene (ADORA2BP) on chrom
NG_000845 Homo sapiens argininosuccinate synthetase pseudogene 2 (ASSP2) on chro
NGJD00846 Homo sapiens argininosuccinate synthetase pseudogene 4 (ASSP4) on chro
NG_000847 Homo sapiens argininosuccinate synthetase pseudogene 5 (ASSP5) on chro
NG_000848 Homo sapiens argininosuccinate synthetase pseudogene 6 (ASSP6) on chro
NG_000849 Homo sapiens ATPase, Na+/K+ transporting, beta 3 pseudogene (ATP1 B3P
NG_000850 Homo sapiens BCL2-like 7 pseudogene 1 (BCL2L7P1) on chromosome 20
NG_000851 Homo sapiens crystallin, beta B2 pseudogene 1 (CRYBB2P1) on chromoson
NG_000852 Homo sapiens cysteine and glycine-rich protein 2 pseudogene (CSRP2P) on
NGJ300853 Homo sapiens cytochrome P450, family 2, subfamily D, polypeptide 8 pseudi
NG_000854 Homo sapiens cytochrome P450, family 2, subfamily D, polypeptide 8 pseudi
NG_000858 Homo sapiens dihydrofolate reductase pseudogene 1 (DHFRP1) on chromos
NG_000859 Homo sapiens eukaryotic translation initiation factor 5A pseudogene 1 (EIF5;
NGJD00861 Homo sapiens glycerol kinase pseudogene 3 (GKP3) on chromosome 4
NG_000862 Homo sapiens quanine nucleotide binding protein (G protein), q polypeptide ]
NG_000863 Homo sapiens hydroxyacyl-Coenzyme A dehydrogenase/3-ketoacyl-Coenzyt
NG J00865 Homo sapiens proliferating cell nuclear antigen pseudogene (PCNAP) on chi
NG_000866 Homo sapiens RNA binding motif, single stranded interacting protein 1 , pseu
NG_000867 Homo sapiens radixin pseudogene 2 (RDXP2) on chromosome X
NG_000868 Homo sapiens ribosomal protein L21 pseudogene 1 (RPL21P1) on chromost
NG_000869 Homo sapiens ribosomal protein L32 pseudogene 1 (RPL32P1) on chromost
NG_000870 Homo sapiens ribonucleotide reductase M2 polypeptide pseudogene 3 (RRI
NG_000871 Homo sapiens ribonucleotide reductase M2 polypeptide pseudogene 4 (RRIV
NG_000872 Homo sapiens tRNA phosphoserine (opal suppressor) pseudogene 1 (TRSP
NG_000873 Homo sapiens makorin, ring finger protein, pseudogene 1 (MKRNP1) on chrc
NG_000874 Homo sapiens heterogeneous nuclear ribonucleoprotein D (AU-rich element
NG_000877 Homo sapiens ribosomal protein L3 pseudogene 1 (RPL3P1) on chromosom
NG_000878 Homo sapiens ribosomal protein L23a pseudogene 3 (RPL23AP3) on chrom'
NG_000880 Homo sapiens arylsulfatase E pseudogene (ARSEP) on chromosome Y
NG_000881 Homo sapiens arylsulfatase D pseudogene (ARSDP) on chromosome Y
NG_000882 Homo sapiens voltage-dependent anion channel 5, pseudogene (VDAC5P) c
NGJ300883 Homo sapiens high-mobility group (nonhistone chromosomal) protein 1-like
NG_000884 Homo sapiens snail homolog 1 like 1 (Drosophila) (SNAI1L1) pseudogene or
NG_000885 Homo sapiens nucleophosmin 1 (nucleolar phosphoprotein B23, numatrin) p
NG_000886 Homo sapiens nucleophosmin 1 (nucleolar phosphoprotein B23, numatrin) p:
NG_000887 Homo sapiens nucleophosmin 1 (nucleolar phosphoprotein B23, numatrin) p.
NG_000889 Homo sapiens ribosomal protein L41, pseudogene 3 (RPL41P3) on chromos
NG_000890 Homo sapiens ribosomal protein L41, pseudogene 2 (RPL41P2) on chromos
NG_000891 Homo sapiens ribosomal protein L41, pseudogene 1 (RPL41P1) on chromos
NG_000892 Homo sapiens high-mobility group (nonhistone chromosomal) protein 17-like
NG_000893 Homo sapiens ribosomal protein S24 pseudogene 1 (RPS24P1 ) on chromos
NG_000894 Homo sapiens ribosomal protein L34 pseudogene 2 (RPL34P2) on chromost
NG_000895 Homo sapiens ribosomal protein L34 pseudogene 1 (RPL34P1) on chromosc
NG_000896 Homo sapiens ribosomal protein S5 pseudogene 1 (RPS5P1) on chromoson
NG_000897 Homo sapiens high-mobility group (nonhistone chromosomal) protein 1-like £
NG_000898 Homo sapiens zinc finger protein 299 pseudogene (ZNF299P) on chromosor
NG_000899 Homo sapiens voltage-dependent anion channel 2 pseudogene (VDAC2P) o
NG_000900 Homo sapiens tubulin, alpha pseudogene (TUBAP) on chromosome 21
NG_000901 Homo sapiens solute carrier family 6, member 6 pseudogene (SLC6A6P) on
NG_000903 Homo sapiens ribosomal protein S5-like (RPS5L) pseudogene on chromosoi
NG_000904 Homo sapiens ribosomal protein S3A pseudogene 1 (RPS3AP1 ) on chromos
NG_000906 Homo sapiens ribosomal protein S20, pseudogene 1 (RPS20P1) on chromos
NG_000907 Homo sapiens ribosomal protein L34 pseudogene 3 (RPL34P3) on chromosc
NG_000908 Homo sapiens ribosomal protein L31 pseudogene 1 (RPL31P1) on chromosc
NG_000909 Homo sapiens ribosomal protein L23 pseudogene 2 (RPL23P2) on chromosc
NG_000910 Homo sapiens ribosomal protein L23a pseudogene 4 (RPL23AP4) on chrom
NG_000911 Homo sapiens ribosomal protein L10 pseudogene 1 (RPL10P1) on chromosc
NG_000912 Homo sapiens ribosomal modification protein rimK-like (E. coli) pseudogene
NG_000913 Homo sapiens protein phosphatase 1 , regulatory (inhibitor) subunit 2 pseudo
NG_000914 Homo sapiens peptidylprolyl isomerase A (cyclophilin A), pseudogene (PPIA
NG_000915 Homo sapiens polymerase (RNA) II (DNA directed) polypeptide C, pseudoge
NG_000916 Homo sapiens poly(rC) binding protein 2, pseudogene 1 (PCBP2P1) on chro
NG_000917 Homo sapiens myosin, light polypeptide 6, pseudogene (MYL6P) on chromo;
NG_000919 Homo sapiens inner membrane protein, mitochondrial (mitofilin) pseudogene
NGJ300920 Homo sapiens heat shock 60kDa protein 1 (chaperonin) pseudogene 7 (HSF
NG_000921 Homo sapiens high-mobility group (nonhistone chromosomal) protein 14 pse
NGJD00922 Homo sapiens H2A histone family, member Z, pseudogene (H2AFZP) on chi
NG_000923 Homo sapiens famesyl diphosphate synthase pseudogene (FDPSP) on chro
NG_000924 Homo sapiens eukaryotic translation initiation factor 4A, isoform 1 , pseudoge
NG_000925 Homo sapiens eukaryotic translation initiation factor 3, subunit 5 epsilon, 47k
NGJD00927 Homo sapiens cytochrome P450, family 4, subfamily F, polypeptide 3-like ps
NG_000929 Homo sapiens complement component 1 , q subcomponent binding protein, f
NG_000930 Homo sapiens F-box and WD-40 domain protein 11 pseudogene 1 (FBXW1 '
NG_000933 Homo sapiens endoplasmic reticulum lumenal protein 28 pseudogene (ERP.
NG_000936 Homo sapiens plakophilin 2 pseudogene 1 (PKP2P1) on chromosome 12
NG_000938 Homo sapiens keratin, hair, basic, pseudogene 1 (KRTHBP1) on chromoson
NG_000939 Homo sapiens keratin associated protein 2 pseudogene 1 (KRTAP2P1) on cl
NG_000940 Homo sapiens keratin, hair, basic, pseudogene 2 (KRTHBP2) on chromoson
NG_000941 Homo sapiens keratin associated protein 3 pseudogene 1 (KRTAP3P1) on cl
NG_000942 Homo sapiens keratin associated protein 9 pseudogene 1 (KRTAP9P1) on cl
NG_000943 Homo sapiens keratin, hair, basic, pseudogene 3 (KRTHBP3) on chromoson
NG_000944 Homo sapiens keratin, hair, basic, pseudogene 4 (KRTHBP4) on chromoson
NG_000945 Homo sapiens ribosomal protein L12-like 3 (RPL12L3) pseudogene on chror
NGJ300946 Homo sapiens ribosomal protein S2-like 1 (RPS2L1) pseudogene on chromo
NG_000948 Homo sapiens 5'-nucleotidase, cytosolic III pseudogene 1 (NT5C3P1) on chr
NG_000949 Homo sapiens pleckstrin homology domain containing, family A member 3 p;
NG_000950 Homo sapiens ribosomal protein S4-like 2 (RPS4L2) pseudogene on chrome
NG_000951 Homo sapiens ribosomal protein S15a pseudogene 1 (RPS15AP1) on chrorr
NG_000952 Homo sapiens ribosomal protein S10-like (RPS10L) pseudogene on chromoi
NG_000953 Homo sapiens ribosomal protein S23 pseudogene 1 (RPS23P1) on chromos
NG_000954 Homo sapiens NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 3, 12
NG_000955 Homo sapiens NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 3, 12
NG_000956 Homo sapiens NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 3, 12
NG_000957 Homo sapiens NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 3, 12
NG_000958 Homo sapiens NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 3, 12
NG_000959 Homo sapiens similar to DNAJ (HEJ1) pseudogene on chromosome 1
NG_000960 Homo sapiens ribosomal protein S27a pseudogene 1 (RPS27AP1) on chrorr
NGJD00961 Homo sapiens RNA, U73B small nucleolar (U73B) pseudogene on chromosc
NG_000962 Homo sapiens ribosomal protein L12 pseudogene 4 (RPL12P4) on chromosc
NG_000963 Homo sapiens ribosomal protein L38 pseudogene 1 (RPL38P1) on chromosc
NG_000964 Homo sapiens ribosomal protein L12-like 2 (RPL12L2) pseudogene on chror
NG_000965 Homo sapiens ribosomal protein L27a pseudogene (RPL27AP) on chromosc
NG_000966 Homo sapiens ribosomal protein L13 pseudogene 2 (RPL13P2) on chromosc
NG_000967 Homo sapiens ribosomal protein S4-like (RPS4L) pseudogene on chromosoi
NGJ300968 Homo sapiens mitochondrial ribosomal protein S16 pseudogene (MRPS16P)
NG_000969 Homo sapiens ribosomal protein L7a like 2 (RPL7AL2) pseudogene on chror
NGJD00970 Homo sapiens ribosomal protein S18 pseudogene 1 (RPS18P1) on chromos
NG_000971 Homo sapiens ribosomal protein L23a pseudogene 6 (RPL23AP6) on chrom'
NG_000972 Homo sapiens ribosomal protein S11 , pseudogene 1 (RPS11 P1 ) on chromos
NG_000973 Homo sapiens ribosomal protein, large, PO pseudogene 1 (RPLP0P1) on chr
NG_000974 Homo sapiens ribosomal protein L7a-like 3 (RPL7AL3) pseudogene on chror
NG_000975 Homo sapiens ribosomal protein L15 pseudogene 1 (RPL15P1) on chromosc
NGJD00977 Homo sapiens mitochondrial ribosomal protein S11 pseudogene 1 (MRPS11
NG_000978 Homo sapiens ribosomal protein L12 pseudogene 3 (RPL12P3) on chromosc
NG_000979 Homo sapiens high-mobility group (nonhistone chromosomal) protein 4-like .
NG_000980 Homo sapiens ribosomal protein L21 pseudogene 4 (RPL21 P4) on chromosc
NG_000981 Homo sapiens ribosomal protein L7 pseudogene 2 (RPL7P2) on chromosom
NGJ0OO982 Homo sapiens ribosomal protein L37 pseudogene 1 (RPL37P1) on chromosc
NG_000983 Homo sapiens ribosomal protein L36 pseudogene 1 (RPL36P1 ) on chromosc
NG_000984 Homo sapiens ribosomal protein S3A pseudogene 3 (RPS3AP3) on chromos
NG_000986 Homo sapiens ribosomal protein L21 pseudogene 2 (RPL21 P2) on chromosc
NG_000987 Homo sapiens ribosomal protein L35a pseudogene (RPL35AP) on chromosc
NG_000988 Homo sapiens ribosomal protein L37a pseudogene 1 (RPL37AP1) on chrom'
NG_000989 Homo sapiens ribosomal protein L31 pseudogene 3 (RPL31 P3) on chromosc
NG_000990 Homo sapiens ribosomal protein L39 pseudogene (RPL39P) on chromosome
NGJD00991 Homo sapiens ribosomal protein L17 pseudogene 1 (RPL17P1) on chromosc
NG_000994 Homo sapiens ribosomal protein L36 pseudogene 2 (RPL36P2) on chromosc
NG_000995 Homo sapiens ribosomal protein S27a pseudogene 2 (RPS27AP2) on chrorr
NG_000996 Homo sapiens ribosomal protein L31 pseudogene 2 (RPL31 P2) on chromosc
NG_000997 Homo sapiens ribosomal protein S3 pseudogene 1 (RPS3P1) on chromoson
NGJ300999 Homo sapiens ribosomal protein L7A-like 4 (RPL7AL4) pseudogene on chroi
NG_001000 Homo sapiens ribosomal protein L24 pseudogene 1 (RPL24P1) on chromosc
NG_001001 Homo sapiens ribosomal protein S10 pseudogene 2 (RPS10P2) on chromos
NGJ301002 Homo sapiens ribosomal protein L19 pseudogene 1 (RPL19P1) on chromosc
NG_001004 Homo sapiens ribosomal protein S3 pseudogene 2 (RPS3P2) on chromoson
NG_001005 Homo sapiens ribosomal protein L7 pseudogene 3 (RPL7P3) on chromosom
NG_001006 Homo sapiens heat shock 10kDa protein 1 (chaperonin 10) pseudogene 1 (h
NG_001007 Homo sapiens cytochrome c oxidase ll-like (MTC02L) pseudogene on chron
NG_001008 Homo sapiens peptidylprolyl isomerase A (cyclophilin A)-like 2 (PPIAL2) psei
NG_001009 Homo sapiens estrogen-related receptor alpha pseudogene (ESRRAP) on cl
NG_001010 Homo sapiens similar to RP42 homolog (L0C153893) pseudogene on chrorr
NG_001012 Homo sapiens TAF2G-like gene (TAF2GL) pseudogene on chromosome 19
NG_001013 Homo sapiens glutaredoxin (thioltransferase) pseudogene (GLI XP) on chror
NG_001014 Homo sapiens peptidylprolyl isomerase A (cyclophilin A) pseudogene 3 (PPI>
NG_001016 Homo sapiens C-reactive protein pseudogene (LOC171422) on chromosome
NG_001017 Homo sapiens FUS1P1 pseudogene (pFUSIPI) on chromosome 20
NG_001019 Homo sapiens immunoglobulin heavy locus (IGH.1@) on chromosome 14
NG_001020 Homo sapiens cystatin pseudogene 1 (CSTP1) on chromosome 20
NG_001021 Homo sapiens cytochrome b-5 pseudogene 4 (CYB5P4) on chromosome 20
NG_001022 Homo sapiens RNA binding motif protein, X-linked pseudogene 1 (RBMXP1)
NG_001023 Homo sapiens eukaryotic translation initiation factor 3, subunit 6 48kDa pseu
NG_001024 Homo sapiens keratin 18 pseudogene 1 (KRT18P1) on chromosome 6
NG_001025 Homo sapiens peptidylprolyl isomerase (cyclophilin) pseudogene 9 (PPIP9) c
NG_001026 Homo sapiens protein phosphatase 1 , regulatory (inhibitor) subunit 2 pseudo
NG_001027 Homo sapiens ribosomal protein L23a pseudogene 1 (RPL23AP1) on chrom'
NG_001028 Homo sapiens eukaryotic translation termination factor 1 pseudogene 1 (ETF
NG_001029 Homo sapiens makorin, ring finger protein, pseudogene 3 (MKRNP3) on chrc
NG_001030 Homo sapiens nuclear fragile X mental retardation protein interacting protein
NG_001031 Homo sapiens ADP-ribosylation factor 4 pseudogene 2 (ARF4P2) on chrome
NG_001032 Homo sapiens cell division cycle 42 pseudogene 1 (CDC42P1) on chromoso
NG_001033 Homo sapiens cytochrome c oxidase subunit Vic pseudogene 2 (COX6CP2)
NG_001035 Homo sapiens endosulfine alpha pseudogene (ENSAP) on chromosome 20
NG_001036 Homo sapiens FAT tumor suppressor homolog 1 (Drosophila) pseudogene 1
NG_001037 Homo sapiens ferritin, light polypeptide pseudogene (FTLP) on chromosome
NG_001038 Homo sapiens glyceraldehyde-3-phosphate dehydrogenase pseudogene 2 (ι
NG_001040 Homo sapiens glutathione S-transferase M3 pseudogene (GSTM3P) on chro
NG_001041 Homo sapiens heterogeneous nuclear ribonucleoprotein A1 pseudogene 3 (I
NG_001043 Homo sapiens keratin 18 pseudogene 3 (KRT18P3) on chromosome 20
NG_001045 Homo sapiens laminin receptor 1 pseudogene 1 (LAMR1P1) on chromosome
NG_001046 Homo sapiens proliferation-associated 2G4 pseudogene 2 (PA2G4P2) on ch
NG__001047 Homo sapiens phosphoglycerate utase 3, pseudogene (PGAM3P) on chroi
NG_001048 Homo sapiens peptidylprolyl isomerase A (cyclophilin A) pseudogene 2 (PPI;
NG_001049 Homo sapiens peptidylprolyl isomerase (cyclophilin) pseudogene 11 (PPIP1 '
NG_001050 Homo sapiens proteasome (prosome, macropain) 26S subunit, non-ATPase,
NG_001051 Homo sapiens prothymosin, alpha pseudogene 6 (PTMAP6) on chromosome
NG_001052 Homo sapiens ring finger protein 11 B, pseudogene (RNF11 B) on chromoson
NG_001053 Homo sapiens small inducible cytokine subfamily E, member 1 (endothelial n
NG_001054 Homo sapiens splicing factor 3a, subunit 3 pseudogene (SF3A3P) on chromi
NG_001055 Homo sapiens synaptosomal-associated protein, 23kDa pseudogene (SNAP
NG_001056 Homo sapiens small nuclear ribonucleoprotein polypeptide F pseudogene 1 i
NG_001057 Homo sapiens spermidine synthase pseudogene 1 (SRMP1) on chromosomi
NG_001058 Homo sapiens suppression of tumorigenicity 13 (colon carcinoma) (Hsp70 in
NG_001060 Homo sapiens tropomyosin 5, pseudogene (TPM5P) on chromosome 20
NG_001061 Homo sapiens ubiquitin-conjugating enzyme E2 variant 1 pseudogene 1 (UB
NG_001062 Homo sapiens exportin, tRNA (nuclear export receptor for tRNAs) pseudoger
NGJ301063 Homo sapiens lysophospholipase l-like (LOC157713) pseudogene on chrom
NGJ301064 Homo sapiens similarto orphan seven transmembrane receptor (RHJI/GuB|
NG__001065 Homo sapiens RH-ll/GuB pseudogene 1 (RH-ll/GuBp1) on chromosome 2
NG_001066 Homo sapiens toll-like receptor 7-like (TLR7-like) pseudogene on chromosor
NG_001067 Homo sapiens alpha-2-macroglobulin pseudogene (A2MP) on chromosome
NG_001068 Homo sapiens actin, gamma pseudogene 1 (ACTGP1) on chromosome 3
NG__001069 Homo sapiens actin, gamma pseudogene 9 (ACTGP9) on chromosome 6
NG_001070 Homo sapiens adenylate kinase 3 pseudogene 1 (AK3P1) on chromosome 1
NG_001071 Homo sapiens aldehyde reductase (aldose reductase) pseudogene (ALDRP]
NG_001073 Homo sapiens S-adenosylmethionine decarboxylase pseudogene 1 (AMDP1
NG_001074 Homo sapiens v-raf murine sarcoma 3611 viral oncogene homolog pseudoge
NG_001075 Homo sapiens ADP-ribosylation factor 4 pseudogene (ARF4P) on chromosoi
NG_001076 Homo sapiens ADP-ribosyltransferase 2 pseudogene (RT6 antigen homolog
NG_001077 Homo sapiens argininosuccinate synthetase pseudogene 1 (ASSP1) on chro
NG_001078 Homo sapiens argininosuccinate synthetase pseudogene 3 (ASSP3) on chro
NG_001080 Homo sapiens activating transcription factor 4 pseudogene (tax-responsive e
NG_001081 Homo sapiens ATPase, Na+/K+ transporting, beta polypeptide-like 1 (ATP1E
NGJ301082 Homo sapiens antiquitin-like 1 (ATQL1) pseudogene on chromosome 5
NG_001083 Homo sapiens antiquitin-like 3 (ATQL3) pseudogene on chromosome 7
NG_001084 Homo sapiens antiquitin-like 4 (ATQL4) pseudogene on chromosome 10
NG_001085 Homo sapiens brain cytoplasmic RNA 1 , pseudogene 2 (BCYR 1 P2) on chn
NG_001086 Homo sapiens basic transcription factor 3, pseudogene 1 (BTF3P1) on chror
NG_001087 Homo sapiens solute carrier family 25 (carnitine/acylcamitine translocase), rr
NG_001088 Homo sapiens calcitonin pseudogene (CALCP) on chromosome 11
NG_001089 Homo sapiens calmodulin 1 (phosphorylase kinase, delta) pseudogene 1 (C/
NG_001090 Homo sapiens calmodulin 1 (phosphorylase kinase, delta) pseudogene 2 (C/
NG_001091 Homo sapiens calmodulin 2 pseudogene 2 (CALM2P2) on chromosome 10
NG_001092 Homo sapiens cyclin D2 pseudogene (CCND2P) on chromosome 11
NG_001093 Homo sapiens cyclin D3 pseudogene (CCND3P) on chromosome 6
NG_001094 Homo sapiens carcinoembryonic antigen-related cell adhesion molecule pse
NG_001095 Homo sapiens carcinoembryonic antigen-related cell adhesion molecule pse
NG_001096 Homo sapiens carcinoembryonic antigen-related cell adhesion molecule pse
NG_001097 Homo sapiens carcinoembryonic antigen-related cell adhesion molecule pse
NG_001098 Homo sapiens carcinoembryonic antigen-related cell adhesion molecule pse
NG_001099 Homo sapiens carcinoembryonic antigen-related cell adhesion molecule pse
NG_001100 Homo sapiens carcinoembryonic antigen-related cell adhesion molecule pse
NGJD01101 Homo sapiens carcinoembryonic antigen-related cell adhesion molecule pse
NG_001102 Homo sapiens carcinoembryonic antigen-related cell adhesion molecule pse
NG_001103 Homo sapiens carcinoembryonic antigen-related cell adhesion molecule pse
NG_001104 Homo sapiens carcinoembryonic antigen-related cell adhesion molecule pse
NGJD01105 Homo sapiens cytochrome c oxidase subunit Via polypeptide 1 pseudogene
NG_001106 Homo sapiens ceruloplasmin (ferroxidase) pseudogene (CPP) on chromosor
NG_001107 Homo sapiens crystallin, gamma F pseudogene 1 (CRYGFP1) on chromosoi
NG_001108 Homo sapiens crystallin, gamma G pseudogene 1 (CRYGGP1) on chromoso
NG_001109 Homo sapiens crystallin, zeta (quinone reductase) pseudogene 1 (CRYZP1)
NG_001110 Homo sapiens catenin (cadherin-associated protein), alpha pseudogene 1 (C
NG_001112 Homo sapiens diazepam binding inhibitor-like 2 (pseudogene) (DBIL2) on ch
NG_001113 Homo sapiens dihydrofolate reductase pseudogene 2 (DHFRP2) on chromos
NG_001114 Homo sapiens ELK2, member of ETS oncogene family, pseudogene 1 (ELK;
NG_001115 Homo sapiens enolase 1 , (alpha) pseudogene (EN01 P) on chromosome 1
NG_001116 Homo sapiens ferredoxin pseudogene 2 (FDXP2) on chromosome 21
NG_001117 Homo sapiens ferredoxin pseudogene 1 (FDXP1) on chromosome 20
NGJD01118 Homo sapiens ferrochelatase pseudogene (FECHP) on chromosome 3
NG_001119 Homo sapiens forkhead box 03B (FOX03B) pseudogene on chromosome 1"
NG_001120 Homo sapiens forkhead box 01 B (FOX01 B) pseudogene on chromosome 5
NG_001121 Homo sapiens ferritin, heavy polypeptide pseudogene 2 (FTHP2) on chromo'
NG_001122 Homo sapiens fucosidase, alpha-L- 1 , tissue pseudogene (FUCA1 P) on chro
NG_001123 Homo sapiens glyceraldehyde-3-phosphate dehydrogenase pseudogene 1 (<
NG_001124 Homo sapiens GDP dissociation inhibitor 2 pseudogene (GDI2P) on chromoi
NG_001125 Homo sapiens glycoprotein, alpha-galactosyltransferase 1 (GGTA1) pseudog
NG_001126 Homo sapiens glycerol kinase pseudogene 1 (GKP1) on chromosome 1
NG_001127 Homo sapiens glycerol kinase pseudogene 6 (GKP6) on chromosome X
NG_001128 Homo sapiens glutamate dehydrogenase pseudogene 2 (GLUDP2) on chron
NG_001129 Homo sapiens glutamate dehydrogenase pseudogene 5 (GLUDP5) on chron
NG_001130 Homo sapiens GM2 ganglioside activator pseudogene (GM2AP) on chromos
NG_001131 Homo sapiens G protein-coupled receptor 32, pseudogene (GPR32P) on chr
NG_001132 Homo sapiens G protein-coupled receptor 33, pseudogene (GPR33) on chro
NG_001133 Homo sapiens G protein-coupled receptor kinase 6 pseudogene (GRK6PS) c
NG_001134 Homo sapiens glutathione peroxidase pseudogene 2 (GPXP2) on chromosoi
NG_001135 Homo sapiens glutathione S-transferase A pseudogene 1 (GSTAP1 ) on chro
NG_001136 Homo sapiens gulonolactone (L-) oxidase pseudogene (GULOP) on chromos
NG_001141 Homo sapiens high-mobility group (nonhistone chromosomal) protein 17 pse
NG_001142 Homo sapiens high-mobility group (nonhistone chromosomal) protein 17 pse
NG_001144 Homo sapiens heat shock 70kDa protein pseudogene 1 (HSPAP1) on chrom
NG_001145 Homo sapiens heat shock 60kDa protein 1 (chaperonin) pseudogene 1 (HSF
NG_001146 Homo sapiens heat shock 60kDa protein 1 (chaperonin) pseudogene 2 (HSF
NG_001147 Homo sapiens heat shock 60kDa protein 1 (chaperonin) pseudogene 3 (HSF
NG_001148 Homo sapiens heat shock 60kDa protein 1 (chaperonin) pseudogene 4 (HSF
NG_001149 Homo sapiens iduronate 2-sulfatase pseudogene 1 (IDSP1) on chromosome
NG_001150 Homo sapiens interferon, omega 15 (pseudogene) (IFNWP15) on chromosoi
NG_001151 Homo sapiens recombining binding protein suppressor of hairless (Drosophil
NG_001152 Homo sapiens IMP (inosine monophosphate) dehydrogenase-like 1 (IMPDHl
NG_001153 Homo sapiens Kallmann syndrome sequence pseudogene (KALP) on chrom
NG_001155 Homo sapiens lactate dehydrogenase B pseudogene (LDHBP) on chromoso
NG_001156 Homo sapiens melanoma antigen, family A, 7, pseudogene (MAGEA7) on ch
NG_001157 Homo sapiens MRE11 meiotic recombination 11 homolog B (S. cerevisiae) (I
NG_001158 Homo sapiens metallothionein 2 pseudogene 1 (processed) (MT2P1) on chrc
NG_001159 Homo sapiens methylenetetrahydrofolate dehydrogenase (NADP+ depender
NG_001160 Homo sapiens metaxin 1 pseudogene (MTX1 P) on chromosome 1
NG_001161 Homo sapiens NADH dehydrogenase (ubiquinone) flavoprotein 2 pseudoger
NG_001162 Homo sapiens nucleophosmin 1 (nucleolar phosphoprotein B23, numatrin) p
NG_001163 Homo sapiens nucleophosmin 1 (nucleolar phosphoprotein B23, numatrin) p
NG_001164 Homo sapiens nucleophosmin 1 (nucleolar phosphoprotein B23, numatrin) p
NG_001165 Homo sapiens nucleophosmin 1 (nucleolar phosphoprotein B23, numatrin) p
NG_001166 Homo sapiens nucleophosmin 1 (nucleolar phosphoprotein B23, numatrin) p
NG_001167 Homo sapiens nucleophosmin 1 (nucleolar phosphoprotein B23, numatrin) p
NG_001169 Homo sapiens ornithine aminotransferase-like 3 (pseudogene) (OATL3) on c
NG_001170 Homo sapiens phosphoglycerate kinase 1 , pseudogene 1 (PGK1 P1 ) on chro
NG_001171 Homo sapiens prohibitin pseudogene 1 (PHBP1) on chromosome 6
NG_001172 Homo sapiens phosphorylase kinase, beta pseudogene 1 (PHKBP1) on chro
NGJD01173 Homo sapiens phosphorylase kinase, beta pseudogene 2 (PHKBP2) on chro
NG_001174 Homo sapiens phosphatidylinositol glycan, class A, pseudogene 1 (PIGAP1)
NG_001175 Homo sapiens phosphatidylinositol glycan, class C, pseudogene 1 (PIGCP1)
NG_001176 Homo sapiens phosphatidylinositol glycan, class F, pseudogene 1 (PIGFP1)
NG_001177 Homo sapiens peptidylprolyl isomerase (cyclophilin) pseudogene 1 (PPIP1) c
NG_001178 Homo sapiens protein phosphatase 1 , regulatory (inhibitor) subunit 8 pseudo
NGJD01179 Homo sapiens protein S pseudogene (beta) (PROSP) on chromosome 3
NG_001180 Homo sapiens prothymosin, alpha pseudogene 3 (gene sequence 34) (PTM/
NG_001181 Homo sapiens prothymosin, alpha pseudogene 4 (gene sequence 112) (PT
NG_001182 Homo sapiens protein tyrosine phosphatase type IVA pseudogene 2 (PTP4A
NG_001183 Homo sapiens RNA, U1 small nuclear pseudogene 1 (RNU1 P1) on chromos*
NGJD01184 Homo sapiens RNA, U1 small nuclear pseudogene 2 (RNU1P2) on chromos*
NG_001185 Homo sapiens RNA, U3 small nucleolar pseudogene 1 (RNU3P1)
NG_001186 Homo sapiens RNA, U4 small nuclear pseudogene 1 (U4/7) (RNU4P1 ) on ch
NGJ001187 Homo sapiens RNA, U4 small nuclear pseudogene 2 (U4/14) (RNU4P2) on c
NG_001188 Homo sapiens RNA, U7 small nuclear pseudogene 1 (RNU7P1) on chromos*
NG_001189 Homo sapiens RNA, U7 small nuclear pseudogene 2 (RNU7P2) on chromos*
NG_001190 Homo sapiens RNA, U7 small nuclear pseudogene 3 (RNU7P3) on chromos*
NG_001191 Homo sapiens RNA, U7 small nuclear pseudogene 4 (RNU7P4) on chromos*
NG_001192 Homo sapiens ribosomal protein L9 pseudogene 1 (RPL9P1) on chromosom
NG_001193 Homo sapiens ribosomal protein L7 pseudogene (RPL7P) on chromosome 5
NG_001194 Homo sapiens sterol-C4-methyl oxidase pseudogene (SC4MOP) on chromos
NG_001195 Homo sapiens SHC (Src homology 2 domain containing) transforming proteii
NG_001196 Homo sapiens steroid-5-alpha-reductase, alpha polypeptide pseudogene 1 (:
NG_001197 Homo sapiens steroid sulfatase (microsomal) pseudogene (STSP) on chrom
NG_001198 Homo sapiens eukaryotic translation termination factor 1 pseudogene 2 (ETF
NG_001199 Homo sapiens TAR (HIV) RNA binding protein 2 pseudogene (TARBP2P) on
NG_001200 Homo sapiens transcription elongation factor A (SU), 1 pseudogene (TCEA11
NG_001201 Homo sapiens thioredoxin-dependent peroxide reductase 2 (thiol-specific anl
NG_001202 Homo sapiens transcription factor Dp-1 pseudogene (TFDP1 P) on chromoso
NG_001204 Homo sapiens tRNA leucine (AAG) pseudogene 1 (TRLP1) on chromosome
NG_001205 Homo sapiens tRNA methionine elongator pseudogene 1 (TRMEP1) on chro
NG_001206 Homo sapiens tubulin, beta polypeptide pseudogene 1 (TUBBP1) on chromo
NG_001207 Homo sapiens ubiquitin A-52 residue ribosomal protein fusion product 1 psei
NG_001208 Homo sapiens ubiquitin A-52 residue ribosomal protein fusion product 1 psei
NG_001209 Homo sapiens ubiquitin-conjugating enzyme E2L 4 (UBE2L4) pseudogene o
NG_001210 Homo sapiens ubiquitin protein ligase E3A pseudogene 2 (UBE3AP2) on chr
NG_001211 Homo sapiens urate oxidase (UOX) pseudogene on chromosome 1
NG_001212 Homo sapiens von Willebrand factor pseudogene (VWFP) on chromosome Σ
NG_001213 Homo sapiens v-yes-1 Yamaguchi sarcoma viral oncogene homolog pseudo*
NG_001214 Homo sapiens zinc finger protein 75b (ZNF75B) pseudogene on chromosom
NG_001215 Homo sapiens interferon-induced protein with tetratricopeptide repeats 1 , psc
NG_001216 Homo sapiens a disintegrin and metalloproteinase domain 1 (fertilin alpha) p
NG_001217 Homo sapiens G protein-coupled receptor 53, pseudogene (GPR53P) on chr
NG_001218 Homo sapiens synaptogyrin 2 pseudogene (SYNGR2P) on chromosome 15
NG_001219 Homo sapiens cytochrome c oxidase subunit Vic pseudogene 1 (COX6CP1 )
NG_001220 Homo sapiens cytochrome c oxidase subunit Va pseudogene 1 (COX5AP1 ) ι
NG_001221 Homo sapiens cytochrome c oxidase subunit Vile pseudogene 1 (COX7CP1
NG_001222 Homo sapiens v-myc myelocytomatosis viral oncogene homolog 3 pseudoge
NG_001223 Homo sapiens voltage-dependent anion channel 1-like pseudogene (VDAC1
NG_001224 Homo sapiens voltage-dependent anion channel 1 pseudogene (VDAC1 P) o
NGJ301226 Homo sapiens H2B histone family, member 0 (H2BFO) pseudogene
NG_001228 Homo sapiens cold shock domain protein A pseudogene 1 (CSDAP1) on chr
NG_001229 Homo sapiens nuclear distribution gene C homolog (A. nidulans) pseudogen
NG_001230 Homo sapiens nuclear distribution gene C homolog (A. nidulans) pseudogen
NG_001231 Homo sapiens nucleophosmin 1 (nucleolar phosphoprotein B23, numatrin) p
NG_001232 Homo sapiens nucleophosmin 1 (nucleolar phosphoprotein B23, numatrin) p
NG_001233 Homo sapiens nucleophosmin 1 (nucleolar phosphoprotein B23, numatrin) p
NG_001234 Homo sapiens macrophage stimulating, pseudogene 8 (MSTP8) on chromos
NG_001235 Homo sapiens macrophage stimulating, pseudogene 7 (MSTP7) on chromos
NG_001236 Homo sapiens macrophage stimulating, pseudogene 6 (MSTP6) on chromos
NG_001237 Homo sapiens macrophage stimulating, pseudogene 5 (MSTP5) on chromos
NGJD01238 Homo sapiens macrophage stimulating, pseudogene 4 (MSTP4) on chromos
NG_001239 Homo sapiens macrophage stimulating, pseudogene 3 (MSTP3) on chromos
NG_001240 Homo sapiens macrophage stimulating, pseudogene 2 (MSTP2) on chromos
NG_001241 Homo sapiens macrophage stimulating, pseudogene 1 (MSTP1) on chromos
NG_001242 Homo sapiens teratocarcinoma-derived growth factor 5, pseudogene (TDGF!
NG_001243 Homo sapiens teratocarcinoma-derived growth factor 4, pseudogene (TDGF'
NG_001244 Homo sapiens teratocarcinoma-derived growth factor 2, pseudogene (TDGF!
NG_001245 Homo sapiens glycosylphosphatidylinositol anchor attachment 1 pseudogene
NG_001246 Homo sapiens ATP-binding cassette, sub-family D (ALD), member 1 , pseudc
NG_001247 Homo sapiens protocadherin alpha 14 pseudogene (PCDHA14) on chromosi
NG_001248 Homo sapiens RNA, U6 small nuclear pseudogene 1 (RNU6P1) on chromos*
NG_001249 Homo sapiens RNA, U4 small nuclear pseudogene 6 (RNU4P6) on chromos*
NG_001250 Homo sapiens RNA, U4 small nuclear pseudogene 5 (RNU4P5) on chromos*
NG_001251 Homo sapiens RNA, U4 small nuclear pseudogene 4 (RNU4P4) on chromos*
NG_001252 Homo sapiens RNA, U4 small nuclear pseudogene 3 (RNU4P3) on chromos*
NG_001253 Homo sapiens RNA, U3 small nucleolar pseudogene 4 (RNU3P4) on chrome
NG_001254 Homo sapiens RNA, U3 small nucleolar pseudogene 3 (RNU3P3) on chrome
NG_001255 Homo sapiens RNA, U3 small nucleolar pseudogene 2 (RNU3P2) on chrome
NG_001256 Homo sapiens RNA, U2 small nuclear pseudogene 3 (RNU2P3) on chromos*
NG_001257 Homo sapiens RNA, U2 small nuclear pseudogene 2 (RNU2P2) on chromos*
NG_001258 Homo sapiens RNA, U2 small nuclear pseudogene 1 (RNU2P1)
NG_001259 Homo sapiens RNA, U1 small nuclear pseudogene 10 (RNU1P10) on chrom
NG_001260 Homo sapiens RNA, U1 small nuclear pseudogene 9 (RNU1P9) on chromos*
NG_001261 Homo sapiens RNA, U1 small nuclear pseudogene 8 (RNU1P8) on chromos*
NG_001262 Homo sapiens RNA, U1 small nuclear pseudogene 7 (RNU1P7)
NG_001263 Homo sapiens RNA, U1 small nuclear pseudogene 6 (RNU1P6) on chromos*
NG_001264 Homo sapiens RNA, U1 small nuclear pseudogene 5 (RNU1P5) on chromos*
NG_001265 Homo sapiens ATP-binding cassette, sub-family D (ALD), member 1 , pseudc
NG_001266 Homo sapiens poly(A) binding protein, cytoplasmic, pseudogene 3 (PABPCF
NG_001267 Homo sapiens poly(A) binding protein, cytoplasmic, pseudogene 2 (PABPCF
NG_001268 Homo sapiens poly(A) binding protein, cytoplasmic, pseudogene 1 (PABPCF
NG_001269 Homo sapiens ATP-binding cassette, sub-family D (ALD), member 1 , pseudc
NG 301270 Homo sapiens ATP-binding cassette, sub-family D (ALD), member 1 , pseudc
NGJJ01271 Homo sapiens molybdenum cofactor synthesis 1 pseudogene 1 (MOCS1P1)
NGJD01272 Homo sapiens G protein-coupled receptor 79 pseudogene (GPR79) on chror
NGJ301273 Homo sapiens mitogen-activated protein kinase kinase 1 pseudogene 1 (MA
NGJD01274 Homo sapiens protein phosphatase 1 , regulatory (inhibitor) subunit 14B psei
NG_001275 Homo sapiens chemokine (C-X-C motif) ligand 1 pseudogene (CXCL1P) on i
NG_001276 Homo sapiens nuclease sensitive element binding protein 1 pseudogene (NS
NG_001277 Homo sapiens capping protein (actin filament) muscle Z-line, alpha 1 pseudc
NG_001278 Homo sapiens UDP glycosyltransferase 2 family, polypeptide B28 pseudogei
NGJD01279 Homo sapiens UDP glycosyltransferase 2 family, polypeptide B27 pseudogei
NGJ0O128O Homo sapiens UDP glycosyltransferase 2 family, polypeptide B26 pseudogei
NG_001281 Homo sapiens UDP glycosyltransferase 2 family, polypeptide B25 pseudogei
NG_001282 Homo sapiens UDP glycosyltransferase 2 family, polypeptide B24 pseudogei
NGJD01286 Homo sapiens fatty acid binding protein 3, pseudogene 2 (FABP3P2) on chrc
NG_001287 Homo sapiens uracil-DNA glycosylase pseudogene 1 (UNGP1) on chromosc
NG_001288 Homo sapiens uracil-DNA glycosylase pseudogene 2 (UNGP2) on chromosc
NG_001289 Homo sapiens Zn-15 related zinc finger protein RLF pseudogene (RLFP)
NG_001290 Homo sapiens 6-pyruvoyltetrahydropterin synthase pseudogene (PTS-P1) or
NG_001291 Homo sapiens recombining binding protein suppressor of hairless (Drosophil
NG_001292 Homo sapiens recombining binding protein suppressor of hairless (Drosophil
NG_001293 Homo sapiens COX17 pseudogene (LOC81993) on chromosome 13
NG_001294 Homo sapiens cytochrome P450, subfamily 51 pseudogene 1 (CYP51 P1 ) on
NG_001295 Homo sapiens ribosomal protein S19 pseudogene 1 (RPS19P1) on chromos
NG_001296 Homo sapiens ribosomal protein S19 pseudogene 2 (RPS19P2) on chromos
NG_001297 Homo sapiens thioredoxin 1 pseudogene 2 (LOC93202) on chromosome 10
NG_001298 Homo sapiens family with sequence similarity 8, member A5 pseudogene (F>
NG_001299 Homo sapiens family with sequence similarity 8, member A6 pseudogene (FJ
NG_001300 Homo sapiens cytochrome c oxidase subunit Vllb pseudogene 1 (COX7BP1;
NG_001301 Homo sapiens ribosomal protein L37 pseudogene 2 (RPL37P2) on chromosc
NG_001302 Homo sapiens ribosomal protein L3 pseudogene 2 (RPL3P2) on chromosom
NG_001303 Homo sapiens thioredoxin 1 pseudogene 4 (LOC124974) on chromosome 1
NG_001305 Homo sapiens mitogen-activated protein kinase kinase 4 pseudogene (LOC1
NG_001306 Homo sapiens FBR-MuSV-associated ubiquitously expressed (fox derived) p
NG_001307 Homo sapiens protein phosphatase 2, regulatory subunit B (B56), gamma isc
NG_001308 Homo sapiens hydroxysteroid (17-beta) dehydrogenase 7 pseudogene 1 (HS
NG_001309 Homo sapiens thioredoxin 1 pseudogene 1 (LOC151276) on chromosome 2
NG_001311 Homo sapiens deleted in split-hand/split-foot 1 pseudogene (DSS1 P1 ) on chr
NG_001313 Homo sapiens lactate dehydrogenase pseudogene (LOC158222) on chromo
NGJ301314 Homo sapiens PHD finger protein 10 pseudogene 1 (PHF10P1) on chromosc
NG_001315 Homo sapiens chromobox homolog 3 gamma pseudogene (LOC159770) on
NG_001316 Homo sapiens ribosomal protein L7a pseudogene 2 (RPL7AP2) on chromosi
NG_001317 Homo sapiens ribosomal protein L7a pseudogene 3 (RPL7AP3) on chromosi
NG_001318 Homo sapiens HSP40 pseudogene (HSP40) on chromosome 2
NG_001319 Homo sapiens ubiquitin-conjugating enzyme-like (UBCH7N2) pseudogene oi
NG_001321 Homo sapiens thioredoxin 1 pseudogene 6 (AF357533) on chromosome 1
NG_001322 Homo sapiens thioredoxin 1 pseudogene 7 (AF357534) on chromosome 2
NG_001323 Homo sapiens glutaredoxin pseudogene 2 (GLRXP2) on chromosome 14
NG_001324 Homo sapiens thioredoxin 1 pseudogene 5 (AF357532) on chromosome 1
NG 301325 Homo sapiens EPF5 pseudogene (EPF5) on chromosome 9
NG_001326 Homo sapiens EPF8 pseudogene (EPF8) on chromosome 16
NG_001328 Homo sapiens sperm autoantigenic protein 17 pseudogene 1 (SPA17P1) on
NG_001329 Homo sapiens high mobility group AT-hook 1 -like 2 (HMGA1 L2) pseudogene
NGJ301330 Homo sapiens thioredoxin 1 pseudogene 3 (AF357530) on chromosome 4
NG_001331 Homo sapiens 3-oxoacid CoA transferase 2 pseudogene (OXCT2P) on chror
NG_001332 Homo sapiens T cell receptor alpha delta locus (TCRA/TCRD) on chromosor
NG_001333 Homo sapiens T cell receptor beta locus (TRB@) on chromosome 7
NG_001334 Homo sapiens growth hormone locus (GH@) on chromosome 17
NG_001335 Homo sapiens genomic large histone family cluster (HFL@) on chromosome
NG_001336 Homo sapiens T cell receptor gamma locus (TRG@) on chromosome 7
NG_001337 Homo sapiens T cell receptor beta variable orphans on chromosome 9 (TRB'
NG_001526 Homo sapiens a disintegrin and metalloproteinase domain 3b (cyritestin 2) (/
NG_001528 Homo sapiens phosphoglycerate kinase 1 , pseudogene 2 (PGK1 P2) on chro
NG_001529 Homo sapiens tyrosinase-like (TYRL) pseudogene on chromosome 11
NG_001531 Homo sapiens similar to tyrosine 3-monooxygenase/tryptophan 5-monooxygι
NG_001532 Homo sapiens similar to casein kinase 1 , alpha 1 (LOC120321) pseudogene
NG_001533 Homo sapiens proteasome (prosome, macropain) subunit, beta type, 3 pseu*
NG_001534 Homo sapiens legumain 2 pseudogene (LGMN2P) on chromosome 13
NG_001535 Homo sapiens brain creatine kinase pseudogene (LOC124144) on chromosc
NG_001537 Homo sapiens coactosin-like, Smith Magenis syndrome chromosome region
NG_001538 Homo sapiens proteasome (prosome, macropain) subunit, beta type, 3 pseu*
NG_001539 Homo sapiens mitochondrial ribosomal protein L9 pseudogene (MRPL9P1) c
NG_001540 Homo sapiens ATP synthase, H+ transporting, mitochondrial F0 complex, su
NG_001541 Homo sapiens synaptogyrin 2 pseudogene (LOC138916) on chromosome 9
NG_001542 Homo sapiens similar to plasmolipin (LOC139061) pseudogene on chromosc
NG_001543 Homo sapiens peptidyl prolyl isomerase H (cyclophilin H) pseudogene (LOC
NG_001544 Homo sapiens eukaryotic translation initiation factor 3, subunit 6 interacting f
NG_001545 Homo sapiens tyrosine 3-monooxygenase/tryptophan 5-monooxygenase actl
NG_001546 Homo sapiens similar to yeast Upf3, variant A pseudogene (LOC147150) on
NG_001547 Homo sapiens Iectin, galactoside-binding, soluble, 9 (galectin 9) pseudogene
NG_001548 Homo sapiens similar to yeast Upf3, variant A pseudogene (LOC147226) on
NGJ301549 Homo sapiens son-pseudogene (LOC148300) on chromosome 1
NG_001550 Homo sapiens u2 small nuclear ribonucleoprotein A' pseudogene (LOC1504'
NG_001551 Homo sapiens u2 small nuclear ribonucleoprotein polypeptide A' pseudogeni
NG_001552 Homo sapiens RAB6C, member RAS oncogene family pseudogene (LOC15(
NG_001553 Homo sapiens tyrosine 3-monooxygenase/tryptophan 5-monooxygenase acti
NG_001554 Homo sapiens RAB6C, member RAS oncogene family pseudogene (LOC15(
NG_001555 Homo sapiens peptidylprolyl isomerase (cyclophilin)-like 1 pseudogene 1 (PF
NG_001556 Homo sapiens protease (prosome, macropain) 26S subunit, ATPase, 1 pseu
NG_001557 Homo sapiens similar to ART-4 protein (LOC152594) pseudogene on chromi
NG_001558 Homo sapiens signal recognition particle 72kD pseudogene (LOC153932) or
NG_001559 Homo sapiens protein phosphatase 2, regulatory subunit B (B56), epsilon isc
NG_001560 Homo sapiens cell division cycle 20 pseudogene (LOC157956) on chromoso
NG_001561 Homo sapiens tyrosine 3-monooxygenase/tryptophan 5-monooxygenase acti
NG_001562 Homo sapiens tyrosine 3-monooxygenase/tryptophan 5-monooxygenase acti
NG_001563 Homo sapiens protein kinase C, iota pseudogene (LOC158948) on chromosi
NG_001564 Homo sapiens proteasome (prosome, macropain) 26S subunit, ATPase, 6 ps
NG_001566 Homo sapiens peptidylprolyl isomerase A (cyclophilin A)-like (PPIAL) pseudc
NG_001567 Homo sapiens ribonuclease H1 pseudogene 3 (RNASEH1 P3) on chromoson
NG_001568 Homo sapiens nitric oxide synthase 2A (inducible, hepatocytes) pseudogene
NG_001569 Homo sapiens similar to high mobility group protein-R (LOC201958) pseudoj
NG_001571 Homo sapiens ribosomal protein L36a pseudogene (dJ507H5.1) on chromos
NG_001572 Homo sapiens cyclin-dependent kinase 7 pseudogene (CDK7PS) on chromo
NG_001573 Homo sapiens TAF13 RNA polymerase II, TATA box binding protein (TBP)-a
NG_001574 Homo sapiens TBP-associated factor 9-like pseudogene (LOC246135) on cr
NG_001576 Homo sapiens deltaNEMO (deltaNEMO) pseudogene on chromosome X
NG_001577 Homo sapiens CDC28 protein kinase regulatory subunit 1A (CKS1A) pseudo
NG_001578 Homo sapiens CDC28 protein kinase regulatory subunit 1 B pseudogene 2 (C
NG_001579 Homo sapiens CDC28 protein kinase regulatory subunit 1B pseudogene 3 (C
NG_001580 Homo sapiens YWHAQ pseudogene 2 (YWHAQP2) on chromosome 22
NG_001581 Homo sapiens transcription elongation factor B (Sill), polypeptide 2 (18kD, el
NG_001582 Homo sapiens transcription elongation factor B (Sill), polypeptide 2 (18kD, el
NG_001583 Homo sapiens ubiquitin carrier protein E2-EPF pseudogene (LOC246719) or
NG_001584 Homo sapiens dynein, cytoplasmic, light polypeptide pseudogene (LOC2467
NG_001585 Homo sapiens ATP synthase, H+ transporting, mitochondrial F0 complex, su
NG_001586 Homo sapiens peptidyl prolyl isomerase H (cyclophilin H) pseudogene 1 (PP
NG_001587 Homo sapiens tyrosine 3-monooxygenase/tryptophan 5-monooxygenase acti
NG_001588 Homo sapiens homolog of C. elegans smu-1 pseudogene (LOC246784) on c
NG_001589 Homo sapiens PTD004 pseudogene (LOC246785) on chromosome 22
NG_001590 Homo sapiens signal recognition particle 68kD pseudogene (LOC252840) or
NG_001591 Homo sapiens signal recognition particle 68kD pseudogene (LOC252841) or
NG_001592 Homo sapiens karyopherin (importin) beta 2 pseudogene (LOC252968) on cl
NG_002151 Homo sapiens olfactory receptor, family 1 , subfamily E, member 3 pseudoge
NG_002153 Homo sapiens olfactory receptor, family 1 , subfamily P, member 1 pseudoge
NG_002154 Homo sapiens olfactory receptor, family 8, subfamily C, member 1 pseudoge
NG_002156 Homo sapiens olfactory receptor, family 8, subfamily B, member 1 pseudoge
NG 002158 Homo sapiens olfactory receptor, family 7, subfamily E, member 41 pseudog
NG_002160 Homo sapiens ifactory receptor, fami ly 7, subfam ly E, member 5 pseudoge NG_002162 Homo sapiens Ifactory receptor, fami ly 7, subfam ly E, member 4 pseudoge NG_002164 Homo sapiens Ifactory receptor, fam ly 7, subfami ly E, member 87 pseudog NG_002166 Homo sapiens factory receptor, fam ly 7, subfam ly E, member 2 pseudoge NG_002168 Homo sapiens ifactory receptor, fam y 7, subfam ly E, member 15 pseudog NG_002170 Homo sapiens Ifactory receptor, fami ly 7, subfam ly A, member 3 pseudoge NG_002172 Homo sapiens Ifactory receptor, fam y 5, subfam ly D, member 3 pseudoge NG_002173 Homo sapiens factory receptor, fam ly 4, subfam ly A, member 1 pseudoge NG_002175 Homo sapiens Ifactory receptor, fami ly 7, subfam ly E, member 14 pseudog NG_002177 Homo sapiens Ifactory receptor, fam ly 7, subfam ly E, member 13 pseudog NG_002179 Homo sapiens Ifactory receptor, fam y 7, subfam ly E, member 12 pseudog NG_002181 Homo sapiens factory receptor, fam ly 7, subfam ly E, member 11 pseudog NG_002183 Homo sapiens ifactory receptor, fami y 7, subfam ly E, member 10 pseudog NG_002184 Homo sapiens Ifactory receptor, fami ly 4, subfam ly C, member 1 pseudoge NG_002185 Homo sapiens Ifactory receptor, fam y 4, subfam ly H, member 8 pseudoge NG_002189 Homo sapiens Ifactory receptor, fam ly 7, subfam ly E, member 16 pseudog NG_002191 Homo sapiens Ifactory receptor, fam ly 7, subfam ly E, member 43 pseudog NG_002195 Homo sapiens Ifactory receptor, fam ly 9, subfam ly A, member 1 , pseudoge NG_002196 Homo sapiens Ifactory receptor, fam ly 12, subfamily D, member 1 pseudog NG_002199 Homo sapiens Ifactory receptor, fam y 51 , subfamily A, member 1 pseudog NG_002200 Homo sapiens Ifactory receptor, fam y 10, subfamily G, member 1 pseudog NG_002201 Homo sapiens Ifactory receptor, fam ly 10, subfamily B, member 1 pseudog NG_002202 Homo sapiens ifactory receptor, fam ly 8, subfamily B, member 7 pseudoge NG_002203 Homo sapiens Ifactory receptor, fam y 8, subfamily B, member 6 pseudoge NG_002204 Homo sapiens Ifactory receptor, fam y 8, subfamily B, member 5 pseudoge NG_002205 Homo sapiens Ifactory receptor, fam y 7, subfamily H, member 1 pseudoge NG_002207 Homo sapiens Ifactory receptor, fam y 7, subfamily E, member 8 pseudoge NG_002211 Homo sapiens Ifactory receptor, fam ly 7, subfamily E, member 33 pseudog NG_002212 Homo sapiens Ifactory receptor, fam y 7, subfamily D, member 1 pseudoge NG_002215 Homo sapiens Ifactory receptor, fam y 7, subfamily A, member 11 pseudog NG_002216 Homo sapiens Ifactory receptor, fam y 2, subfamily N, member 1 pseudoge NG_002218 Homo sapiens vomeronasal 1 receptor 7 pseudogene (VN1 R7P) on chromos NG_002219 Homo sapiens Ifactory receptor, fami y 56, subfamily A, member 5 (OR56AJ NG_002220 Homo sapiens Ifactory receptor, fam y 52, subfamily X, member 1 pseudog NG_002221 Homo sapiens factory receptor, fami y 7, subfamily E, member 94 pseudog NG_002225 Homo sapiens Ifactory receptor, fam ly 7, subfamily E, member 93 pseudog NG_ 002229 Homo sapiens ifactory receptor, fami ly 4, subfamily C, member 7 pseudoge NG_002232 Homo sapiens factory receptor, fam y 51 , subfamily P, member 1 pseudog NG_002233 Homo sapiens factory receptor, fami y 9, subfamily L, member 1 pseudogei NG_002234 Homo sapiens Ifactory receptor, fami ly 52, subfamily J, member 1 pseudogi NG_002235 Homo sapiens ifactory receptor, fami ly 4, subfamily K, member 7 pseudoge NG )02236 Homo sapiens Ifactory receptor, fam y 4, subfamily P, member 1 pseudoge NG_002238 Homo sapiens factory receptor, fam y 1 , subfamily AA, member 1 pseudog NG_002239 Homo sapiens Ifactory receptor, fami ly 2, subfamily AD, member 1 pseudoc NG )02240 Homo sapiens ifactory receptor fam ly 4, subfamily K, member 6 pseudoge NG_002241 Homo sapiens Ifactory receptor, fam y 7, subfamily E, member 91 pseudog NG_002242 Homo sapiens Ifactory receptor, fami ly 4, subfamily K, member 4 pseudoge NG_002243 Homo sapiens Ifactory receptor, fami y 4, subfamily N, member 1 pseudoge NG )02246 Homo sapiens Ifactory receptor, fam y 13, subfamily C, member 1 pseudog NGJ302247 Homo sapiens Ifactory receptor, fam y 4, subfamily C, member 5 (OR4C5) | NG_002250 Homo sapiens factory receptor, fami ly 4, subfamily C, member 2 pseudoge NG 302251 Homo sapiens ifactory receptor, fam y 7, subfamily E, member 83 pseudog NG_002252 Homo sapiens Ifactory receptor, fam y 51 , subfamily J, member 1 (OR51J1 NG_002253 Homo sapiens Ifactory receptor, fam y 5, subfamily BD, member 1 pseudoc NG_002254 Homo sapiens factory receptor, fam y 10, subfamily D, member 5 pseudog NG_002255 Homo sapiens ifactory receptor, fam y 10, subfamily G, member 6 (OR10G NG 002256 Homo sapiens Ifactory receptor, fam y 7, subfamily E, member 97 pseudog
NGJD02257 Homo sapiens olfactory receptor, family 2, subfamily AF, member 1 pseudog
NG_002258 Homo sapiens olfactory receptor, family 7, subfamily L, member 1 pseudogei
NG_002259 Homo sapiens olfactory receptor, family 10, subfamily R, member 1 pseudog
NG_002260 Homo sapiens olfactory receptor, family 10, subfamily T, member 1 pseudog'
NG_002261 Homo sapiens olfactory receptor, family 10, subfamily G, member 5 pseudog
NG_002262 Homo sapiens olfactory receptor, family 52, subfamily S, member 1 pseudog
NG_002263 Homo sapiens olfactory receptor, family 51 , subfamily A, member 5 pseudog
NG_002264 Homo sapiens olfactory receptor, family 4, subfamily C, member 10 pseudog
NG_002265 Homo sapiens olfactory receptor, family 4, subfamily R, member 1 pseudoge
NG_002266 Homo sapiens olfactory receptor, family 52, subfamily J, member 2 pseudogi
NG_002267 Homo sapiens olfactory receptor, family 4, subfamily C, member 9 pseudoge
NG_002268 Homo sapiens olfactory receptor, family 51 , subfamily A, member 3 pseudog
NG_002269 Homo sapiens olfactory receptor, family 52, subfamily E, member 3 pseudog
NG_002270 Homo sapiens olfactory receptor, family 4, subfamily V, member 1 pseudoge
NG_002271 Homo sapiens olfactory receptor, family 7, subfamily E, member 90 pseudog
NG_002272 Homo sapiens olfactory receptor, family 7, subfamily E, member 89 pseudog
NG_002273 Homo sapiens olfactory receptor, family 2, subfamily AL, member 1 pseudog
NG_002274 Homo sapiens olfactory receptor, family 6, subfamily J, member 1 (OR6J1 ) p
NG_002275 Homo sapiens olfactory receptor, family 4, subfamily C, member 4 pseudoge
NG_002277 Homo sapiens olfactory receptor, family 5, subfamily D, member 2 pseudoge
NG_002278 Homo sapiens olfactory receptor, family 7, subfamily A, member 18 pseudog
NG_002279 Homo sapiens olfactory receptor, family 9, subfamily I, member 2 pseudoger
NG_002281 Homo sapiens olfactory receptor, family 5, subfamily M, member 13 pseudog
NG_002282 Homo sapiens olfactory receptor, family 6, subfamily L, member 1 pseudogei
NG_002284 Homo sapiens ubiquitin B pseudogene 1 (UBBP1) on chromosome 2
NG_002285 Homo sapiens ubiquitin B pseudogene 4 (UBBP4) on chromosome 17
NG_002286 Homo sapiens ubiquitin B pseudogene 3 (UBBP3) on chromosome 2
NG_002287 Homo sapiens ubiquitin B pseudogene 2 (UBBP2) on chromosome 1
NG_002288 Homo sapiens suppressor of bimDδ homolog pseudogene (LOC122145) on
NG_002289 Homo sapiens serine hydroxymethyltransferase 1 (soluble) pseudogene (SH
NG_002290 Homo sapiens DKFZP434J193-like pseudogene (LOC259308) on chromoso
NG_002291 Homo sapiens splicing factor 3b, subunit 4, 49kD pseudogene (LOC260329)
NG_002298 Homo sapiens olfactory receptor, family 1 , subfamily D, member 3 pseudoge
NG_002299 Homo sapiens olfactory receptor, family 5, subfamily G, member 1 pseudoge
NG_002302 Homo sapiens olfactory receptor, family 1, subfamily R, member 1 pseudoge
NG_002305 Homo sapiens olfactory receptor, family 5, subfamily B, member 1 pseudoge
NG_002306 Homo sapiens olfactory receptor, family 5, subfamily B, member 10 pseudog
NG_002307 Homo sapiens olfactory receptor, family 4, subfamily H, member 6 pseudoge
NG_002311 Homo sapiens olfactory receptor, family 7, subfamily A, member 15 pseudog
NG_002315 Homo sapiens olfactory receptor, family 7, subfamily E, member 26 pseudog
NG_002316 Homo sapiens olfactory receptor, family 7, subfamily E, member 53 pseudog
NG_002317 Homo sapiens olfactory receptor, family 7, subfamily E, member 62 pseudog
NG_002319 Homo sapiens olfactory receptor, family 2, subfamily H, member 5 pseudoge
NG_002320 Homo sapiens NADH dehydrogenase 2 pseudogene 2 (MTND2P2) on chrorr
NG_002321 Homo sapiens proteasome (prosome, macropain) subunit, alpha type, 6 psei
NG_002322 Homo sapiens olfactory receptor, family 8, subfamily B, member 9 pseudoge
NGJ302323 Homo sapiens high mobility group AT-hook 1-like 3 (HMGA1L3) pseudogene
NG_002324 Homo sapiens high mobility group AT-hook 1-like 1 (HMGA1L1) pseudogene
NG_002325 Homo sapiens COP9 pseudogene (bA345E19.2) on chromosome X
NG_002326 Homo sapiens dihydrolipoamide S-succinyltransferase pseudogene (E2 com
NGJD02327 Homo sapiens HSA12cenp11 beta-tubulin 4Q (TUBB4Q) pseudogene (LOC
NG_002328 Homo sapiens polypyrimidine tract binding protein 1 pseudogene (PTBP1 P) *
NG_002329 Homo sapiens protease, serine, 29 pseudogene (PRSS29P) on chromosome
NG_002331 Homo sapiens apolipoprotein B mRNA editing enzyme, catalytic polypeptide-
NG_002332 Homo sapiens fascin pseudogene (LOC145989) on chromosome 15
NG_002333 Homo sapiens proteasome (prosome, macropain) subunit, alpha type, 7 psei
NG_002334 Homo sapiens HSA16q24 beta-tubulin 4Q pseudogene (LOC197331 ) on chr*
NG_002335 Homo sapiens HSA1 q43-44 beta-tubulin 4Q (TUBB4Q) pseudogene (LOC20
NGJD02336 Homo sapiens tubulin, beta polypeptide pseudogene 5 (TUBBP5) on chromo
NG_002337 Homo sapiens olfactory receptor, family 5, subfamily AK, member 4 pseudog
NG_002338 Homo sapiens HSA1 q42.3 beta-tubulin 4Q pseudogene (LOC255208) on chi
NG_002339 Homo sapiens HSA18p11 beta-tubulin 4Q pseudogene (LOC260334) on chr
NG_002340 Homo sapiens ELF2P2 pseudogene (ELF2P2)
NG_002341 Homo sapiens ELF2P3 pseudogene (AF256221 ) on chromosome 9
NG_002342 Homo sapiens zinc finger protein Np97 pseudogene (LOC260337) on chrom*
NG_002343 Homo sapiens HSA12cenq11 beta-tubulin 4Q (TUBB4Q) pseudogene (LOC.
NG_002348 Homo sapiens sorting nexin 6 pseudogene (LOC126506) on chromosome 1£
NG_002349 Homo sapiens eukaryotic translation elongation factor 1 alpha-like 11 (EEF1ι
NG_002350 Homo sapiens eukaryotic translation elongation factor 1 alpha 1 pseudogene
NG_002351 Homo sapiens sorting nexin 7 pseudogene (LOC203930) on chromosome 1 '
NG_002352 Homo sapiens actin related protein 2/3 complex subunit 1 A pseudogene (LO
NG_002353 Homo sapiens eukaryotic translation elongation factor 1 alpha 1 pseudogene
NG_002354 Homo sapiens actin related protein 2/3 complex subunit 1 A pseudogene (LO
NG_002355 Homo sapiens actin related protein 2/3 complex subunit 1 A pseudogene (LO
NG_002356 Homo sapiens olfactory receptor, family 7, subfamily E, member 19 pseudog
NG_002357 Homo sapiens olfactory receptor, family 5, subfamily AH, member 1 pseudoc
NG_002360 Homo sapiens ribosomal protein S2 pseudogene (LOC125208) on chromosc
NG_002361 Homo sapiens CGI-148 protein pseudogene (CGI-148P) on chromosome 16
NG_002363 Homo sapiens actin related protein 2/3 complex, subunit 3B, 21 kDa (ARPC3
NG_002364 Homo sapiens tumor-associated calcium signal transducer 1 pseudogene (U
NG_002365 Homo sapiens nitric oxide synthase 2A (inducible, hepatocytes) pseudogene
NG_002366 Homo sapiens NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 9, p
NG_002367 Homo sapiens family with sequence similarity 12, member C pseudogene (FJ
NG_002368 Homo sapiens proteasome (prosome, macropain) 26S subunit, ATPase, 3 ps
NG_002369 Homo sapiens Charot-Leyden crystal protein pseudogene 1 (CLCP1 ) on chrc
NG_002370 Homo sapiens NS1 -associated protein 1 pseudogene (LOC149844) on chroi
NG_002371 Homo sapiens dendritic cell protein pseudogene (LOC266683) on chromosoi
NG_002372 Homo sapiens embryonic ectoderm development pseudogene (LOC266694)
NG_002373 Homo sapiens enhancer of zeste homolog 2 (Drosophila) pseudogene (LOC;
NG_002374 Homo sapiens POM121 membrane glycoprotein-like 4 pseudogene (rat) (PO
NG_002375 Homo sapiens oligophrenin 1 pseudogene 1 (OPHN1 P1) on chromosome 25
NG_002376 Homo sapiens fatty acid binding protein 5, pseudogene 1 (FABP5P1) on chrc
NG_002378 Homo sapiens ash2 (absent, small, or homeotic)-like (Drosophila) pseudogei
NG_002379 Homo sapiens proteasome (prosome, macropain) 26S subunit, ATPase, 6 ps
NG_002380 Homo sapiens heat shock 70kDa protein 9B pseudogene (HSPA9BP) on chr
NG_002381 Homo sapiens ubiquitin protein ligase E3A pseudogene 1 (UBE3AP1) on chr
NG_002382 Homo sapiens melanoma antigen pseudogene, family A (psMAGEA) on chrc
NG_002383 Homo sapiens keratin 19 pseudogene (LOC160313) on chromosome 12
NG_002384 Homo sapiens nucleoporin 50 pseudogene (NUP50P) on chromosome 14
NG_002385 Homo sapiens tropomyosin-like (LOC146253) pseudogene on chromosome
NG_002387 Homo sapiens proteasome 26S non-ATPase subunit 2 pseudogene (LOC26I
NG_002388 Homo sapiens nucleoporin 50kDa pseudogene (LOC266785) on chromosorr
NG_002389 Homo sapiens nucleoporin 50 kDa pseudogene (LOC266786) on chromosor
NG_002392 Homo sapiens major histocompatibility complex, class II, DR52 haplotype (D
NG_002393 Homo sapiens adenylate kinase 2 pseudogene (AK2B) on chromosome 1
NG_002394 Homo sapiens calcium-binding tyrosine phosphorylation-regulated protein ps
NG_002395 Homo sapiens proteasome (prosome, macropain) 26S subunit, non-ATPase,
NG_002397 Homo sapiens major histocompatibility complex, class I, BC (HLA-BC) on chi
NG_002398 Homo sapiens major histocompatibility complex, class I, GHKAJ (HLA-GHK/>
NG_002399 Homo sapiens transcription elongation factor B (Sill), polypeptide 1 pseudog
NGJ302400 Homo sapiens piggyBac transposable element derived 3 pseudogene 1 (PGI
NG_002401 Homo sapiens piggyBac transposable element derived 3 pseudogene 2 (PGI
NG_002402 Homo sapiens piggyBac transposable element derived 3 pseudogene 3 (PGI
NG_002403 Homo sapiens piggyBac transposable element derived 3 pseudogene 4 (PGI
NG__002404 Homo sapiens ribosomal protein S9 pseudogene 2 (RPS9P2) on chromoson
NGJD02405 Homo sapiens RNA, U12 small nuclear pseudogene (RNU12P) on chromosc
NG_002406 Homo sapiens olfactory receptor, family 5, subfamily G, member 3 pseudoge
NG_002408 Homo sapiens olfactory receptor, family 13, subfamily C, member 7 pseudog
NG_002409 Homo sapiens olfactory receptor, family 5, subfamily AX, member 1 (OR5AX
NG_002410 Homo sapiens musashi 1 pseudogene (LOC268276) on chromosome 11
NG_002411 Homo sapiens suppressor of cytokine signaling 2 pseudogene 1 (SOCS2P1)
NG_002412 Homo sapiens proteasome 26S non-ATPase subunit 7 pseudogene (LOC28I
NG_002415 Homo sapiens RNA binding motif protein 8B pseudogene (RBM8B) on chron
NGJ302416 Homo sapiens mitochondrial ribosomal protein L11 pseudogene (MRPL11P2
NG_002417 Homo sapiens proteasome 26S non-ATPase subunit 10 pseudogene (LOC2!
NG_002418 Homo sapiens similar to Homeobox protein Meis3 (Meisl -related protein 2) (
NG__002419 Homo sapiens eukaryotic translation initiation factor 1 A pseudogene 1 (EIF1;
NG_002423 Homo sapiens CD8 antigen, beta polypeptide 2, pseudogene (p37) (CD8B2)
NG_002425 Homo sapiens RNA, 7SL, cytoplasmic, pseudogene 1 (RN7SLP1)
NG_002426 Homo sapiens RNA, 7SL, cytoplasmic, pseudogene 2 (RN7SLP2) on chrome
NG_002427 Homo sapiens RNA, 7SL, cytoplasmic, pseudogene 3 (RN7SLP3)
NG_002428 Homo sapiens RNA, 7SL, cytoplasmic, pseudogene 4 (RN7SLP4) on chrome
NG 302429 Homo sapiens RNA, 7SL, cytoplasmic, pseudogene 5 (RN7SLP5) on chrome
NG_002430 Homo sapiens mitochondrial ribosomal protein L48 pseudogene 1 (MRPL481
NG_002431 Homo sapiens nuclear autoantigenic sperm protein pseudogene 1 (NASPP1
NG_002432 Homo sapiens major histocompatibility complex, class II, DR51 haplotype (D
NG_002433 Homo sapiens major histocompatibility complex, class II, DR53 haplotype (D
NG_002434 Homo sapiens pre-B-cell leukemia transcription factor pseudogene 1 (PBXP'
NG_002437 Homo sapiens cytochrome b-5 pseudogene 3 (CYB5P3) on chromosome 14
NG_002438 Homo sapiens LEFTY family pseudogene (LEFTY3) on chromosome 1
NG__002440 Homo sapiens TGFB-induced factor (TALE family homeobox) pseudogene (I
NG_002448 Homo sapiens mitogen-activated protein kinase 6 pseudogene 2 (MAPK6PS
NG_002449 Homo sapiens mitogen-activated protein kinase 6 pseudogene 6 (MAPK6PS
NG_002450 Homo sapiens cytokine receptor-like factor 3 pseudogene (LOC285706) on c
NG_002451 Homo sapiens mitogen-activated protein kinase 6 pseudogene 4 (MAPK6PS
NG_002452 Homo sapiens mitogen-activated protein kinase 6 pseudogene 5 (MAPK6PS
NG_002453 Homo sapiens mitogen-activated protein kinase 6 pseudogene 3 (MAPK6PS
NG_002454 Homo sapiens mitogen-activated protein kinase 6 pseudogene 1 (MAPK6PS
NG_002456 Homo sapiens ribosomal protein L32-like 2 (RPL32L2) pseudogene on chror
NG_002457 Homo sapiens protein tyrosine phosphatase, non-receptor type substrate 1 — li
NG_002458 Homo sapiens actin, beta-like 1 (ACTBL1) pseudogene on chromosome 22
NG_002459 Homo sapiens vomeronasal 1 receptor 9 pseudogene (VN1 R9P) on chromos
NG_002460 Homo sapiens high-mobility group nucleosomal binding domain 2 pseudoger
NG_002461 Homo sapiens ataxin 2 related protein pseudogene (LOC317727) on chrome
NG_002462 Homo sapiens basic leucine zipper nuclear factor 2 pseudogene (BLZF2P) o
NG_002463 Homo sapiens proteasome 26S non-ATPase subunit 12 pseudogene (LOC3
NG_002464 Homo sapiens steroidogenic acute regulator pseudogene 1 (STARP1) on chi
NG_002465 Homo sapiens chromosome 14 open reading frame 55 (C14orf55) pseudoge
NG_002467 Homo sapiens a disintegrin and metalloproteinase domain 21 pseudogene (/
NG_002468 Homo sapiens ribosomal protein L9 pseudogene (LOC254948) on chromoso
NG_002469 Homo sapiens bromodomain containing 7 pseudogene (BRD7P) on chromos
NG_002470 Homo sapiens basic transcription factor 3, pseudogene 2 (BTF3P2) on chror
NG_002471 Homo sapiens ribosomal protein L9 pseudogene (LOC317771) on chromoso
NG_002472 Homo sapiens CDC10 cell division cycle 10 homolog (S. cerevisiae) pseudoc
NG_002473 Homo sapiens cysteine and histidine-rich domain (CHORD)-containing 2 pse
NG_002474 Homo sapiens CCNDBP1 interactor pseudogene (CBPINP) on chromosome
NG_002475 Homo sapiens CDC28 protein kinase regulatory subunit 1 B pseudogene (CK
NG_002476 Homo sapiens coilin pseudogene (COILP) on chromosome 14
NG_002477 Homo sapiens cytochrome c oxidase subunit Va pseudogene 2 (COX5AP2) ι
NG_002478 Homo sapiens ras homolog gene family, member Q pseudogene (RHOQP) c
NG_002479 Homo sapiens cytochrome c oxidase subunit Vila polypeptide 3 pseudogene
NGJD02480 Homo sapiens DEAD (Asp-Glu-Ala-Asp) box polypeptide 18 pseudogene 1 (I
NG_002481 Homo sapiens G protein-coupled receptor 57 (GPR57) pseudogene on chror
NG_002482 Homo sapiens ribosomal protein L21 pseudogene 12 (RPL21 P12) on chrome
NG_002483 Homo sapiens peptidylprolyl isomerase A (cyclophilin A) pseudogene 4 (PPI;
NG_002484 Homo sapiens peptidylprolyl isomerase A (cyclophilin A) pseudogene 5 (PPI;
NG_002485 Homo sapiens Friedreich ataxia pseudogene (FRDAP) on chromosome 14
NG_002486 Homo sapiens laminin receptor 1 pseudogene 4 (LAMR1P4) on chromosome
NG_002487 Homo sapiens ribosomal protein L21 pseudogene 7 (RPL21 P7) on chromose
NG_002488 Homo sapiens ribosomal protein L21 pseudogene 11 (RPL21P11) on chromi
NG_002489 Homo sapiens laminin receptor 1 pseudogene 3 (LAMR1 P3) on chromosome
NG_002490 Homo sapiens ubiquitin-conjugating enzyme E2L 7 pseudogene (UBE2L7) o
NG_002491 Homo sapiens ribosomal protein L21 pseudogene 9 (RPL21 P9) on chromosc
NG_002492 Homo sapiens Rho GTPase activating protein 16 pseudogene (ARHGAP16F
NG_002493 Homo sapiens DnaJ (Hsp40) homolog, subfamily C, member 8 pseudogene
NG_002494 Homo sapiens eukaryotic translation elongation factor 1 alpha 1 pseudogene
NG_002495 Homo sapiens endosulfine alpha pseudogene 2 (ENSAP2) on chromosome
NG_002496 Homo sapiens eukaryotic translation initiation factor 2, subunit 2 beta, pseud
NG_002497 Homo sapiens ATPase, H+ transporting, lysosomal 13kDa, V1 subunit G pse
NG_002498 Homo sapiens eukaryotic translation initiation factor 3, subunit 6 interacting p
NG_002499 Homo sapiens eukaryotic translation initiation factor 4B pseudogene (EIF4BF
NG_002500 Homo sapiens ATP synthase, H+ transporting, mitochondrial F0 complex, su
NG_002501 Homo sapiens tyrosine 3-monooxygenase/tryptophan 5-monooxygenase acti
NG_002502 Homo sapiens tyrosine 3-monooxygenase/tryptophan 5-monooxygenase acti
NG_002503 Homo sapiens uracil-DNA glycosylase pseudogene 3 (UNGP3) on chromosc
NG_002504 Homo sapiens zinc finger protein 405 pseudogene (ZNF405P) on chromosor
NG_002505 Homo sapiens asparaginyl-tRNA synthetase pseudogene (NARSP) on chron
NG_002506 Homo sapiens peptidylprolyl isomerase A (cyclophilin A) pseudogene 6 (PPI;
NG_002507 Homo sapiens laminin receptor 1 pseudogene 5 (LAMR1 P5) on chromosome
NG_002508 Homo sapiens high-mobility group box 1 pseudogene (HMGB1 P) on chromo;
NG_002509 Homo sapiens high-mobility group nucleosome binding domain 1 pseudogen
NG_002510 Homo sapiens high-mobility group nucleosomal binding domain 2 pseudoger
NG_002511 Homo sapiens heterogeneous nuclear ribonucleoprotein C pseudogene (HNI
NG_002512 Homo sapiens heterogeneous nuclear ribonucleoprotein U pseudogene (HNI
NG_002513 Homo sapiens ribosomal protein L10a pseudogene 1 (RPL10AP1) on chrom'
NG_002514 Homo sapiens ATP synthase, H+ transporting, mitochondrial F0 complex, su
NG_002515 Homo sapiens ATP synthase, H+ transporting, mitochondrial F0 complex, su
NG_002516 Homo sapiens BCL2/adenovirus E1 B 19kDa interacting protein 3 pseudogen
NG_002517 Homo sapiens glycine C-acetyltransferase pseudogene (GCATP) on chromo
NG_002518 Homo sapiens ribosomal protein L12 pseudogene 5 (RPL12P5) on chromosc
NG_002519 Homo sapiens ribosomal protein L21 pseudogene 10 (RPL21P10) on chromi
NG_002520 Homo sapiens ribosomal protein L21 pseudogene 13 (RPL21P13) on chromi
NG_002521 Homo sapiens ribosomal protein L21 pseudogene 8 (RPL21 P8) on chromosc
NG_002522 Homo sapiens ribosomal protein S2 pseudogene 4 (RPS2P4) on chromoson
NG_002523 Homo sapiens ribosomal protein S29 pseudogene 1 (RPS29P1) on chromos
NG_002524 Homo sapiens ubiquitin A-52 residue ribosomal protein fusion product 1 psei
NG_002525 Homo sapiens ribosomal protein L24 pseudogene 3 (RPL24P3) on chromosc
NGJ302526 Homo sapiens ribosomal protein, large, P1 pseudogene 1 (RPLP1 P1) on chr
NG_002527 Homo sapiens ribosomal protein L26 pseudogene 4 (RPL26P4) on chromosc
NG_002528 Homo sapiens ubiquitin-conjugating enzyme E2C pseudogene 1 (UBE2CP1)
NG_002529 Homo sapiens tubulin, beta polypeptide pseudogene 3 (TUBBP3) on chromo
NG_002530 Homo sapiens serine/threonine kinase 16 pseudogene (STK16P) on chromo
NG_002531 Homo sapiens abl-interactor 1 pseudogene (ABI1 P) on chromosome 14
NG_002532 Homo sapiens spermidine synthase pseudogene 2 (SRMP2) on chromosomi
NGJD02533 Homo sapiens small nuclear ribonucleoprotein polypeptide G pseudogene (£
NG_002534 Homo sapiens solute carrier family 20 (phosphate transporter), member 1 ps
NG_002535 Homo sapiens ribosomal protein L12 pseudogene 7 (RPL12P7) on chromosc
NG_002537 Homo sapiens SET pseudogene 2 (SETP2) on chromosome 14
NG_002538 Homo sapiens ribosomal protein L13a pseudogene 2 (RPL13AP2) on chrom'
NG_002539 Homo sapiens ribosomal protein L15 pseudogene 2 (RPL15P2) on chromosc
NG_002540 Homo sapiens ribosomal protein L17 pseudogene 3 (RPL17P3) on chromosc
NGJ302541 Homo sapiens ribosomal protein L17 pseudogene 4 (RPL17P4) on chromosc
NG_002542 Homo sapiens ribosomal protein L18a pseudogene 1 (RPL18AP1) on chrom'
NG__002543 Homo sapiens ribosomal protein L18 pseudogene 1 (RPL18P1) on chromosc
NG_002544 Homo sapiens ribosomal protein L22 pseudogene 2 (RPL22P2) on chromosc
NG_002545 Homo sapiens ribosomal protein L23a pseudogene 10 (RPL23AP10) on chrc
NG_002546 Homo sapiens ribosomal protein L23a pseudogene 11 (RPL23AP11 ) on chrc
NG_002547 Homo sapiens eukaryotic translation initiation factor 4E binding protein 1 pse
NG_002548 Homo sapiens heat shock factor binding protein 1 pseudogene 1 (HSBP1P1]
NG_002549 Homo sapiens ribosomal protein L26 pseudogene 2 (RPL26P2) on chromosc
NG_002550 Homo sapiens ribosomal protein L26 pseudogene 3 (RPL26P3) on chromosc
NG_002551 Homo sapiens ribosomal protein L27 pseudogene 1 (RPL27P1) on chromosc
NG_002552 Homo sapiens heat shock 10kDa protein 1 (chaperonin 10) pseudogene 2 (h
NG_002553 Homo sapiens ribosomal protein L36a pseudogene 2 (RPL36AP2) on chrom'
NG_002554 Homo sapiens NEK2 pseudogene (NEK2P) on chromosome 14
NG_002555 Homo sapiens ribosomal protein L36a pseudogene 3 (RPL36AP3) on chrom'
NG_002556 Homo sapiens ribosomal protein L36a pseudogene 4 (RPL36AP4) on chrom'
NG_002557 Homo sapiens molybdenum cofactor synthesis 3 pseudogene (MOCS3P) on
NG_002558 Homo sapiens ribosomal protein L39 pseudogene 2 (RPL39P2) on chromosc
NG_002559 Homo sapiens ribosomal protein L3 pseudogene 4 (RPL3P4) on chromosom
NG_002560 Homo sapiens ribosomal protein L41 pseudogene 4 (RPL41 P4) on chromosc
NG_002561 Homo sapiens ribosomal protein L7a pseudogene 5 (RPL7AP5) on chromosi
NG_002562 Homo sapiens ribosomal protein L7a pseudogene 6 (RPL7AP6) on chromosi
NG_002563 Homo sapiens ribosomal protein L9 pseudogene 6 (RPL9P6) on chromosom
NG_002564 Homo sapiens ribosomal protein S12 pseudogene 1 (RPS12P1) on chromos
NG_002565 Homo sapiens pituitary tumor-transforming 4 pseudogene (PTTG4P) on chro
NG_002566 Homo sapiens ribosomal protein S15a pseudogene 2 (RPS15AP2) on chrorr
NG_002567 Homo sapiens ribosomal protein S15a pseudogene 3 (RPS15AP3) on chrorr
NG_002568 Homo sapiens olfactory receptor, family 11 , subfamily H, member 8 pseudog
NG_002570 Homo sapiens ribosomal protein S18 pseudogene 2 (RPS18P2) on chromos
NG_002571 Homo sapiens ribosomal protein S24 pseudogene 2 (RPS24P2) on chromos
NG_002572 Homo sapiens ribosomal protein S24 pseudogene 3 (RPS24P3) on chromos
NG_002573 Homo sapiens ribosomal protein S27a pseudogene 4 (RPS27AP4) on chrorr
NG_002574 Homo sapiens ribosomal protein S8 pseudogene 1 (RPS8P1) on chromoson
NG_002575 Homo sapiens ribosomal protein S2 pseudogene 2 (RPS2P2) on chromoson
NG_002576 Homo sapiens ribosomal protein S2 pseudogene 3 (RPS2P3) on chromoson
NG_002577 Homo sapiens SSX1 pseudogene (psiSSXI ) on chromosome X
NG_002578 Homo sapiens SSX4 pseudogene (psiSSX4) on chromosome X
NG_002579 Homo sapiens SSX5 pseudogene (psiSSXδ) on chromosome X
NG_002580 Homo sapiens SSX3 pseudogene (psiSSX3) on chromosome X
NG 302581 Homo sapiens mago-nashi homolog, proliferation-associated pseudogene (C
NG_002582 Homo sapiens SSX2 pseudogene (psiSSX2) on chromosome X
NG_002583 Homo sapiens psiSSXδ pseudogene (psiSSXδ) on chromosome X
NGJD02584 Homo sapiens SSX9 pseudogene (psiSSX9) on chromosome X
NG_002585 Homo sapiens SSX6 pseudogene (psiSSX6) on chromosome X
NG_002586 Homo sapiens SSX10 pseudogene (psiSSXIO) on chromosome 6
NG_002587 Homo sapiens SSX7 pseudogene (psiSSX7) on chromosome X
NG_002588 Homo sapiens mortality factor 4 like pseudogene 1 (MORF4LP1 ) on chromos
NG_002589 Homo sapiens mortality factor 4 like pseudogene 2 (MORF4LP2) on chromos
NG_002590 Homo sapiens mortality factor 4 like pseudogene 3 (MORF4LP3) on chromos
NG_002591 Homo sapiens mortality factor 4 like pseudogene 4 (MORF4LP4) on chromos
NG_002592 Homo sapiens MAD2 mitotic arrest deficient-like 1 (yeast) pseudogene (MAC
NG_002593 Homo sapiens cytochrome c oxidase pseudogene 2 (MTC01 P2) on chromos
NG_002594 Homo sapiens nuclear receptor coactivator 4 pseudogene (NCOA4P) on chn
NG_002595 Homo sapiens NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 8, ps
NG_002596 Homo sapiens nicotinamide nucleotide adenylyltransferase pseudogene (NW
NG_002597 Homo sapiens nucleophosmin 1 (nucleolar phosphoprotein B23, numatrin) p.
NG_002598 Homo sapiens 5',3'-nucleotidase, cytosolic pseudogene 1 (NT5CP1) on chro
NGJ302599 Homo sapiens 5',3'-nucleotidase, cytosolic pseudogene 2 (NT5CP2) on chro
NG_002601 Homo sapiens UDP glycosyltransferase 1 family, polypeptide A cluster (UGT
NG_002603 Homo sapiens nuclear transport factor 2 pseudogene 2 (NUTF2P2) on chron
NG_002604 Homo sapiens prostatic binding protein pseudogene 1 (PBPP1) on chromosc
NG_002605 Homo sapiens PCQAP pseudogene (PCQAPP) on chromosome 14
NG_002606 Homo sapiens PDZ and LIM domain 1 pseudogene (PDLIM1P) on chromoso
NG_002607 Homo sapiens proteasome (prosome, macropain) subunit, alpha type, 3 psei
NG_002608 Homo sapiens prothymosin, alpha pseudogene 7 (PTMAP7) on chromosome
NG_002609 Homo sapiens RAN binding protein 20 pseudogene (RANBP20P) on chromo
NG_002610 Homo sapiens replication protein A2 pseudogene (RPA2P) on chromosome
NG_002611 Homo sapiens interleukin 6 signal transducer (gp130, oncostatin M receptor)
NG_002612 Homo sapiens family with sequence similarity 16, member A, Y-linked (FAM1
NG_002613 Homo sapiens family with sequence similarity 16, member B (FAM16B) pseu
NG_002616 Homo sapiens histone 2, H3b (HIST2H3B) pseudogene on chromosome 1
NG_002617 Homo sapiens histone 2, H3a (HIST2H3A) pseudogene on chromosome 1
NG_002618 Homo sapiens histone 3, H2ba (HIST3H2BA) pseudogene on chromosome 1
NG_002619 Homo sapiens histone 2, H2bc (HIST2H2BC) pseudogene on chromosome 1
NG_002620 Homo sapiens histone 2, H2bd (HIST2H2BD) pseudogene on chromosome '
NG_002621 Homo sapiens histone 2, H2ba (HIST2H2BA) pseudogene on chromosome 1
NG_002622 Homo sapiens ELYS transcription factor-like protein TMBS62 pseudogene (L
NG_002623 Homo sapiens ADP-ribosyltransferase (NAD+; poly (ADP-ribose) polymerase
NG_002624 Homo sapiens adaptin, beta 1-like 2 (ADTB1 L2) pseudogene on chromosom
NG_002625 Homo sapiens adaptin, beta 1-like 1 (ADTB1L1) pseudogene on chromosom
NG_002626 Homo sapiens proteasome (prosome, macropain) activator subunit 2 pseudc
NG_002627 Homo sapiens proteasome 26S non-ATPase subunit 10 pseudogene (LOC3:
NG_002628 Homo sapiens proteasome activator subunit 2 pseudogene (LOC338095) on
NG_002629 Homo sapiens proteasome activator subunit 2 pseudogene (LOC338096) on
NG_002630 Homo sapiens proteasome activator subunit 2 pseudogene (LOC338097) on
NG_002631 Homo sapiens proteasome activator subunit 2 pseudogene (LOC338098) on
NG_002632 Homo sapiens proteasome activator subunit 2 pseudogene (LOC338099) on
NG_002633 Homo sapiens cytochrome c oxidase subunit Vb-like 7 (COX5BL7) pseudoge
NG_002634 Homo sapiens serum amyloid A3 pseudogene (SAA3P) on chromosome 11
NG_002636 Homo sapiens YME1-like 2 (S. cerevisiae) (YME1L2) pseudogene on chrome
NG_002637 Homo sapiens argininosuccinate lyase-like (ASLL) pseudogene on chromosc
NG_002638 Homo sapiens colony stimulating factor 2 receptor, beta, 2 (CSF2RB2) pseu
NG_002639 Homo sapiens inorganic pyrophosphatase pseudogene (LOC151842) on chr
NG_002640 Homo sapiens mannan-binding Iectin serine protease 1 pseudogene 1 (MAS
NG_002641 Homo sapiens inorganic pyrophosphatase pseudogene (LOC285591) on chr
NG_002642 Homo sapiens SULT1 D pseudogene (SULT1 D1 P) on chromosome 4
NG_002645 Homo sapiens serine (or cysteine) proteinase inhibitor, clade B (ovalbumin),
NG_002646 Homo sapiens high-mobility group (nonhistone chromosomal) protein 17-like
NG_002647 Homo sapiens vomeronasal 1 receptor 8 pseudogene (VN1R8P) on chromos
NG_002648 Homo sapiens mitochondrial ribosomal protein S16 pseudogene (MRPS16P'
NG_002649 Homo sapiens mitochondrial ribosomal protein S16 pseudogene (MRPS16P:
NG_002650 Homo sapiens mitochondrial ribosomal protein S24 pseudogene (MRPS24P'
NG_002651 Homo sapiens mitochondrial ribosomal protein S25 pseudogene (MRPS25P'
NG_002652 Homo sapiens histone 2, H2bb (HIST2H2BB) pseudogene on chromosome 1
NG_002653 Homo sapiens taste receptor, type 2, member 62 pseudogene (TAS2R62P) c
NG_002655 Homo sapiens ADP-ribosyltransferase (NAD+; poly (ADP-ribose) polymerase
NG_002656 Homo sapiens ADP-ribosyltransferase (NAD+; poly (ADP-ribose) polymerase
NG_002657 Homo sapiens DEAH (Asp-Glu-Ala-His) box polypeptide 9 pseudogene (DH>
NG_002658 Homo sapiens eukaryotic translation initiation factor 4E-like 2 (EIF4EL2) psei
NG_002659 Homo sapiens zinc finger pseudogene (BA393J16.4) on chromosome 10
NG_002660 Homo sapiens KRAB box zinc finger protein pseudogene (BA775A3.1 ) on ch
NG_002661 Homo sapiens hypothetical pseudogene bA291 L22.4 (bA291 L22.4) on chron
NG_002662 Homo sapiens mitochondrial ribosomal protein S5 pseudogene (MRPS5P3) i
NG__002663 Homo sapiens mitochondrial ribosomal protein S21 pseudogene (MRPS21 P'
NG_002665 Homo sapiens mitochondrial ribosomal protein L42 pseudogene (MRPL42P1
NGJD02666 Homo sapiens mitochondrial ribosomal protein S18C pseudogene (MRPS18'
NG__002667 Homo sapiens keratin associated protein 13 pseudogene 1 (KRTAP13P1) or
NG_ 002668 Homo sapiens keratin associated protein 13 pseudogene 2 (KRTAP13P2) or
NG__002669 Homo sapiens keratin associated protein 8 pseudogene 1 (KRTAP8P1) on cl
NG_002670 Homo sapiens keratin associated protein 8 pseudogene 2 (KRTAP8P2) on cl
NG__002671 Homo sapiens keratin associated protein 19 pseudogene 3 (KRTAP19P3) or
NG_002672 Homo sapiens keratin associated protein 19 pseudogene 4 (KRTAP19P4) or
NG_002673 Homo sapiens keratin associated protein 21 pseudogene 1 (KRTAP21 P1) or
NG_002674 Homo sapiens taste receptor, type 2, member 64 pseudogene (TAS2R64P) <
NG_002675 Homo sapiens taste receptor, type 2, member 63 pseudogene (TAS2R63P) <
NG_002676 Homo sapiens taste receptor, type 2, member 65 pseudogene (TAS2R65P)
NG_002679 Homo sapiens potassium large conductance calcium-activated channel, subt
NG__002680 Homo sapiens survival motor neuron pseudogene (SMNP) on chromosome 5
NG_002681 Homo sapiens leukocyte immunoglobulin-like receptor, subfamily A (without '
NG_002682 Homo sapiens MAPRE1 P pseudogene (MAPRE1 P) on chromosome 8
NG_002683 Homo sapiens steroid-5-beta-reductase, beta polypeptide pseudogene 1 (SF
NG_002684 Homo sapiens mitochondrial translational initiation factor 2 pseudogene 1 (M
NG_002685 Homo sapiens histone 1 , H2a, pseudogene 2 (HIST1 H2APS2) on chromosoi
NG_002687 Homo sapiens suppressor of cytokine signaling 2 pseudogene 2 (SOCS2P2)
NG__002688 Homo sapiens keratin associated protein 19 pseudogene 1 (KRTAP19P1) or
NG_002689 Homo sapiens keratin associated protein 19 pseudogene 2 (KRTAP19P2) or
NG__002690 Homo sapiens Prader-Willi/Angelman syndrome region (PWSAS@) on chror
NG_002691 Homo sapiens ATP synthase, H+ transporting, mitochondrial F1 complex, ep
NG_002692 Homo sapiens protein phosphatase 1A pseudogene (LOC137012) on chrom.
NGJ302693 Homo sapiens phosphoribosyl pyrophosphate amidotransferase pseudogene
NG__002694 Homo sapiens mesoderm specific transcript homolog (mouse) pseudogene (
NG__002696 Homo sapiens eukaryotic translation initiation factor 2, subunit 2 beta, pseud
NG_002697 Homo sapiens ALEX2 pseudogene (LOC347674) on chromosome 7
NG_002698 Homo sapiens eukaryotic translation initiation factor 2 beta-like pseudogene
NG_002699 Homo sapiens G protein gamma 5-like subunit (GNG5ps) pseudogene on ch
NG_002700 Homo sapiens endothelin converting enzyme-like 1 , pseudogene 1 (ECEL1 P
NG__002701 Homo sapiens ECEL2 pseudogene 2 (ECEL2) on chromosome 2
NG_002702 Homo sapiens calgizzarin-like (LOC347701) pseudogene on chromosome 7
NG__002703 Homo sapiens mitochondrial ribosomal protein S36 pseudogene (MRPS36P<
NG_002705 Homo sapiens mitochondrial ribosomal protein S36 pseudogene (MRPS36P!
NG__002707 Homo sapiens mitochondrial ribosomal protein S36 pseudogene (MRPS36P!
NG_002709 Homo sapiens mitochondrial ribosomal protein S36 pseudogene (MRPS36P'
NG__002711 Homo sapiens mitochondrial ribosomal protein S36 pseudogene (MRPS36PI
NG_002713 Homo sapiens mitochondrial ribosomal protein S36 pseudogene (MRPS36PI
NG_002716 Homo sapiens calmodulin 2 pseudogene 1 (CALM2P1) on chromosome 17
NG_002717 Homo sapiens nitric oxide synthase 2A pseudogene (LOC284193) on chrom*
NG_002718 Homo sapiens galectin-9 pseudogene (LOC284194) on chromosome 17
NG )02719 Homo sapiens TL132 pseudogene (LOC347716) on chromosome 17
NG_002720 Homo sapiens signal recognition particle 68kD pseudogene (LOC347717) or
NG__002721 Homo sapiens karyopherin (importin) beta 2 pseudogene (LOC347719) on cl
NG_002723 Homo sapiens proteasome (prosome, macropain) 26S subunit, non-ATPase,
NG_002724 Homo sapiens actin, beta pseudogene 7 (ACTBP7) on chromosome 15
NG_002725 Homo sapiens eukaryotic translation initiation factor 5A pseudogene 3 (EIF&
NG__002726 Homo sapiens SRY (sex determining region Y)-box 5 pseudogene (SOX5P) i
NGJ302727 Homo sapiens MHC class I polypeptide-related sequence C (MICC) pseudog
NG__002728 Homo sapiens heterogeneous nuclear ribonucleoprotein A1 pseudogene (hn
NG__002729 Homo sapiens HLA-75 pseudogene (HLA-75) on chromosome 6
NGJ302731 Homo sapiens HLA-90 pseudogene (HLA-90) on chromosome 6
NG_002733 Homo sapiens MHC class I polypeptide-related sequence G pseudogene (Ml
NG_002735 Homo sapiens 3.8-1.5 pseudogene (3.8-1.5) on chromosome 6
NG_002736 Homo sapiens HLA complex P5 pseudogene 12 (HCP5P12) on chromosome
NG_002737 Homo sapiens HLA complex group 2 pseudogene 8 (HCG2P8) on chromoso
NG_002738 Homo sapiens ribosomal protein L7a pseudogene 7 (RPL7AP7) on chromosi
NG_002739 Homo sapiens HLA complex group 4 pseudogene 9 (HCG4P9) on chromoso
NG_002740 Homo sapiens HLA complex P5 pseudogene 13 (HCP5P13) on chromosome
NG_002741 Homo sapiens HLA complex group 4 pseudogene 10 (HCG4P10) on chromo
NG 302742 Homo sapiens HLA complex P5 pseudogene 14 (HCP5P14) on chromosome
NG_002743 Homo sapiens HLA complex group 9 pseudogene 5 (HCG9P5) on chromoso
NG_002744 Homo sapiens HLA complex group 4 pseudogene 11 (HCG4P11 ) on chromo
NG_002745 Homo sapiens HLA complex P5 pseudogene 15 (HCP5P15) on chromosome
NG_002746 Homo sapiens eukaryotic translation initiation factor 5A pseudogene 2 (EIF5/
NG_002747 Homo sapiens hydroxysteroid (17-beta) dehydrogenase pseudogene 1 (HSD
NG_002748 Homo sapiens interleukin 6 signal transducer (gp130, oncostatin M receptor)
NG_002749 Homo sapiens RAN, member RAS oncogene family pseudogene 1 (RANP1)
NG_002750 Homo sapiens SMT3 suppressor of mif two 3 homolog 2 (yeast) pseudogene
NG_002752 Homo sapiens N0D24 pseudogene (NOD24) on chromosome X
NG_002753 Homo sapiens NOD13 pseudogene (NOD13) on chromosome X
NG_002754 Homo sapiens NOD25 pseudogene (NOD25) on chromosome 12
NG_002761 Homo sapiens complement component 4 binding protein, alpha-like 2 (C4BP
NG_002762 Homo sapiens crystallin, gamma E pseudogene 1 (CRYGEP1) on chromosoi
NG_002763 Homo sapiens glutamate dehydrogenase pseudogene 3 (GLUDP3) on chron
NG_002764 Homo sapiens interleukin 9 receptor pseudogene 4 (IL9RP4) on chromosom
NG_002765 Homo sapiens prothymosin, alpha pseudogene 2 (gene sequence 32) (PTM/
NG_002766 Homo sapiens cytochrome P450, subfamily 51 pseudogene 2 (CYP51 P2) on
NG_002767 Homo sapiens glycoprotein, alpha-galactosyltransferase 1 pseudogene (GG"
NG_002768 Homo sapiens beta-lactoglobulin pseudogene (LOC138159) on chromosome
NG_002769 Homo sapiens heat shock 70kDa protein 8 pseudogene (LOC158714) on chi
NG_002770 Homo sapiens pseudogene of IGF-II mRNA-binding protein 3 (LOC346296) e
NG_002771 Homo sapiens major histocompatibility complex, class I, L (HLA-L) pseudoge
NG_002772 Homo sapiens CAP, adenylate cyclase-associated protein, 2 (yeast) pseudoe
NG_002773 Homo sapiens coactosin-like 1 (Dictyostelium) pseudogene 2 (COTL1 P2) on
NG_002775 Homo sapiens keratin pseudogene (LOC147228) on chromosome 17
NG_002776 Homo sapiens keratin pseudogene (LOC284196) on chromosome 17
NG_002777 Homo sapiens keratin pseudogene (LOC339186) on chromosome 17
NG_002778 Homo sapiens keratin pseudogene (LOC339241) on chromosome 17
NG_002779 Homo sapiens keratin pseudogene (LOC339244) on chromosome 17
NG_002780 Homo sapiens keratin pseudogene (LOC339258) on chromosome 17
NG_002781 Homo sapiens keratin pseudogene (LOC353194) on chromosome 17
NG_002782 Homo sapiens keratin pseudogene (LOC353196) on chromosome 17
NG_002785 Homo sapiens interleukin 6 receptor pseudogene (LOC157916) on chromosc
NG_002786 Homo sapiens makorin, ring finger protein, pseudogene 2 (MKRNP2) on chrc
NG_002787 Homo sapiens chromosome 20 open reading frame 189 (C20orf189) pseudo
NG_002788 Homo sapiens bitter taste receptor pseudogene 8 (PS8) on chromosome 12
NG_002790 Homo sapiens pseudogene of origin recognition complex, subunit 1-like (LO(
NG_002791 Homo sapiens selenoprotein W, 1 pseudogene (SEPW1 P) on chromosome
NG_002792 Homo sapiens PC4 and SFRS1 interacting protein 1 pseudogene (PSIP1P) c
NG_002793 Homo sapiens pelota/integrin, alpha 1 region (PELO/ITGA1 @) on chromosoi
NG_002795 Homo sapiens polycystic kidney disease 1 (autosomal dominant) pseudogen
NG_002796 Homo sapiens polycystic kidney disease 1 (autosomal dominant) pseudogen
NG_002797 Homo sapiens polycystic kidney disease 1 (autosomal dominant) pseudogen
NG_002798 Homo sapiens polycystic kidney disease 1 (autosomal dominant) pseudogen
NG_002799 Homo sapiens polycystic kidney disease 1 (autosomal dominant) pseudogen
NG_002800 Homo sapiens polycystic kidney disease 1 (autosomal dominant) pseudogen
NG_002801 Homo sapiens 18S ribosomal RNA pseudogene (LOC359724) on chromosor
NG_002802 Homo sapiens ADP-ribosyltransferase (NAD+; poly (ADP-ribose) polymerase
NG_002803 Homo sapiens apical protein-like (Xenopus laevis) pseudogene (APXLP) on * NG_002804 Homo sapiens arylsulfatase F pseudogene (ARSFP) NG_002805 Homo sapiens calcium/calmodulin-dependent serine protein kinase (MAGUK NG_002806 Homo sapiens adlican pseudogene (ADLICANP) on chromosome Y NG_002807 Homo sapiens cofactor required for Sp1 transcriptional activation, subunit 2, NG_002808 Homo sapiens C-terminal binding protein 2 pseudogene (LOC352905) on ch NG_002809 Homo sapiens eukaryotic translation initiation factor 4A, isoform 1 pseudogei NG_002810 Homo sapiens hypothetical protein FLJ10842 pseudogene (LOC359793) on NG_002811 Homo sapiens glycogenin 2 pseudogene (GYG2P) on chromosome Y NG_002812 Homo sapiens lung cancer candidate FUS1 pseudogene (LOC359794) on ct NG_002813 Homo sapiens neurofilament, light polypeptide 68kDa pseudogene (LOC359 NG_002814 Homo sapiens G protein-coupled receptor 143 pseudogene (GPR143P) on c NG_002815 Homo sapiens 60S ribosomal protein L26 pseudogene (LOC347593) on chrc NG_002816 Homo sapiens splicing factor proline/glutamine rich (polypyrimidine tract bine NG_002817 Homo sapiens solute carrier family 25 (mitochondrial carrier; omithine transp NG_002818 Homo sapiens Smcy homolog, Y chromosome (mouse) pseudogene (SMCYI NG_002819 Homo sapiens transducin (beta)-like 1Y-linked pseudogene (TBL1YP) on chr NG_002821 Homo sapiens ubiquitin-conjugating enzyme E2 variant 1 pseudogene (LOC NG_002822 Homo sapiens ubiquitin specific protease 12 pseudogene 1 (USP12P1) on cl NG_002823 Homo sapiens ubiquitin specific protease 12 pseudogene 2 (USP12P2) on cl NG_002824 Homo sapiens ubiquitin specific protease 12 pseudogene 3 (USP12P3) on cl NG_002825 Homo sapiens voltage-dependent anion channel 1 pseudogene (LOC35980C NG_002826 Homo sapiens Ras-homolog enriched n brain pseudogene 1 (RHEBP1) on c NG_002827 Homo sapiens tochondri ribosomal protein 63 pseudogene 1 (MRP63P1 NG_002828 Homo sapiens tochondri ribosoma protein S5 pseudogene (LOC133332 NG_002829 Homo sapiens tochondri ribosomal protein 63 pseudogene 6 (MRP63P6; NG_002830 Homo sapiens itochondri ribosoma protein S33 pseudogene 1 (MRPS33 NG_002831 Homo sapiens itochondri ribosoma protein L36 pseudogene 1 (MRPL36I NG_002832 Homo sapiens tochondri ribosomal protein S35 pseudogene 1 (MRPS35 NG_002833 Homo sapiens itochondri ribosoma protein S7 pseudogene 2 (MRPS7P2 NG_002834 Homo sapiens itochondri ribosoma protein L51 pseudogene 2 (MRPL51I NG_002835 Homo sapiens itochondri ribosoma protein S18C pseudogene 6 (MRPS1 NG_002836 Homo sapiens itochondri ribosomal protein L49 pseudogene 2 (MRPL49I NG_002837 Homo sapiens itochondri ribosoma protein 63 pseudogene 10 (MRP63P NG_002838 Homo sapiens itochondri ribosoma protein L2 pseudogene 1 (MRPL2P1 NG_002839 Homo sapiens itochondri ribosomal protein S18C pseudogene 4 (MRPS1 NG_002840 Homo sapiens itochondri ribosoma protein S21 pseudogene 8 (MRPS21 NG_002841 Homo sapiens tochondri ribosoma protein L50 pseudogene 1 (MRPL50I NG_002842 Homo sapiens itochondri ribosomal protein S31 pseudogene 1 (MRPS31 NG_002843 Homo sapiens itochondri ribosoma protein L51 pseudogene 1 (MRPL51 I NG_002844 Homo sapiens itochondri ribosomal protein S23 pseudogene 1 (MRPS23 NG_002845 Homo sapiens itochondri ribosoma protein 63 pseudogene 2 (MRP63P2 NG_002846 Homo sapiens itochondri ribosoma protein 63 pseudogene 3 (MRP63P3; NG_002847 Homo sapiens tochondri ribosomal protein 63 pseudogene 7 (MRP63P7 NG_002848 Homo sapiens itochondri ribosoma protein 63 pseudogene 8 (MRP63P8 NG_002849 Homo sapiens itochondri ribosomal protein 63 pseudogene 9 (MRP63P9 NG_002850 Homo sapiens itochondri ribosoma! protein L11 pseudogene 3 MRPL11 I NG_002851 Homo sapiens itochondri ribosoma protein L14 pseudogene 1 MRPL14I NG_002852 Homo sapiens tochondri ribosomal protein L15 pseudogene 1 MRPL15I NG_002853 Homo sapiens tochondri ribosomal protein L20 pseudogene 1 MRPL20I NG_002854 Homo sapiens itochondri ribosomal protein L22 pseudogene 1 MRPL22I NG_002855 Homo sapiens tochondri ribosomal protein L3 pseudogene 1 (MRPL3P1 NG_002856 Homo sapiens itochondri ribosoma protein L30 pseudogene 1 (MRPL30I NG_002857 Homo sapiens tochondri ribosomal protein L32 pseudogene 1 (MRPL32I NG_002858 Homo sapiens tochondri ribosomal protein L35 pseudogene 1 (MRPL35I NG_002859 Homo sapiens itochondri ribosoma protein L35 pseudogene 2 (MRPL35I NG 002860 Homo sapiens tochondri ribosomal protein L35 pseudogene 3 (MRPL35I
NG_002861 Homo sapiens mitochondrial ribosomal protein L35 pseudogene 4 (MRPL35I
NG_002862 Homo sapiens mitochondrial ribosomal protein L42 pseudogene 3 (MRPL42I
NG_002863 Homo sapiens mitochondrial ribosomal protein L45 pseudogene 1 (MRPL45I
NG_002864 Homo sapiens mitochondrial ribosomal protein L49 pseudogene 1 (MRPL49I
NG_002865 Homo sapiens mitochondrial ribosomal protein L50 pseudogene 2 (MRPL50I
NG_002866 Homo sapiens mitochondrial ribosomal protein L50 pseudogene 3 (MRPL50I
NG_002867 Homo sapiens mitochondrial ribosomal protein L50 pseudogene 4 (MRPL50I
NG_002868 Homo sapiens mitochondrial ribosomal protein L53 pseudogene 1 (MRPL53I
NG_002869 Homo sapiens mitochondrial ribosomal protein S10 pseudogene 1 (MRPS10
NG_002870 Homo sapiens mitochondrial ribosomal protein S10 pseudogene 5 (MRPS10
NG_002871 Homo sapiens mitochondrial ribosomal protein S15 pseudogene 1 (MRPS15
NG_002872 Homo sapiens mitochondrial ribosomal protein S15 pseudogene 2 (MRPS15
NG_002873 Homo sapiens mitochondrial ribosomal protein S17 pseudogene 3 (MRPS17
NG_002874 Homo sapiens mitochondrial ribosomal protein S17 pseudogene 5 (MRPS17
NG_002875 Homo sapiens mitochondrial ribosomal protein S17 pseudogene 6 (MRPS17
NG_002876 Homo sapiens mitochondrial ribosomal protein S17 pseudogene 9 (MRPS17
NG_002877 Homo sapiens mitochondrial ribosomal protein S18A pseudogene 1 (MRPS1
NG_002878 Homo sapiens mitochondrial ribosomal protein S18B pseudogene 1 (MRPS1
NG_002879 Homo sapiens mitochondrial ribosomal protein S18B pseudogene 2 (MRPS1
NG_002880 Homo sapiens mitochondrial ribosomal protein S18C pseudogene 3 (MRPS1
NG_002881 Homo sapiens mitochondrial ribosomal protein S18C pseudogene 5 (MRPS1
NG_002882 Homo sapiens mitochondrial ribosomal protein S21 pseudogene 2 (MRPS21
NG_002883 Homo sapiens mitochondrial ribosomal protein S21 pseudogene 3 (MRPS21
NG_002884 Homo sapiens mitochondrial ribosomal protein S21 pseudogene 4 (MRPS21
NG_002885 Homo sapiens mitochondrial ribosomal protein S21 pseudogene 5 (MRPS21
NG_002886 Homo sapiens mitochondrial ribosomal protein S21 pseudogene 6 (MRPS21
NG_002887 Homo sapiens mitochondrial ribosomal protein S21 pseudogene 7 (MRPS21
NG_002888 Homo sapiens mitochondrial ribosomal protein S21 pseudogene 9 (MRPS21
NG_002889 Homo sapiens mitochondrial ribosomal protein S22 pseudogene 1 (MRPS22
NG_002890 Homo sapiens mitochondrial ribosomal protein S29 pseudogene 2 (MRPS29
NG_002891 Homo sapiens mitochondrial ribosomal protein S33 pseudogene 2 (MRPS33
NG_002892 Homo sapiens mitochondrial ribosomal protein S33 pseudogene 3 (MRPS33
NG_002893 Homo sapiens mitochondrial ribosomal protein S33 pseudogene 4 (MRPS33
NG_002894 Homo sapiens mitochondrial ribosomal protein S35 pseudogene 2 (MRPS35
NG_002895 Homo sapiens mitochondrial ribosomal protein S35 pseudogene 3 (MRPS35
NG_002896 Homo sapiens mitochondrial ribosomal protein S6 pseudogene 1 (MRPS6P1
NG_002897 Homo sapiens mitochondrial ribosomal protein S6 pseudogene 2 (MRPS6P2
NG_002898 Homo sapiens mitochondrial ribosomal protein S6 pseudogene 4 (MRPS6P4
NG_002899 Homo sapiens mitochondrial ribosomal protein S7 pseudogene 1 (MRPS7P1
NG_002900 Homo sapiens mitochondrial ribosomal protein S5 pseudogene (MRPS5P4) i
NG_002901 Homo sapiens PCNA pseudogene pF2PCNA (LOC359805) on chromosome
NG_002902 Homo sapiens PCNA pseudogene p1 PCNA (LOC359806) on chromosome 4
NGJ302903 Homo sapiens mitochondrial ribosomal protein S10 pseudogene (MRPS10P:
NG_002904 Homo sapiens mitochondrial ribosomal protein S29 pseudogene 1 (MRPS29
NG_002905 Homo sapiens chloride channel, nucleotide-sensitive, 1 B (CLNS1 B) pseudoc
NGJD02906 Homo sapiens VENT-like homeobox 2 pseudogene 4 (VENTX2P4) on chrom
NG_002907 Homo sapiens mitochondrial ribosomal protein L42 pseudogene 2 (MRPL421
NG_002908 Homo sapiens mitochondrial ribosomal protein L42 pseudogene 4 (MRPL42I
NG_002909 Homo sapiens VENT-like homeobox 2 pseudogene 2 (VENTX2P2) on chrom
NG_002910 Homo sapiens mitochondrial ribosomal protein L10 pseudogene (LOC34895:
NG_002911 Homo sapiens VENT-like homeobox 2 pseudogene 3 (VENTX2P3) on chrom
NG_002912 Homo sapiens mitochondrial ribosomal protein L30 pseudogene 2 (MRPL30I
NG_002913 Homo sapiens mitochondrial ribosomal protein L39 pseudogene (LOC35981!
NG_002914 Homo sapiens mitochondrial ribosomal protein L42 pseudogene 5 (MRPL421
NG_002915 Homo sapiens peroxiredoxin 2 pseudogene 1 (PRDX2P1) on chromosome 1
NG_002916 Homo sapiens MHC class I polypeptide-related sequence E (MICE) pseudog
NG_002917 Homo sapiens PA1-1 mRNA-binding protein pseudogene (LOC359996) on ct
NG_002918 Homo sapiens ATP synthase, H+ transporting, mitochondrial F0 complex, su NG_002919 Homo sapiens CHRNA7 (cholinergic receptor, nicotinic, alpha polypeptide 7, NG_002920 Homo sapiens CHRNA7 (cholinergic receptor, nicotinic, alpha polypeptide 7, NG_002921 Homo sapiens BCL6 co-repressor pseudogene (LOC360000) on chromoson NG_002922 Homo sapiens interferon alpha-L pseudogene (G13P1) on chromosome 9 NG_002923 Homo sapiens glyceraldehyde-3-phosphate dehydrogenase pseudogene (LC NG_002924 Homo sapiens glyceraldehyde-3-phosphate dehydrogenase pseudogene (LC NG_002926 Homo sapiens cytochrome c, somatic pseudogene (HCP46) on chromosome NG_002927 Homo sapiens cytochrome c, somatic pseudogene (LOC360009) on chromo: NG_002928 Homo sapiens RAD17 homolog (S. pombe) pseudogene 2 (RAD17P2) on ch NG_002929 Homo sapiens RAD17 homolog (S. pombe) pseudogene 1 (RAD17P1) on ch NG_002930 Homo sapiens TAF9 pseudogene 1 (TAF9P1) on chromosome Y NG_002931 Homo sapiens TAF9 pseudogene 2 (TAF9P2) on chromosome Y NG_002932 Homo sapiens discs, large homolog 7 (Drosophila) pseudogene (LOC36001< NG_002933 Homo sapiens raft-linking protein pseudogene (LOC360015) on chromosome NG_002934 Homo sapiens capicua homolog (Drosophila) pseudogene (LOC360016) on i NG_002935 Homo sapiens capicua homolog (Drosophila) pseudogene (LOC360017) on ι NG_002936 Homo sapiens HBxAg transactivated protein 2 pseudogene (LOC360018) on NG_002937 Homo sapiens keratin 18 pseudogene 10 (KRT18P10) on chromosome Y NG_002938 Homo sapiens PC4 and SFRS1 interacting protein 2 pseudogene (LOC3600. NG_002939 Homo sapiens protein phosphatase 1 , regulatory (inhibitor) subunit 12B psei NG_002940 Homo sapiens protein phosphatase 1, regulatory (inhibitor) subunit 12B (LOC NG_002941 Homo sapiens ribosomal protein L41 pseudogene (LOC286568) on chromos NG_002942 Homo sapiens ribosomal protein L41 pseudogene (LOC286570) on chromos NG_002943 Homo sapiens intersectin 2 pseudogene (LOC360027) on chromosome Y NG_002944 Homo sapiens intersectin 2 pseudogene (LOC360026) on chromosome Y NG_002945 Homo sapiens intersectin 2 pseudogene (LOC360025) on chromosome Y NG_002946 Homo sapiens intersectin 2 pseudogene (LOC360024) on chromosome Y NG_002947 Homo sapiens tubulin, beta polypeptide 4, member Q pseudogene (LOC140I NG_002948 Homo sapiens tubulin, beta polypeptide 4, member Q pseudogene (LOC349* NG_002949 Homo sapiens hypothetical protein MGC23909 pseudogene (LOC360028) oi NG_002950 Homo sapiens hypothetical protein MGC45134 (MGC45134) pseudogene on NG_002951 Homo sapiens cyclic-AMP-dependent transcription factor ATF-4 pseudogene NG_002952 Homo sapiens hypothetical protein BC016683 pseudogene (LOC360029) on NG_002953 Homo sapiens cytochrome c, somatic pseudogene (HCP1) on chromosome NG )02954 Homo sapiens cytochrome c somatic pseudogene (HCP3) on chromosome NG_002955 Homo sapiens cytochrome c, somatic pseudogene (HCP4) on chromosome NG )02956 Homo sapiens cytochrome c, somatic pseudogene (HCP5) on chromosome NG_002957 Homo sapiens cytochrome c, somatic pseudogene (HCP6) on chromosome ; NG )02958 Homo sapiens cytochrome c somatic pseudogene (HCP7) on chromosome : NG_002959 Homo sapiens cytochrome c somatic pseudogene (HCP8) on chromosome ; NG_002960 Homo sapiens cytochrome c, somatic pseudogene (HCP9) on chromosome ; NG J02961 Homo sapiens cytochrome c, somatic pseudogene (HCP10 on chromosome NG_002962 Homo sapiens cytochrome c, somatic pseudogene (HCP11 on chromosome NG_002963 Homo sapiens cytochrome c somatic pseudogene (HCP12' on chromosome NG_002964 Homo sapiens cytochrome c, somatic pseudogene (HCP13 on chromosome NG_002965 Homo sapiens cytochrome c somatic pseudogene (HCP14 on chromosome NG )02966 Homo sapiens cytochrome c, somatic pseudogene (HCP16 on chromosome NG_002967 Homo sapiens cytochrome c, somatic pseudogene (HCP17; on chromosome NG )02968 Homo sapiens cytochrome c, somatic pseudogene (HCP18 on chromosome NGJD02969 Homo sapiens cytochrome c somatic pseudogene (HCP19 on chromosome NG_002970 Homo sapiens cytochrome c, somatic pseudogene (HCP20 on chromosome NG_002971 Homo sapiens cytochrome c somatic pseudogene (HCP21 on chromosome NG_002972 Homo sapiens cytochrome c, somatic pseudogene (HCP22 on chromosome NG_002973 Homo sapiens cytochrome c somatic pseudogene (HCP23 on chromosome NG_002974 Homo sapiens cytochrome c somatic pseudogene (HCP24' on chromosome NG 002975 Homo sapiens cytochrome c somatic pseudogene (HCP25 on chromosome
NG_002976 Homo sapiens cytochrome c, somatic pseudogene (HCP26) on chromosome
NG_002977 Homo sapiens cytochrome c, somatic pseudogene (HCP27) on chromosome
NG_002978 Homo sapiens cytochrome c, somatic pseudogene (HCP28) on chromosome
NG_002979 Homo sapiens cytochrome c, somatic pseudogene (HCP29) on chromosome
NG_002980 Homo sapiens cytochrome c, somatic pseudogene (HCP30) on chromosome
NG_002981 Homo sapiens cytochrome c, somatic pseudogene (HCP32) on chromosome
NG_002982 Homo sapiens cytochrome c, somatic pseudogene (HCP33) on chromosome
NG_002983 Homo sapiens cytochrome c, somatic pseudogene (HCP34) on chromosome
NG_002984 Homo sapiens cytochrome c, somatic pseudogene (HCP35) on chromosome
NG_002985 Homo sapiens cytochrome c, somatic pseudogene 1 (CYCSP1) on chromosc
NG_002986 Homo sapiens cytochrome c, somatic pseudogene (HCP37) on chromosome
NG_002987 Homo sapiens cytochrome c, somatic pseudogene (HCP38) on chromosome
NG_002988 Homo sapiens cytochrome c, somatic pseudogene (HCP39) on chromosome
NG_002989 Homo sapiens cytochrome c, somatic pseudogene (HCP40) on chromosome
NG_002990 Homo sapiens cytochrome c, somatic pseudogene (HCP41) on chromosome
NG_002991 Homo sapiens cytochrome c, somatic pseudogene (HCP42) on chromosome
NG_002992 Homo sapiens cytochrome c, somatic pseudogene (HCP43) on chromosome
NG_002993 Homo sapiens cytochrome c, somatic pseudogene (HCP44) on chromosome
NG_002994 Homo sapiens cytochrome c, somatic pseudogene (HCP45) on chromosome
NG_002995 Homo sapiens cytochrome c, somatic pseudogene (HCP48) on chromosome
NG_002996 Homo sapiens thymosin-like 5 (TMSL5) pseudogene on chromosome 11
NG_002997 Homo sapiens thymosin-like 7 (TMSL7) pseudogene on chromosome X
NG_002998 Homo sapiens NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 4, 9
NG_002999 Homo sapiens cytochrome c, somatic pseudogene (HCP49) on chromosome
NG_003006 Homo sapiens RNA binding motif protein, Y-linked, family 1, member H (RBΛ
NG_003008 Homo sapiens nuclease sensitive element binding protein 1 pseudogene (b/
NG_003009 Homo sapiens argininosuccinate synthetase pseudogene 8 (ASSP8) on chro
NG_003010 Homo sapiens cytochrome c oxidase subunit Vb-like 1 (COX5BL1) pseudoge
NG_003011 Homo sapiens glyceraldehyde-3-phosphate dehydrogenase-like 4 (GAPDL4]
NG_003012 Homo sapiens glycine dehydrogenase (decarboxylase) pseudogene (GLDCF
NG_003013 Homo sapiens general transcription factor IIF, polypeptide 2-like (GTF2F2L)
NG_003014 Homo sapiens heat shock 90kDa protein 1 , alpha-like 2 (HSPCAL2) pseudoc
NG_003015 Homo sapiens heat shock 90kDa protein 1 , beta pseudogene 1 (HSPCP1 ) oi
NG_003016 Homo sapiens lactate dehydrogenase A-like 1 (LDHAL1) pseudogene on chr
NG_003017 Homo sapiens sorcin-like (SRIL) pseudogene on chromosome 4
NG_003018 Homo sapiens v-raf-1 murine leukemia viral oncogene homolog 1 pseudoger
NG__003019 Homo sapiens actin, beta pseudogene 2 (ACTBP2) on chromosome 5
NG_003020 Homo sapiens actin, beta pseudogene 4 (ACTBP4) on chromosome 5
NG_003021 Homo sapiens argininosuccinate synthetase pseudogene 10 (ASSP10) on cl
NGJ 03022 Homo sapiens argininosuccinate synthetase pseudogene 9 (ASSP9) on chro
NG_003023 Homo sapiens OFD1 pseudogene 1 (OFD1P1) on chromosome 5
NG__003024 Homo sapiens diazepam binding inhibitor-like 1 (DBIL1) pseudogene on chrc
NG__003025 Homo sapiens chemokine ligand 14, chemokine ligand 15 transcription unit (
NG__003026 Homo sapiens endogenous retroviral pol gene-like sequence 2 (ERPL2) psei
NG_003027 Homo sapiens glyceraldehyde-3-phosphate dehydrogenase-like 16 (GAPDL
NG__003028 Homo sapiens ferritin, heavy polypeptide-like 10 (FTHL10) pseudogene on c
NG_003029 Homo sapiens gap junction protein, alpha 1 , 43kDa (connexin 43) pseudogei
NG__003030 Homo sapiens glutamate-ammonia ligase (glutamine synthase)-like 1 (GLUL
NG_003031 Homo sapiens hypoxanthine phosphoribosyltransferase pseudogene 2 (HPR
NG_003032 Homo sapiens moesin-like 1 (MSNL1) pseudogene on chromosome 5
NG__003033 Homo sapiens ribosomal protein S17 pseudogene 2 (RPS17P2) on chromos
NG D03034 Homo sapiens ribosomal protein S20 pseudogene 3 (RPS20P3) on chromos
NG_003035 Homo sapiens ribosomal protein S20 pseudogene 4 (RPS20P4) on chromos
NG_003036 Homo sapiens t-complex 1-like 2 (TCP1L2) pseudogene on chromosome 5
NG_003037 Homo sapiens X-box binding protein pseudogene 1 (XBPP1) on chromosom*
NG_003038 Homo sapiens eukaryotic translation elongation factor 1 beta 3 (EEF1 B3) psi
NG__003039 Homo sapiens actin, gamma pseudogene 10 (ACTGP10) on chromosome X
NG_003040 Homo sapiens brain cytoplasmic RNA 1, pseudogene 1 (BCYRN1P1) on chn
NG_003041 Homo sapiens RNA binding motif protein, Y-linked, family 2, member B pseu
NG_003042 Homo sapiens RNA binding motif protein, Y-linked, family 2, member C pseu
NG_003043 Homo sapiens RNA binding motif protein, Y-linked, family 2, member D pseu
NG_003044 Homo sapiens centromere protein C2, 140kDa (CENPC2) pseudogene on cf
NG_003045 Homo sapiens eukaryotic translation elongation factor 1 beta 1 (EEF1B1) psi
NG_003061 Homo sapiens DNA segment on chromosome 6 (unique, pseudogene) 2723
NG_003062 Homo sapiens ribosomal protein L7a pseudogene 1 (RPL7AP1 ) on chromosi
NG_003063 Homo sapiens zinc finger protein 381 , Y-linked pseudogene (ZNF381 P) on c
NG_003064 Homo sapiens RNA binding motif protein, Y-linked, family 2, member A pseu
NG_003070 Homo sapiens RNA binding motif protein, Y-linked, family 2, pseudogene (LC
NG_003072 Homo sapiens MADS box transcription enhancer factor 2, polypeptide A psei
NG_003073 Homo sapiens protein kinase, cAMP-dependent, regulatory, type I, alpha pse
NG_003074 Homo sapiens RNA binding motif protein, Y-linked, family 2, pseudogene (LC
NG_003075 Homo sapiens RNA binding motif protein, Y-linked, family 2, pseudogene (LC
NG_003076 Homo sapiens ring finger protein 134 pseudogene 1 (RNF134P1) on chromo
NG_003077 Homo sapiens testis specific protein, Y-linked pseudogene 1 (TSPYP1) on cl
NG_003078 Homo sapiens testis specific protein, Y-linked pseudogene 2 (TSPYP2) on cl
NG_003079 Homo sapiens testis specific protein, Y-linked pseudogene 3 (TSPYP3) on cl
NG_003080 Homo sapiens testis specific protein, Y-linked pseudogene 4 (TSPYP4) on cl
NG_003081 Homo sapiens RNA binding motif protein, Y-linked, family 2, pseudogene (LC
NG_003082 Homo sapiens RNA binding motif protein, Y-linked, family 2, pseudogene (LC
NG_003083 Homo sapiens RNA binding motif protein, Y-linked, family 2, pseudogene (LC
NG_003084 Homo sapiens RNA binding motif protein, Y-linked, family 2, pseudogene (LC
NG_003085 Homo sapiens RNA binding motif protein, Y-linked, family 2, pseudogene (LC
NG_003086 Homo sapiens RNA binding motif protein, Y-linked, family 2, pseudogene (LC
NG_003087 Homo sapiens RNA binding motif protein, Y-linked, family 2, pseudogene (LC
NG_003088 Homo sapiens RNA binding motif protein, Y-linked, family 2, pseudogene (LC
NG_003089 Homo sapiens RNA binding motif protein, Y-linked, family 2, pseudogene (LC
NG_003090 Homo sapiens RNA binding motif protein, Y-linked, family 2, pseudogene (LC
NG_003091 Homo sapiens RNA binding motif protein, Y-linked, family 2, pseudogene (LC
NG_003092 Homo sapiens RNA binding motif protein, Y-linked, family 2, pseudogene (LC
NG_003093 Homo sapiens testis specific protein, Y-linked pseudogene 5 (TSPYP5) on cl
NG_003094 Homo sapiens X Kell blood group precursor-related, Y-linked pseudogene 1
NG_003095 Homo sapiens X Kell blood group precursor-related, Y-linked pseudogene 2
NG_003096 Homo sapiens X Kell blood group precursor-related, Y-linked pseudogene 3
NG_003097 Homo sapiens X Kell blood group precursor-related, Y-linked pseudogene 4
NG_003098 Homo sapiens X Kell blood group precursor-related, Y-linked pseudogene 5
NG_003099 Homo sapiens X Kell blood group precursor-related, Y-linked pseudogene (X
NG_003100 Homo sapiens choline kinase-like, carnitine palmitoyltransferase 1 B (muscle^
NG_003101 Homo sapiens ubiquitin-conjugating enzyme E2L 1 (UBE2L1) pseudogene o
NG_003102 Homo sapiens COP9 constitutive photomorphogenic homolog subunit 5 psei
NG_003103 Homo sapiens COP9 pseudogene (LOC375350) on chromosome 3
NG_003104 Homo sapiens zinc finger protein 91 homolog (mouse), ciliary neurotrophic fε
NG_003105 Homo sapiens E2F transcription factor 6 pseudogene (LOC376818) on chror
NG_003106 Homo sapiens E2F transcription factor 6 pseudogene (LOC386610) on chror
NG_003107 Homo sapiens solute carrier family 25 (mitochondrial carrier; adenine nucleo
NG_003108 Homo sapiens similar to v-raf murine sarcoma viral oncogene homolog B1 ps
NG_003109 Homo sapiens osteoclast stimulating factor 1 pseudogene (OSTF1 P) on chrc
NG_003110 Homo sapiens LAG1 longevity assurance homolog 1 (S. cerevisiae), growth i
NG_003111 Homo sapiens serine/threonine kinase 6-like pseudogene (STK6LP) on chroi
NG_003114 Homo sapiens OFD1 pseudogene Y-linked 1 (OFDYP1) on chromosome Y
NG_003115 Homo sapiens OFD1 pseudogene Y-linked 2 (OFDYP2) on chromosome Y
NG_003116 Homo sapiens OFD1 pseudogene Y-linked 3 (OFDYP3) on chromosome Y
NG_003117 Homo sapiens OFD1 pseudogene Y-linked 4 (OFDYP4) on chromosome Y
NG_003118 Homo sapiens OFD1 pseudogene Y-linked 5 (OFDYP5) on chromosome Y
NG_003119 Homo sapiens OFD1 pseudogene Y-linked 6 (OFDYP6) on chromosome Y
NG_003120 Homo sapiens OFD1 pseudogene Y-linked 7 (OFDYP7) on chromosome Y NG_003121 Homo sapiens OFD1 pseudogene Y-linked 8 (OFDYP8) on chromosome Y NG_003122 Homo sapiens OFD1 pseudogene Y-linked 9 (OFDYP9) on chromosome Y NG_003125 Homo sapiens OFD1 pseudogene Y-linked 11 (OFDYP11) on chromosome NG_003126 Homo sapiens OFD1 pseudogene Y-linked 12 (OFDYP12) on chromosome NG_003127 Homo sapiens OFD1 pseudogene Y-linked 13 (OFDYP13) on chromosome λ NG_003128 Homo sapiens OFD1 pseudogene Y-linked 14 (OFDYP14) on chromosome N NG_003129 Homo sapiens OFD1 pseudogene Y-linked 15 (OFDYP15) on chromosome NG_003130 Homo sapiens chromodomain protein, Y-linked 10 pseudogene (CDY10P) or NG_003131 Homo sapiens chromodomain protein, Y-linked 3 pseudogene (CDY3P) on c NG_003132 Homo sapiens chromodomain protein, Y-linked 5 pseudogene (CDY5P) on c NG_003133 Homo sapiens chromodomain protein, Y-linked 9 pseudogene (CDY9P) on c NG_003134 Homo sapiens family with sequence similarity 8, member A7 pseudogene (Fy NG_003135 Homo sapiens family with sequence similarity 8, member A8 pseudogene (F; NG_003136 Homo sapiens family with sequence similarity 8, member A9 pseudogene (F; NG_003137 Homo sapiens family with sequence similarity 8, member A1 pseudogene (L( NG_003138 Homo sapiens chromodomain protein Y-linked 4 pseudogene (CDY4P) on c NG_003139 Homo sapiens chromodomain protein Y-linked 6 pseudogene (CDY6P) on c NG_003140 Homo sapiens chromodomain protein Y-linked 7 pseudogene (CDY7P) on c NG_003141 Homo sapiens chromodomain protein Y-linked 8 pseudogene (CDY8P) on c NG_003142 Homo sapiens chromodomain protein Y-linked 12 pseudogene (CDY12P) or NG_003143 Homo sapiens chromodomain protein Y-linked 13 pseudogene (CDY13P) or NG_003144 Homo sapiens chromodomain protein Y-linked 14 pseudogene (CDY14P) or NG_003145 Homo sapiens chromodomain protein Y-linked 15 pseudogene (CDY15P) or NG_003146 Homo sapiens chromodomain protein Y-linked 16 pseudogene (CDY16P) or NG_003147 Homo sapiens chromodomain protein Y-linked 17 pseudogene (CDY17P) or NG_003148 Homo sapiens chromodomain protein Y-linked 18 pseudogene (CDY18P) or NG_003149 Homo sapiens chromodomain protein Y-linked 19 pseudogene (CDY19P) or NG_003150 Homo sapiens chromodomain protein Y-linked 20 pseudogene (CDY20P) or NG_003151 Homo sapiens chromodomain protein Y-linked 21 pseudogene (CDY21P) or NG_003152 Homo sapiens chromodomain protein Y-linked 22 pseudogene (CDY22P) or NG_003153 Homo sapiens chromodomain protein Y-linked 23 pseudogene (CDY23P) or NG_003154 Homo sapiens chromodomain protein Y-linked 11 pseudogene (CDY11 P) or NG_003155 Homo sapiens ribosomal protein L29 pseudogene 1 (RPL29P1) on chromosc NG_003156 Homo sapiens ribosomal protein L39 pseudogene 3 (RPL39P3) on chromosc NG_003157 Homo sapiens v-raf murine sarcoma 3611 viral oncogene homolog pseudoge NG_003158 Homo sapiens telomeric repeat binding factor (NIMA-interacting) 1 pseudoge NG_003159 Homo sapiens pseudogene of CXYorfl (CXYorfl P) on chromosome 16 NG_003160 Homo sapiens ribosomal protein L24 pseudogene 4 (RPL24P4) on chromose NG_003162 Homo sapiens actin, beta pseudogene 9 (ACTBP9) on chromosome 18 NG_003163 Homo sapiens creatine kinase B pseudogene 1 (CKBP1) on chromosome 16 NG_003164 Homo sapiens transferrin pseudogene (TFP) on chromosome 3 NG_003165 Homo sapiens platelet-activating factor acetylhydrolase, isoform lb, pseudog NG_003166 Homo sapiens olfactory receptor, family 7, subfamily A, member 8 pseudoge NG_003167 Homo sapiens platelet-activating factor acetylhydrolase, isoform lb, pseudog NG_003168 Homo sapiens ubiquitin specific protease 9, Y-linked pseudogene (LOC3873 NG_003169 Homo sapiens ubiquitin specific protease 9, Y-linked pseudogene (LOC3873 NG_003170 Homo sapiens ubiquitin specific protease 9, Y-linked pseudogene (LOC3873 NG_003171 Homo sapiens USP9Y pseudogene 1 (USP9YP1) on chromosome Y NG_003172 Homo sapiens USP9Y pseudogene 2 (USP9YP2) on chromosome Y NG_003173 Homo sapiens ubiquitin specific protease 9, Y-linked pseudogene (LOC3873 NG_003180 Homo sapiens cytochrome P450, family 2, subfamily D, polypeptide 6 genon NG_003183 Homo sapiens BRCA1 pseudogene (LBRCA1) on chromosome 17 NG_003186 Homo sapiens transcription elongation factor A (Sll), 1 pseudogene (LOC395 NG_003187 Homo sapiens deafness dystonia pseudogene (DDPP) on chromosome 2 NG_003188 Homo sapiens chemokine (C-C motif) ligand 3-like 2 (CCL3L2) pseudogene • NG 003189 Homo sapiens ribosomal protein S26 pseudogene 6 (RPS26P6) on chromos
NG_003190 Homo sapiens pseudogene of ribosomal protein L9 (LOC388147) on chrome
NG_003193 Homo sapiens PR/SET domain containing protein 07 pseudogene (SET07p)
NG_003194 Homo sapiens olfactory receptor, family 2, subfamily B, member 8 pseudoge
NG_003195 Homo sapiens olfactory receptor, family 52, subfamily L, member 2 pseudogi
NG_003196 Homo sapiens olfactory receptor, family 10, subfamily D, member 4 pseudog
NG_003197 Homo sapiens olfactory receptor, family 52, subfamily E, member 1 pseudog
NG_003198 Homo sapiens olfactory receptor, family 4, subfamily K, member 3 pseudoge
NG_003200 Homo sapiens ELK2, member of ETS oncogene family, pseudogene 2 (ELKi
NG_003201 Homo sapiens olfactory receptor, family 4, subfamily G, member 2 pseudoge
NG_003215 Homo sapiens eukaryotic translation initiation factor 2, subunit 3 gamma, 52!
NG_003216 Homo sapiens ribosomal protein S25 pseudogene (LOC283114) on chromos
NG_003217 Homo sapiens heat shock 60kDa protein 1 pseudogene (LOC283320) on chi
NG_003218 Homo sapiens synaptogyrin 2 pseudogene (LOC283698) on chromosome 1 J
NG_003219 Homo sapiens transcription elongation factor B (Sill), polypeptide 1 pseudog
NG_003221 Homo sapiens olfactory receptor, family 11 , subfamily I, member 1 pseudoge
NG_003222 Homo sapiens olfactory receptor, family 4, subfamily G, member 3 pseudoge
NG_003230 Homo sapiens olfactory receptor, family 5, subfamily D, member 11 pseudog
NG_003253 Homo sapiens cytochrome c oxidase, subunit 8B pseudogene (COX8B) on c
NG_003254 Homo sapiens immunoglobulin heavy constant epsilon P2 (IGHEP2) pseudo*
NG_003255 Homo sapiens Rhesus blood group cluster (RHD/RHCE@) on chromosome
NG_003256 Homo sapiens gonadotropin-releasing hormone receptor 2 pseudogene (GN
NG_003258 Homo sapiens folate hydrolase 2 (FOLH2) pseudogene on chromosome 11
NG_003259 Homo sapiens ribosomal protein S9 pseudogene 1 (RPS9P1) on chromoson
NG_004075 Homo sapiens laminin receptor 1 pseudogene 14 (LAMR1 P14) on chromoso
NG_004077 Homo sapiens thiopurine S-methyltransferase pseudogene (LOC400650) on
NG_004085 Homo sapiens protein tyrosine phosphatase type IVA pseudogene 1 (PTP4A
NG_004086 Homo sapiens olfactory receptor, family 5, subfamily J, member 1 pseudogei
NG_004087 Homo sapiens olfactory receptor, family 7, subfamily A, member 2 pseudoge
NG_004088 Homo sapiens olfactory receptor, family 2, subfamily E, member 1 pseudoge
NG_004089 Homo sapiens olfactory receptor, family 4, subfamily H, member 12 pseudog
NG_004091 Homo sapiens mitochondrial ribosomal protein S17 pseudogene (MRPS17P'
NG_004092 Homo sapiens mitochondrial ribosomal protein S17 pseudogene (MRPS17P'
NG_004093 Homo sapiens Nanog homeobox pseudogene 8 (NANOGP8) on chromosom
NG_004095 Homo sapiens Nanog homeobox pseudogene 3 (NANOGP3) on chromosom
NG_004096 Homo sapiens Nanog homeobox pseudogene 10 (NANOGP10) on chromosc
NG_004097 Homo sapiens Nanog homeobox pseudogene 9 (NANOGP9) on chromosom
NG_004098 Homo sapiens NANOG homeobox pseudogene 7 (NANOGP7) on chromosoi
NG_004099 Homo sapiens NANOG homeobox pseudogene 2 (NANOGP2) on chromosoi
NG_004100 Homo sapiens NANOG homeobox pseudogene 4 (NANOGP4) on chromosoi
NG_004101 Homo sapiens NANOG homeobox pseudogene 5 (NANOGP5) on chromosoi
NG_004102 Homo sapiens NANOG pseudogene 6 (NANOGP6) on chromosome 10
NG_004103 Homo sapiens NANOG homeobox pseudogene 11 (NANOGP11 ) on chromo:
NGJ304109 Homo sapiens YTH domain family 2 pseudogene (YTHDF2P) on chromosorr
NG_004110 Homo sapiens ribonuclease H1 pseudogene 2 (RNASEH1 P2) on chromoson
NG_004111 Homo sapiens ribonuclease H1 pseudogene 1 (RNASEH1 P1) on chromoson
NG_004112 Homo sapiens endogenous retroviral family W, env(C7), member 1 (syncytin
NG_004113 Homo sapiens CCL3L1-CCL4L1 chemokine gene cluster (CCL3L1-CCL4L1ι
NG_004115 Homo sapiens VKORC1 pseudogene 1 (LOC414355) on chromosome X
NG_ 004116 Homo sapiens VKORC1 pseudogene 2 (LOC414357) on chromosome 1
NG_004122 Homo sapiens olfactory receptor, family 7, subfamily E, member 22 pseudog
NG_004123 Homo sapiens olfactory receptor, family 7, subfamily E, member 21 pseudog
NG_004124 Homo sapiens olfactory receptor, family 5, subfamily E, member 1 pseudoge
NG_004125 Homo sapiens olfactory receptor, family 10, subfamily D, member 3 pseudog
NG_004126 Homo sapiens olfactory receptor, family 10, subfamily D, member 1 pseudog
NG_004127 Homo sapiens olfactory receptor, family 7, subfamily E, member 66 pseudog
NG_004128 Homo sapiens olfactory receptor, family 7, subfamily E, member 47 pseudog
NG_004129 Homo sapiens olfactory receptor, family 7, subfamily E, member 36 pseudog
NG_004130 Homo sapiens factory receptor, fami ly 7, subfamily E, member 29 pseudog NG_004131 Homo sapiens factory receptor, fami ly 7, subfamily E, member 28 pseudog NG_004132 Homo sapiens factory receptor, fami ly 7, subfamily E, member 25 pseudog NG_004133 Homo sapiens factory receptor, fam ly 4, subfamily G, member 1 pseudoge NG_004134 Homo sapiens factory receptor, fami ly 4, subfamily F, member 1 pseudoge NG_004135 Homo sapiens Ifactory receptor, fami ly 4, subfamily E, member 1 pseudoge NG_004136 Homo sapiens Ifactory receptor, fam ly 4, subfamily B, member 2 pseudoge NG_004137 Homo sapiens factory receptor, fami ly 2, subfamily V, member 1 (OR2V1) | NG_004138 Homo sapiens factory receptor, fami ly 1 , subfamily H, member 1 pseudoge NG_004139 Homo sapiens Ifactory receptor, fam ly 5, subfamily P, member 1 pseudoge NG_004140 Homo sapiens factory receptor, fam ly 6, subfamily M, member 3 pseudoge NG_004141 Homo sapiens factory receptor, fami ly 11 , subfamily H, member 2 (OR11 K NG_004142 Homo sapiens Ifactory receptor, fam ly 51 , subfamily B, member 3 pseudog NG_004143 Homo sapiens factory receptor, fam ly 5, subfam ly AL, member 2 pseudog NG_004144 Homo sapiens factory receptor, fam ly 5, subfam ly AL, member 1 pseudog NG_004145 Homo sapiens factory receptor, fam ly 5, subfam ly BM, member 1 pseudoc NG_004146 Homo sapiens factory receptor, fam ly 8, subfam ly K, member 2 pseudoge NG_004147 Homo sapiens ifactory receptor, fam ly 8, subfam ly I, member 1 pseudoger NG_004148 Homo sapiens factory receptor, fam ly 4, subfam ly G, member 4 pseudoge NG_004149 Homo sapiens factory receptor, fam ly 5, subfam ly M, member 7 pseudoge NG_004150 Homo sapiens factory receptor, fami ly 5, subfam ly M, member 6 pseudoge NG_004151 Homo sapiens factory receptor, fam ly 5, subfam ly M, member 5 pseudoge NG_004152 Homo sapiens factory receptor, fam ly 5, subfam ly M, member 4 pseudoge NG_004153 Homo sapiens factory receptor, fami ly 10, subfamily AB, member 1 pseudo NG_004154 Homo sapiens Ifactory receptor, fam ly 5, subfamily M, member 2 pseudoge NG_004155 Homo sapiens factory receptor, fami ly 6, subfamily M, member 2 pseudoge NG_004156 Homo sapiens Ifactory receptor, fam ly 5, subfamily BJ, member 1 pseudog NG_004157 Homo sapiens Ifactory receptor, fam ly 5, subfamily BH, member 1 pseudog NG_004158 Homo sapiens factory receptor, fami ly 5, subfamily AW, member 1 pseudoi NG_004159 Homo sapiens Ifactory receptor, fam ly 4, subfamily W, member 1 pseudoge NG_004160 Homo sapiens Ifactory receptor, fam ly 4, subfamily K, member 12 pseudog NG_004161 Homo sapiens Ifactory receptor, fam ly 4, subfamily K, member 11 pseudog NG_004162 Homo sapiens Ifactory receptor, fam ly 4, subfamily Q, member 1 pseudoge NG_004163 Homo sapiens Ifactory receptor, fam ly 11 , subfamily K, member 1 pseudog NG_004164 Homo sapiens Ifactory receptor, fam ly 11 , subfamily J, member 2 pseudogi NG_004165 Homo sapiens Ifactory receptor, fam ly 11 , subfamily J, member 1 pseudogi NG_004166 Homo sapiens Ifactory receptor, fam ly 11 , subfamily H, member 3 pseudog NG_004167 Homo sapiens Ifactory receptor, fam ly 7, subfamily K, member 1 pseudoge NG_004168 Homo sapiens spondyloepiphyseal dysplasia, late, pseudogene 5 (SEDLP5) NG_004169 Homo sapiens spondyloepiphyseal dysplasia, late, pseudogene 3 (SEDLP3) NG_004170 Homo sapiens Ifactory receptor, fami ily 7, subfami ly E, member 106 pseudo NG_004171 Homo sapiens Ifactory receptor, fam ly 7, subfam ly E, member 105 pseudo NG_004172 Homo sapiens Ifactory receptor, fam ly 4, subfami ly K, member 16 pseudog NG_004173 Homo sapiens factory receptor, fami ly 7, subfami ly E, member 111 pseudo NG_004174 Homo sapiens Ifactory receptor, fam ly 7, subfam ly E, member 104 pseudo NG_004175 Homo sapiens factory receptor fami ly 7, subfam ly E, member 101 pseudo NG_004176 Homo sapiens factory receptor, fami ly 9, subfami ly R, member 1 pseudoge NG_004177 Homo sapiens factory receptor, fami ly 9, subfami ly M, member 1 pseudoge NG_004178 Homo sapiens factory receptor, fami ly 9, subfami ly I, member 3 pseudoger NG_004179 Homo sapiens factory receptor, fam ly 9, subfam ly G, member 3 pseudoge NG_004180 Homo sapiens factory receptor, fami ly 9, subfami ly G, member 2 pseudoge NG_004181 Homo sapiens factory receptor, fami ly 8, subfami ly R, member 1 pseudoge NG_004182 Homo sapiens Ifactory receptor, fam ly 8, subfam ly Q, member 1 pseudoge NG_004183 Homo sapiens factory receptor, fami ly 8, subfam ly L, member 1 pseudogei NG_004184 Homo sapiens [factory receptor, fami ly 8, subfami ly K, member 4 pseudoge NG_004185 Homo sapiens [factory receptor, fam ly 8, subfam ly J, member 2 pseudogei NG 004186 Homo sapiens factory receptor, fami ly 7, subfami ly A, member 128 pseudo
NG_004188 Homo sapiens Ifactory receptor, , fam ly 5, subfam ly W, member 1 pseudoge NG_004189 Homo sapiens Ifactory receptor, , fam ly 5, subfam ly P, member 4 pseudoge NG_004190 Homo sapiens Ifactory receptor, , fami ly 5, subfam ly M, member 12 pseudog NG_004191 Homo sapiens Ifactory receptor, , fam ly 5, subfam ly G, member 5 pseudoge NG_004192 Homo sapiens Ifactory receptor, , fam ly 5, subfam ly G, member 4 pseudoge NG_004193 Homo sapiens Ifactory receptor, , fam ly 5, subfam ly F, member 2 pseudoge NG_004194 Homo sapiens Ifactory receptor, , fam ly 5, subfam ly D, member 17 pseudog NG_004195 Homo sapiens Ifactory receptor, , fam ly 5, subfam ly D, member 15 pseudog NG_004196 Homo sapiens Ifactory receptor, , fami ly 5, subfam ly BR, member 1 pseudoc NG_004197 Homo sapiens Ifactory receptor, , fam ly 5, subfam ly BQ, member 1 pseudoc NG_004198 Homo sapiens Ifactory receptor, , fam ly 5, subfam ly BP, member 1 pseudog NG_004199 Homo sapiens Ifactory receptor, , fam ly 5, subfam ly BN, member 1 pseudog NG_004200 Homo sapiens Ifactory receptor, , fami ly 5, subfam ly BL, member 1 pseudog NG_004201 Homo sapiens Ifactory receptor, , fam ly 5, subfam ly BE, member 1 pseudog NG_004202 Homo sapiens Ifactory receptor, , fam ly 5, subfami ly BC, member 1 pseudoc NG_004203 Homo sapiens Ifactory receptor, , fam ly 5, subfam ly BB, member 1 pseudog NG_004204 Homo sapiens Ifactory receptor, , fam ly 5, subfam ly BA, member 1 pseudog NG_004205 Homo sapiens Ifactory receptor, , fam ly 5, subfami ly B, member 19 pseudog NG_004206 Homo sapiens Ifactory receptor, , fam ly 5, subfami ly B, member 15 pseudog NG_004207 Homo sapiens Ifactory receptor, , fam ly 5, subfami ly AZ, member 1 pseudog NG_004208 Homo sapiens Ifactory receptor, , fam ly 5, subfami ly AQ, member 1 pseudoc NG_004209 Homo sapiens Ifactory receptor, , fam ly 5, subfam ly AP, member 1 pseudog NG_004210 Homo sapiens Ifactory receptor, , fam ly 5, subfami ly AN, member 2 pseudoc NG_004211 Homo sapiens Ifactory receptor, , fam ly 5, subfam ly AM, member 1 pseudoc NG_004212 Homo sapiens Ifactory receptor, , fam ly 5, subfami ly AK, member 3 pseudog NG_004213 Homo sapiens Ifactory receptor, , fam ily 5, subfami ly AK, member 1 pseudog NG_004214 Homo sapiens Ifactory receptor, , fam ly 56, subfam ly A, member 7 pseudog NG_004215 Homo sapiens Ifactory receptor, , fam ly 52, subfam ly Y, member 1 pseudog NG_004216 Homo sapiens Ifactory receptor, , fam ly 52, subfam ly V, member 1 pseudog NG_004217 Homo sapiens Ifactory receptor, , fam ly 52, subfam ly U, member 1 pseudog NG_004218 Homo sapiens Ifactory receptor, , fam ly 52, subfam ly T, member 1 pseudog' NG_004219 Homo sapiens Ifactory receptor, , fam ly 52, subfam ly Q, member 1 pseudog NG_004220 Homo sapiens Ifactory receptor, , fam ly 52, subfam ly P, member 1 pseudog NG_004221 Homo sapiens Ifactory receptor, , fam ly 52, subfam ly N, member 3 pseudog NG_004222 Homo sapiens Ifactory receptor, , fam ly 52, subfam ly H, member 2 pseudog NG_004223 Homo sapiens Ifactory receptor, , fam ly 52, subfami ly E, member 7 pseudog NG_004224 Homo sapiens Ifactory receptor, , fam ly 52, subfam ly B, member 5 pseudog NG_004225 Homo sapiens Ifactory receptor, , fam ly 52, subfam ly B, member 1 pseudog NG_004226 Homo sapiens Ifactory receptor , fam ly 51 , subfami ly K, member 1 pseudog NG_004227 Homo sapiens Ifactory receptor, , fam ly 51 , subfam ly E, member 1 pseudog NG_004228 Homo sapiens Ifactory receptor, , fam ly 51 , subfam ly C, member 1 pseudog NG_004229 Homo sapiens Ifactory receptor , fam ly 51 , subfami ly A, member 10 pseudo NG_004230 Homo sapiens Ifactory receptor, , fam ly 4, subfam ly R, member 3 pseudoge NG_004231 Homo sapiens Ifactory receptor, , fam ly 4, subfam ly R, member 2 pseudoge NG_004232 Homo sapiens Ifactory receptor , fam ly 4, subfam ly D, member 8 pseudoge NG_004233 Homo sapiens Ifactory receptor, , fam ly 4, subfam ly D, member 7 pseudoge NG_004234 Homo sapiens Ifactory receptor, , fam ly 4, subfami ly C, member 14 pseudog NG_004235 Homo sapiens Ifactory receptor, , fam ly 4, subfam ly A, member 8 pseudoge NG_004236 Homo sapiens Ifactory receptor, , fam ly 4, subfam ly A, member 7 pseudoge NG_004237 Homo sapiens Ifactory receptor, , fam ly 4, subfam ly A, member 3 pseudoge NG_004238 Homo sapiens Ifactory receptor, , fam ly 4, subfam ly A, member 21 pseudog NG_004239 Homo sapiens Ifactory receptor, , fam ly 4, subfam ly A, member 19 pseudog NG_004240 Homo sapiens Ifactory receptor, , fam ly 4, subfam ly A, member 18 pseudog NG_004241 Homo sapiens Ifactory receptor, , fam ly 4, subfami ly A, member 14 pseudog NG_004242 Homo sapiens Ifactory receptor, , fam ly 4, subfam ly A, member 13 pseudog NG_004243 Homo sapiens Ifactory receptor, , fam ly 4, subfami ly A, member 12 pseudog NG 004244 Homo sapiens Ifactory receptor, , fami ly 2, subfami ly AH, member 1 pseudog
NG_004245 Homo sapiens Ifactory receptor, , fam iy 10, subfamily Y, member 1 pseudog NG_004246 Homo sapiens Ifactory receptor, , fam ly 10, subfamily W, member 1 (OR10V NG_004247 Homo sapiens Ifactory receptor, , fam ly 10, subfamily V, member 3 pseudog NG_004248 Homo sapiens Ifactory receptor, , fam ly 10, subfamily V, member 2 pseudog NG_004249 Homo sapiens Ifactory receptor, , fam ly 10, subfamily Q, member 2 pseudog NG_004250 Homo sapiens Ifactory receptor, , fam ly 7, subfamily M, member 1 pseudoge NG_004251 Homo sapiens Ifactory receptor, , fam ly 7, subfamily E, member 110 pseudo NG_004252 Homo sapiens Ifactory receptor, , fam ly 6, subfamily L, member 2 pseudogei NG_004253 Homo sapiens Ifactory receptor, , fam ly 7, subfamily E, member 116 pseudo NG_004254 Homo sapiens Ifactory receptor, , fam ly 7, subfamily E, member 108 pseudo NG_004255 Homo sapiens Ifactory receptor, , fam ly 2, subfamily AM, member 1 pseudoe; NG_004256 Homo sapiens Ifactory receptor, , fam ly 7, subfamily A, member 125 pseudo NG_004257 Homo sapiens Ifactory receptor, , fam ly 9, subfamily P, member 1 pseudoge NG_004258 Homo sapiens Ifactory receptor, , fam y 9, subfamily N, member 1 pseudoge NG_004259 Homo sapiens Ifactory receptor, , fam ly 4, subfamily F, member 7 pseudoge NG_004260 Homo sapiens Ifactory receptor, , fam y 2, subfamily W, member 6 pseudoge NG_004261 Homo sapiens Ifactory receptor, , fam y 2, subfamily W, member 4 pseudoge NG_004262 Homo sapiens Ifactory receptor, , fam y 7, subfamily E, member 99 pseudog NG_004263 Homo sapiens Ifactory receptor, , fam y 5, subfamily M, member 14 pseudog NG_004264 Homo sapiens Ifactory receptor, , fam y 7, subfamily A, member 129 pseudo NG_004265 Homo sapiens Ifactory receptor, , fam y 7, subfamily A, member 122 pseudo NG_004266 Homo sapiens Ifactory receptor, , fam y 7, subfamily E, member 100 pseudo NG_004267 Homo sapiens Ifactory receptor, , fam iy 5, subfamily AC, member 1 pseudog NG_004268 Homo sapiens Ifactory receptor, , fam ly 4, subfamily G, member 6 pseudoge NG_004269 Homo sapiens Ifactory receptor, , fam ly 9, subfamily H, member 1 pseudoge NG_004270 Homo sapiens Ifactory receptor, , fam ly 6, subfamily R, member 1 pseudoge NG_004271 Homo sapiens Ifactory receptor, , fam ly 6, subfamily K, member 1 pseudoge NG_004272 Homo sapiens Ifactory receptor, , fam ly 2, subfamily T, member 7 (OR2T7) p NG_004273 Homo sapiens Ifactory receptor , fam ly 2, subfamily L, member 9 pseudogei NG_004274 Homo sapiens Ifactory receptor , fam ly 2, subfamily L, member 6 pseudogei NG_004275 Homo sapiens Ifactory receptor, , fam ly 2, subfamily L, member 5 (OR2L5) p NG_004276 Homo sapiens Ifactory receptor , fam ly 2, subfamily AQ, member 1 pseudog NG_004277 Homo sapiens Ifactory receptor , fam ly 10, subfamily AE, member 1 pseudo NG_004278 Homo sapiens Ifactory receptor , fam ly 10, subfamily AA, member 1 pseudo NG_004279 Homo sapiens Ifactory receptor , fam ly 2, subfamily W, member 2 pseudoge NG_004280 Homo sapiens Ifactory receptor , fam ly 2, subfamily B, member 7 pseudoge NG_004281 Homo sapiens Ifactory receptor, , fam ly 51 , subfamily N, member 1 pseudog NG_004282 Homo sapiens Ifactory receptor, , fam ly 52, subfamily P, member 2 pseudog NG_004283 Homo sapiens Ifactory receptor, , fam ly 2, subfamily AP, member 1 (OR2AP NG_004284 Homo sapiens Ifactory receptor, , fam ly 11 , subfamily M, member 1 pseudog NG_004285 Homo sapiens Ifactory receptor, , fam iy 9, subfamily K, member 1 pseudoge NG_004286 Homo sapiens Ifactory receptor, , fam y 5, subfamily AV, member 1 pseudog NG_004287 Homo sapiens Ifactory receptor, , fami ly 6, subfamily P, member 1 (OR6P1 ) | NG_004288 Homo sapiens Ifactory receptor, , fam y 6, subfamily K, member 4 pseudoge NG_004289 Homo sapiens Ifactory receptor, , fam y 2, subfamily Al, member 1 pseudoge NG_004290 Homo sapiens Ifactory receptor, , fam y 2, subfamily A, member 15 pseudog NG_004291 Homo sapiens Ifactory receptor, , fam y 7, subfamily E, member 136 pseudo NG_004292 Homo sapiens Ifactory receptor, , fam y 2, subfamily A, member 3 pseudoge NG_004293 Homo sapiens Ifactory receptor, , fam: iy 6, subfamily D, member 1 pseudoge NG_004294 Homo sapiens Ifactory receptor, , fam ly 7, subfamily E, member 39 pseudog NG_004295 Homo sapiens Ifactory receptor, , fam ly 4, subfamily C, member 50 pseudog NG_004296 Homo sapiens Ifactory receptor, , fam ly 4, subfamily D, member 12 pseudog NG_004297 Homo sapiens Ifactory receptor, , fam ly 7, subfamily E, member 149 pseudo NG_004298 Homo sapiens Ifactory receptor, , fam ly 10, subfamily AF, member 1 pseudo NG_004299 Homo sapiens Ifactory receptor, , fam ly 8, subfamily V, member 1 pseudoge NG_004300 Homo sapiens Ifactory receptor, , fam ly 5, subfamily BT, member 1 pseudog NG 004301 Homo sapiens Ifactory receptor, , fam ly 8, subfamily T, member 1 pseudoge
NG_004302 Homo sapiens Ifactory receptor, , fam ly 11 , subfamily P, member 1 pseudog NG_004303 Homo sapiens Ifactory receptor, , fami ly 7, subfamily E, member 148 pseudo NG_004304 Homo sapiens Ifactory receptor, , fami ly 52, subfamily Z, member 1 pseudog NG_004305 Homo sapiens Ifactory receptor, , fam ly 52, subfamily M, member 2 pseudog NG_004307 Homo sapiens Ifactory receptor, , fami ly 7, subfamily E, member 1 pseudoge NG_004308 Homo sapiens Ifactory receptor ',, fam ly 10, subfamily J, member 8 pseudogi NG_004309 Homo sapiens Ifactory receptor, , fami ly 7, subfamily E, member 140 pseudo NG_004310 Homo sapiens Ifactory receptor , fam: ly 2, subfamily Q, member 1 pseudoge NG_004311 Homo sapiens Ifactory receptor, , fam ly 52, subfamily B, member 3 pseudog NG_004312 Homo sapiens Ifactory receptor, , fami ly 8, subfamily G, member 3 pseudoge NG_004313 Homo sapiens Ifactory receptor , fami ly 7, subfamily A, member 19 pseudog NG_004314 Homo sapiens Ifactory receptor, , fam ly 7, subfamily A, member 130 pseudo NG_004315 Homo sapiens Ifactory receptor, , fam ly 7, subfamily E, member 161 pseudo NG_004317 Homo sapiens Ifactory receptor , fami ly 55, subfamily B, member 1 pseudog NG_004318 Homo sapiens Ifactory receptor, , fami ly 51 , subfamily R, member 1 pseudog NG_004319 Homo sapiens Ifactory receptor, , fam ly 52, subfamily K, member 3 pseudog NGJ304320 Homo sapiens Ifactory receptor , fami ly 51 , subfamily A, member 9 pseudog NG_004321 Homo sapiens Ifactory receptor, , fam ly 51 , subfamily F, member 5 pseudog NG_004322 Homo sapiens Ifactory receptor, , fam ly 51 , subfamily C, member 4 pseudog NG_004323 Homo sapiens Ifactory receptor, , fam ly 51 , subfamily F, member 3 pseudog NG_004324 Homo sapiens Ifactory receptor, , fam ly 51 , subfamily F, member 4 pseudog NG_004325 Homo sapiens Ifactory receptor, , fam ly 51 , subfamily A, member 6 pseudog NG_004326 Homo sapiens Ifactory receptor, , fam ly 56, subfamily B, member 2 pseudog NG_004327 Homo sapiens Ifactory receptor, , fam ly 4, subfamily A, member 40 pseudog FMG_004328 Homo sapiens Ifactory receptor, , fam ly 4, subfamily A, member 43 pseudog NG_004329 Homo sapiens Ifactory receptor, , fam ly 4, subfamily A, member 6 pseudoge NG_004330 Homo sapiens Ifactory receptor , fam ly 4, subfamily A, member 2 pseudoge NG_004331 Homo sapiens Ifactory receptor, , fam ly 4, subfamily A, member 4 pseudoge NG_004332 Homo sapiens Ifactory receptor, , fam ly 4, subfamily A, member 11 pseudog NG_004333 Homo sapiens Ifactory receptor, , fam ly 4, subfamily A, member 9 pseudoge NG_004334 Homo sapiens Ifactory receptor, , fam ly 4, subfamily A, member 10 pseudog NG_004335 Homo sapiens Ifactory receptor, , fam ly 4, subfamily A, member 17 pseudog NG_004336 Homo sapiens Ifactory receptor, , fam ly 7, subfamily E, member 145 pseudo NG_004337 Homo sapiens Ifactory receptor, , fam ly 2, subfamily AT, member 2 pseudog NG_004338 Homo sapiens Ifactory receptor, , fam ly 2, subfamily AT, member 1 pseudog NG_004339 Homo sapiens Ifactory receptor, , fam ly 10, subfamily N, member 1 pseudog NG_004340 Homo sapiens Ifactory receptor, , fam ly 8, subfamily F, member 1 pseudoge NG_004341 Homo sapiens Ifactory receptor, , fam ly 8, subfamily A, member 2 pseudoge NG_004342 Homo sapiens Ifactory receptor, , fam ly 8, subfamily B, member 10 pseudog NG_004343 Homo sapiens Ifactory receptor, , fam ly 5, subfamily BS, member 1 pseudog NG_004344 Homo sapiens Ifactory receptor, , fam ly 10, subfamily U, member 1 pseudog NG_004345 Homo sapiens Ifactory receptor, , fam ly 6, subfamily C, member 5 pseudoge NG_004346 Homo sapiens Ifactory receptor, , fam ly 6, subfamily C, member 7 pseudoge NG_004347 Homo sapiens Ifactory receptor, , fam ly 6, subfamily C, member 71 pseudog NG_004348 Homo sapiens Ifactory receptor, , fami ly 6, subfamily U, member 2 pseudoge NG_004349 Homo sapiens Ifactory receptor, , fam ly 4, subfamily Q, member 2 pseudoge NG_004350 Homo sapiens Ifactory receptor , fam ly 4, subfamily U, member 1 pseudoge NG_004351 Homo sapiens Ifactory receptor , fami ly 4, subfamily T, member 1 pseudoge NG_004352 Homo sapiens Ifactory receptor, , fam ly 11 , subfamily G, member 1 pseudog NG_004353 Homo sapiens Ifactory receptor, , fami ly 11 , subfamily H, member 5 pseudog NG_004354 Homo sapiens Ifactory receptor, , fam1 ly 11 , subfamily H, member 7 pseudog NG_004355 Homo sapiens Ifactory receptor, , fam ly 4, subfamily N, member 3 pseudoge NG_004356 Homo sapiens Ifactory receptor, , fami ly 4, subfamily F, member 14 pseudog NG_004357 Homo sapiens Ifactory receptor, , fam1 ly 4, subfamily F, member 13 pseudog NG_004358 Homo sapiens Ifactory receptor, , fam ly 4, subfamily F, member 28 pseudog NG_004359 Homo sapiens Ifactory receptor, , fam; ly 4, subfamily F, member 8 pseudoge NG 004360 Homo sapiens Ifactory receptor, , fami ly 7, subfamily E, member 18 pseudog
NG_004361 Homo sapiens olfactory receptor, fam ly 7, subfamily A, member 1 pseudoge NG_004362 Homo sapiens olfactory receptor, fam ly 10, subfamily R, member 3 pseudog NG_004363 Homo sapiens olfactory receptor, fam ly 6, subfamily K, member 5 pseudoge NG_004364 Homo sapiens olfactory receptor, fam ly 10, subfamily J, member 2 pseudogi NG_004365 Homo sapiens olfactory receptor, fam ly 10, subfamily J, member 7 pseudogi NG_004366 Homo sapiens olfactory receptor, fam ly 10, subfamily J, member 9 pseudogi NG_004367 Homo sapiens olfactory receptor, fam ly 10, subfamily J, member 4 pseudogi NG_004368 Homo sapiens olfactory receptor, fam ly 7, subfamily E, member 23 pseudog NG_004369 Homo sapiens olfactory receptor, fam ly 5, subfamily S, member 1 pseudoge NG_004370 Homo sapiens olfactory receptor, fam ly 7, subfamily E, member 55 pseudog NG_004371 Homo sapiens olfactory receptor, fam ly 7, subfamily E, member 35 pseudog NG_004372 Homo sapiens olfactory receptor, fam ly 9, subfamily A, member 3 pseudoge NG_004373 Homo sapiens olfactory receptor, fam ly 2, subfamily R, member 1 pseudoge NG_004374 Homo sapiens olfactory receptor, fam ly 10, subfamily AC, member 1 pseudc NG_004375 Homo sapiens olfactory receptor, fam ly 2, subfamily A, member 13 pseudog NG_004376 Homo sapiens olfactory receptor, fam ly 7, subfamily E, member 158 pseudo NG_004377 Homo sapiens olfactory receptor, fam ly 13, subfamily E, member 1 pseudog NG_004378 Homo sapiens olfactory receptor, fam ly 13, subfamily C, member 6 pseudog NG_004379 Homo sapiens olfactory receptor, fam ly 2, subfamily S, member 1 pseudoge NG_004380 Homo sapiens olfactory receptor, fam ly 13, subfamily D, member 2 pseudog NG_004381 Homo sapiens olfactory receptor, fam ly 11 , subfamily N, member 1 pseudog NG_004382 Homo sapiens olfactory receptor, fam ly 3, subfamily B, member 1 pseudoge NG_004383 Homo sapiens spondyloepiphyseal dyspl asia, late, pseudogene (LOC39259 NG_004384 Homo sapiens oi factory receptor fam ly 7, subfamily E, member 117 pseudo NG_004385 Homo sapiens ol factory receptor, fam ly 7, subfamily E, member 102 pseudo NG_004386 Homo sapiens ol Ifactory receptor, fam ly 7, subfamily E, member 109 pseudo NG_004387 Homo sapiens o Ifactory receptor, fami ly 51 , subfamily A, member 8 pseudog NG_004388 Homo sapiens o factory receptor fam ly 51 , subfamily H, member 1 pseudog NG_004389 Homo sapiens o factory receptor fami ly 51 , subfamily H, member 2 pseudog NG_004390 Homo sapiens Ol factory receptor, fam ly 56, subfamily B, member 3 pseudog NG_004391 Homo sapiens o Ifactory receptor, fam ly 5, subfamily BK, member 1 pseudog NG_004392 Homo sapiens o Ifactory receptor, fami ly 11 , subfamily K, member 2 pseudog NG_004393 Homo sapiens o Ifactory receptor, fam ly 10, subfamily J, member 6 pseudogi NG_004394 Homo sapiens 0 factory receptor, fam ly 7, subfamily E, member 46 pseudog NG_004395 Homo sapiens o Ifactory receptor, fam ly 7, subfamily A, member 121 pseudo NG_004396 Homo sapiens YTH d* omain family 11 pseu dogene (YTHDF1 P) on chromosorr NG_004397 Homo sapiens o Ifactory receptor, fami ly 10, subfamily AH, member 1 pseudc NG_004398 Homo sapiens o Ifactory receptor fam ly 7, subfamily E, member 59 pseudog NG_004399 Homo sapiens 0 factory receptor, fam, ly 7, subfamily E, member 160 pseudo NG_004400 Homo sapiens o Ifactory receptor, fami ly 13, subfamily D, member 3 pseudog NG_004401 Homo sapiens 0 Ifactory receptor fam ly 10, subfamily AE, member 3 pseudo NG_004402 Homo sapiens o factory receptor fam ly 10, subfamily AK, member 1 pseudo NG_004403 Homo sapiens 0 factory receptor, fam ly 11 , subfamily J, member 5 pseudogi NG_004404 Homo sapiens o Ifactory receptor fam ly 11 , subfamily Q, member 1 pseudog NG_004405 Homo sapiens 0 factory receptor, fam ly 13, subfamily Z, member 1 pseudog NG_004406 Homo sapiens 0 factory receptor, fam ly 13, subfamily Z, member 2 pseudog NG_004407 Homo sapiens ol factory receptor, fam Iy 1 , subfamily M, member 4 pseudoge NG_004408 Homo sapiens o factory receptor fam ly 2, subfamily A, member 41 pseudog NG_004409 Homo sapiens 0 factory receptor, fam ly 2, subfamily AO, member 1 pseudog NG_004410 Homo sapiens 0 Ifactory receptor, fam ly 2, subfamily BH, member 1 pseudog NG_004411 Homo sapiens ol Ifactory receptor, fam ly 2, subfamily T, member 32 pseudog' NG_004412 Homo sapiens ol ifactory receptor, fam ly 2, subfamily X, member 1 pseudoge NG_004413 Homo sapiens ol factory receptor, fam ly 4, subfamily A, member 41 pseudog NG_004414 Homo sapiens ol Ifactory receptor, fam ly 4, subfamily A, member 42 pseudog NG_004415 Homo sapiens o! Ifactory receptor, fam ly 4, subfamily A, member 44 pseudog NG_004416 Homo sapiens o Ifactory receptor, fam ly 4, subfamily A, member 45 pseudog NG 004417 Homo sapiens ol factory receptor, fam ly 4, subfamily A, member 46 pseudog
NG_004418 Homo sapiens olfactory receptor, family 4, subfamily A, member 48 pseudog
NG_0O4419 Homo sapiens olfactory receptor, family 4, subfamily A, member 49 pseudog
NG_004420 Homo sapiens olfactory receptor, family 4, subfamily A, member 50 pseudog
NG_0O4421 Homo sapiens olfactory receptor, family 4, subfamily C, member 48 pseudog
NG_0O4422 Homo sapiens olfactory receptor, family 4, subfamily C, member 49 pseudog
NG_0O4423 Homo sapiens olfactory receptor, family 4, subfamily G, member 11 pseudog
NG_0O4424 Homo sapiens olfactory receptor, family 51 , subfamily AB, member 1 pseudc
NG_0O4425 Homo sapiens olfactory receptor, family 51 , subfamily B, member 8 pseudog
NG_0O4426 Homo sapiens olfactory receptor, family 5, subfamily AC, member 4 pseudoc
NG_0O4427 Homo sapiens olfactory receptor, family 5, subfamily AO, member 1 pseudog
NG_0O4428 Homo sapiens olfactory receptor, family 5, subfamily J, member 7 pseudogei
NG_0O4429 Homo sapiens olfactory receptor, family 6, subfamily C, member 64 pseudog
NG_0O4430 Homo sapiens olfactory receptor, family 6, subfamily C, member 66 pseudog
NG_0O4431 Homo sapiens olfactory receptor, family 6, subfamily C, member 69 pseudog
NG_0O4432 Homo sapiens olfactory receptor, family 6, subfamily C, member 72 pseudog
NG_0O4433 Homo sapiens olfactory receptor, family 6, subfamily C, member 73 pseudog
NG_0O4434 Homo sapiens olfactory receptor, family 6, subfamily R, member 2 pseudoge
NG_0O4435 Homo sapiens olfactory receptor, family 7, subfamily E, member 155 pseudo
NG_0O4436 Homo sapiens olfactory receptor, family 7, subfamily E, member 159 pseudo
NG_0O4437 Homo sapiens olfactory receptor, family 7, subfamily G, member 15 pseudog
NG_0O4438 Homo sapiens olfactory receptor, family 7, subfamily H, member 2 pseudoge
NG_0O4439 Homo sapiens olfactory receptor, family 8, subfamily A, member 3 pseudoge
NG_0O4440 Homo sapiens olfactory receptor, family 8, subfamily X, member 1 pseudoge
NG_0O4441 Homo sapiens olfactory receptor, family 4, subfamily C, member 17 pseudog
NG_0O4443 Homo sapiens spondyloepiphyseal dysplasia, late, pseudogene (LOC41475'
NG_0O4444 Homo sapiens spondyloepiphyseal dysplasia, late, pseudogene (LOC41475
NG_0O4445 Homo sapiens spondyloepiphyseal dysplasia, late, pseudogene (LOC41475U
NG_0O4446 Homo sapiens spondyloepiphyseal dysplasia, late, pseudogene 4 (SEDLP4)
NG_0O4625 Homo sapiens olfactory receptor, family 2, subfamily U, member 1 pseudoge
NG_0O4626 Homo sapiens olfactory receptor, family 13, subfamily I, member 1 pseudoge
NG_0O4627 Homo sapiens olfactory receptor, family 1 , subfamily AB, member 1 pseudog
NG_0O4628 Homo sapiens olfactory receptor, family 8, subfamily G, member 7 pseudoge
NG_0O4629 Homo sapiens olfactory receptor, family 7, subfamily E, member 96 pseudog
NG_0O4630 Homo sapiens olfactory receptor, family 1 , subfamily X, member 1 pseudoge
NG_0O4631 Homo sapiens olfactory receptor, family 1 , subfamily X, member 5 pseudoge
NG_0O4632 Homo sapiens olfactory receptor, family 13, subfamily K, member 1 pseudog
NG_0O4633 Homo sapiens olfactory receptor, family 7, subfamily E, member 157 pseudo
NG_0O4634 Homo sapiens olfactory receptor, family 7, subfamily E, member 154 pseudo
NG_0O4635 Homo sapiens olfactory receptor, family 8, subfamily I, member 4 pseudoger
NG_0O4638 Homo sapiens glutathione S-transferase pi pseudogene (GSTPP) on chrome
NG_0O4639 Homo sapiens actin, gamma pseudogene (LOC414754) on chromosome Y
NG_0O4652 Homo sapiens olfactory receptor, family 2, subfamily AJ, member 1 (OR2AJ1
NG_0O4656 Homo sapiens ATP synthase, H+ transporting, mitochondrial F1 complex, ga
NG_0O4658 Homo sapiens MHC class III complement gene cluster, bimodular haplotype
NG_0O4662 Homo sapiens PTPN13-like, Y-linked pseudogene (LOC442865) on chromos
NG_0O4663 Homo sapiens PTPN13-like, Y-linked pseudogene (LOC442866) on chromos
NG_0O4666 Homo sapiens olfactory receptor, family 7, subfamily E, member 31 pseudog
NG_0O4670 Homo sapiens serine/threonine kinase 22A (spermiogenesis associated) (ST
NT_004321 Homo sapiens chromosome 1 genomic contig
NT_004350 Homo sapiens chromosome 1 genomic contig
NT_0O4433 Homo sapiens chromosome 1 genomic contig
NT_004434 Homo sapiens chromosome 1 genomic contig
NT_004487 Homo sapiens chromosome 1 genomic contig
NT_004511 Homo sapiens chromosome 1 genomic contig
NT_004538 Homo sapiens chromosome 1 genomic contig
NT_004547 Homo sapiens chromosome 1 genomic contig
NT_004559 Homo sapiens chromosome 1 genomic contig
NT .004610 Homo sapiens chromosome 1 genomic cont NT_004668 Homo sapiens chromosome 1 genomic cont NT_004671 Homo sapiens chromosome 1 genomic conti NT_004686 Homo sapiens chromosome 1 genomic cont NT_004754 Homo sapiens chromosome 1 genomic cont NT_004836 Homo sapiens chromosome 1 genomic cont NT_004873 Homo sapiens chromosome 1 genomic cont NT_005058 Homo sapiens chromosome 2 genomic cont NT_005079 Homo sapiens chromosome 2 genomic conti NT_00512O Homo sapiens chromosome 2 genomic cont NT_005334 Homo sapiens chromosome 2 genomic cont NT_0054O3 Homo sapiens chromosome 2 genomic conti NT_00541 6 Homo sapiens chromosome 2 genomic cont NT_005535 Homo sapiens chromosome 3 genomic cont NT_005612 Homo sapiens chromosome 3 genomic cont NT .006051 Homo sapiens chromosome 4 genomic cont NT_006081 Homo sapiens chromosome 4 genomic cont NT_00621 6 Homo sapiens chromosome 4 genomic cont NT_006238 Homo sapiens chromosome 4 genomic cont NT .0063O7 Homo sapiens chromosome 4 genomic cont NT_00631 6 Homo sapiens chromosome 4 genomic cont NT_006431 Homo sapiens chromosome 5 genomic cont NT_006576 Homo sapiens chromosome 5 genomic cont NT_00671 3 Homo sapiens chromosome 5 genomic cont NT_007299 Homo sapiens chromosome 6 genomic cont NT_0073O2 Homo sapiens chromosome 6 genomic cont NT_007422 Homo sapiens chromosome 6 genomic cont NT .007583 Homo sapiens chromosome 6 genomic cont NT_007592 Homo sapiens chromosome 6 genomic cont NT_007741 Homo sapiens chromosome 7 genomic cont NT_007758 Homo sapiens chromosome 7 genomic cont NT_00781 9 Homo sapiens chromosome 7 genomic cont NT_007914 Homo sapiens chromosome 7 genomic cont NT_007933 Homo sapiens chromosome 7 genomic cont NT_007995 Homo sapiens chromosome 8 genomic cont NT .008046 Homo sapiens chromosome 8 genomic cont NT_008127 Homo sapiens chromosome 8 genomic cont NT_008183 Homo sapiens chromosome 8 genomic cont NT_008251 Homo sapiens chromosome 8 genomic cont NT_00841 3 Homo sapiens chromosome 9 genomic conti NT_00847O Homo sapiens chromosome 9 genomic cont NT_008476 Homo sapiens chromosome 9 genomic cont NT_008554 Homo sapiens chromosome 9 genomic conti NT_008583 Homo sapiens chromosome 10 genomic contig NT_008705 Homo sapiens chromosome 10 genomic contig NT_00881 8 Homo sapiens chromosome 10 genomic contig NT_008984 Homo sapiens chromosome 11 genomic contig NT_009237 Homo sapiens chromosome 11 genomic contig NT_009487 Homo sapiens chromosome 12 genomic contig NT_009714 Homo sapiens chromosome 12 genomic contig NT_009755 Homo sapiens chromosome 12 genomic contig NT_009759 Homo sapiens chromosome 12 genomic contig NT_009775 Homo sapiens chromosome 12 genomic contig NT_009952 Homo sapiens chromosome 13 genomic contig NT_010194 Homo sapiens chromosome 15 genomic contig NT_010274 Homo sapiens chromosome 15 genomic contig NT 010280 Homo sapiens chromosome 15 genomic contig
NT_010393 Homo sapiens chromosome 16 genomic contig NT_010498 Homo sapiens chromosome 16 genomic contig NT_010505 Homo sapiens chromosome 16 genomic contig NT_010542 Homo sapiens chromosome 16 genomic contig NT_010552 Homo sapiens chromosome 16 genomic contig NT_010641 Homo sapiens chromosome 17 genomic contig NT_010663 Homo sapiens chromosome 17 genomic contig NT_010718 Homo sapiens chromosome 17 genomic contig NT_010755 Homo sapiens chromosome 17 genomic contig NT_010783 Homo sapiens chromosome 17 genomic contig NT_010799 Homo sapiens chromosome 17 genomic contig NT_010859 Homo sapiens chromosome 18 genomic contig NT_010879 Homo sapiens chromosome 18 genomic contig NT_010966 Homo sapiens chromosome 18 genomic contig NT_011109 Homo sapiens chromosome 19 genomic contig NT_011255 Homo sapiens chromosome 19 genomic contig NT_011295 Homo sapiens chromosome 19 genomic contig NT_011333 Homo sapiens chromosome 20 genomic contig NT_011362 Homo sapiens chromosome 20 genomic contig NT_011387 Homo sapiens chromosome 20 genomic contig NT_011512 Homo sapiens chromosome 21 genomic contig NT_011515 Homo sapiens chromosome 21 genomic contig NT_011516 Homo sapiens chromosome 22 genomic contig NT_011519 Homo sapiens chromosome 22 genomic contig NT_011520 Homo sapiens chromosome 22 genomic contig NT_011521 Homo sapiens chromosome 22 genomic contig NT_011522 Homo sapiens chromosome 22 genomic contig NT_011523 Homo sapiens chromosome 22 genomic contig NT_011525 Homo sapiens chromosome 22 genomic contig NT_011526 Homo sapiens chromosome 22 genomic contig NT_011565 Homo sapiens chromosome X genomic contig NT_011568 Homo sapiens chromosome X genomic contig NT_011630 Homo sapiens chromosome X genomic contig NT_011638 Homo sapiens chromosome X genomic contig NT_011651 Homo sapiens chromosome X genomic contig NT_011669 Homo sapiens chromosome X genomic contig NT_011681 Homo sapiens chromosome X genomic contig NT_011726 Homo sapiens chromosome X genomic config NT_011757 Homo sapiens chromosome X genomic contig NT_011786 Homo sapiens chromosome X genomic contig NT_011875 Homo sapiens chromosome Y genomic contig NT_011878 Homo sapiens chromosome Y genomic contig NT_011896 Homo sapiens chromosome Y genomic contig NT_011903 Homo sapiens chromosome Y genomic contig NT_015926 Homo sapiens chromosome 2 genomic contig NT_016297 Homo sapiens chromosome 4 genomic contig NT_016354 Homo sapiens chromosome 4 genomic contig NT_016606 Homo sapiens chromosome 4 genomic contig NT_017696 Homo sapiens chromosome 10 genomic contig NT_017795 Homo sapiens chromosome 10 genomic contig NT_019197 Homo sapiens chromosome 22 genomic contig NT_019273 Homo sapiens chromosome 1 genomic contig NT_019501 Homo sapiens chromosome 9 genomic contig NT_019546 Homo sapiens chromosome 12 genomic config NT_019609 Homo sapiens chromosome 16 genomic contig NT_019686 Homo sapiens chromosome X genomic contig NT 021877 Homo sapiens chromosome 1 genomic contig
NT_021937 Homo sapiens chromosome 1 genomic contig NT_021973 Homo sapiens chromosome 1 genomic contig NT .022052 Homo sapiens chromosome 1 genomic contig NT_022071 Homo sapiens chromosome 1 genomic contig NT_022135 Homo sapiens chromosome 2 genomic contig NT_022139 Homo sapiens chromosome 2 genomic contig NT_022171 Homo sapiens chromosome 2 genomic contig NT_022173 Homo sapiens chromosome 2 genomic contig NT_022184 Homo sapiens chromosome 2 genomic contig NT_022221 Homo sapiens chromosome 2 genomic contig NT_022270 Homo sapiens chromosome 2 genomic contig NT_022327 Homo sapiens chromosome 2 genomic contig NT_022459 Homo sapiens chromosome 3 genomic contig NT_022517 Homo sapiens chromosome 3 genomic contig NT_022778 Homo sapiens chromosome 4 genomic contig NT_022792 Homo sapiens chromosome 4 genomic contig NT_022794 Homo sapiens chromosome 4 genomic contig NT .022853 Homo sapiens chromosome 4 genomic contig NT_023089 Homo sapiens chromosome 5 genomic contig NT_023133 Homo sapiens chromosome 5 genomic contig NT_023148 Homo sapiens chromosome 5 genomic contig NT_023603 Homo sapiens chromosome 7 genomic contig NT_023629 Homo sapiens chromosome 7 genomic contig NT_023666 Homo sapiens chromosome 8 genomic contig NT_023678 Homo sapiens chromosome 8 genomic contig NT_023684 Homo sapiens chromosome 8 genomic contig NT_023736 Homo sapiens chromosome 8 genomic contig NT_023929 Homo sapiens chromosome 9 genomic contig NT_023932 Homo sapiens chromosome 9 genomic contig NT_023935 Homo sapiens chromosome 9 genomic contig NT_024000 Homo sapiens chromosome 9 genomic contig NT_024040 Homo sapiens chromosome 10 genomic contig NT_024133 Homo sapiens chromosome 10 genomic contig NT_024477 Homo sapiens chromosome 12 genomic contig NT_024498 Homo sapiens chromosome 13 genomic contig NT_024524 Homo sapiens chromosome 13 genomic contig NT_024773 Homo sapiens chromosome 16 genomic contig NT .024797 Homo sapiens chromosome 16 genomic contig NT_024812 Homo sapiens chromosome 16 genomic contig NT_024862 Homo sapiens chromosome 17 genomic contig NT_024871 Homo sapiens chromosome 17 genomic contig NT_024972 Homo sapiens chromosome 17 genomic contig NT_025004 Homo sapiens chromosome 18 genomic contig NT_025028 Homo sapiens chromosome 18 genomic contig NT_025215 Homo sapiens chromosome 20 genomic contig NT_025302 Ho o sapiens chromosome X genomic contig NT_025307 Homo sapiens chromosome X genomic contig NT_025319 Homo sapiens chromosome X genomic contig NT_025441 Homo sapiens chromosome Y genomic contig NT_025741 Ho o sapiens chromosome 6 genomic contig NT_025835 Homo sapiens chromosome 10 genomic contig NT_025965 Homo sapiens chromosome X genomic contig NT .025975 Homo sapiens chromosome Y genomic contig NT_026437 Homo sapiens chromosome 14 genomic contig NT_026446 Homo sapiens chromosome 15 genomic contig NT_026943 Homo sapiens chromosome 1 genomic contig NT 026970 Homo sapiens chromosome 2 genomic contig
NT_027140 Homo sapiens chromosome 13 genomic contig NT_028050 Homo sapiens chromosome 1 genomic contig NT_028251 Homo sapiens chromosome 8 genomic contig NT_028392 Homo sapiens chromosome 20 genomic contig NT_028395 Homo sapiens chromosome 22 genomic contig NT_028405 Homo sapiens chromosome X genomic contig NT_028413 Homo sapiens chromosome X genomic contig NT_029289 Homo sapiens chromosome 5 genomic contig NT_029419 Homo sapiens chromosome 12 genomic contig NT_029490 Homo sapiens chromosome 21 genomic contig NT_029860 Homo sapiens chromosome 1 genomic contig NT_029928 Homo sapiens chromosome 3 genomic contig NT_029998 Homo sapiens chromosome 7 genomic contig NT_030008 Homo sapiens chromosome 7 genomic contig NT_030058 Homo sapiens chromosome 9 genomic contig NT_030059 Homo sapiens chromosome 10 genomic contig NT_030187 Homo sapiens chromosome 21 genomic contig NT_030188 Homo sapiens chromosome 21 genomic contig NT_030584 Homo sapiens chromosome 1 genomic contig NT_030737 Homo sapiens chromosome 8 genomic contig NT_030772 Homo sapiens chromosome 10 genomic contig NT_030843 Homo sapiens chromosome 17 genomic contig NT_030871 Homo sapiens chromosome 20 genomic contig NT_030872 Homo sapiens chromosome 22 genomic contig NT_031730 Homo sapiens chromosome 1 genomic contig NT_031847 Homo sapiens chromosome 10 genomic contig NT_032962 Homo sapiens chromosome 1 genomic contig NT_032968 Homo sapiens chromosome 1 genomic contig NT_032977 Homo sapiens chromosome 1 genomic contig NT_032994 Homo sapiens chromosome 2 genomic contig NT_033000 Homo sapiens chromosome 2 genomic contig NT_033172 Homo sapiens chromosome 6 genomic contig NT_033224 Homo sapiens chromosome 10 genomic contig NT_033330 Homo sapiens chromosome X genomic contig NT_033899 Homo sapiens chromosome 11 genomic contig NT_033903 Homo sapiens chromosome 11 genomic contig NT_033927 Homo sapiens chromosome 11 genomic contig NT .033948 Homo sapiens chromosome 6 genomic contig NT_033968 Homo sapiens chromosome 7 genomic contig NT_033985 Homo sapiens chromosome 10 genomic contig NT_034398 Homo sapiens chromosome 1 genomic contig NT_034400 Homo sapiens chromosome 1 genomic contig NT_034401 Homo sapiens chromosome 1 genomic contig NT_034403 Homo sapiens chromosome 1 genomic contig NT_034410 Homo sapiens chromosome 1 genomic contig NT_034471 Homo sapiens chromosome 1 genomic contig NT_034485 Homo sapiens chromosome 2 genomic contig NT_034508 Homo sapiens chromosome 2 genomic contig NT_034772 Homo sapiens chromosome 5 genomic contig NT_034819 Homo sapiens chromosome 5 genomic contig NT_034880 Homo sapiens chromosome 6 genomic contig NT_034885 Homo sapiens chromosome 7 genomic contig NTJ335014 Homo sapiens chromosome 9 genomic contig NT_035036 Homo sapiens chromosome 10 genomic contig NT_035040 Homo sapiens chromosome 10 genomic config NT_035086 Homo sapiens chromosome 11 genomic contig NT 035113 Homo sapiens chromosome 11 genomic contig
NT_035158 Homo sapiens chromosome 11 genomic contig NT_035243 Homo sapiens chromosome 12 genomic contig NT_035325 Homo sapiens chromosome 15 genomic contig NT_035363 Homo sapiens chromosome 16 genomic contig NT_035414 Homo sapiens chromosome 17 genomic contig NT_035608 Homo sapiens chromosome 20 genomic contig NT_037485 Homo sapiens chromosome 1 genomic contig NT_037540 Homo sapiens chromosome 2 genomic contig NT_037622 Homo sapiens chromosome 4 genomic contig NT_037623 Homo sapiens chromosome 4 genomic contig NT_037645 Homo sapiens chromosome 4 genomic contig NT_037704 Homo sapiens chromosome 8 genomic contig NT .037750 Homo sapiens chromosome 10 genomic contig NT_037753 Homo sapiens chromosome 10 genomic contig NT_037852 Homo sapiens chromosome 15 genomic contig NT_037887 Homo sapiens chromosome 16 genomic contig NT_077382 Homo sapiens chromosome 1 genomic contig NT_077383 Homo sapiens chromosome 1 genomic contig NT_077384 Homo sapiens chromosome 1 genomic contig NT_077387 Homo sapiens chromosome 1 genomic contig NT_077388 Homo sapiens chromosome 1 genomic contig NT_077389 Homo sapiens chromosome 1 genomic contig NT_077390 Homo sapiens chromosome 1 genomic contig NT_077402 Homo sapiens chromosome 1 genomic contig NT_077407 Homo sapiens chromosome 2 genomic contig NT_077444 Homo sapiens chromosome 4 genomic contig NT_077451 Homo sapiens chromosome 5 genomic contig NT_077528 Homo sapiens chromosome 7 genomic contig NT_077531 Homo sapiens chromosome 8 genomic contig NT_077567 Homo sapiens chromosome 10 genomic contig NT_077569 Homo sapiens chromosome 10 genomic contig NT_077570 Homo sapiens chromosome 10 genomic contig NT_077571 Homo sapiens chromosome 10 genomic contig NT_077627 Homo sapiens chromosome 13 genomic contig NT_077631 Homo sapiens chromosome 15 genomic contig NT_077661 Homo sapiens chromosome 15 genomic config NT_077677 Homo sapiens chromosome 16 genomic contig NT_077812 Homo sapiens chromosome 19 genomic contig NT_077819 Homo sapiens chromosome X genomic contig NT_077911 Homo sapiens chromosome 1 genomic contig NT_077912 Homo sapiens chromosome 1 genomic contig NT_077913 Homo sapiens chromosome 1 genomic config NT_077914 Homo sapiens chromosome 1 genomic contig NT_077915 Homo sapiens chromosome 1 genomic contig NT_077919 Homo sapiens chromosome 1 genomic contig NT_077920 Homo sapiens chromosome 1 genomic contig NT_077921 Homo sapiens chromosome 1 genomic contig NT_077922 Homo sapiens chromosome 1 genomic contig NT_077930 Homo sapiens chromosome 1 genomic contig NT_077931 Homo sapiens chromosome 1 genomic contig NT_077932 Homo sapiens chromosome 1 genomic contig NT_077933 Homo sapiens chromosome 1 genomic contig NT .077936 Homo sapiens chromosome 1 genomic contig NT_077939 Homo sapiens chromosome 1 genomic contig NT_077941 Homo sapiens chromosome 1 genomic contig NT_077944 Homo sapiens chromosome 1 genomic contig NT 077945 Homo sapiens chromosome 1 genomic contig
NT .077946 Homo sapiens chromosome 1 genon NT .077949 Homo sapiens chromosome 1 geno NT .077962 Homo sapiens chromosome 1 geno NT_077964 Homo sapiens chromosome 1 geno NT.077966 Homo sapiens chromosome 1 geno NT_077967 Homo sapiens chromosome 1 geno NT_077970 Homo sapiens chromosome 1 geno NT .077988 Homo sapiens chromosome 1 geno NT_077995 Homo sapiens chromosome 2 geno NT_077999 Homo sapiens chromosome 2 geno NT_078000 Homo sapiens chromosome 2 geno NT .078003 Homo sapiens chromosome 2 geno NT_078010 Homo sapiens chromosome 4 geno NT .078012 Homo sapiens chromosome 4 geno NT_078016 Homo sapiens chromosome 4 geno NT_078018 Homo sapiens chromosome 5 geno NT.078019 Homo sapiens chromosome 5 geno NT_078020 Homo sapiens chromosome 6 geno NT_078023 Homo sapiens chromosome 6 geno NT_078033 Homo sapiens chromosome 7 geno NT_078037 Homo sapiens chromosome 8 geno NT_078038 Homo sapiens chromosome 8 geno NT .078040 Homo sapiens chromosome 9 geno NT_078041 Homo sapiens chromosome 9 geno NT_078042 Homo sapiens chromosome 9 geno NT_078043 Homo sapiens chromosome 9 geno NT_078044 Homo sapiens chromosome 9 geno NT_078045 Homo sapiens chromosome 9 geno NT_078046 Homo sapiens chromosome 9 geno NT .078047 Homo sapiens chromosome 9 geno NT_078048 Homo sapiens chromosome 9 geno NT_078049 Homo sapiens chromosome 9 geno NT.078051 Homo sapiens chromosome 9 geno NT_078052 Homo sapiens chromosome 9 geno NT_078053 Homo sapiens chromosome 9 geno NT_078055 Homo sapiens chromosome 9 geno NT_078057 Homo sapiens chromosome 9 geno NT .078058 Homo sapiens chromosome 9 geno NT_078059 Homo sapiens chromosome 9 geno NT_078061 Homo sapiens chromosome 9 geno NT_078062 Homo sapiens chromosome 9 geno NT_078063 Homo sapiens chromosome 9 geno NT_078064 Homo sapiens chromosome 9 geno NT_078065 Homo sapiens chromosome 9 geno NT_078066 Homo sapiens chromosome 9 geno NT_078067 Homo sapiens chromosome 9 geno NT_078068 Homo sapiens chromosome 9 geno NT .078069 Homo sapiens chromosome 9 geno NT_078070 Homo sapiens chromosome 9 geno NT_078071 Homo sapiens chromosome 9 geno NT_078072 Homo sapiens chromosome 9 geno NT_078074 Homo sapiens chromosome 9 geno NT.078075 Homo sapiens chromosome 9 geno NT_078076 Homo sapiens chromosome 9 geno NT_078077 Homo sapiens chromosome 9 geno NT_078078 Homo sapiens chromosome 9 geno NT 078079 Homo sapiens chromosome 9 geno

NT_078081 Homo sapiens chromosome 9 genomic contig NT_078083 Homo sapiens chromosome 9 genomic contig NT_078087 Homo sapiens chromosome 10 genomic contig NT_078088 Homo sapiens chromosome 11 genomic contig NT_078092 Homo sapiens chromosome 13 genomic contig NT_078094 Homo sapiens chromosome 15 genomic contig NT_078095 Homo sapiens chromosome 15 genomic contig NT_078096 Homo sapiens chromosome 15 genomic contig NT_078099 Homo sapiens chromosome 16 genomic config NT_078100 Homo sapiens chromosome 17 genomic contig NT_078102 Homo sapiens chromosome 17 genomic contig NT_078103 Homo sapiens chromosome 19 genomic contig NT_078109 Homo sapiens chromosome X genomic contig NT_078110 Homo sapiens chromosome X genomic contig NT_078114 Homo sapiens chromosome X genomic contig NT_078115 Homo sapiens chromosome X genomic contig NT_078116 Homo sapiens chromosome X genomic contig NT_078118 Homo sapiens chromosome X genomic contig NT_079482 Homo sapiens chromosome 1 genomic contig NT_079483 Homo sapiens chromosome 1 genomic contig NT_079484 Homo sapiens chromosome 1 genomic contig NT_079485 Homo sapiens chromosome 1 genomic contig NT_079486 Homo sapiens chromosome 1 genomic contig NT_079487 Homo sapiens chromosome 1 genomic contig NT_079488 Homo sapiens chromosome 1 genomic contig NT_079489 Homo sapiens chromosome 1 genomic contig NT_079490 Homo sapiens chromosome 1 genomic contig NT_079491 Homo sapiens chromosome 1 genomic contig NT_079492 Homo sapiens chromosome 1 genomic contig NT_079493 Homo sapiens chromosome 1 genomic contig NT_079494 Homo sapiens chromosome 1 genomic contig NT_079495 Homo sapiens chromosome 1 genomic contig NT_079496 Homo sapiens chromosome 1 genomic contig NT_079497 Homo sapiens chromosome 1 genomic contig NT_079498 Homo sapiens chromosome 1 genomic contig NT_079499 Homo sapiens chromosome 1 genomic contig NT_079500 Homo sapiens chromosome 1 genomic contig NT_079501 Homo sapiens chromosome 1 genomic contig NT_079502 Homo sapiens chromosome 2 genomic contig NT_079503 Homo sapiens chromosome 2 genomic contig NT_079504 Homo sapiens chromosome 2 genomic contig NT_079505 Homo sapiens chromosome 2 genomic contig NT_079506 Homo sapiens chromosome 3 genomic contig NT_079507 Homo sapiens chromosome 3 genomic contig NT_079508 Homo sapiens chromosome 3 genomic contig NT_079509 Homo sapiens chromosome 3 genomic contig NT_079510 Homo sapiens chromosome 4 genomic contig NT_079511 Homo sapiens chromosome 4 genomic contig NT_079512 Homo sapiens chromosome 4 genomic contig NT_079513 Homo sapiens chromosome 5 genomic contig NT_079514 Homo sapiens chromosome 7 genomic contig NT_079515 Homo sapiens chromosome 7 genomic contig NT_079516 Homo sapiens chromosome 7 genomic contig NT_079517 Homo sapiens chromosome 8 genomic contig NT_079518 Homo sapiens chromosome 8 genomic contig NT_079519 Homo sapiens chromosome 8 genomic contig NT 079520 Homo sapiens chromosome 8 genomic contig
NT_079521 Homo sapiens chromosome 8 genomic contig NT_079522 Homo sapiens chromosome 8 genomic contig NT .079523 Homo sapiens chromosome 8 genomic contig NT_079524 Homo sapiens chromosome 8 genomic contig NT_079525 Homo sapiens chromosome 8 genomic contig NT_079526 Homo sapiens chromosome 8 genomic contig NT_079527 Homo sapiens chromosome 8 genomic contig NT_079528 Homo sapiens chromosome 9 genomic contig NT_079529 Homo sapiens chromosome 9 genomic contig NT_079530 Homo sapiens chromosome 9 genomic contig NT_079531 Homo sapiens chromosome 9 genomic contig NT_079532 Homo sapiens chromosome 9 genomic contig NT_079533 Homo sapiens chromosome 9 genomic contig NT_079534 Homo sapiens chromosome 9 genomic contig NT_079535 Homo sapiens chromosome 9 genomic contig NT_079536 Homo sapiens chro osome 9 genomic contig NT_079537 Homo sapiens chromosome 9 genomic contig NT_079538 Homo sapiens chromosome 9 genomic contig NT_079539 Homo sapiens chromosome 9 genomic contig NT_079540 Homo sapiens chro osome 10 genomic contig NT_079541 Homo sapiens chro osome 10 genomic contig NT_079542 Homo sapiens chromosome 10 genomic contig NT_079543 Homo sapiens chromosome 10 genomic contig NT_079544 Homo sapiens chro osome 10 genomic contig NT_079545 Homo sapiens chromosome 10 genomic contig NT_079546 Homo sapiens chromosome 15 genomic contig NT_079547 Homo sapiens chro osome 15 genomic contig NT_079548 Homo sapiens chro osome 15 genomic contig NT_079549 Homo sapiens chro osome 15 genomic contig NT_079550 Homo sapiens chro osome 15 genomic contig NT_079551 Homo sapiens chro osome 15 genomic contig NT_079552 Homo sapiens chromosome 15 genomic contig NT_079553 Homo sapiens chro osome 15 genomic contig NT_079554 Homo sapiens chro osome 15 genomic contig NT_079555 Homo sapiens chro osome 17 genomic contig NT_079556 Homo sapiens chromosome 17 genomic contig NT_079557 Homo sapiens chro osome 17 genomic contig NT_079558 Homo sapiens chromosome 17 genomic contig NT_079559 Homo sapiens chromosome 17 genomic contig NT_079560 Homo sapiens chromosome 17 genomic contig NT_079561 Homo sapiens chromosome 17 genomic contig NT_079562 Homo sapiens chromosome 17 genomic contig NT_079563 Homo sapiens chromosome 17 genomic contig NT_079564 Homo sapiens chromosome 17 genomic contig NT_079565 Homo sapiens chromosome 17 genomic contig NT_079566 Homo sapiens chromosome 17 genomic contig NT_079567 Homo sapiens chromosome 17 genomic contig NT_079568 Homo sapiens chromosome 17 genomic contig NT_079569 Homo sapiens chromosome 17 genomic contig NT_079570 Homo sapiens chromosome 17 genomic contig NT_079571 Homo sapiens chromosome 17 genomic contig NT_079572 Homo sapiens chromosome 18 genomic contig NT_079573 Homo sapiens chromosome X genomic contig NT_079574 Homo sapiens chromosome X genomic contig NT_079575 Homo sapiens chromosome X genomic contig NT_079576 Homo sapiens chromosome X genomic contig NT 079577 Homo sapiens chromosome X genomic contig
NT_079578 Homo sapiens chromosome X genomic contig
NT_079579 Homo sapiens chromosome X genomic contig
NT_079580 Homo sapiens chromosome X genomic contig
NT_079581 Homo sapiens chromosome Y genomic contig, pseudoautosomal region
NT_079582 Homo sapiens chromosome Y genomic contig, pseudoautosomal region
NT_079583 Homo sapiens chromosome Y genomic contig, pseudoautosomal region
NT_079584 Homo sapiens chromosome Y genomic contig, pseudoautosomal region
NT_079585 Homo sapiens chromosome Y genomic contig, pseudoautosomal region
NT_079586 Homo sapiens genomic contig
NT_079587 Homo sapiens genomic contig
NT_079588 Homo sapiens genomic contig
NT_079589 Homo sapiens genomic contig
NT_079590 Homo sapiens chromosome 7 genomic contig, alternate assembly
NT_079591 Homo sapiens chromosome 7 genomic contig, alternate assembly
NT_079592 Homo sapiens chromosome 7 genomic contig, alternate assembly
NT_079593 Homo sapiens chromosome 7 genomic contig, alternate assembly
NT_079594 Homo sapiens chromosome 7 genomic contig, alternate assembly
NT_079595 Homo sapiens chromosome 7 genomic contig, alternate assembly
NT_079596 Homo sapiens chromosome 7 genomic contig, alternate assembly
NT_079597 Homo sapiens chromosome 7 genomic contig, alternate assembly
NT_079614 Homo sapiens genomic contig
NT_079615 Homo sapiens genomic config
NT_079616 Homo sapiens genomic contig
NT_079617 Homo sapiens genomic contig
NT_079618 Homo sapiens genomic contig
NT_079619 Homo sapiens genomic contig
NT_079620 Homo sapiens genomic contig
NT_079621 Homo sapiens genomic contig
NT_079622 Homo sapiens genomic config
NT_079623 Homo sapiens genomic contig
NT_079624 Homo sapiens genomic contig
NT_079625 Homo sapiens genomic contig
NT_086323 Homo sapiens chromosome 13 sequence, ENCODE region ENr111
NT_086324 Homo sapiens chromosome 2 sequence, ENCODE region EN 12
NT_086325 Homo sapiens chromosome 4 sequence, ENCODE region ENM 13
NT_086326 Homo sapiens chromosome 10 sequence, ENCODE region ENM 14
NT_086327 Homo sapiens chromosome 5 sequence, ENCODE region ENM 15, ALTERN
NT_086328 Homo sapiens chromosome 2 sequence, ENCODE region ENM21
NT_086329 Homo sapiens chromosome 18 sequence, ENCODE region ENM 22
NT_086330 Homo sapiens chromosome 12 sequence, ENCODE region ENM 23
NT_086331 Homo sapiens chromosome 2 sequence, ENCODE region ENM 24, ALTERN
NT_086332 Homo sapiens chromosome 2 sequence, ENCODE region ENM 31
NT_086333 Homo sapiens chromosome 13 sequence, ENCODE region ENM 32
NT_086334 Homo sapiens chromosome 21 sequence, ENCODE region ENM 33
NT_086335 Homo sapiens chromosome 4 sequence, ENCODE region ENM 34, ALTERN
NT_086336 Homo sapiens chromosome 16 sequence, ENCODE region ENr211
NT_086337 Homo sapiens chromosome 5 sequence, ENCODE region ENr212
NT_086338 Homo sapiens chromosome 18 sequence, ENCODE region ENr213
NT_086339 Homo sapiens chromosome 4 sequence, ENCODE region ENr214, ALTERN
NT_086340 Homo sapiens chromosome 5 sequence, ENCODE region ENr221
NT_086341 Homo sapiens chromosome 6 sequence, ENCODE region ENr222
NT_086342 Homo sapiens chromosome 6 sequence, ENCODE region ENr223
NT_086343 Homo sapiens chromosome 4 sequence, ENCODE region ENr224, ALTERN
NT_086344 Homo sapiens chromosome 1 sequence, ENCODE region ENr231
NT_086345 Homo sapiens chromosome 9 sequence, ENCODE region ENr232
NT_086346 Homo sapiens chromosome 15 sequence, ENCODE region ENr233
NTJ386347 Homo sapiens chromosome 17 sequence, ENCODE region ENr234, ALTERI
NT .086348 Homo sapiens chromosome 14 sequence, ENCODE region ENr311 NT_086349 Homo sapiens chromosome 11 sequence, ENCODE region ENr312 NT_086350 Homo sapiens chromosome 16 sequence, ENCODE region ENr313 NT_086351 Homo sapiens chromosome X sequence, ENCODE region ENr314, ALTERN NT_086352 Homo sapiens chromosome 8 sequence, ENCODE region ENr321 NT_086353 Homo sapiens chromosome 14 sequence, ENCODE region ENr322 NT_086354 Homo sapiens chromosome 6 sequence, ENCODE region ENr323 NT .086355 Homo sapiens chromosome X sequence, ENCODE region ENr324 NT_086356 Homo sapiens chromosome 2 sequence, ENCODE region ENr331 NT_086357 Homo sapiens chromosome 7 sequence, ENCODE region ENm001 NT_086358 Homo sapiens chromosome 5 sequence, ENCODE region ENm002 NT_086359 Homo sapiens chromosome 11 sequence, ENCODE region ENm003 NT .086360 Homo sapiens chromosome 22 sequence, ENCODE region ENm004 NT_086361 Homo sapiens chromosome 21 sequence, ENCODE region ENm005 NT_086362 Homo sapiens chromosome X sequence, ENCODE region ENmOOδ NT_086363 Homo sapiens chromosome 19 sequence, ENCODE region ENm007 NT_086364 Homo sapiens chromosome 16 sequence, ENCODE region ENm008 NT_086365 Homo sapiens chromosome 11 sequence, ENCODE region ENm009 NT_086366 Homo sapiens chromosome 7 sequence, ENCODE region ENm010 NT_086367 Homo sapiens chromosome 11 sequence, ENCODE region ENm011 NT_086368 Homo sapiens chromosome 7 sequence, ENCODE region ENm012 NT_086369 Homo sapiens chromosome 7 sequence, ENCODE region ENm013 NT .086370 Homo sapiens chromosome 7 sequence, ENCODE region ENm014 NT_086371 Homo sapiens chromosome 7 sequence, ENCODE region ENm015, ALTERI NT_086372 Homo sapiens chromosome 7 sequence, ENCODE region ENm016, ALTERI NT_086373 Homo sapiens chromosome 7 sequence, ENCODE region ENm017, ALTERI NT_086374 Homo sapiens chromosome 11 sequence, ENCODE region ENr332 NT_086375 Homo sapiens chromosome 20 sequence, ENCODE region ENr333 NT_086376 Homo sapiens chromosome 6 sequence, ENCODE region ENr334 NT 086377 Homo sapiens chromosome 9 sequence, ENCODE region ENr335, ALTERN
|28629867|ref :||NM 000536.11[28629867]
|33859834 |ref| |NM 000601.3|[33859834]
129788995 |ref| |NM 000940.11[29788995]
|24307876 |ref| |NM "000941.1 |[24307876]
|38505191 |ref| |NM 000953.2|[38505191]
|47717124 |ref| |NM 001001132.1 |[47717124]
J48928033 |ref :||NM 001001188.2 |[48928033]
|47933388 |ref| |NM. 001001290.1 | [47933388]
|48255914 |ref :||NM 001001317.1 |[48255914]48255946I |ref| |NM .001001323.1 |[48255946]
|47679100 |ref| |NM 001001324.1 |[47679100]
|47679098 |ref| |NM 001001325.1 |[47679098]
|48255890 |ref| |NM 001001329.1 |[48255890]
|47679088j Jref| |NM 001001330.1 |[47679088]
J48255950 |ref| |NM 001001331.1 | [48255950]
|47778926 |ref| |NM 001001336.1 |[47778926]
|47717128 |ref| |NM 001001342.1 |[47717128]
|47716515 |ref :||NM 001001343.1 |[47716515]
|48255954 |ref| |NM 001001344.1 | [48255954]
J47716518 |ref| |NM 001001346.1 |[47716518]
|47777757 |ref| |NM .001001349.1 |[47777757]
|48255936 |ref| |NM .001001389.1 | [48255936]
|48255938 |ref| |NM .001001390.1 |[48255938]
|48255940| |ref| |NM .001001391.1 |[48255940]
J48255942 |ref| |NM 001001392.1 |[48255942]
|47777311 |ref| |NM 001001394.1 |[47777311]
|47778934 |ref| |NM .001001395.1 I [47778934]
J48255958 jref :||NM 001001396.1 |[48255958]
|48762675 |ref| |NM 001001410.2 j [48762675]
J47777334 |refj |NM 001001411.1 |[47777334]
[47777332 lref||NM .001001412.1 |[47777332]
[47825360 |ref ||NM 001001414.1 |[47825360]
|47777339 |ref| |NM 001001415.1 | [47777339]
|48949871 |ref| |NM .001001417.2 |[48949871]
J48949891 Jref r]|NM .001001418.2 |[48949891]
J47778928 |ref| |NM .001001419.1 |[47778928]
J47778930 |ref ||NM .001001420.1 |[47778930]
|48255878 |ref| |NM .001001430.1 |[48255878]
|48255880 |ref :||NM 001001431.1 |[48255880]
|48255882 |ref| |NM .001001432.1 |[48255882]
|47778942 |ref| |NM .001001433.1 |[47778942]
|47778944| |ref| |NM 001001434.1 H47778944]
J48949791 |ref| |NM .001001435.2 |[48949791]
J47824888 |ref| |NM .001001436.1 |[47824888]
|48949908 |ref| |NM .001001437.2 |[48949908]
|47933396 |ref| |NM .001001438.1 |[47933396]
|47827221 |ref| |NM .001001479.1 |[47827221]
|47933380 |ref| |NM .001001481.1 | [47933380]
J47933382 |ref| |NM .001001482.1 |[47933382]
J47933347 jref| |NM .001001483.1 |[47933347]
|47933340 |ref| |NM .001001484.1 | [47933340]
|48762686 |refj |NM 001001485.1 |[48762686]
J48762688 |ref :||NM 001001486.1 |[48762688]
J48762690 |ref| |NM .001001487.1 j [48762690]
|48255902 |ref| |NM 001001502.1 |[48255902]
J48255925 |ref| |NM 001001503.1 |[48255925]
J48255930 |ref ||NM 001001520.1 | [48255930]
gi|48255967[ref|NM_001001521.1 [[48255967] gi|48255906|ref|NM_001001522.1|[48255906] gi|48255917|ref[NM_001001523.1 [[48255917] gi|48375170|ref|NM_001001524.1 |[48375170] gi|483751 5|ref|NM_001001547.11[48375175] gi|48375179|ref|NM_001001548.1|[48375179] gi[48762692iref|NM_001001549.1 |[48762692] gi|48762694|ref|NM_001001550.11[48762694] gi|48427668[ref|NM_001001551.1 [[48427668] gi[48675833Jref[NM_001001552.2|[48675833] gi|48475059|ref|NM_001001553.1|[48475059] gi|48762696|ref|NM_001001555.1l[48762696] gi|48527956|ref|NM_001001556.1 |[48527956] gi|48475G6ljref|NM_001001557.1|[48475061] gi|48527951[ref|NM_001001560.1 [[48527951] gi|48527953|ref|NM_001001561.1|[48527953] gi|48526508|ref[NM_001001563.1 |[48526508] gi|48762717jref|NM )01001567.lj[48762717] gi|48762719|ref|NM_001001568.1[[48762719] gi|48762721 |ref|NM_00 001569.1 j[48762721] gi!48762723|ref|NM_001001570.1|[48762723] gi)48762725|ref|NM_001001571.11[48762725] gi|48762727[ref|NM_001001572.1 [[48762727] gi|48762729|ref|NM_001001573.1|[48762729] gi|48762731|ref|NM_001001574.1|[48762731] gi|48762733|ref|NM_001001575.1|[48762733] gi|48762735|ref|NM_001001576.1 [[48762735] gi|48762737[ref|NM_001001577.1|[48762737] gi|48762739|ref|NM_001001578.1|[48762739] gij48762741|ref|NM_001001579.1|[48762741] gi|48762743|ref|NM_001001580.1 |[48762743] gi[48762745|ref|NM_001001581.1 ([48762745] gi|48762747|ref|NM_001001582.1|[48762747] gi|48762749iref|NM_001001583.1|[48762749] gi|48762751|ref|NM_001001584.1|[48762751] gi|48762753|ref|NM_001001585.1|[48762753] gi|4876268ljref|NM_001001586.1|[48762681] gi|48762701 |ref|NM_001001651.1 [[48762701] gi|48762703|ref|NM_001001653.1|[48762703] gi(48762705|ref|NM_001001654.1|[48762705] gi|48717225|ref|NM_001001655.1)[48717225] gi]48717230|ref|NM_001001656.1j[48717230] gi|48717233|ref|NM_001001658.1|[48717233] gi|48717235lref]NM_001001659.lj[48717235] gi|48717238|ref|NM_00100166O.1|[48717238] gi|48717240|ref|NM_001001661.1|[48717240] gi|48717243|ref|NM_001001662.1[[48717243] gi|48717248|ref|NM_001001663.1|[48717248] gi|48717262|ref|NM_001001664.1 j[48717262] gi|48717264[ref|NM„001001665.1|[48717264] gi|48717268|ref|NM_001001666.1|[48717268] gi|48717322|ref|NMJ)01001667.1 [[48717322] gi|50582993[ref|NM_001001668.2|[50582993] gi|48717281 |ref|NM_001001669.11[48717281] gi[48717284[ref|NM_00100167O.1([48717284] gi|48717279iref|NM_001001671.1|[48717279] gi|48717327|ref|NM_001001673.1|[48717327]
|48717336 |ref| |NM 001001675.1 |[48717336]
148717446 |ref| |NM 001001676.1 [[48717446]
[48717358 |ref| |NM 001001677.1 |[48717358]
148717360 |ref :||NM 001001678.1 j[48717360]
148717363 |ref||NM 001001679.1 [[48717363]
148717365 |ref| |NM .001001680.1 |[48717365]
J48717367 |ref ]|NM 001001681.1 |[48717367]
J48717369 |ref| |NM .001001682.1 |[48717369]
148717371 |ref| |NM .001001683.1 |[48717371]
148717373 |ref| |NM 001001684.1 |[48717373]
[48717375 |ref| |NM 001001685.1 |[48717375]
148717377 |ref| |NM .001001686.1 |[48717377]
J48717379 |ref :||NM .001001687.1 [[48717379]
J48717381 |ref| |NM 001001688.1 j[48717381]
148717383 |ref| |NM .001001689.1 |[48717383]
148717385 |ref| |NM .001001690.1 |[48717385]
148717387 |ref| |NM 001001691.1 |[48717387]
[48717389 |ref :||NM .001001692.1 |[48717389]
|48717391 |ref| |NM 001001693.1 |[48717391]
[48717393 |refj |NM .001001694.1 |[48717393]
148717395 |ref||NM .001001695.1 |[48717395]
|48717398 lref| |NM .001001696.1 |[48717398]
148717400 |ref| |NM 001001697.1 |[48717400]
J48717402 |ref| |NM .001001698.1 |[48717402]
148717404 |ref| |NM 001001699.1 |[48717404]
J48717406 |ref||NM 001001700.1 |[48717406]
|48717408 |ref ]|NM .001001701.1 |[48717408]
148717410 |ref| |NM .001001702.1 |[48717410]
J48717412| |ref| |NM 001001703.1 [[48717412]
[48717414| |ref| |NM .001001704.1 [[48717414]
[48717416 |ref| |NM .001001705.1 |[48717416]
[48717418 |ref| |NM 001001706.1 |[48717418]
148717420 |ref| |NM .001001707.1 |[48717420]
148717422 |ref| |NM 001001709.1 |[48717422]
148717425 |ref| |NM .001001710.1 |[48717425]
J48717484 |ref| |NM .001001711.1 |[48717484]
148717488 |ref| |NM 001001712.1 |[48717488]
J48762711 |ref| |NM .001001713.1 |[48762711]
|48928037 |ref I|NM .001001715.1 |[48928037]
J48928016 |ref| |NM 001001716.1 |[48928016]
|49355786 |ref| |NM .001001722.1 |[49355786]
|48928030 |ref| |NM .001001723.1 |[48928030]
|48926647 |ref| |NM .001001731.1 |[48926647]
J48926644 |ref| |NM 001001732.1 j[48926644]
J49249976 |ref| |NM .001001734.1 |[49249976]
150428921 |ref| |NM .001001740.2 |[50428921]
149169819 |ref| |NM 001001786.1 |[49169819]
|49574488 |ref| |NM .001001787.1 |[49574488]
J49169832 |ref |NM 001001789.1 |[49169832]
|49169827 |ref| |NM 001001790.1 |[49169827]
|49169836 |ref| |NM 001001791.1 |[49169836]
|49169830 |ref| |NM .001001794.1 |[49169830]
|49169840 |ref| |NM 001001795.1 j[49169840]
|49227243 |ref| |NM 001001802.1 |[49227243]
J49227740 jrefj |NM 001001821.1 |[49227740]
|49227790l |ref| |NM 001001824.1 |[49227790]
J49226829 |ref| |NM 001001827.1 |[49226829]
149258195 ref| |NM 001001850.1|[49258195]
I49355777 ref|[NM 001001851.1 |[49355777]
149258201 ref| [NM .001001852.1 |[49258201]
|49355753| refj |NM 001001870.1 [[49355753]
150312656 ref| |NM 001001871.1 |[50312656]
149402261 ref j|NM 001001872.1 |[49402261]
151036610 ref| |NM 001001873.2[[51036610]
J49574211 ref| |NM .001001874.1|[49574211]
J49574215 ref :j|NM 001001875.1 |[49574215]
150312658 ref| |NM .001001877.1 |[50312658]
J50312660 ref| |NM 001001878.1 [[50312660]
[49574525 ref| |NM .001001887.1|[49574525]
|49619240 ref :||NM .001001888.1 ([49619240]
J49574545 ref| |NM .001001890.1|[49574545]
|49533622 ref| |NM .001001891.1 |[49533622]
1496400101 ref| |NM .001001894.1 |[49640O10]
149640013 ref| [NM .001001895.1 |[49640013]
150054441, ref :||NM .001001912.1 |[50054441]
|50054462 ref :j|NM 001001913.1 |[50054462]
I50054448 ref| |NM .001001914.1 |[50054448]
|50054472 ref| |NM .001001915.1|[50054472]
[50054458| ref ]|NM .001001916.1 [[50054458]
J50054465 ref :||NM .001001917.1|[50054465]
J50054469, ref| |NM .001001918.1 |[50054469]
J500544761 ref| |NM .001001921.1|[50054476]
|50054480 ref| |NM .001001922.1 |[50054480]
150348610 ref| |NM .001001924 ,1|[50348610]
J50348616 ref||NM 001001925, ,1 |[50348616]
|50348621 ref| |NM .001001927. ,1|[50348621]
)50348665| ref| |NM .001001928 ,1 |[50348665]
J50348675 ref| |NM .001001929, ,1 |[50348675]
|50348680 ref| |NM 001001930, ,1 |[50348680]
J50348625 ref| |NM .001001931..1 |[50348625]
J50083286 refj [NM .001001933..1|[50083286]
[50345981 ref| |NM .001001935, ,1 |[50345981]
J50897849 ref||NM .001001936..1 |[50897849]
I50345983 ref| |NM .001001937..1 |[50345983]
150080155 ref :||NM .001001938, ,1|[5008O155]
|50086630 ref :j|NM .001001939. ,1 |[50086630]
J50080192 ref| |NM .001001953, .1|[5008O192]
J50080194 ref| |NM .001001954, ,1 |[5008O194]
150080196 refl |NM .001001956. ,1|[5008O196]
J50080198 ref| |NM .001001957, ,1 |[5008O198]
I50080200 ref| |NM .001001958. ,1 |[5008O200]
|50080202 ref :j|NM 001001959, .1|[5008O202]
J50233855 ref| |NM .001001960..1 |[50233855]
I50233845 ref| |NM .001001961 , .1 |[50233845]
J50233851 refl |NM .001001962..1|[50233851]
I50233863 refl |NM .001001963. ,1|[50233863]
|50233857 refj |NM 001001964, .1|[50233857]
J50233847 ref| |NM .001001965.1|[50233847]
I50233853 ref| |NM .001001966, .1 |[50233853]
J50233861 refl [NM .001001967.1 [[50233861]
|50233849 ref| |NM .001001968..1|[50233849]
|50897853 refl |NM .001001971.1|[50897853]
|50345987 refl |NM .001001973.1|[50345987]
J50234892 refl |NM 001001974..1 |[50234892]
150345990 |ref :||NM .001001975.1 |[50345990] 150345876 |ref| |NM .001001976.1 |[50345876] J50345992 |ref| |NM .001001977.1 |[50345992] J50659099 [refj |NM..001001991.1 |[50659099] [50312663 |ref||NM .001001992.1 |[50312663] |50263052 |ref||NM .001001994.1 |[50263052] J50263047 |ref| |NM .001001995.1 |[50263047] I50263049 |ref| |NM..001001996.1 |[50263049] J50301239 |ref| |NM .001001998.1 |[50301239] J50541951 jref| |NM .001002000.1 [[50541951] 150541953 |ref| |NM..001002001.1 |[50541953] J50541947 |ref| |NM .001002002.1 |[50541947] J50593109 |ref| [NM..001002006.1 | [50593109] J50593509 |ref| [NM .001002014.1 | [50593509] J50593511 |ref| |NM..001002015.1 [[50593511] I50952465 |ref| |NM .001002017.1 |[50952465] J50952467 |ref| |NM .001002018.1 |[50952467] |50346004 |ref| |NM..001002021.1 |[50346004] J50345278 |ref| |NM..001002026.1 |[50345278] I50659068 |ref| |NM 001002027.1 |[50659068] J50345295 |ref| |NM 001002029.1 |[50345295] J50593532 |refj |NM..001002031.1 j[50593532] J50345293 ]ref||NM 001002032.1 |[50345293] 150345291 |ref||NM..001002033.1 |[50345291] I50344743 [ref :||NM .001002035.1 |[50344743] J50345025 |ref [|NM 001002036.1 | [50345025] I50363236 |ref| |NM..001002231.1 |[50363236] J50363234 |ref| |NM..001002232.1 | [50363234] |50363229 |ref| |NM .001002233.1 |[50363229] J50363223 |ref :||NM..001002234.1 |[50363223] |50363220 |ref j|NM .001002235.1 |[50363220] J50363218 |ref| |NM .001002236.1 |[50363218] J50409855 |ref| |NM .001002243.1 |[50409855] J50409809 |ref j|NM. 001002244.1 |[50409809] J50409795 |ref| [NM .001002245.1 |[50409795] J50409803 |ref| |NM .001002246.1 |[50409803] J50409780 |ref| |NM 001002247.1 |[50409780] J50409788 |ref| |NM..001002248.1 |[50409788] J50409749 |refj |NM..001002249.1 |[50409749] I50409737 |ref||NM .001002251.1 |[50409737] |50409690 jref :||NM .001002252.1 |[50409690] J50400080 |ref| |NM..001002255.1 |[50400080] 150659071 |ref| |NM..001002256.1 |[50659071] I50659058 |ref| |NM 001002257.1 |[50659058] J50659073 [refj |NM 001002258.1 |[50659073] I50428932 jref| |NM .001002259.1 |[50428932] J50428926 |ref| |NM .001002260.1 |[50428926] I50557645 |ref| |NM .001002261.1 |[50557645] J50557647 |ref| |NM..001002262.1 |[50557647] 150428916 |ref| |NM 001002264.1 [[50428916] J50539411 |ref| |NM .001002265.1 |[50539411] J50539413 |ref| |NM..001002266.1 |[50539413] 150511938 |ref| |NM..001002269.1 |[50511938] 150511931 |ref| |NM 001002273.1 |[50511931] J50511925 |ref| |NM 001002274.1 |[50511925] J50511927 |ref| |NM 001002275.1 [[50511927] J50541962 jrefj |NM 001002292.1 |[50541962]
150541960 |ref| |NM 001002294.1 |[50541960]
150541958 |ref j|NM "001002295.1 |[50541958]
150541945 jrefj |NM 001002296.1 |[50541945]
J50593020 |ref| |NM "001002755.1 j[50593020]
J50593022 jref| |NM. "001002756.1 |[50593022]
J50593018 jref jjNM .001002757.1 |[50593018]
150959166 jref j|NM. JD01002758.1 j[50959166]
J50593525 jref| [NM..001002759.1 j[50593525]
|50959134| jref| |NM 001002760.1 j[50959134]
J50959142 |ref j|NM. 001002761.1 j[50959142]
|50593536 jref jjNM 001002762.1 j[50593536]
J50582995 Iref j|NM..001002796.1 |[50582995]
J50658066 |ref j|NM 001002799.1 j[50658066]
|50658062| | ref | jNM..001002800.1 j[50658062]
[50658076 jref :||NM .001002810.1 |[50658076]
|50658070 |ref j|NM..001002811.1 |[50658070]
|50658068 jref jjNM..001002812.1 |[50658068]
|50659103 |ref j|NM .001002814.1 [[50659103]
|50878274 |ref :||NM 001002836.1 j[50878274]
[50726959 jrefj |NM..001002837.1 |[50726959]
150845415 |ref jNM .001002838.1 |[50845415]
|50726988 |ref j|NM..001002840.1 j[50726988]
|50845427l | ref : ||NM .001002841.1 |[50845427]
150811872 |ref| INM. 001002843.1 [[50811872]
150811874 |ref| |NM 001002844.1 |[50811874]
J50811876 |ref j|NM. "001002845.1 j[50811876]
150811882 |ref ]|NM .001002847.1 [[50811882]
J50845400 |refj |NM "001002848.1 j[50845400]
|50845404 |ref :||NM. 001002849.1 j[50845404]
J50845385 [ref| |NM. 001002857.1 |[50845385]
I50845387, |ref| |NM 001002858.1 j[50845387]
|50843834 |ref j|NM 001002860.1 |[50843834]
J50959183 jref] |NM..001002861.1 |[50959183]
J50845410 |ref| JNM .001002862.1 |[50845410]
J50845413 jref : ||NM .001002876.1 |[50845413]
150959101 [ref| |NM 001002877.1 j[50959101]
150959109 |ref j|NM .001002878.1 |[50959109]
150959114 |ref jjNM .001002879.1 j[50959114]
150959131 [ref :||NM..001002880.1 |[50959131]
150881949 |ref J|NM .001002881.1 |[50881949]
150878301 jref j |NM 001002901.1 j[50878301]
I50897297 |ref jjNM..001002905.1 |[50897297]
|51093877] [ref||NM..001002906.1 |[51093877]
|50897269 |ref| |NM 001002907.1 j[50897269]
J50962881 |ref j|NM .001002909.1 |[50962881]
J50897271 |refj |NM .001002910.1 j[50897271]
J50897277 jref ]jNM 001002911.1 j[50897277]
J50897283 |ref j|NM..001002913.1 |[50897283]
151036593 jref j|NM .001002914.1 |[51036593]
I50897279 [refj |NM "001002915.1 j[50897279]
|50897287 [ref| |NM 001002916.1 |[50897287]
J50897281 |ref j|NM..001002917.1 |[50897281]
150897291 |ref| |NM 001002918.1 |[50897291]
|50897285 |ref :||NM 001002919.1 j[50897285]
J50897293 |ref| |NM 001002920.1 |[50897293]
[51092298 |ref j|NM 001002921.1 |[51092298]
|50897289 Jref jjNM 001002922.1 j[50897289]
gi|50897295| | ref | |NM_ 001002923.1 |[50897295] gi|50959221 |ref| |NM_ 001002924.1 |[50959221] gi|50979289 jref | |NM_ 001002925.1 |[50979289] gi|50962816 |ref| |NM_ .001002926.1 |[50962816] gi|51093829 [refj |NM_ 001003398.1 j[51093829] gi|50979287 |ref| |NM_ .001003399.1 |[50979287] gi|51093858 |ref| |NM_ 001003406.1 |[51093858] giJ51011128 |ref ]|NM_ 001003443.1 [[51011128] gi|51093852 |ref| |NM_ 001003656.1 |[51093852] gi|51092269 |ref| |NM_ 001003665.1 |[51092269] gi|51093707 |ref j|NM .001003674.1 |[51093707] gi|51093709 |ref| |NM_ 001003675.1 |[51093709] gi|51093378 jref jjNM 001003679.1 |[51093378] gij51093380 jref | |NM_ .001003680.1 |[51093380] gi|46488942 jref] [NM 001079.3 |[46488942] gi|27477067 jref | |NM_ .001132.1 |[27477067] gi|26667190 jref |NM .001222.2 |[26667190] gi|19115953 |ref j|NM 001369.1 |[19115953] gi|33350931 |ref j|NM 001376.2 j[33350931] gi|24307878 jref | |NM_ 001378.1 |[24307878] gi|46195706 [refj |NM_ 001410.1 |[46195706] gi|34222091 |ref| |NM 001547.3 |[34222091] gi|41352711 |ref| |NM 001556.1 j[41352711] gi|27764862| |ref| |NM_ 001636.1 j[27764862] gi|27764864 jref | |NM_ 001763.1 j[27764864] gi|26105977 |ref j|NM 001810.4 j[26105977] gi|31711991 [ref] |NM 001931.2 |[31711991] gi|38348231 |ref| |NM 001947.1 |[38348231] gi|33413399 |ref| |NM 001984.1 j[33413399] gi|24307882 jref | |NM_ .001986.1 |[24307882] gi|38327038 lref j|NM..002154.3 j[38327038] gi|34222089 |ref j|NM..002242.2 [[34222089] giJ28827775 |ref| |NM 002348.1 |[28827775] gi|27502374 jref | |NM.002399.2 |[27502374] gi|23111004 |ref| JNM 002404.1 |[23111004] gi|27764860 jref j |NM .002471.1 |[27764860] gi|23510390 [ref j|NM .002498.1 |[23510390] gi|28195383 ]ref j|NM..002523.1 j[28195383] giJ47078228 |ref| |NM 002596.2 j[47078228] giJ24429565 jref j |NM..002603.1 |[24429565] gi|24429563 |ref j|NM .002604.1 j[24429563] gi|47132535 [refj |NM 002605.2 |[47132535] giJ33300667 jref | |NM..002679.1 |[33300667] gi|38257138 |ref j|NM..002735.1 j[38257138] gi|38257140 lref j|NM .002746.1 j[38257140] gi|23110943 [refj |NM 002791.1 |[23110943] giJ23110924 |ref j|NM _002798.1 |[23110924] gi|37574611 Iref j|NM 002972.1 [[37574611] gi|28076868 jref | |NM _002974.1 |[28076868] gi[40548377j |ref| |NM..002998.2 |[40548377] gi|48475051 jref jjNM. _003013.1 j[48475051] gi|27777631 |ref| |NM _003047.2 |[27777631] giJ29826338 |ref j|NM 003106.2 |[29826338] gi|38373692 jref j |NM 003111.1 j[38373692] gi|27764866 |ref| |NM. _003179.1 |[27764866] gi|41350334| jref | |NM _003196.1 [[41350334] gi|27777635 jref jjNM. 003200.1 |[27777635]
gi|23308730|ref|NM_003302.1|[23308730] gi|23308728|ref[NM 003415.11[23308728] gi|38564321|ref|NM_003444.1|[38564321] giJ31083149Jref|NM_003502.2|[31083149] giJ27436923|ref|NM_003517.2|[27436923] gi|31657108|ref|NM_003575.1 |[31657108] gi|20070102|ref|NM_003598.1 [[20070102] gi|49170033|ref|NM_003638.lj[49170033] giJ41872688)ref|NM_003660.2|[41872688] gi|27501445|ref|NM_003677.2|[27501445] gi|50053005|ref|NM_003700.1|[50053005] gii26006850[ref[NM_003719.1|[26006850] gi|22208998 jref|NM_003724.11[22208998] gi|30089979|ref|NM_003741.2|[30089979] gii34335255|ref|NM_003817.1|[34335255] giJ22035625|ref|NM_003818.2|[22035625] gi|28872760|ref|NM_003828.1|[28872760] gi|28827773|ref|NM_003845.11[28827773] gi|29788999|ref|NM_003848.1j[29788999] gii38176157|ref|NM_003858.2J[38176157] gi|26190607|ref|NM_003898.1|[26190607] gi[33667022|ref|NM_003907.1|[33667022] gi|27501463|ref|NM_003957.1|[27501463] gi|4876294ljref|NM_003959.1|[48762941] gi|50345994|ref|NM_003972.2|[50345994] gi|220276311 ref | N M_004080.11 [22027631] gi|45598368Jref[NM_004097.1[[45598368] gi|34304365Jref|NM_004118.3[[34304365] giJ41352692|ref|NM_004136.1|[41352692] gi|38570145Jref|NM_004200.2|[38570145] gi|32698677|ref|NM_004220.1 |[32698677] gi|32698673|ref|NM_004241.11[32698673] gi|23238229|ref|NM_004242.2|[23238229] gi|46488922|ref|NM_004319.1 j[46488922] gi|32967316Jref|NM_004439.3|[32967316] gi|24307886|ref|NM_004498.1 |[24307886] gi|42415470|ref|NM_004650.1|[42415470] gi|37537693|refJNM_004685.2|[37537693] gi|34335257|ref|NM_004691.3|[34335257] gi|34222090|ref|NM_004764.2i[34222090] gi|22094078|ref|NM_004773.1 |[22094078] gi|37674229|ref|NM_004816.2|[37674229] gi|47078220|ref|NM_004840.2|[47078220] gi|50593006|ref|NM_004884.2|[50593006] gii31377467|ref|NM_004946.1|[31377467] gi|40254811 jref|NM_004947.2|[40254811] gi|19718754|ref|NM_005054.1|[19718754] gi|15812217|ref|NM_005105.2|[15812217] gi|40254809|ref|NM_005126.2|[40254809] gi|42718010|ref|NM_005140.1 j[42718010] gi|22547203|ref|NM_005144.2|[22547203] gi|24307888|ref|NM_005153.1|[24307888] gi|32964829|ref|NM_005202.11[32964829] gi|20270187|ref|NM_005240.1|[20270187] gi[29789001 jref|NM_005241.11[29789001] gi|22779859Jref|NM_005250.1|[22779859] gi|22027523|ref|NM_005272.2|[22027523]
gi|50263051 |ref|NM_005278.3|[50263051] gi|42560226[ref[NM }05349.2|[42560226] gi|34147322|ref|NM_005376.2|[34147322] giJ49087131 jref |NM_005407.1 j[49087131] giJ23199982lref|NM_005482.l j[23199982] gi|29788789|refJNM_005487.2|[29788789] gi|34147320|ref|NM_005533.2|[34147320] gi|38788415|ref|NM_005559.2|[38788415] gi|30840979|ref|NM_005595.1 |[30840979] giJ31652243|ref|NM_005650.l j[31652243] gi|33667050|ref|NM_005669.3|[33667050] gi|21536364|ref|NM_005680.1 |[21536364] gi|24307898Jref|NM 005702.11[24307898] gi|22027540|ref|NM )05707.1|[22027540] giJ32698675|ref|NM_005779.1 |[32698675] gi|44771197|ref|NM_005788.1 [[44771197] gi|31317304|ref|NM_005791.1 |[31317304] gi|27501451 |ref|NM_005840.1|[27501451] gi[40538725|ref|NM_005841.11[40538725] giJ39930312|ref|NM_005848.1 |[39930312] gi|33469918|ref|NM_005914.2|[33469918] gi|22027533|ref|NM_005942.1 |[22027533] gi|22027535Jref|NM_005943.2|[22027535] gi|28866959|ref|NM_005946.1|[28866959] gi|27414494|ref|NM_005947.1 |[27414494] gi|28866946|ref|NM_005949.1 |[28866946] gi|41406063[ref|NM_005964.1 |[41406063] gi|21389314JrefJNM_005984.1 |[21389314] gi|32567783|ref|NM_005995.2|[32567783] giJ49457785Jref|NM_006036.1 |[49457785] gi|48427666|ref|NM_006040.1 |[48427666] gi|50052980Jref|NM_006062.1 [[50052980] gi|24307902[ref|NM_006091.11[24307902] gi|24307904|ref|NM_006108.1 |[24307904] gii27262631 |ref|NM_006133.1 |[27262631] gi|40549417|ref|NM_006151.l i[40549417] gi|38257154Jref|NM_006154.1 |[38257154] gi|23510318|ref|NM_006172.1[[23510318] gi|46049091 |ref|NM_006175.3|[46049091] gi|33354284|ref|NM_006210.1 |[33354284] gi|34147323|ref|NM_006216.2|[34147323] gi|47834321 |ref|NM_006266.2J[47834321] gi|22325384|ref[NM_006277.1 |[22325384] gi|17388802Jref|NM_006452.2|[17388802] gi|37537515|ref|NM_006524.1 |[37537515] gi|38492355|ref|NM_006591.1 [[38492355] gi|38176299|ref|NM_006617.1 |[38176299] gii24307912[refjNM_006630.1 |[24307912] gi|37622348|ref|NM_006631.2|[37622348] giJ46361971 |ref|NM_006635.2|[46361971] gi|28269671 |ref|NM_006642.1 ([28269671] gi|41393186|ref|NM_006647.1 |[41393186] gi|4707822ljref|NM_006673.2|[47078221] gi|34147321 |ref|NM_006714.2|[34147321] gi|38156700|ref|NM_006722.lj[38156700] gi|27901802|ref|NM_006742.1 |[27901802] gi|45827705|ref|NM_006775.l j[45827705]
gi|24307916|ref|NM_006828.1 |[24307916] gi|29789005|refJNM_006832.1 [[29789005] gi|24307918|ref|NM_006857.1|[24307918] gi|37577165|refJNM_006859.2J[37577165] gi|24497546|ref|NM_006897.l j[24497546] gi|38505169|refJNM_006909.1 |[38505169] gi|24307922Jref|NM_006916.1 |[24307922] gi|29893558|ref|NM_006920.2|[29893558] gi|39930603Jref|NM_006939.lj[39930603] gii24307874|refJNM_006955.1 |[24307874] gi|50512287|ref|NM_006956.1 |[50512287] gi|42734301|ref|NM_006959.1 |[42734301] gi|38045955Jref|NM_006961.2|[38045955] gi|46195704|ref|NM_006969.1 |[46195704] gi|24307924Jref|NM_006973.l j[24307924] gi|28274681 jref|NM_006974.1 j[28274681] giJ27734718|ref|NM_006996.1 |[27734718] gi|42734307Jref|NM_007001.1 j[42734307] gi|38569504|ref|NM_007010.2|[38569504] gi|50345874|ref|NM_007041.2|[50345874] gi|45592958|ref|NM_007078.1 |[45592958] gi|23510456[ref|NM_007130.1 |[23510456] gi|41152069|ref|NM_007131.2|[41152069] giJ24307936|ref|NM_007135.1|[24307936] giJ46559734Jref|NM_007137.l j[46559734] giJ37537683[ref|NM_007139.2i[37537683] giJ24307934|ref|NM_007149.1 |[24307934] gi|42476268|ref|NM_007156.2|[42476268] giJ48675875|ref|NM_007157.2|[48675875] gi|24307932|ref|NM_007162.11[24307932] giJ32698687|ref|NM_007174.1 |[32698687] gi|27881505|ref|NM_007189.l j[27881505] gi[39930314iref|NM_007224.1 |[39930314] gi|27754211 jref[NM_007225.1 j[27754211] gi|25282390|ref|NM_007243.1 |[25282390] gil3366707l jref|NM_007261.lj[33667071] giJ33469984|ref|NM_007270.2|[33469984] gi|38148698|ref[NM_007277.3|[38148698] gi[24307928|refJNM_007280.lj[24307928] gi|40804749|ref|NM_007349.2|[40804749] gi|46276865|ref|NM_007356.1 |[46276865] gi|34147324|ref|NM_012073.2|[34147324] gi|29171733|ref|NM_012154.2|[29171733] gi|30061488|refJNM_012156.2|[30061488] gi|30089921|refJNM_012167.1|[30089921] giJ30795120|ref|NM_012174.1|[30795120] gi|45580705|ref|NM_012184.3|[45580705] gi[34222094|refJNM_012212.2[[34222094] gi|41872672|ref|NM_012224.1|[41872672] giJ42734429|refJNM_012232.2J[42734429] gi|24307942|ref|NM_012235.1|[24307942] gi|30410776|ref|NM_012271.1 |[30410776] gi|29789007Jref|NM_012272.1 |[29789007] gi|38569492|ref|NM_012284.1|[38569492] giJ47834347|ref[NM_012292.2|[47834347] gi|27477040|ref|NM_012305.11[27477040] gi|19743793|ref|NM_012309.1 |[19743793]
gi|29366811 |ref|NM_012315.11[29366811] gi|27544940|ref|NM_012335.2|[27544940] gi)46048142Jref|NM_012363.1 |[46048142] gi]50052933|ref|NM_012364.1 |[50052933] gi|45504385|ref|NM_012367.1 |[45504385] gi|50897263|ref|NM 012374.11[50897263] gi|28570169|ref|NM_012378.1 |[28570169] gi|31657128|refjNM_012393.1 |[31657128] giJ31317308|ref[NM_012398.1 |[31317308] gi|45331212Jref|NM_012416.1 |[45331212] gi|24430130|ref|NM_012477.2|[24430130] gi[24430131|ref|NM_012478.2|[24430131] gi|24307962Jref|NM_013304.1 |[24307962] giJ23943857Jref|NM_013321.1 j[23943857] gi|32698691 |ref|NM_013373.1 |[32698691] gi|46488914|ref|NM_014010.3|[46488914] giJ40217846|ref[NM_014014.2|[40217846] gi|34147338|ref|NM_014089.2|[34147338] gi|32483376Jref|NM_014098.2|[32483376] gi|31657139|ref|NM_014215.11[31657139] gi|21265100Jref|NM_014220.1 |[21265100] gi|23943853Jref|NM_014224.1 |[23943853] gi|21265036|ref|NM_014243.1 |[21265036] giJ29789055Jref|NM_014261.1|[29789055] gi|24307946Jref|NM_014282.1 [[24307946] gi|24307948|ref|NM_014284.1 |[24307948] gi[24307950|ref|NM_014290.1|[24307950] gii24307952|ref|NM_014301.11[24307952] gi|22507408|ref|NM_014346.1 |[22507408] giJ24307954Jref|NM_014376.1 |[24307954] gi|7657336|ref|NM_014381.1 |[7657336] giJ24415382|ref[NM_014389.1 |[24415382] gi|50726958|ref|NM_014422.2|[50726958] gi|3079450ljref|NM_014435.1|[30794501] gi|23943855Jref|NM_014441.1 |[23943855] giJ33859667|ref|NM_014455.1 |[33859667] gi|46048172|ref|NM_014460.2|[46048172] gi|50593521 |ref|NM_014472.3|[50593521] gi|21702741 |ref|NM_014494.11[21 02741] gi|27477044Jref|NM_014507.1 |[27477044] giJ22907038Jref|NM_014508.2|[22907038] gi|50726962|ref[NM_014510.l j[50726962] gi|27436930|ref|NM_014562.2[[27436930] gi|32698685|ref|NM_014568.l j[32698685] gi|18959199|ref|NM_014572.1 |[18959199] giJ32698695Jref|NM_014573.11[32698695] gi|30794503|ref|NM_014594.1 |[30794503] gi|2394391 l jref|NM_014602.li[23943911] gi|32698689|ref|NM_014603.1 |[32698689] gi|34222095|ref[NM_014607.2|[34222095] giJ24307968Jref|NM_014608.l j[24307968] giJ24415403|ref|NM_014611.11[24415403] gi|24797105|ref|NM_014613.1 |[24797105] gi|40788002|ref[NM_014614.1|[40788002] giJ44955925Jref|NM_014615.l j[44955925] gi|34878694|refJNM_014647.1 j[34878694] gi|45237192|ref|NM_014655.1 |[45237192]
124307960 |ref J|NM 014657 |[24307960]
142734311 |ref JjNM 014667, |[42734311]
138788371 |ref j|NM 014691, j[38788371]
[31317300 |1ref| |NM 014697 J[31317300]
J45120116 |ref J|NM 014701, |[45120116]
|24307972| |ref| [NM .014756, |[24307972]
|40538727| |ref J|NM .014798, |[40538727]
J29789059 |ref J|NM 014802, |[29789059]
138201615 |ref J|NM .014836, j[38201615]
|45827807| [ref| |NM_.014839, [[45827807]
I24307966 |ref J|NM .014850, |[24307966]
J33469146 |ref J|NM .014854, |[33469146]
|28372520 [ref J|NM 014858, |[28372520]
142734318 |ref J|NM .014881, j[42734318]
|38490530 | ref J |NM .014884, |[38490530]
J19913345 jref J|NM 014919. |[19913345]
J42542404 jref J|NM .014955, J[42542404]
|50345869 jref J |NM 014957. j[50345869]
J44888817 [ref J|NM 014974. j[44888817]
[45120118 |ref J|NM 014975, |[45120118]
121902518 |ref J|NM 014982. ([21902518]
J41054863 |ref J|NM .014989, |[41054863]
131317271 |ref J|NM".014991, |[31317271]
121071076 | ref J |NM 014992. |[21071076]
J44917618 jref JjNM 014997. |[44917618]
J24307970 |ref J|NM 015000, j[24307970]
|39725632 [ref J]NM .015004. [[39725632]
J39930342 jref JjNM .015008. j[39930342]
[30794499 |ref J|NM .015013. |[30794499]
138016126 |ref J|NM .015014. [[38016126]
I45504379 jref J|NM 015015. [[45504379]
[42516566 |ref J|NM .015017. |[42516566]
J34577117 jref J |NM .015018. [[34577117]
[30061508 |ref J|NM .015022. |[30061508]
J39930344 |ref J|NM .015027. |[39930344]
J23097291 |ref J|NM .015029. |[23097291]
138524621 Jref :j|NM 015033. j[38524621]
J40317630 |ref J|NM 015035. |[40317630]
I42734322 |ref j|NM .015037. [[42734322]
125141321 |ref J|NM .015039. |[25141321]
150881947 |ref J|NM 015040. [[50881947]
J42734324 [ref JlNM" 015045. [[42734324]
I22095330 |ref J|NM .015047. [[22095330]
I24307982 jref J |NM .015050. |[24307982]
[24307992 |ref J|NM 015052. |[24307992]
J30911100 |ref J|NM" 015055. |[30911100]
J22035664 |ref J|NM 015059. [[22035664]
J24307986 |ref J|NM .015061, j[24307986]
121359817 |ref j|NM .015065. |[21359817]
J25777691 |ref J|NM" 015066. |[25777691]
[46359074, |ref J|NM 015069. |[46359074]
J49355783 |ref J|NM .015076. |[49355783]
131742504 [ref J|NM .015078. [[31742504]
[40018619 [ref JjNM" 015079. [[40018619]
J46309460; |ref J|NM 015085. [[46309460]
J40806197 [ref J|NM 015087. j[40806197]
J24307990 |ref J|NM 015089. |[24307990]
gi|44888819|ref|NM_015091.1 |[44888819] gi[31657120|ref|NM_015094.l [[31657120] gi|2982634θjref|NM_015099.2|[29826340] gi|46397389|ref|NM_015100.2|[46397389] gi|34304361 |ref[N _015102.2i[34304361] gi|28933450|ref|NM_015103.1 |[28933450] giJ24307994irefJNM_015106.l i[24307994] gi|32698699|ref[NM_015107.1j[32698699] gi|24850455JrefJNM_015110.1 [[24850455] giJ32698693|refJNM_015115.1 {[32698693] gi|33859669|ref|NM_015116.11[33859669] giJ30794493JrefJNM_015117.lj[30794493] gi|27436958|ref|NM_015120.2J[27436958] giJ29789053Jref[NM_015122.lj[29789053] gi|50980306|ref |NM_015134.2|[50980306] gi|34222098|ref|NM_015138.2|[34222098] gi|34222096Jref(NM_015141.2J[34222096] gi|24308008[ref|NM_015143.lj[24308008] gi)21735418|ref|NM_015144.1|[21735418] giJ41872576|ref|NM_015150.1 |[41872576] gi|45827691 |ref|NM_015151.2|[45827691] giJ38424072Jref|NM_015157.l [[38424072] gi|23510374|ref|NM_015158.1|[23510374] giJ24308012Jref|NM_015160.1 [[24308012] gi|24308006|ref|NM_015161.1 |[24308006] gi|23943859|ref|NM_015167.1 |[23943859] giJ29789063Jref[NM_015170.l j[29789063] giJ46049062Jref|NM_015171.1 |[46049062] giJ38016902|ref|NM_015172.2J[38016902] gi|5065806θjref|NM_015173.2[[50658060] gi|34222099|ref|NM_015184.2|[34222099] giJ39930348|ref|NM_015187.1 |[39930348] gii33667083Jref|NM_015190.3|[33667083] gi|38569459[ref)NM_015191.11[38569459] gi|31581523|ref|NM_015198.2[[31581523] gi|46195712|ref |NM_015199.11[46195712] gi|22094120|ref|NM_015200.1 |[22094120] giJ4035377θ[ref|NM_015201.3i[40353770] gi[45433544|refiNM_015203.2J[45433544] gi|47271353Jref|NM_015210.l [[47271353] gi|44889474|ref |NM_015213.2|[44889474] giJ24308034jref|NM_015219.l j[24308034] gi[39930350[ref|NM_015221.1 [[39930350] gii40254858JrefJNM_015229.2J[40254858] giJ44771172|ref JNM_015234.3J[44771172] giJ29789057|refJNM_015238.1 [[29789057] gi|35493724|ref|NM_015243.2|[35493724] giJ38683796JrefJNM_015245.l j[38683796] giJ44917607|ref|NM_015246.l j[44917607] giJ21735416JrefJNM_015250.l j[21735416] gi|44771179|ref|NM_015252.2J[44771179] gi|27597060|ref JNM_015255.1 j[27597060] gi[46255054Jref|NM_015259.3|[46255054] gi|45356150]ref|NM_015261.lj[45356150] gijl9745147JrefJNM_015263.1 J[19745147] giJ38016201 |ref|NM_015265.1 |[38016201] gi|41872702Jref|NM_015266.l j[41872702]
gi|42734330|ref|NM_015268.11[42734330] gi|50659092|ref[NM_015274.l [[50659092] gi|40018628|ref|NM_015275.lj[40018628] giJ45935384|refJNM_015278.3|[45935384] gi|24308026[ref|NM_015281.11[24308026] giJ31563536[ref|NM_015282.1 |[31563536] gι'J38568177Jref|NM_015284.1 [[38568177] gil24586683|ref|NM_015286.4|[24586683] giJ33563285|ref|NM_015289.11[33563285] gi|19526752|ref|NM_015293.1 [[19526752] giJ24308028Jref|NMJj15296.1 j[24308028] giJ34222097|ref|NM_015305.2[[34222097I giJ24308032Jref|NM_015308.1 [[24308032] gi[39725633[refJNM_015315.2|[39725633] gi|18699719Jref[NM_015316.1 [[18699719] giJ38787940|ref|NM_015319.2|[38787940] gi[34732708Jref|NM_015321.1 j[34732708] gi|37059722iref|NM_015323.2|[37059722] gi|24308040|ref|NM_015327.11[24308040] gil24308042lref[NM_015328.1 |[24308042] giJ50726969|ref|NM_015329.2|[50726969] giJ51036581 |ref|NM_015330.1[[51036581] gi|24638432Jref|NM_015331.1|[24638432] gi|47575843|ref|NM_015335.2|[47575843] gi|29244580|ref|NM_015336.11[29244580] gi|29570781 jref[NM_015338.3J[29570781] gi|25121986|ref[NM_015341.2l[25121986] gi[24308048[ref|NM_015342.1|[24308048] giJ40548414[ref|NM_015345.2|[40548414] giJ38202204|ref|NM_015346.2|[38202204] gi|50726971 |ref|NM_015347.2|[50726971] gi|21245133|ref|NM_015350.lj[21245133] gi[27436888iref[NM_015352.1[[27436888] gi|28872811 |ref|NM_015358.11[28872811] gi]47271355|ref|NM_015359.11[47271355] gi|39930352|ref|NM_015360.2|[39930352] giJ31742497|ref|N __015374.11[31742497] gi|40806178|ref|NM_015375.11[40806178] giJ42734332lrefJNM_015378.1 j[42734332] gi|29789067|ref|NM_015381.1 |[29789067] gi|32698701 |ref|NM_015382.1 [[32698701] gi|21070955|ref|NM_015386.1 |[21070955] gi|32698709[ref|NM_015391.11[32698709] gi|32698703[ref|NM_015395.1 [[32698703] giJ22218618Jref|NM_015397.l [[22218618] gi|45827789|ref|NM_015404.11[45827789] gi[24308052Jref [NM_015411.1 [[24308052] gi[24308054[refJNM_015412.l i[24308054] gi[50659097|refiNM_015430.2[[50659097] gi)40353772|refJNM_015431.2J[40353772] gi[45827720|refiNM_015433.2J[45827720] gi|24308060|ref|NM_015436.11[24308060] giJ31083115iref|NM_015439.2|[31083115] gi|40018634JrefiNM_015440.3J[40018634] gi|46195718|ref|NM_015441.1 |[46195718] gi|41152088jref JNM_015443.2j[41152088] gi|22001414JrefJNM_015444.1 |[22001414]
124308066 ref |NM_015446.11[24308066]
140538729 ref |NM_015447.11[40538729]
J3993035 ref[NM_015448.1|[39930354]
(41152071 ref|NM_015457.2|[41152071]
I45827805 ref|NM_015459.3|[45827805]
I23308602 ref[NM_0 5460.1 [[23308602]
J24308068 ref|NM_015461.1|[24308068]
J24308070J ref|NM_015463.11[24308070]
I23943784 refJNM_015464.l[[23943784]
122001416 ref[NM_015465.1 [[22001416]
I24308072 ref |NM_015466.11[24308072]
J22267435 ref|NM_015469.l[[22267435]
I24308074 ref|NM_015470.1 [[24308074]
[46195780 refJNM_015475.2J[46195780]
I24308076 ref|NM_015476.lj[24308076]
[23397665J ref|NM_015477.11[23397665]
I24308078 ref|NM_015481.1 |[24308078]
145120110 ref|NM_015483.1|[45120110]
[37059800 ref|NM_015488.3J[37059800]
J24308080 ref|NM_015503.1|[24308080]
I38524593 ref JNM_015508.2[[38524593]
(39930360 ref|NM_015518.1|[39930360]
[40548417 refJNM_015522.2|[4054841 ]
I24308084 ref |NM_015529.1 [[24308084]
146195720 ref|NM_015531.1 [[46195720]
131377766 ref )NM_015532.2|[31377766]
[40254867 ref|NM_015534.3[[40254867]
I24308086 ref|NM_015541.1 j[24308086]
|25777708j ref|NM_015547.2|[25777708]
J34577048 ref]NM_015548.2|[34577048]
140354211 ref|NM_015553.1|[40354211]
J20127147 ref|NM_015555.lj[20127147]
J24308088 ref JNM_015557.1 [[24308088]
139777611 ref|NM_015558.3J[39777611]
118860828 ref|NM_015560.1 [[18860828]
131657110 ref|NM_015565.1|[31657110]
140217822] ref |NM_015567.1 [[40217822]
J34335276 ref[NM_015568.2J[34335276]
142544242 ref|NM_015569.2|[42544242]
J42476298 refJNM_015575.2([42476298]
134577113 refJNM_015576.1|[34577113]
I24308092 ref |NM_015578.1 [[24308092]
134222103 ref|NM_015585.2|[34222103]
150811833 refJNM_015589.3[[50811833]
J34594663 ref(NM_015597.2[[34594663]
134147330 refJNM_015600.2|[34147330]
J39753956 ref|NM_015602.2[[39753956]
|31317287J ref|NM_015604.2|[3 317287]
[45243553 ref|NM_015605.4J[45243553]
131742483 ref JNM_015608.2J[31742483]
J24308106 ref|NM_015609.1 [[24308106]
130911102 ref]NM_015617.1 [[30911102]
J22027529 ref|NM_015627.11[22027529]
J24308108 ref|NM_015631.11[24308108]
[24308110 ref[NM_015633.1 [[24308110]
J24308112 ref|NM_015634.1|[24308112]
J51093831 ref|NM_015635.2|[51093831]
gi|50052856 |ref J|NM 015639.1 |[50052856] gi|24308114 jref J |NM.015649.1 |[24308114] giJ34147336 |ref| |NM_ 015655.2 [[34147336] gi|46195722 |ref J|NM .015659.1 j[46195722] giJ28416430 |ref J|NM.015660.1 [[28416430] gi|46358427 |ref J|NM .015662.1 [[46358427] gi[31563510 [ref J|NM 015666.2 [[31563510] gi|32698717 |ref J|NM .015667.1 [[32698717] gi|24308118 |ref J|NM .015668.1 |[24308118] giJ46852180 jref JjNM 015687.2 |[46852180] gi|34222107 jref J |NM 015690.2 [[34222107] giJ38570148 |ref J|NM .015691.2 |[38570148] giJ23510326 [ref J|NM.015692.1 |[23510326] gi|44888832 |ref J|NM .015693.2 [[44888832] giJ23821012 jref JjNM.015694.1 |[23821012] gi|42544135 [ref J|NM 015713.3 |[42544135] giJ48675815 |ref J|NM.015723.2 |[48675815] gi|47419910 jref J |NM.015905.2 |[47419910] giJ28558968 |ref J|NM.015979.2 [[28558968] giI31317232 [ref J|NM.016105.2 |[31317232] gi|38327532 |ref J|NM 016133.2 [[38327532] gi|21264364 |ref J|NM 016320.2 j[21264364] gi|23821014 jref J |NM.016544.1 |[23821014] gi|31442760 |ref J|NM 017419.1 [[31442760] gi|34101287 jref J |NM 017437.1 |[34101287] giJ34222109 Jref jjNM.017440.2 [[34222109] gi|39725635 jref JjNM .017510.3 |[39725635] giJ39930370 |ref J|NM..017516.1 [[39930370] gi[40068465 jref J|NM .017519.1 |[40068465] gi|41055988 |ref J|NM..017520.2 |[41055988] gi|50878264 jref J |NM..017525.1 |[50878264] giJ40068041 |ref J|NM 017527.2 |[40068041] gi|24308168 |ref J|NM 017539.1 |[24308168] gi|24475585 jref J |NM..017549.1 |[24475585] gi|45267838 |ref J|NM..017550.1 |[45267838] gi|38708320 lref J|NM .017553.1 |[38708320] gi|50512291 jref J |NM..017554.1 ([50512291] gi|30794491 |ref J|NM 017556.1 [[30794491] gi|24308146 jref JjNM 017563.1 |[24308146] gi[42476034 |ref J|NM..017565.2 |[42476034] gi|48314819 |ref J|NM .017570.1 [[48314819] gi|40316916 jref JjNM 017573.2 |[40316916] gi[30794487 jref J |NM .017576.1 |[30794487] gi[23943913 jref JjNM..017580.1 [[23943913] gi|40353201 [ref jjNM .017602.2 |[40353201] gi[40556377 jref J |NM. 017619.2 |[40556377] gi[40254897 |ref J|NM .017628.2 [[40254897] gi|38569483 jref J|NM. 017641.2 | [38569483] gi|42476026 |ref J|NM .017666.2 [[42476026] giJ29893551 jref J |NM..017672.2 |[29893551] gi|33356553 |ref j|NM .017725.1 [[33356553] giJ46519146 jref JjNM .017747.1 [[46519146] gi|33350929 |ref J|NM..017754.2 |[33350929] gi[40353209 |ref J|NM..017758.2 [[40353209] gi|31543451 |ref J|NM..017771.2 |[31543451] giJ50811884 |ref J]NM..017804.3 |[50811884] gi|23680884 |ref J|NM. 017861.1 |[23680884]
gi|39725639|ref|NM_017871.3|[39725639] giJ33859677[ref[NM_017879.1 [[33859677] giJ24308176Jref|NM_017969.11[24308176] giJ4651915θirefJNM_017978.lj[46519150] giJ24308154|ref|NM_018003.li[24308154] gi|50811886|ref|NM_018069.2J[50811886] gi|22547233|ref|NM_018117.10|[22547233] gi|47059117Jref|NM_018151.1|[47059117] giJ31742491 jref JNM_018177.2|[31742491] gi|42734338|refJNM_018193.11[42734338] giJ41055952|ref|NM_018218.lj[41055952] giJ46852387|ref[NM_018237.2i[46852387] giJ24308156|refJNM_018284.1 [[24308156] giJ37039611 jref|NM_018325.1 [[37039611] gi|37059785Jref|NM_018334.3[[37059785] gi|23510330|ref|NM_018369.11[23510330] gi[38016155|ref|NM_018392.3([38016155] giJ27501455Jref|NM_018397.lj[27501455] gi|24475639)ref|NM_018405.2|[24475639] gi|21359927|ref|NM_018414.2|[21359927] gi|2430816θ[ref|NM_018420.11[24308160] gi[29893546|ref|NM_018424.lj[29893546] gi|21281668|ref|NM_018429.1|[21281668] giJ27544938Jref|NM_018462.3J[27544938] gi|21314681|ref|NM_018646.2|[21314681] gi|38638697|ref|NM_018689.11[38638697] giJ38683863irefJNM_018703.3J[38683863] giJ24308178JrefJNM_018704.1 [[24308178] gi|24308162Jref|NM_018708.11[24308162] gi|24308164|ref[NM_018710.11[24308164] gi|24308166|ref|NM_018711.11[24308166] gi|34222111 |ref[NM_018712.2J[34222111] giJ21237782JrefJNM_018714.1|[21237782] giJ29789089|refjNM_018715.lj[29789089] gi|38158014|ref|NM_018717.2|[38158014] giJ38327656|ref|NM_018837.2J[38327656] gi[2430818θjref|NM_018847.1 [[24308180] gi|24308126[ref|NM_018981.lj[24308126] giJ39930366Jref|NM_018987.1 j[39930366] giJ30749199[ref|NM_018998.2|[30749199] giJ24308130|ref|NM_018999.11[24308130] gi|40254886Jref|NM_019001.2|[40254886] gii34147334Jref|NM_019007.2[[34147334] gi|27894336Jref|NM_019010.lj[27894336] giJ48717494|ref|NM_019015.l[[48717494] gi|3850522ljref|NM_019022.3|[38505221] gi|24308132[ref|NM_019026.l[[24308132] gi|22027515[ref[NM_019029.li[22027515] gi|26553431Jref|NM_019030.1 j[26553431] giJ38016903|ref|NM_019032.2|[38016903] gi[46195724|ref|NM_019036.lj[46195724] gi|21265095[ref|NM_019051.l[[21265095] gi|30410709Jref|NM_019053.2[[30410709] gi[l7511434|ref|NM_019055.4|[17511434] giJ32129213|ref|NM_019065.2J[32129213] gi|24308140|ref|NM_019072.1 |[24308140] gi[45827761[ref|NM_019075.2|[45827761]
gi[41282212|ref|NM_019077.2[[41282212] gi|46249403Jref[NM_0 9078.1 [[46249403] gi|4273433δjref|NM_019085.1 |[42734336] gi|39930376[refJNM_019092.1 [[39930376] gi|28144915Jref|NM_019104.1 |[28144915] giJ50052804|ref|NM )19107.2|[50052804] gi|50843819Jref[NM_019590.2J[50843819] gii34222113JrefJNM_019593.2J[34222113] gi|39930378|ref|NM_019594.1|[39930378] gi|29789101 [ref(NM_019850.1 [[29789101] gi|39930382|ref|NM_020063.1 |[39930382] gi|34222114|ref|NM_020116.2J[34222114] giJ23110975iref(NM_020124.1|[23110975] gi|24308184|ref|NM_020170.1 ([24308184] gi[20514779[ref|NM_020172.1 [[20514779] gi[24308186[ref|NM_020175.1 |[24308186] gi|21281666|ref|NM_020192.1 |[21281666] gi[33569215Jref[NM_020204.2i[33569215] gi[39930392[ref[NM_020207.l i[39930392] gi|42476327|ref|NM_020209.2|[42476327] gi|39777607(ref|NM_020210.2|[39777607] gi|24308188|ref(NM_020211.11[24308188] giJ40556360|reflNM_020212.1 J[40556360] gi|19482155[ref|NM_020214.1|[19482155] giJ45267816iref|NM_020219.2|[45267816] gi|32698727|ref|NM_020223.1 |[32698727] giJ31083343Jref|NM_020311.11[31083343] gi|40789232|refJNM_020312.1 |[40789232] giJ23097298|ref|NM_020318.1 ([23097298] gij28461128jref|NM_020319.1[[28461128] gi|24475643Jref|NM_020320.2|[24475643] giJ34787408|ref|NM_020336.1 |[34787408] gi|50234887|ref|NM_020338.2|[50234887] giJ38569486Jref(NM_020340.2[[38569486] gi|29826330|ref|NM_020341.2J[29826330] gi|34147340Jref|NM_020376.2J[34147340] gi[34147337|ref|NM j20378.2|[34147337] gi|21359928|ref [NM_020383.2|[21359928] gi|29826286|ref|NMJ)20409.2[[29826286] gi|32967587|ref|NM_020416.2[[32967587] gi|31652229|ref)NM_020417.1 |[31652229] gi|32698733 jref |NM_020420.1 [[32698733] gi|31317291 |ref|NM_020429.1 |[31317291] gi[40254932JreflNM_020432.2|[40254932] gi|48976059|ref|NM_020438.3|[48976059] giJ41152505Jref|NM_020440.2|[41152505] gil40353202|ref|NM_020447.2|[40353202] gi|47578098Jref|NM_020451.2|[47578098] gi[40316836[refJNM_020452.1 [[40316836] gi|28466988Jref|NM_020453.2|[28466988] giJ37620168JrefJNM_020455.3|[37620168] gi|20531764[ref|NM_020456.1 |[20531764] gi|40354196|refjNM_020457.2|[40354196] gi[24308198|ref|NM_020462.1 |[24308198] giJ39930396[ref|NM_020463.1 [[39930396] gi|39777615[ref|NM_020468.2|[39777615] giJ41327713Jref[NM_020531.2J[41327713]
gi|47519458|ref|NM_020532.4|[47519458] giJ29570789|ref|NM_020536.2|[29570789] gi|25952130|ref|NM )20546.1 |[25952130] giJ38373681 |ref|NM_020631.2|[38373681] gi|21735574|ref|NM_020647.1 |[21735574] gi|21359934|ref|NM_020693.2|[21359934] gi|37655180Jref|NM_020695.2|[37655180] giJ33468964|refJNM_020696.lj[33468964] gi|45433545Jref|NM_020697.2|[45433545] giJ29789109|ref|NM_020698.l [[29789109] gi|21218437|ref|NM_020699.1 |[21218437] gi|20149303Jref|NM_020701.1 [[20149303] gi|50950116|ref|NM_020702.11[50950116] giJ40789228Jref|NM_020706.lj[40789228] gi[24308206[refJNM_020710.1 |[24308206] gi|34013527|ref|NM_020713.1 |[34013527] giJ29789111 |ref|NM_020714.11[29789111] gi|42415502|ref|NM_020718.2J[42415502] giJ45387944Jref|NM_020728.1|[45387944] gi[40068463|ref|NM_020732.2|[40068463] gi|31621295|ref|NM_020739.1 |[31621295] giJ3131725l iref(NM_020740.l j[31317251] gi|31317257|ref|NM )20742.2|[31317257] gι'J50878291 |ref|NM_020744.2J[50878291] gi|38569416[ref|NM_020745.l [[38569416] gi|42734342|ref|NM_020746.1 |[42734342] giJ2430821θirefJNM_020748.l i[24308210] gi|22748936Jref|NM_020750.1 [[22748936] gi|33457343|ref|NM_020751.1|[33457343] gi[47059124|ref|NM_020752.l [[47059124] giJ45433546|ref|NM_020753.2J[45433546] giJ34222115|ref|NM_020755.2[[34222115] gii22094986JrefJNM_020761.1 j[22094986] gi|44917603|ref|NM_020762.1|[44917603] gi|24416001 |ref|N _020765.11[24416001 ] gi|24308214|ref|NM_020769.1 |[24308214] gi|34222116|refJNM_020771.2J[34222116] gi|32698729JrefjNM_020772.l [[32698729] giJ29789113|refJNM_020773.1 j[29789113] gi|30348953|ref|NM_020774.1 |[30348953] gi[38569481 jref|NM_020775.2|[38569481] gi|24308216|ref[NM_020778.1 [[24308216I giJ34222117Jref(NM_020779.2J[34222117] gi|25777702|refJNM_020781.2|[25777702] gi|34222118[ref|NM_020783.2|[34222118] gi|41349496JrefJNM_020786.11[41349496] gi|47271359|ref|NM_020787.11[47271359] gi|34147342Jref|NM_020789.2|[34147342] giJ45439369Jref|NM_020791.1 j[45439369] giJ4604817δ[ref JNM_020792.2|[46048175] giJ31317254[ref|NM_020795.2|[31317254] gi|33147079|ref|NM_020799.1|[33147079] giJ46409656|ref|NM_020800.1 j[46409656] giJ32698735|refJNM_020801.1 j[32698735] gi|34101267[ref|NM_020803.3|[34101267] giJ47834327[ref|NM_020804.2|[47834327] giJ45237199|refJNM_020808.1 ([45237199]
gi|45433494|ref| |NM_020809.11[45433494] giJ50845423Jrefj |NM_020810.1 |[50845423] giJ40217610jref J|NM_020812.1 [[40217610] gi]32698737Jref ]|NM_020813.1 [[32698737] gi|40385863Jref j[NM_020816.1 [[40385863] gi|24308222Jref J|NM_020817.1 j[24308222] giJ40068514[ref J|NM_020818.1 [[40068514] gi|34452731|ref JjNM_020820.2J[34452731] gi|50345878Jref| |NM_020824.2J[50345878] gi|46195728|ref ]|NM_020825.1 [[46195728] giJ24308232|ref J|NM_020826.11[24308232] gi|22094124|ref| |NM_020828.1 j[22094124] giJ24308226Jref[ |NM_020832.11[24308226] gi[29789117|ref J|NM_020834.1 [[29789117] gi|21314694Jrefj |NM_020839.2J[21314694] gi|4523720l [ref| |NM_020844.11[45237201] gi|24308236|ref |NM_020845.1 [[24308236] giJ21702732Jref ]|NM_020847.1 |[21702732] gi|40538735|ref J|NM_020850.11[40538735] gi|3993040θjref J|NM_020851.1 |[39930400] gi|24475647|ref J|NM_020854.2[[24475647] giJ24308230[ref J|NM_020856.1 [[24308230] gi|24234728|ref J|NM_020858.1 [[24234728] gi[l869972l [ref J|NM_020859.1 [[18699721] gi|21070998|ref J|NM_020860.1 |[21070998] gi|24308240|ref| |NM_020861.1 j[24308240] gi|4648791θ[ref| |NM_020863.2[[46487910] gi|22325365Jref J|NM_020867.11[22325365] gi|24308234|ref J|NM_020868.1 [[24308234] giJ32698739Jref J|NM_020870.1 j[32698739] gi|40068043Jref JjNM_020871.2J[40068043] gi|34222384|ref ]|NM_020873.3|[34222384] gi|45827745|ref J|NM_020875.11[45827745] gi(40254939Jref J[NM_020880.2[[40254939] gi|44917609Jref J|NM_020882.1 [[44917609] gi|30842828Jref ][NM_020889.11[30842828] gi|24308238|ref JjNM_020890.11[24308238] giJ24308252|ref |NM_020892.1 [[24308252] gi|34147343|ref J|NM_020895.2|[34147343] gi|38327036|ref JjNM_020897.1 [[38327036] gi|34222119Jref ]|NM_020899.2J[34222119] gi|5023387l [ref ]|NM_020914.2|[50233871] gi|40254942Jref| |NM_020918.2|[40254942] gi|50845417|ref| |NM_020922.2J[50845417] gi|3269874l[ref J|NM_020925.11[32698741] gi|21071035|ref J|NM_020926.2J[21071035] gi[24308256iref JjNM_020927.11[24308256] gi|20143481 jref jjNM_020932.1|[20143481] gi]32698743|ref J|NM_020935.1 [[32698743] gi|41327778|ref J|NM_020936.1 |[41327778] giJ25141322Jref[ |NM_020939.1 [[25141322] gi[27413907Jref J|NM_020944.2|[27413907] gi|38327643|ref ]|NM_020947.2|[38327643] gi|2430826θjref| |NM_020948.1 j[24308260] gi[24308254|ref J|NM_020951.1 |[24308254] gi|45935356|refj |NM_020952.2[[45935356] gi|39930394|ref ]|NM_020954.11[39930394]
gi|34147341 |refJ|NM 020961.2 [34147341] gi[l9882240|refJ|NM 020962.1 [19882240] gi|46195732JrefJ|NM.020964.1 [46195732] gi|29568112|refJjNM_.020965.2 [29568112] gi|42734346|refJ|NM_020970.1 [42734346] gϊ]40353203|ref||NM_.020971.1 [40353203] gi|48949814|refJjNM_021006.4 [48949814] giJ34304116|ref||NM_021009.2 [34304116] gi[2862652θ[refJ|NM_.021035.1 [28626520] gi|23346635Jref||NM_021044.2 [23346635] giJ32698747|ref NM_021045.1 [32698747] gi|31742502Jref||NM_021059.2 [31742502] gij45387946|ref||NM_021061.1 [45387946] gi|32698745|refJ|NM.021072.1 [32698745] giJ2030409θjrefJ|NM_021088.1 [20304090] gi|33438574[ref||NM_021089.1 [33438574] gϊ|31083192[refJ]NM .021116.1 [31083192] giJ2978912l jref NM_021117.1 [29789121] gi|33667024|refJ|NM..021143.1 [33667024] gi|40538737|ref||NM_021148.1 [40538737] gi|23510452|refJ|NM .021149.2 [23510452] giJ33469959[refJ|NM.021164.2 [33469959] gi|23943865JrefJ|NM .021165.1 [23943865] giJ38049006|refJ|NM 021180.2 [38049006] gi[39930398|refJ|NM 021202.1 [39930398] gi|33438585|refJ|NM .021217.1 [33438585] gi|24308268(refJ|NM .021218.1 [24308268] gi|24308262|ref|NM.021222.1 [24308262] giJ31377724Jref[NM..021224.2 [31377724] gi[40548405JrefJ[NM 021227.2 [40548405] gi|32698749[refJ|NM.021228.1 [32698749] gi|46370091 JrefJ|NM .021237.3 [46370091] gi|32895366|refJ|NM 021250.2 [32895366] gi|30795179|refJ|NM..021260.1 [30795179] gi|39930402JrefjNM 021636.1 [39930402] gi|48675828JrefJ|NM 021649.3 [48675828] giJ41281598|refJ|NM .021652.1 [41281598] giJ50512285|refJ|NM.021915.1 [50512285] gi|31340618JrefJ|NM..021916.2 [31340618] giJ2309730θjrefJ|NM.021936.1 [23097300] gi[39725642Jref|NM..021937.2 [39725642] gi|21630256|ref|NM.022045.2 [21630256] giJ32455257|refJ|NM.022075.2 [32455257] gϊJ44917605JrefJ|NM..022080.1 [44917605] gi|42794774|refJ|NM.022085.3 [42794774] gi|27501457|refJ|NM.022092.1 [27501457] gi|3269875l jrefJ|NM._022106.1 [32698751] gi|46048067|ref|NM._022115.2 [46048067] gi|24308276|refJ|NM._022138.1 [24308276] gi|46195736|refJ|NM_022160.1 [46195736] gi|28269692[refJjNM_022166.1 [28269692] gi|31377716|ref|NM_022351.2 [31377716] gi|20143972]refJ]NM.022475.1 [20143972] gi|32880205|refJ|NM.022478.2 [32880205] gij22538494JrefJjNM .022479.1 [22538494] gi|48093064|refJjNM 022486.3 [48093064] gi|42734348JrefJ|NM 022491.1 [42734348]
gi[39725643|ref|NM_022572.2|[39725643] giJ23943871|ref|NM_022733.1 [[23943871] gi|32171243Jref|NM_022742.2|[32171243] gi[48976052iref|NM_022745.2[[48976052] gi|40255224|ref|NM_022757.3|[40255224] gi[45238579Jref[NM )22824.l i[45238579] gi|51093862|ref|NM_022833.2|[51093862] giJ45237196Jref[NM_022835.1 j[45237196] giJ2887578l jrefJNM_022913.1 [[28875781] gi|30794471 |ref|NM_023002.11[30794471] gi|20302140|ref|NM_023006.1|[20302140] gi|50897299Jref|NM_023939.3|[50897299] giJ4538795θ[ref JNM_023943.1 j[45387950] gi|36054140|refJNM_024007.2J[36054140] gi[31340581 |ref|NM_024019.2J[31340581 ] gi|37674208|ref|NM_024100.2[[37674208] giJ24308288[ref|NM_024316.li[24308288] gi|42544240|ref|NM_024335.2J[42544240] gi|39930458|ref|NM_024336.1 |[39930458] gi|25121937Jref|NM_024342.1 |[25121937] gi|27765071 |ref|NM_024344.11[27765071] gi|23943919JrefiNM_024420.l[[23943919] gi[24308296|ref |NM_024493.11[24308296] giJ38327635|refJNM_024496.2J[38327635] gi|3972571 θ[ref|N _024511.3J[39725710] gi|24797090|ref|NM_024517.1 j[24797090] gi|33469927|ref|NM_024553.2[[33469927] giJ22003857|ref|NM_024621.1 j[22003857] giJ27477137|ref|NM_024625.3J[27477137] gi|34328078|ref|NM_024684.2|[34328078] gi|28875783Jref|NM_024742.1 |[28875783] gii41393588Jref[NM_024769.2J[41393588] gi|47578114|ref[NM_024870.2[[47578114] giJ39653320|ref|NM_024878.lj[39653320] giJ46358069iref|NM_024933.2|[46358069] giJ38679913lref|NM_024953.2|[38679913] gi[47716684[ref|NM_025169.1 [[47716684] giJ37059725|ref|NM_025196.2J[37059725] giJ34147386Jref|NM_025202.2J[34147386] giJ45504381 |ref|NM_025219.1 [[45504381] gi|47679079|ref|NM_025224.1 |[47679079] giJ45592956[refiNM_025248.li[45592956] gi|47132517|ref|NM_025252.3J[47132517] gijl8426878Jref|NM_025256.4J[18426878] gi[33859754[ref|NM_030625.l i[33859754] gi|32698754|ref|NM_030627.1 |[32698754] gi[4477120θ[ref[NM_030628.l j[44771200] gi[29789254iref[NM_030629.1|[29789254] giJ29825822|ref|NM_030630.lj[29825822] giJ30794213|ref|NM_030633.11[30794213] gi|24308298|ref|NM_030634.lj[24308298] gi|2430830θ[ref|NM_030636.1[[24308300] gi|45120112|refJNM_030637.li[45120112] gi|39930462JrefJNM_030639.1 |[39930462] gi|38372910|refJNM_030640.1|[38372910] gi|22035641 |ref|NM_030644.11[22035641] gi|24308302Jref|NM_030645.1|[24308302]
gi|38176150|ref| |NM_030650.1 |[38176150] gi[30581114|ref| |NM_030789.2|[30581114] giJ32698756|ref| |NM_030812.1 |[32698756] gi|34222273jref| |NM_030883.3J[34222273] gi[44890053|ref J|NM_030906.2[[44890053] gi|24308304Jref JjNM_030919.1 |[24308304] gi|42476171|ref| |NM_030922.3[[42476171] gi[34222122Jref| |NM_030923.2J[34222122] gi|19311005|ref[ |NM_030949.1 |[19311005] gi|21265024|ref ]|NM_030957.lj[21265024] gi|30794215Jref ]|NM_030961.1 |[30794215] gi|29788754Jref| |NM_030962.11[29788754] gi|49065663Jref JjNM_031303.11[49065663] gi|50845406Jref| |NM_031444.2|[50845406] gi|34147398|ref J|NM_031448.2J[34147398] gi[32880228|ref jiNM_031454.1 [[32880228] gi|24308313jref| |NM_031467.1|[24308313] gi|31377666|ref J|NM_031490.2|[31377666] gi|23510324|ref ]jNM_031888.1 |[23510324] gi|24586685Jref| |NM_031895.4[[24586685] gi|40549456Jref| |NM_031912.3[[40549456] gi|46195742|ref| |NM_031913.1 |[46195742] giJ32698758Jref JjNM_031914.1 [[32698758] giJ31083305jref J|NM_031935.11[31083305] gi|31543197|ref ]|NM_032017.1 J[31543197] gi|21265074|ref J[NM_032111.1 |[21265074] gi|19882212Jref J|NM_032119.l [[19882212] gi|40353745|ref JjNM_032123.4|[40353745] gi|31542506]ref J|NM_032132.2|[31542506] giJ31542516|ref| |NM_032137.2|[31542516] gi|50428930|ref| |NM_032156.2|[50428930] gi|23943786Jref J|NM_032160.1 [[23943786] gi|50083284|ref| |NM_032165.2|[50083284] gi[29789282[ref J|NM_032168.1 [[29789282] gi[39930468[ref J|NM_032194.1|[39930468] giJ21040313|ref ]|NM_032195.l [[21040313] gi[38683806|ref JjNM_032217.3J[38683806] gi|39752638|ref| |NM_032222.11[39752638] gi|24308321 |ref| |NM_032226.11[24308321] gi[50582992Jref| |NM_032228.4|[50582992] gi|3993047θ[ref J|NM_032230.1 [[39930470] gij46195744jref J|NM_032279.1 |[46195744] giJ42734355jref| |NM_032282.11[42734355] gi|45433498|ref J|NM_032283.1 |[45433498] gi|23943879|ref J|NM_032285.1 j[23943879] gi|49227853Jref J|NM_032286.2|[49227853] gi|29789276|ref J|NM_032422.1 [[29789276] giJ46410932Jref| |NM_032423.2[[46410932] giJ46391093|ref J[NM_032425.3|[46391093] giJ33286443|ref J|NM_032427.1 [[33286443] gi[45331214Jref J|NM_032429.1 [[45331214] gi|24308325|ref jjNM_032430.11[24308325] gi|27436924Jref JjNM_032431.1 [[27436924] gi|40316949|ref ]|NM_032432.2[[40316949] giJ20336723Jref J|NM_032433.1 j[20336723] gi[40217789|ref J|NM_032434.2J[40217789] giJ24308329|ref| |NM_032435.1 j[24308329]
gi|41281611 |ref|NM_032436.1 [[41281611] gi|3346902θjref[NM_032439.11[33469020] gi J39930472Jref|NM_032440.1 [[39930472] gi|24308331 [ref|NM_032444.11[24308331] giJ24308333|ref|NM 032448.11[24308333] gi[33859758|ref|NM j32452.1|[33859758] gi|28557676[ref|NM_032458.l[[28557676] gi|21265092|ref|NM_032477.l[[21265092] gi|26667164|ref|NM_032478.2J[26667164] gi|21265087[ref|NM_032479.2[[21265087] giJ22094134|ref|NM_032482.1|[22094134] gi[23618925[ref(NM_032497.1 [[23618925] giJ28416952|ref|NM_032501.2[[28416952] gij21245131|ref[NM_032505.1j[21245131] gi|47575698|ref|NM_032506.lj[47575698] gi|22296883Jref|NM_032508.l j[22296883] gi(24308335Jref|NM_032511.1 [[24308335] gi|46559760Jref|NM_032512.2J[46559760] gi|37059771|ref|NM_032517.3J[37059771] gi|26190609|ref|NM_032528.1|[26190609] gil26006460|ref|NM_032531.11[26006460] gijl9387853Jref(NM_032536.1([19387853] gii40217818[ref|NM_032539.2[[40217818] gi|50897838JrefJNM_032550.2J[50897838] gi[20070108|ref|NM_032552.1 [[20070108] gi|40556375|ref|NM_032569.2|[40556375] gi[46852144Jref|NM_032590.2[[46852144] giJ29826313|ref|NM_032636.4J[29826313] gi|31377641|ref|NM_032869.2|[31377641] gi[l8699723[ref|NM_032870.1 [[18699723] gi|40805107|ref|NM_032947.3|[40805107] gi(50726961[ref|NM_033026.2[[50726961] gi[l952647θ[ref|NM_033046.li[19526470] gi|2430834ljref|NM_033052.1|[24308341] giJ33438589iref|NM_033053.l[[33438589] gi|39753964|ref|NM_033055.2|[39753964] giJ48375172|ref|NM_033063.lj[48375172] gi|29336042|ref|NM_033064.11[29336042] gii2430835ljref|NM_033067.lj[24308351] gi|23097307|ref|NM_033071.11[23097307] gi|32129198Jref|NM_033082.lj[32129198] gi|32698764|ref|NM_033086.lj[32698764] gi[42734359[ref|NM_033088.1 [[42734359] giJ23397643Jref|NM_033090.lj[23397643] gi|24308345Jref|NM_033107.1|[24308345] gi|3369511 θ[ref |NM_033109.2 [[33695110] gi|24308349Jref|NM_033112.lj[24308349] gi[38683798[ref[NM_033121.1 [[38683798] gi|46430490|ref|NM_033129.lj[46430490] giJ50582976|ref|NM_033141.1 j[50582976] gi[42822888|ref|NM_033160.3|[42822888] gi|19593984|ref|NM_033161.2|[19593984] gi|24308355|ref|NM_033200.1|[24308355] gi|24308353Jref|NM_033201.1 j[24308353] giJ45331210Jref|NM_033206.1|[45331210] gi|50593107|ref|NM_033253.2|[50593107] gi|39930474|refJNM_033267.l j[39930474]
gi|30794221 |ref| |NM_033271.1|[30794221] gi[32698766Jref j[NM_033276.11[32698766] gi|38788287|ref| |NM_033288.2J[38788287] giJ38569418|ref ]|NM_033364.2|[38569418] gi|46485767|ref| |NM_033375.3J[46485767] gi|31324576|ref J[NM_033386.1|[31324576] gi|34222124Jref J|NM_033387.2|[34222124] gi|37674209Jref| |NM_033389.2[[37674209] gi|41350322|ref ]|NM_033392.3|[41350322] gi|49472816lref ]|NM_033393.1 [[49472816] gi|20270211 |ref| |NM_033396.1 |[20270211] gil47271452|ref j[NM_033397.2J[47271452] gi[47604913[ref ]|NM_033402.2|[47604913] gi|46243679|ref| |NM_033404.2[[46243679] giJ34335259Jref jjNM_033405.2[[34335259] giJ42734361 lref| |NM_033407.1 |[42734361] gi|39930478[ref| |NM_033425.1J[39930478] giJ32307174|ref J|NM_033426.2|[32307174] gi|4619575θ[ref J|NM_033429.1 J[46195750] gi|28875785|ref JjNM_033449.1 [[28875785] gi|25914748Jref| |NM_033450.2|[25914748] gi[33457333[ref J|NM_033452.1 [[33457333] gi|50083288Jref J|NM_033505.1J[50083288] giJ25121979|ref| |NM_033510.1 |[25121979] gi[49410494Jref| |NM_033512.2[[49410494] gi|46195752|ref JjNM_033513.l [[46195752] gi|41281602|ref| |NM_033520.l[[41281602] gi|23618847|ref| |NM_033542.1 |[23618847] gi|2430836l jref J[NM_033548.1 j[24308361] gi[38176148|ref| |NM_033553.2[[38176148] gi|24308363]ref J|NM_033557.1 [[24308363] gi[l9924142Jref J|NM_033631.1 [[19924142] gi|32455235Jref ]|NM_033647.2|[32455235] gi]19743814|ref J|NM_033666.1 [[19743814] gi|19743816|ref J|NM_033667.l [[19743816] gi[l 9743818|ref jjNM_033668.1 [[19743818] gi|19743820|ref| |NM_033669.1|[19743820] gi|40255313|ref ]|NM_052843.1 [[40255313] gi|45433500|ref| |NM_052846.11[45433500] gi|32698768Jref J|NM_052847.1 [[32698768] gi[33086946Jref ]|NM_052849.2|[33086946] gi|31543414|ref J|NM_052850.2|[31543414] gi|49472813Jref J|NM_052857.2[[49472813] gi|38202233|ref| |NM_052864.2[[38202233] gi|24119273|ref J|NM_052867.1 [[24119273] gi[39930482Jref| |NM_052878.1|[39930482] gi|3359894l jref JjNM_052892.2J[33598941] giJ38045884Jref J|NM_052896.2|[38045884] giJ46852160|ref ]|NM_052897.3[[46852160] gi[32698770|ref| |NM_052899.1|[32698770] gi[38045885|ref J|NM_052900.2|[38045885] giJ39930484Jref J|NM_052901.1 |[39930484] gi|24308375Jref J|NM_052902.1 [[24308375] gi|48976055Jrefj |NM_052903.2[[48976055] gi|24308379Jref J|NM_052904.11[24308379] gi|38524617|ref JjNM_052905.1 j[38524617] gi|42734363Jref JNM_052909.1 [[42734363]
gi|40217816|ref|NM_052910.1 |[40217816] gι 40804767Jref|NM_052911.1 [[40804767] gi|40538800|refJNM_052913.2[[40538800] giJ42741666jref|NM_052917.1 j[42741666] gi|46195754|ref|NM_052923.1 |[46195754] gi|19882236|ref|NM_052924.l j[19882236] giJ24308381|ref|NM_052925.1|[24308381] giJ24308383Jref|NM_052926.1 [[24308383] gi|46195756JrefJNM_052928.1 |[46195756] gi|24308385|ref|NM_052937.1 |[24308385] gi|42734365Jref|NM_052964.l j[42734365] gi|24308389[ref|NM_052965.l j[24308389] gi|29789284|ref|NM_053041.11[29789284] giJ2447574θjref|NM_053044.2J[24475740] gi|29789292|ref|NM_053051.11[29789292] giJ26024192Jref|NM_053052.2|[26024192] giJ27894377|ref(NM_053277.lj[27894377] gi|32698772|ref|NM_053279.1 [[32698772] gi|2430841 ljref|NM_053282.l j[24308411] gi|50080212[refJN _054104.1 [[50080212] gi|50080218|ref|NM_054105.1 |[50080218] gi|46370074Jref[NM_057163.2[[46370074] gi[24308387[ref(NM_058163.l j[24308387] gi|19718730[ref|NM_058243.1|[19718730] giJ46309852|ref|NM_080574.2l[46309852] gi|32307108|ref|NM_080614.1 |[32307108] giJ29570784|ref[NM_080618.2J[29570784] gii49574583|refJNM_080622.2J[49574583] gi|22129777|ref|NM_080725.l j[22129777] gi[28372502|ref|NM_080747.1 |[28372502] giJ20304092|ref|NM_080751.1 [[20304092] giJ31563538Jref[NM_080753.2|[31563538] gi[50233782|ref|NM_080757.1 j[50233782] giJ38202248|refJNM_080764.2|[38202248] giJ22779935|ref|NM_080827.l [[22779935] gi|24308393|ref|NM_080833.1 |[24308393] gi|38327559|ref|NM_080836.2|[38327559] giJ34878895|ref|NM_080865.2[[34878895] gi|42542385|ref|NM_080866.2|[42542385] gi|27477048|ref[NM_080868.l j[27477048] gi|20069857|ref|NM_080869.1 |[20069857] gi|21264329|ref|NM_080875.1 |[21264329] gi|25014087|ref|NM_080877.1 |[25014087] gi|19718750|ref|NM_080911.lj[19718750] giJ38261966|ref|NM_080928.2J[38261966] gi|18765749|ref|NM_101395.1 |[18765749] gijl8860889|ref|NM_130391.l j[18860889] gijl 8860891 jref|NM_130392.1 [[18860891] gij 18860893|ref|NM_130393.1 j[18860893] gi|40805848JrefJNM_130435.2|[40805848] gi[l8765751 jref|NM_130436.1 |[18765751] gi[l8765753[ref|NM_130437.l j[18765753] gi[l8765755[ref|NM_130438.l j[18765755] gi[l8860895|ref|NM_130440.1 |[18860895] gijl 8765701 |ref|NM_130442.1 [[18765701] gi[l8765745Jref|NM_130444.1 [[18765745] gi|18765747|ref|NM_130445.1 |[18765747]
gi|50511933|ref|NM_130465.2|[50511933] gi|35493958|ref|NM_130466.2|[35493958] gi|1886086ljref|NM_130470.l j[18860861] gijl8860863|ref|NM_ 30471.1 |[18860863] gi|18860865|ref|NM_130472.1 |[18860865] gi|18860867|ref|NM_130473.1 [[18860867] gi[l8860869[ref|NM_130474.l i[18860869] gi|18860872|ref|NM_130475.1 j[18860872] gi|18860874[ref|NM_130476.1 |[18860874] gi|18780276|ref|NM_ 30760.1 |[18780276] gi|1878028θjref|NM_130761.1 |[18780280] gi[l8780284[ref[NM_130762.li[18780284] gi|18765706Jref|NM_130766.1 [[18765706] gi]45580712[ref|NM_130771.2|[45580712] gi|19747272|ref|NM_130775.1 |[19747272] gi|19747268|ref|NM_130776.1|[19747268] gi|19747282[ref|NM_130777.1 |[19747282] gi|18860879 jref|NM_130788.1|[18860879] gi|18860881 jref|NM_130790.1 j[18860881] gi|18860883|ref|NM 30791.1 |[18860883] gi|18860885|ref|NM 30792.1 |[18860885] gi|39777588|ref|NM 30793.3|[39777588] gi[ 19882248|ref|NM_130794.11[19882248] gi|18765697Jref|NM_130797.11[18765697] gi[l876573θ[ref|NM 30798.l j[18765730] gi|18860846|ref|NM_130799.1 |[18860846] gi|18860848Jref|NM_130800.l i[18860848] gijl886085θiref|NM_130801.lj[18860850] gijl8860852Jref]NM_130802.l j[18860852] gi|18860854|ref|NM_130803.11[18860854] gi| 18860856|ref|NM 30804.11[18860856] gi|20070357|ref|NM_130806.2J[20070357] gi|40018625iref|NM_130807.2J[40018625] gijl8677732Jref|NM_130808.1 |[18677732] gi|31377624|ref|NM_130809.2|[31377624] gi| 18677736|ref|NM J 30810.1 j[18677736] gi|18765734[ref|NM_130811.1 [[18765734] gi|40255007|ref|NM_130830.2|[40255007] gijl8860830|ref|NM_130831.1 |[18860830] gi|18860832|ref|NM_130832.1 |[18860832] gi| 18860834Jref|NM J 30833.11[18860834] gi[18860836[ref|NM_130834.l j[18860836] gi| 18860840 jref JNM 30835.11[18860840] gijl8860842Jref|NM_130836.1 |[18860842] gi|18860844|ref|NM_130837.1 |[18860844] gi|19718761 |ref|NMJ30838.1 |[19718761] gi[19718763[ref|N _130839.1 |[19718763] gi| 1991341 θjrefJNM 30840.1 J[19913419] gi[l9913421 (refJNM 30841.1 |[19913421] gi[ 19743911 [ref|NM_130842.1 [[19743911] gi[l9743913Jref|NM_130843.1 |[19743913] gijl8860887Jref|NM 30844.1 j[18860887] gij 18860909 jref|NM_130845.11[18860909] gijl 9743916|ref|NM_130846.1 JH 9743916] gi[22027645[ref|NM J 30847.11[22027645] gi|18677769|ref|NM_130848.1 |[18677769] gi|19115957[ref|NM_130849.1 |[19115957]
gi|19528649|ref[NM_130850.1([19528649] gi|19528651|ref|NM_130851.1|[19528651] gi| 18765704Jref |NM_130852.1 j[18765704] gijl9743920|ref|NM_130853.1|[19743920] gijl9743922|ref|NM_130854.1|[19743922] gi|19743924Jref|NM_130855.lj[19743924] gi|31657106|ref|NM_130896.lj[31657106] gij 18702322 jref |NM_130897.11[18702322] gi|3154209θjref|NM 30898.2|[31542090] gi|18702326|ref|NMJ30899.1|[18702326] gi|18702328|ref|NM_130900.l[[18702328] gi|18702330|ref|NM_130901.1|[18702330] gi| 18702332|ref |NM 30902.11[18702332] gijl9557635|refJNM_130906.l[[19557635] gi|45238855Jref|NM_131915.2|[45238855] gi|19557639|ref|NM_131916.1|[19557639] giil9528654iref[NM_131917.l[[19528654] gi|45580714[ref|NM_133168.2|[45580714] giJ4558071 δjref JNMJ 33169.2|[45580715] gi|48762921|ref|NM_133170.2|[48762921] gi|33469945|ref|NM_133171.2|[33469945] gi[l9528658Jref|NM 33172.lj[19528658] gi|19528660|ref|NM_133173.1|[19528660] gi|19528662|refJNM_133174.lj[19528662] gi|19528664|ref|NM_133175.1|[19528664] gi|19528666|ref|NM_133176.1|[19528666] gi[l974393θjref|NM_133177.li[19743930] gi|19743932|ref|NM_133178.1|[19743932] gijl974727θjrefJNM_ 33179.lj[19747270] gi|21264607|ref|NM_133180.1|[21264607] gi|21071015jref|NM_133181.2|[21071015] gi|31621304JrefJNM_133259.2J[31621304] gi|18959203|ref|NM 33261.1|[18959203] gi|20357541|ref|NM_133262.2J[20357541] giJ3154339ljref|NM_133263.2|[31543391] giJ23199980|ref|NM_133264.2|[23199980] gi[19111149|refJNM_ 33265.1 J[19111149] gi| 19743795Jref |NM_133266.1 [[19743795] gi|18959211|ref|NM_133267.1|[18959211] gi|19718742[ref|NM 33268.1|[19718742] gi| 19743856Jref |NM_133269.11[19743856] gijl9743858|ref|NM_133271.1|[19743858] giil974386θjrefJNM_133272.lj[19743860] gi|19743862Jref|NM_133273.1 J[19743862] gi[l9743864[ref|NM 33274.lj[19743864] gi|19743866|ref|NMJ33277.1|[19743866] gi|19743868|ref|NM_133278.1|[19743868] gi|19743870Jref|NM_133279.1|[19743870] gij 19743872JrefJNM_133280.1 j[19743872] gi|19718798[ref|NM_133282.1|[19718798] gijl9747275|ref|NM_133325.l j[19747275] gi|20357543Jref|NM_133326.11[20357543] gi|1971878liref|N _133327.lj[19718781] giJ31377621|refJNM_133328.2[[31377621] giJ2743699l [refJNM_133329.4J[27436991] gi|19913347|ref|NM_133330.1|[19913347] gijl9913349|ref|NM_133331.lj[19913349]
gi|19913351 |ref|NM_133332.1 |[19913351] gijl 9913353|ref |NM_133333.1 [[19913353] giJ19913355|ref|N _133334.1 |[19913355] gi|19913357|ref|NM_133335.1 |[19913357] gi|1991336θ[ref|NM_133336.l j[19913360] gi|19718758|ref|NM_133337.1 |[19718758] gijl9718783Jref(NM_133338.1 |[19718783] gijl 9718785|ref|NM_133339.1 [[19718785] gij 19718787|ref |NM_133340.11[19718787] gijl9718789Jref|NM_133341.l[[19718789] gi|19718791 |ref|NM_133342.1 |[19718791] gi|19718793|ref|NM_133343.1 |[19718793] gi|19718795|ref|NM_133344.11[19718795] gi J19115959 jrefjNM_133367.1 j[19115959] giJ45387948[ref|NM_133368.1 [[45387948] giJ21166354Jref|NM_133370.1 [[21166354] gi|31377622|ref|NM_133371.2|[31377622] gi|48675819|ref|NM_133373.2[[48675819] gi[34147483|ref|NM_133375.2|[34147483] gi|19743822[ref(NM_133376.l [[19743822] gijl 971880θ[ref|NM_133377.1 [[19718800] giJ50659087|ref|NM_133378.2[[50659087] gi[20143915|ref|NM_133379.l [[20143915] gi|19747278iref[NM_133430.l i[19747278] gijl9747280|ref|NM_133431.1 |[19747280] gi|20143917|ref|NM_133432.1 |[20143917] gi[47578104[ref|NM_133433.2i[47578104] gi|19718773|ref(NM_133436.11[19718773] giJ20143921 |ref|NM_133437.l [[20143921] gi|19743890|ref|NM_133439.11[19743890] gi|19263339|ref|NM_133443.1 j[19263339] gi|45331216Jref|NM_133444.1 ][45331216] gi|20143963|ref|NM_133445.l [[20143963] gi|19263342|ref|NM_133446.11[19263342] giJ42734372Jref|NM_133448.1 [[42734372] giJ42734374Jref|NM_133450.11[42734374] giJ3345733l jref|NM_133452.1 j[33457331] giJ50511940|ref|NM_133455.2J[50511940] gi|31442419|ref|NM_133456.1 |[31442419] gijl 9263346 jref|NM_133457.1 j[19263346] giJ3993051 Olref|NM_133459.1 [[39930510] gi[33457329JrefJNM_133462.1 [[33457329] gi|28274685|ref|NM_133466.11[28274685] gi[38455425[ref|NM_133467.2|[38455425] gi|38327519[ref|NM_133468.2|[38327519] gi[32698774Jref|NM_133473.11[32698774] giJ51036585|ref|NM_133474.li[51036585] gi|36054193[ref|NM_133476.2|[36054193] gi[l9743826|ref[NM_133478.l j[19743826] gi[l9743829Jref|NM_133479.l [[19743829] gi|19743893|ref|NM_133480.l [[19743893] gijl 9743895|refJNM_133481.1 [[19743895] gi|19924130|ref|N _133482.1 j[19924130] gi|19311007Jref [NM_133483.1 [[19311007] gi|19743570|ref|NM_133484.1 |[19743570] gi|30315648Jref|NM_133486.1 j[30315648] gi|19924134|ref|NM_133487.1 |[19924134]
gi|19424121|ref|NM_133489.1 |[19424121] gi|27436994|ref[NM_133490.2|[27436994] gi|34147485Jref|NM_133491.2J[34147485] gi|19424127|ref|NM_133492.1|[19424127] gi[l9424129[reflNM_133493.1|[19424129] gijl 9424131 [ref |NM_133494.11[194241311 gi[41529827[ref|NM_133496.3|[41529827] gi|28329446|ref|NM_133497.2J[28329446] gi|33563295|ref|NM_133498.2J[33563295] gijl9924096|refJNM_133499.1 [[19924096] gi|19743800|ref|NM_133502.1 |[19743800] gi[47419926Jref[NM_133503.2|[47419926] gi J47419924|ref |NM_ 33504.2 j[47419924] gi|47419923Jref|NM_133505.2)[47419923] gi|47419922|ref|NM 33506.2|[47419922] gi|47419921 jref |NM_133507.2J[47419921] gi|46255038[ref|NM_133509.2|[46255038] gi|46255037|ref|NM_133510.2|[46255037] giJ46399195[ref|NM_133625.2J[46399195] gij 19924120|ref|NM_133627.1 [[19924120] gi|19924122|ref|NM_133628.1|[19924122] gi[l9924124|ref|NM_133629.1|[19924124] gijl9924126Jref|NM_133630.l[[19924126] gi|19743805|ref|NM_133631.1|[19743805] gijl9924106Jref|NM_133632.l j[19924106] gi[l9924108|ref|NM_133633.1|[19924108] gi|49574586|ref|NM_133634.2|[49574586] giJ49574587Jref|NM_133635.3i[49574587] gi|19525732Jref|NM_133636.1 j[19525732] gijl9525734[ref|NM_133637.1|[19525734] gi|19525736|ref|NM_133638.1|[19525736] gi|20070359iref|NM_133639.2J[20070359] gi[31652219|ref|NM_133640.3|[31652219] gi|1992414θ[ref|NM_133642.1|[19924140] giJ41281652|ref |NM_133644.1 j[41281652] gijl9882216Jref|NM_133645.lj[19882216] gi|19526766|ref|NM_133646.1 [[19526766] gi[2277993l[ref|NM_133650.li[22779931] gi|19913364Jref|NM_134258.l[[19913364] gij 19913366[ref JNM_134259.11[19913366] gijl974390θjref|NM_134260.l[[19743900] gi| 19743902|ref|NM_134261. 1[19743902] gi| 19743904|ref |NM_134262.11[19743904] giJ20336276[ref|NM_134263.1|[20336276] gi|20143909[ref|NM_134264.1 [[20143909] giJ20143911 |ref|NM_134265.11[20143911] gi|20336281|ref|NM_134266.lj[20336281] gi[38454323JrefJNM_134268.3|[38454323] gijl9913395Jref|NM_134269.l[[19913395] giil9913397|ref|NM_134270.1j[19913397] gi|19743839|ref|NM_134323.lj[19743839] gi[l974384ljrefJNM_134324.1|[19743841] gi|20336287|ref[NM_134325.1|[20336287] gij 19913442|ref JNM_134421.11[19913442] gi|20143953[ref|NM_134422.1|[20143953] gi|20143955|ref|NM_134423.1|[20143955] gi|20143957|ref|NM_134424.11[20143957]
gi|20336273|ref|NM_134425.1 |[20336273] gi[20336278iref[NM_134426.lj[20336278] gi| 19913391 |ref|NM_134427.1 |[19913391] gi|19743883|ref|NM_134428.1|[19743883] gi|19913402|ref|NM_134431.1 |[19913402] gijl9743878|ref|NM_134433.l j[19743878] gi[20143929|ref|NM_134434.1 |[20143929] gijl9924155JrefJNM_134440.1|[19924155] gi|20143932|ref|NM_134441.1|[20143932] gii22219460Jref[NM_134442.2i[22219460] gijl9745161 |ref|NM_134444.1 |[19745161] gi[41281655(ref|NM_134445.1 |[41281655] giJ41281669Jref|NM_134446.1 |[41281669] gi| 19924160|ref |NM_134447.1 j[19924160] gi|24430221 |ref|NM_134470.2|[24430221] giJ20336204|ref|NM_138270.l j[20336204] gi|20336206Jref|NM_138271.1 [[20336206] gi|19913374JrefJNM_138272.l j[19913374] gijl9913376Jref|NM_138273.1|[19913376] gi|19913378Jref|NM_138274.1 [[19913378] gi|19913380|ref|NM_138275.11[19913380] gijl9913382Jref|NM_138276.lj[19913382] gij 9913384Jref|NM 38277.1 |[19913384] gi|19923079[ref|NM_138278.1 |[19923079] gijl 9923712Jref|NM_138279.11[19923712] gi|45580744|ref|NM 38280.3|[45580744] gi[2014396l irefJNM_138281.1 [[20143961] gi|20357538[ref|NM_138282.1 |[20357538] gij 19882258 jref JNM_138283.1 j[19882258] gijl 9923714|ref|NM_138284.1 [[19923714] gi|31982903|ref|NM 38285.2|[31982903] gijl9923077irefJNM_138286.lj[19923077] gi|31377615Jref|NM_138287.2J[31377615] gi|46358345Jref|NM_138288.2J[46358345] gi[ 19923075|ref|NM J 38289.1 [[19923075] gi|19923089|ref|NM_138290.11[19923089] giJ20336198|ref|NM_138292.1 |[20336198] gi|20336200Jref|NM_138293.1 |[20336200] gi|19923081 |refJNM_138294.1 j[19923081] giJ33359209Jref|NM_138295.2J[33359209] gi|19923720|ref|NM 38296.1 [[19923720] gi|20143940|ref|NM_138297.l [[20143940] gi|20143919|ref|NM_138298.l[[20143919] gi[20143923JrefJNM_138299.l j[20143923] gi|34147514|ref|NM_138300.2|[34147514] giJ23510397Jref|NM_138316.2j[23510397] gi|20143943|ref|NM_138317.1 [[20143943] giJ20143945|ref[NM_138318.1 |[20143945] gi|20336179Jref|NM_138319.l j[20336179] gi|20336181 jrefJNM 38320.1 [[20336181] gi|20336183[ref|NM_138321.1 |[20336183] gi[20336185|ref|NM_138322.1 |[20336185] gi[20336187|ref|NM_138323.1 [[20336187] giJ20336189|ref[NM_138324.l j[20336189] gi|27894284|refJNM_138325.2J[27894284] gi|21264327|ref|NM 38326.l j[21264327] gi|21264323JrefJNM 38327.1 |[21264323]
gi[21264325|ref|NM_138328.1|[21264325] gi|21264319|ref|NM_138329.1|[21264319] gi|21264321|ref|NM_138330.1|[21264321] gi|21687142|ref|NM_138331.1|[21687142] gi|49472829|ref|NM_138333.3|[49472829] gi|19923878|ref|NM_ 38334.1|[19923878] gij 1992388θ[ref|NM_138335.1 j[19923880] gi| 19923882Jref|NM_138336.11[19923882] giJ42490739|ref|NM_ 38337.3|[42490739] giJ24475813|ref|NM_138338.1|[24475813] gi|34304337[ref|NM_138340.3[[34304337] gi|24308397[ref|NM_138341.lj[24308397] giJ31543092Jref|NM_138342.2J[31543092] gi|41871964|ref|NM_138343.2J[41871964] gi|31543094|ref |NM_138344.2|[31543094] gi|25286702Jref|NM_138346.lj[25286702] gi|34147525|reflNM_138347.2|[34147525] gi[34222357iref|NM_138348.3[[34222357] giJ34147526[ref|NM_138349.2[[34147526] gi|42734378|ref|NM_138350.2J[42734378] gi|39930516Jref|NM_138352.11[39930516] gi|24308405Jref|NM_138355.l[[24308405] gi|46195760|ref|NM_138356.1|[46195760] gi[24308399Jref|NM_138357.1 j[24308399] giJ34328079|ref|NM_138358.2J[34328079] gi|33457327|ref|NM_138360.1 ([33457327] gi|31543096Jref|NM_138361.2[[31543096] giJ30794245|ref|NM_138362.lj[30794245] gi|19923898|ref|NM_138363.1)[19923898] gi|4053877liref|NM_138364.2J[40538771] gi|38679899|ref|NM_138368.2|[38679899] gi[34147528[refJNM_138369.1|[34147528] giil992390θ[ref|NM_138371.l[[19923900] gi|2168726ljrefJNM_138372.lj[21687261] gi|31377612|ref|NM_138373.2|[31377612] gi|24308407|ref[NM_138375.lj[24308407] gi|24308431|ref|NM_138376.1|[24308431] gi|19923904|ref|NM_138379.1|[19923904] gij 19923906Jref|NM_138381.1 [[19923906] gi|46195762|ref|NM_138383.1|[46195762] gi|24308425|ref|NM_138384.11[24308425] gi[l9923908[refJNM_138385.li[19923908] gij 19923910|ref|NM_138386. lj[19923910] giJ46852181 jref|NM_138387.2|[46852181] gi|29789372|ref|NM_138389.11[29789372] gi|31982893|ref|NM_138390.2|[31982893] gi(34222351 [ref(NM_138391.3J[34222351] gi|19923916|ref|NM_138392.li[19923916] gi|19923918|ref|NM_138393.lj[19923918] giJ20149708JrefJNM_138394.2J[20149708] gi|24308435|ref|NM_138395.1 [[24308435] gi[40255015|ref|NM_138396.3J[40255015] gi|19923924[ref|NM_138397.1|[19923924] giJ38679901|ref|NM_138399.2J[38679901] gi[24308439[ref|NM_138401.1|[24308439] gi|2014971θ[ref|NM_138402.2[[20149710] gi[40286635(ref[NM_138403.3|[40286635]
gi|50355976|ref|NM_138408.2[[50355976] giJ24308441 |ref|NM_138409.1 [[24308441] gi|31657097|refJNM_138410.2|[31657097] gi|34147533Jref|NM_138412.2|[34147533] gi|31543059Jref|NM_138413.2|[31543059] gi|19923934|ref|NM 38414.11[19923934] gi(19923936|ref|NM_138415.l[[19923936] gi[34147534|ref|NM_138416.1|[34147534] gi|49472811|ref|NM_138417.2|[49472811] gi|33300669Jref|NM_138418.2|[33300669] gi|39653314|ref|NM_138419.2|[39653314] gi|19923942[ref|NM_138421.11[19923942] gij 19923944|ref|NM_138422.11[ 9923944] gi|29826288Jref|NM_138423.2J[29826288] gi| 19923948|ref |NM_138424.11[19923948] gi|34147536|ref|NM_138425.2|[34147536] gi|31377613Jref|NM_138428.2|[31377613] gi|20149319|ref|NM_138429.1|[20149319] gi|40316948Jref|NM_138430.3J[40316948] gi|19923956|ref|NM_138431.1|[19923956] gi|34147537|ref|NM_138432.2|[34147537] gi(34330156|ref|NM_138433.2|[34330156] gi[49574496irefiNM_138434.2|[49574496] gi)19923962|ref|NM_138435.11[19923962] gi[34147538Jref|NM_138436.2|[34147538] gi|45238856|ref|NM_138437.2|[45238856] giJ34147539|ref|NM_138439.lj[34147539] gi|39930520|ref|NM_138440.1|[39930520] gi|31581597|ref|NM_138441.1 [£31581597] gi|34147541|refJNM_138442.2J[34147541] gi|34147542|ref|NM_138443.2|[34147542] gi|40255011|refJNM_138444.2J[40255011] gi|41349503Jref[NM_138445.2|[41349503] gi| 19923976|ref |NM_138446.11[19923976] gi|34147543JrefJNM_138447.1|[34147543] gi|20336764|ref|NM_138448.2)[20336764] giJ30348973[ref|NM_138450.3J[30348973] gi| 19923980|ref |NM_138451.11[19923980] gi| 19923982 jref|NM_138452.11[19923982] gi(34147545|ref[NM_138453.2|[34147545] gi|19923986|ref|NM_138454.1|[19923986] gi|34147546|refJNM_138455.2|[34147546] gi|45238853|ref|NM_138456.3|[45238853] gi|29789366Jref|NM_138457.11[29789366] gi[24308443(ref[NM_138458.l[[24308443] gii34147547(ref|NM_138459.2|[34147547] gi|32130531 |ref|NM_138460.2|[32130531] gijl9923994iref[NMJ38461.lj[19923994] gi|37594445|ref|NM_138462.2|[37594445] gi[34147548JrefJNM_138463.2[[34147548] gi|41152252|ref|NM_138465.3|[41152252] gi|42734380jref|NM_138467.11[42734380] giJ22095387|ref|NM_138468.3J[22095387] gi|39930522|ref|NM_138471.1|[39930522] gi|38372900JrefJNM_138473.2J[38372900] gi[33457310|ref|NM_138476.2|[33457310] gi|33147083jref|NM_138477.1 [[33147083]
gi|42734382|ref|NM_138479.11[42734382] gi[l9924014JrefJNM_138482.lj[19924014] gi|24308447Jref|NM_138484.1 j[24308447] giJ24308449JrefiNM_138487.1|[24308449] giJ45505153|ref|NM_138492.4|[45505153] gi|28416426|refJNM_138494.1|[28416426] gi|19924022|ref[NM_138497.1 [[19924022] gi|46358349JrefJNM_138499.2|[46358349] gi|50726974|refJNM_138501.4J[50726974] giJ42544204|ref|NM_138551.2|[42544204] gi|2033631θjref|NM_138553.1|[20336310] gi|19924148[ref|NM_138554.1|[19924148] gi|20143966[ref|NM_138555.1|[20143966] gi| 19924150|refJNM_138556.1 [[19924150] gi|19924152|ref|NM_138557.1|[19924152] gi|20336238|ref|NM_138558.1|[20336238] gi|20336312(ref|NM_138559.1[[20336312] giJ20302144Jref|NM_138563.l[[20302144] giJ20302146|ref|NM_138564.1|[20302146] giJ20357555|ref|NM_138565.1|[20357555] gi|20336215|ref|NM_138566.1|[20336215] gi|32528307|ref|NM_138567.2|[32528307] gi[29788775Jref|NM_138568.2|[29788775] giJ20070373|ref|NM_138569.1|[20070373] giJ20070375Jref|NM_138570.1|[20070375] gi|21359981|ref|NM_138571.3|[21359981] gi J20070377|ref (NMJ 38572. lj[20070377] giJ20070379Jref|NM_138573.1|[20070379] giJ20070381|ref|NM_138574.lj[20070381] giJ20070383|ref|NM 38575.11[20070383] gi|49574493Jref|NM_138576.2|[49574493] gi|20336334Jref[NM_138578.lj[20336334] giJ41281674iref|NM_138608.li[41281674] gi|20336745|ref|NM_138609.11[20336745] gi|20336747|ref|NMJ 38610.11[20336747] gi|20302154|ref|NM_138612.1|[20302154] giJ20336291 jrefJNM_138614.1 j[20336291] gi|20336293|ref|NM_138615.1|[20336293] gi|20336218Jref|NM_138616.lj[20336218] gi|20336220|ref|NM_138617.1|[20336220] gi|20336222Jref|NM_138618.11[20336222] gi|20336266JrefJNM_138619.lj[20336266] gi|20336297|ref|NM_138620.1|[20336297] giJ46276872Jref|NM_138621.2J[46276872] gi|46276873|ref|NM_138622.2J[46276873] giJ46276874iref|NMJ38623.2|[46276874] gi|46276875|ref|NM_138624.2|[46276875] gii46276876JrefiNM_138625.2i[46276876] gi|46276877|ref|NM_138626.2J[46276877] giJ46276878Jref|NM_138627.2J[46276878] gi|20336762|ref|NM_138632.11[20336762] gi|20336195|ref|NM_138633.1|[20336195] gi|20302157Jref JNM_138634.11[20302157] giJ41406072|ref|NM_138635.2J[41406072] gi|45935389JrefJNM_138636.2|[45935389] gi|41281671 jref|NM_138637.11[41281671] gi|33946277|ref|NM_138638.11[33946277]
gi[20336329(ref[NM_138639.1|[20336329] giJ20336263Jref|NM_138640.1|[20336263] giJ24797109|ref|NM 38643.1 [[24797109] gi|24797111 jref|NM_138644.11[24797111] gi|20336255Jref |NM_138687.11[20336255] giJ20302170|ref|NM_138688.11[20302170] giJ21071069JrefJNM_138691.2J[21071069] gi|20162553|ref|NM_138693.1|[20162553] gi|25777664Jref|NM_138694.2i[25777664] gi|29294614|refJNM_138697.2|[29294614] gi|38016157|ref|NM_138698.2|[38016157] gi|45433512|ref|NM_138699.2|[45433512] giJ33469990Jref|NM_138700.2[[33469990] gi]20162565|ref|NM_138701.11[20162565] gi|20162567Jref[NMJ 38702.1 [[20162567] gi|49574499|ref|NM_138703.3[[49574499] gi|29826297|ref|NM_138704.2|[29826297] giJ50897265[ref|NM_138705.2J[50897265] gi|47271458|refJNM_138706.2|[47271458] giJ20336474Jref |NM_138707.11[20336474] gi|41281682|ref|NM_138709.1|[41281682] gi|20336232Jref|NM_138711.1 [[20336232] gi|20336234|ref|NM_138712.1|[20336234] giJ27886522JrefJNM_138713.2|[27886522] gi|27886519|ref|NM_138714.2[[27886519] gi|20357511 jrefJNM_138715.1 [[20357511] gi|20357514(ref|NM_138716.1|[20357514] giJ20336250|ref|NM_138717.lj[20336250] gi)20336284|ref|NM_138718.1|[20336284] gi|20336751|ref|NM_138720.1 [[20336751] gi|21040327|ref|NM_138722.1|[21040327] gi(21040329Jref(NM_138723.li[21040329] giJ2104033ljref|NM_138724.lj[21040331] gi|25952067|ref|NM_138726.2J[25952067] gi[38201634[ref|NM_138727.2|[38201634] gi|38201635|ref|NM_138728.2|[38201635] gi|38201636|ref|NM_138729.2[[38201636] giJ23238230|ref|NM_138730.1|[23238230] gi|40255033|ref|NM_138731.2|[40255033] gi[21166379[ref|NMJ38732.li[21166379] gi|31543396|ref|NM_138733.2|[31543396] gi|21166381 jrefJNM 38734.1 j[21166381] gi|21070966|ref|NM_138735.1|[21070966] giJ41281684JrefJNM_138736.1 [[41281684] giJ21166384[refjNM_138737.1 j[21166384] gi|20270262|ref |NM_138738.11[20270262] giJ20270264JrefiNM_138739.11[20270264] gi|41281687|ref|NM_138740.lj[41281687] gi|34335117|ref|NM_138761.2|[34335117] gi|34335120|ref|NM_138762.2|[34335120] gi|34335121|ref|NM_ 38763.2|[34335121] gi|34335124|ref|NM_138764.2|[34335124] giJ34335125Jref|NM_138765.2J[34335125] gi[21070973|ref|NM_138766.11[21070973] gi|4279461 liref[NM_138768.2J[42794611] gi|20270302|refJNM_138769.1 j[20270302] gi|20270306(ref[NM_138770.1[[20270306]
gi[40255028|ref|NM_138771.2|[40255028] gi|20270310|ref|NM_138773.1 |[20270310] giJ3422214θjref|NM_138774.2J[34222140] gi|20270314|ref|NM_138775.1 |[20270314] gi|40317613|ref|NM_138777.2|[40317613] gi|24308451|ref|NM_138778.11[24308451] gi|34222136|ref|NM_138779.2|[34222136] gi|20270304JrefJNM_138780.1 [[20270304] gi|49574540jref|NM_138781.2|[49574540] gi|20270322irefJNMJ 38783.1 [[20270322] gi|20270324|ref|NM_138784.1 [[20270324] gi|20302037|ref|NM_138785.1 |[20302037] gi|20270326Jref JNM_138786.11[20270326] gi|42476215|ref|NM_138787.2|[42476215] gi|31543061 |ref|NM_138788.2|[31543061] gi|37059737|ref|NM 38789.2|[37059737] gi|20270334|ref|NM_138790.1 [[20270334] giJ20302039Jref[NM_138791.11[20302039] giJ37059738|ref|NM_138792.2J[37059738] gii31543621 |ref|NM_138793.2J[31543621] gi|20270340|ref|NM_138794.11[20270340] giJ38679903|ref|NM_138795.2|[38679903] gi|37059739Jref|NM_138796.2|[37059739] giJ20270346Jref|NM_138797.1 |[20270346] gi|20270348|ref|NM_138798.11[20270348] gi|40548386|ref|NM_138799.2|[40548386] gi|20270352|ref|NM_138800.1 |[20270352] giJ20270354Jref|NM_138801.1 j[20270354] giJ20270356Jref|NM_138802.11[20270356] gi|3137761 OJref |NM_138803.2|[31377610] gi|21314782|ref|NM 38804.2|[21314782] gi|38455417|ref [NM_138805.2J[38455417] gi[41327722Jref|NM_138806.3i[41327722] gi|34147617|ref|NM_138807.2J[34147617] gi[20270368Jref|NM_138808.1 [[20270368] gi|31982874Iref|NM_138809.2J[31982874] giJ23199968|ref|NM_138810.2|[23199968] gi|38570063|ref|NM_138811.3|[38570063] giJ20270376|ref|NM_138812.l j[20270376] giJ44888834[ref|NM_138813.1 [[44888834] gi|20304126|ref|NM_138814.1 |[20304126] gi|31982875Jref|NM_138815.2J[31982875] gi|20270382|ref|NM_138817.1 |[20270382] gi[34147618Jref|NM_138818.2|[34147618] giJ20270386[ref|NM_138819.1 [[20270386] gi|20270388|ref|NM_138820.11[20270388] gi|21070979Jref|NM_138821.1 |[21070979] gi[2107098l [refJNM_138822.1 [[21070981] giJ20357587|ref|NM_138923.11[20357587] gi|20336211 [ref|NM_138924.1 [[20336211] gi|21040319[ref|NM_138925.1 |[21040319] gi|2104032l jrefJNM_138926.1 [[21040321] gi|21040325|ref|NM_138927.l [[21040325] gi|22027531 Jref|NM_138928.11[22027531] giJ42544196|refJNM_138929.2|[42544196] giJ42544194irefJNM_138930.2|[42544194] gi|21040335|ref|NM_138931.1|[21040335]
gi|20357574|ref[NM_138932.11[20357574] gi|20357577Jref|NM_138933. lj[20357577] giJ20336252Jref|NM_138934.1 [[20336252] gi|21070992|ref|NM_138937.1 |[21070992] gi|21070994Jref|NM_138938.l[[21070994] gi|25777654|ref|NM_138939.1|[25777654] gi|25777656Jref|NM_138940.li[25777656] gi[20986530|ref|NM_138957.1|[20986530] gi|21071002Jref|NM_138958.11[21071002] gi|20373170|ref|NM_138959.1|[20373170] gi|3432894θjref|NM_138960.3i[34328940] gi|20452463|ref|NM_138961.1 [[20452463] giJ2512199ljref|NM_138962.2J[25121991] gi]20373176|ref|NM_138963.lj[20373176] giJ44921613Jref|NM_138964.2|[44921613] gi|23510394|ref|NMJ38966.2J[23510394] gi[42544128Jref(NM_138967.2J[42544128] gi|40807362|ref|NM_138969.2[[40807362] giJ41350304Jref|NM_138970.2|[41350304] gi|46255012Jref|NM_138971.2[[46255012] gi|46255013|ref|NM_138972.2|[46255013] gi|46255014|ref(NM_138973.2|[46255014] gi|20986505[ref[NM_138980.lj[20986505] giJ20986507Jref|NM_138981.1|[20986507] gi|20986509[ref|NM_138982.1 [[20986509] gi|41281694|ref|NM_138983.1|[41281694] gi|21040359(ref(NM_138991.1|[21040359] giJ2104036l[ref|NM_138992.lj[21040361] gi|47519715|ref|NM_138993.2][47519715] gi[20544136iref|NM_138994.1|[20544136] gi|20428778|ref|NM_138995.11[20428778] gi[2054414l[ref|NM_138996.1|[20544141] giJ21040372Jref|NMJ38998.1|[21040372] gi|20452469|ref|NM_138999.11[20452469] giJ21040338Jref|NM_139002.1|[21040338] gi|21040340|ref|NM_139003.1|[21040340] gi|21040342JrefJNM_139004.lj[21040342] gi|21040344|ref|NM_139005.l[[21040344] gi|21040346|refJNM_139006.lj[21040346] giJ21040348Jref |NM_139007.1 j[21040348] giJ21040350|ref|NM_139008.1 |[21040350] gi|21040352|ref|NM_139009.1 j[21040352] gi|21040354Jref|NM_139010.1|[21040354] gi|21040356|ref|NM_ 39011.11[21040356] gi|20986511 |ref |NM_139012.1 j[20986511 ] gi|20986513Jref|NM 39013.1 j[20986513] gi|20986515Jref|NM_139014.li[20986515] gi[3753769θjref|NM_139015.3|[37537690] giJ31982870|ref|NM_139016.2|[31982870] gi|38455420|ref|NM_139017.3|[38455420] gi|38455397|ref JNMJ 39018.2|[38455397] giJ20502985[ref|NM_139021.1 j[20502985] gi|3759553l[ref|NM_139022.2J[37595531] gi|37595532|ref|NM_139024.2|[37595532] gi|21265033|ref|NMJ39025.1|[21265033] gi|21265042|ref|NM_139026.11[21265042] gi|21265045|refJNM_139027.1|[21265045]
gi|21265048|ref|NM_139028.1 |[21265048] gi|34328914Jref|NM_139030.2J[34328914] gi|20986498|ref|NM 39032.11[20986498] giJ2098650θjrefJNM_139033.1 j[20986500] gi|20986502|ref|NMJ 39034.11[20986502] gi|21071045Jref|NM_139035.1 j[21071045] gi|48255897|ref|NM_139045.2J[48255897] gi|20986518|ref|NM_139046.1|[20986518] giJ2098652θjref|NM_139047.1 j[20986520] gi|21071053|ref|NM_139048.11[21071053] gi|20986522Jref|NM_139049.1 j[20986522] gi|21071017|ref|NM_139053.1 |[21071017] gi|40806184|ref|NM_139054.2|[40806184] giJ21265057JrefJNM_139055.lj[21265057] gi|2126506θ[ref|NM_139056.1 |[21265060] gi|21265063|ref|NM_139057.1 |[21265063] giJ24497588Jref(NM_139058.li[24497588] gi|20544144|ref|NM_139062.1 [[20544144] gi|21237807JrefJNM_139067.1[[21237807] gi|21237738|ref[NM_139068.1 |[21237738] gi|21237741 |ref|NM_139069.1 |[21237741] gi|21237744JrefJNM_139070.1 |[21237744] giJ21264347Jref|NM_139071.1 |[21264347] gi|31542542|ref|NM_139072.2|[31542542] gi|31712019|ref|NM_139073.2|[31712019] gi|30061486|ref|NM_139074.2|[30061486] giJ20589957|ref|NM_139075.1|[20589957] gi|20589960|ref|NM_139076.lj[20589960] giJ21237767|refJNM_139078.lj[21237767] gi|20986487|ref|NM_139118.1 |[20986487] gi|20986489|refJNM_139119.1 |[20986489] giJ2098649l iref|NM_139120.lj[20986491] gi|20986494|refiNM_139121.lj[20986494] giJ21536354Jref|NM_139122.1J[21536354] gi|21536356Jref|NM_139123.1 J[21536356] gi|21237774|ref|NM_139124.1 j[21237774] gi|21264358|ref|NM_139125.l [[21264358] gi|22538483|ref|NM_139126.2|[22538483] gi|2126436δjrefJNM_139131.1J[21264368] giJ2126437θjref|NMJ 39132.11[21264370] gi|21264574|ref|NM_139135.1 |[21264574] giJ24497456Jref[NM_139136.2i[24497456] gi[24497455|ref|NM_139137.2|[24497455] gi|21265054|refJNM_139155.lj[21265054] gi|3414762θjref|NM_139156.1 |[34147620] gi|47132530|ref|NM_139157.2|[47132530] giJ21040234|refJNM_139158.1 j[21040234] giJ26080432|ref|NM_139159.2|[26080432] gi|21040238Jref|NM_139160.1 [[21040238] gi|28144891 |ref|NM_139161.2J[28144891] gi|22538487|ref|NM_139162.2|[22538487] giJ21040244|ref|NM_139163.lj[21040244] gi[21040246JrefJNM_ 39164.l [[21040246] giJ21040248JrefJNM_139165.1 j[21040248] gi|31543660|ref |NM_139166.2|[31543660] gi|21040252|ref|NM_139167.1 |[21040252] gi|21040254Jref|NM_139168.1|[21040254]
gi|34303921 |ref|NM_139169.3|[34303921] gi|21040258|ref|NM_139170.1|[21040258] giJ2104026θjref|NM_139171.1 j[21040260] giJ21040262|ref|NM_139172.1 j[21040262] gi|21040264|ref |NM_139173.11[21040264] gi|31543071 [ref |NM_139174.2|[31543071] gi|21040268|ref|NM_139175.1|[21040268] gi|46049099|refJNM_139176.2J[46049099] gi|31542247JrefJNM_139177.2|[31542247] gi|21040274|refJNM_139178.1|[21040274] gi|21040276|ref|NM_139179.1|[21040276] giJ21264596|ref|NM_139181.lj[21264596] gi|21264593JrefJNM_139182.1|[21264593] giJ34452706|ref|NM_139199.1|[34452706] gi|21237793[ref|NM_139201.1 [[21237793] gi|21237731 |ref|NM_139202.1 |[21237731] gi|21264609|ref|NM 139204.11[21264609] gi|21237798|ref|NMJ39205.1|[21237798] gi|21327707|ref|NM 139207.1 j[21327707] gi|21264360|ref|NMJ39208.1 |[21264360] gi|21166358|ref|NMJ39209.1 |[21166358] gi|47717121 |ref|NMJ39211.2|[47717121] gi|47717120|ref|NM_139212.2|[47717120] gi|34328942[ref|NM_139214.2|[34328942] gi[2132770θjref|NMJ39215.1 |[21327700] gi|39777589Jref|NMJ39235.3|[39777589] gii21327692Jref|NM_139238.1|[21327692] gi(41281708 [ ref | NMJ 39239.1|[41281708] giJ31543098Jref|NMJ39240.2|[31543098] gi|21245127|ref|NMJ39241.1|[21245127] gi|31543267|ref|NMJ39242.2|[31543267] giJ21245123JrefJNMJ39243.1|[21245123] gi|31652246irefJNMJ39244.2|[31652246] gi|21245119Jref|NMJ39245.1|[21245119] gi|3137760θjref[NMJ39246.3i[31377600] gi|24497586|ref|NMJ39247.2|[24497586] gi[21245105[ref[NMJ39248.1([21245105] giJ23110998iref|NMJ39249.2J[23110998] gi|21281684|ref|NMJ39250.1|[21281684] gi|21327694|ref|NMJ39264.1 |[21327694] gi|34147619|ref|NMJ39265.2|[34147619] giJ2153630θjref|NMJ39266.1|[21536300] gi[21450855|ref|NMJ39267.1|[21450855] gi|21269876JrefJNMJ39273.li[21269876] gi|2126987ljref|NMJ39274.l[[21269871] gi|21493034|ref|NMJ39275.1 [[21493034] gi|47080104|ref|NM J 39276.2|[47080104] giJ21327704[ref|NMJ39277.lj[21327704] giJ45643139|ref|NMJ39278.2|[45643139] gi|46852158[refJNMJ39279.3J[46852158] gi|27544926Jref|NMJ 39280.1 [[27544926] gi|21281676Jref|NMJ39281.1 |[21281676] gi[23943885Jref|NMJ39282.li[23943885] giJ21281678Jref|NMJ39283.lj[21281678] giJ21281672|ref|NMJ39284.1|[21281672] gi|21281674[ref|NMJ39285.lj[21281674] gi|38683841 |ref|NMJ39286.3|[38683841]
121493038 | refl JNM 139289.11(21493038] |21328450 [ref JjNM. 139290.1 [[21328450] 121327684 jref J |NM 139312.1 |[21327684] |21327686 |ref J|NM 139313.1 [[21327686] 121536397 jref J |NM 139314.11(21536397] 121536358 |ref J|NM .139315.1 |[21536358] J21536422 |ref :||NM 139316.11(21536422] 121536420 |ref| |NM 139317.1 |[21536420] J27886644 jref J |NM..139318.3 [[27886644] 121322233 |ref J|NM 139319.1 |[21322233] J23312387 |ref j|NM 139320.11(23312387] [21450860 |ref J|NM..139321.1 j[21450860] 121450862 jref JjNM .139322.1 |[21450862] [31742480 |ref j|NM .139323.2 |[31742480] 121536399 |ref J|NM..139343.1 [[21536399] 121536401 | ref | JNM 139344.11(21536401] 121536403 |ref J|NM..139345.1 |[21536403] 121536406 |ref J|NM 139346.1 [[21536406] 121536408 |ref| |NM 139347.1 [[21536408] 121536410 |ref J|NM..139348.1 |[21536410] 121536412 |ref J|NM 139349.1 |[21536412] 121536414 |ref| |NM .139350.1 |[21536414] 121536416 |ref J|NM 139351.11(21536416] 121536361 |ref J|NM..139352.1 [[21536361] 121536366 jref JjNM 139353.1 |[21536366] 121450843 |ref J|NM 139354.1 [[21450843] [21450845 | refl JNM. 139355.11(21450845] [21464133 |ref JjNM 144488.11(21464133] J21464135 [ref JjNM..144489.1 |[21464135] J21464141 jref JjNM. 144490.11(21464141] |21536293 [ref J|NM 144492.1 |[21536293] 141281711 [ref J|NM..144494.1 |[41281711] [41281714 jref J|NM..144495.11(41281714] 121493023 Jref JjNM 144497.1 [[21493023] J21450852 |ref J|NM .144498.11(21450852] 122027521 jref J|NM..144499.1 [[22027521] [21464106 |ref J|NM 144501.11(21464106] 121464108 |ref J|NM .144502.1 |[21464108] [21464110 | ref J|NM .144503.11(21464110] [21464112 | ref J|NM. 144504.11(21464112] J21464126 lref J|NM .144505.1 |[21464126] 121464128 | ref JjNM .144506.1 |[21464128] 121464130 [refj |NM 144507.1 [[21464130] [24476009 (ref| |NM 144508.1 [[24476009] 121389336 |ref| |NM 144563.11(21389336] 147080101 |ref| |NM 144564.4 [[47080101] [46485464 |ref J|NM. 144565.2 [[46485464] |36030945 |ref J|NM. 144567.2 |[36030945] 121389334 |ref| [NM 144568.11(21389334] [47271474 | ref j|NM .144569.3 |[47271474] |46361989 | ref J|NM .144570.2 |[46361989] [21687267 jref J |NM..144571.1 |[21687267] [21389338 jref J |NM..144573.11(21389338] J31317279 [ref J|NM. 144574.2 |[31317279] 141152100 [ref J|NM 144575.2 j [41152100] [31542761 jref J|NM .144576.2 |[31542761] [21389346 |ref J|NM. 144577.11(21389346]
134147697 |ref j|NM .144578 |[34147697] [21389350 |ref J|NM 144579 [[21389350] J243O7870 |ref J|NM 144580 |[24307870] [21389360 |ref j|NM 144581 j[21389360] |4251 8075 |ref J|NM. 144582 1(42518075] |4771 7097 |ref J|NM 144583 |[47717097] [21389358 [ref J|NM..144584 j[21389358] [24497484 [ref J|NM .144585 1(24497484] [40255056 jref jjNM..144586 j[40255056] I50345832 | ref | |NM 144587 1(50345832] [50557643 jref| |NM .144588 [[50557643] 131542746 (ref| |NM .144589 |[31542746] [21389370 jref] |NM .144590 [[21389370] [21389380 |refj |NM..144591 [[21389380] [21389384 jref | |NM .144593 [[21389384] [21389378 jref] |NM .144594 [[21389378] 131542757 |ref| |NM .144595, [[31542757] [21389382 |ref JjNM..144596 1(21389382] J21389392 |ref JjNM..144597, [[21389392] [33285014 [ref JjNM .144598, 1(33285014] |41406090 [ref J|NM..144599, |[41406090] [21389390 |ref J|NM .144600, [[21389390] 132130524 |ref J|NM..144601 , |[32130524] J37059750 |ref J|NM .144602, j[37059750] 134222190 |ref J|NM..144603, j[34222190] [31377594 jref J |NM .144604, |[31377594] |21389408 |ref J|NM..144605, j[21389408] J34335183 jref JjNM 144606. j[34335183] J40255059 |ref J|NM..144607. |[40255059] [21389406 | ref J |NM .144608. j[21389406] [21389416 |ref J|NM .144609. [[21389416] [21389410 | ref J |NM 144610. |[21389410] 140255061 |ref j|NM .144611. |[40255061] I42734440 |ref J|NM..144612. 1(42734440] J40538807 |ref j|NM 144613. [[40538807] [28144903 jref J |NM .144614. |[28144903] J21389428 [ref J|NM .144615. [[21389428] 139725701 |ref j|NM 144616. |[39725701] [21389432 jref J |NM .144617. |[21389432] [21389426 |ref J|NM .144618. |[21389426] J21389430 |ref J|NM .144620. [[21389430] J31377596 |ref j|NM .144621. 1(31377596] 121389434 jref J |NM ,144622. |[21389434] J21389444J |ref J|NM .144623. 1(21389444] J21389438 [ref jjNM 144624. [[21389438] J40255062I [ref JjNM .144625. j[40255062] J21389442; [ref JjNM 144626. [[21389442] [21389452 jref JjNM .144627. [[21389452] [21389446 jref JjNM..144628. 1(21389446] [21389456 Jref JjNM 144629. [[21389456] J4741 9890 jref JjNM .144631 , 1(47419890] J40255064 |ref J|NM..144632. 1(40255064] J27886666 Jref J|NM..144633, [[27886666] 134222163 jref J |NM..144634, [[34222163] J40255250 jref J|NM..144635, |[40255250] J21389468 jref J |NM..144636, 1(21389468] 134222165 |ref J|NM. 144637, [[34222165]
gi|2 389472|ref[NMJ44638.1 ([21389472] gi|21389466JrefJNMJ44639.1 [[21389466] gi|24430202|ref|NMJ44640.2|[24430202] gi|21389470[refJNMJ44641.1 [[21389470] gi|32401419|ref|NMJ44642.3|[32401419] gi|21389474JrefJNM J44643.1 [[21389474] gi[31543824|ref(NMJ44644.2|[31543824] gι'J21389478JrefJNMJ44645.1|[21389478] gi|32189367|ref|NMJ44646.2|[32189367] giJ34222166|ref|NMJ44647.2|[34222166] gi[21389482|ref|NMJ44648.11[21389482] gi|21389524|ref|NMJ44649.l[[21389524] gi J21389518 jref JNM 44650.1 [[21389518] gi[47271461|ref|NMJ44651.2|[47271461] gi|21389522|ref|NMJ44652.11[21389522] gi|42716306Jref]NMJ44653.3J[42716306] gi[21389526Jref|NMJ44654.li[21389526] gi|42476008|ref|NMJ44657.2|[42476008] gi|4O068508| ref | NM J 44658.2J[40068508] gi|48995169[ref[NMJ 44659.3|[48995169] gi|21389544|ref|NMJ44660.11[21389544] gi|3422217θ[ref|NMJ44661.2J[342221 0] giJ21389548[ref[NMJ 44662.1 [[21389548] gi|21389542|ref|NMJ44663.1|[21389542] gi|46195786[ref|NMJ44664.3[[46195786] gi|31377590|ref|NMJ44665.2|[31377590] giJ21389554(ref|NMJ44666.1|[21389554] gi|21389550|ref|NM J44667.1 j[21389550] gi|34222172|ref|NMJ44668.2|[34222172] gi|21389558[ref|NMJ44669.lj[21389558] gi|21389560Jref|NM J 44670.1 [[21389560] gi[24432055Jref|NMJ44671.2J[24432055] gi|24638434|ref|NMJ44672.2[[24638434] gi|31563435[ref|NMJ44673.2|[31563435] gi[21389568JrefJNMJ44674.11[21389568] gi|21389570|ref|NMJ44675.1 [[21389570] giJ21389572[ref|NMJ44676.1|[21389572] giJ3981218θ[ref|NMJ44677.2|[39812180] giJ33186853|ref|NMJ44678.2|[33186853] giJ21389576[reflNMJ44679.li[21389576] giJ21389578Jref|NMJ44681.1|[21389578] gi|4O255066|ref(NMJ44682.2J[40255066] giJ3414770θ[ref|NMJ44683.2|[34147700] gi|21389584|ref|NMJ44684.lj[21389584] gi[34222353|refJNMJ44685.3|[34222353] gi|2138958δjref |NM J 44686.11[21389588] gi|21955153[ref(NMJ44687.1|[21955153] gi|4O255068iref|NMJ44688.2J[40255068] gi|34303923|ref|NMJ44689.3J[34303923] gi[21389592[refiNMJ44690.li[21389592] gi|46852396|ref|NMJ44691.3|[46852396] gi[21389596|ref|NMJ44692.1([21389596] gi|21687263|ref|NMJ44693.l[[21687263] gi|21389598|ref|NMJ44694.1|[21389598] gi|21389600|ref|NMJ44695.1 [[21389600] gi|51093835|ref|NMJ44696.3|[51093835] gi|31543068JrefINMJ44697.2|[31543068]
giJ24432064|ref|NMJ44698.2|[24432064] gi|49249974|ref|NMJ44699.2|[49249974] gi|24430211 jref|NMJ44701.2[[24430211] gi|21389614|ref|NMJ44702.1|[21389614] gi|21389608[ref|NMJ44703.l[[21389608] gi|21389616|ref|NMJ44704.1|[21389616] gi|34222169|ref|NMJ44705.2[[34222169] giJ3414770liref|NMJ44706.2|[34147701] gi|21389622|ref|NMJ44707.11[21389622] gi|21389306[ref|NMJ44709.lj[21389306] gi|30795194|ref|NMJ44710.21(30795194] giJ40255070Jref|NM J44711.3J[40255070] gi|38198666|ref|NMJ44712.2|[38198666] giJ21389490|ref|NMJ44713.lj[21389490] gi|21389492|ref|NMJ44714.1|[21389492] gi|3252689ljref|NMJ44715.2[[32526891] gi|21389496[ref|NMJ44716.11[21389496] gi|40255072|ref|NMJ44717.2|[40255072] gi|40255074|ref|NMJ44718.2[[40255074] gi[40255075|ref|NMJ44719.2|[40255075] gi|46852162|ref|NMJ44720.2|[46852162] gi|37059751|ref|NMJ44721.2|[37059751] giJ46411157|ref|NMJ44722.2J[46411157] giJ21389510|ref|NMJ44723.1|[21389510] gi|21687269[ref|NMJ44724.l[[21687269] gi|40255077|ref|NMJ44725.2|[40255077] giJ21389514Jref|NMJ44726.l j[21389514] gi|21389516|ref|NMJ44727.1|[21389516] giJ2153633θjref|NMJ44728.1|[21536330] gi|21536332|ref|NMJ44729.lj[21536332] gi|21536319|ref|NMJ44732.1|[21536319] giJ21536321 jref|NMJ44733.11[21536321] gi|21536323|ref|NMJ44734.11[21536323] gi[47524169iref|NMJ44736.3[[47524169] gi|21536336|ref|NMJ44765.l[[21536336] gi[21464138[ref|NMJ44766.11[21464138] gi|31563332|ref|NMJ44767.3|[31563332] gi J21618328|ref|NM J 44769.1 [[21618328] gi|48762928|ref|NMJ44770.2|[48762928] gi|21426826[refJNMJ44772.1|[21426826] gi|30581162|ref|NMJ44773.2[[30581162] gi|21426844|ref|NMJ44775.11[21426844] gi|21614514|ref|NMJ44776.1|[21614514] gi|21536307|ref|NMJ44777.1 [[21536307] gi|46411178|ref|NMJ44778.2[[46411178] gi|47778937|ref|NMJ44779.1|[47778937] gi|21614500|ref|NMJ44780.11[21614500] gi|21735593|ref|NM J 44781.11[21735593] giJ21618335iref|NMJ44782.l[[21618335] gi[21618356Jref|NM J 44947.1 [[21618356] gi|41349445|ref|NMJ44949.2|[41349445] gi|2161453θ[ref|NMJ44956.1|[21614530] gi[21614532[ref|NMJ44957.1 [[21614532] gi[21450646[ref|NMJ44962.li[21450646] gi|21450642Jref|NMJ44963.l[[21450642] gi[21450650|ref|NMJ44964.11[21450650] gi[21450644iref|NMJ44965.l[[21450644]
gi|21450654|ref|NMJ44966.1J[21450654] gi|40255079|ref| NMJ 44967.2 j[40255079] gi|21450658Jref|NMJ44968.lj[21450658] giJ21450652|ref|NMJ44969.1|[21450652] giJ21450662|ref|NMJ44970.1|[21450662] gi|47271496|ref|NMJ44972.3|[47271496] giJ32307180Jref|NMJ44973.2J[32307180] gi|21699053|ref|NMJ44974.lj[21699053] giJ50086625|ref|NMJ44975.2|[50086625] gii21699059JreflNMJ44976.1|[21699059] gi|21450670|ref|NMJ44977.1|[21450670] giJ21450664Jref|NMJ44978.l[[21450664] giJ34222176|ref|NMJ44979.2J[34222176] giJ21450668Jref|NMJ44980.lj[21450668] giJ21450678[ref |NMJ44981.11[21450678] gi|34787414|ref|NMJ44982.3|[34787414] giJ21450676Jref[NMJ44984.l[[21450676] gi[32880202|ref|NMJ44985.2|[32880202] gi|21450690Jref|NMJ44987.1|[21450690] gi|41349504Jref|NMJ44988.2|[41349504] gi|45267820|ref|NMJ44990.2|[45267820] gi(22001419JrefiNMJ44991.2|[22001419] gi|5035598l[ref|NMJ44992.3|[50355981] gi|4132775ljref|NMJ44994.6|[41327751] gi[3977758ljref|NMJ44995.2|[39777581] gi|33598957|ref|NMJ44996.2|[33598957] gi[34335184Jref|NMJ44997.3J[34335184] giJ2145071θiref|NMJ44998.1|[21450710] gi|34147703|ref|NMJ44999.2|[34147703] gi|31377592[ref|NMJ45000.2[[31377592] gi|21450708|ref|NMJ45001.1|[21450708] gi|21450718iref|NMJ45003.l[[21450718] gi|4031695ljref|NMJ45004.4J[40316951] giJ37039614JrefJNMJ45005.3J[37039614] giJ37059752[ref|NMJ45006.2|[37059752] gi|21450724|ref|NMJ45007.lj[21450724] giJ21450726Jref|NMJ45008.li[21450726] gi|24432063Jref|NMJ45010.2|[24432063] giJ24462252Jref|NMJ45011.2J[24462252] gi)32171246[ref|NMJ45012.3i[32171246] giJ21450728Jref|NMJ45013.1|[21450728] gi|2145073θjrefJNMJ45014.lj[21450730] gi|3154316θjref|NMJ45015.2([31543160] gi|31542212Jref|NMJ45016.2|[31542212] giJ21450742Jref|NMJ45017.lj[21450742] gi|25072198Jref|NMJ45018.2[[25072198] gi|31542759|ref|NMJ45019.2|[31542759] gi[34222173|ref|NMJ45020.2|[34222173] gi|50539409[refJNMJ45021.4J[50539409] gi|37059791|ref|NMJ45023.3[[37059791] gi|21450748|ref|NMJ45024.1|[21450748] gi|34222175|ref|NMJ45025.2|[34222175] gi|38570144(ref|NMJ45026.2[[38570144] gi|47271465|ref|NMJ45027.3J[47271465] gi|37059793JreflNMJ45028.3[[37059793] gi|21450766Jref|NMJ45029.1|[21450766] gi|21450760|ref|NM 145030.1 [[21450760]
124432071 | ref | |NM 145032. |[24432071] 121450764 |ref J|NM .145033. |[21450764] |21450774 [ref JjNM. 145034. |[21450774] |40255082 |ref J|NM 145035. |[40255082] J21450778 | ref | |NM..145036. |[21450778] [21450780 |ref| JNM 145037. [[21450780] 121450776 | ref J |NM .145038. |[21450776] J49619233 |ref J|NM..145039. [[49619233] 147132586 |ref| JNM 145040. |[47132586] 121450795 |ref J|NM .145041. [[21450795] [34147705 |ref j|NM..145042. [[34147705] [21450799 |ref J|NM .145043. |[21450799] [21699063 [ref J|NM 145044. |[21699063] [40068494 | ref | |NM 145045. |[40068494] J34222181 jref J|NM .145046. |[34222181] J45643132 | ref J |NM .145047. |[45643132] 134222178 |ref J|NM. 145048. [[34222178] [21450801 | ref J |NM .145049. [[21450801] 147271469 |ref J|NM..145050. |[47271469] J34147709 |ref J|NM .145051. 1(34147709] 121450815 | ref J |NM .145052. [[21450815] |50086627 |ref J|NM .145053. |[50086627] 131543066 jref J|NM .145054. 1(31543066] J21450813 Jref J|NM. 145055. |[21450813] J21450823 [ref J|NM .145056. [[21450823] J30089965 jref JjNM 145057. |[30089965] 121450827 [ref J|NM..145058. 1(21450827] J21450821 | ref JjNM .145059. 1(21450821] J21450831 |ref J|NM..145060. [[21450831] J47419927 jref J |NM..145061. 1(47419927] 121450835 jref JjNM 145062. |[21450835] 134147711 jref| JNM 145063. j[34147711] 121450837 |ref J|NM .145064. |[21450837] 121450833 |ref J|NM 145065. 1(21450833] [22027467 jref j |NM 145068. |[22027467] [21614506 |ref J|NM .145071. 1(21614506] [21614537 |ref J|NM 145074. |[21614537] [39725702 |ref J|NM..145080. 1(39725702] 121483181 [ref J|NM 145102. j[21483181] 121618348 |ref j|NM .145109. 1(21618348] 121618350 jref J |NM 145110. [[21618350] 121495177 | ref J|NM..145111 , 1(21495177] 121704262 jref j |NM 145112. [[21704262] 121704264 |ref J|NM..145113. [[21704264] [21704266 | ref J|NM 145114, [[21704266] |21536373 | refl JNM 145115. [[21536373] 121704268 | ref j|NM 145116, |[21704268] J38044281 jref J |NM .145117, |[38044281] J41281724 jref JjNM 145119, |[41281724] [21704278 | refl JNM. 145159, 1(21704278] [21729894 | ref JlNM..145160, |[21729894] 121729896 jref J |NM..145161 , |[21729896] 121729898 |ref J|NM..145162, 1(21729898] [21553312 |ref J|NM..145165, |[21553312] [38569485 |ref J|NM..145166, [[38569485] [21553314 |ref J|NM 145167, |[21553314] [21553310 |ref J|NM 145168, 1(21553310]
gi|21553316 Jref) NM 145169 1(21553316] gi|40255086 [refj NM .145170 |[40255086] giJ39753962 jref] NM 145171 j[39753962] gi|21553320 |ref] NM..145172 |[21553320] gi|21553322 |ref] NM .145173 1(21553322] giJ21553334 |ref| NM 145174 1(21553334] gi|21553328 jrefj NM .145175 j[21553328] gi[21553330 [ref| NM .145176 |[21553330] gi|21553324 |ref| NM .145177 1(21553324] gi|38327523 [ref] NM 145178 j[38327523] gi|21553338 |ref| NM .145179 |[21553338] gi|24475827 |ref| NM .145180 |[24475827] gi|22035619 |ref| NM 145182 j[22035619] gi|22035621 [ref| NM..145183 [[22035621] gi|40806198 (refj NM 145185 [[40806198] gi|21729875| [refl NM..145186 |[21729875] giJ21729885] [refj NM..145187 [[21729885] giJ21729887 [ref| NM..145188 [[21729887] giJ21729889 |ref| NM 145189 1(21729889] gi|21729891 [refj NM..145190 |[21729891] giJ21729877 |ref| NM .145196 [[21729877] giJ21729879 | ref | NM 145197 [[21729879] giJ21729881 |ref| NM..145198 |[21729881] giJ21729883 |ref| NM .145199 [[21729883] gi[46852395 |ref| NM..145200 [[46852395] giJ40255088 |refj NM .145201 |[40255088] giJ48976058 [ref| 145202 j[48976058] giJ34222183 | ref | NM .145203 [[34222183] gi[33942065 [ref| NM .145204 |[33942065] gij34222180 jrefj NM..145205 [[34222180] gi|21624647 |ref| NM .145206 1(21624647] giJ21624653 |ref| NM..145207 [[21624653] gi|21624655 |ref| NM..145208 j[21624655] gi|21735603 [ref| NM .145212 [[21735603] gi|21735605 [ref| NM .145213 |[21735605] giJ24497621 jrefj NM..145214 [[24497621] gi[50593527| |ref| NM .145230 |[50593527] giJ21687227 |ref| NM..145231 1(21687227] gi|37059754 |ref| NM .145232 [[37059754] giJ34147713 | ref | NM..145233 [[34147713] gi|34147714 [ref| NM .145234 |[34147714] giJ34222185 |ref| NM..145235 1(34222185] giJ21687138 jref | NM .145236. |[21687138] giJ21955121 [ref| NM..145237 [[21955121] gi|34147716 jref | NM 145238 |[34147716] giJ21687111 [ref| NM 145239 |[21687111] gi|21687047 [ref| NM ,145241. |[21687047] giJ21687049 [refj NM 145242 [[21687049] gi|33186904 jrefj NM..145243 |[33186904] giJ34222182 jrefj NM 145244 [[34222182] gi|34147717 |ref| NM 145245, |[34147717] giJ50409631 |ref| NM..145246 1(50409631] gi|50593523 jref | NM 145247 j[50593523] giJ37059756] |ref| NM 145248. 1(37059756] giJ21699069 |ref| NM.145249 |[21699069] gi[50355979 |ref| NM 145250 j[50355979] gi|34222188 |ref| NM 145251 |[34222188]
gi|21687059|ref|NMM 45252.11[21687059] gi|21687061|ref|N J45253.1|[21687061] gi|21687071|ref|NMJ45254.lj[21687071] giJ22547113Jref|NMJ45255.2|[22547113] gi|45593137|ref|N J45256.2|[45593137] gi|4867582ljref|NMJ45257.2|[48675821] gi|21687083|ref|N J45258.1|[21687083] gi|21687097|ref|N J45259.1|[21687097] gi|21687099Jref JNM J 45260.1 [[21687099] gi[41152082|ref|NMJ45261.2|[41152082] gi[31543062JrefJNMJ45262.2([31543062] gi|21687118|ref|N J45263.11[21687118] gi|34222184|ref|N J45265.2|(34222184] gi[4055627θ[refiNMJ45266.4i[40556270] gi|46275838|ref|NMJ45267.2|[46275838] gi|24432075(ref|NMJ45268.2|[24432075] gi|34222189|ref|NMJ45269.2|[34222189] gi|21687187|ref|NM J 45270.11[21687187] giJ34147718Jref|NMJ45271.2|[34147718] gi[37059757|ref|NMJ45272.2|[37059757] gi|37059758(ref[NMJ45273.2J[37059758] gi|21687219|ref|N J45274.1|[21687219] gi|21687234JrefiNMJ45275.1|[21687234] gi(21687236(ref(NMJ45276.1|[21687236] gi|44662822|ref|NMJ45277.3|[44662822] gi|21687148|ref|NMJ45278.1[[21687148] giJ41406058|refJN J45279.3|[41406058] gi|31377537|ref|NMJ45280.3|[31377537] giJ2846069ljrefJNMJ45282.1([28460691] giJ21699083|ref|NMJ 45283.1 [[21699083] gi|40255093|ref|N J45284.3|[40255093] gi|21699077|ref|NMJ45285.l[[21699077] gi|21686994|ref[NIVlJ45286.1|[21686994] gii21686996Jref(NMJ45287.l[[21686996] gi|2168725l[ref|NMJ45288.1|[21687251] gi|40255095|ref|NMJ45291.2|[40255095] gi|34222191|ref|NMJ45292.2J[34222191] gi|21686972|ref|NMJ45293.1|[21686972] giJ37574045Jref|NMJ45294.3|[37574045] gi|42734386|ref|N J45295.2|[42734386] gi|21686976|ref|NIVIJ45296.1|[21686976] gi[21699081|ref|NIVtJ45297.li[21699081] gi|31083201|ref|NMJ45298.3|[31083201] gi|21686978Jref|NIVIJ45299.1|[21686978] gi[2168698θ[ref[NMJ45300.1|[21686980] gi|21945057|ref|NIVlJ45301.1|[21945057] giJ21687067[ref|NIVIJ45303.1|[21687067] gi|34147719|ref|NIVlJ45304.2|[34147719] gi|2168715θ[ref|NIVlJ45305.1|[21687150] gi|21687152|ref|NMJ45306.1|[21687152] gi|26190613|ref|NMJ 45307.2|[26190613] giJ3414772θiref[NMJ45308.2[[34147720] gi|21687174|ref|NMJ45309.1|[21687174] gi|2194506θ[ref|N J45310.1|[21945060] gi|22202613|ref|M M 45311.1 [[22202613] gi|3414772ljref|NMJ45312.2|[34147721] gi[21687176Jref(NJ J45313.lj[21687176]
gi|21687122|ref|NMJ45314.1|[21687122] gi|2191887l jref|NMJ45315.2|[21918871] gi|31543075|ref|NMJ45316.2|[31543075] gi[21735579|ref|NMJ45320.1|[21735579] gi|21735581 |ref | NMJ 45321.1|[21735581] giJ21735583|ref|NMJ45322.1J[21735583] gi|21735585|ref|NMJ45323.1|[21735585] gi|21735587|ref|NMJ45324.1|[21735587] gi[21704273[ref|NMJ45325.lj[21704273] gi|21687265|ref|NMJ45326.1|[21687265] giJ22035566Jref|NMJ45328.lj[22035566] gi|21735608Jref|NMJ45330.lj[21735608] gi|2173556l[ref|NMJ45331.1|[21735561] gi|21735563|ref|NMJ45332.1|[21735563] gi|21735565|ref|NMJ45333.1|[21735565] gi[34304341 |ref|NM J45341.2|[34304341] giJ21735558|ref|NMJ45342.1 [[21735558] gi|21735615Jref|NMJ45343.lj[21735615] gi|21735617jref|NMJ45344.1|[21735617] gi|21703361 |ref|NMJ45345.1 [[21 03361] gi|41281730Jref|NMJ45346.1|[41281730] gi|27437003|ref|NMJ45347.1|[27437003] gi|27437005Jref|NMJ45348.1[[27437005] gi|33598926Jref|NMJ45349.lj[33598926] giJ3359893θ[ref|NMJ45350.1|[33598930] gi|33598932|ref|NMJ45351.11[33598932] gi|33598934[ref|NMJ45352.1|[33598934] gi|22035652Jref|NMJ45637.1|[22035652] giJ22035608JrefJNMJ45638.lj[22035608] gi|22035643|ref|NMJ45639.1|[22035643] gi|22035645Jref|NMJ45640.1|[22035645] giJ22035647[ref|NMJ45641.1|[22035647] giJ22035649|ref| NMJ 45642. lj[22035649] gi|22035593|ref|NMJ45644.1|[22035593] gi[21717802Jref|NMJ45645.lj[21717802] gi|39777583|ref| NMJ 45646.2|[39777583] gi|21717806|ref|NMJ45647.lj[21717806] gi[21717815|ref(NMJ45648.1[[21717815] gi[30061503|ref|NMJ45649.2J[30061503] gi|40255097[ref|NMJ45650.2|[40255097] gi|21717813|ref|NMJ45651.11[21717813] giJ2323824l jref|NMJ45652.2J[23238241] gi|21717823|ref|NMJ45653.11[21717823] giJ31563532Jref| NMJ 45654.2|[31563532] giJ30061505[ref|NMJ45655.2J[30061505] gi[33457325|ref|NMJ45657.1|[33457325] gi[2171783l[ref|NMJ45658.lj[21717831] gi|28416912Jref|NMJ45659.2J[28416912] gi|22035656|ref| NMJ 45660.1|[22035656] gi[49619235Jref|NMJ45662.2[[49619235] gi|22035681 |ref| NMJ 45663.11[22035681] gi[22027491Iref|NMJ45664.lj[22027491] giJ22027495|ref|NMJ45665.lj[22027495] gi|22035557Jref|NMJ45685.1|[22035557] gi[4624936θ[ref|NMJ45686.2|[46249360] gi|46249362|ref|NMJ45687.2|[46249362] gii22035553|refJNMJ45689.1[[22035553]
gi|21735624|ref|NMJ45690.11[21735624] gi|22538424Jref|NMJ45691.2J[22538424] gi|22027647|ref|NMJ45693.1|[22027647] gi|22027633Jref|NMJ45695.1|[22027633] giJ22035559|ref|NM J45696.11[22035559] gi|22027506Jref|NMJ45697.li[22027506] giJ21735486Jref|NMJ45698.1|[21735486] giJ22907036Jref|NMJ45699.2[[22907036] giJ2202751θjrefJNMJ45701.lj[22027510] giJ22209000|ref|NMJ 45702.1|[22209000] gi|27262644|ref|NMJ45714.1|[27262644] gi|23312391|refJNMJ45715.1|[23312391] giJ21955169Jref[NMJ45716.lj[21955169] gi|23312393|ref|NMJ45719.l[[23312393] gi[28416947Jref|NMJ45720.2J[28416947] gi|22027617|ref|NMJ45725.lj[22027617] giJ22027619iref|NMJ45726.1|[22027619] giJ22035662[ref|NMJ 45727.1 [[22035662] giJ22027637Jret]NMJ45728.lj[22027637] giJ22035589Jref|NMJ45729.1|[22035589] gi|22027652|ref|NMJ45730.1|[22027652] giJ39777618|ref(NMJ45731.2J[39777618] gi|22035571|ref|NMJ45733.1|[22035571] gi|22035573JrefJNMJ45734.1[[22035573] gi|22027527[ref|NMJ45735.1|[22027527] giJ22035699JrefJNMJ 45738.1 [[22035699] gi[22035611 jreflNMJ 45739.11[22035611] gi(48375174|refJNMJ45740.2J[48375174] gi|22035667|ref|NMJ45747.1|[22035667] gi|22035669[ref|NMJ45748.1|[22035669] gi|22027623|ref|NMJ45751.1|[22027623] gi|22027474Jref|NMJ45752.1j[22027474] giJ2195517ljrefJNMJ45753.l[[21955171] giJ34147722]ref|NMJ45754.2J[34147722] gi|21955236irefJNMJ45755.1|[21955236] gi|21955238|ref|NMJ45756.1|[21955238] gi|22027626Jref[NMJ45759.li[22027626] giJ22091447JrefJNMJ45762.1|[22091447] gi|22091449Jref|NMJ45763.lj[22091449] gi|22035633iref|NMJ45764.1|[22035633] gi|22035635|refJNMJ45791.11[22035635] giJ22035637|ref|NMJ45792.1|[22035637] gi|2203569ljref|NMJ45793.lj[22035691] gi|22035583Jref|NMJ45794.1|[22035583] gi[22035585iref|NMJ45795.li[22035585] gi|46397391|ref|NMJ45796.2|[46397391] giJ47132623Jref|NMJ45798.2[[47132623] gi|33624782|ref|NMJ45799.2|[33624782] gi|33624799|ref|NMJ45800.2| [33624799] gi[3362482θiref|NMJ45802.2[[33624820] gi|22027629|ref|NMJ 45803.11 [22027629] gi|21956638Jref(NMJ45804.1|[21956638] giJ2195664θjref|NMJ45805.1|[21956640] gi[2827470θ[refJNMJ45806.2|[28274700] gi|24308064|ref|NMJ45807.11 [24308064] gi|21956644|ref |NM J 45808.11 [21956644] gi|21956646Jref|NMJ45809.1|[21956646]
gi|22027513|ref|NMJ45810.1|[22027513] giJ22027550|ref|NM~'l45811.1 [[22027550] gi|22202628|ref|NMJ45812.1|[22202628] giJ22202630Jref|NMJ45813.1 [[22202630] gi|22027553|ref|NMJ45814.1|[22027553] gi|22027556|refJNM 145815.1 [[22027556] gi|41281739|ref|NMJ45818.1|[41281739] gi|22202615|ref|NMJ45858.1|[22202615] gi|2253879ljref|NMJ45859.1 [[22538791] gi|22538793|ref|NMJ45860.1 |[22538793] gi|22325374|ref|NMJ45861.1 [[22325374] gi|22209008JrefJNMJ45862.1|[22209008] giJ22208950Jref|NMJ45863.lj[22208950] giJ2220899lirefJNMJ45864.lj[22208991] gi[22004646Jref|NMJ45865.1 [[22004646] giJ22035628|ref|NMJ45867.1|[22035628] gi[22165430JrefJNMJ45868.l[[22165430] gi|22165432)refJNMJ45869.1|[22165432] gi|22202623|ref|NMJ45870.1|[22202623] gi|22202625[ref|NMJ45871.11[22202625] giJ22208963|ref|NMJ45872.1 |[22208963] gi|22325359|ref|NMJ45886.l[[22325359] gi|22325361 [ref|NMJ45887.11[22325361] giJ22208983Jref|NMJ45888.1J[22208983] gi[22538404[ref|NMJ45891.1 [[22538404] gi|22538406|ref|NMJ45892.11[22538406] giJ22538408|ref[NMJ45893.lj[22538408] giJ22208986|ref|NMJ45894.1|[22208986] giJ22208988|ref|NMJ45895.1|[22208988] gi[22202634[ref|NMJ45896.1 [[22202634] gi|22202636|ref|NMJ45897.1 |[22202636] gi(22538807Jref[NMJ45898.1[[22538807] giJ22208966|ref|NM J45899.11[22208966] giJ2220897θjref|NMJ45901.11[22208970] gil22208972|ref|NMJ459O2.11[22208972] gi|22208974Jref|NMJ45903.1|[22208974] gi|22208976|ref|NMJ45904.1|[22208976] giJ22208978Jref|NMJ45905.1|[22208978] gi[22325378|ref|NMJ45906.1|[22325378] gi[41281748[ref|NMJ45909.1 |[41281748] giJ41281752Jref|NMJ45910.11[41281752] gi[23308736Jref|NMJ45911.1 j[23308736] giJ31543287[ref[NMJ45912.3|[31543287] giJ33942075Jref|NMJ45913.2J[33942075] giJ37537685[ref[NMJ45914.2|[37537685] giJ22202618|ref|NMJ45918.1 [[22202618] gi|22547137|ref|NMJ46387.11[22547137] gi(22547139[ref|NMj46388.1|[22547139] gi|23065546|ref|NMJ46421.11[23065546] gi|31377586|ref|NMJ47127.2)[31377586] gi|40316919Jref|NM J47128.3|[40316919] gi|33359214|ref|NMJ47129.2|[33359214] gi|2447583l[ref|NMJ47130.11[24475831] gi|22165417|ref|NMJ47131.11[22165417] gi[22165419[refJNMJ47132.1 [[22165419] giJ22212924Jref|NMJ47133.1 [[22212924] gi|22212926|ref|NMJ47134.11[22212926]
|22202605 [ref J|NM .147147.11(22202605] [23065556 [ref J|NM 147148.1 |[23065556] J23065559 Jref j|NM .147149.11(23065559] [22325355 [ref J|NM. .147150.1 |[22325355] [22325382 |ref J|NM .147152.1 j[22325382] [41350331 [ref J|NM .147156.3 j[41350331] [22212933 [ref J|NM 147157.1 |[22212933] J22212935 |ref J|NM .147158.1 j[22212935] 122212937 |ref j|NM 147159.1 j[22212937] J22325389 |ref J|NM .147160.2 j[22325389] I25777709 |ref J|NM 147161.2 |[25777709] 122212921 jref J |NM .147162.11(22212921] J22212917 |ref J|NM. 147164.11(22212917] J22538388 |ref J|NM .147166.11(22538388] J22219468 [ref JjNM .147168.1 |[22219468] [22219470 [ref J|NM .147169.1 j[22219470] [22538390 |ref J|NM 147171.1 |[22538390] [22219464 jref J|NM .147172.1 j[22219464] 122219466 | ref j |NM .147173.1 |[22219466] J27597081 [ref J|NM .147174.2 j[27597081] J24432101 jref J |NM 147175.2] j[24432101] J34303920 [ref J|NM .147180.2] j[34303920] J28373060 | ref JjNM. 147181.2 |[28373060] J28373061 jref JjNM .147182.2 j[28373061] |28373062 |ref J|NM .147183.2 j[28373062] J22538445 jref J |NM. 147184.11(22538445] |22538392 jref J |NM. 147185.1 j[22538392] 122547118 jref J|NM .147187.11(22547118] 122547148 [ref JjNM .147188.1 j[22547148] J22218338 [ref JjNM .147189.1 |[22218338] J22218344 [ref JjNM. 147190.1 |[22218344] J22218340 jref JjNM .147191.1 j[22218340] |27436934 |ref J|NM. 147192.2 j[27436934] [22218342 jref J |NM .147193.1 j[22218342] J22218352 |ref J|NM. 147194.1 j[22218352] J22507406 jref J |NM ,147195.1 j[22507406] 122218346 |ref J|NM. 147196.1 j[22218346] 131563541 jref | |NM ,147197.2] [[3 563541] J23238189 |ref| |NM .147198.2 j[23238189] [22218356 |ref| |NM_ ,147199.11(22218356] |22538425 jrefj |NM .147200.1 j[22538425] [22218620 |ref J|NM. 147202.11(22218620] 142544197] jref | |NM ,147203.2] [[42544197] J22547179 |ref J|NM. 147204.11(22547179] J22538456 jref j |NM ,147223.1 j[22538456] |22538458 [ref| |NM_ 147233.1 j[22538458] [22538427] |ref J|NM ,147686.1 j[22538427] |46370088 |ref| |NM ,147777.2 j[4637O088] I22538430 |ref J|NM ,147780.1 j[22538430] [22538432 jref JjNM ,147781.1 [[22538432] |22538434 |ref j|NM .147782.1 |[22538434] |22538436 jref J |NM .147783.1 [[22538436] |22325387 |ref J|NM 148169.1 |[22325387] |22538439 jref JjNM .148170.1 [[22538439] |22325367 |ref j|NM .148171.1 |[22325367] I22538477 |ref J|NM .148172.11(22538477] J22538479 |ref| JNM 148173.1 j[22538479]
gi|22538417[ref|NM J48174.11[22538417] gi|22547211|ref|NMJ48175.1[[22547211] giJ22547214|ref|NMJ48176.lj[22547214] gi|22547143[ref|NMJ48177.lj(22547143] gi|22325369|ref|NMJ48178.lj[22325369] gι'J22325371 [refJNM J48179.11[22325371] gi|27262648Jref|NMJ48414.lj[27262648] giJ2726265θjrefJNMJ48415.lj[27262650] gi|27262652|ref|NMJ48416.lj[27262652] gi|22547128Jref|NMJ48570.lj[22547128] gi(22547130(ref(NMJ48571.1([22547130] gi|22538810[refJNMJ48672.lj[22538810] gi|48475049|ref|NMJ48674.2J[48475049] gi|45433549[ref|NMJ48675.2|[45433549] giJ22380642|ref[NMJ48676.1|[22380642] giJ22538492lref|NMJ48842.lj[22538492] gi|22538489|ref|NMJ48886.l[[22538489] gi|22547124|ref]NMJ48887.1|[22547124] gi|22538797|ref|NMJ48888.l[[22538797] gi|22507398|ref|NMJ48894.1|[22507398] giJ22507402Jref[NMJ48896.1[[22507402] giJ22507404|ref|NMJ48897.li[22507404] giJ2253841 θ[ref|NM J 48898.1 j[22538410] gi|22538412|ref|NMJ48899.1|[22538412] gi|22538414|ref|NMJ48900.1|[22538414] giJ23238193iref|NMJ48901.l[[23238193] gi|23238196[ref|NMJ48902.1|[23238196] gi|23397645Jref|NMJ48903.l[[23397645] gi|34335284[refiNMJ48904.2|[34335284] giJ34335285|ref|NMJ48905.2J[34335285] gi|22547168|ref|NMJ48906.1[[22547168] gi[2254717θ[ref|NM J 48907.11[22547170] gi|22547172Jref[N J 48908.1 j[22547172] gi|22547175|ref|NMJ 48909.1 [[22547175] gi|22547218|ref|NMJ48910.1|[22547218] gi|23312389Jref|NMJ48911.l[[23312389] gi|23200007|ref[NMJ48912.1([23200007] gi|23200011 |ref|NMJ48913.11[23200011] giJ23200013iref[NMJ48914.1|[23200013] gil23200015|ref|NMJ48915.1|[23200015] gi|23200017|ref|NMJ48916.1|[23200017] gi|22547188|ref|NMJ48918.1|[22547188] giJ34335278Jref[NMJ48919.2|[34335278] gi|22538452Jref|NMJ48920.1|[22538452] gi[41327739Jref|NMJ48921.2|[41327739] giJ41281767|ref|NMJ48923.11[41281767] giJ2320000ljrefJNMJ48936.1 [[23200001] gi|23110931 |ref|NMJ48954.lj[23110931] gi[23111031 jref|NM_148955.11 [23111031] giJ23199997Jref|NMJ48956.11[23199997] gi|31652245|ref|NMJ48957.2|[31652245] giJ31077214[ref|NMJ48959.2|[31077214] gi|2250737ljref|NMJ48960.1|[22507371] gi|42544244|ref|NMJ48961.3j[42544244] gi|40255258|ref|NMJ48962.3|[40255258] gi|22507379|ref|NMJ 48963.1 j [22507379] giJ23110951 |ref[NMJ48964.11[23110951]
gi|23200020|ref|NMJ48965.1 |[23200020] gi|23200022|ref|NMJ48966.1 |[23200022] gi[23200024[ref|NMJ48967.l [[23200024] gi|23200026|ref|NMJ48968.1 |[23200026] gi[23200028|ref|NMJ48969.l [[23200028] gi|23200030|ref|NMJ48970.1 |[23200030] giJ23200032Jref[NMJ48971.1 [[23200032] gi|23200034|ref|NMJ48972.1 [[23200034] giJ23200036|refJNMJ48973.l j[23200036] giJ23200038Jref|NMJ48974.l j[23200038] gi|23110994Jref|NMJ48975.1 [[23110994] giJ23110934JrefJNMJ48976.l j[23110934] gi|23510399|ref|NMJ48977.l j[23510399] gi[23510401 jref[NMJ48978.11[23510401] gi|23110956|ref|NMJ48979.11[23110956] giJ23200003Jref|NMJ48980.1 |[23200003] giJ23200005[ref|NMJ49379.l i[23200005] gi|23110938|ref|NMJ52132.11[23110938] gi|2319997θ[ref|NMJ52133.l i[23199970] gi[44889478[ref|NMJ52219.2|[44889478] gi|4054940θjref|NMJ52221.2J[40549400] gi|23238199|ref[NMJ52222.l i[23238199] gi|23312367Jref|NMJ52223.1 |[23312367] gi[23312373[refJNMJ52224.11[23312373] gi|23312375|ref|NMJ52225.1 [[23312375] gi[23312377iref|NMJ52226.l [[23312377] gi|23111046Jref|NMJ 52227.11[23111046] giJ42476079Jref|NMJ52230.2J[42476079] gi|22726202|ref|NMJ52232.1 |[22726202] gi|2311105θjrefJNMJ52233.1 |[23111050] gi|23111063[ref|NMJ 52235.1 [[23111063] gi|23065525|ref|NMJ 52236.11[23065525] gi|23065528Jref|NMJ52237.1 |[23065528] giJ23111054[ref|NMJ52238.1 |[23111054] gi|23199977Jref|NMJ52240.1 |[23199977] gi[23238225[ref|NMJ 52243.1 [[23238225] gi|23111027|ref JNM J 52244.1 [[23111027] gi|23238253Jref|NMJ52245.1 |[23238253] gi|23238255|ref|NMJ 52246.1 [[23238255] gi[23238257[ref|NMJ 52247.1 [[23238257] gi|22748616|ref|NMJ52250.1 |[22748616] gi[34098963Jref|NMJ 52251.2J[34098963] gi|23238260|ref|NMJ 52253.11[23238260] giJ23110947Jref|NM_152255.11[23110947] gi|23308686Jref|NMJ 52257.11[23308686] gi|31543194Jref|NMJ52259.2|[31543194] gi|23308688|ref|NMJ 52260.1 [[23308688] gi[22748614Jref|NMJ52261.1 [[22748614] giJ22748624|ref|NMJ 52262.11[22748624] giJ22748618|ref|NMJ52263.1 |[22748618] giJ4025510θjrefJNMJ52264.2[[40255100] gi|22748622Jref|NMJ52266.11(22748622] giJ31542782Jref|NMJ52267.2J[31542782] gi|34303925|ref|NMJ52268.2|[34303925] gi[22748626[ref|NMJ52269.1 |[22748626] gi[42476006[ref|NMJ52270.2|[42476006] gi[40217795Jref|NMJ52271.2J[40217795]
gi|22748640|ref|NMJ52272.1|[22748640] gi|3432808θiref[NMJ52274.2i[34328080] gi[38683852[refJNMJ52275.2J[38683852] gi|22748634|ref|NMJ 52277.1 [[22748634] gi|23397408|ref|NMJ52278.1|[23397408] gi[25014092iref|NMJ52279.2[[25014092] gi|31377588lref|NM J 52280.2|[31377588] gi|22748638[ref[NMJ 52281.1 [[22748638] gi|42476016|ref|NM J 52282.2[[42476016] gi|23097314|ref|NMJ52283.1|[23097314] gi[40548376|ref|NMJ52284.2[[40548376] gi|22748652|ref|NMJ52285.1|[22748652] gi[38524588|ref|NMJ52286.2|[38524588] gi|4080510liref[NMJ52287.2J[40805101] gi[22748650|ref|NMJ52288.1|[22748650] gi|2274866θ[ref|NMJ 52289.1 [[22748660] giJ22748654lref|NMJ52290.lj[22748654] gi|22748664|ref|NMJ 52291.1 j[22748664] gi|34303922iref|NMJ52292.2J[34303922] gi|38202254|ref|NMJ52295.3|[38202254] gi[49249971|ref|NMJ52296.3[[49249971] gi|27262629Jref|NMJ52298.2|[27262629] gi[23097284[ref|NMJ 52299.1 [[23097284] gi[38569502[ref[NMJ52300.2i[38569502] gi|31559785Jref|NMJ 52301.3|[31559785] gi[23097278[ref|NMJ52302.li[23097278] gi|22748680|ref|NMJ 52303.11(22748680] giJ22748674[ref|NM J 52304.11[22748674] gi|31982952|ref|NMJ52305.1|[31982952] giJ23312361|ref|NMJ52306.2|[23312361] gi[22748678Jref |NM J 52307.11[22748678] gi|24308244[ref|NMJ52308.1l[24308244] giJ45505138|ref JNM J 52309.2J[45505138] gi[23097309[ref|NMJ 52310.1 [[23097309] gi[22748684|ref|NMJ52311.1 j[22748684] gi|33285007|ref|NMJ52312.2[[33285007] gi[4080735θiref|NMJ52313.2J[40807350] gi|22748688[ref|NMJ52314.1|[22748688] gi[22748690|ref|NMJ52315.1|[22748690] gi[22748692[ref[NM J 52316.1 [[22748692] gi[22748694[ref|NMJ52317.li[22748694] gi|22748696|ref|NMJ 52318.1 [[22748696] gi|34303924[ref[NMJ52319.2|[34303924] gi[22748700|refJNMJ 52320.1 [[22748700] gi|22748702|ref|NMJ 52321.11[22748702] gi[22748704|refJNMJ52322.l[[22748704] gi]22748706|ref|NMJ 52323.1 [[22748706] gi[22748708Jref[NMJ52324.1|[22748708] gi|22748710|ref|NMJ 52325.11[22748710] gi|50355986[ref|NMJ52326.2[[50355986] gi|22748714|ref|NMJ52327.1 |[22748714] gi|40316947[ref|NMJ52328.3[[40316947] gi|45439341 |ref|NMJ52329.3[[45439341] gi|34303929[ref|NMJ52330.2J[34303929] gi|22748722|ref|NMJ 52331.1 [[22748722] gi|31377583|ref|NMJ52332.2J[31377583] gi|22748726|ref|NMJ 52333.11[22748726]
gi|38570110|ref J|NM_ 152334.2] |[385701 10] giJ22748730|ref| |NM_ 152335.1 |[22748730] gi|22748732|ref J|NM_ 152336.1 [[22748732] gi|22748734|ref J|NM_ 152337.1 |[22748734] gi[22748736Jref| |NM_ 152338.1 [[22748736] gi|31377584|ref| |NM_ .152339.2 1(31377584] giJ23097312|ref JjNM_ 152340.1 |[23097312] gi|31542755|ref JjNM. ,152341.2 [[31542755] gi|22748742|ref J|NM_ 152342.1 |[22748742] gi|22748744|ref| [NM_ .152343.1 [[22748744] gi|22748746|ref| |NM_ 152344.1 |[22748746] gi|33636763|ref J|NM.152345.3 [[33636763] giJ2274875θjref J|NM .152346.1 [[22748750] gi|34222281 |ref J|NM.152347.3 |[34222281] giJ22748754|ref J|NM .152348.1 [[22748754] gi|22748756|ref J|NM..152349.1 |[22748756] giJ22748758jref J|NM .152350.1 [[22748758] gi|31982944|ref J|NM 152351.2 1(31982944] gi|22748762|ref J|NM.152352.1 [[22748762] giJ22748764|ref J|NM .152353.1 1(22748764] gi|3856670l [ref J|NM .152354.2 |[38566701] gi|22748768|ref J|NM.152355.1 |[22748768] gi|31542771 [ref J|NM 152356.2 1(31542771] giJ22748772Jref J|NM .152357.1 1(22748772] gi|22748774Jref] |NM .152358.1 1(22748774] gi[22748776iref J|NM. 152359.1 1(22748776] giJ40789269|ref j|NM .152360.2 [[40789269] gi|24308367|ref J|NM. 152361.1 [[24308367] giJ22748780|ref J|NM .152362.1 1(22748780] giJ40255103[ref J|NM. 152363.2 j[40255103] giJ22748786Jref J|NM 152365.1 1(22748786] gi|24432073jref J|NM..152366.2 [[24432073] gi|22748790|ref JjNM 152367.1 [[22748790] gi|32698778Jref J|NM..152369.2 [[32698778] gi|22748798|ref| |NM 152371.1 j[22748798] gi|40255105Jref| |NM. 152372.2 |[40255105] gi|38678533|ref J|NM .152373.2 1(38678533] gi[22748804|ref JjNM 152374.1 [[22748804] gi|22748806Jref J|NM..152375.1 1(22748806] gi|31377582|ref J|NM 152376.2 [[31377582] giJ22748810|ref JjNM..152377.1 [[22748810] gi[22748812|ref| |NM..152378.1 1(22748812] gi|22748814jref J|NM .152379.1 |[22748814] gi|22748818|ref JjNM .152382.1 |[22748818] giJ32964831 [ref J|NM 152383.2 [[32964831] giJ22748822[ref J|NM .152384.1 [[22748822] gi|22748824Jref JjNM..152385.1 |[22748824] gi[23510288|ref j|NM .152386.2 1(23510288] gi|45387952[ref J[NM 152387.2 j[45387952] giJ2274883θiref J|NM..152388.1 |[22748830] gi[22748832Jref J|NM .152389.1 [[22748832] gi[22748834|ref J|NM 152390.1 [[22748834] gi|4044540θjref JjNM. 152391.2 [[40445400] gi[22748838|ref J|NM..152392.1 |[22748838] giJ4559314l jref j|NM .152393.2 [[45593141] gi|31543187|ref J|NM..152394.2 |[31543187] giJ24308369Jref JjNM 152395.1 1(24308369]
|22748844 | ref| |NM .152396.1 |[22748844] 122748846 |ref ]|NM 152397.1 |[22748846] 122748848 |ref| |NM..152398.1 j[22748848] [22748850 |ref| |NM..152399.1 |[22748850] J22748852 |ref :||NM 152400.1 j[22748852] [22748854 |ref| |NM .152401.11(22748854] [28460693 jref :j|NM .152402.1 j[28460693] [33469928 |ref| |NM 152403.2 j[33469928] J24432082 |ref J|NM .152404.2 j[24432082] J22748860 |ref JjNM. 152405.1 j[22748860] [40255108 [ref JjNM .152407.2 |[40255108] J22748864 [ref J|NM..152408.1 j[22748864] 140255110 |ref J|NM .152409.2 j[40255110] [22748868 jref J |NM. 152410.1 j[22748868] |22748870 | ref J|NM..152411.1 j[22748870] [22748872 | ref J|NM .152412.1 j[22748872] J22748874 | ref J|NM..152413.1 |[22748874] [44680137 |ref J|NM..152414.31(44680137] J22748878 |ref J|NM .152415.1 |[22748878] |22748880 [ref J|NM .152416.1 j[22748880] J22748882 |ref J|NM. 152417.11(22748882] |45243529 |ref| |NM .152418.2 j[45243529] [22748888 |ref J|NM..152420.1 j[22748888] J34303930 jref JjNM .152421.2 j[34303930] [30089951 jref j|NM .152422.31(30089951] [24432076 | ref J |NM..152423.2 |[24432076] [22748896 | ref J |NM..152424.1 j[22748896] |22748898 |ref J|NM .152425.1 j[22748898] [22748900 |ref J|NM .152427.1 j[22748900] 140255112 |ref J|NM..152428.21(40255112] J31377576 jref JjNM .152429.2 |[31377576] J22748910 |ref JjNM..152430.1 [[22748910] J22748904 jref J INM .152431.1 [[22748904] 138327515 |ref J|NM..152433.2 [[38327515] J22748918 [ref| |NM .152434.1 [[22748918] J22748912 Iref J|NM .152435.1 [[22748912] [22748922 |ref J|NM 152436.1 [[22748922] 122748916 | ref J |NM .152437.1 [[22748916] [22748926 |ref J|NM..152439.1 |[22748926] [47458824 I ref J|NM 152440.3 j[47458824] [22748930 | ref J |NM..152441.1 |[22748930] [41393615 I ref J|NM 152442.2 [[41393615] [22748934 | ref J |NM .152443.1 |[22748934] J22748928 [ref J|NM 152444.1 j[22748928] I22748942 |ref J|NM .152445.1 |[22748942] [31542243 | ref J |NM .152447.2 j[31542243] [22748940 I ref J|NM .152448.11(22748940] J40288200 I ref J|NM 152449.2 |[40288200] I22748944 jref J |NM 152450.11(22748944] I50582990 | ref J |NM .152451.2 [[50582990] 134303931 |ref| [NM .152453.21(34303931] J22748952 jref JjNM .152454.1 [[22748952] 131542776 I ref J|NM .152455.2 j[31542776] [22748956 I ref J|NM .152456.11(22748956] I22748966 |ref J|NM .152457.1 j[22748966] |34222277 |ref J|NM .152458.41(34222277] J31543192 I ref JjNM 152459.2 |[31543192]
gi(48255961|ref[NMJ52460.2|[48255961] gi|50345998|ref|NMJ52461.2|[50345998] giJ22748974Jref|NMJ52462.lj[22748974] gi|22748968Jref|NMJ 52463.1 [[22748968] gi|22748978|ref|NMJ 52464.11[22748978] gi|22748972[ref[NMJ52465.1|[22748972] gi|22748982|ref|NMJ52466.lj[22748982] giJ22748976|refJNMJ52467.l[[22748976] gi|31377553lref|NMJ52468.3[[31377553] gi|22748990|refJNMJ 52470.11[22748990] gi|22748994|ref|NMJ52472.1[[22748994] gi|22748988Jref|NMJ52473.lj[22748988] gi|34303933|ref|NMJ52474.2|[34303933] gi|22748992|ref|NMJ52475.1|[22748992] gi|22749002|ref|NMJ52476.1 |[22749002] gi(46358359|ref|NMJ52477.2J[46358359] gi|22749006Jref|NMJ52478.1|[22749006] gi|46391095|ref|NMJ52479.3J[46391095] giJ2274901θjref|NMJ52480.11[22749010] gi|22749004Jref|NMJ 52481.1 |[22749004] gi|22749014|ref|NMJ52482.1([22749014] gi|22749008|ref|NMJ 52483.1|[22749008] giJ3857011 δjrefJNMJ 52484.2|[38570116] gi|31542750|ref|NMJ52485.2|[31542750] gi|22749012|ref|NMJ52486.1|[22749012] gi|22749022|ref|NMJ52487.1|[22749022] gι'J22749016 jref|N J 52488.1 j[22749016] gi|22749026|ref|NMJ 52489.11[22749026] gi|22749020|ref|NMJ52490.1|[22749020] gi J47458826|ref|NM J 52491.3|[47458826] gi|22749024|ref|NMJ 52492.11[22749024] gi[38570114iref|NMJ52493.2[[38570114] gi|22749028Jref|NMJ 52494.11[22749028] gi|22749038|ref|NM J 52495.11[22749038] gi|22749032Jref|NMJ52496.1|[22749032] gi|22749042|ref|NMJ52497.1|[22749042] gi|22749036(ref|NMJ 52498.1|[22749036] gi|22749046Jref|NMJ 52499.1 [[22749046] gi|22749040|refjNMJ 52500.11[22749040] gi|39540513Jref |NM J 52501.2|[39540513] gi|47578108|ref|NM 152503.2J[47578108] gi|31542748|ref|NMJ52504.2|[31542748] giJ34303934[ref|NMJ52505.2|[34303934] giJ22749058|ref|NMJ 52506.1 [[22749058] gi|22749052Jref|NMJ52507.1|[22749052] gi|22749056|ref|NMJ 52509.11[22749056] gi|22749066Jref[NMJ52510.1|[22749066] gii51093844iref|NMJ52511.3J[51093844] gi|34222283Jref|NMJ52512.3|[34222283] gi|32526889[ref|NMJ52515.2|[32526889] gi|22749078[ref |NM J 52516.11[22749078] giJ22749072Jref|NMJ52517.1|[22749072] gi|40217793|ref|NMJ 52519.2|[40217793] gi|31542751|ref|NMJ52520.2|[31542751] giJ37059759Jref|NMJ52522.2J[37059759] gi|22749084|ref|NM 152523.11[22749084] gi[22749094|ref|NMJ52524.1|[22749094]
gi|37059794 jrefj |NM_ 152525.31(37059794] gi|45505181 [ref J|NM_ 152526.3 [[45505181] gi|42415495 |,ref J|NM_ 152527.3 [[42415495] gi[22749102| |ref| |NM_ 152528.1 [[22749102] gi|40255146| |ref J|NM.152529.31(40255146] gi[31542773| |ref J|NM.152531.3 |[31542773] gi|22749104J |ref JjNM 152533.11(22749104] gi|449176 " |ref J|NM.152534.2 [[44917614] gi|500801 61 jref JjNM.152536.21(50080161] gi|22749122 |ref J|NM..152538.1 [[22749122] giJ22749116, |ref J|NM.152539.11(22749116] gi|40255253] |ref J|NM..152540.3 [[40255253] gi|31542532 jref JjNM .152542.2 [[31542532] giJ22749124 | ref J |NM.152543.1 |[22749124] gi|22749134 [ref J|NM .152544.1 [[22749134] giJ22749128 |ref J|NM.152545.11(22749128] gi|22749138 [ref J|NM .152546.1 |[22749138] giJ22749132 jref J |NM .152547.11(22749132] gi|22749142] |ref J|NM 152548.1 [[22749142] giJ22749136 jref J |NM..152549.1 |[22749136] giJ47578102; |ref J|NM 152550.2 [[47578102] giJ33457354| jref JjNM..152551.2 [[33457354] gi|22749150' Jref J]NM..152552.1 j[22749150] gi|34303935 jref J]NM .152553.2 |[34303935] gi[22749154 |ref J]NM 152554.11(22749154] gi|22749158 j,ref J |NM .152556.1 |[22749158] giJ46358365| jref J |NM..152557.3 j[46358365] giJ48675824' |ref J|NM..152558.2 [[48675824] giJ30795189 |ref J|NM..152559.2 |[30795189] giJ44681483 |ref J|NM..152562.2 j[44681483] gi|24211014 |ref J|NM. 152563.11(24211014] gi|35493700 jref J |NM. 152564.3 [[35493700] gi|22749164 |ref j|NM 152565.1 [[22749164] giJ22749178 |ref J|NM 152568.11(22749178] gi|22749172 jref J |NM..152569.1 j[22749172] gi|22749182 |ref J|NM .152570.11(22749182] gi|22749176 |ref J|NM .152571.1 j[22749176] gi|31377569 jref J |NM..152572.2 j[31377569] giJ40255118 |ref j|NM 152573.2 |[40255118] gi|22749190 jref J]NM..152574.1 [[22749190] gi|22749188 [ref J|NM .152577.1 [[22749188] gi|22749198 jref J |NM .152578.1 [[22749198] giJ50053828 |ref J|NM. 152579.2 |[50053828] gi|22749196 jref JjNM 152581.1 [[22749196] gi|37537697 I ref J |NM..152582.3 |[37537697] giJ22749204 |ref J|NM..152583.1 [[22749204] gi|24211016, |ref J|NM 152584.1 [[24211016] giJ34303936 jref JjNM..152585.1 |[34303936] giJ40255120 jref J]NM. 152586.21(40255120] giJ40255122 |ref J|NM..152587.2 [[40255122] gi|22749210, |ref J|NM .152588.1 |[22749210] gi|22749212] |ref j|NM 152589.1 [[22749212] giJ22749214 [ref J]NM..152590.1 |[22749214] gi|22749216 jref J |NM..152591.11(22749216] giJ38570051 |ref J|NM .152592.2 |[38570051] gi|22749220, jref J |NM .152594.1 [[22749220] gi|25777745| jref JjNM. 152595.21(25777745]
gi|34303938|ref|NMJ52596.2|[34303938] gi|34222280Jref|NMJ52597.3|[34222280] giJ22749228Jref|NMJ52598.lj[22749228] gi|46048468|ref|NMJ52599.2|[46048468] gi|22749232|ref|NMJ52600.lj[22749232] gi|22749234|ref|NMJ 52601.11[22749234] gi|22749236Jref|NM J 52602.11[22749236] gi|34303940|ref|NMJ52603.2J[34303940] gi|23097320|ref| N M J 52604.1 J[23097320] giJ46094067|refJNMJ52605.2i[46094067] gi|32306517|ref|NMJ 52606.2|[32306517] gi[22749242Jref(NM J 52607.1 ([22749242] gi|40255124Jref|NMJ52608.2i[40255124] giJ22749246|ref)NMJ52609.1|[22749246] gi|22749248|ref|NMJ 52610.1 j[22749248] gi|34303942|ref |NM J 52611.2|[34303942] giJ29501808iref[NMJ52612.2i[29501808] gi|22749254Jref|NMJ52613.1|[22749254] gi|22749256|ref|NMJ52614.l[[22749256] giJ22749258|ref|NMJ 52615.11[22749258] giJ29029527|ref|NMJ52616.3|[29029527] gi[31377565Jref(NMJ52617.2|[31377565] giJ40217787|ref|NMJ52618.2[[40217787] giJ22749266irefJNMJ 52619.11[22749266] gi|37622897|ref|NMJ52620.2[[37622897] gi|37059795Jref|NMJ 52621.3|[37059795] giJ40255126irefiNMJ52622.2J[40255126] gi[22749274Jref!NMJ52623.1|[22749274] gi|37537718|ref|NMJ 52624.3|[37537718] giJ2274927δ[ref |NM J 52625.1 j[22749278] gi|37537682|ref|NMJ52626.2|[37537682] gi|24432084Jref|NMJ52628.2J[24432084] gi|34303943Jref|NMJ52629.2J[34303943] gi|34303945|ref|NMJ52630.2|[34303945] gi[22749288[reflNMJ 52631.1 [[22749288] giJ2274929θ[ref|NM J 52632.11[22749290] gi|22749292|ref|NMJ52633.1|[22749292] gi[22749296[ref|NMJ52635.1|[22749296] gi|22749298|ref|NMJ 52636.11[22749298] giJ2274930θjref|NMJ 52637.11[22749300] giJ34303946Jref|NMJ52638.2J[34303946] gi|40548402|ref|NMJ52640.3|[40548402] gi[50845421(ref|NMJ 52643.5|[50845421] giJ22749312Jref|NMJ52644.lj[22749312] giJ22749318|refJNMJ52647.1|[22749318] gi[22749322iref[NMJ52649.li[22749322] gi|22749324|ref|NMJ 52652.1| [22749324] gi|22749326|ref|NMJ52653.lj[22749326] giJ22749328Jref JNM J 52654.1 j[22749328] gi|40217797|refJNMJ52655.2J[40217797] gi[33286435[ref|NMJ52657.3i[33286435] gi|22749336[ref|NM J 52658.11[22749336] gi[22749340|ref|NMJ52660.1|[22749340] giJ22749344Jref|N J 52662.11[22749344] gi[32441282|refJNMJ52663.2|[32441282] gi|2274935θ[ref|NMJ52665.lj[22749350] gi|22749352|ref|NMJ52666.1|[22749352]
gi|23308748[ref|NMJ 52667.11[23308748] gi|22749356Jref[NMJ 52670.11[22749356] giJ50881948|ref[NMJ52671.2J[50881948] gi]47271484|ref|NMJ52672.3|[47271484] gi|2309733θiref[NMJ52673.li[23097330] gi|23097324|ref|NMJ52675.1|[23097324] giJ22749362Jref[NMJ 52676.1 [[22749362] gi|22749364[ref|NMJ52677.1|[22749364] giJ32698776|ref|NMJ52678.1|[32698776] gi|34328082|ref|NMJ52679.2|[34328082] gi|22749366|ref|NM J 52680.1 [[22749366] giJ22749368[ref[NMJ52681.lj[22749368] giJ2274937θiref|NMJ52682.lj[22749370] gi|22749372|ref)NMJ 52683.1 j[22749372] giJ22749374Jref|NMJ 52684.1 [[22749374] gi|44680142|ref|NMJ52685.2|[44680142] gi(22749376|ref(NMJ52686.1|[22749376] giJ22749378Jref|NMJ 52687.1 ([22749378] gi|22749380|ref|NMJ52688.1|[22749380] gi[40255128[ref|NMJ52689.2J[40255128] gi|24497590|ref|NMJ 52690.11[24497590] giJ31542248iref|NMJ52692.2J[31542248] gi|31543182|ref|NMJ52693.2|[31543182] giJ22749386Jref|NMJ 52694.1 j[22749386] gi[4727144θiref[NMJ52695.3|[47271440] gi|38201637|ref(NMJ 52696.31 [38201637] giJ34303947Jref[NMJ52697.2J[34303947] gi|22749392Jref(NMJ 52698.1|[22749392] giJ34303948Jref|NMJ52699.2i[34303948] gi[34303949[ref|NMJ52700.2|[34303949] gi|31657091|ref|NMJ52701.2|[31657091] gi|22749400|refJNMJ52702.lj[22749400] giJ40288202iref(NMJ52704.2J[40288202] giJ22749406(refJNMJ 52705.1 ([22749406] gi|3430395θirefJNMJ52706.2p4303950] gi(31652216Jref|NM J 52707.2|[31652216] gi|22749412|ref|NMJ 52710.11[22749412] gi[34303951|ref|NMJ52713.2|[34303951] gi|22749416|ref|NMJ52715.1|[22749416] gi|22749426|ref|NMJ 52716.11[22749426] gi|2274942θ(ref|NMJ 52717.11[22749420] giJ2274943θ[refiNMJ52718.1|[22749430] gi[22749424iref|NMJ52719.lj[22749424] giJ23510392|ref|NMJ 52720.1|[23510392] giJ47575848[ref|NMJ 52721.2J[47575848] gi[40795672|ref|NMJ52722.3|[40795672] gi|22749428|ref|NM J 52723.1 j[22749428] gi|34577082[ref|NMJ52724.2[[34577082] gi|22749432|ref|NMJ52725.1|[22749432] giJ22749442Jref|NMJ 52726.1 [[22749442] gi|30181235Jref|NMJ52727.4J[30181235] gi|22749444|refJNMJ 52728.11[22749444] gi(38570155(refiNMJ52729.2i[38570155] gi|41529833|ref|NMJ52730.3|[41529833] gi|22749448[refJNMJ 52731.11[22749448] gi|32964824Jref|NMJ52732.2|[32964824] gi|23308690|ref|NM J 52733.1 [[23308690]
gi|47271470 |ref J|NM. 152734.2 |[47271470] gi[24475836 jrefj JNM 152735.2 ([24475836] gi(34303952 |ref J|NM. 152736.2 [[34303952] gi|22749454 jref J]NM 152737.11(22749454] gi[22749456 [ref JjNM. 152738.1 ([22749456] gi[24497557 | ref JjNM 152739.2 [[24497557] gi[41393565 [ref J|NM 152740.2] |[41393565] gi|22749458 |ref J|NM 152742.1 ([22749458] giJ25092657 | ref J |NM. 152743.1 |[25092657] giJ32880200 |ref J|NM 152744.21(32880200] gi[23097337 | ref J |NM. .152745.1 |[23097337] gi|22749464 jref J |NM 152747.11(22749464] gi|33356135 [ref J|NM. .152748.2 |[33356135] gi(37059760 jref JjNM .152749.2 [[37059760] gi|40255132 [ref| NM .152750.2 |[40255132] giJ22749470 |ref| NM 152751.1 j[22749470] gi|31377567 |ref J|NM .152753.2 ([31377567] giJ41406085 | ref JjNM. 152754.2 j[41406085] gi|22749478 |ref J|NM. .152755.1 j[22749478] gi[22749482 jrefj JNM. .152757.11(22749482] gi|40255134 |ref (jNM .152758.2 |[40255134] giJ40255136 |ref J|NM .152759.31(40255136] gi|22749488 |ref J|NM. 152760.1 ([22749488] giJ22749496 jref J |NM .152761.11(22749496] giJ22749492 |ref J|NM .152762.1 |[22749492] gi(34303954 [ref JjNM .152763.21(34303954] gi[22749494 |ref J|NM .152764.1 |[22749494] gi|34303955 |ref j|NM .152765.2 [[34303955] giJ37059761 |ref J|NM .152766.2 |[37059761] gi|22749502 |ref] |NM ,152769.1 ([22749502] gi(22749508 jref J |NM ,152770.11(22749508] gi|22749510 Jref JjNM 152771.1 ([22749510] giJ22749516 |ref J|NM ,152772.11(22749516] gi|34303956 |ref J|NM ,152773.2 P4303956] gi|22749518 [ref J|NM 152774.1 ([22749518] giJ22749514 jref J |NM 152775.1 j[22749514] gi|22749520 jref J|NM. 152776.11(22749520] giJ34303957| I ref J |NM 152777.2 j[34303957] gi[22749524 |ref J|NM. 152778.1 ([22749524] gi(22749526 |ref J|NM 152779.1 j[22749526] gi|31343502 I ref ( |NM 152780.21(31343502] giJ22749528 [ref| |NM~ 152781.1 [[22749528] gi(34303958 |ref J|NM. 152782.2 ([34303958] gi(40255138 |ref J|NM. 152783.2 |[40255138] gi|38678531 jrefj NM. 152784.21(38678531] gi|33186855 |ref| NM_ 152785.2 [[33186855] gi|22749538 |ref J|NM. 152786.11(22749538] giJ38146004 |ref J|NM 152787.2 |[38146004] giJ50511944 |ref J|NM. 152788.31(50511944] gi|22749542 jref JjNM. 152789.1 [[22749542] giJ40255144 |ref J|NM. 152791.31(40255144] gi|22758145 |ref J|NM. 152792.1 |[22758145] giJ23618862 jref JjNM 152793.1 j[23618862] giJ23065534 |ref| |NM_ 152794.1 [[23065534] gi|23065537 jref J]NM. 152795.1 ([23065537] gi|23065540 [ref J]NM. 152796.1 j[23065540] gi|23111035 [ref JjNM. 152826.11(23111035]
gi(23111040[ref|NMJ52827.1([23111040] gi|34304375|ref|NMJ52828.2|[34304375] giJ23238187|ref|NMJ52829.1|[23238187] gi[23238213iref(NMJ52830.li[23238213] gi|23238215|ref|NMJ52831.1|[23238215] gi(22779872|ref|NMJ52832.1|[22779872] giJ4054839θiref|NMJ52834.2[[40548390] gi|22779869|ref|NMJ 52835.1|[22779869] giJ23238245|ref|NMJ 52836.11[23238245] gi|23238247|ref|NMJ 52837.11[23238247] giJ33469953Iref|NMJ52838.2|[33469953] gi|23110967[ref|NMJ52840.1|[23110967] gi|23110969Jref|NMJ 52841.1J[23110969] giJ23110971 jrefJNMJ52842.1|[23110971] gi|23110973lref|NMJ52843.1|[23110973] gi|38045916|ref|NMJ52850.2J[38045916] giJ23238235iref[NMJ52851.lj[23238235] giJ23238237|ref|NM J 52852.11[23238237] gi|23312370|ref|NMJ52854.1|[23312370] giJ23110979|ref|NM J 52855.1 ([23110979] gi|23111017|ref|NMJ 52856.11[23111017] gi|23199973[ref|NMJ52857.li[23199973] gi[23199975|ref|NM J 52858.1 j[23199975] giJ22902135JrefJNMJ52860.lj[22902135] gi|23238210|ref|NMJ52862.l([23238210] gi|42476063|ref|NMJ52864.2|[42476063] gi|23110988|refJNMJ52866.li[23110988] gi(2311099θiref(NMJ52867.lj[23110990] giJ23110983|ref|NMJ52868.lj[23110983] gii23111020|refJNM_152869.li(23111020] gi|23397656|ref|NMJ 52870.11[23397656] giJ2351042θ[ref|NMJ52871.1|[23510420] gi)23510422|ref|N J 52872.1 [[23510422] gi|23510424|ref|NM J 52873.1 [[23510424] gi[23510426|ref|NMJ 52874.11[23510426] gi|23510428|ref|NMJ52875.1|[23510428] gi[2351043θjref(NMJ52876.1([23510430] gi[23510433|ref)NMJ52877.lj[23510433] giJ23111002JrefJNM_152878.1 j[23111002] gi(25777597|ref|NMJ52879.2J[25777597] gi|27886605|ref|NMJ52880.2|[27886605] gi|27886606[ref(NMJ 52881.2[[27886606] gi|27886607iref|NMJ52882.2J[27886607] giJ27886608JrefJNMJ52883.lj[27886608] gi|40805822|ref|NMJ52888.1|[40805822] gi|23097237|ref|NMJ52889.1|[23097237] gi[32964828[ref(NMJ52890.4|[32964828] gi(23097243|ref|NMJ52891.l[[23097243] gi|23097239Jref|NMJ52892.1|[23097239] gi|23312363[ref|NMJ52896.lj[23312363] gi|23510349|ref|NMJ52897.1|[23510349] giJ40217806|ref|NMJ52898.2([40217806] gi|23821022[ref|NMJ52899.1l[23821022] gi|23097339JrefJNM J 52900.1 j[23097339] gi[23097245[ref(NM J 52901.1 [[23097245] gi(33239373|ref|NMJ52902.2|[33239373] gi|45333882Jref|NMJ52903.3i[45333882]
|23097259 |ref J|NM 152904.1 |[23097259] (34303960 [ref J|NM .152905.21(34303960] (38488711 |ref J|NM 152906.2 [[38488711] [38261959 jref JjNM 152908.2 ([38261959] (34303961 jref JjNM 152909.2 [[34303961] [32307168 [ref J|NM .152910.31(32307168] 146397315 jref JjNM .152911.2 j[46397315] (32306535 |ref J|NM .152912.3 ([32306535] |23097273 |ref j|NM 152913.1 ([23097273] |23097269 (ref J|NM..152914.1 j[23097269] [23397682 |ref J|NM 152916.1 |[23397682] |23397684 |ref j|NM..152917.11(23397684] J23397686 | ref J |NM 152918.1 |[23397686] [23397688 |ref J|NM .152919.1 |[23397688] |23397690 [ref| |NM..152920.1 ([23397690] (23397692 [refj JNM .152921.1 |[23397692] [38026872 jref J|NM..152924.2 [[38026872] (23397695 jref JjNM..152925.1 |[23397695] [23397697 |ref J|NM .152926.1 ([23397697] J23397699 |ref J|NM ,152927.1 |[23397699] 123397701 |ref J|NM. ,152928.1 [[23397701] J23397703 jref J |NM ,152929.1 [[23397703] [23397705 |ref j|NM. ,152930.11(23397705] |23397707 | ref J |NM 152931.11(23397707] J23510345 |ref J|NM ,152932.1 ([23510345] 123312381 jref J |NM. 152933.11(23312381] J23312383 |ref J|NM ,152934.1 |[23312383] |23397638 jref J|NM. ,152939.11(23397638] (23510436 jref JjNM 152942.1 ([23510436] 141281777] jref j |NM 152943.1 [[41281777] J23308700 [ref J|NM 152945.1 |[23308700] J41281781 |ref J|NM. 152988.11(41281781] J30061560 |ref J|NM 152989.2 |[30061560] J24475838 |ref j|NM 152990.2 [[24475838] J24041022 |ref| |NM_ 152991.1 ([24041022] (23510405 jref J |NM 152992.1 [[23510405] J31343495 |ref| |NM_ 152994.2 ([31343495] 137674231 (ref| JNM 152995.3 j[37674231] (23308723 |ref] |NM 152996.1 |[23308723] I50428928 |ref| |NM 152997.21(50428928] [23510383 | ref J]NM.152998.1 ([23510383] (25092600 [ref J|NM 152999.21(25092600] [30387616 |ref J|NM 153000.3 [[30387616] J24430154 [ref JjNM 153001.11(24430154] [23308584 jref JjNM 153002.1 ([23308584] (23308598 jref J)NM 153003.11(23308598] [23510357 |ref J|NM 153005.1 [[23510357] J23308586 |ref J|NM 153006.11(23308586] [31559788 [ref J|NM.153007.3 ([31559788] 131559787 jref JjNM 153008.3 [[31559787] J30410025 jref JjNM 153010.3 |[30410025] 151036604 |ref J|NM 153011.2 |[51036604] [23510440 |ref (|NM 153012.1 |[23510440] (40255107 |ref J|NM 153013.2 ([40255107] J23308520 jref j|NM 153014.1 |[23308520] J23308544 |ref J|NM 153015.1 [[23308544] J23308510 jref] JNM 153018.1 |[23308510]
gi|29824429|ref|NMJ 53019.2|[29824429] gi|2330856θ[ref|NMJ 53020.1 [[23308560] gi(23308506|ref|NMJ 53022.11[23308506] gi[23308552[ref|NMJ53023.1 ([23308552] gi|23308558[ref|NMJ53024.1|[23308558] giJ23308500|ref|NM J 53025.1 j[23308500] giJ23308518Jref|NMJ53026.1 [[23308518] giJ23308540|ref[NM J 53027.11[23308540] gi|23308516Jref|NMJ53028.11[23308516] gi|4892801 βjref |NM 53029.3J[48928018] gi|23308528(ref|NMJ 53031.11[23308528] gi|23308548|ref|NMJ53032.1|[23308548] gi|2330855θ(ref|NM J 53033.1 ([23308550] gii40255102(ref|NMJ53034.2[[40255102] gi[23308504(ref|NM J 53035.11[23308504] gi(23308562[ref|NMJ53036.1 |[23308562] giJ23308536[ref|NMJ53038.11[23308536] gi(23308512|ref(NMJ53040.1 j[23308512] gi[23308538|ref|NMJ 53041.1 [[23308538] gi[45433553iref[NMJ53043.3i[45433553] gi[23308524Jref |NM J 53044.1 ([23308524] gi|33356141|ref|NMJ 53045.2|[33356141] giJ42734387|ref|NMJ 53046.11[42734387] gi|23510361 |ref|NMJ 53047.11[23510361] gi|23510363(ref|NMJ53048.lj[23510363] gi(23510386[ref|NMJ 53050.1 [[23510386] giJ23510388|ref |NM J 53051.1 [[23510388] gi[23510395[ref|NMJ53181.2|[23510395] gi|41281787|ref|NMJ53182.1 [[41281787] giJ41393548JrefjNMJ53183.1|[41393548] giJ23346423|ref|NMJ53184.1|[23346423] gi|47157333Jref|NM J 53186.2J[47157333] gi|23510409Jref|NM J 53187.1 [[23510409] gi|23510380|ref|NMJ 53188.11[23510380] gi|23510417|ref|NMJ53189.l [[23510417] gi|23510414|ref|NMJ53191.1|[23510414] gi|24497600|ref |NM J 53200.11[24497600] gi[24234685|ref|NMJ 53201.11(24234685] gij24041039|ref|NMJ 53202.11[24041039] gi(23397448|ref|NMJ53204.1 |[23397448] gi|23397450|reflNM J 53206.1 j[23397450] gi|49472839|ref|NMJ53207.3([49472839] gi[23397455|ref(NMJ 53208.1 [[23397455] giJ23397457|ref|NMJ 53209.11(23397457] giJ23397461 [refJNMJ 53211.1 [[23397461] giJ23397463Jref|NMJ 53212.1 ([23397463] gi(40255148|ref[NMJ53213.2i[40255148] gi|2339747θ[ref|NMJ 53214.11[23397470] gii23397472[ref[NMJ53215.li[23397472] giJ23463325Jref|NMJ53216.1|[23463325] gi[23397474(ref[NMJ53217.1 ([23397474] gi[23397476(ref|NMJ53218.1|[23397476] gi(44921601 [ref |NM J 53219.2|[44921601] gi|23397480|ref|NMJ 53220.11[23397480] gi|23510284|ref|NMJ 53221.11[23510284] gi|23397482|ref|NMJ 53223.1 j[23397482] gi|4884373θjref|NMJ53225.2|[48843730]
gi|23397491[ref|NMJ 53226.11[23397491] giJ23397495|ref|NMJ53228.1[[23397495] gi|23397497Jref|NMJ53229.1|[23397497] gi|23397499|ref|NMJ 53230.11[23397499] gi|4025515θjref|NMJ53231.2|[40255150] gi|34222202|ref|NMJ53232.2J[34222202] giJ23397509Jref|NMJ 53233.1 ([23397509] giJ34222285(ref|NMJ53234.3J[34222285] gi(28416944Jref|NMJ53236.2J[28416944] gi[23397519[ref|NMJ 53238.1 [[23397519] giJ28875802|ref|NMJ53239.2|[28875802] gi|34304359Jref|NMJ53240.3|[34304359] gi|2339753ljref|NMJ53244.lj[23397531] giJ23397533Jref|NMJ53246.lj[23397533] gi|23397535iref[NMJ 53247.1 [[23397535] giJ31377558|ref|NM J 53248.2|[31377558] gi|4945785θ[ref|NMJ53251.2|[49457850] giJ45120095JrefJNMJ53252.2J[45120095] gi|24497628JrefJNMJ53253.28|[24497628] gi|23397543Jref|NMJ53254.1|[23397543] gi|33469926|ref|NMJ53255.2|[33469926] giJ31377559|ref|NMJ53256.2|[31377559] gi J46371194 jref|N J 53257.2([46371194] giJ23397553Jref|NMJ 53260.11[23397553] giJ37059796Jref|NMJ 53261.3J[37059796] gi|23397557|ref|NMJ53262.1|[23397557] gii23397559Jref|NMJ53263.l[[23397559] gi[40255154|ref|NMJ53264.2|[40255154] giJ23397563JrefJNMJ 53265.1 j[23397563] gi|34222205|ref|NMJ53266.2|[34222205] gi|40538801|ref|NMJ53267.3|[40538801] gi(23397569|ref|NMJ53268.l([23397569] gi|23943927|ref|NMJ53269.l([23943927] giJ2339757ljref|NMJ53270.1|[23397571] gi|23397573[ref|NMJ53271.1 ([23397573] gi|23510334|ref|NMJ53273.1|[23510334] gii23397575irefJNMJ53274.li[23397575] giJ24497475|ref|N J 53276.1 ([24497475] glJ24497477JrefJNMJ53277.lj[24497477] gi|24497479|ref|NMJ53278.l([24497479] gi|24497481 |ref|NMJ 53279.11[24497481] gi(23510339Jref|NMJ53280.1|[23510339] gi|24497563|ref|NM 153281.1|[24497563] gi|24497565Jref|NMJ53282.1|[24497565] giJ24497567|ref|NMJ 53283.1 [[24497567] gi|24497569|ref|NMJ 53284.11[24497569] giJ2449757ljrefJNMJ53285.1|[24497571] gi|24497573(ref|N J 53286.11[24497573] giJ37704381 jrefJNM J 53289.2J[37704381] giJ23463298[ref|NMJ 53290.1 ([23463298] gi|23463288|ref|NMJ 53291.1|[23463288] gi(24041031|ref(NMJ53292.1|[24041031] gi|24497494|ref|NMJ53320.1|[24497494] gi|24430 62|ref|NMJ 53321.1 j[24430162] gi|24430164|ref|NMJ 53322.1 [[24430164] gi|24475840|ref|NMJ53324.2|[24475840] gii24233573Jref|NMJ53325.1|[24233573]
gi|24497576|ref|NMJ53326.1|[24497576] gi|24119167|ref|NM_153328.11[24119167] gi|34222200|ref|NMJ53329.2|[34222200] giJ46094077Jref[NMJ53330.2|[46094077] gi|40255152|ref|NMJ 53331.2|[40255152] gi|31543183|ref|NMJ53332.2|[31543183] gi|23503246(ref[NMJ 53333.1 j[23503246] gi[33598938|ref|NMJ53334.3p3598938] gi|33569217|ref|NMJ 53335.3|[33569217] gi(2350326θ[ref[NMJ 53336.1 j[23503260] gi|23503254|ref|NMJ 53337.11[23503254] gi|23503264Jref|NMJ53338.l[[23503264] gi|23503258[ref|NMJ 53339.11[23503258] gi(31377554[ref|NMJ53340.2J[31377554] gi[23503262|ref|NMJ 53341.1 j[23503262] gi[23503272|ref|NMJ53342.1|[23503272] giJ37059767[ref|NMJ53343.2i[37059767] gi[23503276|ref|NMJ 53344.1 ([23503276] gi(23503270|ref|NMJ53345.1|[23503270] gi[34222211|ref|NMJ53346.2J[34222211] gi|23503274|ref|NMJ 53347.11 [23503274] gii30795122(ref|NMJ53348.2[[30795122] gi(34222207Jref|NMJ53350.2p4222207] gi|37059768JrefiNMJ53353.2i[37059768] gi|34222213[ref|NMJ53354.2([34222213] gi|34222210|ref|NMJ53355.2|[34222210] gi[23503282|ref|NMJ53356.1|[23503282] gi|23503292|ref|NMJ 53357.11[23503292] giJ23503286|ref|NMJ 53358.1 [[23503286] gi|23503298|ref|NMJ53359.lj[23503298] giJ23503290|ref|NMJ 53360.1 [[23503290] gi J37059769Jref|NM J 53361.2[[37059769] gi(23503296|ref|NMJ53362.1|[23503296] gi|2350330δ[ref|NMJ 53363.1 j[23503306] gi|2350330θ[ref|NM J 53364.11[23503300] giJ23503310|ref|NM J 53365.11[23503310] gi|23503314|ref|NMJ53367.1|(23503314] gi[23503308|ref|NMJ53368.1|[23503308] gi|24158475iref|NMJ53369.l[[24158475] gi|23503318|ref|NMJ 53370.11[23503318] giJ34222215iref[NMJ 53371.2|[34222215] gi|24119276|ref|NM J 53373.1 ([24119276] giJ23503312Jref|NMJ53374.1|[23503312] gi(23503320|ref|NMJ53375.l([23503320] gi|23503316JrefJNMJ53376.1|[23503316] gi(40255156iref|NMJ53377.3|[40255156] gi|24497486|ref|NMJ 53378.11[24497486] gii24041013[ref(NMJ53379.li[24041013] gi|23510454|ref|NMJ 53380.11[23510454] gi|41281793Jref|NMJ 53381.1 [[41281793] gi|24234725(ref[NMJ 53425.11[24234725] gi(24234707[ref|NMJ53426.1 ([24234707] gi|24234710|ref|NMJ53427.1 |[24234710] gi|24430182|refJNMJ53437.lj[24430182] gi|23592219|ref(NMJ53442.1|[23592219] gi[46488945(refiNMJ53443.2i[46488945] gi[2359222l(ref|NMJ 53444.1 [[23592221]
123592229 |ref J|NM.153445. |[23592229]
(23592223 jref J|NM 153446. 1(23592223]
132481210 I ref J |NM 153447. [[32481210]
[38455418 jref j|NM .153448. 1(38455418]
J24475843 |ref J|NM 153449. |[24475843]
J23592231 |ref J|NM. 153450. 1(23592231]
(23943891 |ref (|NM .153451. ([23943891]
[23592241 jref J |NM 153453. |[23592241]
(23592235 |ref J|NM .153454. [[23592235]
[45580706 |ref J|NM..153456. ([45580706]
(24430194 |ref J|NM..153460. 1(24430194]
|24430196 |ref J|NM..153461. |[24430196]
J24430198 [ref JjNM .153462. 1(24430198]
|24430200 |ref J|NM..153463. j[24430200]
(24234755 |ref J|NM .153464. ([24234755]
124041017 |ref J|NM..153477. |[24041017]
[24234759 (ref J|NM 153478. 1(24234759]
|24234762 jref J |NM .153479. |[24234762]
(24430203 (ref J|NM. 153480. j[24430203]
J24430205 jref J |NM .153481. j[24430205]
|24430207 |ref J|NM..153482. ([24430207]
|24430209 (ref J|NM .153483. j[24430209]
124430148 jref J INM .153485. 1(24430148]
|37595753 jref J |NM..153486. [[37595753]
J24475844 (ref J|NM 153487. 1(24475844]
J34335237 |ref J|NM .153488. P4335237]
J24234695 [ref JjNM 153490. [[24234695]
147717113 |ref J|NM .153497. ([47717113]
J23943849 [ref JjNM 153498. [[23943849]
J27437016 [ref JjNM..153499. ([27437016]
[27437018 |ref| |NM 153500. [[27437018]
I23957689 jref J|NM .153603. |[23957689]
123957691 |ref j|NM. 153604. ([23957691]
(23957695 | ref J |NM .153606. |[23957695]
|23957697 |ref j|NM .153607. j[23957697]
|23957699 |ref J|NM .153608. |[23957699]
J23957701 jref J |NM. 153609. 1(23957701]
I32698779 jref j |NM .153610. |[32698779]
(48976062 [ref J|NM..153611. ([48976062]
|45267823 | ref J |NM 153612. |[45267823]
[23957707 lref j|NM .153613. |[23957707]
139204546 [ref JjNM..153614. [[39204546]
J23957679 jref J |NM 153615. |[23957679]
|24234734 [ref J|NM .153616. 1(24234734]
I24234737 |ref J|NM..153617. |[24234737]
I24234740 jref J |NM 153618. ([24234740]
[24234743 |ref J|NM .153619. [[24234743]
[24497508 |ref J|NM .153620. 1(24497508]
124497512 jref J |NM..153631. 1(24497512]
J24497514 Iref JjNM .153632. |[24497514]
I24497539 |ref J|NM .153633. [[24497539]
[38683845] jref J |NM .153634. P8683845]
(25121973 |ref J|NM .153635. 1(25121973]
125141327 [ref J|NM..153636. ([25141327]
J24430168 [ref J|NM..153637, [[24430168]
J24430170 [ref J|NM..153638, |[24430170]
(24430172 [ref J|NM. 153639, ([24430172]
gi|24430174lref |NM J 53640.11[24430174] giJ24430176|ref|NMJ53641.1|[24430176] gi[24497446[ref|NMJ 53645.11[24497446] gi|24111227|ref|NMJ 53646.11[24111227] gi|29788984|ref |NM J 53647.11[29788984] gi[29788986(ref|N J 53648.1 ([29788986] gi|39725631 |ref[NM J 53649.2J[39725631] gi[24497503|ref[NMJ 53675.1 [[24497503] gi|41281807|ref|NMJ53676.1|[41281807] gi|24497596|ref|NMJ53681.11[24497596] gi|24497598Jref|NMJ53682.li[24497598] gil24497615lref|NMJ 53683.1 [[24497615] giJ24497448lref |NM J 53684.1 ([24497448] giJ31559779[ref|NMJ 53685.2|[31559779] gi|34916053|ref|NMJ53686.4|[34916053] gi|2423351 e(ref[NM J 53687.11(24233516] gi|2423353l[ref[NMJ 53688.1 [[24233531] gi|31581540|refJNMJ53689.3i[31581540] gi|45333897|ref|NMJ53690.4|[45333897] gi|46049072(ref|NMJ53691.3|I46049072] gil30410026JrefiNMJ53692.2i(30410026] gi|24497543|ref |NM J 53693.11[24497543] gi[50345249|ref|NMJ53694.3[[50345249] gi|46195795|ref|NMJ 53695.2|[46195795] gi|30410031|ref|NMJ53696.2|[30410031] gi(24233529(ref|NMJ 53697.11[24233529] gi|24308513|ref|NMJ 53699.1 [[24308513] gi[31559780[ref|NMJ53700.2J[31559780] gi|24497439Jref(NMJ 53701.1 [[24497439] giJ24308455|ref|NMJ 53702.11[24308455] gi|45505136|ref|NMJ 53703.3|[45505136] gi[42476194JrefJNMJ53704.3[[42476194] gi|33859778[ref|NMJ53705.2|[33859778] giJ31559786|ref|NMJ53706.2([31559786] gi|24308463Jref|NMJ 53707.11[24308463] gi|24308421|ref|NMJ53708.l([24308421] gi(24308423(ref|NM J 53709.1 j[24308423] gi[24308453[ref|NMJ 53711.1 [[24308453] gi|42734436[ref|NMJ53712.3([42734436] gi|24308457|ref|NMJ53713.1|[24308457] giJ24308459|ref|NMJ 53714.11(24308459] gi(2449755θ(ref(NMJ 53715.1 [[24497550] gi|32526912|ref|NMJ53716.l([32526912] gi|2449753θ(ref[NMJ 53717.1 ([24497530] gi|34335247|ref|NMJ53718.2J[34335247] gi|34335248|ref|NMJ53719.2|[34335248] gi|24430134|ref|NMJ 53741.1 [[24430134] gi[34328938|ref|NMJ53742.3[[34328938] gi(24371271 jrefJNM J 53746.11[24371271 ] gi|24430185|refJNMJ53747.11[24430185] gi|24497457|ref|NMJ 53748.11[24497457] gi|24371249|ref |NM J 53750.1 ([24371249] gi|24371253Jref [N J 53752.11[24371253] gi|24371255|refJNMJ 53754.11[243712551 gi|24371259|ref|NMJ53756.11[24371259] gi|24371267|ref|NMJ53757.1|[24371267] giJ30795209Jref|NMJ 53758.1 [[30795209]
gi[28559067[ref(NMJ53759.2|[28559067] gi|24497463|ref|NMJ53763.1|[24497463] giJ24497466JrefJNMJ53764.1J[24497466] giJ24497468[refJNMJ53765.lj[24497468] gi|24497470|ref|NMJ53766.1|[24497470] gi(24497472|ref[NMJ53767.li[24497472] gi|24797113|ref|NMJ53768.1J[24797113] gi[24797115|ref|NMJ53769.1|[24797115] gi|24797117|ref|NMJ53770.1|[24797117] gi|24415991|ref|NMJ53773.1 |[24415991] gi|2442957l[ref|NMJ53809.1|[24429571] gi J34222217[ref[NM J 53810.2|[34222217] gi|24429573|ref|NM J 53811.11[24429573] gi(24432092|ref|NMJ53812.lj[24432092] gi|24429581|ref|NMJ53813.11[24429581] gi|24797100|ref|NMJ53815.1|[24797100] gi|39777616|ref|NMJ53816.2[[39777616] gi|24797088JrefJNMJ53818.1J[24797088] gi|24797102|ref|NMJ53819.1j[24797102] giJ25121957Jref|NMJ 53822.1 [[25121957] gi|24432099|ref|NMJ53823.1|[24432099] gi|24797094|ref[NMJ53824.1|[24797094] giJ24432103[ref[NM J 53825.1 j[24432103] gi|27502403JrefJNMJ53826.2J[27502403] gi|27436916|ref|NMJ53827.2|[27436916] gi|47519507|ref|NMJ53828.2|[47519507] giJ2788659l[ref|NMJ 53831.2J[27886591] gi)24476015|ref|N J 53832.1 ([24476015] gi|24475862|ref|NMJ53833.1|[24475862] gi|24475864|ref|NMJ53834.1 |[24475864] gi|24475866|ref|NMJ53835.1|[24475866] gi[24475868(ref(NMJ53836.1|[24475868] gi(24475870Jref|NMJ53837.1)[24475870] gi|24475872Jref|NMJ53838.1|[24475872] gi|24475874|ref|NMJ 53839.1 [[24475874] gi|24475876|ref|NM J 53840.11[24475876] gi(24797134Jref|NMJ56036.1[[24797134] giJ24797136iref[NMJ56037.l([24797136] giJ27437041 |ref|NM J 56038.2|[27437041] gi|27437043|ref|NMJ56039.2[[27437043] gi|41281804|ref|NMJ 70587.11[41281804] giJ24496784(ref[NM J 70589.1 j[24496784] gi|4128182θiref|NMJ70600.lj[41281820] giJ47458051 jreflNM J 70601.3J[47458051] gi|26051253|ref|NMJ 70602.11[26051253] gi|26051255|ref|NMJ 70603.11[26051255] gi|26051257|ref|NMJ70604.lj[26051257] gi|29029543|ref|NMJ70605.2|[29029543] giJ24586652Jref[NM J 70606.1 j[24586652] giJ38201610lref|NMJ70607.2J(38201610] gi|25121983|ref|NMJ 70609.11[25121983] gi[28558999[refiNMJ70610.2J[28558999] gii34222218(ref|NMJ70662.2J[34222218] gi|27436918|ref|NMJ70663.2J[27436918] giJ24638451 [ref|NMJ70664.1 j[24638451] gi|27886537|ref[NMJ70665.2|[27886537] gi[24762238|ref|NMJ70672.lj[24762238]
gi|27502376|ref|NMJ70674.2|[27502376] gi[27502377(ref|NMJ70675.2i[27502377] gi|27502378|ref|NMJ70676.2J[27502378] giJ27502379[ref|NMJ70677.2J[27502379] gi|24762247|ref|NMJ 70678.11[24762247] gi|25777712|ref|NMJ70679.lj[25777712] gii25306282[ref|NMJ70681.l([25306282] gi|28416917|ref|NMJ70682.2J[28416917] giJ28416918Jref|NM J 70683.2J[28416918] gi|26449156|ref|NM J 70685.1 [[26449156] gil25777703Jref|NMJ 70686.11[25777703] gi[25306286|ref (NM J 70691.1 ([25306286] gi|25121935|ref|NMJ 70692.11[25121935] gi|2516826θjref|NMJ70693.lj[25168260] giJ2500653liref(NMJ70694.1 [[25006531] gi|28178842|ref|NMJ 70695.2|[28178842] giJ25777725|ref|NMJ 70696.1 [[25777725] gi|25777727(ref |N J 70697.1 [[25777727] gi|24850130|ref|NMJ70698.1|[24850130] gi[24850108iref|NMJ70699.li[24850108] gi|24797155|ref|NMJ70705.1|[24797155] giJ34335244|ref|NMJ70706.2J[34335244] gi|27436945|ref|N J 70707.11[27436945] gi|27436947Jref|NMJ 70708.1 j[27436947] giJ25168266iref[NMJ 70709.1 [[25168266] gi|31317310|ref|NMJ70710.2|[31317310] giJ25470889(ref[NM J 70711.1 j[25470889] gi|25777679Jref|NMJ 70712.1 [[25777679] gi|25777681 |ref|NM J 70713.11[25777681] gi[25777683[ref|NMJ70714.li[25777683] giJ25777685|ref|NMJ 70715.1 ([25777685] gi[25777687|ref(NMJ 70716.11[25777687] giJ25777689Jref|N J 70717.1 ([25777689] gi|25777605|ref|NMJ 70719.11(25777605] gi(25777634[ref|NM J 70720.1 ([25777634] gi|25121992jref|NMJ 70721.1|[25121992] giJ25777609(ref|NMJ 70722.11[25777609] gi|2545348θ(ref|NMJ 70723.1 j[25453480] gi|25777666|ref|NMJ 70724.11(25777666] gi|2577774l(ref|NMJ 70725.1 [[25777741] gi(25777735[ref|NMJ70726.lj[25777735] gi|34106706|ref|NMJ70731.2[[34106706] gi[34106707Jref|NMJ70732.2i[34106707] gi|34106708|ref|NMJ70733.2|[34106708] gi|34106709|ref|NMJ70734.2|[34106709] giJ34170263|refJNMJ70735.3|[34170263] gi|25777637|ref|NM J 70736.11[25777637] gi|25777639(ref|NMJ 70737.1|[25777639] giJ2530627ljref[NMJ 70738.1J[25306271] gi|25306274Jref|NMJ 70739.1 [[25306274] gi|25777720|ref|NMJ70740.1|[25777720] gi|25777627|ref|NM J 70741.11[25777627] giJ25777629(ref|NM J 70742.1 [[25777629] giJ28416903Jref|NMJ70743.2|[28416903] giJ32261317|ref|NMJ 70744.2|[32261317] gi(29171724|ref|NMJ 70745.2|[29171724] giJ4637009θjrefJNMJ70746.2J[46370090]
128605136 | ref J |NM .170750.1 |[28605136]
125777618 |ref J|NM 170751.1 ([25777618]
I25777620 jref J |NM 170752.1 |[25777620]
I25777743 |ref J|NM..170753.1 [[25777743]
|38787956 [ref (jNM .170754.2 |[38787956]
I25777700 jref J |NM 170768.11(25777700]
J25777715 jref jjNM .170769.1 |[25777715]
125777717] | ref J |NM. 170770.11(25777717]
[25952151 |ref J|NM 170771.1 j[25952151]
I25777674 |ref j|NM..170773.11(25777674]
|25777676 jref JjNM. 170774.1 [[25777676]
I25777646 |ref J|NM .170775.1 j[25777646]
(40538803 |ref j|NM 170776.3 |[40538803]
141327723 |ref J|NM..170780.21(41327723]
|25777672 |ref J|NM .170781.1 [[25777672]
I25777649 |ref J|NM..170782.11(25777649]
|25777706 |ref J|NM .170783.1 [[25777706]
125914753 |ref J|NM. 170784.11(25914753]
125952117 jref JjNM .171825.1 [(25952117]
|27886641 |ref J|NM..171827.1 [[27886641]
|25952098 jref J |NM .171828.1 ([25952098]
125952101 |ref J|NM .171829.1 [[25952101]
125952104 |ref J]NM. 171830.1 ([25952104]
126051232 jref JjNM .171846.1 [[26051232]
I25777693 |ref JjNM. 171982.1 [[25777693]
|28565284 jref J |NM ,171997.1 j[28565284]
125188192 |ref J|NM. 171998.1 [[25188192]
[25188194 jref (jNM ,171999.1 ([25188194]
(25188188 | ref J |NM 172000.1 |[25188188]
[50301233 |ref J|NM..172002.3 j[50301233]
(38016203 |ref J|NM. ,172003.2 [[38016203]
|40548404 |ref J|NM 172004.2 |[40548404]
125188180 jref JjNM. ,172005.1 ([25188180]
131563539 jref J |NM 172006.21(31563539]
J25952146 jref J|NM ,172014.1 [[25952146]
J25777697 |ref J|NM. 172016.1 ([25777697]
126051277 |ref J|NM 172020.11(26051277]
|25952070 jref JjNM 172024.11(25952070]
I25952073 [ref JjNM. ,172025.1 [[25952073]
J25952076 | ref J |NM 172026.11(25952076]
J25777623 [ref J|NM. 172027.11(25777623]
|25777625 jref J |NM 172028.1 j[25777625]
146048189 [ref J|NM ,172037.2 |[46048189]
J26051270 |ref J|NM. ,172056.1 j[26051270]
126051272 |ref (|NM 172057.1 j[26051272]
126667215 |ref J|NM. ,172058.1 j[26667215]
126667218 | ref ( |NM ,172059.1 |[26667218]
(26667221 |ref J|NM ,172060.11(26667221]
[45120120 |ref J|NM. ,172069.1 [[45120120]
140255162 jref (jNM ,172070.21(40255162]
J26051205 |ref J|NM 172078.1 [[26051205]
126051207 |ref J|NM. ,172079.11(26051207]
126051209 [ref J|NM ,172080.1 |[26051209]
126051211 |ref J|NM 172081.1 [[26051211]
126051213 |ref J|NM ,172082.1 |[26051213]
[26051215 |ref J|NM 172083.1 [[26051215]
J26051217 |ref J|NM 172084.1 |[26051217]
gi|26051245|ref|NMJ72087.1[[26051245] gi(26051247|ref[NMJ 72088.1 j[26051247] gi|47519573|ref[NMJ72089.2J[47519573] giJ26051222Jref|NMJ72095.l[[26051222] giJ26051224|ref|NMJ72096.1 [[26051224] gi|26051226|ref|NMJ 72097.1 ([26051226] gi|26667245|ref|NMJ 72098.1 [[26667245] gi|2788663θjref[NMJ 72099.11[27886630] giJ27886632Jref|NM J 72100.1 [[27886632] gi|27886634|ref|NMJ 72101.1 |[27886634] gi|27886636|ref|NMJ72102.1|[27886636] gi|2666725ljref|NMJ72103.li[26667251] gi|26667254|ref|NMJ72104.1|[26667254] gi|26667256|ref|NMJ72105.1|[26667256] gi|26051261|ref|NMJ72106.lj[26051261] gi|26051263Jref|NMJ 72107.11[26051263] giJ26051265[ref|NMJ72108.1|[26051265] gi(26051267|ref|NMJ72109.1|[26051267] giJ26667229|ref|NMJ72110.l([26667229] gi|26667231 |ref|NM J 72111.11[26667231] giJ26667234|ref|NMJ72112.1|[26667234] gi|26667239|ref[NMJ 72113.1|[26667239] giJ26667182JrefJNMJ72115.li[26667182] gi|26667185|ref|NMJ72127.1|[26667185] gi|26667188[ref|NMJ72128.1|[26667188] giJ27436968|ref JNM J 72130.1 j[27436968] gi|31563540|ref|NMJ72131.2[[31563540] gi|26024320|ref|N J 72138.11[26024320] giJ28144900|ref|NMJ72139.2|[28144900] gi|26024324|ref|NMJ 72140.11[26024324] gi|46255024Jref |NM J 72159.2|[46255024] gi|27436965|ref|NMJ72160.1|[27436965] gi|26638654|ref|NMJ 72163.1|[26638654] giJ27262633Jref|NM J 72164.1 J[27262633] gi(26638663Jref|NMJ 72165.1|[26638663] gi|26638665|ref|NMJ72166.11[26638665] gi|4128181θ[ref|NMJ72167.1|[41281810] gi|41281827JrefJNMJ72168.1|[41281827] giJ26667195|ref|NMJ72169.lj[26667195] gi|40354202Jref |NM J 72170.2|[40354202] gi|26667202|ref|NMJ72171.1 |[26667202] giJ26667205Jref|NM J 72172.1 j[26667205] gi|26667210|ref|NMJ 72173.1|[26667210] gi|26787983Jref|NMJ72174.lj[26787983] giJ26787985|ref|NMJ72175.1|[26787985] gi|2666717θ(ref|NM J 72177.11 [26667170] gi|26667173|ref|NMJ72178.1|[26667173] giJ26190615|ref|NMJ72193.1|[26190615] gi(41281816Jref|NM_172195.1 j[41281816] giJ26787965Jref|NMJ 72196.11[26787965] gi|26787961|ref|NMJ72197.11[26787961] gi|27436985|refJNMJ 72198.11[27436985] giJ2666589θ(refiNMJ 72199.1 [[26665890] gi|26787981|ref|NMJ 72200.11[26787981] gi|27436977|ref|NMJ 72201.11[27436977] gi|27437010|ref|NMJ72206.1|[27437010] gi|27437012|ref|NMJ72207.1|[27437012]
gi|27436894 |refJ|NM 172208 1(27436894] gi|27436896 |refJ|NM 172209 ([27436896] gi|27262662 I refJjNM 172210 j[27262662] gi|27262664 |refJ|NM. 172211 |[27262664] gi(27262666 Iref (jNM 172212 |[27262666] giJ27886638 [ref J|NM .172213 ([27886638] gi(27437020 [refJ|NM 172214 [[27437020] gi|27437022 | refJ |NM .172215 ([27437022] gi|27437024 |refJ|NM 172216 [[27437024] gi|27262656 |refJ|NM. .172217 ([27262656] gi|27262637 |ref (|NM 172218 1(27262637] giJ27437048 |ref J|NM. .172219 |[27437048] gi|27437050 |refj|NM. .172220 j[27437050] gi|27436935 jref J |NM .172225 |[27436935] gi(27437026 [refj|NM. .172226 1(27437026] gi|27437007 |ref J|NM. 172229 ([27437007] gi[27436926 jref (jNM 172230 1(27436926] gi|33469963 |ref(|NM .172231 |[33469963] gi(27262625 |ref J|NM .172232 1(27262625] gi|27477075 |ref J|NM. .172234 |[27477075] gi(27436890 |ref J|NM .172236 j[27436890] gi|27363485 |ref (jNM. .172238 |[27363485] gi|26665874 |ref J|NM 172239 [[26665874] giJ26665868 |ref J|NM. .172240 |[26665868] giJ26665878 |ref J|NM .172241 [[26665878] gi(27262640 Jref JjNM. .172242 ([27262640] giJ46249399 | ref J |NM .172244 1(46249399] gi|27437031 |ref J|NM. .172245 j[27437031] giJ27437033 jref J |NM .172246 j[27437033] gi[27437035 jref JjNM .172247 |[27437035] gi|27437037 jref JjNM 172248 [[27437037] gi(27437039 jref JjNM .172249 |[27437039] gi|26892294 |ref J|NM. ,172250. 1(26892294] giJ27436907 jref ( |NM 172251 |[27436907] giJ29553943 jrefj |NM 172311 |[29553943] gi|27502372 |ref J|NM. 172312 ([27502372] gi(27437044 jref J |NM 172313 [[27437044] gi[27477080 jref (|NM 172314. ([27477080] gi(27502380 |ref J|NM 172315. [[27502380] gi[27502382 |ref J|NM 172316 ([27502382] gi|27436989 |ref J|NM 172318 |[27436989] gi|27436932 |ref J|NM 172337 1(27436932] gi|28144919 |ref J|NM 172341 [[28144919] gi(27477083 |ref J|NM 172343 ([27477083] gi[27436992 Jref J|NM 172344 |[27436992] giJ27436921 |ref J|NM 172345. [[27436921] gi|27436954 jref JjNM 172346, ([27436954] gi[27436995 |ref J|NM 172347, [[27436995] gi|27477091 |ref J|NM 172348, ([27477091] gi(27477094 |ref J|NM 172349, 1(27477094] giJ27502404 jref JjNM 172350, |[27502404] gi|27502406 |ref J|NM 172351, 1(27502406] gi|27502408 |ref J|NM 172352, |[27502408] gi|27502410 jref J |NM. 172353, 1(27502410] giJ27502412 jref J |NM. 172354, ([27502412] giJ27502414 jref JjNM. 172355. 1(27502414] gi[27502416 jrefjjNM. 172356 [[27502416]
127502418 [ref JjNM 172357 1(27502418] |27502420 |ref j|NM .172358 1(27502420] J27502422 jref J|NM .172359 [[27502422] I27502424 |ref J|NM 172360 |[27502424] J27502426 |ref j|NM..172361 [[27502426] J27437000 |ref J|NM 172362 1(27437000] J33589828 jref J |NM 172364 ([33589828] (27312028 jref| |NM .172365 [[27312028] [30089920 jref JjNM. 172366 |[30089920] (27312030 |ref J|NM .172367 |[27312030] [27363487 |ref J|NM..172369 j[27363487] (48843726 jref JjNM 172370 1(48843726] |34222093 [ref JjNM .172373 j[34222093] J27477088 jref J|NM. 172374 1(27477088] J27886645 |ref (|NM .172375 j[27886645] J27886647 jref JjNM..172376 ([27886647] J50428920 |ref (|NM 172377 [[50428920] (27477116 |ref (|NM .172386 1(27477116] |27502385 |ref J|NM..172387, ([27502385] |27502387 |ref J|NM. 172388, |[27502387] J27502390 [ref J|NM. 172389, j[27502390] |27502392 |ref J|NM .172390, ([27502392] I27370564 |ref J|NM..173039, 1(27370564] |27502395 jref J |NM 173042, |[27502395] |27502397 |ref J|NM..173043, j[27502397] J27502399 jref JjNM 173044, |[27502399] J27413157 [ref J|NM..173050, 1(27413157] J27436939 jref jjNM 173054, |[27436939] 127881485 |ref J|NM .173055, 1(27881485] 127881487 |ref J|NM. 173056, 1(27881487] J27881489 jrefj |NM .173057. j[27881489] (27881491 jref JjNM .173058. j[27881491] 127881493 |ref J|NM. 173059. |[27881493] J27765084 jref JjNM .173060. j[27765084] J27765086 jrefj (NM 173061. [[27765086] J27765088 |ref J|NM..173062. 1(27765088] J28416908 |ref (|NM. 173064. ([28416908] J28416910 |ref J|NM .173065. 1(28416910] I34335286 |ref J|NM..173073. 1(34335286] J27894290 |ref (|NM .173074. 1(27894290] I50083290 jref J |NM .173075. [[50083290] J30795237 |ref J|NM 173076. ([30795237] J27436870 jref J|NM. 173077. |[27436870] J40217821 jref ( |NM .173078. 1(40217821] J27436874 |ref J|NM .173079. j[27436874] J27436868 |ref J|NM .173080. ([27436868] I27436878 |ref J|NM..173081. ([27436878] I27436872 jref JjNM .173082. 1(27436872] J32996736 |ref J|NM .173083. j[32996736] |27436876 [ref JjNM .173084. j[27436876] [31343353 jref J |NM .173086. |[31343353] J27765073 [ref JjNM .173087. 1(27765073] J27765075 Jref JjNM..173088. |[27765075] J27765077 jref J |NM. 173089. ([27765077] J27765079 jref JjNM .173090. ([27765079] J27886540 jref JjNM..173091. j[27886540] [27886650 |ref J|NM. 173092. j[27886650]
gi|42475557|ref J|NMJ73156.1 |[42475557] giJ27894343Jref J|NMJ73157.11[27894343] giJ27894345|ref ([NMJ 73158.1 ([27894345] gi|41281830|ref J|NMJ 73159.11[41281830] giJ5065910ljrefj |NMJ73160.2([50659101] gi|27894306|ref ]|NMJ73161.1 J[27894306] giJ27886664Jref J|NMJ 73162.1 j[27886664] gi|27886554|refj |NMJ73163.1 |[27886554] gi|27886558(ref| |NMJ73164.l j[27886558] giJ2788656θjref JjNMJ 73165.11[27886560] gi|27477060|ref JjNMJ73167.1 [[27477060] giJ27894309|ref J|NMJ73170.1 |[27894309] gi|27894348|ref J|NMJ 73171.1|[27894348] gi|27894350|ref| NMJ73172.l [[27894350] giJ27894352Jrefj NMJ 73173.11[27894352] gi|27886585|ref| |NMJ73174.l j[27886585] giJ27886587|ref J[NMJ 73175.11[27886587] gi|27886589|ref J|NMJ 73176.1 [[27886589] gi|27894372|ref| |NMJ73177.1 |[27894372] gi|27894312|ref J|NMJ 73178.11[27894312] gi[34335288Jref||NMJ73179.2J[34335288] gi|50557653|ref J|NMJ73191.2([50557653] gi|50557649|ref J|NMJ 73192.2J[50557649] gi|50557655|refj [NMJ73193.2([50557655] gi|50557651 Jref J|NMJ73194.2J[50557651] giJ50557650|ref J|NMJ 73195.2J[50557650] giJ50557652|ref JjNMJ 73197.2([50557652] gi|27894356|ref| JNMJ 73198.1 j[27894356] giJ27894358|ref J|NMJ73199.1 |[27894358] gi(2789436θjref J|NM J 73200.1 j[27894360] gi|47132613|ref J|NMJ 73201.2|[47132613] gi|27894295|ref J|NMJ 73202.11[27894295] giJ27894297Jref J|NMJ 73203.11[27894297] gi|27894299|ref J|NMJ73204.1 ([27894299] giJ2789430l jref J|NMJ 73205.11[27894301] gi|27532983|ref J|NMJ 73206.11[27532983] gi|28178844Jrefj |NMJ73207.l ([28178844] giJ28178848|ref J|NMJ 73208.1 j[28178848] giJ28178850|ref (|NMJ 73209.11[28178850] giJ28178852|ref J|NMJ73210.l [[28178852] giJ28178854Jref JiNMJ 73211.1 ([28178854] gi|27894340|ref| [NMJ73213.1 |[27894340] gi|27886523|ref J|NMJ 73214.11[27886523] giJ27886525|ref JjNMJ73215.1 |[27886525] gi|2776509θjref| |NMJ73216.l ([27765090] giJ27765092|ref J|NMJ73217.1 |[27765092] gij27894286Jref JjNMJ 73341.1 |[27894286] giJ50593104Jref JjNMJ73342.2J[50593104] giJ27894333Jref J|NMJ73343.1 [[27894333] gi|27765095|ref J|NMJ 73344.1 |[27765095] gi|50051734Jref J|NMJ73351.1|[50051734] giJ27597103|ref J|NMJ 73352.11[27597103] gi|31795562|ref J]NMJ73353.2|[31795562] giJ48762713(ref JjNMJ 73354.2J[48762713] giJ34222223Jref JiNMJ 73355.2|[34222223] giJ28559018Jref JJNMJ73357.2|[28559018] giJ28559019Jref JjNMJ73358.2|[28559019]
gi|50659063|refJ|NM..173360.2 [50659063] gi|40255164|refJ|NM.173362.2 [40255164] gi|27754777|refJjNM..173452.1 [27754777] gi|47132536Jref| NM. 173454.1 [47132536] giJ47132538Jrefj NM.173455.1 [47132538] giJ4713254θjrefJ|NM.173456.1 [47132540] giJ47132542JrefJ|NM. 173457.1 [47132542] giJ27894325|ref (|NM.173459.1 [27894325] gi|37704383|ref J|NM. 173460.1 [37704383] gi|50083294|ref| [NM 173462.2 [50083294] gi[33186919Jref J|NM .173463.2 [33186919] gi|28212273JrefJ|NM 173464.1 [28212273] giJ29725623JrefJ|NM .173465.2 [29725623] gi|27735032|refJ|NM. 173466.1 [27735032] gi|29648316|ref (|NM .173467.2 [29648316] giJ41406062|ref J|NM. 173468.2 [41406062] gi|29789400|refJ|NM .173469.1 [29789400] gi|27735036|refj|NM .173470.1 [27735036] giJ27735034JrefJ|NM..173471.1 [27735034] gi|27735040|refJ|NM 173472.1 [27735040] gi|50234894JrefJ|NM .173473.2 [50234894] gi|40789088JrefJ|NM 173474.2 [40789088] giJ27735046JrefJ[NM .173475.1 [27735046] gi|40255165|refJ|NM 173476.2 [40255165] giJ34304382JrefJjNM..173477.2 [34304382] gi|27735056JrefJ|NM .173478.1 [27735056] gi|47271480|refJ|NM .173479.2 [47271480] giJ28315872JrefJ|NM..173480.1 [28315872] gi|34222226|refJ|NM .173481.2 [34222226] gi|27735062JrefJ|NM .173482.1 [27735062] gi|27735072|refJ|NM 173483.1 [27735072] giJ31377545JrefJJNM. .173484.2 [31377545] giJ40255167JrefJ|NM. .173485.2 [40255167] gij42734389jref||NM_ 173486.1 [42734389] giJ27735074|ref||NM_ .173487.1 [27735074] giJ30520362|ref||NM_ 173488.2 [30520362] gi|38564324|ref||NM_ .173489.2 [38564324] gi|42476211 |ref||NM_ 173490.4 [42476211] gi|27735088|ref||NM_ .173491.1 [27735088] giJ27735084|ref||NM_ .173492.1 [27735084] gi|27735094|ref||NM_ 173493.1 [27735094] giJ33859792Jref|NM_ .173494.1 [33859792] giJ2773509θ[ref|NM_ 173495.1 [27735090] giJ34222227|refJ|NM_ .173496.21 [34222227] giJ27735098|refj|NM_ 173497.1 [27735098] gi|47564113|ref||NM_ .173499.2 [47564113] giJ28466990|ref||NM_ 173500.2 [28466990] gi|27735108|ref||NM_ .173501.1 [27735108] giJ27735116Jrefj|NM_ ,173502.1 [27735116] giJ27735112Jref||NM_ ,173503.1 [27735112] giJ27735120|ref||NM_ ,173505.1 [27735120] giJ40255169|ref|NM_ ,173506.3 [40255169] giJ27735128Jref[NM_ 173507.1 [27735128] giJ27735126Jref[NM_ 173508.1 [27735126] giJ34222229|refJ|NM_ 173509.2 [34222229] gi|27735132Jrefj|NM_ 173510.1 [27735132] giJ2773513θjref||NM_ 173511.1 [27735130]
gi|27735136 |ref J|NM..173512.1 [[27735136] giJ34222230 |ref J|NM 173513.2 [[34222230] gi|27735148 |ref J]NM. 173514.1 j[27735148] giJ27735138 |ref (]NM .173515.1 |[27735138] gi|27735154 [ref JjNM .173516.11(27735154] giJ47717107 jref jjNM 173517.3 ([47717107] giJ40255171 jref J|NM 173518.2 ([40255171] gi|27734848 jref J|NM..173519.11(27734848] giJ27734942 jref (|NM 173521.1 [[27734942] gi|27734962 |ref J|NM 173522.1 |[27734962] gi|29337289 [ref J|NM..173523.2 ([29337289] giJ40255173 |ref J|NM..173524.2 [[40255173] gi|27734960 jref JjNM..173525.11(27734960] gi|27734950 jref J |NM. 173526.1 j[27734950] gi[27734952 |ref (|NM..173527.11(27734952] gi(27734946 jref J|NM. 173528.1 |[27734946] gi[40255175 |ref (|NM..173529.31(40255175] gi|27734940 [ref J|NM. 173530.1 |[27734940] gi|27734944 | ref JjNM..173531.1 ([27734944] gi|27734936 |ref J|NM. 173532.1 [[27734936] gi|40255177 | ref | |NM_ .173533.21(40255177] gi(47778939 jref| |NM_ .173535.2 j[47778939] gi|31742489 |ref| |NM_ .173536.21(31742489] gi|48762699 |ref J|NM_ 173537.2 [[48762699] gi|27734928 jref JjNM_ 173538.1 ([27734928] giJ27734918 jref | |NM_ .173539.1 |[27734918] giJ27734920 |ref| |NM_ .173540.11(27734920] gi|27734914 jref| |NM_ 173541.1 |[27734914] gi[37059775 jrefj |NM_ 173542.21(37059775] gi(27734910 jrefj |NM_ .173543.1 |[27734910] gi|31542206 |ref| |NM_ 173544.2 j[31542206] gi|27734904 jref J |NM_ .173545.1 [[27734904] gi|27734908 |ref] |NM_ ,173546.1 |[27734908] gi|38679904 |ref| |NM_ 173547.21(38679904] gi|27734902 jref J |NM_ ,173548.1 |[27734902] gi[27734896 [ref (|NM_ 173549.11(27734896] gi|38348728 [ref J|NM_ 173550.2 [[38348728] giJ27734888 jref| |NM_ 173551.1 j[27734888] gi|34222231 jref JjNM_ 173552.2 [[34222231] gi|27734882 |ref| [NM_ 173553.11(27734882] gi[27734884 |ref J|NM_ 173554.11(27734884] gi|27734876 |ref J|NM_ 173555.1 ([27734876] gi|40255179 |ref (|NM_ 173556.2 |[40255179] giJ27734872 |ref J|NM_ 173557.11(27734872] gi|34222232 | ref | JNM_ 173558.2 |[34222232] gi[27734868 Iref JjNM_ 173559.1 [[27734868] gi|27734870 jref JjNM_ 173560.1 |[27734870] gi(27734864 Irefj |NM_ 173561.11(27734864] gi|40255181 [ref J|NM_ 173562.3 |[40255181] gi|27734860 Iref JjNM_ 173563.11(27734860] gi|37537553 (ref| |NM_ 173564.2 |[37537553] gi|27734854 |ref| |NM_ 173565.11(27734854] gi|27734856 |ref| |NM_ 173566.1 |[27734856] gi|31542788 |ref| |NM_ 173567.21(31542788] gi|27734852 Irefj |NM_ 173568.1 |[27734852] gi|50234885 |ref J|NM_ 173570.21(50234885] gi|27734834 |ref| |NM_ 173571.11(27734834]
gi|27734836 refl NM. 73572.1 |[27734836] gi[27734828 ref| NM .1 73573.1 |[27734828] gi|27734832 ref| NM..1 73574.1 1(27734832] gi|32455268 ref| NM .173575.2 P2455268] giJ27734826 ref| NM .1 73576.1 [[27734826] gi[27734820 ref| NM. 173578.1 |[27734820] gi|27734812 ref| NM .1 73579.1 ([27734812] giJ27734816 refl NM _1 73580.1 ([27734816] gi|27734806 refl NM .1 73581.1 1(27734806] gi[31377547] ref| NM .1 73582.2 1(31377547] gi|27734800 ref| NM .1 73583.1 |[27734800] giJ27734804 ref| NM .1 73584.1 [[27734804] giJ27734796 ref| NM .1 73586.1 |[27734796] gi|34222235 refl NM 1 73587.2 1(34222235] giJ27734790 ref| NM..1 73588.1 j[27734790] gi|42761478 refj NM .1 73589.2 [[42761478] gi|27734786 refj NM .1 73590.1 |[27734786] gi|27734788 ref| NM .1 73591.1 1(27734788] giJ38566691 ref| NM .1 73593.2 1(38566691] gi|27734776 ref| NM..1 73596.1 1(27734776] gi[27734768 refl NM..1 73597.1 |[27734768] giJ34222392 refj NM..1 73598.2 ([34222392] gi|27734764 refj NM .1 73599.1 j[27734764] giJ27734696 refl NM..1 73605.1 [[27734696] gi|31377542 ref| NM 1 73607.2 |[31377542] gi|27734692 ref| NM .1 73608.1 |[27734692] gi|27734694 ref| NM .1 73609.1 [[27734694] gi|27734706 refl NM..1 73610.1 |[27734706] gi(27734702 refl NM..1 73611.1 j[27734702] giJ28212275 refl NM 1 73613.1 ([28212275] gi|27734708 refj NM 1 73614.1 |[27734708] gi|27734712 ref| NM .1 73616.1 |[27734712] gi|27734731 ref| NM 1 73617.1 j[27734731] giJ27734726 ref| NM .1 73618.1 |[27734726] gi|27734735 ref| NM .1 73619.1 j[27734735] giJ27734733 refl NM. 1 73620.1 |[27734733] giJ27734741 refl NM..1 73621.1 1(27734741] gi|27734737 refl NM .1 73622.1 P7734737] gi|27734745 refj NM .1 73623.1 ([27734745] giJ27734743 ref| NM.1 73624.1 [[27734743] gi|45580703 ref| NM .1 73625.3 |[45580703] giJ27734747 ref| NM .1 73626.1 j[27734747] gi|31542774 ref| NM .1 73627.2 |[31542774] gi[27734756 ref| 1 73628.1 [[27734756] gi|27734982 ref| NM_ .1 73629.1 |[27734982] giJ38201695 refj NM_ 1 73630.2 ([38201695] gi|38044285 ref| NM_ .1 3631.2 P8044285] giJ40255187 refj NM_ 1 73632.2 1(40255187] gi|27734988 ref| NM_ .1 73633.1 [[27734988] giJ27735026 ref| NM_ .1 73635.1 1(27735026] giJ50080220 refj NM_ 173636.3 ([50080220] gi[27735042 refj NM_ 1 3637.1 [[27735042] gi|31341145 ref| NM 1 73638.2 |[31341145] gi(27735030 ref| NM_ 1 3639.1 j[27735030] gi|27735060 refl NM_ 173640.1 |[27735060] gi|27735058 ref| NM_ 173641.1 |[27735058] gi|27735068 refl NM 1 3642.1 |[27735068]
gi|27735064|ref|NMJ73643.1|[27735064] gi|27735086|ref|NMJ 73644.1|[27735086] gi[27735076|ref|NMJ 73645.11[27735076] giJ27735096Jref|NMJ 73646.1 ([27735096] gi|31543079JrefJNMJ73647.2J[31543079] giJ39725716|ref|NMJ73648.2|[39725716] gii27735114|refJNMJ73649.l[[27735114] gi|277351 OδjrefJNMJ 73650.1 ([27735106] giJ27734956Jref|NMJ 73651.1 j[27734956] gi[27734974|ref[NMJ73652.lj[27734974] gi|27734934|ref|NMJ 73653.11[27734934] giJ39930530|refJNMJ 73654.1 [[39930530] giJ27734906|ref|NMJ 73656.11[27734906] giJ27734924JrefJNMJ 73657.1 [[27734924] gi[28212277(ref[NMJ 73658.1 ([28212277] gi|27734886|ref|NMJ 73659.11[27734886] gi|31542769Jref|NMJ73660.2i[31542769] gi|27734842Jref|NMJ 73661.11[27734842] giJ27734858JrefJNMJ 73662.1 j[27734858] gi|2839504θ[ref|NMJ 73663.1 [[28395040] gi|46195790|ref|NMJ 73664.3|[46195790] gi[27734814Jref|NMJ 73665.11[27734814] gi|27734778Jref(NM J 73666.1 ([27734778] gi|27734794|ref|NM J 73667.1 [[27734794] gi|27734690|ref|NM J 73669.11[27734690] gi|27734739|ref|NMJ 73670.11[27734739] giJ27734722Jref|NM J 73671.1 ([27734722] gi|27735044Jref|NMJ 73672.11[27735044] giJ27735001 |ref|NM J 73673.1 j[27735001] gi[27735142iref|NMJ73674.lj[27735142] gi|27735140|ref|NMJ 73675.11[27735140] giJ27734926Jref JNM J 73676.1 [[27734926] giJ27734968|ref|NM 73677.1 ([27734968] gi|27734978JrefJNMJ 73678.1 [[27734978] giJ34222238|ref|NMJ73680.2[[34222238] gi|27734980|ref|NMJ 73682.11[27734980] giJ34222239|ref|NMJ73683.2|[34222239] gi|27734760|ref|NMJ 73685.11[27734760] gi|27735122Jref|NMJ 73687.1 j[27735122] gi|27734999Jref|NMJ 73688.11[27734999] gi|46391097|ref|NMJ73689.3J[46391097] gi|27735124Jref|NMJ73690.1|[27735124] gi|27734964|ref|NM J 73691.1 [[27734964] gi|40316838|refJNMJ 73694.2|[40316838] giJ2773489θjref|NMJ73695.lj[27734890] gi|27734774|ref|NM J 73698.11[27734774] gi|27734780|ref|NM J 73699.1 [[27734780] giJ28316807|ref|NMJ73700.1|[28316807] gi|47419915Jref|NMJ73701.lj[47419915] giJ32307165|ref|NMJ 73728.2|[32307165] gi|31341089 jref JNM 73791.2J[31341089] gi|31341095|ref|NMJ 73793.2|[31341095] giJ31341096|ref|NMJ73794.2i(31341096] gi|31341099|ref|NM J 73795.2|[31341099] gi|31341107|refJNMJ73797.2|[31341107] giJ31341108|ref|NMJ73798.2J[31341108] gi|31341110|ref|NM J 73799.2|[31341110]
gi|46397360|ref|NM..173800.31(46397360] gi|42476002|refJNM .173801 1(42476002] gi|31341117|ref|NM .173802.2 |[31341117] gi|31341122|ref|NM .173803.21(31341122] gi|31560865|ref(NM .173804.3 |[31560865] giJ31341123[ref|NM 173805.2 j[31341123] gi|31341127|ref|NM 173806.2 |[31341127] gi|31341128|ref|NM .173807.21(31341128] gi(27754173|ref|NM. 173808.1 ([27754173] gii46852397Jref[NM .173809.2 j[46852397] gi(34222234|ref|NM. 173810.3 j[34222234] gi|31341135JrefJN 173811.2 j[31341135] gi[40255183|ref|NM. 173812.3 j[40255183] giJ31341139(ref[NM .173813.2 [[31341139] giJ40255185|ref|NM .173815.31(40255185] gi[28376647(ref|NM..173821.1 j[28376647] giJ28376657[ref|NM..173822.1 j[28376657] giJ34222236[refJNM .173824.2 j[34222236] gi[28376659|refJNM .173825.1 |[28376659] gi|28376649|ref|NM 173826.11(28376649] gi|28395048|ref|NM..173827.1 j[28395048] gi[40255189(ref[NM .173828.3 j[40255189] gi[31341149|ref[NM_ .173829.2 |[31341149] giJ34222237|ref|NM 173830.3 j[34222237] gi|31341152|ref|NM_ .173831.21(31341152] giJ31341155[ref|NM .173832.2 |[31341155] gi|47271476|ref|NM 173833.3 |[47271476] gi(31341159|ref|NM .173834.21(31341159] gi|27894316|ref|NM .173841.1 |[27894316] gi[27894318iref[NM_ .173842.11(27894318] gi|2789432θjref|NM .173843.11(27894320] giJ27886565|ref|NM_ 173844.1 j[27886565] gi|3422224θ(ref|NM .173846.31(34222240] gi[31341185|refJNM .173847.2 j[31341185] gi|40255191 |ref|NM_ 173848.3 j[40255191] giJ29171761 |ref|NM_ .173849.21(29171761] gi|31341191 |ref|NM_ 173850.21(31341191] gi|27777666|ref|NM_ .173851.1 j[27777666] gi|31341196|ref|NM_ .173852.21(31341196] gi(31341197|ref|NM_ 173853.2 j[31341197] gi|40255193|ref[NM_ .173854.3 ([40255193] gi(50355989[ref(NM .173855.3 [[50355989] gi|27777672|ref|NM .173856.1 |[27777672] gi|27777668|ref|NM_ 173857.1 j[27777668] gi[27777674|refJNM_ .173858.1 j[27777674] gi[28372506(ref|NMl 173859.1 ([28372506] gi[27804314|ref(NM_ 173860.1 [[27804314] gi|41281845|ref|NM_ .173872.11(41281845] gi[28178815|ref(NM_ 174855.1 |[28178815] gi|281 8818[ref|NM_ ,174856.1 [[28178818] gi|28144896(ref|NM_ ,174858.1 |[28144896] giJ28178837iref|NM_ ,174869.1 |[28178837] gi|28872764|ref|NM_ ,174871.1 ([28872764] gi[28416920|ref|NM_ 174872.1 j[28416920] gi(28416922[ref|NM_ 174873.1 [[28416922] gi(37622908[ref|NM_ 174878.2 [[37622908] gi|28144911 jref JNM_ 174880.1 ([28144911]
gi|28144892|ref|NMJ74881.11[28144892] giJ28144894JrefiNMJ74882.li[28144894] gi|28178856|ref|NMJ74886.1|[28178856] gi|34222241|ref|NMJ74887.2J[34222241] gi|31341340|ref|NMJ74889.2|[31341340] gi|28376663|ref(NMJ74890.1 [[28376663] giJ31341349|ref|NMJ74891.2|[31341349] gi|28461132JrefJNMJ74892.1|[28461132] gi|31341350|ref|NMJ 74896.2|[31341350] gi|28372524|ref|NM J 74897.11[28372524] gi|28372526Jref|NMJ 74898.11[28372526] gi|31343457|refJNM J 74899.3|[31343457] gi|4247627θ(ref|NMJ74900.2|[42476270] gi|31341354JrefJNMJ74901.2|[31341354] giJ31341355|ref|NMJ74902.2|[31341355] gi|37537698|refJNMJ74905.2|[37537698] gi|28376665Jref|NMJ74906.1|[28376665] gi|2837253θ[ref|NM J 74907.1 [[28372530] gi|33186926JrefJNM J 74908.2J[33186926] gi(28372532[ref|NMJ74909.1 ([28372532] gi|28372534|ref|NMJ 74910.11[28372534] gi|40255195[ref[NMJ74911.3[[40255195] gi|30410024|ref|NM J 74912.2J[30410024] giJ2821227ljref|NMJ74913.lj[28212271] gi|31341368(ref|NMJ74914.2([31341368] gi|28372496|ref|NMJ74916.1 |[28372496] gi(28372536(ref[NMJ 74917.1 ([28372536] giJ27885012|refJNMJ74918.lj[27885012] gi|31341380|ref|NMJ 74920.2|[31341380] gi[28372538|ref|NMJ 74921.1|[28372538] giJ41393592|ref|NMJ 74922.3|[41393592] giJ28316809Jref|NMJ 74923.1 [[28316809] gi|28372542|ref|NM J 74924.11[28372542] gi|28372544|ref|NMJ74925.1|[28372544] giJ28376667[ref|NM J 74926.1 [[28376667] gi|28376651|ref|NMJ74927.11[28376651] gi[28372546Jref|NMJ74928.1|[28372546] giJ31341385Jref|NMJ74930.2i(31341385] gi|2839505θ(ref|NMJ74931.11[28395050] gi|28372550|ref|NMJ74932.l[[28372550] gi|34222242(ref|NMJ74933.2J[34222242] gi|28372554Jref|NMJ74934.1|[28372554] gi|31317306|ref|NMJ74936.2|[31317306] gi|28372558JrefJNMJ 74937.1|[28372558] gi|34222248|ref|NMJ74938.3|[34222248] giJ31341399 jrefJNM J 74939.2|[31341399] gi|28372560|ref|NMJ 74940.11[28372560] gi|5065909θjref[NMJ 74941.3|[50659090] gi|28372562|ref|NMJ 74942.1|[28372562] gi|31341411|ref|NMJ74943.2J[31341411] gi|42734400|ref|NMJ74944.2|[42734400] gi|28372566|ref|NMJ74945.1|[28372566] gi|29126235|ref|NMJ74947.2|[29126235] gi|31340828|ref|NMJ74950.2|[31340828] gi[3134334θ(ref[NMJ 74951.2|[31343340] gi|33457323|ref|NMJ 74952.11[33457323] gi|28373104|ref|NMJ74953.1|[28373104]
gi|28373106|ref|NM_ 174954.1 |[28373106] gi(28373108Jref|NM_ 174955.1 ([28373108] gi|28373110|ref|NM_ 174956.1 [[28373110] gi(28373112[ref|NM_ 174957.1 [[28373112] gi(28373114[ref|NM_ 174958.1 [[28373114] gi|33457321 |ref|NM_ 174959.1 [[33457321] gi(31581528[ref|NM_ 174961.2 1(31581528] gi|31581525|ref|NM_ 174962.2 j[31581525] gi(28373067|ref|NM_ 174963.1 1(28373067] gi|28373069|ref|NM_ 174964.1 [[28373069] gi|2837307l [reflNM_ 174965.1 |[28373071] giJ28373073[ref|NM_ 174966.1 j[28373073] gi|28373075|ref|NM_ 174967.1 1(28373075] gi|28373077|ref|NM_ 174968.1 [[28373077] gi|28373081 |ref|NM_ 174969.1 [[28373081] giJ28373083|ref(NM_ 174970.1 |[28373083] gi|28373085|ref|NM_ 174971.1 1(28373085] gi[28373087(ref|NM_ 174972.1 |[28373087] gi[48255895(ref|NM_ 174975.3 j[48255895] giJ2837251θ[ref|NM_ 174976.1 |[28372510] gi|30410718[ref|NM_ .174977.2 |[30410718] gi(28372512[ref|NM_ 174978.1 1(28372512] gi(27923946[ref|NM_ 174980.1 |[27923946] gi|31343345|ref|NM_ 174981.2 [[31343345] gi|34222225[ref|NM_ .174983.2 |[34222225] gi|28373118|ref|NM_ .175038.1 ([28373118] gi|2837309l [ref|NM_ 175039.1 ([28373091] gi|28373093|ref[NM_ .175040.1 1(28373093] gi|30179912|ref|NM_ 175047.2 ([30179912] gi|28373098(ref[NM. 175052.1 [[28373098] giJ31341312[refJNM_ .175053.2 [[31341312] gi|28416433Jref|NM_ .175054.2 |[28416433] gi|28872748|ref|NM..175055.2 [[28872748] giJ28372568(ref[NM..175056.1 1(28372568] giJ49574494Jref|NM..175057.2 ([49574494] gi|48675826[ref|NM_ .175058.3 |[48675826] gi|28269706|ref|NM..175060.1 |[28269706] gi|31341302|ref|NM..175061.2 [[31341302] gi[40255196(ref|NM. 175062.2 [[40255196] gii45580693Jref|NM..175063.3 [[45580693] gi(30795187[ref[NM. 175064.2 |[30795187] gi[29171728|ref|NM. 175065.2 1(29171728] gi(37059776|ref|NM. 175066.2 |[37059776] gi|28173557|ref|NM..175067.1 ([28173557] gi|31341282[ref|NM. 175068.2 1(31341282] giJ28329429Jref[NM. 175069.1 ([28329429] gi[28329426[refiNM..175071.1 j[28329426] giJ28329432[ref|NM. 175072.1 1(28329432] gi(28329435(ref[NM..175073.1 [[28329435] giJ31341283Jref|NM..175075.2 [[31341283] gi[28416423(ref(NM..175077.1 1(28416423] giJ45597457[ref(NM..175078.1 [[45597457] gi(28416934Jref|NM..175080.1 |[28416934] gi[28416936[ref(NM..175081.1 1(28416936] giJ28416898iref[NM..175085.1 |[28416898] gi[28373128Jref|NM..175566.1 1(28373128] giJ28416928|ref[NM. 175567.1 [[28416928]
gi[28416930[ref|NMJ75568.1|[28416930] gi[28212219|ref|NMJ75569.1|[28212219] gi|2841695δjref jNM J 75571.11[28416955] giJ28373193[ref|NMJ 75573.11[28373193] gi|40806203|ref|NMJ75575.4|[40806203] giJ28558992Jref|NMJ75605.2|[28558992] gi|2837312ljref|NMJ75607.lj[28373121] gi|28416435|ref |NM J 75609.11[28416435] gi|28416401 (ref|NMJ 75610.1 [[28416401] gi|28416443|ref |NM J 75611.11[28416443] giJ28373123[ref|NMJ 75612.1 [[28373123] giJ28373125(ref|NMJ75613.lj[28373125] giJ46370098Jref|NMJ75614.2|[46370098] gi|31342253|ref |NM J 75616.2|[31342253] gi|31581520|ref|NMJ75617.2|[31581520] gi|28269686Jref|NMJ75619.1|[28269686] giJ31342255[ref|NMJ 75622.2J[31342255] gi|41281855Jref|NMJ 75623.1 J[41281855] gi|4128187θjrefJNMJ75624.1|[41281870] gi|41281861|ref|NMJ75625.1|[41281861] gi|41281876|ref|NMJ75626.1|[41281876] giJ41281867jrefJNMJ75627.1|[41281867] gi)28559068|ref |NMJ 75629.11[28559068] giJ2855907θ(ref|NMJ75630.1|[28559070] gi|28329415|ref|NMJ75634.1|[28329415] gi|28329418|ref|NMJ75635.1|[28329418] gi|28329421|ref|NMJ75636.l[[28329421] giJ28559005|ref|NMJ 75698.1|[28559005] gi(46852393Jref|NMJ75709.2J[46852393] gi|28559008|ref|NMJ 75711.1 ([28559008] gi|28558983|ref|NMJ75719.1|[28558983] gi[28558985irefJNMJ 75720.1 [[28558985] giJ28558987JrefJNMJ75721.lj[28558987] gi|28558989lrefJNMJ75722.lj[28558989] giJ28559016|ref|NMJ75723.1|[28559016] giJ28559022|ref|NMJ 75724.11[28559022] gi|28559024Jref| NMJ 75725.1 [[28559024] giJ28559026JrefJ NMJ 75726.1|[28559026] gi|28559028JrefJNMJ75727.1J[28559028] giJ28559030|reflNMJ 75728.1 ([28559030] gi|28559012Jref|NMJ 75729.11[28559012] gi|31342257[ref|NMJ75733.2J[31342257] gi|31342258Jref|NMJ75734.2|[31342258] giJ31652254Jref JNM J 75735.3|[31652254] gi|40217838Jref |NM J 75736.3|[40217838] gi|31343329|ref |NM J 75737.2|[31343329] giJ31343330[ref|NMJ 75738.2|[31343330] gi|31343337jref|NMJ75739.2|[31343337] gi|50233786Jref|NMJ75741.1|[50233786] giJ29029617|ref|NMJ75742.1 ([29029617] gi|29029619|ref|NMJ75743.1|[29029619] giJ34222243[ref|NMJ75744.3|[34222243] giJ45505176Jref |N J 75745.3J[455051 6] giJ31343439Jref|NMJ75747.2J[31343439] giJ28411949|ref|NMJ75748.li[28411949] gi|28610148|ref|NMJ75767.1|[28610148] gi|28559002|ref |NMJ 75768.11[28559002]
gi|39753955|ref|NMJ75834.2[[39753955] gi|28559073|refJNMJ 75839.1 [[28559073] gi|28559075|ref |NM J 75840.11[28559075] giJ28559077|refJNMJ 75841.1J[28559077] giJ28559079Jref|NMJ 75842.1 ([28559079] gi|28558997|ref|NMJ 75847.11[28558997] giJ2855906θjref|NM J 75848.1 ([28559060] giJ28559062|ref|NM J 75849.1 [[28559062] giJ28559064JrefJNMJ 75850.1 ([28559064] giJ41281881|ref[NMJ75851.1|[41281881] gi|39725959|ref|NMJ75852.3|[39725959] gi|40353730|ref(NMJ75854.4J[40353730] gi|48762943Jref[NMJ75857.3J[48762943] giJ31343637Jref|NMJ75858.2J[31343637] giJ28559084iref[NMJ 75859.1 ([28559084] gi|28466998|ref|NMJ 75861.1|[28466998] gi|29029571 [ref(NM J 75862.2[[29029571] giJ40068461 jref|NM J 75863.2J[40068461] giJ2861O155|ref|NMJ75864.1|[28610155] gi|41281873(ref[NMJ75865.li[41281873] gi|28467002|ref|NMJ75866.1|[28467002] gi|28872779|ref[NMJ75867.1|[28872779] gi[29029626JrefJNMJ 75868.1 j[29029626] gi|39725709Jref|NMJ75870.3J[39725709] gi|3134361θ(ref|NMJ75871.2|[31343610] gi|38570118|ref|NMJ 75872.3|[38570118] gi|44921609(ref|NMJ75873.3|[44921609] giJ31343594|ref|NMJ75874.2|[31343594] gι'J40354215|ref|N J 75875.3J[40354215] giJ44921614|ref|NMJ75876.2|[44921614] giJ40255200|ref|NMJ75877.3|[40255200] gii31343579Jref(NMJ75878.2J[31343579] gi[31343527|ref|NMJ75881.2J[31343527] gi|38524590|ref|NM J 75882.11[38524590] gi(31343528|ref|NMJ75884.2[[31343528] gi|31343498|ref|NMJ75885.2J[31343498] gi|31343499|refjNMJ75886.2[[31343499] gi(3134348θ(ref |NM J 75887.2|[31343480] gi|40255202|ref|NMJ75892.3|[40255202] gi[31343331|ref|NMJ75895.2|[31343331] giJ31343323|ref|NMJ75898.2|[31343323] giJ3134226θiref|NMJ75900.2J[31342260] gi|31342241|ref|NMJ75901.2|[31342241] gi|34222251 jrefJNM J 75902.3J[34222251] giJ31342242|ref|NMJ75903.2|[31342242] gi|31342232|ref|NMJ75904.2J[31342232] gi|31342225|ref|NMJ75906.2|[31342225] giJ51036599|ref|NMJ75907.3J[51036599] giJ40255204|refJNMJ75908.3|[40255204] giJ40255231|ref|NMJ75910.4J[40255231] gi[3134221θiref|NMJ75911.2([31342210] gi|29893811 |ref|NMJ75913.2|[29893811] gi(31341870[ref|NMJ75918.2|[31341870] gii31341799Jref|NMJ75920.2J[31341799] gi|51036606|refJNMJ75921.3|[51036606] giJ31341688 jref|NM J 75922.2J[31341688] gi|31341689|ref |NM J 75923.2|[31341689]
gi|31341675|ref|NM_J75924.2|[31341675J gi|28872755|ref|NM_175929.11[28872755] giJ28872804JrefJNM_175931. l j[28872804] gi|28872729|ref|NM_175932.1 [[28872729] gi|28872750|ref|NM_175940.11[28872750] gi|28872742|refJNM_1 6071.1 j[28872742] gi j28872744)ref [NM_1 6072.11[28872744] gi|28872767|ref|NM_176081.1 J[28872767] gi|28872769Jref|NM_176083.1 j[28872769] gi|28872771 |ref|NM_176084.11[28872771] gi|28872773|ref|NM_176085.11[28872773] gi J28872775Jref|NM_176086.1 ([28872775] giJ28872789JrefJNM_176095.1 ([28872789] gi|28872791 |ref|NM_176096.11[28872791] gi|28603817|ref|NM_176782.1 |[28603817] gi(3058114θjref[NM_176783.1 [[30581140] gi|29171683JrefJNM_176786.1 ([29171683] gi|34328904|ref|NM_J 76787.2|[34328904] gi|28872762Jref|NM_176789.11[28872762] gi|31343584|ref|NM_176791.2|[31343584] giJ28872731 jref|NM_176792.1 [[28872731] gi|28872735|ref|NM_176793.11[28872735] giJ28872737Jref|NM_176794.1 j[28872737] gi|47117696|ref|NM_176795.2|[47117696] gi|29029607|ref|NM_176796.11[29029607] gi|29029609|ref|NM_176797.1 |[29029609] giJ29029611 jref|NM_J 6798.11[29029611] gi|29171686|ref|NM_176799.11[29171686] giJ28872758JrefJNM_176800.1 J[28872758] gi|29826324|ref|NM_176801.1 J[29826324] gi|28872739|ref|NM_176805.1 [[28872739] gi|35493763iref|NM_176806.2|[35493763] gi|28827788|refJNM_176810.1 |[28827788] giJ33667039JrefJNM_176811.2J[33667039] gi|40549398|ref|NM_176812.3|[40549398] gi|38455428|ref|NM_176813.3|[38455428] gi|39753952|ref(NM_176814.3[[39753952] gi|31341213Jref|NM_176815.2|[31341213] gi[31341214|refJNM_176816.2|[31341214] giJ28827796|ref|NM_176817.11[28827796] gi|50978623|ref|NM_J 76818.11[50978623] gii33519449JrefJNM_176820.2|[33519449] gi|33519448|ref|NM_176821.2|[33519448] gi|31543282Jref|NM_J 76822.2|[31543282] gi|31341192|ref|NM_J 6823.2|[31341192] gi|29029556|ref|NM_176824.11 [29029556] gi|45935388|ref|NM_176825.2|[45935388] giJ29171691 jref|NM_J 76826.1 ([29171691] gi|29029563|ref|NM_176853.11[29029563] gii30410795Jref|NM_176863.l j[30410795] gi|29171697|ref|NM_176866.1 |[29171697] gi|31881619|refjN _176867.2|[31881619] gi|2917170ljref|NM_176869.1|[29171701] gi|28866965|ref|NM_176870.11[28866965] gi|40288187|ref|NM_176871.2|[40288187] gi|29029533|ref|NM_176874.11 [29029533] giJ33356159[refJNM_176875.2|[33356159]
gi|29029604|ref|NM J 76876.11[29029604] giJ29029545Jref JNM J 76877.1 j[29029545] gi|29029547|ref|NMJ 76878.11[29029547] giJ40255206Jref|NMJ76880.3J[40255206] gi|28882025|ref|NMJ 76881.1 |[28882025] gi|28882032Jref|NMJ 76882.11[28882032] giJ28882027|ref|N J 76883.1 j[28882027] gi|28882036JrefJNMJ76884.1|[28882036] gi|28882030|ref|NMJ76885.11[28882030] gi|29126244|ref|NMJ76886.11[29126244] gi[28882040|ref|NMJ 76887.11[28882040] gi|28882034|ref|N J 76888.11[28882034] gi|28882046Jref|NMJ 76889.11[28882046] gi(28882038[refJNMJ 76890.1 [[28882038] gi|31341119|ref|NMJ 76891.2J[31341119] giJ29171722Jref|NM J 76894.1 ([29171722] giJ29171737Jref|NM 76895.1 ([29171737] gi|28893580|ref)NMJ 77398.11[28893580] gi|41281894(refJNMJ77399.1 |[41281894] gi|28912911 |ref|NMJ 77400.11[28912911] giJ31560862JrefiNMJ77402.3pi 560862] gi|38490536|refJNMJ77403.3|[38490536] gi|29171708JrefJNMJ77404.1|[29171708] gi|29029551 |ref|NMJ77405.11[29029551] gi|29171741|ref|NMJ77414.11[29171741] giJ2917171θiref|NMJ77415.li[29171710] giJ29501806[ref|NMJ77417.1|[29501806] gi|29337285)refJNMJ 77422.11[29337285] giJ29171752|ref|NMJ77423.1|[29171752] gi|28933464|ref|NMJ 77424.1 j[28933464] gi|34335274|ref|NMJ77427.2|[34335274] gi|29171704Jref(NMJ 77433.1 ([2917 704] gi|29029588|refJNMJ 77434.1 ([29029588] gi|29171749|ref |NM J 77435.1 ([29171749] giJ29029560|ref|NMJ 77436.11[29029560] gi|28973794|ref|NMJ77437.lj[28973794] gi|29294650[ref|NMJ 77438.1|[29294650] gi|2902959θjref|NMJ 77439.1J[29029590] giJ28973798[ref|NMJ 77441.11[28973798] giJ29294652|ref|NM J 77442.11[29294652] gi[29294628(ref|NMJ77444.1|[29294628] gi[31343344Jref|NMJ77452.2|[31343344] gi|31343354JrefJNMJ77453.2|[31343354] gi|40255207(refJNMJ77454.2|[40255207] gi|31343355|ref|NMJ77455.2|[31343355] gi|29294654|ref|NM J 77456.1 [[29294654] gi|50346002|ref|NMJ77457.2[[50346002] giJ29294642Jref(N J 77458.1 j[29294642] giJ29337282JrefJNMJ77476.li(29337282] gi|29294644|ref|NMJ 77477.11[29294644] gi|29126240|ref|NMJ77478.1|[29126240] giJ29171718Jref|NMJ77483.1|[29171718] gi|29294634|ref|N J 77524.11[29294634] gi|29294636)ref|NMJ77525.1|[29294636] giJ29171744Jref|NMJ77526.1|[29171744] gi|29550892|ref|NMJ77528.1 |[29550892] gi[29540534[ref[NMJ 77529.1 j[29540534]
gi|29540536|ref|NMJ77530.1[[29540536] giJ31377831|ref|NMJ77531.2|[31377831] gij41393612Jref|NMJ77532.3|[41393612] giJ37594466Jref|NMJ77533.2|[37594466] gi|29540538Jref|NMJ 77534.11[29540538] gi(29337293(ref[NMJ 77535.11[29337293] giJ29540542jref[NM J 77536.11[29540542] gi)29337295JrefJNMJ 77537.1 j[29337295] gij29171729|ref|NMJ77538.1|[29171729] gi|29294616|ref|NMJ77539.1|[29294616] gi[29294618(ref|NMJ77540.1|[29294618] giJ29294620|ref|N J77541.1|[29294620] gi)29294623Jref|NMJ 77542.11[29294623] gi|29171746|refJNMJ77543.l[[29171746] gi|31343531[ref|NMJ77549.2|[31343531] giJ31343532[ref|NMJ77550.2|[31343532] gi[41152145[ref|NMJ77551.3[[41152145] giJ2955092θ[ref|NMJ 77552.11[29550920] gi|29540558|ref|NMJ 77553.11[29540558] gi|30090002|ref|NMJ 77554.11[30090002] gi|29540532|ref|NMJ 77555.11[29540532] gi|29540540 jref[NM J 77556.1 [[29540540] gi|29540546JrefJNMJ 77557.11[29540546] giJ29540548[ref(NMJ 77558.1 ([29540548] gi|47419901|ref|NMJ77559.2J[47419901] gi|47419903|ref|NMJ77560.2|[47419903] gi|30089927|ref|NMJ 77924.11[30089927] gi J29553969Jref JNM 77925.1 j[29553969] gi|29570787|ref|NMJ 77926.11[29570787] gi|29550849(ref|NMJ 77937.11[29550849] gi|34222253|ref|NM J 77938.1 ([34222253] giJ29570770|ref|NM J 77939.1 ([29570770] giJ47578120|ref|NMJ77947.2|[47578120] giJ47578121|refJNMJ77948.2|[47578121] gi|29540561 |ref|NMJ 77949.11[29540561] gi|29557854|ref|NMJ 77951.1 ([29557854] gi[29557938|ref|NM J 77952.1 [[29557938] gi[29570774[ref[NM J 77953.1 J[29570774] giJ29570776JrefJNMJ 77954.1 J[29570776] gi|29544739|ref|NMJ 77959.11[29544739] gi|31342626|ref|NMJ77963.2|[31342626] gi[40255209[ref(NMJ77964.3([40255209] gi|31342620|ref|NMJ77965.2[[31342620] giJ34222255|ref|NMJ77966.3J[34222255] giJ31342615|ref|NMJ77967.2|[31342615] giJ2955802ljref|NMJ 77968.11[29558021] gi|29558098|refJNMJ 77969.1 j[29558098] giJ29826278Jref[N J 77972.1 j[29826278] gi|31563385|ref|NMJ 77973.11[31563385] gi[29826290|ref|NMJ77974.l i[29826290] gi|29826298|ref|NMJ 77976.1 j[29826298] gi[30061493|ref(NMJ 77977.1 [[30061493] gi|30089975|refJNMJ 77978.1|[30089975] gi|30089977Jref|NMJ 77979.1 j[30089977] gi|29570799|ref|NMJ 77980.11[29570799] gi|29826281|ref|NMJ 77983.11[29826281] gi(29542730|ref|NMJ 77985.1 j[29542730]
gi|31342522 ref JlNM 177986.2|[31342522] giJ42558278 ref J|NM 177987.1 |[42558278] gi|29826284 ref J|NM. 177988.1 1[29826284] giJ30089996 ref (|NM. _177989.1 |[30089996] giJ29826328 ref (|NM 177990.1 ([29826328] giJ29826316 ref J|NM. 177991.11[29826316] giJ30089949 ref JlNM 177995.11(30089949] giJ30061490 ref j|NM 177996.11[30061490] giJ29725634 ref J|NM 177998.1 1[29725634] gi|30089931 ref J|NM. 177999.1 |[30089931] gi[30065642 ref JjNM 178000.1 |[30065642] gi|30065644 ref JjNM. 178001.1 |[30065644] giJ30065646 ref (|NM 178002.1 |[30065646] gi|30065648, ref JjNM..178003.11(30065648] giJ29788782 ref| [NM .178004.1 ([29788782] gi|41281897 ref JjNM..178006.1 |[41281897] giJ41281 19"0"6" ref jjNM .178007.1 j[41281906] giJ4128 11901 refl JNM .178008.1 |[41281901] giJ323071 69 ref JjNM .178009.2|[32307169] giJ3006 115557 ref J|NM .178010.1 ([30061557] giJ40255215 ref J|NM 178011.2|[40255215] gi|42476191 ref J|NM..178012.3|[42476191] giJ29788777 ref (jNM 178013.1 |[29788777] giJ34222261 ref JjNM .178014.2|[34222261] giJ29789450 ref J|NM 178019.1 j[29789450] gi|30061497 ref J|NM .178025.1 1[30061497] giJ30061499 ref JlNM .178026.1 |[30061499] giJ30089936 ref J|NM 178031.1 |[30089936] gi|29837647 ref J|NM..178033.1 1[29837647] gi|29837661 ref JjNM .178034.1 1 [29837661] gi|38045891 refl JNM .178037.1 |[38045891] giJ38045893 ref JlNM..178038.1 |[38045893] gi|38045895 ref J|NM .178039.1 1[38045895] gi|38045897 refl JNM 178040.1 j[38045897] giJ30089998 ref J]NM .178042.1 1[30089998] giJ30061564 ref JjNM.178043.1 1[30061564] giJ30089944 ref (|NM .178044.1 1[30089944] gi|31342420, ref J|NM.178120.2|[31342420] giJ49472644] ref J|NM.178121.2|[49472644] giJ31342415 ref JlNM.178122.2|[31342415] giJ31342410 ref| |NM_ .178123.2J[31342410] gi|31377534 refl JNM..178124.3|[31377534] gi(31342404 ref (|NM 178125.2|[31342404] gi[31342405 ref| NM_ .178126.2|[31342405] giJ31342398 refl NM.178127.2|[31342398] giJ31342399 ref| NM_ .178128.21(31342399] giJ38373667 ref NM ,178129.3|[38373667] giJ31342394 ref J]NM_ 178130.2|[31342394] gi|31083135 ref] NM_ 178134.2|[31083135] gi|31342377 ref JjNM_ 178135.2|[31342377] giJ30089918 ref J|NM_ 178136.1 |[30089918] gi|30089941 ref J|NM_ 178138.2|[30089941] giJ30061506 ref| |NM_ ,178140.1 |[30061506] giJ30474872] ref) |NM_ ,178145.1 1[30474872] gi|30026033 ref| |NM_ 178148.1 1[30026033] giJ30795118 ref| |NM_ ,178150.1 1[30795118] giJ30181239| refl |NM_ 178151.1 |[30181239]
gi|30181241|ref|NMJ78152.1|[30181241] gi|30181243|ref|NMJ78153.11[30181243] giJ30410723|ref|NMJ78154.lj[30410723] giJ30410725Jref(NMJ78155.1|[30410725] gi|30410727|ref|NMJ78156.11[30410727] gi|30410729|ref|NMJ78157.lj[30410729] giJ3041078θ(ref|NMJ78159.lj[30410780] gi[30039713[ref|NMJ78160.11[30039713] gi|30039709|ref|NMJ78161.1 j[30039709] giJ31342353|ref(NMJ78167.2i[31342353] gi|30039687|ref|NMJ78168.1 |[30039687] gi(31342347|refJNMJ78169.2i[31342347] giJ31342341 jref|NMJ 78170.2J[31342341] gi|31342342JrefJNMJ78171.2i[31342342] gi|31342336JreflNM J 78172.2|[31342336] gi|31342337|ref|NMJ78173.2|[31342337] giJ3134233θ[ref|NMJ 78174.2|[31342330] gi|31342331|ref|NMJ78175.2|[31342331] gi|31342324Jref|NMJ78176.2|[31342324] gi|31342325Jref|NM J 78177.2J[31342325] gi|30410802|ref|NMJ78181.1 |[30410802] gi|30260189Jref|NMJ78190.1|[30260189] gi|30260191|ref[NMJ78191.1|[30260191] giJ30410845Jref|NMJ78221.l j[30410845] gi|30795256|ref|NMJ 78225.1 ([30795256] giJ30749197|ref|NMJ78226.1 ([30749197] gi(46397356Jref|NMJ78228.2[[46397356] gi(3975396θiref|NMJ78229.3J[39753960] giJ30102943|ref|NMJ78230.lj[30102943] gi(30102945|ref|NMJ78231.1 [[30102945] gi[32455240|ref|NMJ78232.2J[32455240] gi(30102939Jref|NMJ78233.1|[30102939] gi(30410789iref(NMJ78234.l j[30410789] gi|30410715|ref|NMJ78237.1|[30410715] gi[3017991θ[ref|NMJ78238.li[30179910] gi|30795247|ref|NMJ 78270.11(30795247] giJ30795249Jref(NM J 78271.1 [[30795249] gi|30179904|refJNMJ78272.1|[30179904] gi|30179906Jref|NMJ78273.1|[30179906] gi[3817582θ(ref[NMJ 78275.3|[38175820] gi|31342170|ref |NM J 78276.2|[31342170] gi|30581154Jref|NMJ78311.11[30581154] giJ30581156Jref|NMJ78312.l([30581156] giJ30315657iref|NMJ78313.l[[30315657] gi|31342079iref|NMJ78314.2([31342079] giJ3047487θ(ref|NMJ 78324.1 [[30474870] gi|47132612Jref|NMJ78326.2([47132612] giJ30581169JrefJNMJ78329.1 ([30581169] gi|30581149|ref|NMJ78331.1|[30581149] giJ3058115liref(NMJ78332.li[30581151] gi(4128191 ojrefJNMJ 78335.11[41281910] gi|31083034|refJNMJ78336.1|[31083034] gi[31341973|ref|NMJ78338.2J[31341973] giJ30350205Jref|NMJ 78339.11[30350205] gi|30350211 lref|NMJ 78340.1 ([30350211] giJ30350207Jref|NMJ 78341.1|[30350207] gi[30350213iref|NMJ78342.1|[30350213]
gi|30350209 jref j|NM. 178343.1 |[3035O209] gi[30350215 jref ( |NM 178344.11(30350215] giJ30387653 Iref JjNM..178348.1 j[30387653] gi|30387655 jref J]NM. 178349.1 |[30387655] giJ30387651 |ref J|NM 178351.11(30387651] gi|30387647 |ref J|NM ,178352.1 |[30387647] gi|30387649 jref J |NM. 178353.1 ([30387649] giJ30387645 jref JjNM. 178354.1 j[30387645] gi|30387641 |ref (|NM .178356.1 [[30387641] giJ42476188 [ref J|NM. 178422.3 |[42476188] gi|30795201 |ref| JNM .178423.11(30795201] gi|30581116 |ref J|NM. 178424.11(30581116] gi(30795203 |ref J|NM .178425.1 [[30795203] gi|30795239 |ref J|NM 178426.11(30795239] giJ30795241 | ref J |NM..178427.1 |[30795241] gi|30410044 |ref (|NM .178428.11(3041 0044] giJ30410046 [ref J|NM 178429.1 [[3041 0046] giJ30410042 jref JjNM. 178430.1 [[3041 0042] gi[30410038 |ref J]NM .178431.1 |[3041 0038] giJ30795254 |ref J|NM. 178432.1 [[30795254] gi|30410040 jref J |NM .178433.1 |[3041 0040] gi]30410036 |ref J|NM. 178434.1 |[3041 0036] gi|30410032 |ref (|NM 178435.1 ([3041 0032] gi|30410034 jref J |NM .178438.1 [[3041 0034] gi(34222268 |ref J|NM. 178439.31(34222268] gi|30795181 jref (jNM .178441.1 |[30795181] giJ41281904 |ref J|NM 178443.11(41281904] gi|30795216 jref J |NM. 178445.1 |[30795216] giJ31341967 [ref j|NM..178448.2 ([31341967] gi|31341960 jref J|NM .178449.2 |[31341960] gi|31341961 [refj JNM 178450.2] 1(31341961] gi[31341954 |ref J|NM 178451.2] |[31341954] gi|40255219 |ref J|NM .178452.3] ([40255219] gi|31341948 jref J |NM .178453.2 [[31341948] gi[30425377 |ref J|NM..178454.11(30425377] gi|45333915 jref J |NM..178456.2 |[45333915] giJ30425381 |ref (|NM .178460.11(30425381] gi|30425385 jref J]NM..178463.1 [[30425385] giJ30425387 |ref J|NM 178465.1 |[30425387] gi|30425389 jref J|NM 178466.11(30425389] giJ30425391 jref JjNM..178467.1 ([30425391] g 0425393 |ref (|NM 178468.1 j[30425393] gi|31341942 jref J |NM..178470.2 [[31341942] gi|30425399 |ref J|NM 178471.1 ([30425399] gi|30425401 |ref J|NM. 178472.11(30425401] gi(30425403 [ref (|NM 178477.1 [[30425403] gi|50263054 |ref J|NM..178483.2 |[50263054] giJ30425409 |ref J]NM 178491.11(30425409] gi|40255213 |ref JNM 178493.3 [[40255213] gi|31341935 jref J |NM 178494.2 [[31341935] gi(34222262 |ref J|NM 178495.3 [[34222262] gi|31341922 Iref JjNM 178496.21(31341922] giJ31341915 |ref J|NM 178497.2 |[31341915] gi|31341916 |ref J|NM 178498.211(31341916] giJ31341910 lref J|NM 178499.2 |[31341910] giJ31341911 I ref J |NM..178500.2 ([31341911] gi|31341899 jref JjNM. 178502.2 |[31341899]
140255222] |ref J|NM 178504. 1(40255222]
[50233825] [ref J|NM .178505. |[50233825]
131341888 |ref J|NM.'178507. j[31341888]
(31341881 jref (jNM 178508. [[31341881]
|34222266 [ref J|NM 178509. |[34222266]
[30425443 jref jjNM 178510. [[30425443]
(31341849 jref JjNM .178514. |[31341849]
131341844 jref JjNM 178516. j[31341844]
131341845 |ref J|NM .178517. ([31341845]
[31341836 |ref j|NM.178518. |[31341836]
(31341837 jref (jNM .178519. ([31341837]
131341830 |ref J|NM.178520. 1(31341830]
J49574542 jref ( |NM 178523. [[49574542]
131341825] |ref J|NM..178525. ([31341825]
J31341818 |ref J|NM.178527. |[31341818]
131341815 [ref J|NM..178530. 1(31341815]
J31341781 |ref J|NM..178532. [[31341781]
(31341771 jref J |NM 178536. 1(31341771]
[40789264 jref J |NM.178537. |[40789264]
131341765] |ref J|NM .178538. 1(31341765]
(31341766 (ref J|NM 178539. |[31341766]
131341752] jref J |NM..178540. ([31341752]
J31341753 |ref J|NM 178542. |[31341753]
[45545420 |ref J|NM 178543. [[45545420]
131341741 |ref J|NM..178544. ([31341741]
131341734] [ref (|NM 178545. 1(31341734]
J31341735] jref J |NM 178546. 1(31341735]
131341729 |ref J|NM .178547. [[31341729]
131341730 jref J |NM..178548. 1(31341730]
[31341723 |ref J|NM .178549. ([31341723]
J31341724 |ref J|NM .178550. |[31341724]
131341712 |ref J|NM..178552. |[31341712]
(31341713 |ref JjNM 178553. 1(31341713]
J45505170 |ref J|NM .178554. [[45505170]
J33356133 |ref JjNM..178555. 1(33356133]
131560863 |ref| (NM .178556. ([31560863]
J31341708] [ref J|NM .178557. ([31341708]
(31341703 jrefJ |NM..178558. |[31341703]
[36413606 jrefJjNM.178559. [[36413606]
131341698 |ref (|NM .178562. [[31341698]
[42558242 |ref J|NM .178563. 1(42558242]
J31341683 IrefJjNM.178564. [[31341683]
[40255217 jrefJ |NM .178565. 1(40255217]
131341670 |refJNM 178566. [[31341670]
I30425552 jrefJ |NM..178568. ([30425552]
[31340723 |ref|JNM 178569. j[31340723]
I30425562 [refJ|NM .178570. j[30425562]
J31340718 |refj|NM 178571. |[31340718]
130581142 jrefJjNM .178578. 1(30581142]
(30581144 [refjjNM 178579. 1(30581144]
[30581108 |refJ|NM .178580. [[30581108]
J30581110 |refJ|NM .178581. |[30581110]
(30581112| |refJ|NM .178582, ([30581112]
J31317267] |refJ|NM .178583 |[31317267]
J30795192 |refJ|NM .178584, [[30795192]
[31317269 |refJ|NM .178585 |[31317269]
[31083235] jrefJjNM 178586 1(31083235]
131083242 |ref JjNM 178587.1 |[31083242]
)31083249 [ref J|NM.178588.11(31083249]
131343604 jref JjNM.178812.2 j[31343604]
[45433550 |ref J|NM.178813.4 j[45433550]
(31341423 [ref J|NM 178814.2 |[31341423]
(31341468 |ref J|NM_ 178815.2 |[31341468]
[31341470 |ref J]NM 178816.21(31341470]
(45827702 Iref JjNM 178817.3 |[45827702]
[31343534 jref J]NM.178818.21(31343534]
J31343375 |ref J|NM 178819.2 |[31343375]
|34222267 |ref J]NM 178820.31(34222267]
[32189424 |ref| JNM 178821.1 |[32189424]
|38490687 |ref J|NM 178822.31(38490687]
131343459 |ref J|NM. 178823.2 |[31343459]
J34222272 |ref J|NM 178824.3 j[34222272]
131341259 |ref J|NM 178826.2 |[31341259]
|32526893 jref J |NM .178827.31(32526893]
[40255226 |ref J|NM. ,178828.3 j[40255226]
131341424 jref J]NM ,178829.2 j[31341424]
131341376 (ref j|NM 178830.21(31341376]
131340688 [ref J|NM. ,178831.2 |[31340688]
(31341691 jref jjNM ,178832.2 j[31341691 ]
147271478 [ref J|NM 178833.3 [[47271478]
[31343533 jref JjNM ,178834.2 ([31343533]
131343491 jref J|NM. ,178835.2 |[31343491 ]
|31343395 jref (jNM 178836.2 [[31343395]
131340687 |ref J|NM. 178837.2 |[31340687]
[50080216 jref JjNM. ,178838.3 ([50080216]
140255221 |ref (|NM 178839.3 |[40255221 ]
131341936 |ref J|NM ,178840.2 [[31341936]
131343485 jref J |NM. ,178841.2 |[31343485]
(31341477 jref J |NM. 178842.21(31341477]
[30524927 |ref J|NM 178844.1 ([30524927]
131077206 |ref JNM ,178849.11(31077206]
131077208 jref J |NM 178850.1 |[31077208]
|40255277 |ref J|NM. 178857.41(40255277]
(34222270 |ref J|NM 178858.3 |[34222270]
131341260 Jref J|NM. ,178859.21(31341260]
131341319 jref JjNM. ,178860.2 |[31341319]
J37059780 [ref jjNM 178861.3 |[3705978O]
J30578409 |ref J|NM 178862.11[30578409]
J31341469 |ref J|NM ,178863.21(31341469]
131341471 |ref J|NM ,178864.2 |[31341471 ]
131340713 jref J |NM..178865.21(31340713]
J42542387 |ref J|NM 178867.3 |[42542387]
(32130535 |ref J|NM .178868.3 [[32130535]
J31077197 jref J |NM 180699.1 |[31077197]
J31077199 |ref JjNM..180703.1 [[31077199]
131083279 |ref J|NM 180976.1 ([31083279]
131083287 |ref J|NM..180977.1 ([31083287]
131083044 jref J |NM..180981.11(31083044]
[31083059 |ref J|NM..180982.1 |[31083059]
(31657146 Iref J|NM. 180989.3 [[31657146]
J31340935] |ref j|NM 180990.2 |[31340935]
(38679889 [ref J|NM 180991.4 j[38679889]
J41281926 [ref J|NM 181041.1 j[41281926]
J41281916 |ref j|NM. 181042.1 |[41281916]
gi|31083143 |ref J|NM. 181050 1(31083143] gi|31077210 [ref J]NM 181054 ([31077210] gi|31083090 jref
:||NM 181076. [[31083090] gi|31083098 |ref J|NM 181077 [[31083098] gi|31083173 |ref j|NM 181078 [[31083173] gi|31083179 |ref J|NM .181079 1(31083179] gi|41281932 jref JjNM 181093 ([41281932] gi|31317281 jref JjNM
,181265 [[31317281] gi|31563389 |ref j|NM
,181268 j[31563389] gi|31563391 Iref JjNM 181269 ([31563391] gi|31563393 |ref J|NM. 181270 [[31563393] gi|31563395 jref ( |NM .181271 1(31563395] gi|31563397 |ref J|NM 181272 ([31563397] gi|31563401 | ref J |NM .181283 |[31563401] gi|31563403 |ref j|NM .181285 [[31563403] gi|31563405 | ref J |NM. .181286 ([31563405] gi|31563407 (ref J|NM. 181287 ([31563407] gi|31563409 |ref J|NM .181288 [[31563409] gi|31563411 |ref J|NM .181289 [[31563411] gi|31563413 jref J |NM. 181290 J[31563413] gi|31317273 jref J |NM. 181291 1(31317273] gi|31563415 |ref J|NM. .181292 1(31563415] gi|31563417 jref J|NM .181293 ([31563417] gi|31563419 |ref J|NM 181294 ([31563419] gi|31563421 jref JjNM. .181295 |[31563421] gi|31563423 Iref JjNM 181296 1(31563423] gi|31563425 jref J]NM. .181297 ([31563425] gi|31563427 |ref J|NM 181298 1(31563427] gi|31563429 jref J]NM .181299 |[31563429] gil31563431 jref J]NM .181300 1(31563431] gi|31563433 jref |NM. .181301 [[31563433] gi|31317275 |ref| |NM .181302 1(31317275] gi|31083065 |ref JNM. .181304 ([31083065] gi|31083071 |ref J|NM 181305 [[31083071] gi|31083076 |ref JNM. .181306 ([31083076] gi|31083084 jref J |NM 181307 1(31083084] gi|31317277 jref (jNM .181308 [[31317277] gi|31317240 |ref j|NM. .181309 ([31317240] gi|31317242 jref J |NM .181310 1(31317242] gi|31317258 [ref J|NM .181311 [[31317258] gi|31317260 jref J |NM 181312 ([31317260] gi|31317262 [ref J|NM .181313 |[31317262] gi|31317264 |ref J|NM. .181314 ([31317264] gi|31317255 jref J |NM 181332 [[31317255] gi|31317217 |ref J|NM 181333 1(31317217] gi|31317219 |ref J|NM .181334 1(31317219] gi|31317221 jref J]NM. .181335 ([31317221] gi|31341428 jref J|NM 181336 |[31341428] gi|31560860 |ref j|NM 181337 [[31560860] gi|31317245 jref JjNM. 181339 ([31317245] gi|31317283 | ref J|NM. 181340 ([31317283] gi|31317285 |ref J|NM 181341 1(31317285] gi|31317230 |ref J|NM .181342 [[31317230] gi|31317289 jref J |NM 181349 [[31317289] gi|41281936 |ref J|NM. 181351 [[41281936] gi|31317296 jref J|NM 181353 1(31317296] gi|34222269 |ref (|NM 181354 1(34222269]
gi[31317233(ref|NMJ81355.1|[31317233] giJ41281950jref|NM J 81356.11[41281950] gi|31657100|ref|NMJ81357.1|[31657100] giJ38201643JrefiNMJ81358.lj[38201643] gi|31317248|ref|NMJ81359.1|[31317248] giJ31317294|ref)NMJ81361.1][31317294] gi|31742515|ref|NMJ81425.li(31742515] gi|32483403|ref|NMJ81427.1|[32483403] giJ31088849|refJNMJ81428.l([31088849] gi|31088851|ref|NMJ81429.1|[31088851] gi|31563333JrefJNMJ81430.l([31563333] gi|31563335|ref|NMJ81431.1|[31563335] gi|31341 94|ref)NMJ81435.2|[31341794] giJ31563351|ref|NMJ81441.1|[31563351] gi|31563502Jref|NMJ81442.1|[31563502] gi|31317208|ref|NMJ81443.lj[31317208J giJ31657135(ref|NMJ81446.1|[31657135] gi|31324529|ref|NMJ81449.1j[31324529] gi[34335126(ref(NMJ81453.2|[34335126] gi|31563353|ref|NMJ81454.1|[31563353] gi|31563355|ref|NMJ81455.1|[31563355] gi(31563357[ref|NMJ81456.1|[31563357] gi|31563339|ref|NMJ81457.1|[31563339] gi|31563341|ref|NMJ81458.lj[31563341] gi|31563343[ref|NMJ81459.1|[31563343] gi|31563345|ref|NMJ81460.1|[31563345] gi|31563347|ref|NMJ81461.1|[31563347] gi|31563359|ref|NMJ 81462.1|[31563359] gi|31563361|ref)NMJ81463.1|[31563361] giJ31563363|ref|NMJ81464.1|[31563363] gi|31563365|ref|NM J 81465.11[31563365] giJ31563373Jref|NMJ81466.1|[31563373] giJ31563375|ref|NMJ81467.1|[31563375] gi|31563377|ref|NMJ81468.1([31563377J giJ31563379Jref|NMJ81469.1|[31563379] gi|31563533|ref|NMJ81471.1|[31563533] gi|31657098Jref|NMJ81472.1|[31657098] gi|51093725|refJNMJ 81481.2|[51093725] gi|51093715|ref|NMJ81482.2|[51093715] gi|51093722|ref|NMJ81483.2|[51093722] gi|31563369|ref|NMJ81484.11[31563369] gi|31563371[ref(NMJ81485.1([31563371] gi|31652231|ref(NMJ81486.1|[31652231] gi|32129210Jref|NMJ81489.3|[32129210] gi|31652217Jref(NMJ81491.1|[31652217] gi|31652241|refJNMJ81492.1|[31652241] gi|31657143|ref|NMJ81493.l([31657143] gi[31652237|ref[NM 181500.11[31652237] gi|31657141|ref|NMJ81501.1|[31657141] gi|31563527|ref|NMJ81502.l[[31563527] gi|31415881|ref|NMJ81503.l[[31415881] gi|32455251(ref|NMJ81504.2([32455251] giJ31415879(ref|NMJ81505.1I[31415879] gi|42476317Jref|NMJ81506.3|[42476317] gi[31657122Jref(NMJ81507.11[31657122] gi|31657124|ref|NMJ81508.11[31657124] gi[31563517Jref|NMJ81509.1([31563517]
gi|31657104|ref|NMJ81510.1|[31657104] gi|3165222ljref|NMJ81512.l([31652221] gi|31652223|ref|NMJ81513.1|[31652223] gi[31652225|ref|NMJ81514.l([31652225] giJ31652227|ref|NMJ81515.1|[31652227] gii40549457[ref|NMJ81519.2([40549457] gi|31657095|ref|NMJ81521.l([31657095] giJ32307102[ref|NMJ81522.li[32307102] gi|32455247|ref(NMJ81523.l[[32455247] giJ32455249Jref|NMJ 81524.11[32455249] gi(32307104Jref|NMJ 81525.1 ([32307104] giJ31563523|ref|NMJ81526.1|[31563523] giJ3156351 ljref|NMJ81527.l[[31563511] giJ31563513[ref|NMJ81528.l[[31563513] gi|32307106|ref|NMJ81530.l[[32307106] giJ31881692|ref|NMJ81531.1[[31881692] gi[31581555|ref(NMJ81532.2i[31581555] giJ3245149l(ref|NMJ81533.3|[32451491] gi[31581553|ref|NMJ81534.2|[31581553] giJ3158155l[ref(NMJ81535.2[[31581551] gi[31559824|ref|NMJ81536.1|[31559824] giJ3158155θ[refJNMJ81537.2i[31581550] gi[3155982θjref|NMJ81538.l[[31559820] gi(31581548[ref|NMJ81539.2|[31581548] gi|31652239|ref|NMJ81552.1|[31652239] giJ32130525(ref|NMJ81553.1|[32130525] gi[32130527(ref|NMJ81554.li[32130527] gi|32130529|ref|NMJ81555.1|[32130529] gi(31795537iref|NMJ81558.l([31795537] gi|34335185|ref|NMJ81571.1 [[34335185] gi[31795590|ref|NMJ81572.2|[31795590] giJ31881686Jref|NM_181573.l([31881686] giJ32313574[ref|NMJ81575.2i[32313574] gi|31712023irefJNMJ81576.li[31712023] gi|31795539|ref|NMJ81578.1|[31795539] gii31742495Jref|NMJ81581.li[31742495] gi|31742507(ref|NMJ81597.l[[31742507] gi|31742512Jref|NMJ81598.lj[31742512] gi[31791007Jref|NM_181599.lj[31791007] gi|31791003JrefJNMJ81600.l([31791003] gi|31791037|ref|NMJ81602.1|[31791037] gi(31791039 jref|NM_181604.1 j[31791039] gi|31791033Jref|NMJ81605.lj[31791033] gi|31791029Jref|NMJ81607.1|[31791029] gi|31791031|refJNMJ81608.l[[31791031] gii31791025|ref|NMJ81609.lj[31791025] giJ31791027Jref JNM_181610.11[31791027] giJ3179102l jref|NMJ81611.l[[31791021] giJ31791023|ref|NM_181612.1 j[31791023] giJ31791017|ref|NM_181614.1j[31791017] gi|31791019|ref|NMJ81615.l([31791019] giJ31791013(ref|NM_181616.1|[31791013] giJ31791015|ref|NMJ81617.1|[31791015] giJ32130532|ref|NMJ81618.1|[32130532] gi[31791009iref|NMJ81619.l([31791009] gi|31791011|ref|NMJ81620.1|[31791011] gi[38569478|refiNMJ81621.2|[38569478]
gi|31791001 |ref JNM 181622.11(31791001] giJ31790997|ref| |NM_ 181623.1 |[31790997] gi|31790999|ref |NM_ 181624.1 1(31790999] giJ32307153Jref (NM_ 181640.1 |[32307153] giJ32307155|ref |NM_ 181641.1 j[32307155] gi]32313598Jref| |NM_ 181642.1 |[32313598] gi|31791054Jref |NM_ 181643.1 j[31791054] giJ37059798(ref J]NM_ 181644.2 |[37059798] gi|31791050|ref |NM_ 181645.1 ([31791050] gi(37202115Jref J|NM_ .181646.2] |[37202115] giJ31791046|ref |NM_ 181647.1 [[31791046] giJ32455259(ref JNM. .181651.1 |[32455259] giJ3245526l [ref JNM_ 181652.1 ([32455261] gij3179556θjref JNM. 181654.1 |[31795560] giJ31795554Jref JNM. 181655.1 ([31795554] gi|31795556|ref JNM. 181656.1 1(31795556] giJ3188179l jref J|NM .181657.1 [[31881791] gi|32307125|ref J|NM 181659.1 ([32307125] gi(34556190|ref J|NM. .181661.1 ([34556190] gi|50511946|ref jNM 181670.2 1(50511946] giJ32307141 |ref JjNM. .181671.1 1(32307141] giJ32307147Jref [NM .181672.1 |[32307147] gi[32307149|ref JNM 181673.1 1(32307149] giJ32307112Jref JNM. .181674.1 [[32307112] gi|32307114|ref J|NM .181675.1 1(32307114] giJ32307116Jref JNM. 181676.1 j[32307116] giJ32307118Jref JNM .181677.1 |[32307118] gi|32307120|ref JNM 181678.1 1(32307120] gi[32307129Jref J|NM .181679.1 |[32307129] giJ48717493Jref J|NM. 181684.2 [[48717493] giJ32140179Jref JNM. .181686.1 1(32140179] gi|32140181 |ref JNM 181688.1 ([32140181] gij32307135Jref J|NM 181689.1 |[32307135] giJ32307162Jref JNM .181690.1 1(32307162] giJ32455263Jref JNM 181696.1 [[32455263] gi(32455265|ref J]NM. 181697.1 j[32455265] giJ32171183(ref J|NM. .181698.1 |[32171183] gi|32455243|ref (NM .181699.1 [[32455243] gi|32171251 |ref JNM 181701.1 j[32171251] giJ32483370|ref JNM .181702.1 1(32483370] gi|32483411 |ref J|NM 181703.1 |[32483411] giJ32699146Jref JNM .181704.1 1(32699146] gi|32171235|ref |NM 181705.1 |[32171235] gi|42476213Jref JNM. 181706.3 [[42476213] giJ32698787|ref (|NM 181707.1 |[32698787] giJ32171232Jref JNM 181708.1 j[32171232] giJ40255256Jref JNM. 181709.2 1(40255256] giJ3422236l jref (NM 181710.2 |[34222361] giJ3217122θ[ref J|NM 181711.1 1(32171220] gi|44917612Jref J|NM 181712.2 [[44917612] giJ36030972Jref J|NM. .181713.3 |[36030972] gi|32171218Jref JNM 181714.1 1(32171218] giJ32171214|ref J|NM 181715.1 1(32171214] gi|32171195Jref JNM 181716.1 |[32171195] gi|32171210|ref JjNM 181717.1 1(32171210] giJ38708312Jref JNM 181718.3 1(38708312] gi|42476209Jref J|NM 181719.2 1(42476209]
132171208 |ref| |NM..181720.1 |[32171208] I42734392 |ref JNM .181721.11(42734392] 132171202 |ref| |NM .181722.1 [[32171202] J32171204 |ref] |NM..181723.1 ([32171204] 132171198 | ref J |NM .181724.1 j[32171198] 132401415 |ref J|NM..181725.1 |[32401415] J32171200 jref JjNM .181726.11(32171200] (32171193 jref JjNM..181727.1 |[32171193] [32481217 |ref J|NM..181733.11(32481217] (33188451 |ref J|NM .181737.1 |[33188451] [33188453 jref J |NM .181738.11(33188453] (32454730 jref J |NM..181739.1 |[32454730] |32454732 jref J |NM..181740.11(32454732] |32454745 |ref J|NM .181741.1 [[32454745] |32454747 j ef J INM .181742.11(32454747] I38678523 jref J |NM..181744.1 |[38678523] J32698783 jref JjNM..181745.1 [[32698783] [32455255 Iref JjNM 181746.1 ([32455255] |32454753 [ref J|NM..181747.11(32454753] (32455238 jref j |NM 181755.1 [[32455238] 132308168 [ref J|NM..181756.11(32308168] [32967275 jref jjNM .181762.1 ([32967275] 133356156 |ref J]NM 181773.2 |[33356156] 142476321 [ref J|NM..181774.2 ([42476321] [34222359 |ref JjNM. 181775.2 |[34222359] J32401452 Jref (jNM .181776.1 [[32401452] J32967277 [ref J|NM..181777.1 |[32967277] 132401472 jref J |NM..181780.1 ([32401472] |33946292 jref J |NM .181781.2 |[33946292] (42476174 (ref J|NM .181782.2 [[42476174] J32401450 |ref J|NM. 181783.1 |[32401450] [32401444 [ref JjNM .181784.1 [[32401444] 132401446 |ref J|NM .181785.1 ([32401446] 134330189 |ref j|NM. 181786.21(34330189] 132401442 |ref J|NM..181787.1 [[32401442] (32401436 jref jjNM .181788.11(32401436] 132401438 |ref J|NM .181789.1 ([32401438] 132401432 |ref J|NM..181790.11(32401432] 132401434 jref J |NM .181791.1 |[32401434] J32483389 I ref j|NM .181794.11(32483389] 132483391 I ref J|NM..181795.1 |[32483391] |32479522 jref ( |NM .181797.11(32479522] J32479524 |ref J|NM. 181798.1 [[32479524] |32967282 jref j |NM .181799.11(32967282] (32967284 [ref J|NM. 181800.1 ([32967284] [32967286 jref | JNM..181801.11(32967286] J32967288 I ref J |NM .181802.1 [[32967288] J32967290 |ref J|NM .181803.11(32967290] |32483380 jref JjNM..181804.1 |[32483380] J32483382 jref J |NM .181805.11(32483382] |45580729 [ref J|NM .181806.2 |[45580729] J32441270 Irefj JNM .181807.1 j[32441270] J32698791 |ref J|NM .181808.11(32698791] J32441272 |ref J|NM..181809.11(32441272] J32528309 |ref J|NM .181814.1 |[32528309] (32967253 |ref J|NM..181825.1 [[32967253] J32967255 jref JjNM. 181826.1 j[32967255]
gi|32967257|ref[NMJ81827.1[[32967257] giJ32967259JrefJNMJ81828.1J[32967259] gi(32967261 |ref(NM J 81829.1 J[32967261] gi|32967263Jref|NMJ81830.lj[32967263] gi|32967513|ref|NMJ81831.1|[32967513] gii32967265|ref|NMJ81832.1|[32967265] giJ32967267[ref|NMJ81833.1|[32967267] giJ32967269Jref[NM J 81834.1 j[32967269] gi(3296727l[ref|NMJ 81835.11[32967271] gi|40255262Jref|NMJ81836.3|[40255262] gi|32483368|ref|NMJ 81837.11[32483368] gi[33188455iref[NMJ81838.lj[33188455] giJ32483385Jref|NM J 81839.11[32483385] giJ32469494JrefJNM J 81840.1 j[32469494] gi|32469500|ref|NMJ 81841.1 ([32469500] gi|32469508|ref|NMJ 81842.11[32469508] gi|32469514|ref|NMJ81843.1|[32469514] giJ38327514Jref|NMJ81844.2J[38327514] gi|32813444|ref|NMJ81846.1|[32813444] gii40556374Jref|NMJ81847.2i[40556374] gi|32483358|ref|NMJ 81861.1|[32483358] gi|32528277|ref|NMJ81862.1|[32528277] gι
'J32528279Jref|NM J 81863.1 j[32528279] giJ3252828ljrefJNMJ81864.lj[32528281] gi[32528283iref|NMJ81865.l([32528283] gi|32528285|ref|NMJ81866.1|[32528285] giJ3248336θjref|NMJ81868.lj[32483360] giJ32483362Jref(NMJ81869.lj[32483362] giJ32479518|ref|NMJ81870.1|[32479518] gi[32967596[ref|NMJ 81871.1 ([32967596] gi|32490573|ref|NMJ81872.1|[32490573] giJ32490578irefJNMJ81873.li[32490578] gi(32528267|ref |NM J 81874.11[32528267] giJ32528269Jref|NM J 81875.1 j[32528269] gi|32967592|ref|NMJ 81876.11[32967592] gi|37675274|ref |NM J 81877.2|[37675274] giJ3289536θjref|NMJ81879.1|[32895360] gi|49355827|ref|NMJ81880.1|[49355827] gi|32490588JrefJNMJ81882.1|[32490588] giJ32490566(ref|NMJ81885.li[32490566] gi|33149309|ref|NMJ81886.1|[33149309] giJ33149311 (ref[NMJ81887.1 [[33149311] gi|33149313[ref|NMJ81888.1|[33149313] gi|33149315|ref|NM_181889.1 j[33149315] gi|33149317|ref|NMJ81890.1|(33149317] giJ33149319|ref|NMJ81891.1|[33149319] gi(3314932ljref|NMJ81892.li[33149321] gi|33149323Jref|NMJ81893.1|[33149323] giJ32528272JrefJNMJ81894.1|[32528272] giJ32967585|ref|NMJ 81897.11[32967585] gi|38569455Jref|NM J 81900.2|[38569455] gi|32895362(refJNMJ81985.li[32895362] gi|32895364|ref |NM J 81986.11[32895364] giJ32528290Jref|NMJ82314.li[32528290] giJ32528296Jref |NM J 82398.11[32528296] gi|33286419|ref|NMJ82470.1|[33286419] gi|3328642liref(NMJ82471.li[33286421]
132967318 |ref J|NM. 182472.1 |[32967318] I32967304 jref (jNM 182476.11(32967304] I33286437 (ref J|NM. 182477.11(33286437] |32967306 jref j |NM 182480.1 [[32967306] 132699148 |ref J|NM .182481.1 ([32699148] (32699150 I ref J |NM 182482.1 |[32699150] J33286433 |ref J|NM 182483.1 |[33286433] J32699144 |ref J|NM. 182484.1 [[32699144] J33188438 jref JjNM .182485.1 |[33188438] [32967299 |ref J|NM 182486.1 ([32967299] |32698794 |ref J|NM 182487.1 [[32698794] 132698814 |ref J|NM .182488.1 [[32698814] |32698789 |ref J|NM. 182489.1 j[32698789] (32698785 |ref J|NM 182490.1 ([32698785] 132698821 |ref JjNM .182491.1 [[32698821] [32698823 jref J|NM .182492.1 ([32698823] J32698825 |ref J|NM..182493.1 [[32698825] J32698827 |ref J|NM..182494.1 ([32698827] J41349679 |ref J|NM..182495.31(41349679] J32698829 |ref J|NM..182496.1 j[32698829] [32698831 |ref J|NM .182497.11(32698831] (38788218 |ref J|NM..182498.2 j[38788218] |32698834 |ref J|NM .182499.11(32698834] I32698836 |ref J|NM .182500.1 JJ32698836] J42740902 jref J |NM .182501.21(42740902] (32698840 |ref J|NM .182502.1 |[32698840] I32698844 |ref J|NM..182503.1 [[32698844] [34222370 |ref j|NM .182504.2 |[34222370] |38327628 |ref J|NM..182505.31(38327628] |32698850 jref ] |NM_ .182506.1 JJ32698850] |32698852 jref JjNM .182507.1 JJ32698852] I32698854 |ref J|NM..182508.1 |[32698854] (40805853 jref J]NM
,182509.21(40805853] [32698858 |ref J|NM 182510.1 [[32698858] I40255263 |ref J|NM
,182511.2 |[40255263] (32698865 |ref J|NM.
,182513.1 j[32698865] [32698869 |ref J|NM .182516.1 |[32698869] |32698863 |ref J|NM 182517.1 j[32698863] (32698873 |ref J|NM .182518.1 |[32698873] |32698867 jref J |NM
,182519.1 j[32698867] [32698877 |ref J|NM
,182520.1 j[32698877] J32698871 |ref J|NM
,182521.1 [[32698871] J40255264 |ref J|NM
,182522.2 |[40255264] (32698875 jref J |NM.
,182523.1 |[32698875] J32698885 |ref J|NM
,182524.1 j[32698885] (32698879 jref J |NM 182525.1 [[32698879] |32698889 |ref J|NM
,182526.11(32698889] J32698883 |ref J|NM 182527.1 [[32698883] [32698893 jref JjNM
,182528.1 ([32698893] J32698887 I ref J]NM .182529.1 |[32698887] J32698897 |ref J|NM .182530.11(32698897] J32698891 jref JjNM 182531.1 |[32698891] 132698901 [ref J|NM 182532.11(32698901] J32698895 jref JjNM .182533.1 ([32698895] J32698905 |ref J|NM 182534.1 j[32698905] I32698899 |ref J|NM .182535.1 ([32698899] J32698947 jref J|NM 182536.1 ([32698947]
147519876 |ref J|NM .182537.2 |[47519876] 147271488 Iref JjNM. 182538.3 |[47271488] (32698909 jref J |NM .182539.11(32698909] [32698913 |ref J|NM..182541.1 j[32698913] (32698917 jref JjNM 182543.1 JJ32698917] I32698925 |ref J|NM..182546.1 j[32698925] |33457307 |ref J|NM .182547.21(33457307] I34222377 jref JjNM..182548.2 j[34222377] [33438593 |ref J|NM .182549.11(33438593] (50659060] |ref J|NM..182551.3 |[50659060] (33519460 jref J]NM .182552.21(33519460] [32698937] |ref J|NM 182553.1 |[32698937] 132698931 Iref JjNM .182554.11(32698931] 132698941 jref JjNM .182556.1 [[32698941] I32698935] |ref J|NM .182557.1 [[32698935] |32698945| jref JjNM. 182558.11(32698945] J32698939 |ref J|NM .182559.1 |[32698939] |32698951 |ref JjNM .182560.1 j[32698951] J32698943 |ref J|NM..182561.1 j[32698943] |32698955] |ref J|NM..182562.1 j[32698955] I34222374] |ref J|NM 182563.2 |[34222374] J32698959 |ref (jNM .182564.1 j[32698959] J34222379 jref J |NM..182565.21(34222379] J32698963 Iref (jNM .182566.1 |[32698963] |50979285| |ref JjNM..182568.2 [[50979285] 132698961 Iref JjNM .182569.1 |[32698961] 132698971 I ref J |NM..182570.11(32698971] I40255265] Irefj JNM .182572.2 j[40255265] |32698975 I ref J |NM..182573.1 j[32698975] I32698969 |ref| JNM .182574.1 [[32698969] I32698979 |ref J|NM..182575.11(32698979] [32698983 jref JjNM. 182577.1 [[32698983] (32698977] |ref| JNM .182578.11(32698977] [32698987] |ref JjNM..182579.1 IJ32698987] (32698981 jref JjNM .182580.11(32698981] 132698991 |ref JjNM..182581.1 P2698991] I32698995 J]ref] JNM .182583.1 [[32698995] |32698989 Iref JjNM..182584.1 IJ32698989] [32698993 |ref J|NM 182585.11(32698993] (32699002] |ref J|NM..182586.1 j[32699002] J32698997] |ref J|NM. 182587.11(32698997] 147519912 |ref J|NM .182589.2 j[47519912] J32699000 |ref J|NM .182590.11(32699000] [32699010 |ref J|NM..182591.1 |[32699010] (32699004 jref JjNM 182592.1 ([32699004] [32699014 |ref J|NM..182594.1 [[32699014] (32699008 jref (jNM..182595.1 ([32699008] (32813448 |ref (|NM 182596.1 |[32813448] 132699018 |ref J|NM .182597.11(32699018] 132699012 jref JjNM..182598.11(32699012] J32699016 jref JjNM .182600.1 [[32699016] J32699025 [ref (jNM 182603.1 |[32699025] I32699029 Iref JjNM .182605.1 [[32699029] (47271482 |ref JjNM..182606.2 |[47271482] |34222376 |ref J|NM 182607.21(34222376] I40255267 |ref JjNM 182608.2 |[40255267] J33438597 |ref J|NM 182609.11(33438597]
gi|32699037|ref|NMJ82610.1[[32699037] gi|32699041|ref|NMJ82611.1|[32699041] giJ32699044|ref(NMJ82612.lj[32699044] giJ33504572Jref|NMJ82613.11[33504572] gi[34222380|ref|NMJ82614.2|[34222380] gi|33859820|ref(NMJ 82615.1 j[33859820] gi[32699049JrefiNMJ82616.1|[32699049] gi|32699051 |ref|N J 82617.11[32699051] giJ33504574Jref|NMJ 82619.1 ([33504574] gi|32699053|ref|NMJ 82620.11[32699053] gi|32699055|ref|NMJ 82621.11[32699055] gi|32699057|ref|NMJ82623.lj[32699057] giJ47271492 jref |NMJ 82625.2 j[47271492] giJ3269906ljref|NMJ82626.lj[32699061] gi|32699063Jref|NMJ82627.1 ([32699063] gi|32699065|ref|NMJ82628.1|[32699065] giJ32699069[reflNMJ 82631.1J[32699069] giJ32699071 |ref [NMJ 82632.11[32699071] giJ33438599Jref)NMJ 82633.11[33438599] gi|32699073|ref|NMJ82634.1 ([32699073] gi|32699075|ref|NMJ 82635.11[32699075] gi|33286409|ref|NMJ82637.lj[33286409] gi|33286411 |ref|N J 82638.11[33286411] giJ33286413Jref|NMJ 82639.1 j[33286413] giJ33188462|ref|NMJ82640.1 ([33188462] gi|38788273|ref|NMJ 82641.2|[38788273] giJ32813442Jref|NMJ82642.li[32813442] gi|33188432[ref|NMJ82643.1J[33188432] giJ32967313[ref|NMJ 82644.1 ([32967313] giJ3362072ljref|NMJ82645.2J[33620721] gi|33188440|ref|NMJ82646.1|[33188440] gi|33286423|ref[NMJ82647.1[[33286423] gi|32967604|ref|NM J 82648.1 [[32967604] giJ33239450|refJNMJ 82649.1 [[33239450] giJ32964822Jref|NMJ 82658.1|[32964822] gi|33188426|ref|NMJ82659.1|[33188426] giJ33188428(ref|NMJ82660.li[33188428] gi|32967302|ref|NMJ82661.1|[32967302] giJ33469969Jref|NMJ 82662.1 j[33469969] gi|3299673θjref|NMJ 82663.1 j[32996730] gi|32996732|ref|NMJ 82664.1 ([32996732] gi[32996734|ref|NMJ 82665.1 j[32996734] gi|33359690[ref|NMJ 82666.1 [[33359690] giJ3335654θjref[NMJ 82676.1|[33356540] gi|33359693|ref|NMJ 82678.11[33359693] gi|33356549|ref|NM J 82679.11[33356549] gi|33356555|ref|NMJ82680.1|[33356555] gi|33356557|ref|NMJ82681.1|[33356557] gi|33359696|ref JNM J 82682.1 j[33359696] gi|33149303[refJNMJ82683.lj[33149303] gi(33149305|refJNMJ82684.11[33149305] gi|33359679Jref|NMJ 82685.1 j[33359679] giJ33359218|refJNMJ82686.1|[33359218] gi[33383238Jref|NM J 82687.1 j[33383238] giJ33359700|ref|NMJ 82688.1 j[33359700] gi(33359685(ref|NMJ82689.lj[33359685] gi|33359687|ref|NMJ82690.1|[33359687]
[33188446 [refJ|NM 182691. 1(33188446] 133188448 jref J |NM 182692. j[33188448] J33356153 jref J|NM 182697. 1(33356153] [33186883 jref J |NM 182699. 1(33186883] J39812495 Jref J|NM 182700. 1(39812495] [33186886 | ref J |NM 182701. |[33186886] [33186881 iref JjNM 182702. 1(33186881] J38176293 |ref J|NM.182703. |[38176293] 133186894 jref J |NM.182704. ([33186894] 133186900 jref J |NM.182705. |[33186900] [45827730 Iref (jNM 182706. 1(45827730] (36287059 |ref JjNM
.182709. [[36287059] J36287068 jref | JNM 182710. 1(36287068] (33239446 Iref JjNM.182712. |[33239446] J33239442 jref ( |NM 182715. 1(33239442] (34335187 Iref JjNM.182717. j[34335187] 134335189 |ref J|NM 182718. 1(34335189] 134335191 Iref JjNM.182719. |[34335191] J34335193 Iref JjNM.182720. [[34335193] (34335195 jref JjNM
.182721. |[34335195] [34335197 |ref J|NM 182722. 1(34335197] (34335199 Iref JjNM.182723. |[34335199] [34335201 |ref (|NM 182724. 1(34335201] |34335203 Iref JjNM.182725. |[34335203] J33286429 |ref (|NM 182728. ([33286429] [33519425 Iref JjNM.182729. |[33519425] (33356543 [ref J|NM
.182734. [[33356543] [33519468 |ref J|NM.182739. |[33519468] (33598939 jref JjNM.182740. ([33598939] [33300664 jref J |NM.182741. |[33300664] 133519427 |ref J|NM 182742. 1(33519427] |33519429 [ref J|NM 182743. [[33519429] 133519446 [ref J|NM 182744. 1(33519446] 133469916 |ref J|NM.182746. |[33469916] (33519451 jref JjNM.182749. ([33519451] [33383236 |ref (|NM.182751. [[33383236] (38679906 |ref JjNM.182752. ([38679906] |33300650 jref J]NM 182755. |[33300650] (33300652 |ref J]NM.182756. |[33300652] I50284695 jref
: ||NM 182757. |[50284695] I33300656 |ref| |NM_ 182758. 1(33300656] J33300648 jref ]jNM 182759. 1(33300648] |38202249 |ref J|NM .182760. [[38202249] J33300654 |ref J|NM.182761. 1(33300654] 147271497 [ref (|NM
,182762. ([47271497] J33519457 |refj|NM ,182763, [[33519457] J33469946 jref J |NM 182764, |[33469946] J33342279 jref JjNM ,182765, j[33342279] J51093833 jref J |NM.
,182766, |[51093833] (47716688 jref J |NM.182767, |[47716688] J34335209 jref J |NM 182769, 1(34335209] [34335211 Jref J|NM 182770 [[34335211] (34335213 |ref J|NM 182771, ([34335213] [34335215 |ref J|NM 182772 [[34335215] J33359210 jref J |NM.182774 ([33359210] [33359216 jref J |NM .182775 [[33359216] [33469921 jref JjNM 182776 1(33469921]
M 182777.21(34222375] M 182779.1 |[33354235] M
"182789.21(34452727] M 182790.1 [[33386694] M 182791.11(33943100] M. 182792.1 ([33386698] M .182793.11(33386702] M. 182794.1 j[33386700] M..182795.1 [[33391149] M .182796.1 ([33519454] M 182797.1 ([33469938] M..182798.1 ([33469930] M .182799.1 [[33469934] M..182800.1 ([33383232] M .182801.1 ([33469936] M..182802.1 j[33469923] M .182804.1 ([33413401] M 182810.11(33469973] M..182811.1 ([33598945] M 182812.11(33469961] M .182826.1 ([33598921] M..182827.11(33469942] M 182828.1 |[33457296] M..182829.11(33469144] M 182830.2 [[38158012] M..182831.1 ([33457298] M 182832.1 ([33457305] M..182833.1 ([33457300] M 182835.1 |[33469977] M..182836.1 ([33469950] M..182838.1 |[33469136] M 182847.1 [[33519443] M..182848.2 |[38570071] M..182849.1 j[33519435] M..182850.1 j[34335217] M .182851.1 [[33519437] M..182852.1 |[33519439] M. 182853.1 |[34335219] M .182854.11(33504570] M. 182894.1 [[34365782] M .182895.1 [[33598936] M..182896.11(33598955] M 182898.1 ([33589858] M..182899.21(41351543] M..182901.1 [[33667100] M .182902.2 j[34222381] M .182903.2 |[33620743] M..182904.21(40255269] M 182905.1 |[39573706] M..182906.11(33667102] M 182907.1 j[33946273] M..182908.3 |[40548401] M..182909.1 |[33667104] M..182910.1 ([33624860] M .182911.1 ([41281983] M 182912.1 |[33624866]

M. 182913.11(33624872]
gi|33624878 Jref J|NM. 182914.11(33624878] gi|41281973 jref (jNM 182915.1 |[41281973] gi|41281989 jref JjNM. 182916.1 ([41281989] giJ38201620 |ref (]NM. 182917.21(38201620] gi|46255022 jref J |NM 182918.2 |[46255022] giJ41281980 |ref J|NM. 182919.1 [[41281980] giJ33624895 |ref (|NM. 182920.11(33624895] giJ33624901 jref J |NM. 182921.1 j[33624901] gi|41281995 |ref J|NM 182922.1 ([41281995] gi[41055203 |ref J|NM 182923.2 j[41055203] gi|33667116 jref J |NM. 182924.1 j[33667116] giJ33667110 |ref J|NM 182925.11(33667110] gi(33620774 I ref ( |NM 182926.1 |[33620774] gi|33636767 jref J |NM. .182931.1 ([33636767] gi|33946303 I ref J|NM 182932.1 |[33946303] gi|33946305 jref J|NM. 182933.11(33946305] gi[33636774 jref JjNM
,182934.1 [[33636774] gi|33636770 |ref JjNM 182935.1 ([33636770] gi(33946307 j ef J INM.
,182936.11(33946307] gi|33636741 |ref J|NM 182943.1 [[33636741] giJ33946316 jref J |NM.
,182944.1 ([33946316] gi|33946318 |ref J|NM 182945.1 j[33946318] gi(33946320 |ref J|NM 182946.1 |[33946320] giJ33667114 jref J |NM.
,182947.11(33667114] gi|46909585 [ref (|NM 182948.21(46909585] gi(45545422 | ref j |NM.
,182960.2 |[45545422] gi|41281986 [ref (|NM .182961.1 ([41281986] gi|33946284 [ref JlNM 182962.1 |[33946284] gi[38044283 | ref J |NM.
,182964.31(38044283] giJ41282001 |ref (|NM 182965.1 [[41282001] gi|33667052 |ref j|NM.
,182966.1 j[33667052] gi|36054149 jref J |NM .182970.2 H36054149] giJ33667031 jref JjNM 182971.1 [[33667031] gi[33667027 |ref J|NM. .182972.1 j[33667027] gi|33667062 |ref j|NM. .182973.11(33667062] gi|33667064 jref J |NM. .182974.1 [[33667064] gi|33946294 jref J |NM 182975.11(33946294] gi|33946296 Iref JjNM .182976.1 [[33946296] gi|33695085 |ref J|NM 182977.11(33695085] gi|33695152 |ref J|NM 182978.1 ([33695152] gi(33695098 |ref J|NM. .182980.1 j[33695098] gi|33695100 jref J |NM 182981.1 |[33695100] gi|33695160 [ref J|NM. .182982.1 ([33695160] gi|33695154 jref J |NM 182983.1 |[33695154] gi[34222388 |ref J|NM 182984.2 |[34222388] gi(37595542 |ref J|NM .182985.2 [[37595542] gi[46358067 jref JjNM .183001.21(46358067] gi|33946310 jref JjNM .183002.1 |[33946310] giJ33859846 |ref J|NM .183003.11(33859846] gi|37537715 jref j |NM. 183004.3 ([37537715] giJ33859836 Iref JjNM 183005.1 |[33859836] gi|34335250 [ref J|NM .183006.1 [[34335250] giJ41281992 [ref J|NM .183008.11(41281992] gi[33946279 jref J |NM .183009.1 j[33946279] gi|41327159 |ref J|NM. .183010.1 [[41327159] gi|34335221 jref JjNM .183011.1 |[34335221] gi|34335223 jref JjNM 183012.11(34335223]
134335225 Iref JjNM .183013.1 |[34335225] (34304367 |ref J|NM .183040.11(34304367] 134304369 |ref J|NM 183041.1 [[34304369] [34305292 |ref J|NM .183043.1 ([34305292] 134305294 |ref J|NM .183044.1 [[34305294] 134305296 |ref J|NM 183045.1 ([34305296] (34335261 |ref J|NM 183047.1 [[34335261] I34335263 jref J |NM .183048.1 ([34335263] 134013529 |ref J|NM..183049.1 |[34013529] [34101271 jref ( |NM .183050.1 [[34101271] |34452692 |ref J|NM..183057.1 ([34452692] (34098967 |ref J|NM .183058.1 [[34098967] |34098969 jref J |NM..183059.1 |[34098969] I34335227 |ref J|NM .183060.1 ([34335227] J38176298 jref J |NM .183062.2 [[38176298] |34304330 jref J |NM..183063.1 ([34304330] 134101277 |ref (|NM 183065.1 |[34101277] |34098958 |ref J|NM .183075.11(34098958] J34304335 jref J |NM..183078.1 [[34304335] (34335269 jref ( |NM .183079.11(34335269] |34485715 |ref (|NM .183227.1 |[34485715] |38045958 |ref J|NM..183228.11(38045958] [38045960 |ref J|NM..183229.1 ([38045960] |38045962 |ref (|NM 183230.1 j[38045962] J38045964 |ref J|NM..183231.1 |[38045964] [38045966 |ref J|NM 183232.1 ([38045966] (34734072 |ref J|NM..183233.1 [[34734072] |34485705 |ref (|NM .183234.1 ([34485705] |34485708 jref J |NM 183235.1 [[34485708] 134485710 |ref J|NM..183236.1 ([34485710] |34304332 (ref (|NM .183237.1 [[34304332] [34222390 |ref J|NM..183238.1 ([34222390] 138016130 jref J |NM 183239.1 [[38016130] (34222394 |ref J|NM..183240.1 ([34222394] |39930540 jref J |NM 183241.1 [[39930540] 134304112 jref JjNM 183242.1 j[34304112] |34328927 |ref J|NM .183243.1 |[34328927] J34304353 |ref J|NM..183244.11(34304353] |34304378 jref J |NM 183245.1 |[34304378] (34304355 |ref j|NM 183246.1 j[34304355] J34304346 |ref J|NM..183247.1 |[34304346] J34452729 jref JjNM 183323.11(34452729] I34452687 |ref| JNM .183337.1 j[34452687] 134335133 |ref J|NM .183352.11(34335133] I34452685 jref J |NM 183353.1 |[34452685] |34452702 |ref J|NM .183356.1 [[34452702] (34486091 |ref J|NM .183357.1 ([34486091] I34452708 |ref J|NM. 183359.11(34452708] J37577094 jref jjNM 183360.11(37577094] I37577096 |ref J|NM .183361.1 [[37577096] J41582235 jref jjNM. 183372.2 [[41582235] |42476062 jref JjNM 183373.21(42476062] (34419648 [ref JjNM..183374.1 j[34419648] [34419638 jref | [NM 183375.11(34419638] J34419646 |ref J|NM 183376.1 ([34419646] [34452694 jrefj JNM 183377.1 [[34452694] [34419640 |ref J|NM 183378.1 ([34419640]
gi|34419642|ref|NMJ 83379.11[34419642] giJ34577046irefJNMJ83380.lj[34577046] giJ34577086JrefJNMJ 83381.1 [[34577086] gi[34577088|ref|NMJ83382.1|[34577088] giJ34577090|refJNMJ 83383.11[34577090] giJ34577092|ref|NMJ 83384.11[34577092] gii3457707θjref[NM 183385.1 [[34577070] gi[34577072|ref|NMJ 83386.1|[34577072] gi|34485719|ref|NMJ 83387.1 H34485719] giJ3445271θjref|NMJ 83393.11[34452710] gi|34452712|ref|NMJ83394.1|[34452712] gi|34878689|ref(NMJ83395.1[[34878689] giJ34452735Jref|N 83397.1 [[34452735] gi|34577095|ref|NMJ83398.1|[34577095] giJ34577097|ref|NMJ 83399.1 ([34577097] gi|34577099|ref|NMJ 83400.11[34577099] gi|34577101(ref[NMJ83401.11[34577101] giJ34577076 jrefJNM J 83404.1 j[34577076] gi|34878748|ref|NMJ 83412.1 j[34878748] gi(34878752(ref|NMJ83413.1|[34878752] gi|35493944|ref|NMJ 83414.11[35493944] gi(35493951|ref(NMJ 83415.11[35493951] gi|41393558Jref[NMJ83416.2J[41393558] giJ35493753JrefJNMJ83418.lj[35493753] gi|3549380θ(ref|NMJ 83419.11[35493800] gi|34878764|ref|NMJ 83420.11[34878764] giJ34878768Jref|NMJ83421.1|[34878768] giJ34556206(ref|NMJ 83422.1J[34556206] gi|34577104Jref JNM J 83425.1 j[34577104] giJ34577109(ref|NMJ 84041.1 [[34577109] gi|35493718|ref|NMJ84042.1 |[35493718] giJ34577111 jref(NMJ 84043.1 j[34577111] gi|34878835|ref|NMJ84085.1|[34878835] giJ34878843Jref|NMJ 84086.1 j[34878843] gi[34878851|ref|NMJ84087.1 ([34878851] gi|37577147|ref|NMJ 84231.11[37577147] giJ35493810|ref|NM 184234.l([35493810] gi|35493816Jref[NMJ 84237.1|[35493816] giJ35493821 |ref|NM J 84241.11[35493821] gi|35493828Jref|NM J 84244.1 ([35493828] gi|34878869Jref|NMJ 87841.1|[34878869] gi|34740336Jref|NMJ 94071.1|[34740336] giJ34850056|refJNMJ 94072.1 [[34850056] giJ34740328|ref|NMJ 94247.1 j[34740328] gi|34740330|ref|NMJ 94248.11[34740330] giJ34740338|ref|NMJ 94249.11[34740338] gi|34740326(ref|NMJ94250.li[34740326] gi|34740332|ref|NM J 94251.11[34740332] gi|34740324|ref|NMJ 94252.11 [34740324] gi(34808709|refJN J 94255.1 [[34808709] giJ35493986Jref|NMJ94259.1|[35493986] gi|35493995|ref|NMJ94260.l([35493995] gi|35494002JrefJNM J 94261.1 ([35494002] gi)34915991|ref|NMJ 94270.11[34915991] gi|37595538|ref|NM J 94271.1 ([37595538] gi|34915997|ref|NMJ 94276.1 [[34915997] gi|34915999Jref|NMJ94277.1[[34915999]
M .194278.21(44890061] M
,194279.1 |[34996486] M. ,194281.2 ([42822883] M 194282.1 |[34996482] M
,194283.1 [[34916003] M 194284.1 ([34916005] M 194285.11(39930546] M
,194286.2 [[50083280] M.
,194287.11(34916009] M
,194288.1 |[34916011] M
,194289.11(34916013] M
,194290.1 |[34916017] M.
,194291.1 j[35038578] M
,194292.1 P5038600] M.
,194293.21(40255271] M
,194294.1 |[34916023] M. ,194295.11(34916015] M
,194298.1 |[34916027] M 194299.11(34916021] M. 194300.1 j[34916031] M 194302.11(34916025] M. 194303.1 |[34916035] M 194309.1 ([34916043] M. 194310.1 |[34916037] M.194312.11(34916041] M 194313.1 [[34916049] M.194314.11(34916045] M 194315.1 |[37577125] M.194316.11(37577127] M 194317.2 |[50355992] M.194318.2 ([46358363] M 194319.11(34996524] M.194320.1 |[34996534] M 194322.1 [[35493852] M.194323.11(35493859] M 194324.1 ([34996532] M.194325.1 |[34996520] M.194326.1 |[34996526] M 194327.1 [[34996518] M.194328.1 ([37577176] M 194329.1 [[37577178] M. 194330.1 |[37577180] M 194331.11(37577182] M 194332.1 |[37577184] M. 194352.1 ([37588855] M 194356.1 [[37577161] M 194358.1 ([37588858] M 194359.11(37588860] M. 194428.1 [[38158019] M. 194429.1 [[36287109] M. 194430.1 |[37577169] M. 194431.11(37577171] M. 194434.1 |[37588849] M 194435.11(37588852] M.
,194436.1 |[37595755] M 194439.11(37059809]
M 194441.1 |[37221188]
M 194442.1|[37595751] M.
"194447.11(37577114] M 194448.1[[37577116] M 194449.1|[37221174] M..194450.11(37577118] M
"194451.1|[37577163] M. 194452.11(37588864] M..194453.1|[37588866] M 194454.11(37221186] M..194455.1|[37221181] M 194456.11(37221183] M..194457.1|[37577129] M 194458.11(37577131] M..194460.1j[37622893] M 194463.11(37588872] M..197939.1|[37655168] M..197941.1j[37577167] M .197947.1 |[37675372] M..197948.1 [(37675374] M 197949.1 |[37675376] M..197950.11(37675378] M 197951.1 |[37675380] M..197952.1 |[37675382] M. 197953.11(37675384] M .197954.1|[37675386] M..197955.11(37694066] M 197956.1|[37537557] M..197957.11(37577141] M..197958.1|[37537705] M..197960.1[[37577088] M..197961.1|[37577090] M 197962.11(37537703] M .197964.1[[37537547] M. 197965.1j[37537551] M
..197966.1 |[37574725] M 197967.11(37574727] M..197968.1|[37574605] M. 197970.11(37574730] M .197972.1|[37574613] M..197973.11(37574156] M 197974.1j[37574627] M .197975.11(37574623] M 197976.1|[37595550] M..197977.1([37574607] M 197978.1 [[37574617] M..198038.1 ([37594456] M..198039.1 [[37594458] M .198040.1 |[37595527] M .198041.1 |[37594460] M..198042.2|[40288186] M..198043.1 ([37594450] M..198044.11(37594452] M..198045.1|[37594446] M..198046.11(37594448] M..198047.1J[37594468] M..198053.11(37595564]
M. 198055.1[[37622340]
|37622906 |ref J|NM 198056.1 |[37622906]
137622902 jref JjNM 198057.1 j[37622902]
(37622342 lref J|NM 198058.1 |[37622342]
|46049104 |ref J|NM 198060.2 [[46049104]
[37622886 |ref J|NM 198061.1 |[37622886]
|41584203 | ref J |NM 198066.2 ([41584203]
|46391098 |ref J|NM 198074.3 |[46391098]
(39930570 | ref J |NM. 198075.1 [[39930570]
(40255279 jref J |NM 198076.2 ([40255279]
[37620205 |ref J|NM 198077.11(37620205]
137620213 [ref J|NM 198078.1 |[37620213]
J38016132 jref J |NM. 198079.1 j[38016132]
J37620215 |ref (jNM .198080.1 [[37620215]
J37620211 jref J]NM. 198081.1 |[37620211]
|37620217 [ref J|NM .198082.1 [[37620217]
J37693520 jref JjNM. ,198083.1 P7693520]
[37675276 |ref| NM ,198085.1 j[37675276]
|37675279 |ref| NM ,198086.1 P7675279]
[37675267 |ref| NM. ,198087.1 ([37675267]
(37675269 |ref| NM. ,198088.1 |[37675269]
|37655172 |ref J|NM 198089.1 [[37655172]
[37674288 |ref| NM. ,198097.1 |[37674288]
137694061 |ref| NM 198098.11(37694061]
(37694064] |ref| NM. ,198120.11(37694064]
[38045887 |ref| NM 198123.1 ([38045887]
(38045889 jref J |NM. .198124.1 |[38045889]
[38158004 |ref (|NM 198125.11(38158004]
(38045933 jref | NM .198128.11(38045933]
|38045909 |ref| JNM 198129.1 [[38045909]
[38149909 jref J |NM..198138.1 [[38149909]
[38049013 |ref| JNM. 198139.1 |[38049013]
I39930590] |ref J|NM .198141.11(39930590]
139930568 |ref J|NM. .198147.1 |[39930568]
|39930572 jref J |NM. .198148.11(39930572]
|39930574 |ref J|NM. 198149.1 |[39930574]
|39930576 |ref| JNM .198150.1 [[39930576]
[37700248 |ref J|NM. .198151.1 ([37700248]
(37700251 jref] NM. 198152.1 |[37700251]
|37700253 [ref] |NM .198153.1 [[37700253]
J37700255 [ref J]NM. .198154.1 j[37700255]
J38026959 jref | NM. .198155.1 |[38026959]
|38045905 jrefj NM .198156.1 ([38045905]
[38157975 |ref| NM. .198157.1 |[38157975]
(38156696 jref J |NM 198158.11(38156696]
[38156698 |ref J|NM .198159.1 [[38156698]
(38049008 jref j |NM 198173.1 [[38049008]
[38049010 jref JjNM. .198174.1 [[38049010]
[38045912] Iref JjNM 198175.1 |[38045912]
138156702 |ref J|NM. .198177.11(38156702]
138156704 jref | JNM. .198178.1 [[38156704]
138016136 |ref J|NM 198179.1 |[38016136]
J38016139 |ref J|NM 198180.11(38016139]
J38044109 |ref J|NM .198181.1 |[38044109]
[38045902 |ref J|NM .198182.11(38045902]
138157985 jref JjNM 198183.1 ([38157985]
(38044107 Iref JjNM 198184.11(38044107]
(38044105 jref] |NM 198185.1 [[38044105]
gi|46488916]ref|NM J 98186.2|[46488916] gi|46488918jref JNM J 98187.2|[46488918] giJ38016948Jref|NMJ 98188.11[38016948] gi|40805829|ref|NM J 98189.2|[40805829] gi|38016906|ref|NM J 98194.1 [[38016906] giJ38045943|ref|NMJ 98195.11[38045943] gi|38683839|ref|NMJ 98196.11[38683839] gi[38045945[ref[NMJ 98197.1 ([38045945] giJ38016925Jref|NM J 98201.1 [[38016925] gi|38016921 |ref |NM J 98202.1 [[38016921] gi|38201611 |ref JNM J 98204.11[38201611] gi|38201613Jref|NM J 98205.1 ([38201613] gi|38176295|ref [NM J 98207.11[38176295] gi[38176291|ref(NMJ98212.li[38176291] gi|38016929|ref|NMJ98213.1|[38016929] gi|50897855|ref|NMJ98215.2|[50897855] gi|38150006(refjNMJ 98216.1 ([38150006] gi[38201662|refJNMJ98217.l[[38201662] gi|38201664iref|NMJ98218.1 ([38201664] gi|38201666Jref|NMJ98219.11[38201666] gi|38149980|ref|NMJ98220.11[38149980] gi|38201616[ref|NM J 98225.1 ([38201616] gi[38327598[ref|NMJ 98227.11[38327598] gi[38327600[ref|NMJ 98229.1 ([38327600] gi|38327602|ref|NMJ 98230.11[38327602] gi|38201681|ref|NMJ98232.l([38201681] giJ38201683[ref(NM J 98234.1 j[38201683] gi[38201685irefJNMJ98235.lj[38201685] gi|38026933|ref|NMJ 98236.11[38026933] gi|3820224θ[ref|NMJ 98239.11[38202240] gi|38044111 |ref|NM J 98240.11[38044111] gil38201622|ref|NM J 98241.1 [[38201622] gi[38201624Jref |NM J 98242.1 [[38201624] gi[38176282|ref|NMJ 98243.11[38176282] giJ38201626(refJNMJ 98244.1 ([38201626] gi|38044287|ref|NMJ 98252.11(38044287] gi]38201697[refJNM J 98253.1 ([38201697] gil38201699|ref(NMJ 98254.1 ([38201699] gi[38201701|ref|NMJ98255.1|[38201701] giJ38201646|ref|NMJ 98256.11[38201646] gi|38201648|ref|NMJ 98257.11[38201648] gi|3820165θ(ref|NM J 98258.1 [[38201650] gil46397365|reflNMJ 98261.2)[46397365] gi|46397367[ref[NMJ98262.2|[46397367] gi|46397363[ref[NMJ98263.2|[46397363] gi|38146117|ref|NMJ98264.1|[38146117] gi|38201679|ref|NMJ98265.1|[38201679] gi(38201656(ref(NMJ 98266.1 [[38201656] gi[38201658[ref|NMJ98267.l[[38201658] gi[38201639(ref|NMJ 98268.11(38201639] gi|38201641 |ref|NMJ 98269.11[38201641] gi(40549455|ref|NMJ98270.2|[40549455] gi(38093638iref(NMJ 98271.1 ([38093638] gi[38093642[ref[NMJ 98274.1 [[38093642] gi|38093644Jref|NMJ 98275.11[38093644] gi|38093646|ref|NMJ 98276.11[38093646] gi|38093648|ref|NMJ 98277.11[38093648]
138093650 |ref J|NM. 198278.1 |[38093650] J38093652 jref ( |NM .198279.1 ([38093652] J38093656 [ref J|NM J 98281.1 |[38093656] J38093658 I ref J|NM "198282.1 [[38093658] J38093660 jref J |NM 198283.1 |[38093660] J38093664 jref J |NM .198284.1 [[38093664] J38093662 |ref J|NM. "198285.1 ([38093662] J38201671 |ref J|NM "198287.11(38201671] 138158010 |ref JjNM 198289.1 ([38158010] J38202216 jref J |NM..198291.1 ([38202216] [38146011 jref J |NM "198309.11(38146011] (38146007 Iref JjNM 198310.1 |[38146007] 141282024 Iref JjNM 198312.1 [[41282024] [47498549 jref JjNM..198315.2 |[47498549] J38787969 Irefj [NM 198316.1 j[38787969] [38194228 jref J |NM..198317.1 [[38194228] (38195086 |ref JjNM 198318.11(38195086] [38195083 |ref JjNM..198319.1 |[38195083] |38327525 jref J INM..198320.1 [[38327525] (38788159 Iref JjNM. 198321.21(38788159] I38327626 |ref J|NM 198324.1 IJ38327626] J38201652 |ref J|NM..198325.1 j[38201652] 138201629 jref J |NM 198327.1 |[38201629] 138201631 |ref J|NM .198328.1 ([38201631] J38327031 jref J |NM 198329.1 ([38327031] 138327541 |ref J|NM. 198330.11(38327541] |38327543 |ref J|NM .198331.1 |[38327543] J38327547 | ref J |NM 198333.1 ([38327547] I38202256 [ref J|NM..198334.11(38202256] (38230584 jref JjNM 198335.1 |[38230584] I38327528 |ref JjNM .198336.1 j[38327528] [38327530 |ref JjNM..198337.1 j[38327530] (38198662 jref J |NM 198353.1 j[38198662] [38505267 [ref j|NM..198376.1 |[38505267] (38505269 jref J |NM..198377.1 ([38505269] [38505271 |ref j|NM 198378.1 j[38505271] I38505273 |ref J|NM .198379.1 |[38505273] (38505275 |ref J|NM..198380.1 j[38505275] [38201716 jref ( |NM .198381.1 [[38201716] (38505277 jref jjNM .198382.1 |[38505277] |38505279 Iref JjNM 198383.11(38505279] J38505281 |ref J|NM .198384.1 [[38505281] |38505283 |ref J|NM .198385.11(38505283] J38505285 |ref J|NM..198386.1 j[38505285] (38505287 |ref J|NM .198387.1 ([38505287] J38505289 |ref J|NM .198388.1 j[38505289] 138202231 [ref J|NM. 198389.11(38202231] (38229312 |ref J|NM .198390.1 j[38229312] [38202221 |ref JjNM..198391.1 |[38202221] (38202238 |ref JjNM..198392.1 [[38202238] J38327619 jref JjNM 198393.11(38327619] (38327629 |ref J|NM .198394.1 [[38327629] 138327551 |ref J|NM..198395.11(38327551] J38505291 |ref J|NM..198396.1 [[38505291] J38505293 jref JjNM..198397.11(38505293] 138327614 |ref J|NM..198398.1 |[38327614] (38261964 |ref J|NM. 198399.1 [[38261964]
J38257150 |ref J|NM..198400.11(38257150 150053931 |ref (|NM 198401.2|[50053931 J42476153 [ref J|NM .198402.2[[42476153] J46048200 |ref J|NM.198403.2([46048200] 138257143 |ref J|NM .198404.1([38257143 141282030 Jref J|NM..198406.1|[41282030] J38455409 Irefj [NM 198407.1[[38455409 [38372932 jref JjNM..198426.11(38372932 J38372930 |ref (|NM 198427.1 ([38372930 J38569433 |ref J|NM..198428.1 [[38569433 |38327604 |ref JjNM 198430.1 |[38327604 [38327040 |ref JjNM..198431.1 |[38327040; (38327606 [ref JjNM 198432.1 ([38327606 J38327563 |ref JjNM .198433.1 |[38327563] (38327565 Iref JjNM 198434.1 [[38327565 (38327567 |ref JjNM .198435.1 |[38327567 [38327569 jref JjNM..198436.1 ([38327569 138327571 |ref J|NM .198437.1 |[38327571 138327517 |ref J|NM .198439.1 [[38327517 I50845408 |ref JjNM..198440.2|[50845408 (38348197 |ref JjNM..198441.1 ([38348197 [38348199 jref J INM .198442.1 [[38348199 (38348201 jref JjNM 198443.1 |[38348201 I38348203 Iref JjNM .198444.1 |[38348203 J38348205 | ref J |NM..198445.1 ([38348205 I38348207 |ref (jNM .198446.1 |[38348207 (38348210 |ref JjNM.198447.1 [[38348210 138348212 |ref (jNM..198448.1 ([38348212 [38348214 |ref JjNM 198449.1 [[38348214 138348216 IrefJjNM..198450.1 |[38348216 (38348218 jrefJ |NM 198451.11(38348218 I38348220 |refJ|NM .198452.11(38348220] [39930584 |refJjNM..198457.11(39930584 I38348235 jrefjJNM 198458.1 ([38348235 (45356146 |refJ|NM..198459.2|[45356146 J38348239 jrefJ |NM 198460.1 [[38348239 (38348241 IrefJjNM .198461.1 |[38348241 139653312 |refJjNM.198462.1 ([39653312 I38348243 irefjJNM .198463.1 [[38348243 [38348247 IrefJjNM .198464.1 |[38348247 |38348249 jrefJ|NM 198465.1 |[38348249 J38348251 IrefJjNM .198466.11(38348251 J38348253 |refJ|NM .198467.1 |[38348253 |38348255 JrefJ|NM .198468.1|[38348255' J38348257 [refJ|NM .198469.1IJ38348257 J38348259 |refJ|NM .198471.1j[38348259 J38348261 jrefJ |NM .198472.1|[38348261 |40353240 |refJjNM 198473.1 [[40353240' [38348263 |refJ]NM .198474.1 |[38348263; J38348267 |refJ]NM..198476.1 ([38348267] J38348269 jrefJ|NM .198477.1 |[38348269] (38348271 [refJNM 198478.11(38348271 (38348273 |refJ|NM .198479.1|[38348273; [47271375 |refJ|NM .198480.2j[47271375] J38348277 |refJ|NM 198481.11(38348277 [38348279 |refJ|NM 198482.1|[38348279 [38348281 jrefJjNM 198483.11(38348281
|38348283|ref|NMJ 98484.11[38348283] [38348285iref[NMJ 98485.1 ([38348285] 150053871 jrefjNMJ 98486.2|[50053871] (38348289iref(NMJ 98488.1 [[38348289] 138348291 |ref|NMJ 98489.1 [[38348291] [50234888Jref[NMJ 98490.1 ([50234888] |38348293|ref|NMJ 98491.1|[38348293] [38348295iref[NMJ 98492.1 j[38348295] i38348297(ref|NMJ98493.l[[38348297] |38348299|ref|NMJ 98494.11[38348299] J38348301|refJNMJ 98495.1 [[38348301] J38348303[ref|NMJ 98496.1 ([38348303] (38348309[ref|NMJ 98498.11[38348309] (38348311 |refJNM J 98499.11(38348311] [38348315|ref|NMJ98501.lj[38348315] i38348319(ref|NMJ98502.li[38348319] (41349442|ref|NMJ98503.2|[41349442] J38348323Jref|NMJ 98504.1 [[38348323] J38348327Jref|NMJ 98506.11[38348327] J38348321|ref|NMJ 98507.11[38348321] [38348331 |ref|NMJ 98508.1 j[38348331] i38348335|ref|NMJ98510.lj[38348335] |38348329[ref|NMJ 98511.11[38348329] |38348339|ref|NMJ98512.1|[38348339] J39653324|ref|NMJ 98513.1|[39653324] 142476012|ref|NMJ 98514.2|[42476012] |38348337|ref|NMJ98516.1|[38348337] [48762676(ref|NM J 98517.2|[48762676] |3834834lirefJNMJ98519.1|[38348341] [38348351 (ref|NMJ 98520.1 [[38348351] |38348345|ref|NMJ 98521.1|[38348345] |38348355|ref|NMJ 98524.11[38348355] [38348349Jref|NMJ98525.1|[38348349] J42734394Jref|NM J 98526.11[42734394] |38348359Jref|NMJ 98527.1 j[38348359] |38348353|ref|NMJ 98529.11[38348353] J41327759(ref|NM J 98531.3J[41327759] |38348367|ref|NMJ 98532.11[38348367] 138348361 jref|NMJ98533.1|[38348361] i40254803[ref|NMJ 98534.1 j[40254803] (38348371|ref|NMJ98537.1|[38348371] [38348365Jref[NMJ 98538.11[38348365] |39930586|ref|NMJ 98539.11[39930586] J42821106(ref|NM J 98540.2J[42821106] |38348369(ref|NMJ 98541.1J[38348369] |38524613|ref|NMJ98542.lj[38524613] |38348377|ref|NMJ 98543.1 j[38348377] [41327699iref|NMJ98544.2|[41327699] |40255273iref(NMJ98545.2|[40255273] |38348381|ref|NMJ98546.l[[38348381] (39753967|ref|NMJ 98547.1 j[39753967] (38348383[ref|NMJ 98549.11[38348383] (38348385[ref|NM J 98550.11[38348385] (38348389|ref[NMJ 98552.11[38348389] 138348391 jref|NMJ 98553.11[38348391] |38348732JrefJNMJ 98554.1 [[38348732] i38348393[ref|NMJ 98557.1 ([38348393]
gi|38348395|ref|NMJ 98559.11[38348395] giJ38348397|ref|NMJ 98560. lj[38348397] gi [38348399|ref|NM J 98562.11[38348399] gi|38348401 jrefJNM J 98563.1 j[38348401] giJ38348403|ref|NM J 98564.11[38348403] gi|38348405|ref|NM J 98565.11[38348405] giJ38348407|ref|NMJ 98566.1|[38348407] giJ38348409Jref(N J 98567.1 j[38348409] gi|38348411 JrefjNMJ 98568.11[38348411] gi(5035594θjref[NMJ 98569.1 [[50355940] gi|38348413Jref|NMJ98570.1|[38348413] gi|38348415[ref|NMJ98571.1|[38348415] gi|38348417[ref|N J 98572.1 j[38348417] giJ3834841 θjref |NM J 98573.1 j[38348419] gi|38348427|ref|NMJ 98577.1 ([38348427] gi|38524615|ref|NMJ98580.1|[38524615] gi|38348431|ref|NMJ 98581.1 ([38348431] gi|38348433Jref|NMJ 98582.1|[38348433] gi|38348435|ref|NMJ98584.lj[38348435] giJ38348437|ref|NMJ98585.lj[38348437] gi|40255282|ref|NMJ98586.2|[40255282] gi|38327608(ref(NMJ 98587.1|[38327608] giJ38372924 jref |N J 98589.1 j[38372924] gi|38372920|ref|NMJ98590.1|[38372920] gi|38372922|ref|NMJ 98591.1|[38372922] gi|38372916|ref |NM J 98593.11[38372916] gi|38372912|ref[NMJ98594.1[[38372912] gi|40807503Jref[NMJ 98595.1|[40807503] giJ38327657|ref|NMJ 98596.1 j[38327657] gi|38373670|ref|NMJ 98597.11[38373670] gi|38373676|ref|NMJ 98679.11[38373676] gi|38373683|ref[NMJ98681.l[[38373683] gi[38373679|ref(NMJ 98682.1|[38373679] gi|38371738|ref|NM J 98686.1 j[38371738] gi|38490567|ref|NMJ 98687.11[38490567] giJ38490574|ref|NMJ 98688.11(38490574] gi[38490570|ref[NMJ98689.lj[38490570] gi[38490576|ref|NMJ 98690.1|[38490576] gi(38490572Jref|NMJ 98691.1 j[38490572] gi|38490578|ref|NMJ98692.1|[38490578] gi(46250734|ref|NMJ 98693.11[46250734] gi|38490580|ref|NMJ98694.l[[38490580] gi(38490582Jref JNM J 98695.1 ([38490582] gi|38490584Jref JNM J 98696.11[38490584] gi|38371736|ref|NMJ98697.1|[38371736] gi|38371743iref[NMJ98698.li[38371743] gi|38490586|ref|NMJ98699.1|[38490586] giJ3857008θjrefJNMJ98700.lj[38570080] gi J38424080|refJN J 98704.1 [[38424080] gi|38424074|ref|NMJ98705.1|[38424074] giJ38424084JrefJNMJ98706.lj[38424084] gi|38424078|ref|NMJ 98707.11[38424078] gi J38424082Jref JNM J 98708.1 j[38424082] gi|38569406JrefJNMJ 98709.1 [[38569406] gi|38505173|ref|NMJ 98712.11(38505173] giJ38505175Jref |NM J 98713.1 [[38505175] gi|38505177|ref|NMJ98714.1|[38505177]
138505179 |refJ|NM.198715.11(38505179] J38505181 |refJjNM 198716.1[[38505181] 138505183 |refJjNM .198717.1([38505183] J38505185 (refJ|NM.198718.11(38505185] 138505187 |refJjNM .198719.1[[38505187] J38505189 IrefJjNM 198720.1([38505189] I38570074 jrefJjNM 198721.1[[38570074] J38454321 |refJ|NM.198722.11(38454321] (38505155 jref (jNM 198723.11(38505155] |38683835 |refj|NM 198793.1 |[38683835] [38570134 I ref J |NM..198794.1 |[38570134] 138505160 |ref J|NM 198795.11(38505160] 138505197 jref J|NM .198797.1 |[38505197] (38569427 | refj |NM..198798.11(38569427] [38569439 jref JjNM..198799.1 |[38569439] |38488756 |ref J|NM..198822.11(38488756] 145433551 jref J|NM .198827.2|[45433551] |38488766 |ref J|NM..198828.11(38488766] 138505164 jref J |NM.198829.1|[38505164] |38569422 jref JjNM .198830.1j[38569422] J38504668 |ref J|NM 198833.1|[38504668] I38679959 |ref J[NM 198834.11(38679959] |38679963 jref JjNM .198835.1|[38679963] (38679966 |ref J|NM 198836.1|[38679966] [38679970 |ref J|NM .198837.1[[38679970] I38679973 | ref J |NM .198838.1 |[38679973] (38679976 |ref J|NM .198839.1([38679976] I38502322 |ref JjNM..198841.11(38502322] [38504666 jref J INM.198843.11(38504666] [38502315 jref J |NM..198844.1j[38502315] |38504664 (ref J|NM .198845.1([38504664] J38504660 [ref J|NM..198846.1[[38504660] (38505210 jref JjNM 198847.1|[38505210] [38502311 jrefj|NM .198849.11(38502311] 147271490 |ref J|NM 198850.2([47271490] (38502313 jrefjjNM.,198851.1|[38502313] 138524611 |ref J|NM.198853.1[[38524611] |38569446 jref JjNM ,198855.1|[38569446] [38570108 jrefj|NM ,198856.11(38570108] (38524586 [ref JjNM.,198857.11(38524586] J38524619 jref J |NM ,198859.1([38524619] J38570139 [ref JjNM.,198867.1[[38570139] J45597174 |ref J|NM..198868.2|[45597174] J38570098 |ref JjNM 198880.1([38570098] J38570102 |ref J|NM 198881.11(38570102] I38569476 jref J |NM .198883.1|[38569476] (38605732 |ref J|NM 198887.11(38605732] [38683815 |ref J|NM..198889.1j[38683815] |38683869 |ref J|NM .198890.11(38683869] (38787934 jref J |NM 198892.1 [[38787934] 138788301 |ref J|NM .198893.1 ([38788301] J38679887 |ref J|NM .198896.1 |[38679887] (38683848 |ref J|NM .198897.11(38683848] [40217840 jref J |NM..198900.11(40217840] (38679883 |ref J|NM .198901.11(38679883] [38787990 |ref J|NM .198902.11(38787990] J38788149 IrefjjNM 198903.11(38788149]
138788154 | ref J |NM .198904.1 |[38788154] 138708316 jref JjNM 198907.11(38708316] I42734396 | ref J |NM 198920.1 |[42734396] (42794264 jref JjNM 198923.21(42794264] 138708010 |ref JjNM 198924.1 |[38708010] |39777609 jref J|NM 198925.1 j[39777609] J39545574 |ref J|NM 198926.1 |[39545574] J41351540 |ref J|NM .198928.21(41351540] J41282055 |ref J|NM 198929.1 |[41282055] 141282050 |ref J|NM .198930.1 |[41282050] 139777613 (ref j|NM..198935.1 [[39777613] J40068468 |ref J|NM 198938.1 |[40068468] [40068470 jref J |NM .198939.1 ([40068470] (40068472 |ref J|NM .198940.11(40068472] [39812105 [ref J|NM..198941.1 [[39812105] (39573729 jref J |NM..198943.1 |[39573729] [39812354 jref J |NM .198944.1 ([39812354] (39573712 |ref J|NM..198945.1 [[39573712] [46852170 jref J |NM .198946.21(46852170] 139573717 |ref JjNM .198947.1 ([39573717] |40288275 jref JjNM 198948.11(40288275] (40288277 |ref JjNM .198949.1 ([40288277] [40288279 |ref J|NM 198950.11(40288279] J39777598 jref j |NM..198951.1 [[39777598] (40288281 |ref J|NM 198952.1 j[40288281] J40288283 |ref J|NM .198953.1 |[40288283] |40288285 |ref j|NM 198954.1 |[40288285] [39812196 [ref J|NM .198955.11(39812196] (39812500 |ref J|NM. 198956.1 |[39812500] [39777585 |ref J|NM .198963.1 ([39777585] (39995088 |ref J]NM..198964.1 |[39995088] [39995090 jref J]NM ,198965.11(39995090] I39995092 [ref J|NM. ,198966.1 |[39995092] |39725956 I ref J |NM 198968.11(39725956] J39812018 (ref J|NM ,198969.1 |[39812018] 139812026 |ref J|NM. ,198970.1 [[39812026] J39725943 |ref J|NM. ,198971.1 JJ39725943] J39752688 [ref J|NM..198973.1 j [39752688] J40068476 jref J |NM 198974.1 [[40068476] 139812491 jref J |NM .198976.11(39812491] [39777603 |ref JjNM. ,198977.1 j[39777603] |39752662 | ref J]NM ,198988.11(39752662] I39752666 |ref J|NM 198989.1 ([39752666] J48675829 jref J]NM .198990.3 [[48675829] [40217616 jref J]NM .198991.1 |[40217616] (39752670 jref J |NM 198992.1 ([39752670] I39752672 |ref| |NM .198993.1 P9752672] J39752674 jref J |NM. 198994.1 [[39752674] I39752676 |ref| JNM .198995.1 ([39752676] J39752678 |ref J|NM .198996.1 |[39752678] J40538879 jref J |NM. 198998.1 [[40538879] J39752682 |ref J|NM 198999.11(39752682] J39752684 |ref (|NM .199000.11(39752684] J39752686 |ref j|NM .199001.11(39752686] I39777605 |ref J|NM .199002.1 |[39777605] J40068499 |ref J|NM .199003.1 JJ40068499] J39812054 |ref J|NM 199004.1 JJ39812054]
gi|39841070|ref|NMJ99005.1[[39841070] gi|4132770θ[ref|NMJ99006.1|[41327700] gi|47157335|ref|NMJ 99037.2|[47157335] gi|39930611 jrefJNM J 99039.1 [[39930611] gi|40317633|ref|NMJ 99040.11[40317633] gi|39932580Jref|NMJ 99043.1 j[39932580] gi[40316917|ref|NMJ99044.2J[40316917] giJ40804769JrefJNMJ 99046.1 j[40804769] giJ3993060lirefJNMJ99047.li[39930601] giJ40018637(ref|NMJ 99050.1 ([40018637] gi|39979637|ref |NM J 99051.1 [[39979637] gi(40018635 jref |NM J 99052.1 ([40018635] gi|39995077|ref|NMJ 99053.1 ([39995077] giJ40018626|refJNM J 99054.1 ([40018626] giJ41327780Jref|NMJ99069.lj[41327780] gi|41327782|ref|NMJ99070.1 |[41327782] gi|47080097|ref|NMJ 99071.2([47080097] gi|40068513|ref|NMJ99072.2|[40068513] giJ41327784Jref|NMJ99073.lj[41327784] gi|41327786|ref|NM J 99074.11[41327786] gi|40068056|ref |NM J 99075.11[40068056] gi[40068054(ref|NMJ99076.1|[40068054] gi|4006805θjref|N J 99077.11(40068050] giJ40068046[ref|NM J 99078.1 j[40068046] gi[40068486Jref[NMJ99121.li[40068486] gi|40217809|ref|NMJ99122.1|[40217809] giJ40068482Jref|NMJ99123.l([40068482] gi|40217800[ref|NMJ99124.1|[40217800] giJ40217791|refJNMJ99126.1|[40217791] gi[40353747[ref|NMJ99127.lj[40353747] gi|40806192|ref|NMJ99129.1|[40806192] gi[4021762ljref|NMJ99131.lj[40217621] gi|40217625|ref[NMJ 99133.11[40217625] giJ40353244(ref|NM J 99135.11[40353244] gi|40217629|ref|NMJ99136.1|[40217629] gi|40288198|ref|NMJ99138.l([40288198] gi|40288192|ref[NMJ99139.1|[40288192] gi|40288287[ref|NMJ99141.lj[40288287] gi[40806166|ref|NMJ99144.lj[40806166] gi[40549414Jref|NMJ 99160.11[40549414] gi|40316909|ref |NM J 99161.11[40316909] gi|40316945|ref|NMJ99162.1|[40316945] gi|40353752|ref(NMJ99163.2|[40353752] giJ40316943Jref|NMJ99165.1j[40316943] gi|40316938|ref|NM J 99166.11[40316938] gi|40316925|ref|NMJ99167.1|[40316925] gi|40316923|ref(NMJ99168.lj[40316923] gi|40317615|ref|NMJ99169.1|[40317615] giJ40317617|ref|NMJ99170.lj[40317617] gil40317619|refJNMJ99171.lj[40317619] gii41152108iref|NMJ99173.2J[41152108] giJ40316842|refJNMJ99175.1|[40316842] gi|40317621|ref|NMJ99176.1|[40317621] gi|40317623|ref|NMJ 99177.1 ][40317623] gi|40353743|ref|NMJ99179.1|[40353743] gi|40353741 |ref|NMJ99180.1|[40353741] gi|40353242JrefJNMJ99181.1|[40353242]
140353749 I ref J |NM 199182.11(40353749] 140353238 [ref J|NM..199183.1 ([40353238] J40354200 jref J |NM 199184.11(40354200] 140353733 (ref J|NM. 199185.1 j[40353733] J40353763 jref J |NM .199186.1 j[40353763] J40354194 (ref J|NM. 199187.1 ([40354194] [40353739 jref J |NM .199188.1 [[40353739] (40353735 |ref J|NM. 199190.1 |[40353735] |40353760 jref J |NM 199191.1 ([40353760] |40353758 |ref J|NM 199192.1 |[40353758] I40353754 |ref J|NM..199193.11(40353754] [40353756 jref JjNM 199194.1 |[40353756] 140549421 jref J|NM 199202.1 ([40549421] 140806189 jref JjNM..199203.1 ([40806189] J40548396 Iref JjNM 199204.11(40548396] J40548407 Iref JjNM..199205.1 |[40548407] |40548374 j ef J INM..199206.1 |[40548374] |40385866 Iref JjNM 199227.1 [[40385866] [40805872 |ref J|NM 199228.1 [[40805872] J40385882 |ref J|NM..199229.1 j[40385882] 140549410 jref JjNM..199231.1 j[40549410] |40548424 jref J |NM..199232.1 j[40548424] [40549412 |refj JNM 199234.1 j[40549412] 140548419 |ref JjNM..199235.1 j[40548419] J46195764 |ref (jNM .199242.1 [[46195764] J40385872 |ref JjNM..199243.1 [[40385872] J40445386 jref jjNM 199244.11(40445386] J40549445 (ref J|NM..199245.1 [[40549445] [40805831 jref J |NM 199246.1 [[40805831] (40804469 |ref (|NM .199247.11(40804469] 140804471 |ref J|NM..199248.1 [[40804471] [40548381 jref j |NM .199249.11(40548381] |40548383 |ref J|NM..199250.1 j[40548383] |40549424 | ref J |NM .199254.1 ([40549424] I40549426 [ref j|NM..199255.1 IJ40549426] |40549432 [ref J|NM .199259.1 ([40549432] (40549434 |ref J|NM .199260.1 [[40549434] |40549436 Jref J|NM..199261.1 j[40549436] [42490765 |ref JjNM 199262.2 [[42490765] 140805851 jref J|NM..199263.1 |[40805851] J40805855 |ref j|NM..199265.1 [[40805855] [40548323 jref ( |NM .199280.1 |[40548323] 140548321 [ref J|NM .199282.1 j[40548321] J51093873 |ref J|NM. 199285.2 |[51093873] I40548325 jref ( |NM .199286.1 [[40548325] J42476196 Iref jjNM. 199290.2 |[42476196] J40786541 jref J |NM .199292.1 ([40786541] I40786543 |ref J|NM..199293.1 |[40786543] 141327702 jref J|NM .199294.1 |[41327702] J41327704 |ref J|NM .199295.11(41327704] |40805857 |ref J|NM .199296.1 j[40805857] [40806218 |ref J|NM..199297.11(40806218] (40806220 jref J |NM..199298.1 |[40806220] [40556392 jref J |NM. 199320.1 ([40556392] (40556388 |ref J|NM .199321.1 [[40556388] [40807495 |ref J|NM 199324.11(40807495] [41349490 iref JjNM 199326.1 ([41349490]
140788000 |ref JjNM. 199327.1 IJ40788000]
140788010 Iref (jNM 199328.11(40788010]
141056258 jref JjNM 199329.1 |[41056258]
(46249350 jref | |NM. 199330.1 ([46249350]
[46249352 jref JjNM 199331.1 |[46249352]
J46249354 |ref J|NM. 199332.1 [[46249354]
I46255056 jref J |NM 199334.2 |[46255056]
142476114 |ref j|NM .199335.21(42476114]
J40786393 jref J|NM. 199336.1 ([40786393]
140786401 jref j |NM ,199337.11(40786401]
I40786395 jref J|NM. 199338.1 [[40786395]
|40786405 |ref J|NM 199339.11(40786405]
|40786409 |ref J|NM ,199341.1 ([40786409]
I40786403 |ref j|NM 199342.1 [[40786403]
140786413 jref J |NM. ,199343.1 |[40786413]
(40786407 |ref (|NM 199344.1 ([40786407]
[48374080 jref J |NM. ,199345.2 |[48374080]
(40786417 |ref J|NM. ,199346.1 [[40786417]
140786421 |ref| JNM ,199348.1 |[40786421]
J40786415 |ref J|NM 199349.11(40786415]
|40786425 jref J |NM. ,199350.1 |[40786425]
140786419 jref J|NM. ,199351.1 j[40786419]
(50355687 I ref ( |NM 199352.2 |[50355687]
[41349483 jref J |NM. .199353.1 |[41349483]
141349485 (ref J|NM .199354.1 |[41349485]
(40806186 |ref J|NM 199355.1 |[40806186]
J40805874 jref j |NM. .199356.11(40805874]
J40788017 jref J |NM ,199357.1 IJ40788017]
|40786537 |ref J|NM. ,199358.11(40786537]
J40805861 jref J |NM. ,199359.1 |[40805861]
I40805863 |ref J|NM 199360.11(40805863]
|40805865 |ref J|NM. .199361.1 |[40805865]
(40805867 jref J |NM 199362.11(40805867]
I40805869 |ref J|NM. .199363.1 [[40805869]
[40806172 jref J |NM 199367.11(40806172]
141872442 |ref J|NM. .199368.1 ([41872442]
I40807442 jref ( |NM 199413.11(40807442]
|40807444 |ref J|NM. .199414.1 [[40807444]
140806195 |ref J|NM 199415.1 ([40806195]
|40807449 |ref J|NM. .199416.1 |[40807449]
J41327788 jref JjNM. 199417.1 |[41327788]
140805840] |ref (|NM 199418.11(40805840]
140805846 |ref J|NM .199420.1 |[40805846]
140807479 |ref J|NM. .199421.1 [[40807479]
|40806208 jref j |NM 199423.1 ([40806208]
J40806210 jref JjNM 199424.1 [[40806210]
[40806215 |ref J|NM. .199425.1 |[40806215]
(40806226 jref J |NM .199426.1 j[40806226]
J40806228 |ref J|NM .199427.11(40806228]
|40806169 [ref J|NM. .199436.11(40806169]
141349459 |ref J|NM .199437.1 [[41349459]
141349461 |ref JjNM. .199438.11(41349461]
[41349463 |ref| |NM. .199439.1 |[41349463]
(41399284 jref JjNM. .199440.1 j[41399284]
[40807458 jref JjNM. .199441.1 [[40807458]
J40805824 jref JjNM. .199442.1 [[40805824]
|40795666 [ref J|NM. 199443.1 j[40795666]
|40805826 Iref JjNM .199444, 1(40805826] [40789309 Iref JjNM 199450, 1(40789309] (40789305 [ref JjNM 199451, ([40789305] 140789307 jref jjNM .199452, |[40789307] [41282073 |ref J|NM..199453, 1(41282073] 141349471 |ref (|NM 199454, j[41349471] 140804751 |ref J|NM..199456, |[40804751] I49574497 |ref (|NM.199459, ([49574497] |40804754 |ref (|NM .199460, j[40804754] |40804760 |ref J|NM .199461, |[40804760] (40806180 |ref j|NM..199462, |[40806180] [40807367 |ref J|NM..199464, ([40807367] 141349500 |ref J|NM .199478, ([41349500] (41349450 [ref j|NM..199482, j[41349450] 141327693 jref J |NM 199483, [[41327693] 141327695 jref J]NM..199484, 1(41327695] |41327697 jref J |NM .199485, [[41327697] 141327686 Iref JjNM..199487, 1(41327686] |41152073 jref J|NM .199511, j[41152073] [41152075 (ref JjNM.199512, ([41152075] (41327688 [ref J|NM .199513, j[41327688] 141350319 |ref J|NM..201222, 1(41350319] J41327775 |ref J|NM..201224, |[41327775] (41152111 [ref J|NM .201252, |[41152111] 141327707 |ref J|NM..201253, j[41327707] 141152083 |ref J|NM .201259. ([41152083] 141152078 |ref J|NM..201260. |[41152078] 141152080 |ref J|NM .201261. 1(41152080] [41406093 |ref J|NM..201262, |[41406093] 141352699 jref J |NM.201263. [[41352699] 141872556 |ref J|NM .201264. ([41872556] (41152106 |ref j|NM .201265. ([41152106] 141872561 |ref J|NM..201266. 1(41872561] 141872566 jref J |NM.201267. ([41872566] [41350326 |ref J|NM .201268. ([41350326] (41152092 |ref J|NM..201269. ([41152092] |50980300 jref J |NM 201274. j[50980300] 141327729 |ref J|NM .201277. [[41327729] 141350315 jref J |NM 201278. [[41350315] [41872571 [ref J|NM .201279. [[41872571] (41152236 jref J |NM 201280. |[41152236] [41350317 |ref J|NM..201281. [[41350317] (41327731 jref J |NM 201282. |[41327731] [41327733 |ref J|NM..201283. ([41327733] 141327735 jref J |NM.201284. |[41327735] (41152234 |ref (|NM .201286, [[41152234] [41349453] jref J |NM..201348. |[41349453] 141406051 jref ( |NM .201349. 1(41406051] (41282244 [ref J|NM .201377. ([41282244] [41322911 I refJ |NM..201378, [[41322911] (41322907 jrefJjNM..201379, ([41322907] I47607493 jrefJjNM..201380, 1(47607493] [41322909 jrefJ]NM..201381, j[41322909] (41322918 jrefJ]NM..201382, [[41322918] [41322913 jrefJ]NM..201383, |[41322913] (41322922 IrefJ]NM .201384, 1(41322922] (41406081 jrefJ]NM 201397, |[41406081]
gi|41872402|ref|NM_201398.1 |[41872402] giJ41352702Jref|NM -01399.1 |[41352702] gi|42476336|refJNM_201400.lj[42476336] giJ4134993ljref|NM_201402.1|[41349931] giI4140606θiref(NM_201403.l i[41406060] gi|41393553|ref|NM_201412.11[41393553] giJ41406054JrefJNM_201413.1|[41406054] giJ41406056Jref|NM_201414.lj[41406056] gi|41393607Jref|NMJ?01428.lj[41393607] gii41393603Jref(NM_201429.1 [[41393603] gi|41393605|refJNM_201430.1 J[41393605] giJ41393609|ref[NM_201431.1 [[41393609] gi|41406077[ref|NM_201432.1|[41406077] giJ41406079|refJNM_201433.lj[41406079] giJ41393613JreflNM_201434.l i[41393613] gi|42475943|ref|NM_201435.1 |[42475943] gi|41406066[ref[NM_201436.l[[41406066] gi|45439354|ref|NM_201437.1[[45439354] giJ41872338|refJNM_201438.11[41872338] giJ41872343Jref|NM_201439.1 ([41872343] gi|41872348(ref|NM_201440.11[41872348] gi|46411155[ref|NM_201441.1 j[46411155] gi|4139360l irefJNM_201442.lj[41393601] giJ46411158JrefJNM_201443.1 [[46411158] gil41872487[ref|NM_201444.2J[41872487] gi|41872493|ref|NM_201445.11[41872493] giJ41393582|ref|NM_201446.l [[41393582] gi|4255828θ(ref|NM_201453.1|[42558280] giJ41872367JrefjNM_201515.1 j[41872367] gi[41406068Jref[NMJ201516.1 |[41406068] gi|41406070|ref|NM_201517.1 |[41406070] gi|41462411 |ref|NM_201520.1 [[41462411] giJ41871945|ref|NM_201521.1|[41871945] gi|41871954Jref|NM_201522.11[41871954] gi|41871959Jref|NMJ201523.1 |[41871959] gi|41584201 |ref|NM_201524.1 |[41584201] giJ41584197Jref[NMJ>01525.1j[41584197] gi[41582238|ref(NM_201526.l i[41582238] giJ41872521|ref|NM_201532.1 |[41872521] giJ41872526|ref[NM_201533.l i[41872526] gi|4254421θ(ref|NM_201535.1 |[42544210] gi|42544212|ref|NM_201536.1 |[42544212] gi|42544215[refJNM_201537.11[42544215] gi|42544217|ref|NM_201538.11[42544217] giJ42544219Jref|NM_201539.l ([42544219] giJ4254422l (ref|NM_201540.1 ((42544221] gi|42544223|ref|NM_201541.11[42544223] giJ48762707iref[NM_201542.2i[48762707] giJ42544186[ref|NM_201543.1 j[42544186] gi|4254419θjref|NM_201544.1 |[42544190] gi(42544192Jref|NM_201545.l j[42544192] gi|41680634|ref|NM_201546.1 ([41680634] giJ41680687|ref[NM 01548.l i[41680687] gi|41680693Jref|NM_201550.lj[41680693] gi|42544199Jref|NM_201552.1 j[42544199] gi|42544201|ref|NM_201553.1 ([42544201] gi|41872499|ref|NM_201554.11[41872499]
142403574 | ref J |NM .201555.11(42403574] J42403563 Iref JjNM 201556.1 [[42403563] J42403569 Iref JjNM..201557.11(42403569] [42519915 jref J |NM..201559.1 |[42519915] (42415532 |ref (|NM .201563.11(42415532] [42415530 jref J |NM .201564.1 j[42415530] J42415487 [ref J|NM 201565.1 [[42415487] J42475554 jref J |NM 201566.1 |[42475554] I42490759 jref JjNM .201567.1 |[42490759] J42476069 |ref J|NM..201568.1 ([42476069] (42476065 (ref J|NM 201569.1 [[42476065] [45359833 jref j |NM .201570.1 ([45359833] (45359835 |ref J|NM .201571.11(45359835] |45359837 |ref J|NM 201572.1 [[45359837] J42415528 |ref j|NM..201574.11(42415528] 142491357 |ref| NM .201575.1 j[42491357] J42415526 I ref| NM..201589.11(42415526] I45359839 | ref | NM .201590.1 JJ45359839] 142476107 |ref| NM..201591.11(42476107] 142476104 |ref J|NM..201592.1 |[42476104] J45359827 |ref J|NM .201593.1 ([45359827] (42491353 |ref J]NM .201594.1 |[42491353] 142476100 |ref J|NM. 201595.1 [[42476100] [45359831 |ref (|NM .201596.1 |[45359831] 145359841 I ref J |NM. 201597.11(45359841] (42475945 |ref (|NM..201598.1 ([42475945] I42475940 I ref J |NM. 201599.11(42475940] 142491361 |ref| NM..201612.1 j[42491361] [42476084 |ref| JNM. 201613.11(42476084] 142491355 |ref J|NM .201614.1 |[42491355] 142490741 jref JjNM .201623.11(42490741] J42516564 Iref JjNM..201624.1 [[42516564] (42490743 [ref J|NM..201625.1 ([42490743] [42516560 jref J |NM. 201626.1 |[42516560] (42516573 jref J |NM .201627.1 j[42516573] [42491369 Iref JjNM..201628.1 |[42491369] (42518064 jref JjNM. 201629.1 JJ42518064] |42544232 |ref J|NM .201630.1 |[42544232] [42518071 Jref J|NM. 201631.1 ([42518071] 142518077 jref J |NM .201632.1 |[42518077] 142518073 Jref j|NM 201633.11(42518073] (42714656 |ref J|NM .201634.1 j[42714656] 142518081 I ref | |NM .201636.11(42518081] (42519911 |ref J|NM .201647.1 ([42519911] 142519908 |ref j|NM..201648.1 j[42519908] I50233798 jref | NM 201649.11(50233798] |42542395 |ref| NM 201650.1 j[42542395] [42542382 |ref| NM..201651.1 ([42542382] I42542399 jref | NM .201653.1 |[42542399] |42544208 |ref| NM .201994.1 ([42544208] 142544124 |ref J|NM .201995.1 [[42544124] 142544126 |ref J|NM 201997.1 ([42544126] 142544122 |ref J|NM .201998.1 |[42544122] 142544171 jref J |NM..201999.1 [[42544171] 147458817 jref JjNM 202000.2 |[47458817] 142544168 Iref JjNM 202001.1 ([42544168] 142544166 jref J]NM 202002.1 [[42544166]
142544160 |ref J|NM .202003.11(42544160] |44829052 jref JjNM .202004.2 [[44829052] 142544148 |ref J|NM 202467.1 j[42544148] 142544143 jref J |NM .202468.11(42544143] 142544145 |ref J|NM..202469.1 |[42544145] [42544139 |ref ]|NM .202470.1 [[42544139] (42544141 jref :j|NM .202494.1 ([42544141] [42544116 [ref J|NM. 202758.11(42544116] 142544181 |ref J|NM 203281.1 ([42544181] |42543993 |ref J|NM. 203282.1 |[42543993] (42560224 |ref J|NM..203283.1 ([42560224] [42560222 jref JjNM..203284.1 j[42560222] (42560230 jref JjNM..203285.11(42560230] |42560232 |ref (|NM 203286.1 |[42560232] [42714663 |ref J|NM..203287.11(42714663] 142718019 jref J |NM..203288.11(42718019] 142560241 |ref J|NM 203289.11(42560241] J42560245 |ref J|NM..203290.1 |[42560245] 142718014 jref J |NM..203291.11(42718014] 142718016 | ref J|NM .203292.11(42718016] 144680121 [ref J]NM 203293.11(44680121] 144680123 |ref J|NM..203294.1 [[44680123] 144680125 jref JjNM..203295.11(44680125] J44680127 |ref| JNM 203296.1 |[44680127] J44680129 I ref J |NM..203297.11(44680129] 142558251 jref JjNM 203298.1 j[42558251] [42558253 I ref J|NM. 203299.1 [[42558253] I42558257 |ref J|NM .203301.1 [[42558257] |45238852 | ref J |NM 203302.2 |[45238852] (42558259 jref J|NM .203303.1 [[42558259] 142558261 |ref JjNM 203304.1 ([42558261] I42558263 Iref JjNM..203305.11(42558263] |42558267 |ref JjNM 203306.1 [[42558267] |42627890 |ref JjNM .203307.1 ([42627890] |42558265 |ref J|NM .203308.1 |[42558265] |42558269 |ref J|NM..203309.1 [[42558269] [42600568 jref J |NM. 203311.1 ([42600568] 144680135 iref JjNM .203314.11(44680135] 144680132 |ref J|NM .203315.1 |[44680132] (42794010 jref J |NM. 203316.11(42794010] I42794778 |ref J|NM .203318.11(42794778] J42716304 |ref J|NM..203326.1 |[42716304] 144680147 jref JjNM. 203327.11(44680147] 142761473 |ref J|NM .203329.1 [[42761473] 142716301 iref J|NM .203330.11(42716301] 142716298 jref ( |NM .203331.1 ([42716298] (42740906 |ref JjNM..203339.1 j[42740906] [42741649 jref J |NM 203341.1 |[42741649] 142716290 |ref J|NM .203342.1 ([42716290] J42716288 | ref J |NM..203343.1 |[42716288] J42741651 |ref J|NM 203344.11(42741651] 142716279 |ref J|NM..203346.1 [[42716279] 142714610 |ref J|NM..203347.1 ([42714610] [42733591 |ref J|NM 203348.1 |[42733591] (44680158 |ref J|NM .203349.21(44680158] 142741683 jref JjNM 203350.11(42741683] J42794766 |ref J|NM 203351.1 [[42794766]
gi|42741674|ref[NM_203352.1 |[42741674] gi|42741676[refJNM_203353.l j[42741676] giJ42740904|ref|NM_203354.1 |[42740904] gi[42740898JrefJNM -03355.lj[42740898] gi|42740900|ref|NM_203356.1 |[42740900] gi|42740894|ref(NM_203357.1 |[42740894] gi(42734502irefJNM_203364.li[42734502] gi|47132516|ref|NM_203365.2|[47132516] giJ42766423Jref|NM_203370.1|[42766423] gi|42766421 |refJNM_203371.11[42766421] gi|42794753|ref|NM_203372.1 |[42794753] gi|42794272|ref|NM_203373.1 |[42794272] gi|42794621 |ref|NM_203374.1 j[42794621] gi|42794615|ref|NM_203375.11[42794615] gii42794617ireflNM_203376.l ([42794617] gi|44955884|ref|NM_203377.1 J[44955884] giJ44955887iref|NM_203378.l [[44955887] gi|42794757|ref|NM_203379.11[42794757] gi|42794759(ref|NM_203380.1 |[42794759] gi|42794274|reflNM_203381.11[42794274] gi|42822892|ref|NM_203382.1 |[42822892] giJ42822871 [ref|NM_203383.1 ([42822871] gi[42794607|ref|NM_203384.1 |[42794607] gi|42822873|ref|NM 203385.lj[42822873] gi|42822869|ref|NM_203386.l j[42822869] gi|42822867|ref|NM_203387.1 |[42822867] giJ42822863(refJNM_203388.l j[42822863] gi|42822865|ref JNM_203389.1 j[42822865] gi|42794262JrefJNM_203390.1 |[42794262] gi|42794762|ref|NM_203391.1 [[42794762] gi|42794268Jref|NM_203392.1 |[42794268] gi|42794613|ref|NM_203393.11[42794613] giJ44955909(ref[NM_203394.lj[44955909] giJ42794270jref|NM J>03395.1 j[42794270] giJ44890049JrefJNM_203399.l [[44890049] gi|42821107|ref|NM_203400.1 ([42821107] gi|44890051 |ref|NM_203401.11[44890051] gi[4282288l (ref|NM_203402.li[42822881] giJ42822875|ref JNM_203403.1 J[42822875] gi|42822877|ref|NM_203405.1|[42822877] gi(42822879Jref|NM_203406.l i[42822879] gi|42821109(ref|NM_203407.11[42821109] giJ42821113Jref|NM_203408.11[42821113] giJ44662812Jref(NM >03411.1|[44662812] gi|44662818|ref|NM_203412.l j[44662818] gii4466283l [refJNM_203413.11[44662831] gi|44662825JrefiNM_203414.l i[44662825] gi|44662827[ref|NM_203415.1 [[44662827] gi[44889962|ref|NM_203416.1 ([44889962] gi|44680107lref|NM_203417.1|[44680107] gi|44680109|ref|NM_203418.11[44680109] gi[44662814|ref|NM_203419.1 |[44662814] gi|44662820|ref|NM_203422.1 |[44662820] gi|44662802|ref|NM_203423.1 j(44662802] gi(44662816|ref|NM_203424.1 |[44662816] gi|44662806|ref|NM_203425.1 |[44662806] gi|44680152|ref|NM_203426.1 |[44680152]
gi|44680116 refl JNM 203428.11(44680116] giJ44680118 ref J|NM.203429.1 1(44680118] gi|45439310 ref (|NM 203430.1([45439310] gi|45439312 ref J|NM .203431.11(45439312] gi|44680113 ref J|NM .203433.11(44680113] giJ44681485 ref J]NM.203434.1|[44681485] giJ44771205 ref| NM .203436.11(44771205] giJ50409938 ref| NM .203437.2][[50409938] gi|50409606 ref| NM 203438.2([50409606] gi[50409576 ref J|NM .203439.2[[50409576] giJ50408561 ref j|NM 203440.2|[50408561] gi[50409592 ref JjNM.203441.2([50409592] gi|45243523 refj NM 203444.1|[45243523] giJ45243526 refl NM.203445.11(45243526] gi|44889476 refl JNM.203446.1([44889476] gi[44889959 ref J|NM..203447.1([44889959] giJ44888813 ref J|NM 203448.1([44888813] gi[44888824 ref J|NM..203451.1[[44888824] gi|44888826 ref J|NM..203452.1([44888826] gi|44888828 ref J|NM .203453.1|[44888828] gi(44888830 ref J|NM .203454.1|[44888830] gi|45439315 ref J|NM.203456.1[[45439315] gi|45439317 ref JjNM .203457.1j[45439317] gi|46397352 ref JlNM .203458.2([46397352] gi|44955928 ref JjNM.203459.11(44955928] gi|44921607 ref J|NM .203462.1|[44921607] gi(45007001 ref J|NM 203463.1([45007001] gi|45439343 refj NM..203466.1[[45439343] gi|45439345 ref| NM..203467.1IJ45439345] gi|45827717 refj NM..203468.11(45827717] giJ45439324 ref JjNM 203471.1|[45439324] giJ45439348 ref J|NM..203472.11(45439348] gi|45439330 ref (jNM 203473.1([45439330] gi|45439332 refl NM..203474.1j[45439332] gi|45439334 ref JjNM 203475.1|[45439334] gi(45439336 ref J|NM..203476.11(45439336] gi[45238850 ref J|NM 203477.1|[45238850] gi|45120103 ref J|NM .203481.11(45120103] gi|45243560 ref J|NM..203486.11(45243560] gi|45243533 refl JNM 203487.1j[45243533] gi|45243546 refl JNM.203488.1j[45243546] gi|45267834 refj NM 203494.11(45267834] giJ45333905 refl NM.203495.1[[45333905] gi|45333908 ref JjNM .203497.11(45333908] gi[45446746 ref J|NM..203499.1[[45446746] giJ45269144| ref J|NM .203500.1[[45269144] gi|45580691 ref J|NM 203503.1([45580691] gi|45359845 ref] |NM..203504.1|[45359845] gi|45359848 ref J|NM 203505.11(45359848] gi|45359858 ref JjNM 203506.1|[45359858] gi|48375168 ref JjNM 203510.1|[48375168] gi|45387928 ref J|NM.205543.1[[45387928] giJ45387924 ref (|NM..205545.1 |[45387924] giJ45387930 ref J|NM.205548.1|[45387930] gi|45387954 refJjNM..205767.1|[45387954] giJ45439301 refJ|NM..205768.11(45439301] giJ45505168 refJlNM.205833.1|[45505168]
147777760 ref|NM..205834. 1(47777760]
147777678 ref|NM.205835, |[47777678]
145545408 ref|NM 205836, 1(45545408]
J45505160 ref|NM .205837, [[45505160]
[45505156 ref|NM 205838. ([45505156]
I45580739 ref|NM..205839, [[45580739]
J45580735 refjNM.205840, [[45580735]
[45505158 ref|NM 205841. 1(45505158]
145545410 ref|NM 205842, 1(45545410]
J45505150 ref|NM 205843, 1(45505150]
[45446744 ref|NM .205845. [[45446744]
(45593131 ref|NM 205846, ([45593131]
J45447089 ref|NM 205847. 1(45447089]
I45504375 ref|NM 205848. 1(45504375]
|45504370 ref|NM 205849. 1(45504370]
(45504368 ref|NM 205850. 1(45504368]
J45504360 reflNM .205852. |[45504360]
I45504354 ref|NM 205853. 1(45504354]
I45504356 reflNM .205854. 1(45504356]
J45504350 ref|NM 205855. j[45504350]
|45504352 reflNM .205856. 1(45504352]
I45504347 reflNM 205857. 1(45504347]
145505144 refjNM 205858. 1(45505144]
J45504345 ref|NM_J205859. 1(45504345]
J45545404 ref| NM 205860. [[45545404]
J45580737 ref|NM 205861. 1(45580737]
J45827766 ref|NM.205862. 1(45827766]
145505183 ref|NM .205863. 1(45505183]
(45505177 ref|NM 205864. 1(45505177]
145580695! ref|NM 206538. 1(45580695]
J45593146 ref|NM 206539. j[45593146]
I45545426 ref|NM 206594. |[45545426]
I45545428 ref|NM 206595. 1(45545428]
I45545436 ref|NM 206808. |[45545436]
145580731 ref|NM 206809. |[45580731]
145545418 ref|NM 206810. 1(45545418]
145545412 ref|NM 206811. 1(45545412]
J45580733 ref|NM 206812. 1(45580733]
145545414 ref|NM 206813. 1(45545414]
145593133 ref|NM 206814. 1(45593133]
J45580716 ref|NM 206817. 1(45580716]
145580718 ref|NM 206818. 1(45580718]
146049124 reflNM 206819. j[46049124]
146049118 ref|NM 206820. |[46049118]
J46049109 reflNM 206821. 1(46049109]
I45827738 ref|NM 206824. 1(45827738]
145643126 ref|NM 206825. [[45643126]
(45643128 reflNM 206826. |[45643128]
I45592962 reflNM .206827. 1(45592962]
J45593127 reflNM 206828. |[4559327]
J45592951 ref|NM 206831. ([45592951]
(45592948 reflNM 206832. |[45592948]
I45592953 reflNM.206833. [[45592953]
J45597461 ref|NM 206834. |[45597461]
145594313 refjNM 206835. |[45594313]
145643120 ref|NM 206836. 1(45643120]
(45643130 reflNM 206837. |[45643130]
gi|45594507|ref|NM..206838.1 [45594507] gi|45643134JrefJNM 206839.1 [45643134] giJ45643124|ref|NM' .206840.1 [45643124] gi|45827749JrefiNM 206841.1 [45827749] gi|45827775|ref|NM" 206852.1 [45827775] gi|45827707(ref|NM 206853.1 [45827707] gi|45827709|ref|NM 206854.1 [45827709] gi|45827711 |ref|NM .206855.1 [45827711] gi|45827777|ref|NM .206857.1 [45827777] gi|46049056|refJNM .206858.1 [46049056] gi|45827752[ref(NM" .206860.1 [45827752] gi|45827754|ref|NM..206861.1 [45827754] giJ45827756Jref|NM .206862.1 [45827756] giJ45827689iref(NM .206866.1 [45827689] gi|45827797|ref|NM" 206873.1 [45827797] gi|46249375|ref|NM 206876.1 [46249375] gi|46249377|ref|NM .206877.1 [46249377] gi|46047428JrefJNM 206880.1 [46047428] gi|45827799|ref|NM" .206883.1 [45827799] gi|45827801 |ref|NM _206884.1 [45827801] gi|45827803|ref|NM _206885.1 [45827803] gi|46047434|ref|NM _206886.1 [46047434] gi|45827725|ref|NM _206887.1 [45827725] gi|45827693|ref|NM _206889.1 [45827693] gi|45827695[ref|NM .206890.1 [45827695] gi|45827697|ref|NM _206891.1 [45827697] gi|4604743l (ref|NM_ .206892.1 [46047431] gi|46195766|ref|NM .206893.1 [46195766] gi|46047454[ref(NM .206894.1 [46047454] gi|46047466|ref|NM .206895.1 [46047466] gi|45827703|ref|NM .206898.1 [45827703] gi|46255004|ref|NM_ .206900.1 [46255004] gi|46255006|ref|NM .206901.1 [46255006] gi|46255008|ref|NM_ .206902.1 [46255008] gi|47458814|ref|NM .206907.2 [47458814] gi|45827779|ref|NM .206908.1 [45827779] gi|45827715|ref|NM .206909.1 [45827715] gi|45827781 |ref|NM .206910.1 [45827781] gi|45827783|ref|NM .206911.1 [45827783] gi|45827785|ref|NM 206912.1 [45827785] gi|45827722|ref|NM..206914.1 [45827722] gi|46094059Jref(NM .206915.1 [46094059] gi|46094061 |refJNM" .206917.1 [46094061] giJ46048444(ref[NM .206918.1 [46048444] gi|51011132|ref|NM" 206919.1 [51011132] giJ46048447irefJNM .206920.1 [46048447] gi|4604845ljref|NM~ .206921.1 [46048451] gi|46048454JrefJNM 206922.1 [46048454] gi|46048457|ref|NM~ 206923.1 [46048457] giJ45935382[ref|NM .206925.1 [45935382] gi|47578100|ref|NM 206926.1 [47578100] giJ46255061 |ref|NM 206927.1 [46255061] gi|46255063|ref|NM_ .206928.1 [46255063] gi|46255065iref[NM. 206929.1 [46255065] giJ46255067|ref|NM .206930.1 [46255067] gi|46249405Jref|NM 206933.1 [46249405] gi|46255051 |ref|NM 206937.1 [46255051]
gi|46249346|ref|NM l._206938.11[46249346] giJ46249356|ref[NM l_206939.1 ([46249356] gi|46249358|ref|NM I_206940.1J[46249358] gi|46249413|ref|NM .206943.1 [[46249413] giJ45935359Jref|NM L.206944.1 [[45935359] gi|45935361|ref|NM 206945.1 j[45935361] gi|45935363Jref|NM l._206946.1 |[45935363] giJ45935365(refJNM 206947.1|[45935365] gi(45935367|ref|NM. 206948.1 j[45935367] giJ46048519irefJNM 206949.1 |[46048519] giJ46249366[ref|NM 206953.1 ([46249366] gi|46249368Jref|NM _206954.1 |[46249368] gi|46249370|refJNM _206955.11[46249370] gi[46249372(ref[NM _206956.11[46249372] gi|46249415|ref|NM" _206961.11[46249415] gi|46255048Jref|NM 206962.1|[46255048] gi|46255042|ref|NM 206963.1|[46255042] gi|46255031|ref|NM 206964.1|[46255031] gi|46255034|ref|NM 206965.11[46255034] gi|46195432Jref[NM. 206966.11[46195432] giJ47894448irefJNM" 206967.1|[47894448] gi|46370076|ref[NM 206994.1|[46370076] gi|46240863[refJNM 206996.1|[46240863] gi|4624367θjref|NM~ 206997.11[46243670] gi|46240865|ref|NM _206998.1|[46240865] gi|46275836Jref|NM 206999.11[46275836] gi|46276879|ref|NM 207002.11[46276879] gi|4627688ljref|NM 207003.1|[46276881] gi|4687710ljref|NM 207005.11[46877101] gi|46255016Jref|NM 207006.1|[46255016] gi|48949836Jref|NM_ 207007.2|[48949836] giJ4635841θjrefJNM 207009.2|[46358410] gi|46370094|ref|NM 207012.1 |[46370094] gi|46276892|ref|NM 207013.1|[46276892] giJ46276884|ref|NM 207014.11[46276884] gi|47777356|ref|NM 207015.11[47777356] gi|46370059Jref|NM_ 207032.1|[46370059] gi|46370061|ref|NM_ 207033.11[46370061] gi|46370063|ref|NM 207034.1|[46370063] gi|46309850|ref|NM_ .207035.11(46309850] giJ46370079|ref|NM 207036.1 [[46370079] giJ4637008l[ref|NM_ 207037.1|[46370081] gi|46370083Jref|NM_ 207038.1 j[46370083] gi|46370085Jref|NM_ 207040.11[46370085] gi|46389549|ref|NM 207042.1|[46389549] gi(4638955l [ref|NM_ 207043.1|[46389551] gi|46389553|ref|NM_ 207044.1 [[46389553] gi|46389555|ref|NM_ 207045.11[46389555] gi|46389557|ref|NM_ 207046.1|[46389557] gi|46389559|ref|NM_ 207047.1 [[46389559] giJ46358079(ref[NM_ 207102.1 [[46358079] gi|46358065(ref|NM_ 207103.11[46358065] gi|47578094|ref|NM_ 207106.1|[47578094] gi|47578096Jref|NM_ .207107.1 j[47578096] gi|46488920|ref|NM_ 207108.1 ([46488920] gi|46370055Jref|NM. 207111.2([46370055] giJ46361986|ref|NM 207112.1 [[46361986]
gi|46361975|ref|NM_207113.11(46361975] giJ46361969|ref|NM_207115.lj[46361969] giJ46370056|ref|NM_207116.11[46370056] gi|46397376|ref|NM_207117.2|[46397376] gi|46359854|ref|NM_207118.11[46359854] gi|46397368|ref|NM_207119.11[46397368] giJ46402320|ref|NM_207121.2|[46402320] gi|46370068|ref|NM_207122.1|[46370068] gi|46370070|ref|NM_207123.1|[46370070] gi|4639731 ljref|NM_207125.11[46397311] giJ46397313|ref|NM_207126.11[46397313] gi[46397307|ref|NM_207127.11[46397307] gi|46397309|ref|NM_207128.1|[46397309] gi|46397305|ref|NM_207129.1|[46397305] gi|46389561 |ref|NM_207168.11[46389561] gi|46371997|ref|NM_207170.1|[46371997] gi|46397393|ref|NM_207171.11[46397393] gi|46395495|ref|NM_207172.11[46395495] gi[46391084|ref|NM_207173.1 |[46391084] gi[46592955|ref|NM_207174.lj[46592955] gi|46397399|ref|NM_207181.11[46397399] gi(46395478|ref|NM_207186.l[[46395478] gi|46399199|ref|NM_207189.11[46399199] gi|46909591 |ref|NM_207191.11[46909591] gi|46909593|ref|NM_207194.1|[46909593] gi|46909595|ref|NM_207195.11[46909595] gi|46909597Jref|NM_207196.11 [46909597] gi|46909599|ref|NM_207197.1|[46909599] giJ46402495|ref|NM_207283.1|[46402495] gi|46402493|ref|NM_207284.1|[46402493] giJ46402501 jref|NM_207285.1 j[46402501] gi|46402497|ref|NM_207286.1|[46402497] giJ46402505Jref|NM_207287.lj[46402505] gi|46402499|ref|NM -07288.1|[46402499] gi|46402508|ref|NM_207289.1|[46402508] giJ46402503|ref|NM_207290.1|[46402503] gi|46877104|ref|NM_207291.11[46877104] gi|46411163Jref|NM_207292.11[46411163] gi|46411165|ref|NM_207293.11[46411165] gi|46411167|ref|NM_207294.11[46411167] gi|46411169|ref|NM_207295.11[46411169] gi|46411171 |ref|NM_207296.11[46411171] gi|46411173|ref|NM_207297.11[46411173] gi|46877065|ref|NM_207299.1|[46877065] giJ46410930|refJNM_207303.1|[46410930] giJ46411179|ref|NM_207304.11[46411179] gi|46409253|ref|NM_207305.1|[46409253] gi|46409257|ref|NM_207306.1|[46409257] gi|46409259|ref|NM_207307.1|[46409259] gi|46409261 |ref|NM_207308.11[46409261] gi|46409263|ref|NM_207309.1 |[46409263] gi|46409265|ref|NM_207310.11[46409265] giJ46409267JrefJNM_207311.11[46409267] gi|46409269|ref|NM 207312.1|[46409269] gi|46560554[refJNM_207313.1 j[46560554] gi|46409271 |refJNM_207314.11[46409271] gi|46409273|ref|NM 207315.11[46409273]
gi|46409275|ref|NM_207316.1|[46409275] gi[46409277Jref(NM_207317.1 ([46409277] giJ47271499|ref)NM_207319.2J[47271499] gi|46409281 jrefJNM_207320.1 j[46409281] gi|46409283(refJNM_207321.1 ([46409283] gi|46409285[ref|NM_207322.1|[46409285] gi|46409287|ref|NM_207323.lj[46409287] gi|46409289Jref JNM_207324.1 j[46409289] gi|46409291 |ref|NM_207325.1 ([46409291] gi|46409293iref[NM_207326.li[46409293] gi|49533620|ref|NM_207327.3|[49533620] gi|46409297JrefJNM_207328.l([46409297] gi|46409299|ref[NM_207329.11[46409299] gi)4640930ljrefJNM_207330.1|[46409301] gi|4657593l[ref|NM_207331.1|[46575931] gi|46409303|ref|NM_207332.1|[46409303] gi|46559736[ref[NM_207333.lj[46559736] giJ46409305Jref JNM_207334.1 j[46409305] gi(46409307Jref|NM_207335.1|[46409307] gi|46409309|ref|NM_207336.1|[46409309] gi|46559738|ref|NM_207337.l[[46559738] giJ46409311 jrefJNM_207338.1 [[46409311] gi|46409313(ref|NM_207339.1 ([46409313] giJ46409315JrefJNM_207340.lj[46409315] gi|46409317|ref|NM_207341.11[46409317] gi|46409321 |ref|NM_207343.1 ([46409321] gi|46409323Jref|NM_207344.1|[46409323] gi|46409325(ref|NM_207345.11[46409325] gi|46559740|ref JNM_207346.11[46559740] gi|46409327IreflNM_207347.l([46409327] giJ46409329Jref|NM_207348.1|[46409329] gi|46409331 jref|NM_207349.11[46409331] gi(46409333|ref|NM_207350.1[[46409333] gi|46409335Jref|NM_207351.11[46409335] gi|46409337|ref|NM_207352.lj[46409337] gi|46575927|ref|NM_207353.1|[46575927] gi[46409339|ref|NM_207354.1|[46409339] gi|46409341 (ref(NM_207355.11[46409341] gi|46409343Jref|NM_207356.1 [[46409343] gi|46409345JrefJNMJ.07357.lj[46409345] gi[46409347|ref|NM_207358.1|[46409347] gi|46409349|ref|NM_207359.1|[46409349] gi|46409355Jref(NM_207362.1|[46409355] gi|46409385JrefJNM_207363.li[46409385] gi|46409359Jref(NMJ207364.1|[46409359] gi|46409353Jref|NM_207365.li[46409353] gi|46409363|ref|NM_207366.1|[46409363] gi[46409357(ref(NM_207367.1[[46409357] gi|46409367|ref|NM_207368.1|[46409367] gi|46409371 |ref|NM_207370.11[46409371] gi(50726976[ref|NM_207371.2i[50726976] gi|46409365|ref|NM_207372.1|[46409365] gii46409375[ref|NM_207373.l([46409375] gi[46409369JrefJNM_207374.1|[46409369] giJ46409379|ref|NM_207375.1|[46409379] gi[46409373Jref|NM_207376.1|[46409373] gi|46409383|ref|NM_207377.1|[46409383]
gi|46409377|ref|NM_207378.11[46409377] gi|46409389|ref|NM_207379.11[46409389] gi|46409381 |ref|NM_207380.11[46409381] gi|46409393|ref|NM_207381.11[46409393] gi|46409387|ref|NM_207382.11[46409387] giJ46409397|ref|NM_207383.11[46409397] gi|46409391 (ref|NM_207384.11[46409391] gi|46409401 jref(NM_207385.1 ([46409401] gi|46409395|ref|NM_207386.1 |[46409395] gi|46409405|ref|NM_207387.11[46409405] gi|46409399|ref|NM_207388.1 ([46409399] giJ46409409|ref|NM_207389.11[46409409] gi|46409403|ref|NM_207390.11[46409403] gi|46409413|ref|NM_207391.11[46409413] gi|46409407|ref|NM_207392.1 |[46409407] gi|46409417|ref|NM_207393.11[46409417] gi|46409411 jref|NM_207394.1 j[46409411] gi|46409421 |ref|NM_207395.11[46409421] gii46409415irefJNM_207396.l i[46409415] gi|46409425|ref|NM_207397.1|[46409425] gi|46409419|ref|NM_207398.11[46409419] gi|46409429Jref|NM_207399.11[46409429] gi|46409423|ref|NM_207400.11[46409423] giJ46409433(ref(NM_207401.1 ([46409433] gi|46409437|ref|NM_207403.1 |[46409437] giJ46559762Jref|NM_207404.2|[46559762] gi|46409441 Jref|NM_207405.1 [[46409441] gi|46409435|ref|NM_207406.11[46409435] gi|46409445[ref|NM_207407.1 ([46409445] gi|46409439|ref|NM_207408.11[46409439] giJ46409449|refJNM_207409.l j[46409449] gi|46409443|ref|NM_207410.1 |[46409443] gi|46409453|ref|NM_207411.11[46409453] gi|46409447|ref|NM_207412.11[46409447] gi|46409457|ref|NM_207413.11 [46409457] giJ46409451 [refJNM_207414.11[46409451] gi|46409455|ref|NM_207416.11[46409455] gi|46409465|refJNM_207417.1 ([46409465] gi|48675831 |ref|NM_207418.2|[48675831] gi|46409469JrefiNM_207419.1 |[46409469] gi|46409463|ref|NM_207420.11[46409463] giJ46409561 (ref[NM_207421.11[46409561] gi(46409467|ref|NM_207422.l j[46409467] gi|46409471 |ref|NM_207423.11[46409471] gi|46409473|ref|NM_207424.1 |[46409473] gi|46409477|ref|NM_207426.11[46409477] gi|46409479|ref|NM_207427.11(46409479] gi|46409481 |ref|NM_207428.11[46409481] gi(46409483|ref|NM_207429.1 |[46409483] gi|46430765[ref(NM_207430.1 [[46430765] gi|46409487|ref|NM_207432.1 |[46409487] gij46409489|ref|NM_207433.11[46409489] gi|46409491 |ref|NM_207434.11[46409491] gi[46409493[ref|NM_207435.1 [[46409493] gi[46409495irefiNM_207436.l j[46409495] gi|46409497|ref|NM_207437.1 |[46409497] gi|46409499|ref|NM_207438.11[46409499]
gi|46559770|ref|NM_207439.2|[46559770] giJ46409503[ref JNM_207440.1 ([46409503] gi|46409505|ref|NM_207441.11[46409505] giJ46409507|ref|NM_207442.l[[46409507] gi(46409509irefJNM_207443.li[46409509] gi|46409511 |ref|NM_207444.1 ([46409511] gi|46409513(ref|NM_207445.1|[46409513] gi(46409515|ref|NM_207446.11[46409515] gi|46409517|ref|NM_207447.1 j[46409517] gi|46409519|ref|NM_207448.11[46409519] gi|46409521 |ref|NM_207449.11[46409521] gi|46409523(ref|NM_207450.1|[46409523] giJ46409525|ref |NM_207451.1 [[46409525] gi|46409527jref|NM_207452.1|[46409527] gi|46409529|ref|NM_207453.1|[46409529] gi|46409531 jref|NM_207454.11[46409531] gi|46409539|refJNM_207458.1|[46409539] giJ46409541 (refJNM_207459.1 [[46409541] gi|46409547|refJNM_207460.lj[46409547] gi|46409543|ref(NM_207461.11[46409543] gi|46409551 |ref|NM_207462.11[46409551] gi|46409545|ref|NM_207463.lj[46409545] gi|46409555(ref|NM_207464.1 [[46409555] gi|46409549|refJNM_207465.lj[46409549] gi|46409559Jref|NM_207466.1|[46409559] gi[46409553|ref|NM_207467.1|[46409553] gi|46409565Jref|NM_207468.1 ([46409565] gi|46409557Jref JNM_207469.11[46409557] gi|46409569Jref|NMJ207470.1 [[46409569] giJ46409563|refJNM_207471.11[46409563] gi|46409573Jref|NM_207472.1|[46409573] giJ46409567[ref|NM_207473.1[[46409567] gi J46409577|ref [NM_207474.1 [[46409577] gi|46409571 Jref JNM_207475.11[46409571] gi|46409575|ref|NM_207477.1|[46409575] giJ46409581 |ref|NM_207478.11[46409581] gi|46409583|ref|NM_207479.1|[46409583] giJ46409585 jref JNM_207480.1 j[46409585] gi|46409587|ref|NM_207481.11[46409587] gi|46409589|refJNM_207482.1|[46409589] gi|46409591|ref|NM_207483.11[46409591] gi|46409593[ref|NM_207484.li[46409593] gi|46409595Jref|NM_207485.lj[46409595] gi|46409597|ref |NM_207486.11[46409597] gi|46409599Jref|NM_207487.l([46409599] gi|46409605|ref|NM_207488.1|[46409605] giJ46409601 |ref|NM_207489.1 ([46409601] gi|46409609|ref|NM_207490.1|[46409609] gi|46575933Jref|NM_207491.1 [[46575933] gi|46409603|ref|NM_207492.1J[464096b3] gi|46409613|ref|NM_207493.1|[46409613] gi|46409607irefJNM_207494.li[46409607] giJ46559764|ref|NM_207495.2j[46559764] gi|46409611 |ref|NM_207496.11[46409611] gi|46409621 jref JNM_207497.1 [[46409621] gi|46409615(ref|NM_207498.1 [[46409615] gij46409623|ref|NM_207499.1|[46409623]
gi|46409619|ref|NM_207500.1|[46409619] gi[46409629JrefiNM_207501.1 ([46409629] gi|46409625Jref|NM_207502.1 ([46409625] gi|46409633|ref|NM_207503.1|[46409633] gi|46409627|ref|NM_207504.1 ([46409627] giJ46409637|ref|NM_207505.l j[46409637] gi|46409631 [ref(NM_207506.1 j[46409631] giJ46409641|ref|NM_207507.1 [[46409641] gi|46409635|refJNM_207508.1 ([46409635] gi|46409643|ref|NM_207509.1 [[46409643] gi|46559773|ref|NM_207510.2[[46559773] gi|46409645|ref|NM_207511.1 ([46409645] gi|46409647JrefJNM_207512.1 ([46409647] gi|46409649|ref|NM_207513.1|[46409649] gii46447819|refJNM_207514.1|[46447819] gi|46447826|ref|NM_207517.1 |[46447826] giJ46909583|ref|NM_207518.1 |[46909583] gi|46488943|ref|NM_207519.1 [[46488943] gi|47519489|ref|NM_207520.1 ([47519489] gi|4751956l(ref|NM_207521.1|[47519561] gi|48375166jref|NM_207577.11[48375166] gi|46909586|ref|NM_207578.1|[46909586] gi|46485770|ref|NM_207581.11[46485770] gi|46485772|ref|NM_207582.1 |[46485772] gi|46488934|ref|NM_207584.1|[46488934] gi|46488936|ref|NM_207585.11[46488936] gi[46592963(ref(NM_207627.11[46592963] gi|46592970|ref|NM_207628.1|[46592970] gi|46592977|ref|NM_207629.1|[46592977] gi|46592983|ref|NM -07630.1|[46592983] giJ46518521|ref|NM_207644.lj[46518521] gi|46559746|ref|NM_207645.1|[46559746] gi|46518523|ref|NM_207646.1|[46518523] gi|46518525|ref|NM_207647.1|[46518525] gi[5054197θjrefJNM_207660.2[[50541970] gi|50541969|ref|NM_207661.2|[50541969] gi|50541966|ref|NM_207662.2|[50541966] gi|46592999JrefiNM_207672.11[46592999] gi[47078226|ref|NM_212460.11[47078226] gi|47132574|ref|NM_212461.11[47132574] gi[47078244|ref|NM_212464.11[47078244] gi|47078246|ref|NM_212465.11[47078246] giJ47078248|ref|NM_212467.11[47078248] giJ47078277|ref|NM_212469.11[47078277] giJ47132580|ref|NM .12471.11[47132580] gi|47132582|ref|NM_212472.1|[47132582] gi|47132548|ref|NM_212474.1|[47132548] gi|47132550|ref|NM_212475.11[47132550] gi|47132552|ref|NM_212476.1|[47132552] gi(47132554|ref|NM_212478.11[47132554] gi|47078242|ref|NM_212479.1|[47078242] gi[47078223[ref[NM_212481.1 ([47078223] gi|47132556|ref|NM .12482.1|[47132556] gi|47078237|ref|NM -12492.1 |[47078237] gi|47078230|ref|NM_212502.1 |[47078230] gi|47078232|ref|NM_212503.1|[47078232] gi|47078254|ref|NM_212530.1|[47078254]
|47078217 ref J|NM 212533 |[47078217] 147157321 ref j|NM .212535 [[47157321] (47157324 ref J|NM 212539 |[47157324] [47078263 ref J|NM 212540 1(47078263] I47078257 ref J|NM 212543 |[47078257] (47086494 ref J|NM 212550 1(47086494] 151036605 ref J|NM 212551 ([51036605] [47086484 refl JNM..212552 1(47086484] I47086474 refl (NM. 212553 [[47086474] |47086468 refl [NM 212554 1(47086468] I47086458 ref JlNM 212555 J[47086458] I47086442 ref j|NM .212556 1(47086442] I47086438 ref J|NM .212557 j[47086438] I47086434 refl |NM.212558 |[47086434] I47086426 refl |NM_ .212559 1(47086426] 147132590 refl JNM 213560 |[47132590] (47132599 ref J]NM 213566, |[47132599] 147080102 refl JNM .213568, |[47080102] 147087156 refl JNM .213569, 1(47087156] 147132518 ref J|NM .213589, |[47132518] J47132523 ref J|NM 213590, 1(47132523] (47132602 refl JNM 213593, ([47132602] 147132526 refl JNM 213594, 1(47132526] [47106056 refl JNM 213596, |[47106056] 147106060 ref J|NM .213597, ([47106060] 147106062 ref J|NM.213598, 1(47106062] J47106047 ref (|NM 213599, j[47106047] (47106064 ref J|NM.213600, 1(47106064] 147106066 ref (|NM 213601 , j[47106066] 147106068 ref jlNM 213602. 1(47106068] [47106070 refl JNM 213603. |[47106070] 147106072 ref JlNM 213604. [[47106072] J47106074 ref J|NM 213605. ([47106074] J47106049 ref J|NM 213606. j[47106049] 147106051 ref (jNM 213607. 1(47106051] 147106053 ref| |NM .213608. ([47106053] J50962798 ref| |NM .213609. [[50962798] 147132594 ref| |NM 213611. 1(47132594] 147132596 ref| |NM .213612. |[47132596] 147132598 refl |NM 213613. |[47132598] 147132531 ref jjNM 213618. [[47132531] [47717099 ref ]jNM 213619. j[47717099] 147717101 refl |NM 213620. ([47717101] (47519840 refl |NM 213621. 1(47519840] J47132533 reflNM 213622. |[47132533] (47578110 refl JNM 213631 , 1(47578110] [47578112 refl JNM 213632, 1(47578112] 147419899 refl JNM 213633, ([47419899] (47157327 ref J]NM.213636, |[47157327] [47419917 refl JNM .213645, ([47419917] 147419919 refl JNM 213646, 1(47419919] J47524174 refl JNM 213647, j[47524174] [47419937 ref J|NM .213648, IJ47419937] J47458810 ref (jNM 213649, 1(47458810] I47458808 ref J|NM..213650, |[47458808] (47458040 ref (|NM.213651 , |[47458040] I47458049 ref J|NM 213652, |[47458049]
|47458047 ref|NM_213653.11[47458047]
[47458042 ref|NM_213654.11[47458042]
(47458035 ref(NM_213655.1 [[47458035]
147458038 ref|NM_213656.11[47458038]
[47717091 ref|NM_213657.11[47717091]
147717093 ref|NM_213658.1 ([47717093]
(47458819 ref JNM_213662.1 ([47458819]
147519615 ref|NM_213674.11[47519615]
[47497975 ref|NM_213720.11[47497975]
(47523379 ref|NM_213723.11[47523379]
[47523373 ref|NM_213724.11[47523373]
J47524166 ref[NM_213725.1|[47524166]
|47523377j ref|NM_213726.11[47523377]
[47551355 ref|NM_214461.1 ([47551355]
(47551353 ref(NM_214462.11[47551353]
[47607494 ref|NM_214675.1 [[47607494]
I47607496 ref|NM_214676.1 ([47607496]
(47607498 ref|NM_214677.1 [[47607498]
I47607500 ref|NM_214678.1|[47607500]
J47607502 ref|NM_214679.11[47607502]
147551346 ref|NM_214710.11[47551346]
J47551348 ref|NM_214711.11[47551348]
114728171 ref|XM_001279.4|[14728171]
115294418 ref |XM_001290.5|[15294418]
114720227 ref |XM_001296.4|[14720227]
114720376 ref|XM_001322.2([14720376]
J14721336 ref|XM_001393.2|[14721336]
118546180 ref JXM_001442.4|[18546180]
114742707 ref|XM_001463.2|[14742707]
J14726320 ref|XM_001527.4|[14726320]
114726884 ref|XM_001541.3|[14726884]
[14734600 refJXM_001607.2|[14734600]
(14725410 ref|XM_001644.4|[14725410]
114729031 ref|XM_001654.4|[14729031]
[14729023 ref|XM_001655.4J[14729023]
I29727539 ref|XM_001677.3|[29727539]
(14735909 ref|XM_001690.4|[14735909]
141204894 ref|XM_007651.13|[41204894]
[37546668 ref|XM_010658.8|[37546668]
I42659898 ref|XM_012219.7|[42659898]
(42656511 ref|XM_015334.6|[42656511 ]
J42655785 ref|XM_015717.7|[42655785]
J42659340 ref|XM_016093.7|[42659340]
I37539642 ref|XM_016113.3|[37539642]
J42657382 ref|XM_016532.6|[42657382]
J37546615 ref|XM_016548.4J[37546615]
I42660439 ref|XM_016713.5|[42660439]
[42656229 ref|XM_017374.8|[42656229]
(37540277 ref|XM_017661.3|[37540277]
142656981 ref[xM_017966.10|[42656981]
[42660185 ref|XMJ318399.7|[42660185]
J42660716 ref|XM_018432.5|[42660716]
[42662446 ref|XM_018487.5|[42662446]
(42660088 ref|XM_027045.111[42660088]
[41196212 ref|XM_027074.6|[41196212]
I42662509 ref |XM_027105.4|[42662509]
I42655789 ref|XM_027162.3|[42655789]
142660312 ref|XM 0272364|[42660312] |41203734 ref||XM 027237 7|[41203734] 118562991 reflXM 027307 ,2 j[18562991] |42660315[ reflXM .027330 ,13|[42660315] |42657548 reflXM 027658 7|[42657548] J42661856 ref|XM .028067 7|[42661856] |42656936 reflXM .028217 3J[42656936] 137552100 ref|XM..028253 3|[37552100] 141146627 ref|XM .028413 7J[41146627] J42660130 reflXM 028522 5|[42660130] 142662215 reflXM 028810 6|[42662215] |29742588 ref|XM .029084 ,5|[29742588] |42657171 ref|XM 029101 8|[42657171] [41194246 ref|XM .029323 4|[41194246] 141203774] ref|XM 029353 3|[41203774] 142661384] reflXM .029429 7|[42661384] [42661064] ref|XM .029438 6|[42661064] 142660024 ref|XM 029805 10|[42660024] 141057799 reflXM 029962 4|[41057799] 141146958 reflXM .030300 6|[41146958] J42661610 reflXM .030378 4|[42661610] I42659359 reflXM .030445 7J[42659359] I42662233 ref|XM .030559 4|[42662233] I42662234] ref|XM .030577 8J[42662234] 141058288] ref|XM .030665 4|[41058288] I42656830] ref|XM .030669 9|[42656830] J41149078] reflXM .030729 5|[41149078] 142661688 reflXM .030892 4|[42661688] 137552114 reflXM 030893 3|[37552114] 127485351 reflXM .030896 3|[27485351] |42661697| refl XM 030958 8|[42661697] |42656397| ref] XM .031009 7|[42656397] |42660307| ref] XM 031102 4|[42660307] I42660309 refl XM .031104 7|[42660309] I42656653 ref|XM.031246 10|[42656653] 114758391 ref| XM 031342 1 |[14758391] 142661350] ref| XM 031357 6|[42661350] 141107703] refl XM .031401 7|[41107703] I37550558 reflXM.031553 8|[37550558] 141150031 refl XM .031561 7|[41150031] |37540832] ref]XM 031689 7|[37540832] 141150034] ref|XM 031706 8|[41150034] 141204883 ref|XM .031744 .5J[41204883] I42659366 reflXM .031975 7|[42659366] 141208847 ref|XM .032059 3J[41208847] 122056189 refjXM .032181 .3|[22056189] 142661621 ref|XM .032278 .10|[42661621] I42655755 ref|XM 032397 4|[42655755] 142656433 ref|XM .032542 ,5|[42656433] |42657389 ref|XM .032571 .5[[42657389] J42661979 ref|XM .032678 7|[42661979] 137541812 ref|XM 032693 .2J[37541812] 142661970 reflXM .032812 4|[42661970] |41144879 reflXM .032901 .7J[41144879] 141209336 ref|XM .032945 5|[41209336] I42662389 ref|XM.032996 5|[42662389] 141151370 refjXM 032997 5|[41151370]
gi|42659913|ref|XM_033113.5|[42659913] gi[3754622θjrefixM_033173.7([37546220] gii37545893irefjXM_033370.9J[37545893] giJ42660237|ref|XM_033371.4J[42660237] gi|42660236[ref|XM_033391.10|[42660236] gi|42655898Jref|XM_033704.5|[42655898] gi|42662548(ref|XM_033853.3[[42662548] gi|4265566l jref|XM_034086.5|[42655661] gi|37540070|ref|XM_034262.7[[37540070] gi|41148717|ref|XM_034274.12|[41148717] gi|27479970JrefixM_034594.7|[27479970] gi|42659159|ref|XM_034623.3|[42659159] gi|42659160|ref|XM_034640.6|[42659160] gi|37546832|ref|XM_034717.9|[37546832] gi|37542014(ref|XM_034819.4|[37542014] gi|41147048|ref|XM_034872.8|[41147048] gi[4266045θjref|XM_034904.7|[42660450] gi|37541632|ref|XM_035037.3|[37541632] gi|42657309|ref|XM_035299.5|[42657309] gi(42657054iref|XM_035371.7[[42657054] gi|41207713Jref|XM_035405.7|[41207713] giJ4265994θ(ref|XM_035497.12|[42659940] gi|42660453(ref|XM_035527.10|[42660453] giJ41146833|ref|XM_035572.9|[41146833] gi|37549734|ref|XM_035601.6([37549734] gi|4265894l [ref|XM_035825.5|[42658941] gi(42659563[ref|XM_035863.7i[42659563] gi|41146641 jref|XM_035946.8|[41146641] gi|41148968|ref|XM_035953.3|[41148968] gi|42661038(ref|XM_036115.5i[42661038] gi|41151126|ref|XM_036218.5|[41151126] gi|37547941|ref|XM_036299.7|[37547941] gi|42655598(reflXM_036612.9i[42655598] gi|42659237[ref|XM_036708.12|[4265923η gi|42662413|ref |XM_036729.2|[42662413] gi|41117169|ref|XM_036740.5|[41117169] gi|41151435|ref|XM_036936.3|[41151435] gi|41209794[ref|XM_036942.3|[41209794] gi|42656790|ref|XM_036988.10|[42656790] gi|42662525[ref|XM_037493.4|[42662525] gi|41149775(ref|XM_037523.9[[41149775] gi|18580213(ref|XM_037557.2|[18580213] gi|37563738|ref|XM_037759.5|[37563738] gi|41146746[ref(xM_037817.10|[41146746] gi|42659052|ref|XM_038063.9|[42659052] gi|37552547|ref|XM_038150.3|[37552547] gi|41107444|ref|XM_038288.9|[41107444] gi|18599776|ref|XM_038291.5|[18599776] gi|41126376|ref|XM_038298.5|[41126376] gi|42660052|ref|XM_038436.111[42660052] giJ42662459(ref[xM_038520.9|[42662459] gi[42656296|ref|XM_038567.8|[42656296] gi|42656295Jref|XM_038576.7|[42656295] gi(42661999|ref|XM_038604.7|[42661999] gi|37545373|ref|XM_038664.7|[37545373] gi[42657420JrefixMJ038920.8i[42657420] gi(l6159362[refixM_038999.2|[16159362]
1426571 12 ref]XM 039169.4|[42657112] 142659816 reflXM 039218. 11[[42659816] J27501258 reflXM 039385. 2|[27501258] 1426581 14] reflXM .039393. 5|[42658114] J2206881 refjXM.039495. 3|[22068817] I42659832, ref|XM 039515. 12|[42659832] J42656339I ref)XM 039548, ,4J[42656339] I42656337, refl|XM 039570. 7[[42656337] [411371 13 refjXM .039627, 91(41137113] [41123798 refl|XM 039676. 5|[41123798] 142659156 reflXM .039698. 9|[42659156] [42660465 refl|XM 039702. 4J[42660465J I42655797 ref||XM .039721, 5|[42655797] I42656267 ref|]XM .039733, 4|[42656267] (42656367 refl[XM 039762, ,5|[42656367] I29728546 reflXM 039796. 9|[29728546] |42659787 reflXM .039877, ,10|[42659787] 141208812 reflXM 039908. ,3|[41208812] |42661137 refl|XM 039922. 4J[42661137] 142656541 reflXM 040149, ,4J[42656541] 137551016 refl]XM 040265. 6|[37551016] |42662664 refl;XM .040383, ,8|[42662664] J37546226 reflIXM 040486. 3|[37546226] |41188087 ref|XM 040527. ,6|[41188087] [41150292] ref)XM 040592. ,5J[41150292] |42660358 ref|XM .040910. 5|[42660358] J42659016 reflXM 041018, 5|[42659016] 141148771 ref|XM 041020, 5J[41148771] J42660327] reflXM .041116. 7|[42660327] |41191478 reflXM 041126, 6|[41191478J 142660201 reflXM .041162. 5J[42660201] 141206442 reflXM 041191. 6|[41206442] 142657103 refjXM .041221. 7[[42657103] [411371 10] refjXM 041363. 111(41137110] 141150611 refjXMJ041964. 9J[41150611] |37550278 refl XM 042066. 8|[37550278] 142656815 reflXM 042178. 7|[42656815] J42660712 refjXM 042234, 5J[42660712] 141146674 reflXM .042301, ,6|[41146674] 141058225] reflXM 042323. ,9|[41058225] (42656140 reflXM 042500, ,5|[42656140] J4114661 ref|XM_l042635. 5|[41146617] J42657049 ref) XM 042661 ,6|[42657049] 137546881 refl]XM 042685, 3|[37546881] J37544667 refjIXM 042698, ,6|[37544667] 137540871 reflXM 042833, ,81(37540871] I42656684 refj|XM 042936, ,5|[42656684] 141210290 reflXM .042978 .31(41210290] I42659964 ref]IXM .043118, 7|[42659964] |42661088 ref|XM .043272 .8|[42661088] |42656527 ref|(XM 043492 5|[42656527] 127479150 ref|XM 043493 .5|(27479150] i|42656525 reflXM 043500 4J[42656525] [27482185 reflXM .043613 8|[27482185] J42656715; ref|!XM .043624, 7|[42656715] [42662596 reflXM 043653 .8|[42662596] (42655948 ref]IXM 043739 5|[42655948]
127481852 Iref JjXM .0438634|[27481852] (42660940 Iref JjXM .043979 5[[42660940] [41 147199 Irefj JXM .043989 8|[41147199] (41 149835 I ref J|XM .044062 8|[41149835] 142656142 |ref J|XM .044166 .7|[42656142] (41 146806 I ref J|XM .044178 9J[41146806] I42659260 |ref J|XM .044196 .7J[42659260] [42657971 |ref J|XM .044212 5|[42657971] (41 149839 (ref J|XM 044334 5|[41149839] [42657086 |ref J|XM 044434 111(42657086] [42657077 |ref :||XM .044461 11 [[42657077] 142660515 |ref J|XM 044580 .6|[42660515] J42658931 |ref| JXM .044622 5J[42658931] |42660753 lref J|XM 044632 5|[42660753] |42658865 [ref J|XM 044727 7|[42658865] [41202066 |ref J|XM 044836 .91(41202066] |27485586 |ref :||XM .044921 4|[27485586] |42658892 |ref| |XM 045086 .5|[42658892] I42655863 [ref J|XM 045113 .5|[42655863] 141200895 [ref J[XM .045271 8|[41200895] 142656310 Iref JjXM 045283 .4J[42656310] |41 146539 |ref J|XM .045290 3|[41146539] 141222695 |ref J|XM 045308 ,7J[41222695] (42662263 |ref J|XM .045421 3|[42662263] J42659902 [ref J|XM 045423 4|[42659902] |37544582 |ref J|XM .045581 7|[37544582] (42662653 Iref JjXM 045705 7|[42662653] |29745945 [ref J|XM 045712 .5J[29745945] (42660102 [ref J|XM 045787, ,10|[42660102] [42660104 |ref||XM 045792, ,10|[42660104] 141199222 |ref J|XM 045907, ,4|[41199222] 141146545] |ref J|XM 045911, ,8|[41146545] J37552368 |ref J|XM .046099, 2|[37552368] (42656722 |ref J|XM 046264, ,8|[42656722] 137551981 |ref J|XM .046305, 3|[37551981] |42661672 [ref J|XM 046390, 7|[42661672] 142662271 |ref J|XM .046437, δ([42662271] J42655873 |ref J|XM 046531, 4|[42655873] I42656444 |ref J|XM .046570, 6|[42656444] [37547050 |ref J|XM 046581, 6|[37547050] I37556088 |ref J|XM 046600, 91(37556088] |41148426 |ref J|XM 046677 ,8|[41148426] I42656570 |ref J|XM 046685 .9J[42656570] |42655962| |ref J|XM .046751 6|[42655962] |42655822] |ref J|XM 046808 12|[42655822] J42659307] [ref J|XM 046861 3|[42659307] J42662549 jref (jXM .047025 7|[42662549] J42658904 |ref J|XM 047083, 7|[42658904] J42662502] |ref J|XM .047214 .7|[42662502] J42662718 [ref J|XM 047325 8|[426627i8] |41144342] |ref J|XM 047355 .7|[41144342] J37550037] |ref J|XM 047357 .4|[37550037] (42660842 [ref J|XM .047462 8|[42660842] |42655922j |ref J|XM .047499 7|[42655922] J42662024 |ref J|XM .047550 5|[42662024] J42662018| |ref J|XM .047554 8|[42662018] J37552328 Iref JjXM 047610 2|[37552328]
gij42661115 reflXM.0476177|[42661115] giJ42656550 refj XM 047707, 6|[42656550] gi|42660976 ref| XM 047734, 10|[42660976] giJ42659920 ref| XM .047770 ,7|[42659920] gi |42657199 ref| IXM..047995, 7|[42657199] gi|41147344 ref]jXM .048070, 3J[41147344] gi|42655726 ref| XM .048104, ,4J[42655726] gi|42660273 ref| XM 048128 ,9|[42660273] gi|37546575 ref] XM 048235, 2p7546575] gi|42662073 ref| XM 048362 ,8[[42662073] gi|42659190 ref| |X 048462 ,6|[42659190] gil42659186 ref] XM .048592, ,4J[42659186] gi|42660003 ref| XM .048675 ,7|[42660003] gi| 16157523 ref| |XM 048721 ,2|[16157523] gij41195095 ref| XM .048747, ,9|[41195095] gi|42656062 ref| XM 048774, ,5J[42656062] gi|22049345 ref| |XM 048786.5|[22049345] gi|42656006 ref| XM 048825, ,10|[42656006] gi|42659387 ref| XM .048898, ,7|[42659387] gi|42660770 ref| XM 049037 ,8|[42660770] gi 141146656 ref| XM 049078, 4J[41146656] gi|41150978 ref| XM 049237, .6[[41150978] gi|42659384 ref| XM 049349, .13|[42659384] gij42659383 refl XM 049351, ,4|[42659383] gi|42660965 refl XM 049380, 3|[42660965] gi|42657717 refl XM 049384 ,8J[42657717] gi|42656765 reflXM.049575, ,11 |[42656765] gi|37550230 refl XM .049619, ,6|[37550230] gi|41188800 ref|]XM 049695.6J[41188800] gi|37546955 ref|XM .0499522J[37546955] giJ41150752 ref]XM 050041 ,91(41150752] gi|41195082 ref|]XM .050219 ,81(41195082] gi|37546014 ref|jXM 050278 ,7|[37546014] gi|42658841 ref]XM 050325.61(42658841] giJ27501317 reflXM .0504782J[27501317] gi|42661994 reflXM 050561, 9|[42661994] gi|42661996 ref| XM 0505643([42661996] gi|42656988 refl XM 050625.4|[42656988] gi|42661998 reflXM 050644, 3|[42661998] gi|42656576 ref| XM .050846.6|[42656576] giJ37549382 ref]XM 0510173|[37549382] gi|42659353 ref|jXM .051081.10|[42659353] gi|37542913 ref|XM 051091.9|[37542913] giJ42660773 ref|XM .051197.8J[42660773] gi|42660774] ref||XM 0512006|[42660774] gi|41191485 ref|IXM .051221.5J[41191485] gi|37550534 ref|XM 051264111[37550534] gi|42658942] ref||XM 051271.7|[42658942] gij42660281 ref]IXM .0516994|[42660281] gi|42660545 ref|XM 051862 ,5|[426605'45] gi|42659501 refjXM 051956 ,10|[42659501] gi|42655635 refl|XM .0525616|[42655635] giJ42656950 ref|XM 052597 ,7|[42656950] gi|42657064 ref|XM 052620 ,8|[42657064] gi|41151001 refl[XM 053074.7J[41151001] giJ42656469 ref|XM 0531777|[42656469] gi|42661986 refjXM 053966.8J[42661986]
gi|42656432|ref|XM_054284.7|[42656432] gi[42657998)ref|XM_054313.5|[42657998] gi|42659250|ref|XM_054983.7J[42659250] gi|42660053Jref|XM_055095.5l[42660053] giJ4110746θjref|XM_055481.7J[41107460] gi|41190421 |ref|XM_055636.3|[41190421] gi|42657832|ref|XM_055725.4|[42657832] gi|37551979|ref |XM_055866.5|[37551979] giJ4266075θjref|XM_056254.6|[42660750] gi|42662468Jref|XM_056282.4|[42662468] gi|41148706Jref|XM_056298.6|[41148706] giJ29735852[ref|XM_056434.2|[29735852] giJ42656369|ref|XM_056455.4|[42656369] gi|37542922|ref|XM_056680.5|[37542922] giJ42659969irefixM_056681.9|[42659969] gi|42656063|ref|XM_057040.4|[42656063] gi|42656009JrefjXM_057107.5|[42656009] gi|37550516|ref|XM_057296.2|[37550516] gi|29739598|ref|XMJD58332.3|[29739598] gi|41149338|ref|XM_058335.8|[41149338] gi|42659386Jref|XM_058404.5|[42659386] gi|41149540|ref|XM_058426.8|[41149540] gi|42659921|ref|XM_058513.8J[42659921] giJ42660085JrefjXM_058581.11|[42660085] gi|42660202|ref|XM_058611.9|[42660202] gi|42660341|ref|XM_058628.7|[42660341] giJ42660291 jref|XM_058661.7J[42660291] gi|42659885|ref|XMJ)58677.5|[42659885] gi|41204989Jref|XM_058719.8|[41204989] gi|41150166[ref|XM_058720.6I[41150166] gi|27483117|ref|XM_058721.6|[27483117] giJ42660724[refIXMJ358743.9|[42660724] gi|42661183|ref|XM_058857.2|[42661183] gi|37543396Jref|XM_058879.7|[37543396] gi|42661425|ref|XM_058931.7|[42661425] gi|42662074|ref|XM_058956.7|[42662074] gi|42662075|ref|XM_058961.6|[42662075] giJ41208833Jref|XM_058964.6|[41208833] gi|42661984|ref|XM_058967.10|[42661984] gi|42661606Jref|XM_058997.7|[42661606] gi|42661724|ref|XM_058999.9|[42661724] gi|41150973|ref|XM_059037.5|[41150973] giJ37552320Jref[XM_059047.4|[37552320] giJ41151025Jref[XM_059051.3|[41151025] giJ42655753JrefIXM_059061.4l(42655753] gi|37549519Jref|XM_059074.2|[37549519] gi|4265559θ(ref|XM_059095.9|[42655590] gi|42655582[ref[XM_059104.5|[42655582] gi|37548619jref|XM_059132.3J[37548619] giJ42655737JrefJXMJ359140.4|[426557'37] gi|42655916|refIXM_059166.81(42655916] gi|37555943|ref|XM_059256.5|[37555943] gi|37556080|ref|XM_059267.5|[37556080] gi(42662438(ref|XM_059318.7i[42662438] gi|42656344Jref|XMJ359341.5J[42656344] gi|42656497|ref|XM_059368.6J[42656497] gi|42656530|ref|XM_059384.8J[42656530]
gi|17437350Jref|XM_059396.11[17437350] gi(42656269JrefixM_059399.4|[42656269] gi|42656194|ref|XM_059438.5|[42656194] gi|41191473|ref|XM_059461 j[41191473] gi|30148215|ref|XM_059462.3|[30148215] gi|18553091|ref|XM_059473.2J[18553091] gi|37549556iref|XM_059482.6J[37549556] gi(42656805|ref|XM_059492.6|[42656805] gi|41194207|ref|XM_059548.3|[41194207] gi|37550095|ref|XM_059578.4|[37550095] gi|37541042|ref |XM_059598.6|[37541042] gi|17462608[ref|XM_059608.1|[17462608] gi|29729032|ref|XM_059617.4|[29729032] gi|41147021 Jref|XM_059669.5|[41147021] gi|42657204|ref|XM_059672.5|[42657204] gi|42657174[ref|XM_059689.4|[42657174] gi|41146992|ref|XM_059702.8|[41146992] giJ27498358[ref|XM_059729.3|[27498358] giJ42657522|ref|XM_059730.3|[42657522] gi|41147314|ref|XM_059776.5|[41147314] gi(42657955iref(xM_059830.5|[42657955] gi|42657920jref|XM_059832.6[[42657920] gijl7466805Jref(xM_059909.1|[17466805] gi|37539506|ref|XM_059923.3|[37539506] giJ42658954[ref|XM_059929.6|[42658954] giJ42659015irefjXM_059954.5|[42659015] gi[42659014(ref|XM_059956.4|[42659014] gi|42659266|ref|XM_059972.8J[42659266] gi|42659306|ref|XM_059987.8|[42659306] gi(42662604|ref|XM_060020.7[[42662604] gi|29743757Iref|XM_060054.3([29743757] gi|41103603|ref|XM_060087.9[[41103603] giJ37546836|ref|XM_060104.4|[37546836] giJ37548317[ref|XM_060171.3|[37548317] gi|37546776|ref|XM_060278.3|[37546776] gi|42655702[ref|XM_060301.2|[42655702] gi|37546800|ref|XM_060303.6|[37546800] gi(37546802|ref|XM_060305.2[[37546802] gi|17437014|ref|XM_060307.1|[17437014] gijl 7437026|refJXM_060309.1 j[17437026] gi|17437031 jref|XM_060310.11[17437031] gi|17437058Jref|XM_060315.1 [[17437058] gi(37546808[ref|XM_060316.3|[37546808] giJ41114875|ref|XM_060318.10|[41114875] gi|17437334|ref|XM_060328.11[17437334] gi|17441007|ref|XM_060417.11[17441007] gi(37549793Jref|XM_060458.4|[37549793] gi|17443792|ref|XM_060509.11[17443792] gi|17444700|ref|XM_060535.11[17444700] gi(37548017|ref|XM_060537.8|[375480l7] gii42655698JrefixM_060563.8J[42655698] gi|42655695|ref|XM_060569.4J[42655695] gi|17445393(ref|XMJ360572.11[17445393] gi|20534826|ref|XM_060580.4|[20534826] giJ41188099|ref|XM_060597.2|[41188099] gi[37547099[ref[XM_060880.9l[37547099] gi|41107481 |ref|XM_060887.3|[41107481]
117451118 reflXM..060943, .11(17451118] I37549775 rer IXM .060945.2|[37549775] J37549781 reflXM .060951, ,3|[37549781] (l8549724 refl XM .060952 ,2|[18549724] 118549722 reflXM .060953, ,2|[18549722] J37549785 refl XM .060955 ,3J[37549785] 117451521 ref| (XM .060956 ,11(17451521] 118549712 refl (XM 060957, ,2|[18549712] I37550467 ref) (XM .060970 ,9|[37550467] |37550988 refl JXM 061055 ,8J[37550988] I37550868 refl XM .061222 ,3J[37550868] 117471846 reflXM 061427, ,11[17471846] (30154419 refl XM .061542, ,6|[30154419] {41201024 reflXM .061562, ,7J[41201024] |29743494 refl XM .061610, ,2|[29743494] I29743488] reflXM 061611.2|[29743488] (17456752 refl XM 061614, .11(17456752] 137541675 reflXM .061619, ,2|[37541675] j17456788 reflXM. 061624.1 ([17456788] (30156200 refl XM .061626, ,3|[30156200] 122061836 refl XM .061627 ,2|[22061836] (37541669 refl XM 061628, .31(37541669] 137541350 ref) XM 061656, ,8|[37541350] (42659879, ref] XM 061666 ,3J[42659879] (30156125 refl XM 061674 ,5J[30156125] (30156121 refl XM 061676, ,2|[30156121] I42659857 reflXM .061677, ,3J[42659857] I42659785 refl XM .061849, 8|[42659785] 141149619] refl XM .061864, 4|[41149619] 141149621 refl XM 061871, ,7|[41149621] 137541464] refl IXM 061880, ,2|[37541464] |37541472 refl XM 061888 ,2|[37541472] (37541444 refj |XM 061890, ,8|[37541444] J37541737 refj |XM 061930 ,6|[37541737] J30156406 refl |XM .062025, ,3|[30156406] {174601021 refl |XM 062162, ,1|[17460102] |17461543 refl XM 062263, ,11(17461543] 137541711 refl |XM .062269, ,8|[37541711] 137541713] refj IXM 062272. ,91(37541713] 117461456 reflXM .062285. ,1|[17461456] 118605326 refj IXM .062286, ,2|[18605326] 141201045] reflXM 062300 ,4|[41201045] |41149734| refl XM 062437 ,8|[41149734] 117474308] reflXM 062467, ,11[17474308] J37542979! refl XM 062468 ,4([37542979] 141201746 refl |XM 062520, ,3|[41201746] 117474608 refl |XM 062553 ,11(17474608] I37542963 refl XM 062594 ,2|[37542963] I37542965] refl XM 062598.3|[37542965] [42659900 refl XM 062645.5|[42659900] 141149870 refl XM..062735 ,7|[41149870] 117475183 refl |XM..062788 ,11[17475183] J41149910 refl XM 062871 ,5|[41149910] 142660124 refl XM .062872 ,4|[42660124] 141202709 reflXMJ .062890 ,3J[41202709] 117475713] ret XM. 062912 ,11(17475713] 141202714 reflXM. 062966 ,3|[41202714]
141149926 |ref|XM 063084.3 |[41149926] J37546112 (ref|XM 063123.4 [[37546112] I37546094 IreflXM 063138.2 ([37546094] j17454506 jref|XM 063202.1 |[17454506] 141149959 (ref|XM 063287.7 |[41149959] |37546044 |ref|XM 063308.31(37546044] (17476687 jref|XM 063310.11(17476687] |37546046 |ref|XM 063315.2 ([37546046] j17476953 |ref|XM 063336.1 |[17476953] J42660384 (ref|XM 063481.3 |[42660384] 137540881 |ref|XM 063630.51(37540881] J41206619 |ref|XM 063871.5 [[41206619] 130158712 jref|XM .063919.2 |[30158712] 141150479 |ref|XM 064003.71(41150479] 141150253 (ref|XM 064062.6 ([41150253] J42660862 [ref|XM 064152.71(42660862] 141222836 (ref|XM 064177.4 |[41222836] 142661206 |ref|XM 064190.31(42661206] I37544075] [ref|XM 064257.5 IJ37544075] 117490807 |ref[XM 064265.11(17490807] J42661091 |ref|XM 064298.6 |[42661091] [42661255] |ref|XM .064333.6 [[42661255] I37545854 |ref|XM 064689.5 IJ37545854] 142661698 |ref|XM .064856.71(42661698] (17482322 |ref|XM 064859.1 |[17482322] J42661730 (ref|XM 064865.61(42661730] [42662030 [ref|XM 064879.61(42662030] I37552580 (ref|XM .064883.9 [[37552580] 117482451 |ref|XM 064884.1 |[17482451] J41208817| |ref|XM .064903.91(41208817] 137552052 |ref|XM 065006.3 |[37552052] J41208717 |ref|XM 065026.31(41208717] (42662045 |ref|XM 065050.31(42662045] 141151146 |ref|XM 065124.6 |[41151146] (17483458 [ref|XM 065153.1 |[17483458] [20546504 |ref|XM 065166.2 |[20546504] (41125738 |ref]XM 065237.61(41125738] [37539788 |ref|XM 065278.2 |[37539788] 117435003 |ref|XM 065316.1 |[17435003] I37547008 |ref|XM 065332.71(37547008] 117435576 (ref|XM .065348.1 |[17435576] 117440039 [ref|XM 065416.11(17440039] 137547218 |ref|XM 065445.2 j[37547218] I37538529 (ref|XM 065555.3 IJ37538529] (41191523 |ref|XM 065722.31(41191523] I37546984 |ref|XM 065743.6 IJ37546984] I37539466 |ref|XM 065750.41(37539466] j17446851 |ref|XM .065828.1 j[17446851] 117447611 |ref|XM_ .065899.1 |[17447611] j17483582 [ref|XM 065998.1 ([17483582] J17483803 |ref|XM 066003.11(17483803] 117484248 |ref|XM 066040.11(17484248] J37555965 jref|XM .066058.31(37555965] I37555988 |ref|XM 066069.2 j[37555988] j17484446 |ref|XM 066102.1 |[17484446] 117484617 |ref|XM 066139.11(17484617] 117484710 |ref|XM 066162.1 [[17484710]
gi| 17484771 [ ref JXM 066176 ([17484771] gi|17484775 IrefjXM 066177 1(17484775] gi|17484794] |ref|XM .066189 |[17484794] gi|37559466 | refJXM .066243 [[37559466] gij 17484973 | reflXM .066339. ([17484973] gi| 17484984 | reflXM .066350, [[17484984] gi| 17484986 |ref|XM 066351, |[17484986] giJ37546343 | reflXM 066443, |[37546343] gi[l7459234 |ref|XM .066452, ([17459234] gi(42662689 | reflXM 066457 1(42662689] gi|17459334, |ref|XM 066469. 1(17459334] gi|17459487J |ref|XM 066484 1(17459487] gi|37546196 |ref|XM .066534 P7546196] gi|22055468 Jref|XM 066585 |[22055468] giJ37546245 |ref|XM 066621 1(37546245] gi|42662668 |ref|XM 066685 |[42662668] gi|41151610 |ref|XM 066690, |[41151610] gi[37546398 |ref|XM .066695 1(37546398] gi|30157519 |ref|XM 066701. 1(30157519] gi| 17486064 |ref|XM .066752 |[17486064] gi|41151542 |ref|XM 066765. j[41151542] gi|37546482 Jref|XM 066859, P7546482] giJ42662610 |ref|XM 066946, 1(42662610] gi|37546653 |ref|XM 067076. 1(37546653] gi]29730274 |ref|XM 067176, |[29730274] gi| 17438568 |ref|XM 067193, [[17438568] giJ37550693, |ref|XM 067228. |[37550693] gi|37550025| |ref|XM 067369, ([37550025] giJ37550121 |ref|XM 067448. 1(37550121] gi|41193310 |ref|XM 067503 [[41193310] gi(42656838 |ref|XM 067585 [[42656838] gi|41146527 |ref|XM..067605 |[41146527] gi|37539782 jrefjXM 067904 1(37539782] gi|17436546 |ref|XM 067994 |[17436546] gi|37540583 |ref|XM 068121 1(37540583] gi|17446452 |ref|XM 068229 |[17446452] gijl7442118 |ref|XM 068430 ([17442118] gi|27481868 |ref|XM 068602, [[27481868] giJ27498253 |ref|XM 068632 1(27498253] giJ37550426 |ref|XM 068681 |[37550426] giJ37550428 |ref|XM 068682 ([37550428] gi| 17449884 |ref|XM 068889 |[17449884] giJ41146962 |ref|XM .068903 1(41146962] gi(42657452 |ref|XM 069035 1(42657452] gi|37549598 |ref|XM 069595 ([37549598] gi|41147579 |ref|XM .069609 ([41147579] giJ37538553 | reflXM 069612 |[37538553] gi|37538544 | reflXM .069616 ([37538544] gi|37538538 | reflXM 069619 [[37538538] giJ37538535 | reflXM .069621 [[37538535] gi|37538533 |ref|XM .069623 |[37538533] giJ37538248 | reflXM 069728 1(37538248] giJ37538751 |ref|XM 069734 1(37538751] gi|37538755 I reflXM .069743 j[37538755] gi| 17449530 | reflXM.069842, |[17449530] gi|37574709 |ref|XM .070233, |[37574709] giJ37555926 | reflXM 070277 1(37555926]
141148450 [ref JjXM 070619. |[41148450] [37540610 [ref j|XM 071013. 1(37540610] I37540345 [ref JjXM 071061. [[37540345] J42659281 [ref JjXM 071093. [[42659281] 137540519 jref JjXM .071096. [[37540519] 137540517 jref JjXM .071097. |[37540517] 137540515 |ref JjXM.071098. 1(37540515] I29733995 [refj JXM 071099. j[29733995] 117470104 jrefl JXM 071150. 1(17470104] J37540553 [ref JjXM 071151. ([37540553] J37539657 jrefl JXM 071173. ([37539657] I37539655 [ref JjXM 071201. |[37539655] J42659590 Jrefl JXM 071712. 0|[42659590] |42660270 Jref I|XM .071793. 1(42660270] |42660738 jref JjXM 071866. |[42660738] [42657526 Jref J|XM 072402. JJ42657526] 142659191 Irefl JXM .072554. IJ42659191] 141207711 Jref| |XM .084000. 1(41207711] J42659354 |ref j|XM 084357. 1(42659354] I42659352 |ref :||XM 084377. 1(42659352] J41149211 |ref| |XM .084445. [[41149211] I42659528 |ref ]|XM 084467. 1(42659528] I42659504 |ref J|XM .084482. 1(42659504] J41200884 jref JjXM 084514. [[41200884] 141149677] |ref J|XM 084529. |[41149677] J42659801 |ref J|XM 084530. ([42659801] I42659799 Iref J|XM .084578. 1(42659799] I42659605] jref JjXM 084672. |[42659605] 142660041 Iref J|XM 084845. 1(42660041] 141149777] |ref J|XM 084852. |[41149777] [42659889 jrefl JXM 084868. 1(42659889] [41149920 |ref J|XM .084990. |[41149920] I37545842 jref J|XM 085028. 1(37545842] I37546090 |ref J|XM 085127. |[37546090] J37545946 |ref J|XM 085138. 1(37545946] I42660338 |ref J|XM 085175. |[42660338] I22054302 |ref J|XM 085200. 1(22054302] I42660459 |ref J|XM .085231. |[42660459] |42660456 [ref J|XM 085234. |[42660456] 141204917 J]ref J|XM 085236. 1(41204917] 137540961 jref JjXM .085261. 1(37540961] J42660424] |ref J|XM .085290. 0|[42660424] I42660436 |ref J|XM .085316. |[42660436] [37541851 jrefl JXM 085347. 1(37541851] I42660790 |ref J|XM 085367. |[42660790] 141206428 |ref| |XM .085375. 1(41206428] I42660792 jrefj |XM 085383. IJ42660792] I42660856 |ref| |XM .085463. 1(42660856] I42660868 iref JjXM 085507. |[42660868] I22070290 jref JjXM .085517. [[22070290] 142661086 Jref jlXM .085578. |[42661086] (41150644 Iref J|XM 085596. |[41150644] 141150647 Iref j|XM 085606. ([41150647] [41222805 jref JjXM 085634. [[41222805] J42661082 jref JjXM 085689. j[42661082] 141150671 lref J|XM 085722. |[41150671] J20558545 lref j|XM 085724. 1(20558545]
gi|42659136|ref|XM_085775.4|[42659136] gi|37545635|ref|XM_085777.2|[37545635] gi|42661682|ref|XM_085824.6J[42661682] gi|18589705|ref|XM_085830.1|[18589705] gi|42661679|ref|XM_085831.8|[42661679] giJ42661680|ref|XM_085833.4|[42661680] gi|42661736|ref|XM_085836.9([42661736] gi|42661651 |ref|XM_085851.5|[42661651] gi|42661961|refJXM_085870.3|[42661961] gi|41208766|ref|XM_085929.9|[41208766] gi|41208760|ref|XM_085932.7j[41208760] gi|18590348[ref|XM_O85967.1|[18590348] gi|42661628|ref|XM_086001.4([42661628] gi|42661583|ref|XM_086046.3|[42661583] gi|42662009|ref|XM_O86095.5|[42662009] gi|42655653|ref|XM_086186.4|[42655653] giJ42655651|ref|XM_086188.9|[42655651] gi|42655854|ref|XM_086257.3|[42655854] gi|18546481|refJXM_086287.1|[18546481] gi|37549242|ref|XM_086308.2|[37549242] giJ42659515|ref|XMJ 86343.6|[42659515] gi|42655649Jref|XM_086344.5i[42655649] giI42655628|ref|XM_O86360.4|[42655628] giJ42655728|ref|XM_086402.7|[42655728] gi|42655860|ref|XM_O86409.7([42655860] gi|42660780|ref|XM_O86494.6|[42660780] gi|41057824|ref|XM_086604.4|[41057824] gi|42655609|ref|XM_O86616.3|[42655609] gi|37552857|ref|XMJ386622.2|[37552857] gi| 18591841 jref |X D86637.11[18591841 ] gi!42662240|ref|XM_O86648.6|[42662240] gi|37555961|ref|XM_086650.5|[37555961] gi|18592730|ref|XM_O86725.1|[18592730] gi|18592785|ref|XM_086732.1|[18592785] gi|29745755|ref|XM_086761.3l[29745755] gi|41209881|ref|XM_086826.5|[41209881] gi|42662414|ref|XMJ386876.5|[42662414] gi|42662479|ref|XM_086879.3|[42662479] gi|37563666Jref|XMJ386894.3|[37563666] gi|42662507|ref|XM_O86905.7|[42662507] gi|41125769|ref|XMJ)86931.6|[41125769] giJ42656424|refjXMJD86937.6i[42656424] gi|42656393Jref|XMJ386996.4|[42656393] gi|29731736|ref|XMJD87056.4|[29731736] gi|42656539Jref|XM_087062.3|[42656539] giJ41123844|ref|XMJ387089.4|[41123844] gi|37546904|ref|XM_O87O97.2|[37546904] gi[4265654θiref[xM_087137.7J[42656540] gi|37549348|ref|XMJ387141.2[[37549348] gi|41119823|ref|XM_087167.5|[41119823] gi(41190366JrefjXM_O87171.3J[41190366] gi|4265657ljref|XM_087182.4|[42656571] gi|18553076|ref|XM_O87200.11[18553076] gi[l8553104(ref[xM_O87208.1 [[18553104] gi(41126909|ref(xM_087225.4J[41126909] gi|42656885|ref|XM_087254.4j[42656885] gi|42656762|ref|XM_087353.7|[42656762]
|4265671Oi |ref J|XM .087384.9 |[42656710] 141146547 Jref J|XM 087386.61(41 146547] J42657084 jref J|XM 087483.4] ([42657084] J30147807 Iref JjXM .087490.4] 1(30147807] 141146790 iref J|XM .087499.7 |[41 146790] [42656987 Jref J|XM .087500.91(42656987] J27477586 jref JjXM .087553.61(27477586] I42657026 jref JjXM .087593.9 j[42657026] [42657273 Iref J|XM 087671.4 ([42657273] 142657194 Jref J|XM .087672.3 j[42657194] (42657242 jref JjXM .087761.3 |[42657242] I29735634 |ref J|XM 087762.2 j[29735634] 129739190 jref JlXM 087800.21(29739190] |42657546 jref J|XM 087804.3 |[42657546] [37552150 jref JlXM 087859.4 ([37552150] (37551363 jref JjXM .087901.2 |[37551363] [42657563 [ref JjXM .087928.31(42657563] 142657761 jref J]XM .088066.6 |[42657761] 118566487J |ref J|XM 088072.1 [[18566487] |37538767| jref JjXM .088118.5 ([37538767] (l8600760 |ref J|XM 088140.11(18600760] (37538513 jref JjXM 088142.5 j[37538513] J37538517 jJref JlXM 088143.31(37538517] [42658839 |ref J|XM 088315.5 j[42658839] |41148443 jref JjXM .088331.6 [[41 148443] |41148702 |ref j|XM 088367.3 |[41 148702] (l8570944 jref JjXM .088376.1 |[18570944] 142659041 jref| |XM 088459.9 j[42659041] J18572392 jrefj |XM .088491.11(18572392] 118572749 Irefj JXM 088516.1 j[18572749] J42659262 jref J]XM 088525.5 |[42659262] [41148953 jref JjXM 088551.8 |[41 148953] [42659291 |ref J|XM .088566.7] |[42659291] I27478473 Iref JjXM 088567.51(27478473] I42659224 [ref JjXM 088578.7 (J[42659224] J42662573 |ref J|XM .088636.5 |[42662573] 137546511 Iref JjXM 088677.2 |[37546511] [18595830 Iref JjXM 088679.11(18595830] J18595832 jrefl JXM .088680.1 [[18595832] J37546267J |ref| |XM 088683.21(37546267] 120554311 jrefj |XM .088684.21(20554311] 142662618 jref JlXM 088686.6 ([42662618] [37546271 jref jjXM .088691.3 |[37546271] |29746315| |ref J|XM 088726.21(29746315] (42662600 jref jjXM 088735.5 ([42662600] 142656420 Iref JjXM 088768.61(42656420] 142655633 jref JjXM 088797.6 [[42655633] 137549515 jref J|XM..088817.5 |[37549515] [42655678 lref J|XM .088951.51](426556.78] J37539862 Iref J|XM 089081.51 [[37539862] [41111279 [ref J|XM 089243.8 [[41 111279] (37548666 |ref J|XM .089281.21(37548666] I37548778 [ref J|XM 089307.4 ([37548778] 137549771 |ref J|XM .089384.71(37549771] I42659374 jref JjXM .089747.4 |[42659374] J37541657 jref J|XM .089858.6 |[37541657] I42659859 jref J|XM 089863.5 [[42659859]
gi|22061786|ref|XM_089866.3|[22061786] gijl8605333[ref(xM_090203.1 [[18605333] giJ3754191θjrefIXM_090294.4|[37541910] gi|27479432Iref|XM_090844.2[[27479432] gi|42660316|ref |XM_090885.4|[42660316] gi|4266044θjref|XM_091156.10|[42660440] gi|42660719|ref|XM_091331.4|[42660719] gii37545693|ref[xM_091809.2([37545693] giJ42661353|ref|XM_091830.7|[42661353] gi|41208797(ref|XM_091886.9|[41208797] gi[27485419(refixM_091914.2i[27485419] gi|42661633[ref|XM_092019.6([42661633] giJ37549566iref[xM_092241.7i[37549566] gi|37549413|ref|XM_092267.2|[37549413] gi[42656556(ref|XM_092342.6|[42656556] gi|37539780|ref[xM_092553.2([37539780] gi|18553506|ref|XM_092681.1|[18553506] gi|4115130θ[refIXM_092778.4I[41151300] gi[4115149θ(reflXM_092995.4I[41151490] gil42662558|ref|XM_093024.5([42662558] giJ37546384|ref|XM_093087.3|[37546384] gi|18555449|ref|XM_093644.1|[18555449] gi|42656817|ref|XM_093813.6|[42656817] giJ42657087JrefiXM_093839.9|[42657087] gi[29728071|ref|XM_093895.6|[29728071] gi|42657051|ref|XM_094066.6|[42657051] giJ41146694 jref (XM_094074.6[[41146694] gi|37550191 |ref|XM_094581.4|[37550191] gi|42657535|ref|XM_094794.3|[42657535] gi|37555895(ref|XM_095568.6|[37555895] gi|41148939[refixM_095746.8|[41148939] gi|37540472|ref|XM_095965.3|[37540472] gi|29731922|ref|XM_095991.6|[29731922] giJ42659319(refjXM_096317.δ([42659319] gi|18575382|ref|XM_096376.11[18575382] gi|18577864|ref|XM_096472.1|[18577864] giI42659835|ref|XM_096516.3|[42659835] giJ18580675|ref|XM_096642.1|[18580675] gi[29738271 |ref(xM_096676.2J[29738271] gi|42660126|ref|XM_096688.5|[42660126] gi|42660353|ref|XM_096733.3|[42660353] gi[l8582778|ref|XM_096734.1|[18582778] gi(22054399iref|XM_096852.2|[22054399] gi|37540868Jref|XM_096864.2|[37540868] gi J18584496(ref|XM_096883.11[18584496] gi[29740242(ref|XM_096885.6|[29740242] gi|37540911 |ref|XM_096919.2|[37540911] giJ37544571 |ref|XM_097065.5|[37544571] gi|37552028|ref|XM_097265.7([37552028] gi[41208738irefixM_097278.5i[41208738] gi[l8590346Jref|XM_097347.1 [[18590346] gi|29741281 [ref|XM_097351.2|[29741281] gi|42655887|ref|XM_09758O.4J[42655887] gi|37548657Iref|XM_097622.6|[37548657] gi|29744642|ref|XM_097725.2|[29744642] gi|18591824|ref|XM_097729.1|[18591824] gi[37556106|refIXM_097736.4([37556106]
gi|37556090 refl JXM 097753.4] |[37556090] gi|27501092 ref JjXM 097792.2] 1(27501092] giJ42662426 ref J|XM 097886.6 j[42662426] giJ41123824 refl JXM .097977.3 [[41123824] gij 18552042 ref JjXM 098008.1 ([18552042] gi|29731277 refl JXM .098030.2 |[29731277] giJ41146532 ref J|XM 098117.4 [[41146532] gi|42656856 refl JXM 098163.2 JJ42656856] gi|20532905 ref j|XM 098164.4 |[20532905] gi|42656985 refl JXM .098238.8 [[42656985] giJ20540354 refl |XM .098317.2 [[20540354] gi|18560129 ref JjXM 098350.1 1(18560129] gi|42657240 refl JXM .098368.5 ([42657240] gi(29736547] ref JjXM .098406.5 J[29736547] gi| 18562551 refl JXM .098450.1 [[18562551] gij 18563960 refl JXM 098512.1 1(18563960] gi|29737065 refl JXM 098625.2 [[29737065] gi|42658958 ref j|XM 098762.1 1|[42658958] gi|27479090 refl JXM .098828.2 P7479090] gi|20537481 refl JXM 098847.4 ([20537481] gi|42659230 ref J|XM .098940.6 ([42659230] gi|42662674 ref J|XM 098980.5 [[42662674] gi|30156011 refl JXM 099034.5 1(30156011] gi|37564886 refl JXM. 104657.3 1(37564886] gi|42657109 refl jXM .106386.8 1(42657109] giJ42659600 refl JXM .113228.3 IJ42659600] gi|20468179 ref JjXM .113596.1 |[20468179] gi|42659472 refl JXM 113625.5] 1(42659472] gi|42659390 ref JjXM 113641.4] j[42659390] giJ37541634 refl JXM 113678.4] 1(37541634] gi|41149670 ref J|XM .113696.3 1(41149670] gi|42660050 refl jXM 113706.6 1(42660050] giJ42660040 refl IXM 113708.5 [[42660040] gi|42659927 ref JjXM 113743.6 |[42659927] giJ42660247 refl JXM .113763.5 1(42660247] gij20543268] refl JXM 113776.1 1(20543268] giJ42660487] refl JXM 113796.4 1(42660487] giJ42660444 refl JXM 113825.3 1(42660444] gi|37541794 refl JXM .113871.4 1(37541794] gi|42661098 refl JXM 113912.3 1(42661098] giJ37543738 ref JjXM 113916.4] 1(37543738] gi|42661118 refl JXM .113947.6 1(42661118] giJ41150884 refl JXM 113967.4 1(41150884] giJ37545685 refl JXM 113971.2 1(37545685] gi|41208895 refl |XM 113978.5 1(41208895] gi|42661868 refl IXM 114000.3 1(42661868] gi|42655712 refl ;|XM 114047.4 ([42655712] gi|42655632 refl XM 114067.8 1(42655632] giJ41107442 refl JXM ,114087.5 |[41107442] gi|42655815| refl JXM 114090.2 1(42655815] gi|37549492 ref JjXM. ,114129.3 ([37549492] giJ20479073 refl JXM 114152.1 1(20479073] gi|37552860 ref JjXM 114156.3 1(37552860] gi|41151221 refl JXM.114158.3 ([41151221] gi(37555950 refl |XM_ 114166.3 [[37555950] giJ42656212 refl JXM 114222.3 |[42656212] gi|42656272 ref JXM 114272.4 1(42656272]
gi|42656519|ref|XM_114297.6|[42656519] giJ42656813JreflXM_114301.5J[42656813] gi|37549667|reffXM_114303.2J[37549667] gi|37550602Jref|XM_114317.6J[37550602] gi[20477602|ref|XM_114355.1 ([20477602] gi|42656954|ref|XM_114415.4|[42656954] gi|42657101|ref|XM_114418.4|[42657101] giJ42657288irefixM_114430.7([42657288] gi|42657274|ref|XM_114432.7[[42657274] gi|41147173(ref|XM_114447.6|[41147173] gi|42657398|ref|XM_114456.6l[42657398] giJ41147232Jref(XM_114481.6|[41147232] gi|42658032|ref[XM_114560.4|[42658032] gi|41148559|refJXM_114611.6J[41148559] gi|42658768|ref|XM_114618.5|[42658768] gi|42658886|ref|XM_114621.4|[42658886] gi[30151559Iref|XM_114685.3J[30151559I gii27486035|ref|XM_114723.2|[27486035] giJ42655596|ref|XM_114735.6|[42655596] gi(27479781|ref|XM_114973.3[[27479781] gi|37541649|ref|XM_114987.3|[37541649] gi|42659827|ref|XM_115009.6|[42659827] gi|29746573|ref|XM_115092.2|[29746573] gi|37541778JreflXM_115100.2J[37541778] giJ37547158Jref JXM_115715.4J[37547158] gi|41190802Jref JXM_115760.4J[41190802] gi|37549449|ref |XM 15769.2|[37549449] gi|3754647ljref|XM_115897.2i(37546471] gi|37546504Jref|XM_115925.5|[37546504] gi|22041538Jref|XM_115974.2p2041538] gi|41194218 jref |XM_116036.5 j[41194218] gi|20543127Jref|XM_116384.1|[20543127] gi|20543398|ref|XM_116396.l[[20543398I gi|42657525|ref|XM_116497.4|[42657525] gi|4265817ljrefjXM_116623.4J[42658171] giJ41149924|ref|XM 16936.3|[41149924] gi|37540886|ref|XM_116970.2p7540886] gi|42660482|ref|XM_116971.6|[42660482I gi J37545338 jref|XM_116980.3J[37545338] gi|22068066|ref|XM 117014.2J[22068066] gi|42660896(ref|XM 117030.2J[42660896] gi|42661267|ref|XM 117044.2|[42661267] gi|20559322|ref|XM 117056.1 [[20559322] gi|22060692Jref|XM 117100.2J[22060692] gi|20485835|ref|XM_117112.1 j[20485835] gi|42661573|ref|XM_117117.5J[42661573] gi|2047132θjref|XM 17152.11[20471320] gi|42655766|ref|XM 17174.5|[42655766] gi|37558992|ref|XM_117213.2|[37558992] gi|42662490Jref|XM_117224.7|[42662490] gi|20534043Jref|XM 117236.1 |[2O534043] gi|37547148|ref|XM_117239.5|[37547148] gi[20535783(ref|XM 17257.1 j[20535783] gi|42656562|ref|XM_117266.3J[42656562] giJ20537324Jref|XM_117268.1 j[20537324] gi|42656764|ref|XM_11 294.6|[42656764] gi|20552934Jref|XM_117408.11[20552934]
142657958 |ref J|XM 117451.41(42657958] (20478094 |ref J|XM 117514.1 [[20478094] 142662535 |ref J|XM 117548.21(42662535] J42659910 |ref J|XM. 165401.5 |[42659910] I42656287] |ref J|XM 165448.4 j[42656287] |37538525 |ref J|XM 165511.4 j[37538525] (42662665 [ref j|XM 165534.61(42662665] [42660679 jref JjXM 165921.4 ([42660679] I42655772 |ref J|XM 165973.61(42655772] |42659343 |ref J|XM 166086.5 ([42659343] I42659338 Iref JjXM 166090.6 [[42659338] J41149312 Iref JjXM 166103.4 ([41149312] [29740938 [refj JXM 166110.3 |[29740938] 142659510 |ref| |XM 166125.31(42659510] J42659553 Irefj |XM 166132.4 |[42659553] (42659449 |ref| |XM 166138.61(42659449] I42659456 |ref J|XM 166140.3 [[42659456] [42659509 |ref J|XM 166160.61(42659509] (42659623 [ref J|XM 166164.6 |[42659623] I42659654 [ref J|XM 166203.51(42659654] J42659659 [ref j|XM .166213.4 |[42659659] (42659661 |ref J|XM 166227.41(42659661] 141149566 jref JjXM 166254.4 |[41149566] |42659739 [ref J|XM 166256.71(42659739] I37545360 [ref :||XM 166270.2 |[37545360] |42657441 |ref J|XM ,166300.41(42657441] 141147283 [ref j|XM 166320.4 |[41147283] I27498545 [ref J|XM 166346.21(27498545] [37552180 jref JjXM ,166372.21(37552180] |42657609 [ref J|XM 166376.31(42657609] 130155180 |ref J|XM 166420.5 j[30155180] 141197117 jref JjXM. ,166432.51(41197117] I42657655 |ref| JXM 166443.4 IJ42657655] I42657634 Jref JlXM 166450.2 ([42657634] I42657666 |ref J|XM 166451.3 [[42657666] 141197125 Irefj JXM 166453.5 |[41197125] 127498571 jref JjXM 166479.21(27498571] I42657854 Iref JjXM .166508.5 |[42657854] I37538326 jref J|XM 166523.2 |[37538326] 142657812 jref JjXM 166527.61(42657812] [42657839 |ref J|XM. .166529.4 |[42657839] 141222005 |ref J|XM 166532.31(41222005] 137538415 jrefj |XM 166571.2 |[37538415] (42657897 |ref J|XM 166573.41(42657897] [42659350 |ref J|XM 166630.71(42659350] (41149360 |ref J|XM 166659.4] |[41149360] J20472483 Jref J|XM 166707.1 |[20472483] 137551324 Jrefl JXM 166720.41(37551324] J20481373 jref JjXM 166747.1 |[20481373] I20558728 |ref J|XM 166757.11(20558728] 141149497 jref JjXM 166767.61(41149497] [42659648 |ref J|XM 166776.3 ([42659648] (20558763 jref j|XM .166777.1 |[20558763] [22063015 jref J|XM .166780.21(22063015] I20558775 jref J|XM .166781.1 |[20558775] I37540789 |ref J|XM 166782.4 |[37540789] (37541091 jref JjXM 166805.4 |[37541091]
141200403 |ref J|XM .166808.3 |[41200403]
[41200412 |ref J|XM ,166813.3 j[41200412]
J37541081 ]ref J|XM ,166818.2 |[37541081]
(42659702 |ref J|XM ,166820.31(42659702]
[41200421 |ref J|XM 166823.3 j[41200421]
(20482069 |ref J|XM 166825.11(20482069]
[30156899 |ref J|XM 166829.21(30156899]
(20482090 |ref J|XM ,166831.1 j[20482090]
[22063727] Jref J|XM ,166834.21(22063727]
(37541095 [ref J|XM ,166835.4 j[37541095]
120482131 |ref ]|XM ,166845.1 ([20482131]
[42659652 Jrefj |XM ,166856.4 |[42659652]
137541062 [ref J[XM. ,166868.3 [[37541062]
(37541064 [ref| |XM 166869.3 |[37541064]
141200383 Jrefj |XM ,166871.31(41200383]
141149513 [ref| |XM ,166872.61(41149513]
137541072 [ref J|XM 166877.61(37541072]
120481991 Jref J|XM ,166878.1 |[20481991]
[20481995 Jrefl JXM 166879.11(20481995]
I20482422 Jrefl JXM ,166898.1 |[20482422]
137541141 jref J]XM 166899.31(37541141]
137541145 |ref J|XM 166900.3 |[37541145]
137541153 jref JjXM ,166903.31(37541153]
137541115 jref JjXM 166910.3 ][37541115]
J20482337 jref JjXM 166912.11(20482337]
137541119 |ref JjXM 166914.3 |[37541119]
137541121 |ref J|XM.166915.31(37541121]
|20482348 Jref J|XM 166916.11(20482348]
I20482350 Jref JlXM 166917.11(20482350]
I20482352 |ref J|XM 166918.11(20482352]
137541125 Jrefl JXM 166923.4 [[37541125]
141200480 jrefl JXM 166926.2 |[41200480]
142660172 |ref J|XM 166966.51(42660172]
I37545377 jref JjXM 166971.31(37545377]
J20558392 Iref JjXM.167001.11(20558392]
142657451 jref JjXM 167044.4 |[42657451]
I27498554 |ref J|XM 167072.31(27498554]
I42657597 Iref JjXM.167147.4 ([42657597]
141197075 |ref J|XM 167149.41(41197075]
I27498465 |ref J|XM 167152.31(27498465]
I37538454 Jref J|XM 167254.3 j[37538454]
130152203 Jref J|XM 167275.41(30152203]
[42659470 Jref J|XM 167709.31(42659470]
137551030 jref (|XM 167711.21(37551030]
142660135 |ref J|XM 167908.31(42660135]
J20561649 |ref J|XM 168030.1 j[20561649]
J37551106 Iref JjXM 168053.41(37551106]
137551107 |ref J|XM 168055.3 ([37551107]
I42657650 |ref J|XM 168060.3 IJ42657650]
|20554839 |ref J|XM 168073.11(20554839]
137551273 jref J|XM ,168101.41(37551273]
141222052 Iref JjXM 168223.3 [[41222052]
J42658069 |ref J|XM ,168302.41(42658069]
I37546494 |ref J|XM 168354.21(37546494]
J37538594 |ref J|XM 168521.31(37538594]
|41147653 |ref J|XM 168578.4 [[41147653]
I42658073 jrefl IXM 168583.5 j[42658073]
142658072 | ref J |XM 168585.41(42658072]
142658078 Jref J|XM 168590.5] 1(42658078]
[27499115 |ref J|XM 169057.2] 1(27499115]
(42659771 jref J|XM 169227.2] 1(42659771]
141200907 Jref J|XM 169258.5] [[41200907]
130158339 |ref j|XM 169434.5] 1(30158339]
141149301 Jref J|XM 170525.3 ([41149301]
142660014 jref JjXM 170536.4] 1(42660014]
I42656842 |ref J|XM 170597.2 [[42656842]
J42659551 jref JjXM 170620.61(42659551]
|41149239 [ref J|XM 170637.41(41149239]
|22062495 |ref J|XM 170658.1 |[22062495]
I42659682 jref J|XM 170659.2 ([42659682]
|42659681 jref j|XM 170667.51(42659681]
141149757] jref J|XM 170708.4 |[41149757]
142660171 |ref jjXM 170736.4 [[42660171]
I42660254 Jref J|XM 170749.7 |[42660254]
|37545993 Jref J|XM 170754.3 |[37545993]
J37540845] ]ref J|XM 170777.21(37540845]
J37544656 | ref J|XM 170840.2 |[37544656]
142661126 Iref JjXM 170842.41(42661126]
141188805] jrefl JXM 170909.3 |[41188805]
137548610 |ref J|XM 170950.21(37548610]
I37563637] jref JjXM 170994.21(37563637]
(37546950 Jref J|XM 171008.2 |[37546950]
I42656404] Jref J|XM 171013.31(42656404]
I37550242] I ref J |XM 171032.4 ([37550242]
141146635] |ref J|XM 171040.4 |[41146635]
I37550029 jref JjXM 171054.21(37550029]
142656713 Jref J|XM 171060.51(42656713]
J42657004 jref JjXM 171068.31(42657004]
141146836 |ref J|XM 171078.5 [[41146836]
|42657083 jref J|XM 171081.4 [[42657083]
J22047411 |ref J|XM 171094.11(22047411]
|42657270 |ref J|XM 171105.7 [[42657270]
137538173 |ref J|XM 171150.2 P7538173]
J42657745 |ref J|XM 171154.21(42657745]
|22062795| |ref J|XM 171156.1 |[22062795]
141222074] |ref J|XM 171158.31(41222074]
I42657855] |ref J|XM 171163.41(42657855]
137538271 I ref J |XM ,171165.211(37538271]
|42658070 [ref J|XM 171171.3 |[42658070]
I27478465 jref J|XM 171207.2] 1(27478465]
(41148948 |ref J|XM 171224.3] ([41148948]
[27499039 |ref J|XM 171410.21(27499039]
J22061830 jref jjXM 171424.1 [[22061830]
J22063112 I refl JXM 171447.11(22063112]
137541762 jref JjXM 171489.41(37541762]
142659881 [ref JjXM 171490.31(42659881]
[22064848 |ref J|XM 171495.11(22064848]
I22065074 [ref J|XM 171503.11(22065074]
(22066626 |ref J|XM 171528.11(22066626]
137541642] |ref j|XM 171536.41(37541642]
I37542967 J]ref JlXM 171578.3 ([37542967]
141201765 |ref J|XM .171581.4| [[412017651
|37543887j |ref J|XM 171590.211(37543887]
J22067232] jref JjXM 171766.11(22067232]
M .171855.31(29736979] M 171892.1 j [22066239] M 171973.4 |[41151186] M. 172230.11(22040879] M 172341.4 |[41146585] M .172389.1 ([22042250] M 172751.3 |[37540561] M 172801.2] j[37546509] M 172852.3 |[41200346] M 172855.2] j[42659613] M 172860.2 |[37541004] M 172861.41](42659663] M 172874.3 |[42659905] M 172889.61(42659966] M 172917.2 J]J42660342] M .172929.5] |[42660437] M 172931.5] |[42660567] M .172968.3 |[37544031] M .172995.2 |[42661963] M 173012.2] 1(30150520] M 173015.11(22043462] M 173036.2 ([27481296] M 173063.4 |[42656216] M 173068.11(22042868] M .173083.5 |[42656773] M 173084.3 ([42656880] M .173087.3 [[42656763] M 173105.2 |[29730062] M 173119.21(27480047] M .173120.1 |[22042061] M 173132.2 ([27485459] M 173135.1 |[22044796] M 173140.11(22047801] M 173160.2 |[29735169] M .173164.11(22054253] M 173166.11(22054273] M .173173.3 |[41147399] M 175125.41(41148813] M 208028.5 [[41149277] M 208035.4 |[42659547] M 208042.31(42659831] M 208043.21(42659848] M 208058.4 [[42659849] M 208060.3 |[42659853] M 208061.3 ([30157573] M .208072.4 [[42659358] M 208080.21(41201710] M .208097.3 ([41149930] M 208108.3 j[37540765] M 208116.2 j[37540831] M 208142.3 |[41150433] M .208145.4 JJ42660979] M..208151.2 |[42661224] M 208185.31(42661356] M 208200.4 |[42661966] M 208203.31(37552350]

M 208204.4 j[42662008]
J41107432] |ref|XM .208213.31(41107432]
J42655966 |ref|XM .208234.3 [[42655966]
I42662290 |ref|XM 208250.3 |[42662290]
137563621 |ref|XM 208261.2 |[37563621]
I42656208 |ref|XM .208270.4 [[42656208]
I42656304 [ref|XM .208281.4 | [42656304]
J42656870 IrefjXM 208300.4 ([42656870]
J37549681 |ref|XM .208312.2 |[37549681]
I42656709 (ref|XM 208313.4 [[42656709]
J41194209 lref|XM .208316.3 |[41194209]
137550719 lref|XM 208317.21(37550719]
142656941 lref|XM 208319.4 |[42656941]
142657031 |ref|XM 208320.41(42657031]
J41147202 |ref|XM .208333.3 |[41147202]
141147323 |ref|XM 208352.3 ([41147323]
I42657638 |ref|XM .208356.4 [[42657638]
I42657744 |ref|XM 208361.41(42657744]
J42658040 |ref|XM. 208373.4 |[42658040]
I42659056 |ref|XM .208403.31(42659056]
141210565 jref|XM 208423.3 ([41210565]
|42662655 |ref|XM 208431.41(42662655]
J42662633 |ref|XM 208438.41(42662633]
I42662595 |ref|XM 208443.41(42662595]
I42659467 |ref|XM 208502.21(42659467]
141200370 |ref|XM 208522.31(41200370]
|42659672 |ref|XM .208524.21(42659672]
I42659602 (reflXM 208529.5 |[42659602]
130156512 |ref|XM .208541.21(30156512]
130156506 [ref|XM 208543.21(30156506]
142659612 IreflXM 208545.4 |[42659612]
141149570 (ref|XM 208554.3 ([41149570]
141149538 [reflXM .208563.21(41149538]
I27499430 |ref|XM 208604.1 [[27499430]
J41149787 |ref|XM .208612.3 ([41149787]
I42659996 |ref|XM 208613.21(42659996]
J42659906 |ref|XM 208635.4 [[42659906]
142659961 |ref|XM 208647.41(42659961]
141149701 |ref|XM 208658.41(41149701]
141149812 |ref|XM 208667.31(41149812]
|37540307] |ref|XM .208690.2 |[37540307]
J41203806 jreflXM 208731.31(41203806]
I42660328 IreflXM .208746.4 ([42660328]
I42660375 IrefJXM 208766.41(42660375]
I42660493 |ref|XM 208778.41(42660493]
I42660399 |ref|XM 208798.5 ([42660399]
I42660523 |ref|XM 208809.31(42660523]
I42660664 |ref|XM 208835.4 [[42660664]
130155904 (reflXM 208847.21(30155904]
141206141 IreflXM 208859.31(41206141]
142660710 |ref|XM .208863.5] |[426607'10]
142661008 [ref|XM .208887.4 |[42661008]
J42660944 |ref|XM 208889.51(42660944]
J42660926 |ref|XM 208891.6 j[42660926]
[42660899 |ref|XM 208908.5 |[42660899]
J42661001 |ref|XM .208927.51(42661001]
I29747594 IreflXM 208930.21(29747594]
142661037 IreflXM 208944.6 ([42661037]
gi|42661164 reflXM 208990.3 |[42661164] giJ42661169 ref|XM .208993.4] 1(42661169] gi|42661246 ref|XM 209012.5 ([42661246] gi(27500473 ref|XM .209041.1 ([27500473] giJ42661062 reflXM 209073.5 |[42661062] giJ37545066 reflXM .209076.3 j[37545066] giJ41150895 reflXM 209083.3 ([41150895] gi|37545072 reflXM 209097.2 1(37545072] gi|27485378 reflXM .209104.1 |[27485378] giJ41208859 reflXM 209111.2| ([41208859] gi|42661649 reflXM .209138.2! 1(42661649] gi|41151009 reflXM 209140.4 [[41151009] gi|27485318 reflXM .209145.1 1(27485318] gi|27485201 reflXM .209149.1 1(27485201] gi|42661686 reflXM .209155.3 j[42661686] giJ42661706 reflXM .209163.2 |[42661706] gi]42661980 reflXM 209178.4 |[42661980] giJ42657464 reflXM .209180.4 ([42657464] gi|41208827 reflXM 209187.3 |[41208827] gi|30155334 reflXM .209196.2 [[30155334] gi|41151123 reflXM .209204.3 ([41151123] gi(37549578 reflXM .209227.4 |[37549578] gi|37539870 reflXM.209234.3 1(37539870] giI42655768 reflXM 209252.3 ([42655768] gi|42656013 reflXM 209337.4 1(42656013] gi|41151256 reflXM 209363.3 1(41151256] gi|42662401 reflXM .209384.4 1(42662401] gi[42662403 reflXM 209394.3 ([42662403] gi|27501210 reflXM .209408.1 j[27501210] gi|41118733 reflXM 209423.3 |[41118733] gi|41125720 reflXM 209429.5 1(41125720] giJ27477986 reflXM 209489.1 [[27477986] gi|42656508 reflXM 209490.5 ([42656508] gi|42656569 reflXM 209500.3 j[42656569] gi(41191862 reflXM 209505.3 1(41191862] gi|42656623 reflXM 209509.3 1(42656623] gi|41145035 reflXM 209550.4 |[41145035] gi|42656646 reflXM 209554.2 ([42656646] gi|41145038 reflXM 209559.3 1(41145038] gi|27477654 reflXM 209563.1 ][27477654] gi|41146601 reflXM 209569.4 [[41146601] gi|37550058 reflXM 209579.2 j[37550058] gi|42656833 reflXM 209597.5 1(42656833] gi|42656984 reflXM 209604.2 ([42656984] gi|42657081 reflXM 209607.6 1(42657081] gi|41146768 reflXM 209612.3 j[41146768] gi|42657040 reflXM 209616.4 [[42657040] gil42657021 reflXM .209640.2 1(42657021] gi|27480622 reflXM .209643.1 |[27480622] gi[42657097 reflXM .209655.5 j[42657097] gi|426571.11 reflXM. 209656.3 [[42657111] g37549920 reflXM 209668.2 ([37549920] giJ41147001 reflXM .209695.3 1(41147001] gi|42657402 reflXM 209700.6 j[42657402] gi|42657418 reflXM.
"209704.5] [[42657418] gi[37550409 reflXM
"209719.3 |[37550409] giJ37550204 ref|XM 209728.2] 1(37550204]
gi|42657483|ref|XM_209741.3|[42657483] gi|27483643|refJXM_209753.1|[27483643] gi(42657836(ref|XM_209824.3|[42657836] giJ37538447|ref|XM_209827.4J[37538447] giJ42658815irefiXM_209870.5i(42658815] gi|42658784Iref|XM_209889.2([42658784] giJ41148491 |refjXM_209893.4J[41148491] gi|41148565Jref|XM_209902.3([41148565] giJ42658778irefjXM_209910.5[[42658778] gi|41148759|ref|XM_209913.3|[41148759] gi|42658920|ref|XM_209918.3|[42658920] gi|42658970|ref|XM_209920.5|[42658970] gi|41148719(ref|XM_209936.2J[41148719] giJ37540569Jref|XM_209941.3l[37540569] gi(37539565iref|XM_209955.2|[37539565] gi|41148969|ref|XM_209967.3J[41148969] gi|41149985|refjXM_210022.4[[41149985] gi(42662550|ref|XM_210035.3|[42662550] giJ41151686JreflXM_210042.3|[41151686] gi|42662601|ref|XM_210048.3|[42662601] gi J41210269|ref |XM_210054.3|[41210269] gi|42662684|ref|XM_210062.4J[42662684] giJ37546386|ref[XM_210082.2|[37546386] giJ37546308(refjXM_210094.2J[37546308] gi|27479851|ref|XM_210119.1|[27479851] gi|37541786|ref|XM_210168.3|[37541786] gi|37541788|ref |XM_210169.2|[37541788] gij3754168l[ref|XM_210180.2|[37541681] gi|37541683|ref|XM_210181.4|[37541683] gi|41201047|refJXM_210184.4J[41201047] gi|2749914θ[ref |XM_210186.1 [[27499140] gi|42659706|ref|XM_210193.3|[42659706] gi|37541113[ref|XM_210196.3|[37541113] gi|27484970|ref|XM_210242.lj[27484970] giJ41203869[ref|XM_210282.4I[41203869] gi|37543400|ref|XM_210324.2([37543400] gi|27482991 |ref|XM_210334.11[27482991] gi|27485362|ref|XM_210365.1|[27485362] gi|27485409|ref|XM_210382.1|[27485409] giJ27479896|ref|XM_210394.1|[27479896] gi|37547220|ref|XM_210400.2|[37547220] gi|37547422|ref|XM_210411.3|[37547422] giJ37546496|ref|XM_210501.3|[37546496] gi|37546632|refjXM_210515.2|[37546632] gi|3755026θ(ref|XM_210543.4J[37550260] gi|27477792|ref|XM_210557.1 |[27477792] gi|27477860|ref|XM_210559.1|[27477860] giJ41146535(ref|XM_210562.4|[41146535] gi|41193269|ref|XM_210576.4i(41193269] gi|37540418Jref|XM_210581.2J[375404
'l8] gi|27498077|ref|XM_210613.1|[27498077] gi|27479022|ref|XM_210642.1 |[27479022] gi|42659285[ref|XM_210752.3J[42659285] gi|42659287|ref|XM_210755.4|[42659287] gi|42661990|ref|XM_210787.3j[42661990] gi|41151484|ref|XM_210826.4|[41151484] gi|37550959|ref|XM_210856.2|[37550959]
gi|42659402|ref|XM_210860.3|[42659402] gi|42659520|ref|XM_210876.2|[42659520] gi J27499154|ref|XM_210906.11[27499154] gi|42659616Jref|XM_210908.4|[42659616] gi|42659937|ref|XM_211009.5[[42659937] gi(42659950|ref|XM_211028.2|[42659950] gi|27499637|ref|XM_211040.1 [[27499637] gi|27482549|ref|XM_211079.11[27482549] giJ42660278|ref|XM_211086.3 j[42660278] gi|37546007|ref|XM_211088.2|[37546007] gi|27479552|ref|XM_211089.1 [[27479552] gi|42660333|ref|XM_211090.2|[42660333] gi(27479546Iref|XM_211092.1 j[27479546] gi|27479548|ref|XM_211108.11[27479548] gi|27483466|ref|XM_211174.11[27483466] gi|27484664(ref|XM_211197.11[27484664] gi|27500051 |ref|XM_211244.1 [[27500051] gi|42660938JreflXM_211246.4|[42660938] gi|42660810|ref|XM_211251.3|[42660810] gi|42661052[ref|XM_211287.4|[42661052] gii2750029θ(ref|XM_211291.1 [[27500290] gi|42661215|ref |XM_211305.5|[42661215] gi|42661054|ref|XM_211339.2|[42661054] gi|42661148|ref|XM_211345.2|[42661148] gi|42661278Jref|XM_211367.2|[42661278] giJ42661391 jref|XM_211403.5|[42661391] gi|42661386|ref|XM_211408.3|[42661386] gi|27485087|ref|XM_211413.11[27485087] gij27485049|ref|XM_211422.1 j[27485049] giJ27485209|ref|XM_211432.11[27485209] gi|42661608Jref|XM_211447.3|[42661608] giJ42661993[ref|XM_211460.4|[42661993] gi(42655855|ref|XM_211509.2|[42655855] gi|30148114|reflXM_211518.2|[30148114] giJ42655831 |reflXM_211529.4([42655831] gi|27478973|ref|XM_211557.11[27478973] giJ42655875Jref|XM_211573.2J[42655875] gi|42662238|ref|XM_211627.3|[42662238] gi|27486285|ref|XM_211694.11[27486285] gi|4265621 θ[ref|XM_211707.2J[42656210] gi|37547147Jref|XM_211736.4J[37547147] gi|41190383|reflXM_211749.3|[41190383] gi|29732583Jref|XM_211764.2([29732583] giJ27477614|ref|XM_211768.11[27477614] gi|42656789|ref|XM_211805.2J[42656789] gi|37550613|ref[xM_211816.3|[37550613] gi|27477609|ref|XM_211837.1 ([27477609] gi|42656708[ref|XM_211843.2J[42656708] gi|27478065|ref|XM_211853.1 J[27478065] gi|37550635|ref|XM_211858.4|[37550δ
'35] gi|27481474|ref|XM_211871.11[27481474] gi|27481242Jref|XM_211896.11[27481242] gi|37540652Jref|XM_211908.3J[37540652] gi|27480565|refJXM_211923.11[27480565] gi|27479013|ref|XM_211983.1 [[274790131 gi|4265739θ(ref|XM_211988.2|[42657390] giJ27479310|ref|XM_211995.1 [[27479310]
gi|27483664|ref|XM_212013.11[27483664] gi[42657530|reflXM_212022.2J[42657530] giJ42657721 (reflXM_212061.4J[42657721] gi|27481114|ref|XM_212067.11[27481114] giJ27478393JrefjXM_212094.11[27478393] gi|2747924θjref|XM_212123.11[27479240] gi|37552419|ref|XM_212162.2|[37552419] gi|2973429θ[ref|XM_212170.2|[29734290] giJ41148791 [ref|XM_212238.3([41148791] giJ41148794Iref|XM_212241.3J[41148794] gi|27486196Jref|XM_212319.1 [[27486196] gi|27501414|ref|XM_212326.11[27501414] gi|42657602|ref|XM_212581.3|[42657602] gi|42659396|ref|XM_290185.2|[42659396] gii4266063l [ref|XM_290225.4i[42660631] gi|42662394Jref|XM_290331.4J[42662394] gii37547242Jref|XM_290342.3|[37547242] gil42656309(ref|XM_290345.4|[42656309] giJ42656776Jref|XM_290351.4|[42656776] gi|42657679(ref|XM_290385.3|[42657679] gi[42657734Jref|XM_290389.3J[42657734] gi|42658025Jref|XM_290401.4|[42658025] gi|42659329[ref|XM_290462.4J[42659329] gi[42659349ireflXM_290463.3|[42659349] gi|42659474(ref|XM_290482.3|[42659474] gi[42659618JreflXM_290501.2|[42659618] gi[42659642(ref|XM_290502.2|[42659642] gi(4265968θirefjXM_290506.3J[42659680] gi|42659684Jref|XM_290509.4|[42659684] gi|42659671 |ref|XM_290516.5J[42659671] gi|41200343|ref|XM_290517.3|[41200343] giJ42659747|ref|XM_290527.3|[42659747] giJ30157437[ref|XM_290536.2|[30157437] gi|41149629|ref|XM_290546.2|[41149629] gi|29746157|ref(xM_290547.1 ([29746157] gij41149666|ref|XM_290552.3|[41149666] gi|29741968(ref|XM_290558.1 [[29741968] gi|42659977|ref|XM_290559.4|[42659977] gi|42660067JreflXM_290579.3J[42660067] gi[29744337|ref|XM_290592.1 |[29744337] gi|42659924|reflXM_290597.4|[42659924] giJ42659939|ref|XM_290598.2|[42659939] gi|37545374|ref|XM_290600.2|[37545374] gi|29736037|ref|XM_290609.1 j[29736037] gi|42660283|ref|XM_290615.3|[42660283] gi|42660362iref|XM_290626.2J[42660362] gii42660377irefixM_290629.4J[42660377] gi|37540877|ref|XM_290631.2J[37540877] gi(42660522iref|XM_290645.3|[42660522] giJ42660668[ref|XM_290660.5|[42660668] gi|4266070θ(ref|XM_290667.3|[42660700] gii42660956|ref|XM_290670.4J[42660956] gi|42660717|ref|XM_290671.5|[42660717] gi(42660789|ref|XM_290684.2|[42660789] gi|37541953Jref|XM_290702.2|[37541953I gi(42660905irefixM_290704.3J[42660905] giJ42660966(ref|XM_290712.3|[42660966]
gi|42656825|ref|XM_290714.4|[42656825] giJ42661002Jref|XM_290722.3J[42661002] gi|37542517|ref|XM_290732.3J[37542517] gi(42661055[ref|XM_290734.4|[42661055] giJ41150605Jref|XM_290737.3|[41150605] gil42661294Jref|XM_290743.4[[42661294] gi|42661172|ref|XM_290755.3|[42661172] gi|42661170|ref|XM_290758.4([42661170] gi|42661271|ref|XM_290768.4([42661271] gi|42661282|refjXM_290777.2|[42661282] giJ37543367|ref|XM_290780.2([37543367] gi|42661125|ref|XM_290781.2J[42661125] gi|29739479|ref|XM_290782.1|[29739479] gi|41150812|ref|XM_290786.3[[41150812] gi[42661239irefjXM_290793.4([42661239] gi|42661247|ref|XM_290795.4[[42661247] gi|41150707|ref|XM_290799.4|[41150707] gi|42661375|ref|XM_290809.3|[42661375] gi|37545637|ref|XM_290811.2|[37545637] gi|42661351|refjXM_290817.2i[42661351] gi|37545112|ref|XM_290818.2|[37545112] gi|42661625|ref|XM_290820.3|[42661625] gi|41208783|ref|XM_290822.3([41208783] gi|42661626|ref|XM_290829.3[[42661626] gi|29741265Jref|XM_290831.11[29741265] gi|41150974|ref|XM_290835.2|[41150974] giJ42661597|ref|XM_290838.4|[42661597] gi|37551907|ref |XM_290842.2|[37551907] gi|42661654|ref|XM_290848.2[[42661654] gi|37552312|ref|XM_290850.3|[37552312] gi(42661878|ref[xM_290854.3|[42661878] gi|41151134Jref|XM_290865.3|[41151134] gi|29742605|ref|XM_290866.1j[29742605] gi|42661964|ref|XM_290867.2J[42661964] giJ42655588JrefjXM_290872.4i[42655588] gi|42655844|ref|XM_290902.3([42655844] giJ4111010ljref]XM_290922.2J[41110101] gi|37548529|ref|XM_290923.2|[37548529] gi|42658307|ref|XM_290925.4|[42658307] giJ42656768Jref|XM_290927.4|[42656768] gi|42655993|ref|XM_290936.2[[42655993] gi|42655747|ref|XM_290941.3|[42655747] gi|37549539|ref|XM_290944.2|[37549539] gi(42656017[refixM_290948.2i[42656017] gi|41114572Jref|XM_290949.3|[41114572] giJ3756375ljref|XM_290972.2|[37563751] gi|42662458|ref|XM_290973.3J[42662458] gi|42656625|ref|XM_290985.4|[42656625] giJ2972934θiref[xM_290994.l([29729340] giJ42656458|ref|XM_291001.4|[42656458] gi|42656456iref[xM_291005.4i[42656456] gi[42656476iref|XM_291007.3[[42656476] gii42656506JrefiXM_291015.3J[42656506] gi|37546921 |ref|XM_291016.2|[37546921] gi|29731718|ref|XM_291017.1|[29731718] gi|42656317|ref|XM_291019.4|[42656317] gi|29732251 |ref|XM_291020.11[29732251]
gi|42656908|ref|XM_291028.3|[42656908] gi(42656642Jref[XM_291054.2|[42656642] giJ42656807Jref|XM_291055.2J[42656807] giJ42656711 jref|XM_291057.41(42656711] gi|41144315Jref|XM_291059.3|[41144315] gi[42656798|ref|XM_291062.4|[42656798] giJ42656721|ref|XM_291063.4|[42656721] gi|41193248|ref|XM_291064.3|[41193248] gi|29731560|ref|XM_291074.1 j[29731560] gi|29731972Jref|XM_291075.1 ([29731972] gi|375507111ref|XM_291077.2|[37550711] giJ42656887lref|XM_291080.2|[42656887] gi|29732565(ref|XM_291085.1 |[29732565] gi|42656835|refjXM_291088.5|[42656835] gi|42657009|ref|XM_291095.4|[42657009] gi|41146802|ref|XM_291099.3|[41146802] gi|29729561 (ref|XM_291105.1 ([29729561] giJ29729553|ref|XM_291106.11[29729553] giJ42657113Jref|XM_291111.4|[42657113] giJ4265716θjref JXM_291117.2J[42657160] gi|42657157|ref|XM_291120.5|[42657157] gi|41146989Jref|XM_291128.3J[41146989] giJ42657143|ref|XM_291137.3|[42657143] gi|41146996|ref|XM_291139.3|[41146996] giJ37550318|reflXM_291141.3J[37550318] gi|42657379|ref|XM_291142.4|[42657379] gi|42657428|ref|XM_291144.5|[42657428] gi|42657488|ref|XM_291154.4|[42657488] giJ42657514|ref jXM_291159.4|[42657514] giJ29740697JrefjXM_291161.1 j[29740697] gi|29742395|ref|XM_291169.11[29742395] gi|29742336|ref|XM_291170.11[29742336] gi|42657654|ref|XM_291181.2|[42657654] gi|42657677|ref|XM_291200.4|[42657677] giJ42657680JrefjXM_291202.2[[42657680] gi|37538155|ref(XM_291204.2|[37538155] giJ29733770|ref(XM_291208.11(29733770] gi|41222000|ref(XM_291222.4|[41222000] gij41222117|ref|XM_291223.3|[41222117] gi|42658087|ref|XM_291241.4|[42658087] gi|42658036|ref|XM_291247.3|[42658036] giJ42658944|ref|XM_291253.5|[42658944] gi(29734346|ref[XM_291261.1 ([29734346] gi|42658868Jref|XM_291262.3|[42658868] giJ37552425JrefjXM_291266.3J[37552425] giJ41148477|ref|XM_291268.4|(41148477] gi|37552427JrefjXM_291269.2i[37552427] gi|42658867|ref|XM_291270.2|[42658867] gi|37552665|ref|XM_291277.2|[37552665] gi|42658799JrefJXM_291282.4|[42658799] gi[42658988(ref[xM_291291.3i[42658988] gi|42659205|ref|XM_291314.3|[42659205] gi|42659157|ref|XM_291315.4|[42659157] gi|41151552(refjXM_291321.2|[41151552] giJ37546309|ref|XM_291322.2|[37546309] gi[42662632iref|XM_291326.2i[42662632] gi|29744085Jref|XM_291334.1 ([29744085]
gi|42662590|ref|XM_291335.2|[42662590] gi|42662575|ref|XM_291339.3|[42662575] gi|41151657|ref|XM_291344.31(41151657] gi|37546165|ref|XM_291345.2|[37546165] gii42662657Jref|XM_291346.4|[42662657] gi|42655594JrefjXM_291378.3|[42655594] gi|41106691 Jref|XM_291387.2|[41106691] gi[37539873Jref|XM_291392.2|[37539873] gi|37539876|ref|XM_291394.2([37539876] giJ41058243|ref|XM_291395.2|[41058243] gi|41058244|ref|XM_291396.3J[41058244] gi|41187873Jref|X _291400.3|[41187873] gi|3753986θ(ref|XM_291419.2|[37539860] gi(29728788[reflXM_291428.1 j[29728788] gi[37546790Jref|XM_291435.2|[37546790] gi[29728998Jref|XM_291436.1 |[29728998] gi|29729000Jref|XM_291437.1 |[29729000] gi|29729002Jref|XM_291438.11[29729002] gi|29729004|ref|XM_291439.11[29729004] gi|37546812Jref|XM_291441.2|[37546812] giJ37540095ireflXM_291464.2|[37540095] gi|41057797|ref|XM_291485.3|[41057797] gi|42655878(ref|XM_291508.5|[42655878] gi|3754855l [ref|XM_291533.2|[37548551] gi|37548627|ref|XM_291543.2|[37548627] giJ37548628[reflXM_291544.2|[37548628] gi|37549777|refJXM_291548.2j[37549777] gi|42656136|ref|XM_291569.3|[42656136] gi|37548786|ref|XM_291577.2|[37548786] gi|41188544|ref|XM_291584.2|[41188544] gi|41221595|ref|XM_291607.4|[41221595] gi|29733317|ref|XM_291623.1 |[29733317] gi|41114575|ref|XM_291625.3|[41114575] gi|37549625|ref|XM_291627.2|[37549625] gi|41114486|ref|XM_291638.3|[41114486] gi|30148316|ref|XM_291643.2|[30148316] gi(42656029|ref|XM_291645.3|[42656029] gi|37550870|ref|XM_291663.2|[37550870] gi|42659375|ref|XM_291671.4|[42659375] gi|41149112|ref|XM_291697.2|[41149112] gi|3755086l jreflXM_291698.2|[37550861] gi|29739316|ref|XM_291704.11[29739316] gi|41149235|ref|XM_291716.4|[41149235] gi|41149253|ref|XM_291723.3|[41149253] gi|41149259|ref|XM_291725.3|[41149259] gi|37551034|ref|XM_291726.2|[37551034] gi|42659457|ref|XM_291729.4|[42659457] gi|42659534|ref|XM_291741.4|[42659534] gi[29740609|ref|XM_291745.11[29740609] gi|37551288|ref|XM_291757.2|[37551288] gi|41149391 |ref|XM_291763.3|[41149391] gi|37551328|ref|XM_291767.2|[37551328] gi|37551336[ref|XM_291770.3|[37551336] gi|29740945|ref|XM_291771.11[29740945] gi|37541322|ref|XM_291786.2|[37541322] gi|37541647|ref|XM_291793.3|[37541647] gi|37541689Jref|XM_291806.21(37541689]
gij42659820 ref|XM 291816.41(42659820] gi|37540787 ref|XM 291838.3 |[37540787] giJ42659650 ref|XM .291857.4 1(42659650] giJ41200399 reflXM 291859.3 [[41200399] gi|42659704 reflXM 291862.3 1(42659704] giJ37541135 reflXM 291885.2 1(37541135] giJ37541311 reflXM .291892.2] 1(37541311] giJ37550438 reflXM 291924.2] ([37550438] gi|41149641 reflXM 291943.4 ■]1(41149641] gi|41149625 reflXM .291947.4 1(41149625] gi|37541727 reflXM 291974.2 1(37541727] gi|37541709 reflXM 291977.2 ([37541709] gi|37541715 reflXM 291980.3 [[37541715] gi|29746586 reflXM 291981.1 j[29746586] gi|37541721 reflXM .291986.3 [[37541721] gi|37541926 reflXM 291989.2 [[37541926] gi|37541876 reflXM .291991.2 [[37541876] giJ42659970 reflXM 292012.5 1(42659970] gi|42660035 reflXM .292021.3 [[42660035] gi]29742308 reflXM 292023.1 |[29742308] gi]37543804] reflXM .292027.2 [[37543804] gi|37543771 reflXM 292029.3 |[37543771] gi|41149783 reflXM .292035.3 1(41149783] gi|29743247] reflXM 292046.1 ([29743247] gi(37542898 reflXM .292048.3 [[37542898] gi|37542975| reflXM .292049.3 1(37542975] gi[37542977 reflXM 292051.3 1(37542977] gi|37543902 reflXM .292064.2! 1(37543902] gi|29743830 reflXM 292085.1 1(29743830] gi|42660026 reflXM 292093.4] |[42660026] gi|29744101 reflXM .292098.1 1(29744101] giJ29744569 reflXM 292109.1 1(29744569] giJ42662724 reflXM 292122.4] 1(42662724] gi|37542949 reflXM 292136.2 1(37542949] gi|42660182 reflXM 292160.5 ([42660182] gi|41202674 reflXM 292184.3 |[41202674] gi|42660161 reflXM 292193.4 [[42660161] gi|41202627 reflXM 292197.4 |[41202627] gi|29738932 reflXM 292210.1 1(29738932] gil37546050 reflXM 292225.2 1(37546050] gi|37546054 reflXM 292227.2 (1[37546054] gi|42660292] reflXM 292260.3 ([42660292] gi|37546100| reflXM 292301.21 1(37546100] gi|41204964 reflXM 292357.3 j[41204964] gi|37540971 reflXM .292384.2 1(37540971] gi|37540983 reflXM 292389.3 [[37540983] giJ41150007 reflXM .292394.4 1(41150007] gi|37541427 reflXM 292468.3 |[37541427] gi|29747214 reflXM 292503.1 1(29747214] gi|42660749 reflXM .292504.4 1(42660749] gi[29747247 reflXM .292512.1 1(29747247] gi|37541859 reflXM .292527.2 (][37541859] gi|37542210 reflXM .292562.3 j[37542210] gil37542034 reflXM..292573.3 P7542034] gi|29737769 reflXM .292596.1 j[29737769] gi|42661197 reflXM .292624.4 1(42661197] giJ42661204 reflXMJ 292627.4 ([42661204]
gi|37544488|ref|XM_292664.2|[37544488] gi|37543752[ref|XM_292674.2|[37543752] gi|37545084(ref|XM_292678.2|[37545084] gi|29743632|ref|XM_292700.l([29743632] gi|37545877|ref|XM_292707.2|[37545877] gi[42661354Jref|XM_292717.5|[42661354] gi|37552026|ref|XM_292723.3|[37552026] gi|37552123[ref|XM_292724.3|[37552123] gi(41208938(ref|XM_292729.3|[41208938] gi(37551888JrefixM_292740.2J[37551888] gi|41150997|ref|XM_292745.3|[41150997] gi|41208721 jref|XM_292765.3([41208721] gi|37551986|ref|XM_292778.2|[37551986] giJ42661676JrefixM_292779.4[[42661676] gi|37551992Jref |XM_292784.2p7551992] gi(37551872|ref|XM_292785.3[[37551872] giJ30155117|ref(xM_292796.2J[30155117] gi|37552337|ref|XM_292803.2|[37552337] gi|41151164Jref|XM_292810.3|[41151164] giJ37552519|reflXM_292813.2|[37552519] gi|41151158Jref|XM_292817.3l(41151158] gi(2974291θ[ref|XM_292819.1|[29742910] gi|41151098|ref |XM_292820.3J[41151098] gi|4115116θ(ref |XM_292824.4|[41151160] gi|42662049|ref|XM_292832.5J[42662049] gi|42662051|ref|XM_292836.2[[42662051] gi|37539464(ref|XM_292850.2|[37539464] gi[42656223(ref|XM_292873.4|[42656223] gi|30147787|ref|XM_292889.2[[30147787] gi|37547182|ref |XM_292895.2J[37547182] gi(41125757lref|XM_292943.3([41125757] gi|37546973|ref|XM_292957.2|[37546973] gi|37547028|ref|XM_292958.3J[37547028] gi|41191507|ref|XM_292963.3|[41191507] gi|41191513|ref|XM_292968.3|[41191513] gi|42656472JrefixM_292982.4J[42656472] giJ37549394|ref|XM_293018.2|[37549394] gi|37546996|ref|XM_293026.3|[37546996] giJ42656288JrefixM_293029.3i(42656288] gi|37547014|ref|XM_293034.2|[37547014] gi|29731856|ref|XM_293042.11[29731856] gi|42656254(ref|XM_293090.3|[42656254] gi[42656426|ref|XM_293092.4i[42656426] gi|29744194[ref|XM_293097.1 ([29744194] giJ37555969|ref|XM_293104.2J[37555969] gi|42662246[ref|XM_293106.4|[42662246] gi|37555984|ref|XM_293121.2|[37555984] gi|41151216Jref|XM_293123.3|[41151216] giJ29745634[ref|XM_293157.11[29745634] gi|41151301|ref|XM_293160.4|[41151301] giJ41202683(ref|XM_293177.4|[41202683] gi[4266243θ(ref|XM_293225.4|[42662430] gi|41209907Jref|XM_293226.3|[41209907] gi|29743323|ref|XM_293276.l([29743323] giJ42662546ireflXM_293284.4i[42662546] gi|29743718Jref|XM_293293.11[29743718] gi(30156583|ref|XM_293312.2|[30156583]
gi|37546258|ref|XM_293320.3[[37546258] giJ37546580[refixM_293325.2[[37546580] giJ29744221 jref|XM_293332.11[29744221] gi|42662614Jref|XM_293334.3|[42662614] gi|37546247|ref|XM_293342.2|[37546247] giJ37546382|ref|XM_293352.3|[37546382] giJ42662670|ref|XM_293354.5|[42662670] giJ37546402Jref|XM_293360.3|[37546402] gi(37546435jref|XM_293366.3([37546435] gi|29745490|ref|XM_293380.1|[29745490] gi|29745444|ref|XM_293387.1|[29745444] giJ29746068|ref|XM_293396.1|[29746068] gi|37546273Jref|XM_293398.2|[37546273] gij37546461 )ref|XM 293401.2|[37546461] gi|42662701|ref|XM_293405.3|[42662701] giJ41151604|ref|XM_293407.3J[41151604] gi|29746316Jref|XM_293412.1|[29746316] gi|41151525|refJXM_293416.3J[41151525] gi|37546637|ref|XM_293449.2|[37546637] gi|41151762Jref|X _293460.4J[41151762] gi|2972799l[ref|XM_293514.1|[29727991] gi|37550598|ref|XM_293529.2|[37550598] gi|37550133|ref|XM_293542.3|[37550133]
' giJ42660001 jreflXM_293565.4J[42660001] gi|29730079|ref|XM_293570.lj[29730079] gi|42656852|reflXM_293577.3J[42656852] gi[37550583|ref|XM_293580.2|[37550583] gi|41194243|ref|XM_293581.3|[41194243] gi|3755014l(refjXM_293596.3|[37550141] gi)41146533Jref|XM_293599.4|[41146533] gi|37550617|ref|XM_293633.2|[37550617] gi|29727605|refjXM_293656.1|[29727605] gi|42656942|ref|XM_293669.4|[42656942] giJ37539816|refjXM_293671.3|[37539816] gi|37540053)reflXM_293680.2|[37540053] gi|41146661 |ref|XM_293687.2|[41146661] gi|41146794Jref|XM_293715.4J[41146794] gi|37540623|ref|XM_293745.2|[37540623] gii37540593|ref|XM_293801.3|[37540593] gi|41146900Jref|XM_293802.3|[41146900] gi|29734520|ref|XM_293821.11[29734520] gi|42657409|ref|XM_293828.4|[42657409] gi|29734534[ref|XM_293829.1|[29734534] gi|29735241|ref|XM_293868.1|[29735241] gi|42657341|ref|XM_293875.4|[42657341] gi|41147015[ref|XM_293886.3|[41147015] gi|37549991|ref|XM_293893.2|[37549991] gi|42657201|ref|XM_293903.4|[42657201] gi|41147189|ref|XM_293911.3|(41147189] gi|41147080|ref|XM_293918.3|[41147080] gi|37550304(ref|XM_293923.2|[37550304] gi|42657303|ref|XM_293924.3|[42657303] gi|41147085[ref|XM_293927.3|[41147085] gi|37549904Jref|XM_293937.3|[37549904] gi|37549908Jref|XM_293943.2|[37549908] gi|41147215|ref|XM_293971.3|[41147215] gi|37550397|ref|XM_293976.3|[37550397]
M 293984.31(41196238] M 294017.4 ([41196213] M 294019.21(37551137] M 294070.31(41147377] M 294077.3 [[42657592] M .294093.3 |[42657604] M 294139.4 |[42657665] M .294165.4 j[42657697] M 294209.41(41147843] M .294219.3 j[42657808] M 294247.1 |[29735275] M 294249.31(37538435] M .294261.2 [[37538519] M..294265.31(42657999] M 294310.1 P9735560] M .294311.11(29735568] M 294316.3 [[37538540] M 294318.21(37539408] M .294319.2 ([37538555] M 294328.2 j[37538725] M 294353.11(29737062] M 294354.3 j[41199207] M .294357.4 |[42658105] M 294365.21(37538771] M 294370.21(37538717] M 294383.51(42658860] M 294387.21(37555922] M 294438.3 [[37552398] M 294450.31(41148655] M 294456.2 [[37552464] M 294468.1 |[29735973] M .294473.11(29735831] M 294476.11(29735919] M 294480.21(37556129] M .294521.31(41148775] M 294531.4 ([42659295] M 294533.3 [[41222701] M .294534.1 |[29732655] M 294540.31(42659166] M .294567.31(42659212] M .294568.2! [[37540355] M 294574.2 ([37540311] M .294581.11(29734086] M 294584.21(37540451] M .294590.11(29734032] M 294592.21(41149037] M 294634.4 ([42657840] M .294666.1 [[29740943] M 294675.4 |[42659628] M .294680.1 |[29745032] M .294688.41(41149602] M 294692.1 |[29746497] M .294723.2 JJ42660059] M .294743.1 |[29736049] M. 294750.1 j[29740257] M 294751.3 |[42660491]

M 294765.41(42660642]
|42660623|ref|XM_294775.3|[42660623] (29741962|ref|XM_294778.1 [[29741962] |29747019|ref|XM_294794.1|[29747019] 141114101 (ref|XM_294802.4J[41114101] J29738586iref|XM_294854.1|[29738586] |41150776|ref|XM_294867.4([41150776] |29739848|ref|XM_294894.lj[29739848] |29743450|ref|XM_294906.1|[29743450] |4266168l[ref|XM_294910.2J[42661681] |29741463[ref|XM_294914.li[29741463] 137552318|ref|XM_294919.2|[37552318] 129742513|ref|XM_294922.1 ([29742513] 137549618|ref|XM_294960.2p7549618] |42656059iref(xM_294963.3|[42656059] J41114106|ref|XM_294993.3([41114106] |37547044|ref|XM_294997.2|[37547044] [29744653[ref|XM_295007.1 [[29744653] |29745744|ref|XM_295017.1 ([29745744] ]42656201|ref|XM_295034.2|[42656201] |37547393(ref|XM_295058.2i[37547393] 129730133Iref|XM_295059.1 ([29730133] J29731086ireflXM_295062.1 [[29731086] 142656641 Jref|XM_295091.2([42656641] |42656846Jref|XM_295096.4|[42656846] |42656746|ref|XM_295097.2|[42656746] |41146819|ref|XM_295120.3|[41146819] i29730466iref[xM_295126.li[29730466] |29735426|ref|XM_295146.1|[29735426] 142657169|ref|XM_295155.3J[42657169] |37551141 jref|XM_295166.2|[37551141] |29739184|ref|XM_295169.lj[29739184] |29740706|ref|XM_295178.1|[29740706] i42658208|ref|XM_295195.4|[42658208] |42657898|ref|XM_295200.2J[42657898] 129737091 |ref|XM_295205.11[29737091] |42658103|ref|XM_295213.5|[42658103] |41148640|ref|XM_295252.3|[41148640] |29732623|refJXM_295257.1 |[29732623] 129733991 |ref|XM_295261.1 [[29733991] |41222673(ref|XM_295263.3|[41222673] J42662680|ref|XM_295270.2[[42662680] |37549764|ref|XM_295309.2([37549764] J4111411 θjref(xM_295598.3i[41114110] |3755129θ(ref|XM_295865.2|[37551290] |41201734|ref|XM_296117.3|[41201734] |37544742|ref|XM_296315.3|[37544742] |41150330|ref|XM_296817.4[[41150330] [41151003(ref |XM_297205.3 j[41151003] 142656616|ref|XM_297687.4|[42656616] |42662252Jref|XM_297816.4|[426622
'52] [41151624(refixM_298045.3i[41151624] 137546417|ref|XM_298053.2|[37546417] |42656785|ref|XM_298151.3[[42656785] |37549693iref|XM_298233.3J[37549693] (42656099[ref[xM_301210.3J[42656099} |42659408|ref|XM_350780.3|[42659408] (42659967|ref|XM_350880.3|[42659967]
gi]37549654|ref|XM_351335.11[37549654] gii42662297(ref[xM_351366.3i[42662297] gi|42656398|ref|XM_351473.3|[42656398] gi|37550244[refIXM_351574.11[37550244] gi[42657293[ref|XM_351617.2J[42657293] gi|37538596[ref|XM_351723.11[37538596] gi|42659053[ref|XM_351854.2|[42659053] gi|41151745Iref|XM_351948.2|[41151745] gi|42660714Jref|XM_352159.3|[42660714] gi(42656402[ref|XM_352463.2|[42656402] gi(37546315|ref|XM_352847.1 ([37546315] giJ37539071 [ref|XM_353628.11[37539071] gi[42659538iref|XM_370532.2J[42659538] giI4114935θ[ref|XM_370533.11[41149350] giJ41149356(ref|XM_370534.11[41149356] gi|42659541|ref|XM_370536.2[[42659541] giJ41149361 [ref|XM_370537.11[41149361] gi|42659543|ref|XM_370538.2([42659543] gi[41149371 jref[xM_370541.11[41149371] gi|42659556Jref|XM_370542.2|[42659556] gi(42659562|ref|XM_370543.2|[42659562] gi[42659564|refjXM_370545.2J[42659564] gi[42659414iref|XM_370554.2|[42659414] gi[42659395|ref|XM_370555.2|[42659395] gi|41149221 (ref|XM_370557.11[41149221] gi|41149306|ref|XM_370560.11[41149306] gi|41149320|ref|XM_370562.1 [[41149320] gi|41149102Jref|XM_370564.1|[41149102] giJ42659341 |ref|XM_370565.2([42659341] giJ4114911 θ[ref|XM_370566.1 ([41149110] gi|42659347|ref|XM_370567.2[[42659347] gi|41149124Jref|XM_370569.1|[41149124] gi|41149128Jref|XM_370570.1 [[41149128] gi|41149130|refJXM_370571.1 )[41149130] gi|42659356|ref|XM_370573.2|[42659356] gi[42659363irefjXM_370575.2i[42659363] gi|42659388|ref|XM_370577.2|[42659388] giJ42659433Jref|XM_370580.2[[42659433] gi|42659424|ref|XM_370582.2|[42659424] gi|42659571Jref|XM_370584.2J[42659571] gi(41149409|ref|XM_370585.11[41149409] giJ41149413|ref|XM_370586.1 [[41149413] giJ42659475|ref|XM_370589.2|[42659475] giI42659482Jref|XM_370591.2|[42659482] gi|41149289|ref|XM_370593.11[41149289] gi(41149588|ref|XM_370597.11[41149588] gi[42659691 |ref|XM_370603.2|[42659691] gi|41149664|ref|XM_370605.1 ([41149664] gi|41149668(reflXM_370606.1 ([41149668] gi|42659797|ref|XM_370607.2|[426597
'97] giJ42659802JrefjXM_370610.2[[42659802] gi(42659803Jref|XM_370611.2J[42659803] gi|41200855|ref|XM_370612.1|[41200855] gi[4265981θ(ref|XM_370613.2|[42659810] gi[41200868irefjXM_370615.1 j[41200868] gi(42659828iref(xM_370616.2|[42659828] gi|41200912(refjXM_370618.1 [[41200912]
gi|42659850|ref|XM_370619.2|[42659850] giJ41149514|ref|XM_370621.11[41149514] giJ41149518|ref|XM_370622.11[41149518] gi|42659657|ref|XM_370623.2|[42659657] giJ41149536|ref|XM_370629.11[41149536] giJ41200335Jref|XM_370630.1 j[41200335] gi|41200349|ref|XM_370631.11[41200349] gi|42659675|ref|XM_370632.2|[42659675] giJ41200354|ref|XM_370633.11[41200354] gi(41200356[ref|XM_370634.1 |[41200356] giJ42659728|ref|XM_370635.2|[42659728] gi|41149547Jref|XM_370636.1 j[41149547] gi|41149555|ref|XM_370638.11[41149555] gi|41149557|ref|XM_370639.11[41149557] giJ41149568|ref|XM_370642.11[41149568] gi|41149613|ref|XM_370644.11[41149613] giJ41149623|ref|XM_370648.11[41149623] gi|41149627|ref|XM_370649.11[41149627] gi|42659575|ref(xM_370651.2|[42659575] gi|42659581 |ref|XM_370652.2|[42659581] gi|42659583|refjXM_370653.2|[42659583] gi|42659586|ref|XM_370654.2|[42659586] giJ41149433|ref|XM_370657.11[41149433] gi|41149437|ref|XM_370658.11[41149437] gi[42659617|ref|XM_370660.2|[42659617] gi|41149451 |ref|XM_370661.11[41149451] gi|41149453|ref|XM_370662.11[41149453] giJ42659622|ref|XM_370663.2|[42659622] gi|41149457Jref|XM_370664.11[41149457] gi[42659633|ref|XM_370665.2|[42659633] giJ41149469Jref|XM_370666.1 ([41149469] gi|42660071 |ref|XM_370667.2|[42660071] gi|41149855(ref|XM_370668.1 j[41149855] gi|41149785Jref(xM_370669.1 j[41149785] gi|42659998|ref|XM_370672.2|[42659998] gi|42660005|ref|XM_370674.2|[42660005] gi|42660007|ref|XM_370675.2|[42660007] gi|41149808Jref|XM_370678.11[41149808] gi|41202037|ref|XM_370681.l j[41202037] gi|42660021 |ref|XM_370682.2J[42660021] gi|41202045|ref|XM_370684.1 |[41202045] gi|42660034|ref|XM_370685.2|[42660034] gi|41202063(ref|XM_370686.1 |[41202063] gi[41202068[ref[xM_370687.1 j[41202068] giJ42660039[ref|XM_370688.2|[42660039] gi|42659930|ref|XM_370690.2|[42659930] gi|42659938|ref|XM_370691.2([42659938] giJ42659946iref|XM_370692.2J[42659946] gi|42659947Jref|XM_370693.2|[42659947] gi|41149769Jref|XM_370695.1 j[41149769] giJ41201711 |ref|XM_370696.11[41201711] gi|42659983Jref|XM_370697.2|[42659983] gi|41149681 |ref|XM_370699.1 j[41149681] gi|41149703|ref|XM_370702.11[41149703] giJ41149713JrefjXM_370704.l j[41149713] giJ41149719Jref|XM_370705.1 [[41149719] gi|4114988δ(ref|XM_370707.1 [[41149886]
gi|41149831|ref|XM_370709.1|[41149831] giJ42660139[ref|XM_370710.2|[42660139] gi|42660140|ref|XM_370711.2|[42660140] gi|4266014liref|XM_370713.2J[42660141] gi|42660142|ref|XM_370714.2|[42660142] gi|42660145|ref|XM_370715.2|[42660145] giJ42660203irefixM_370716.2i[42660203] gi|41202601|ref|XM_370718.1|[41202601] gi|42660156Jref|XM_370721.2J[42660156] giJ42660162Jref|XM_370722.2[[42660162] gii41202624[ref|XM_370723.li[41202624] gi|42660168|ref|XM_370724.2|[42660168] gi|41202637|ref|XM_370725.l([41202637] gi|41202645|refjXM_370726.1|[41202645] gi|41202655|ref|XM_370727.1|[41202655] gi|4266019θiref|XM_370728.2J[42660190] gi|42660193Jref JXM_370729.2J[42660193] gi|41149912|ref|XM_370731.l[[41149912] gi|42660129|ref|XM_370732.2|[42660129] gi|42660215|ref|XM_370733.2J[42660215] gi|42660223|ref|XM_370734.2|[42660223] gii42660232Jref|XM_370737.2[[42660232] gi|41149957|ref|XM_370738.11[41149957] gi|42660240|ref|XM_370754.2|[42660240] gi|42660245|ref|XM_370756.2J[42660245] giJ41149971 jref|XM_370758.11[41149971] gi|42660248|ref|XM_370759.2|[42660248] gi|42660251|ref|XM_370760.2|[42660251] gi|42660294|ref|XM_370762.2[[42660294] gi|41203713|ref|XM_370763.1|[41203713] gi|41203742|ref|XM_370765.1|[41203742] gi|4266032θiref|XM_370767.2J[42660320] gi|41203750|ref|XM_370768.1|[41203750] gi|41203761 |ref|XM_370769.1|[41203761] gi|41203790|ref|XM_370772.1|[41203790] gi|41203813|ref|XM_370776.l([41203813] gi|42660363|ref|XM_370777.2|[42660363] gi|41203834|ref|XM_370778.1|[41203834] gi|42660371|ref|XM_370779.2|[42660371] gi|42660381 (ref|XM_370781.2|[42660381] gi|42660382|reflXM_370782.2J[42660382] giJ41203856Jref|XM_370785.1 ([41203856] gi|42661362(ref|XM_370826.2J[42661362] giJ41150115Jref[X _370829.11[41150115] gi|41150119iref|XM_370830.11[41150119] giJ41150123JrefjX _370831.1|[41150123] giJ42660618|ref|XM_370832.2|[42660618] gi|41150139|ref|XM_370833.1|[41150139] gi|41150141|ref|XM_370834.1|[41150141] gi|41150143|ref|XM_370835.1|[41150143] gi|41150145|ref|XM_370836.1|[41150145] gi|42660662|ref|XM_370837.2|[42660662] gii42660669ireflXM_370838.2J[42660669] giJ42660594ireflXM_370839.2J[42660594] gi(42660680|refjXM_370840.2i[42660680] gi|42660592iref|XM_370843.2([42660592] gi[42660675|refjXM_370844.2i[42660675]
gi|41150003|ref|XM_370845.11[41150003] gi|41150005Jref|XM_370846.1 j[41150005] gi|42660413|ref|XM_370848.2([42660413] gi|4115001 θjref|XM_370849.1 [[41150010] gi|41150014JreflXM_370851.l j[41150014] gi|41150016|ref(XM_370852.1 j[41150016] gi|42660425Jref|XM_370853.2J[42660425] gi|42660435|ref(XM_370855.2[[42660435] gi|41150032Jref|XM_370856.1 [[41150032] gi|41204880|ref|XM_370858.11[41204880] gi[41204903|ref|XM_370863.11[41204903] gi(41204905|ref|XM_370864.1 j[41204905] gi|41204910JreflXM_370865.1 [[41204910] gi|4120491 δjref |XM_370866.1 j[41204918] gi|42660458|ref|XM_370867.2|[42660458] gi|41204930|ref|XM_370868.11[41204930] gi|42660479|ref|XM_370871.2|[42660479] gi|42660486JreflXM_370872.2|[42660486] giJ4120496ljreflXM_370873.l i[41204961] giJ41204972[ref|XM_370876.1 ([41204972] gi|41204979|ref|XM_370878.11[41204979] gi|42660512|ref|XM_370879.2|[42660512] gi|4266052θ[ref|XM_370880.2|[42660520] gi|42660654(ref|XM_370881.2|[42660654] giJ42660526iref|XM_370882.2i[42660526] gi(41205009|ref|XM_370883.11[41205009] gi|41150155|ref|XM_370886.1 j[41150155] gi|41150159|ref|XM_370887.1 j[41150159] gi|42660648|ref|XM_370893.2([42660648] gi[41150173|ref|XM_370894.1 j[41150173] giJ41150036Jref|XM_370895.1 [[41150036] gi|42660551 [ref|XM_370897.2J[42660551 ] gi|41150042(ref|XM_370898.11[41150042] giJ4266056l (ref|XM_370899.2J[42660561] gi|4266O571|ref|XM_3709O4.2|[42660571] giJ41205604|ref|XM 370905.11[41205604] gi|41205607|ref|XM_370906.11[41205607] gi[42660572Jref|XM_370908.2J[42660572] gi|41205617|ref|XM_370909.1 |[41205617] gi|42660587Jref|XM_370910.2|[42660587] giJ41150078|ref|XM_370911.11[41150078] gi|42660682|ref|XM 370917.2J[42660682] gii42660686iref|XM_370918.2J[42660686] gi|41150493|ref|XM_370924.1 ([41150493] gi|41150497[ref|XM_370925.11[41150497] giJ41150501 jreflXM_370926.1 j[41150501] giJ41150503Jref|XM_370927.11[41150503] gi|42661011 |ref|XM_370928.2|[42661011] gi|41150317|ref|XM_370930.11(41150317] giJ41150321 |ref|XM_370931.1 ([41150321] gi[41150323ireflXM_370932.1 ([41150323] gi|42660702|ref|XM_370934.2J[42660702] gi|42660704|ref|XM_370935.2|[42660704] gi|41150249Jref|XM_370938.1 j[41150249] gi|42660942(ref|XM_370939.2|[42660942] gi|41150267|ref|XM_370942.11[41150267] gi|42660734|ref|XM_370943.2|[42660734]
gi|42660736 |ref|XM_.370944.21(42660736] gi[42660744: |ref|XM_370946.21(42660744] gi(41206115 (reflXM 370947.1 |[41206115] gi|41206124 (reflXM 370948.11(41206124] gi|41206126 |ref|XM_370949.1|[41206126] gi|41206138 |ref|XM 370952.11(41206138] gi|42660928 |ref|XM_370958.21(42660928] gi|41150395 IreflXM 370959.1 |[41150395] giJ41150405 J]reflXM 370965.11(41150405] giJ41150409 |ref|XM 370966.1 j[41150409] gi[42660967 J]reflXM 370967.2([42660967] gi|42660969 JreflXM 370968.2([42660969] gi(41150429 |ref|XM 370972.11(41150429] gi|41150431 [reflXM 370973.1 |[41150431] giJ41150435 |ref|XM 370974.11(41150435] gi|41150439 [ref|XM 370975.1 |[41150439] gi|41150522 IreflXM .370977.1 |[41150522] gi[42660995 IreflXM .370980.21(42660995] giJ41150483 IreflXM .370981.11(41150483] gi|42660766 |ref|XM 370982.2([42660766] gi|42660794 |ref|XM 370984.2|[42660794] gi|41206439 IreflXM 370986.11(41206439] gi|41206445 [reflXM .370987.1j[41206445] gi|41150359 IreflXM .370988.1|[41150359] gi|41150375 IreflXM .370991.11(41150375] gi|41150377 IreflXM 370992.1|[41150377] gi|41150379 IreflXM 370993.1 |[41150379] gi|42660825 IreflXM 370995.21(42660825] gi|41150297] IreflXM 370997.11(41150297] gi|41150588 IreflXM 371001.1([41150588] giJ41150601 IreflXM 371006.1([41150601] gi|42661076 (reflXM 371008.21(42661076] gi|42661079] |ref|XM .371009.21(42661079] gi|41150616 IreflXM 371010.1([41150616] gi|41150622] IreflXM 371012.1|[41150622] gi|41150630 IreflXM 371013.1|[41150630] gi|41150634 |ref|XM 371014.1|[41150634] gi|42661096 IreflXM 371015.2IJ42661096] gi|42661099 IreflXM 371016.21(42661099] gi|41150642 IreflXM 371017.11(41150642] gi|41150645| |ref|XM .371018.1|[41150645] gi|42661109 IreflXM .371019.2[[42661109] gi|42661114 (reflXM 371020.2|[42661114] giJ42661122 JreflXM .371023.2|[42661122] giJ42661128 IreflXM 371024.2|[42661128] gi|41150788 |ref|XM .371026.11(41150788] gi|41150794 IrefjXM .371028.11(41150794] gi|41150802 |ref|XM 371032.1|[41150802] gi|41150718 IreflXM .371034.1|[41150718] giJ42661245] JreflXM .371035.2|[42661245] gi|41150726 IreflXM .371036.1j[41150726] giJ42661257 |ref|XM 371039.2|[42661257] gi|42661302 |ref|XM .371043.2|[42661302] giJ42661304 IreflXM .371046.21(42661304] giJ42661141 Jref|XM 371052.2[[42661141] gi|41150711 [reflXM 371053.1([41150711] gi|42661147 |ref|XM 371054.2IJ42661147]
gij41222803 |ref|XM 371056.11(41222803] gi|42661174 (refJXM 371057.2 ([42661174] gi|41222810 |ref|XM 371058.1 |[41222810] gi|41222812 (reflXM 371059.1 ([41222812] giJ41222819 |ref|XM .371062.1 ([41222819] gi|41222821 JreflXM. 371063.1 ([41222821] gi|41222827 |ref|XM 371066.1 |[41222827] gi|42661186 |ref|XM 371067.2 j[42661186] gi|42661195 IreflXM 371068.21(42661195] gi|41222833 JreflXM. 371069.11(41222833] gi|41222839 IreflXM 371070.1 ([41222839] giJ41222851 |ref|XM 371074.1 [[41222851] gi|41150538 IreflXM 371077.11(41150538] gi(41150550 IreflXM 371078.11(41150550] gi|42661040 IreflXM .371079.21(42661040] gi|41150758 IreflXM 371081.1 |[41150758] gi|41150760 IreflXM 371082.1 |[41150760] gi|41150770 |ref|XM 371083.1 |[41150770] giJ41150778 IreflXM 371084.11(41150778] gi|41150782 jrefjXM 371085.11(41150782] gij41150579 IreflXM 371086.1 [[41150579] gi|41150830 |ref|XM .371088.1 |[41150830] gi|42661312 IreflXM 371089.2 |[42661312] giJ42661316 IreflXM 371092.21(42661316] gi|42661323 jref|XM_ .371097.2 j[42661323] gi|42661328 IreflXM 371098.2 j[42661328] giJ42661337 |ref|XM 371105.2 j[42661337] gi|41150874 IreflXM 371106.11(41150874] gi|42661346 |ref|XM_ .371107.21(42661346] gi|42661347 JreflXM 371108.2 |[42661347] gij41150885 |ref|XM 371109.11(41150885] gi|41150887 |ref|XM 371110.1 |[41150887] gi|42661358 IreflXM 371111.2 |[42661358] gi|42661374 IreflXM .371113.2 |[42661374] gi|42661382 IreflXM 371114.21(42661382] gi|42661403 IreflXM 371115.2 |[42661403] gi|42661399 IreflXM 371116.2 |[42661399] gi|41150938 JreflXM 371117.11(41150938] gi|41150942 JreflXM 371118.1 [[41150942] gi|41150918 IreflXM 371120.11(41150918] gi|41151015 IreflXM 371121.1 |[41151015] gi|41151023 |ref|XM 371122.11(41151023] gl|41151032 |ref(XM 371125.11(41151032] gi|41151049 IreflXM 371130.11(41151049] gi|41151066 |ref|XM. 371132.1 |[41151066] gi)42661959 IreflXM 371134.21(42661959] gi|41151084 J|reflXM 371138.1 |[41151084] gi|42661974 IreflXM 371139.2 |[42661974] gi|42661975 |ref|XM. 371140.2 |[42661975] gi|41151090 IreflXM 371141.1 ([41151090] gi|41151092 IreflXM 371142.11(41151092] gi|42661987 |ref|XM .371143.2 |[42661987] gi|41151106 |ref|XM 371145.1 |[41151106] gi|42662001 |ref|XM 371146.21(42662001] gi|41151119 IreflXM 371147.1 |[41151119] giJ42662007 IreflXM 371150.21(42662007] gi|41151129 jrefjXM 371151.11(41151129]
gi|41151131|ref|XM_371152.1|[41151131] giJ41151136JrefjX _371153.11[41151136] gi|42662012Jref|XM_371154.2|[42662012] gi|42662026JreflXM_371155.2|[42662026] giJ42661556JrefjXM_371157.2J[42661556] gi|41150966|ref|XM_371158.1 j[41150966] gi|42661571 |reflXM_371159.2|[42661571] gi|42661575ireflXM_371160.2([42661575] giJ42661588JrefjXM_371161.2|[42661588] gi|41150990|ref|XM_371164.lj[41150990] giJ41150993JreflXM_371165.1 j[41150993] gi|42661624|ref|XM_371167.2J [42661624] gi|41208707|ref|XM_371170.l[[41208707] gi|41208718|ref|XM_371174.1|[41208718] gi|41208728|ref|XM_371175.1|[41208728] gi|41208736Jref|XM_371176.1|[41208736] gi|41208740|ref|XM_371177.1|[41208740] gi|41208742Jref|XM_371178.11[41208742] gi|42661652Jref|XM_371179.2|[42661652] gi|42661655|ref|XM_371181.2|[42661655] gi|41208755|ref|XM_371182.1|[41208755] gi|41208757|ref|XM_371183.1|[41208757] gi|4266166θiref|XM_371184.2|[42661660] gi|42661677|ref|XM_371187.2|[42661677] gi|41208805|ref|XM_371190.11[41208805] gi|4266170ljreflXM_371191.2|[42661701] giJ41208814|ref|X _371192.lj[41208814] gi|42661712|ref|XM_371195.2|[42661712] gi|42661713|ref|XM_371196.2|[42661713] gi|41208836|ref|XM_371197.1|[41208836] gi|42661725|ref|XM_371198.2|[42661725] gi|41208856JreflXM_371200.1|[41208856] gi|41208865|ref|XM_371201.1|[41208865] gi|42659146|ref|XM_371202.2J[42659146] gi|42661310|reflXM_371204.2|[42661310] gi|42657412iref|XM_371205.2J[42657412] gi|42655827|ref|XM_371206.2|[42655827] gi|41107521|ref|XM_371207.1|[41107521] giJ41115074(reflXM_371208.1 ([41115074] gi|42656049|ref|XM_371210.2|[42656049] gi|42655845Jref|XM_371214.2|[42655845] gi|41107719JreflXM_371215.1|[41107719] gii4265562θjreflXM_371216.2J[42655620] giJ41114495|reflXM_371221.1|[41114495] gi|41114514|ref|XM_371222.11[41114514] gi|41114517|ref|XM_371223.lj[41114517] gi|41115267|ref|XM_371225.11[41115267] giJ42655691|ref|XM_371227.2|[42655691] gi|41188261|ref|XM_371230.1|[41188261] gi|41114740|ref|XM_371232.11[41114740] gi|42656067|ref|XM_371234.2|[42656067] gi|41114776(ref|XM_371235.11[41114776] gi|41114779|ref|XM_371236.11[41114779] gi|41107685|ref|XM_371237.11[41107685] giJ41107687|ref|XM_371238.11[41107687} gi|42655835|ref|XM_371239.2J[42655835] gi|42656107|ref|XM_371241.2|[42656107]
gi|42655738|ref|XM_371243.2|[42655738] gi|41107413|ref|XM_371244.11[41107413] giJ41107415(reflXM_371245.1 ([41107415] gil4265575θ(ref|XM_371246.2|[42655750] gi|42655760|reflXM_371248.2l(42655760] gi|42655763|ref|XM_371249.2|[42655763] gii42655764(refixM_371250.2i[42655764] gi|41107446[ref|XM_371252.11[41107446] gi[4110744δ(ref JXM_371253.1 ([41107448] gi|42655773[ref|XM_371254.2|[42655773] gi|41107466|ref|XM_371257.11[41107466] gi|41107468Jref|XM_371258.11[41107468] gi|41107475|ref|XM_371259.11[41107475] gi[4105827θ(ref|XM_371260.11[41058270] gi|4265565θ[ref|XM_371261.2|[42655650] gi|41058281 [ref|XM_371262.1 ([41058281] gi|41058285|ref |XM_371263.1 J[41058285] giJ41113031 jrefJXM_371265.1 j[41113031] gi|42655660|ref|XM_371267.2|[42655660] giJ42655662|ref|XM_371268.2[[42655662] giJ42655663|ref|XM_371269.2|[42655663] giJ41103632(reflXM_371273.1 [[41103632] giJ41057791|reflXM_371276.1|[41057791] gi|42655580|ref|XM_371277.2|[42655580] gi|41057801|ref|XM_371278.1 [[41057801] gi|41057805|ref|XM_371279.1|[41057805] gi[41057816iref[xM_371280.li[41057816] gi|42655584Jref|XM_371281.2|[42655584] gi|42655585|ref|XM_371283.2J[42655585] gi|42655979|ref|XM_371284.2|[42655979] gi|41221590|ref|XM_371285.l([41221590] gi|4265598θ[reflXM_371286.2l[42655980] giJ41115410|reflXM_371288.11[41115410] gi|41115452Jref|XM_371291.1 [[41115452] giJ42656074|reflXM_371292.2J[42656074] gi|42655803|ref|XM_371299.2|[42655803] gi|42656078|refJXM_371301.2|[42656078] gi|41116299|ref|XM_371302.11[41116299] gi[41107502|ref|XM_371304.1 j[41107502] gi|41107504|ref|XM_371305.11[41107504] gi|42655811 |ref|XM 371306.2|[42655811] gi|42655714|ref|XM_371310.2|[42655714] gi|42655715|ref|XM_371311.2|[42655715] gi|42655717|ref|XM_371312.2|[42655717] giJ41106710|ref|X _371313.1|[41106710] gi|41106713|ref|XM_371314.1|[41106713] giJ41117172|ref |XM_371315.1 [[41117172] gi|41117178|ref|XM_371316.11[41117178] gi[41188797lref|XM_371320.1 ([41188797] gi|41108615|ref|XM_371326.1|[411086
'15] gi|42655858Jref|XM_371328.2|[42655858] giJ4118810θjref|X 371329.1|[41188100] gi|42655955Jref|XM_371330.2|[42655955] gi|42655956|ref|XM_371331.2|[42655956] gi|41112865 jref |XM_371332.11[41112865} gi|41112868|ref|XM 371333.1 |[41112868] gi|41112871 |ref|XM_371334.11[41112871]
141188447 |refj|XM .371335 1(41188447]
J41188453 |ref J|XM 371336 [[41188453]
I42655824 jrefl JXM .371338 1(42655824]
J42655603 Iref JjXM 371339 ([42655603]
I42655604 Iref JjXM 371340 |[42655604]
142655613 Iref JjXM 371343 [[42655613]
(41110084 Iref JjXM.371344 1(41110084]
142655915 |ref J|XM .371346 1(42655915]
[41107724 |ref J|XM 371350 |[41107724]
I42655985 |ref J|XM.371351 ([42655985]
J42655988 |ref J|XM 371352 1(42655988]
142655991 |refj|XM 371353 |[42655991]
141114118 |ref J|XM 371354 JJ41114118]
|41114123 |ref J|XM .371355 |[41114123]
141104552 jref J|XM 371356 1(41104552]
141104561 |ref J|XM .371357 1(41104561]
141104564 jref JjXM 371358 1(41104564]
141117236 |ref J|XM 371359 1(41117236]
[41115793 jref JjXM 371369 1(41115793]
I42656098 |refj|XM 371374 1(42656098]
J41117409 |ref J|XM 371380 1(41117409]
142656153 |ref J|XM 371384 1(42656153]
J42656155 jref JjXM 371385 1(42656155]
142656150 |ref JjXM 371388 1(42656150]
I42662266 jref J|XM .371390 1(42662266]
(41151267 |ref J|XM 371391 1(41151267]
141151298 |ref J|XM 371395 1(41151298]
|41209275 jrefl JXM 371397 1(41209275]
I42662293 |ref J|XM 371398 1(42662293]
J41151219 jrefl JXM 371399 [[41151219]
J42662236 Iref JjXM.371401 1(42662236]
J42662237 |ref J|XM 371402 1(42662237]
I42662243 jref JjXM .371403 ([42662243]
J41151206 |ref J|XM 371405 |[41151206]
141151304 |ref J|XM 371407 1(41151304]
142662310 |ref J|XM.371409 [[42662310]
J42662322 |ref J|XM 371411 1(42662322]
I42662328 jref JjXM 371413 1(42662328]
141209339 |ref J|XM 371416 1(41209339]
|42662366 |ref J|XM.371417 1(42662366]
I42662357 JJrefl JXM 371418 |[42662357]
J41151366 jref JjXM 371421 1(41151366]
141151376] Jref J|XM 371423 1(41151376]
141151378] jref JjXM 371424 1(41151378]
141151391 Jref J|XM 371429 |[41151391]
141151393 |refj|XM 371430 j[41151393]
141151413 Jrefl JXM .371431 |[41151413]
I42662425 Jref JjXM 371436 1(42662425]
J41151431 |ref J|XM 371437 1(41151431]
|42662427 |ref J|XM 371455 |[42662427]
I42662435 |ref J|XM 371459 1(42662435]
|42662437! Jref J|XM 371460 1(42662437]
I42662442 jref JjXM 371461 1(42662442]
141209839 Jref J|XM 371463 |[41209839]
J41209868 Jref J|XM.371466 1(41209868}
141209879 [ref J|XM 371468 1(41209879]
I42662478 jrefl JXM.371469 |[42662478]
giJ42662482|ref|XM_371470.2|[42662482] giJ42662483Jref(XM_371471.2|[42662483] giJ41151474|ref|XM_371474.1|[41151474] giJ41118879Jref|X _371476.11[41118879] giJ41117712|ref|X _371477.1[[41117712] gi|42656191 |ref(xM_371478.2J[42656191] giJ42656197JrefjXM_371479.2[[42656197] giJ41123791' jref|XM_371480.1 [[41123791] gi|42656264Jref|XM_371481.2|[42656264] gi|42656268|ref|XM_371484.2J[42656268] giJ4112383θjrefjXM_371485.1|[41123830] gi|42656276|ref|XM_371486.2|[42656276] gi|42656278|ref|XM_371487.2|[42656278] gii42656280|ref|XM_371488.2|[42656280] gi|41190376|ref|XM_371489.1|[41190376] gi|41190388JrefjXM_371490.1|[41190388] giJ41190397|ref|XM_371491.11[41190397] gi|41190406|ref|XM_371492.11[41190406] gi|41190411 jref|XM_371493.1 [[41190411] gi|41190413|ref|XM_371494.11[41190413] gi|41190434JreflXM_371495.1 [[41190434] gi|42656319|ref|XM_371496.2|[42656319] gi|4265633l[ref|XM_371497.2|[42656331] giJ41190460|ref|XM_371500.11[41190460] gi|42656340|reflXM_371501.2|[42656340] gi|42656341|ref|XM_371502.2J[42656341] gi|41190469Jref|XM_371503.1|[41190469] gi|41190474|ref|XM_371504.11[41190474] gi|42656347|ref|XM_371505.2|[42656347] gi|42656348|ref|XM_371506.2|[42656348] gi|42656355|ref|XM_371511.2|[42656355] giJ41190499JreflXM_371513.1 ([41190499] gi|41190501 |ref|XM_371514.11[41190501] gi|42656357JreflXM_371515.2J[42656357] gi|41190507|ref|XM_371517.11[41190507] gi|42656413|ref|XM_371534.2|[42656413] gii42656382ireflXM_371535.2([42656382] gi|42656384|ref|XM_371536.2|[42656384] gi|41124491 |ref|XM_371537.11[41124491] giJ41124501Jref|XM_371539.1|[41124501] giJ41190797|ref|XM_371540.11[41190797] gi|42656219|ref|XM_371542.2|[42656219] gi|42656232|ref|XM_371543.2|[42656232] gi|42656236|ref|XM_371544.2|[42656236] giJ42656248|ref|XM_371546.2|[42656248] gi|42656249|ref|XM_371547.2[[42656249] gi|41118730|reflXM_371552.1 [[41118730] giJ4112636θ(ref|XM_371555.1 [[41126360] gi|42656430|ref|XM_371558.2|[42656430] giJ41125704|ref|XM_371561.11[41125704] gi|41125714|ref|XM_371564.11[41125714] gi|41125731 |ref|XM_371567.11[41125731] gi|42656435|ref|XM_371568.2|[42656435] gi|42656436|ref|XM_371569.2|[42656436] gi|42656439|ref|XM_371571.2|[42656439] gi|41125761 |ref|XM_371572.11[41125761] gi|42656448|ref|XM_371573.2|[42656448]
gi|42656520|ref|XM_371575.2|[42656520] gi(4265652l iref(xM_371576.2J[42656521] giJ42656526iref|XM_371577.2|[42656526] giJ41134014Jref|XM_371581.11[41134014] gi|41191407|ref|XM_371583.11[41191407] giJ41191413Jref|XM_371584.11[41191413] gi|41191422Jref|XM_371585.11[41191422] gi|42656546|ref|XM_371586.2J[42656546] gi|41191437|ref|XM_371588.l[[41191437] gi|42656560Jref|XM_371590.2|[42656560] giJ41191456|ref|XM_371591.1|[41191456] gi|42656566JrefixM_371592.2J[42656566] gi|41191468[ref|XM_371593.11[41191468] gi(42656577|ref[xM_371594.2[[42656577] gi|42656585|ref|XM_371595.2|[42656585] gi[41191497|ref|XM_371600.1|[41191497] gi|41126904Jref|XM_371603.1 ([41126904] gi[42656485(ref|XM_371604.2I[42656485] gi[41126925 jref JXM_371605.1 [[41126925] giJ42656611 [ref |XM_371606.2J[42656611] gi|41144256Jref|XM_371614.1 )[41144256] gi|42656679|ref |XM_371617.2|[42656679] gi|41144279Jref|XM_371618.1 [[41144279] gi|41144284[ref|XM_371619.11[41144284] gi(41144299|ref|XM_371621.11[41144299] gi|41144319|ref|XM_371622.11[41144319] gi[41144322Jref|XM_371623.11[41144322] g|J41144347|ref|XM_371625.11[41144347] gi|4114435θjref|XM_371626.1 [[41144350] gi|41144364|ref|XM_371629.11[41144364] giJ42656714Jref|XM_371630.2[[42656714] gi|41193238|ref|XM_371631.11[41193238] gi|42656723|ref|XM_371632.2J[42656723] gi(41193259|ref |XM_371638.11[41193259] gi|41193262|ref|XM_371639.11[41193262] gi|41193266|ref|XM_371641.11[41193266] gi|41193278|ref|XM_371643.11[41193278] giJ41193286(ref |XM_371645.11[41193286] giJ41137106JrefjXM_371647.l j[41137106] gi(42656643|ref|XM 371649.2|[42656643] gi|41137141 |ref|XM_371653.11[41137141] giJ41146519Iref|XM_371655.1|[41146519] gi|41146521 |ref|XM_371656.1 [[41146521] giJ41146530|ref|XM 371658.11[41146530] gi|42656808|ref|XM_371660.2J[42656808] giJ41146558|ref|XM_371662.11[41146558] gi[4265682l (ref|XM_371663.2|[42656821] gi|42656822[ref|XM_371664.2|[42656822] giJ41146581 |ref|XM_371665.11[41146581] gi[42656845|ref|XM 371666.2|[42656845] giJ41146591 |ref|XM_371668.11[41146591] giJ41146595|ref|XM_371670.11[41146595] gi|42656848|ref|XM_371671.2|[42656848] gi|42656857|ref[XM_371672.2|[42656857] gi|41146621 |ref|XM_371674.11[41146621] gi|41146631 JreflXM 371677.1 [[41146631] giJ41146633|ref|XM 371678.1|[41146633]
141146639 |ref|XM .371679.11(41146639] g J42656876 IreflXM .371680.2 j[42656876] [41146644 |ref|XM 371681.1 1(41146644] g 141146646 jref|XM .371682.1 |[41146646] |41144857 |ref|XM 371683.1 |[41144857] g J41144883 JreflXM .371684.1 1(41144883] [41146823 IreflXM 371687.1 j[41146823] J41146867 |ref|XM 371690.1 ([41146867] J42657118 IreflXM 371691.2 1(42657118] J42656916 JreflXM 371692.2 |[42656916] 141146841 [reflXM .371693.1 ([41146841] g |41146843 JreflXM 371694.1 [[41146843] 142657010 IreflXM .371695.2 j[42657010] 141146664 IreflXM 371697.1 1(41146664] (4 146666 IreflXM..371698.1 1(41146666] 141146680 [reflXM .371701.1 ([41146680] 141195078 JreflXM 371702.1 1(41195078] J42656955 IreflXM 371705.2 |[42656955] J42656961 JreflXM 371706.2 1(42656961] I42657000 IreflXM .371709.2 j[42657000] 141146690 IreflXM 371710.1 [[41146690] 141146692 IreflXM .371711.1 |[41146692] J41146720 (reflXM 371714.1 ([41146720] J42656999 (reflXM .371715.2 1(42656999] [41146758 IreflXM 371717.1 1(41146758] 141146762 IreflXM .371718.1 |[41146762] 141146902 IreflXM 371719.1 |[41146902] J41146906 JreflXM.371722.1 [[41146906] (41146914 [reflXM 371725.1 [[41146914] (41146925 JreflXM.371726.1 1(41146925] J41147105 JreflXM .371728.1 1(41147105] 141147107 IreflXM .371729.1 j[41147107] [41147111 IreflXM 371731.1 1(41147111] J41147113 IreflXM .371732.1 j[41147113] 141147115 IreflXM..371733.1 1(41147115] J42657340 IreflXM .371736.2 j[42657340] I42657342 IreflXM .371738.2 |[42657342] [42657308 IreflXM 371740.2 |[42657308] 142657311 |ref|XM 371741.2 [[42657311] 141147253 IreflXM .371743.1 1(41147253] 141147263 |ref|XM_ 371749.1 1(41147263] (41147177 IreflXM 371754.1 1(41147177] I42657385 IreflXM .371755.2 |[42657385] 142657391 (reflXM 371757.2 1(42657391] 141147203 IreflXM .371758.1 1(41147203] (41147205 jref|XM_ 371759.1 1(41147205] I42657395 IreflXM .371760.2 1(42657395] I42657227 IreflXM .371761.2 [[42657227] I42657235 IreflXM 371762.2 ([42657235] 141147007 |ref|XM_ 371763.1 |[41147007] J41147011 IreflXM .371764.1 [[41147011] 141147013 |ref|XM_ 371765.1 ([41147013] J41147032 |ref|XM_ .371768.1 1(41147032] 141147036 |ref|XM_ .371769.1 1(41147036] 141147038 |ref|XM_: 371770.1 1(41147038] 141147040 |ref|XM_ .:371771.1 [[41147040] J42657283 |refjXM_: 371772.2 j[42657283]
gi|41146994 reflXM 371776.1 |[41146994] gi|42657191 reflXM 371777.2 1(42657191] giJ41146936 reflXM 371778.1 [[41146936] gi|42657203 reflXM .371781.2] |[42657203] gi[41146952 reflXM 371782.1 |[41146952] giJ41146954 reflXM 371783.1 [[41146954] gi|41147230 reflXM 371790.1 |[41147230] gi|41147233 reflXM 371791.1 ([41147233] gi|42657416 reflXM 371793.2 1(42657416] gi|41147306 reflXM .371796.1 J[41147306] giJ41147308 reflXM 371797.1 1(41147308] gi|42657574 reflXM 371798.2 1(42657574] gi|42657579 reflXM 371801.2 [[42657579] gi|42657617 reflXM 371809.2 1(42657617] gi|41197088 reflXM 371812.1 1(41197088] gi|41197093 reflXM 371813.1 1(41197093] gi|41197097 reflXM 371814.1 1(41197097] gi|41197099 reflXM 371815.1 1(41197099] gi|42657635 reflXM 371816.2] 1(42657635] gi|41197105 reflXM 371817.1 |[41197105] gi|41197107 reflXM .371818.1 1(41197107] gi|41197110 reflXM 371819.1 1(41197110] gi|42657643 reflXM 371820.2 1(42657643] gi|42657651 reflXM 371822.2 ([42657651] gi|41197139 reflXM 371823.1 1(41197139] gi|42657659 reflXM 371824.2 ([42657659] gi|41197144 reflXM 371825.1 1(41197144] gi|41197153 ref|XM 371826.1 [[41197153] giJ41147317 reflXM 371829.1 1(41147317] gi|41147326 reflXM 371832.1 1(41147326] gi|41147336 reflXM 371835.1 1(41147336] gi|42657436 reflXM 371837.2 1(42657436] gi|42657437 reflXM 371838.2] ([42657437] gi|41147275 reflXM 371841.1 |[41147275] gi|41147279 reflXM 371842.1 |[41147279] gi|41196188 reflXM 371843.1 [[41196188] gi|42657450 reflXM 371844.2 1(42657450] gi|41196203 reflXM 371845.1 ([41196203] gi|41196206 reflXM 371846.1 |[41196206] gi|42657457 reflXM 371847.2 1(42657457] gi[42657473 reflXM .371848.21 1(42657473] gi|41196223 reflXM 371849.1 1(41196223] gi|41196228 reflXM 371850.1 |[41196228] gi|41196231 reflXM 371851.1 ([41196231] gi|42657492 reflXM 371853.2 [[42657492] gi|41147367 reflXM 371856.1 1(41147367] gi|42657549 reflXM 371857.2 1(42657549] gi(42657552 ref|XM 371858.2 ([42657552] gi|41147427 reflXM .371863.1 |[41147427] gi|42657931 reflXM 371873.2] |[426579'31] giJ41222034 reflXM 371874.1 [[41222034] gi|42657848 reflXM 371877.2 ([42657848] gi(42657850 reflXM .371878.2 [[42657850] gi|41222049 reflXM .371879 |[41222049] gi|42657859 reflXM 371884.2 JJ42657859] gi|41222071 reflXM .371885.1 ([41222071] gi|41222092 reflXM 371889.1 |[41222092]
gi|41222100|reflXM_371891.1|[41222100] gi|41222114|ref|XM_371897.11(41222114] gi|41147431 [ref |XM_371901.11[41147431] gi|42658325|ref|XM_371904.2[[42658325] gi|42658028|ref|XM_371917.2|[42658028] gi|41147511 |ref|XM_371923.1 [[41147511] gi|42657785|ref|XM_371925.2|[42657785] giI4265803θiref|XM_371930.2|[42658030] gi|41147634|ref|XM_371933.1|[41147634] gi|42658058|ref|XM_371936.2[[42658058] gi|42658071|ref|XM_371939.2|[42658071] giJ42658074(ref|XM_371943.2|[42658074] gi|41199202[ref|XM_371948.11[41199202] gi|42658586|ref|XM_371949.2[[42658586] gi|42658119|ref|XM_371953.2|[42658119] gi|42658120|ref|XM_371954.2|[42658120] gi|42658128iref|XM_371956.2|[42658128] gi|41147550|ref|XM_371960.11[41147550] gi|41147554|ref|XM_371995.11[41147554] gi|41147572|ref|XM_372002.11[41147572] gi|41147455[reflXM_372004.11[41147455] gi|41147457|ref|XM_372005.11[41147457] gi|41147530|ref|XM_372006.11[41147530] gi|41148485|ref|XM_372009.11[41148485] gi|42658797|ref|XM_372010.2|[42658797] gi|42658809[ref|XM_372011.2J[42658809] gi|42658816|ref|XM_372013.2|[42658816] gi[42658813|ref|XM_372017.2|[42658813] gi|41148571 |ref|XM_372018.1 [[41148571] gi|42658874Jref|XM_372019.2J[42658874] gi|42658881|ref|XM_372024.2|[42658881] gi|41148589|ref|XM_372025.11[41148589] gi|41148591 |ref|XM_372026.1 ([41148591] gi|41148593|ref|XM_372027.1 j[41148593] gi|42658889|ref|XM_372028.2J[42658889] gi|42658854|ref|XM_372030.2|[42658854] giJ42658866|ref|XM_372031.2[[42658866] gi|41148444|ref|XM_372035.1|[41148444] giJ41148446Jref JXM_372036.11[41148446] giJ42658989|ref|XM_372037.2|[42658989] gi|4265899l[ref|XM_372038.2|[42658991] gi|41148766[ref|XM_372039.1 [[41148766] gi|41148455|ref|XM_372040.11[41148455] gi|42658948|ref|XM_372041.2|[42658948] giJ41148704|ref|XM_372042.11[41148704] gi|41148710|ref|XM_372045.11[41148710] gi|41148723|ref|XM_372046.11[41148723] gi|41148725|ref|XM_372047.11[41148725] gi|42658972|ref|XM_372048.2|[42658972] gi|42658977|ref|XM_372050.2|[42658977] gi|41148647|ref|XM_372054.lj[41148647] gi|41148651 |ref|XM_372055.11[41148651] gi|42658935|ref|XM_372058.2|[42658935] giJ41148533|ref|XM_372060.11[41148533] gi|41148469Jref|XM_372062.11[41148469} gi|41148475|ref|XM_372063.1 [[41148475] gi|42658895|ref|XM_372069.2|[42658895]
142659158 refjXM 372074.211(42659158]
142659161 reflXM 372075.21(42659161]
141148960 ref|XM .372076.11(41148960]
142659171 ref|XM 372077.2] [[42659171]
(41148973 refjXM 372078.11(41148973]
J41148978 refl[XM .372083.1 [[41148978]
[42659209 ref|XM .372086.2] 1(42659209]
|41148825 ref|XM 372088.11(41148825]
J41148827 ref||XM .372089.11(41148827]
[42659125 refl[XM 372090.21 |[42659125]
141148833 reflXM 372092.1 |[41148833]
J42659071 ref|XM .372094.2] 1(42659071]
142659128 ref||XM 372097.2 (1[42659128]
[42659082 refl[XM 372099.2 |[42659082]
J41148849 reflXM 372100.11(41148849]
[41148855 ref|XM 372102.11(41148855]
(41148857 ref|XM .372103.1 |[41148857]
|42659092 refl[XM .372104.2] 1(42659092]
J41148869 ref|XM .372108.11(41148869]
J42659101 ref|XM .372109.21(42659101]
142659130 ref||XM 372110.21 ([42659130]
J41148917 refl[XM 372111.11(41148917]
142659111 reflXM 372112.2] 1(42659111]
141148877 ref|XM .372114.11(41148877]
142659117 ref||XM 372116.2 |[42659117]
[41148768 refl[XM 372117.11(41148768]
141148777 reflXM 372118.11(41148777]
141148779 reflXM .372119.11(41148779]
(41148781 ref|[XM 372120.11(41148781]
|42659023 ref|[XM 372121.2! 1(42659023]
J41148785 reflXM 372122.11(41148785]
141148787 reflXM .372123.11(41148787]
[42659025 ref|[XM 372124.21 |[42659025]
141148795 ref||XM .372125.1 j[41148795]
[42659301 reflXM .372128.21](42659301]
J41149029 reflXM 372132.1 |[41149029]
141149031 ref|[XM 372133.11(41149031]
[42659240 reflIXM .372137.2] 1(42659240]
142659251 reflXM 372138.2 |[42659251]
(42659257 ref|XM 372140.2 [[42659257]
(41222645 reflIXM 372141.11(41222645]
J42659269 ref|XM 372142.2 [[42659269]
142659271 ref|XM 372143.21(42659271]
(41148815 reflXM 372148.1 |[41148815]
[41148819 ref|XM 372150.11(41148819]
142659310 ref|XM 372154.21(42659310]
J41199565 reflXM 372157.1 ([41199565]
(41199571 ref|XM 372159.11(41199571]
J41199577 ref|XM 372160.11(41199577]
J41199582 refXM 372161.11(41199582]
J41199585] ref|XM .372162.1 [[41199585]
(42659118 ref|XM 372163.21(42659118]
J41148891 reflXM .372168.11(41148891]
J41148893 ref|XM..372169.11(41148893]
[41148923 reflXM 372175.1 |[41148923}
142659137! reflXM .372177.2 |[42659137]
J41148931 ref| XM 372180.1 [[41148931]
gi[42662766[ref(XM_372181.2([42662766] gi|42662767|ref|XM_372182.2|[42662767] giJ41151802|ref|XM_372186.11[41151802] gi|41151807[ref|XM_372190.11[41151807] giJ41151809|ref|XM_372191.1 [[41151809] giJ42662777JreflXM_372192.2J[42662777] gi|42662781 [ref|XM_372193.2|[42662781] gi(41151823Jref JXM_372194.1 j[41151823] giJ42662789|ref|XM_372195.2|[42662789] gi|42662654|ref|XM_372197.2|[42662654] gi|41151590|ref|XM_372198.11[41151590] gi|4266266θiref[XM_372199.2J[42662660] gi|42662667Jref|XM_372200.2|[42662667] giJ42662672|refjXM_372201.2[[42662672] gi|41151727|ref|XM 372202.1 j[41151727] gi|41210566[ref|XM_372203.1 |[41210566] gi|42662536|ref|XM_372204.2|[42662536] gi|42662545|ref|XM_372205.2j[42662545] giJ41151494(reflXM_372208.1 j[41151494] gi|41151496|ref|XM_372209.11(41151496] gi|41151498|ref|XM_372210.1 [[41151498] gi|41151500|ref|XM_372212.1 |[41151500] giJ42662553JreflXM_372213.2J[42662553] gi|41151526Jref|XM_372214.11[41151526] gi[42662555|ref|XM_372223.2|[42662555] gi|42662556|ref|XM_372224.2|[42662556] gi|41151518|ref|XM_372226.11[41151518] giJ42662617|ref|XM_372227.2|[42662617] giJ42662627|ref)XM_372231.2|[42662627] gi|42662629|ref|XM_372233.2|[42662629] gi|42662605|ref|XM_372239.2|[42662605] gi|41151689Iref(XM_372245.1 |[41151689] gi|41151626|ref|XM_372247.1 j[41151626] gi|4266268ljref|XM_372248.2J[42662681] gi|41151634|ref|XM_372253.11[41151634] gi|41151637|ref jXM_372254.1 j[41151637] giJ41151641 jref|XM_372255.1 ([41151641] gi|42662690|ref|XM_372257.2J[42662690] gi|41151663|ref|XM_372258.1 [[41151663] gi|41151575|ref|XM_372261.11[41151575] gi|42662640|reflXM_372262.2J[42662640] gi|42662645iref|XM_372267.2([42662645] gi|41151674Jref|X _372268.1 j[41151674] gi|41151701 jref|XM_372272.11[41151701] giJ42662719|ref|XM_372273.2([42662719] gi|41151707|ref|XM_372274.1 ([41151707] gi|41151715 jref |X _372275.11[41151715] gi|42662734Jref|XM_372282.2|[42662734] giI42662736Jref|XM_372286.2|[42662736] giJ4266274l jref|XM_372289.2i[42662741] giJ42662744iref|XM_372292.2|[42662744] gi|41151768lref|XM_372295.1 j[41151768] gi[41151770|ref|XM_372296.1 |[41151770] gi|41149251 |ref|XM_372302.1 J[41149251] gi|41149263Jref|XM_372303.11[41149263] giJ41149265|ref|XM_372304.11[41149265] gi|42659471 |ref|XM_372305.2|[42659471]
(41149399 |ref J|XM 372306.11(41149399]
J41149401 Jref J]XM 372307.1 ([41149401]
J41149403 Irefj |XM 372308.1 ([41149403]
|41149405 [ref J|XM 372309.1 j[41149405]
[41149215 |ref| |XM 372310.1 |[41149215]
[41149217 J]refl JXM 372311.1 ([41149217]
I42659406 Jref :jJXM 372312.2 ([42659406]
J41149334| |ref :||XM 372314.1 [[41149334]
141149336 Irefl IXM 372315.1 [[41149336]
141149340 jref ]1XM 372316.11(41149340]
|41149342 Irefl JXM 372317.11(41149342]
J41149174 |ref| |XM 372319.1 |[41149174]
[41149176 |ref J|XM 372320.11(41149176]
141149178 Jref JXM 372321.11(41149178]
J41149180 Iref IjXM 372322.1 |[41149180]
141149182 Iref jlXM 372323.11(41149182]
(41149184 Irefl JXM 372324.1 ([41149184]
(41149186, |ref J|XM 372325.11(41149186]
J41149192 Irefl IXM 372328.1 j[41149192]
141149194 [ref J|XM 372329.11(41149194]
141149237 Irefl |XM 372330.1 |[41149237]
J41149227 Irefl JXM 372331.11(41149227]
(41149281 [refl JXM 372334.1 j[41149281]
J41149283 Irefl JXM 372335.1 |[41149283]
J41149271 Irefl JXM 372337.1 [[41149271]
J41149295 Iref JjXM 372340.1 |[41149295]
[41149299 Irefl [XM 372341.1 [[41149299]
J41200927 Irefl JXM 372343.11(41200927]
141200932 Irefl JXM 372345.11(41200932]
[41200935 Irefl JXM 372346.11(41200935]
|41200939 Irefl JXM 372347.11(41200939]
J41200942 [refl JXM 372348.1 ([41200942]
|41200945 jrefl |XM 372349.11(41200945]
]41200947 J|refl |XM 372350.1 j[41200947]
142659867 Irefl |XM 372351.2 ([42659867]
141200959 Irefl JXM 372352.1 |[41200959]
[41200962 Irefl JXM 372353.11(41200962]
J41200965 Irefl IXM 372354.1 |[41200965]
J41200967 Irefl JXM 372355.1 [[41200967]
(41200969 |ref JXM 372356.11(41200969]
J41200972 Irefl IXM 372357.1 |[41200972]
141200974 Irefl |XM 372358.11(41200974]
J41200976 Irefl JXM 372359.1 ([41200976]
141200979 |ref J|XM 372360.11(41200979]
[41200983] |ref J|XM 372361.1 ([41200983]
141200985 Irefl JXM .372362.1 |[41200985]
|42659873 Irefl JXM 372364.21(42659873]
|41200992 [ref JjXM 372365.11(41200992]
141200994 |ref J|XM 372366.11(41200994]
J41200997 Irefl JXM .372367.11(41200997]
141200999 Jref :]|XM .372368.11(41200999]
J41201001 |ref J|XM 372369.11(41201001]
141201004 jref JjXM 372370.1 [[41201004]
141201006 |ref J|XM 372371.11(41201006]
[41201015 Iref JjXM .372372.1 |[41201015]
J41201018 Iref JjXM .372373.1 |[41201018]
141201027 Jref JjXM 372374.1 j[41201027]
gi|41201031 |ref|XM_372375.11[41201031] gi|41201035|ref|XM_372376.1|[41201035] gi|4120104θ[ref|XM_372377.1 ([41201040] gi[41201042|ref|XM_372378.1|[41201042] gi|41201050Iref|XM_372379.1j[41201050] gi|41201053Jref|XM_372380.li(41201053] giJ41201056 jrβf|XM_372381.11[41201056] gi|41149607|ref|XM_372382.1 ([41149607] giJ41200377|ref|XM_372384.lj[41200377] gi(41200379iref|XM_372385.1|[41200379] gi|41200386|ref|XM_372386.1|[41200386] giJ41200391 |reflXM_372387.1|[41200391] gii41200394|ref|XM_372388.1J[41200394] gi|42659698|ref|XM_372389.2|[42659698] gi|41200407|ref|XM_372390.1|[41200407] giJ41200409|refjXM_372391.lj[41200409] gi|41200416|ref|XM_372393.l[[41200416] gi|41200418|ref|XM_372394.11[41200418] gi|41200424(ref|XM_372395.1|[41200424] gi|41200427|ref|XM_372396.11[41200427] gi|41200431 |ref|XM_372397.lj[41200431] gi|4l200440|ref|XM_372399.1|[41200440] gi|41200443|ref|XM_372400.l[[41200443] gi|41200446|ref|XM_372401.l([41200446] gi|41200448|ref|XM_372402.1|[41200448] gi|41200452|ref|XM_372403.l[[41200452] gi|41200455|ref|XM_372404.1|[41200455] gii41200460|ref|XM_372405.lj[41200460] gi|41200470|ref|XM_372406.1|[41200470] giJ41200477|ref|XM_372409.1|[41200477] gi|41200487|ref|XM_372410.l([41200487] gi|41200489iref|XM_372411.1|[41200489] gi|42659710|ref|XM_372412.2|[42659710] gi|41200497|ref|XM_372413.1|[41200497] gii41200499JreflXM_372414.lj[41200499] gi|41200501|ref|XM_372415.1|[41200501] gi|41200503|ref|XM_372416.l([41200503] gi|41200509|ref|XM_372418.1|[41200509] gi|42659696|ref|XM_372420.2|[42659696] gi|41149631 |ref|XM_372423.1 ([41149631] gi|41149633|ref|XM_372424.1|[41149633] gi|41149635|ref|XM_372425.11[41149635] gi|41149637|ref|XM_372426.11[41149637] gi|41149639|ref|XM_372427.l[[41149639] gi|41149643|ref|XM_372428.11[41149643] gi[42659783|ref|XM_372429.2|[42659783] giJ41149477|ref|XM_372430.11[41149477] giJ41149479|ref|XM_372431.11[41149479] gi|41149481 |ref|XM_372432.11[41149481] gi|41149483[ref|XM_372433.1 [[41149483] gi|41149485|ref|XM_372434.11[41149485] gi|41149487|ref|XM_372435.1 [[41149487] gi|41149489|ref|XM_372436.1 j[41149489] gi|41149491 |ref|XM_372437.11(41149491] gi|41149493|ref|XM_372438.1|[41149493] giJ42659621 [reflXM_372441.2J[42659621] gi|41149501 (ref|XM_372442.1|[41149501]
gi|41149503|ref|XM 372443.1 [41149503] gi|41149505|ref|XM 372444.1 [41149505] gi|41149507|ref|XM 372445.1 [41149507] gi|41149861 |ref|XM .372447.1 [41149861] gi|41149863Jref|XM 372448.1 [41149863] gi|41202077[ref|XM .372449.1 [41202077] gi|41202079|ref|XM _372450.1 [41202079] gi|41202088(ref|XM _372452.1 [41202088] gi|41202091 IreflXM _372453.1 [41202091] gi|41201751 IreflXM 372456.1 [41201751] gi|4265999θ[reflXM _372457.2 [42659990] gi|41201762|ref|XM _372459.1 [41201762] gi|41201767|ref|XM .372460.1 [41201767] gi|41201770|ref|XM 372461.1 [41201770] gi[41201772Jref|XM .372462.1 [41201772] gi|41201774(ref|XM .372463.1 [41201774] gi|41201777|reflXM _372464.1 [41201777] gi|41201779|ref|XM .372465.1 [41201779] gi|41201784|ref|XM .372466.1 [41201784] gi|41201790|ref|XM .372468.1 [41201790] gi|41201793Jref|XM 372469.1 [41201793] gi|41201795|ref|XM .372470.1 [41201795] gi|41201797|ref|XM .372471.1 [41201797] gi|41149726|ref|XM .372472.1 [41149726] giJ42659918|reflXM 372473.2 [42659918] gi|4114973θ(ref|XM .372474.1 [41149730] gi|41149736Jref|XM 372476.1 [41149736] gi|41149845Jref|XM .372480.1 [41149845] gi|41149851 IreflXM 372482.1 [41149851] gi|41149773|ref|XM 372483.1 [41149773] gi|41202688Jref|XM .372485.1 [41202688] gi|41202691 JreflXM 372486.1 [41202691] gi|41202694[ref|XM .372487.1 [41202694] gi|41202696 JreflXM.372488.1 [41202696] gi|41202701 [reflXM .372490.1 [41202701] gi|41202705|ref|XM 372491.1 [41202705] gi|41202711 IreflXM 372493.1 [41202711] gi|41202716|ref|XM 372494.1 [41202716] gi|41149922|ref|XM 372496.1 [41149922] gi|41149943|ref|XM 372497.1 [41149943] gi|41149928|ref|XM_ 372498.1 [41149928] gi|41203861 JreflXM .372499.1 [41203861] gi|41203864|ref|XM_ 372500.1 [41203864] gi|41203866|ref|XM .372501.1 [41203866] gi|41203872Jref|XM_ 372502.1 [41203872] gi|41203875|reflXM .372503.1 [41203875] gi|41203877|reflXM .372504.1 [41203877] gi|41203880|ref|XM_ .372505.1 [41203880] gi|41203882|ref|XM_ 372506.1 [41203882] gi|41203885|ref|XM_ 372507.1 [41203885] gi|41203888|refjXM_ 372508.1 [41203888] gi|41203891|ref|XM_ 372509.1 [41203891] gi|41203895[ref|XM_ 372521.1 [41203895] gi|41203897|ref|XM 372522.1 [41203897] gi|41203900|ref|XM_ 372524.1 [41203900] giJ41203903(reflXM_ .;372525.1 [41203903] gi|41203909iref|XM_; 372527.1 [41203909]
gi|42660323|ref|XM .372528.2 |[42660323] gi|41203916Jref|XM .372532.1 1(41203916] gi|41203921 IreflXM .372534.1 [[41203921] gi|41203924|ref|XM .372535.1 [[41203924] gi|41203926|ref|XM .372536.1 j[41203926] gi|41150127|ref|XM .372542.1 1(41150127] gi|41150129|ref|XM .372543.1 |[41150129] gi|41150131 JreflXM .372544.1 1(41150131] gi|41150137|ref|XM 372547.1 |[41150137] gi|41150147|ref|XM .372548.1 1(41150147] gi|41150149|ref|XM .372549.1 j[41150149] gi|41150151 [reflXM .372550.1 1(41150151] gi|41150072Jref|XM 372553.1 [[41150072] gi|41150076|ref|XM .372555.1 |[41150076] gi|41150064|ref|XM 372556.1 ([41150064] gi|41205014Jref|XM..372559.1 ([41205014] gi|42660407|refjXM .372560.2 |[42660407] gi|41205021 IreflXM 372562.1 [[41205021] gi|41205026|ref|XM .372563.1 [[41205026] gi|41205033lref|XM .372565.1 |[41205033] gi|41205035(ref|XM .372566.1 1(41205035] gi|42660537|ref|XM 372568.2 |[42660537] gil41205042JreflXM .372569.1 |[41205042] gi|41205044|ref|XM .372570.1 1(41205044] gi|41205051 JreflXM 372573.1 ([41205051] gi]42660472|ref|XM 372574.2 [[42660472] gi|41205056(ref|XM 372575.1 1(41205056] gi|41205058|ref|XM 372576.1 [[41205058] gi|41205061 [reflXM 372577.1 j[41205061] gi|42660492|ref|XM.372578.2 1(42660492] gii42660494JreflXM .372579.2 j[42660494] gi|42660499|ref|XM~ 372580.2 [[42660499] giJ42660509|ref|XM 372581.2 |[42660509] gi(41205075 JreflXM 372583.1 j[41205075] gi|42660513|ref|XM .372584.2 |[42660513] gi|41205080|ref|XM 372585.1 1(41205080] gi|41150177|reflXM .372586.1 1(41150177] giJ41205627|reflXM_ 372588.1 ([41205627] gi|41205629Jref|XM 372589.1 1(41205629] gi|41205631 IreflXM.372591.1 [[41205631] giJ4266b598[reflXM 372592.2 1(42660598] giJ41150092|ref|XM~ 372593.1 |[41150092] gi|41150098Jref|XM .372596.1 [[41150098] gi|41150100|ref|XM 372597.1 1(41150100] gi|41150102|ref|XM .372598.1 [[41150102] gi|41150104|ref|XM_ 372599.1 ([41150104] gi|41150209Jref|XM .372601.1 |[41150209] gi|42661004|ref|XM_ 372606.2 (][42661004] gi|41150516|ref|XM_ 372607.1 j[41150516] gi|41150518|reflXM_ 372608.1 1(41150518] gi|41150520|ref|XM_ 372609.1 ([41150520] gi|41206146|ref|XM_ .372611.1 |[41206146] gi|41206154[ref|XM_ 372614.1 [[41206154] gi|4266074θjref|XM_ .:372615.2 j[42660740] gi|41150449|ref|XM_ .:372616.1 [[41150449] gi|41150451 [ref|XM_: 372617.1 |[41150451] gi|41150453|ref|XM_: 372618.1 |[41150453]
gi|41150461 |ref|XM .372622.11(41150461] gi|41150467|ref|XM .372625.1 |[41150467] gi|41150469|ref|XM .372626.1 |[41150469] gi|41150473Jref|XM .372628.1 1(41150473] gi|41150475|ref|XM .372629.1 |[41150475] gi|41150477|ref|XM .372630.1 j[41150477] gi|41150530|ref|XM .372631.1 |[41150530] gi|41150532|ref|XM 372632.1 1(41150532] gi|41150534|ref|XM 372633.1 ([41150534] gi|41150536|ref|XM .372634.1 1(41150536] gi|41150485Jref|XM 372637.1 ([41150485] gi|41150290|ref|XM .372638.1 ([41150290] gi]41206449|ref|XM 372639.1 1(41206449] gi|41206452(ref|XM .372640.1 ([41206452] giJ41206454(ref|XM 372641.1 |[41206454] gi|41206457|ref|XM 372642.1 [[41206457] gi|41206459|ref|XM 372643.1 1(41206459] gi|41206626[ref|XM 372644.1 |[41206626] giJ41150345|ref|XM. 372646.1 1(41150345] gii42660826[ref|XM 372647.2 1(42660826] gi|41150309|ref|XM 372648.1 j[41150309] gi|41150679|ref|XM 372650.1 j[41150679] gi|41150681 (reflXM 372651.1 1(41150681] gi|41150687|ref|XM 372654.1 j[41150687] gi|41150689Jref|XM .372655.1 |[41150689] gi|41150693|ref|XM 372657.1 ([41150693] gi[42661116Jref|XM .372658.2 1(42661116] gi|41150697|ref|XM .372660.1 ([41150697] gi|41150806|ref|XM 372662.1 1(41150806] gi|42661288|ref|XM 372663.2 |[42661288] gi|41150754Jref|XM .372666.1 j[41150754] gi|41206826|ref|XM .372668.1 j[41206826] gi|4120683θ[ref|XM 372669.1 [[41206830] gi|41206832|ref|XM 372670.1 1(41206832] gi|4120684θ(reflXM 372673.1 j[41206840] gi|41222862|ref|XM 372675.1 ([41222862] gi|41222865Jref|XM .372676.1 |[41222865] gi|42661207|ref|XM .372677.2 |[4266120η gi|41222869|ref|XM .372678.1 1(41222869] gi|41150571 IreflXM 372679.1 1(41150571]. gϊ|41150573Jref|XM .372680.1 |[41150573] gi[41150577|reflXM .372682.1 1(41150577] giJ41150836|ref|XM .372685.1 1(41150836] giJ41150904|ref|XM_ 372687.1 [[41150904] gi|41150908|ref|XM .372689.1 1(41150908] gi|41207734|ref|XM_ 372692.1 1(41207734] gi|41207737|ref|XM_ .372693.1 ([41207737] gi|41150948|ref|XM_ 372695.1 |[41150948] gi|4115095θ[ref|XM 372696.1 ([41150950] giJ41150934|ref|XM_ 372697.1 [[41150934] gi|41150936|ref|XM_ 372698.1 1(41150936] gi|41150924Jref|XM 372700.1 j[41150924] gi|41151051 IreflXM .372701.1 |[41151051] gi|41151053 JreflXM .372702.1 1(41151053] gi|41151055|ref|XM_ .372703.1 1(41151055} gi|41151057|ref|XM_ .372704.1 1(41151057] giJ41151059fref JXM_ 372705.1 |[41151059]
gi|41151152|ref|XM_372707.11[41151152] gi|41151154Jref|XM_372708.11[41151154] giJ42662028|ref|XM_372709.2J[42662028] giJ41151166Jref|X _372711.11[41151166] gi|41151168|ref|XM_372712.11[41151168] giJ41151172|reflXM_372714.l[[41151172] giJ41151174[ref|XM_372715.11[41151174] giJ42661988|ref|XM_372716.21(42661988] giJ41151184|ref|XM_372719.1 j[41151184] giJ41151188|ref|XM_372720.11[41151188] gi|41151190|ref|XM_372721.11[41151190] gi[42661572|ref|XM_372723.2|[42661572] gi|42661741 |ref|XM_372724.2([42661741] gil41208878|ref|XM_372726.1 |[41208878] giJ42661745Jref|XM_372727.2J[42661745] gi|42661747|ref|XM_372728.2l(42661747] giJ42661750|ref|XM_372730.2|[42661750] giJ41208888|ref]XM_372731.l [[41208888] gi|41208890|ref|XM_372732.1|[41208890] gi|41208893|ref|XM_372733.lj[41208893] gii41208898iref|XM_372735.lj[41208898] gi|41208903|ref|XM_372737.1 |[41208903] giJ41208906[ref|XM_372740.1 |[41208906] gi|41208909|ref|XM_372741.1|[41208909] giJ42661702(ref|XM_372745.2|[42661702] gi|41208923|ref|XM_372746.11[41208923] gi|41208928|ref|XM_372748.1|[41208928] gi|41208930|ref|XM_372749.1|[41208930] gi(41208933|ref|XM_372750.1 |[41208933] gi|41208936|ref|XM_372751.1|[41208936] gi|41208941 |ref|XM_372753.1|[41208941] gi[41208943ireflXM_372754.1 |[41208943] gi|41107709|ref|XM_372755.1|[41107709] gi]41058248JreflXM_372757.1|[41058248] gi|41058250|ref|XM_372758.11[41058250] gi|42655640|ref|XM_372759.2|[42655640] giJ41058258Jref|X _372760.1 j[41058258] gi|41114579|ref|XM_372761.11[41114579] giJ41114582|ref|XM_372762.1 [[41114582] gi|41114520|ref|XM_372763.11[41114520] gi|41114524|ref|XM_372764.11[41114524] gi|41114528|ref|XM_372765.11[41114528] giJ41104470|refJXM_372767.11[41104470] gi|41115343[ref|XM_372768.11[41115343] giJ41188267Jref|XM_372769.11[41188267] giJ41188270|ref|XM_372770.1 [[41188270] giJ41107695|ref|XM_372771.11[41107695] gi|41107699Jref|XM_372773.l[[41107699] gi|41107701 |ref|XM_372774.11[411077.01] gi|41109192JrefjXM_372775.1l[41109192] gi J41116237|ref|XM_372776.11[41116237] giJ41107477|ref|XM_372777.11[41107477] gi|41107479Jref|XM_372778.1 j[41107479] gi|41107483|reflXM_372779.11[41107483] gi|41107487|ref|XM_372780.1 |[41107487] gi|41107489|ref|XM_372781.11[41107489] gi|42655674|ref|XM_372784.2|[42655674]
gi|41103645 IreflXM 372785.11(41103645] giJ41115391 IreflXM .372788.1 |[41115391] gi[41115429 JreflXM 372789.11(41115429] gi|41107500 IreflXM 372792.1 [[41107500] giJ41106723 [reflXM 372795.1 ([41106723] giJ41188815 IreflXM .372796.11(41188815] giJ41188817 J]ref|XM 372797.11(41188817] gi|42656134 IreflXM 372798.2 |[42656134] gi|41188823 IreflXM 372799.11(41188823] gi|41188827 J|reflXM 372800.11(41188827] giJ41188832 IreflXM 372801.1 j[41188832] gi|41188836 IreflXM 372802.11(41188836] gi|42655953 IreflXM 372803.2 |[42655953] giJ41188108 IreflXM 372804.11(41188108] gι'J41188110 IreflXM 372805.1 j[41188110] giJ41188113 IreflXM .372806.11(41188113] gi!41188116 IreflXM .372807.11(41188116] giJ41117211 JreflXM. 372809.11(41117211] gi|41187877 JreflXM .372811.11(41187877] gi(41187879 IreflXM 372812.1 ([41187879] gi|41187882 IreflXM 372813.11(41187882] giJ41187887 IreflXM .372815.11(41187887] gi|41110139 IreflXM .372816.11(41110139] giJ41110142] IreflXM 372817.1 j[41110142] gi|41114126 JreflXM 372818.11(41114126] gi|42655696 IreflXM 372820.21(42655696] giJ41104582 IreflXM 372821.11(41104582] giJ41104585 IreflXM 372822.11(41104585] gi|41104588 IreflXM 372823.1 j[41104588] gi|41104591 JreflXM 372824.11(41104591] gi|42655704 (reflXM 372825.21(42655704] gi|41104601 IreflXM 372826.1 [[41104601] gi|41104605 IreflXM..372827.1 ([41104605] giJ41117239 [reflXM .372833.11(41117239] gi|41117281 IreflXM 372834.11(41117281] giJ41115747 IreflXM .372838.1 |[41115747] giJ41117459 IreflXM 372840.11(41117459] gi|41117528 IreflXM 372841.1 ([41117528] giJ42656159 [reflXM 372842.21(42656159] gi|41117534 (]reflXM .372843.1 [[41117534] gi|41117540 IreflXM 372845.11(41117540] gi|42662250 IreflXM .372858.2 ([42662250] giJ41151289 IreflXM 372859.1 [[41151289] gi|41151293 IreflXM .372863.11(41151293] giJ41209284 IreflXM 372864.11(41209284] giJ42662223 IreflXM 372866.2 ([42662223] gi|41151246 (reflXM 372869.1 |[41151246] giJ41151250 IreflXM..372870.11(41151250] gi|41151252 IreflXM.372871.11(41151252] giJ41151208 [reflXM.372873.11(41151208] gi|41151210 IreflXM 372874.11(41151210] gi(41151334 IreflXM.372875.1 [[41151334] gi|41151336 IreflXM .372876.1 ([41151336] gi|41151340 |ref|XM_ .372878.11(41151340] gi|41151354 JreflXM .372879.1 [[41151354] giJ41151362] IreflXM..372880.11(41151362] gi|42662381 IreflXM.372882.2 ([42662381]
gi|42662387|ref|XM_372884.2|[42662387] gi|41151407(ref|XM_372887.1|[41151407] gil41151411 [ref|XM_372889.11[41151411] gi|4120989liref|XM_372890.1|[41209891] gi|41209893Jref|XM_372891.lj[41209893] gi|41209895|ref|XM_372892.1|[41209895] gil41209898[reflXM_372893.1|[41209898] gi|4120990ljreflXM_372894.li[41209901] gi|41209903|ref|XM_372900.1|[41209903] gi|41209910|ref|XM_372901.l((41209910] gi|41209917|ref|XM_372905.1 [[41209917] giJ41151462|reflXM_372907.1|[41151462] gi|41151464|ref|XM_372908.1 [[41151464] gi|41124044|ref|XM_372910.1 [[41124044] gi|42656376 jref|XM_372911.2|[42656376] gi|41123925|ref|XM_372913.lj[41123925] giJ41118885JreflXM_372914.1 ([41118885] gi|41127715|ref|XM_372916.1 )[41127715] gi|41117724JreflXM_372917.11[41117724] gi[41117727Jref |XM_372918.11[41117727] gi|42656266|ref|XM_372919.2|[42656266] gi|41190512|ref|XM_372920.11[41190512] gi|41190514|ref |XM_372921.11(41190514] gi|41190516|ref |XM_372922.11[41190516] gi|41190519Jref|XM_372923.1 j[411905191 gi|41190521 |ref|XM_372924.11[41190521] gi|41190526|ref|XM_372926.1 [[41190526] gi|42656361|ref|XM_372927.2|[42656361] gi|41190531 |ref|XM_372928.11[41190531] gi|41190543|ref|XM_372933.1 ([41190543] gi|41124889Jref|XM_372940.11[41124889] giJ41124892(reflXM_372941.11[41124892] gi|41190805|ref|XM_372942.1 ][41190805] gi|41119886JreflXM_372945.11[41119886] giJ41119889Jref |XM_372946.1 [[41119889] gi|411 4831 jref|XM_372947.1 [[41124831] gi|42656214|ref|XM_372950.2|[42656214] gi|41118768|reflXM_372952.1 ([41118768] giJ41118771 |ref|XM_372953.11[41118771] gi(41118774[ref|XM_372954.1 [[41118774] gi|41125783Jref|XM_372957.1 j[41125783] gi|41125789|ref|XM_372958.11[41125789] giJ41125793(reflXM_372959.1 ([41125793] gi|41125803|ref |XM_372963.11[41125803] gi|41191502Jref|XM_372964.1|[41191502] giJ41 91505|ref[XM_372965.11[41191505] gi|41191510|ref|XM_372966.1|[41191510] giJ41191516|refJXM_372967.1 j[4119 516] gi[42656605ireflXM_372969.2i[42656605] gi|41191526 jref |XM_372970.11[41191526] gi|41191533|ref|XM_372973.11[41191533] gi|41126929|ref|XM_372974.11[41126929] gi]41126932|reflXM_372975.1 j[41126932] gi|42656621|ref|XM_372977.2[[42656621] gil41191872|ref|XM_372978.1|[41191872] gi|41191875|ref |XM_372979.1 [[41191875] gi|41191877|ref [XM_372980.11[41191877]
gi|41134364 ref|XM_ 372981.11(41134364] giJ41134572 ref|XM_ 372982.1 1(41134572] gi|41134599 ref|XM_ 372983.1 |[41134599] gi[41193290 reflXM..372984.1 ([41193290] giJ41193293 ref|XM_ 372985.1 ([41193293] gi|41193295 reflXM.372986.1 ([41193295] gi|41193297 reflXM.372987.1 1(41193297] giJ41193302] reflXM..372988.1 1(41193302] giJ41193313 reflXM.372991.1 1(41193313] gi|41193317 reflXM..372992.1 1(41193317] gi[42656658 reflXM 372993.2 ([42656658] gi|41137154 reflXM 372995.1 1(41137154] gi|41137157 reflXM .372996.1 [[41137157] gi|42656896 reflXM 372997.2 ([42656896] giJ41194224 reflXM .372998.1 j[41194224] gi|41194232 reflXM .373001.1 1(41194232] giJ41194237| ref|XM 373002.1 1(41194237] gi|41194240] reflXM.373003.1 |[41194240] giJ41194251 reflXM.373004.1 1(41194251] gi|41194254] reflXM.373005.1 [[41194254] giJ41194257 reflXM.373006.1 1(41194257] giJ41144894 reflXM..373007.1 |[41144894] giJ41146811 reflXM 373009.1 1(41146811] gi|41146880 reflXM 373016.1 j[41146880] gi|41146890 reflXM..373021.1 1(41146890] giJ41146892 reflXM .373022.1 |[41146892] gi|41146821 reflXM .373025.1 1(41146821] giJ41146809] reflXM 373026.1 [[41146809] gi|41146742 reflXM .373027.1 1(41146742] giJ41146744] reflXM. 373028.1 [[41146744] giJ41195099 reflXM .373029.1 1(41195099] gi|42656935 reflXM 373030.2 |[42656935] gi|41195109 reflXM .373031.1 |[41195109] gi|41195115 reflXM 373033.1 ([41195115] gi|42657002 reflXM. 373035.2 j[42657002] giJ41146728 reflXM .373036.1 1(41146728] gi|41146730 reflXM .373037.1 [[41146730] giJ41146732 reflXM .373038.1 1(41146732] gi|41146734 reflXM .373039.1 ([41146734] giJ41146736 reflXM .373040.1 1(41146736] gi|41146782 reflXM. 373041.1 ([41146782] gi|41146784 reflXM .373042.1 [[41146784] gi|41146786 reflXM 373043.1 [[41146786] gi|41195532] reflXM 373053.1 1(41195532] giJ41195541 reflXM. 373056.1 [[41195541] giJ41195544 reflXM..373057.1 |[41195544] giJ41195547 reflXM .373058.1 1(41195547] gi|41195551 reflXM .373059.1 |[41195551] giJ41147133 reflXM .373061.1 [[41147133] giJ41147157] reflXM .373073.1 [[41147157] gi|41147161 reflXM .373075.1 [[41147161] gi|41147163 reflXM .373076.1 1(41147163] giJ41147165 reflXM .373077.1 ([41147165] gi|41147167 reflXM .373078.1 1(41147167] giJ41147169 reflXM _373079.1 ([41147169] gi|41147171 reflXM _373080.1 [[41147171] giJ41147097 reflXM. 373082.1 ([41147097]
141147101 Irefl jXM .373084.1 |[41147101]
[41147257 |ref J|XM 373085.1 |[41147257]
141147261 iref J|XM .373087.1 |[41147261]
J41147265 Iref JjXM..373088.1 1(41147265]
[41147209 iref J|XM .373089.1 1(41147209]
141147211 Irefl JXM 373090.1 j[41147211]
(41147213 iref j|XM .373091.1 1(41147213]
[41147217 jref JjXM 373092.1 j[41147217]
(41147219 iref J|XM..373093.1 1(41147219]
141147068 Irefl JXM .373094.1 1(41147068]
[41147070 Irefl JXM..373096.1 1(41147070] .
(41147003 Irefl JXM .373097.1 |[41147003]
J41146960 Irefl JXM .373098.1 1(41146960]
141146964 Irefl JXM 373099.1 |[41146964]
141146966 Irefl JXM..373100.1 1(41146966]
141146968 Iref J|XM .373101.1 |[41146968]
|41146974 Irefl |XM..373103.1 1(41146974]
141147247 j]ref JjXM .373106.1 |[41147247]
J41147249 Irefl JXM .373107.1 j[41147249]
(42657506 Iref JjXM..373109.2 1(42657506]
J41147433 Iref JjXM .373156.1 j[41147433]
J41147419 jref JjXM..373157.1 |[41147419]
[41199267 jref J|XM 373170.1 1(41199267]
(41199274 iref J|XM .373171.1 |[41199274]
[41199289 [ref J|XM..373178.1 j[41199289]
141199294 jref J|XM .373180.1 ([41199294]
J41199320 jref JlXM .373190.1 1(41199320]
141147581 Iref JjXM 373209.1 1(41147581]
141147583 jrefl JXM .373210.1 [[41147583]
141147589 Irefl JXM..373214.1 [[41147589]
|41147599 | refl JXM .373219.1 |[41147599]
141147462] iref JlXM .373223.1 |[41147462]
141147534 Irefl JXM..373224.1 ([41147534]
J41148509 jref J|XM 373227.1 1(41148509]
J41148523 Irefl JXM .373233.1 |[41148523]
(41148525 jref JjXM 373234.1 1(41148525]
[41148601 jref JjXM 373236.1 |[41148601]
J41148605 iref JlXM .373238.1 j[41148605]
J41148607 Irefl JXM 373239.1 1(41148607]
(41148611 iref J|XM .373241.1 |[41148611]
[41148613 | ref J |XM 373242.1 1(41148613]
141148615 Irefl JXM .373243.1 |[41148615]
(41148557] Irefl jXM 373245.1 1(41148557]
[41148453 jref J|XM 373246.1 ([41148453]
(41148638 jref j|XM 373248.1 [[41148638]
141148461 Irefl JXM .373249.1 ([41148461]
J41148741 Irefl JXM 373250.1 j[41148741]
|41148745] lref J|XM 373252.1 1(41148745]
141148747] |ref J|XM .373253.1 |[41148747]
J42658968 jrefl JXM .373255.2 ([42658968]
J41148675 Irefl JXM 373257.1 [[41148675]
141148679 lref J|XM 373259.1 ([41148679]
J42658917 Irefl JXM .373260.2 |[42658917]
[41148687 Irefl JXM 373263.1 1(41148687]
(41148479 Irefl JXM 373265.1 |[41148479]
[41148481 Irefl JXM 373266.1 1(41148481]
[41148622 Iref JjXM 373270.1 1(41148622]
gi|41148998 IreflXM 373275.11(41148998] gi|41149000 |ref|XM_ 373276.1 ([41149000] gi|41149002 (reflXM..373277.1 [[41149002] gi|41149010 [reflXM 373281.11(41149010] giJ41149014] IreflXM..373283.1 |[41149014] giJ41149016 JreflXM 373284.1 [[41149016] giJ42659197 IreflXM..373287.2 |[42659197] giJ41149020 [reflXM 373288.1 [[41149020] giJ41149022 IreflXM 373289.11(41149022] giJ41149024 IreflXM. 373290.11(41149024] gi|41148845 (reflXM 373295.11(41148845] gi|41148867j IreflXM 373297.1 [[41148867] giJ41148797] |ref|XM 373298.1 ([41148797] giJ41148801 ]ref|XM 373300.11(41148801] gi|41148803 IreflXM 373301.11(41148803] giJ41148805 IreflXM. 373302.1 |[41148805] gi|41148807] [reflXM 373303.11(41148807] gi|41148809 IreflXM..373304.1 |[41148809] giJ42659029 [reflXM .373305.21(42659029] gi|42659283 IreflXM. 373308.2 j[42659283] gi|41222683 IreflXM .373309.11(41222683] gi|41222686 IreflXM .373310.1 |[41222686] gi|41222688 IreflXM. 373311.11(41222688] gi|41222692 |ref|XM..373312.1 ([41222692] gi[42659256 JreflXM..373313.21(42659256] giJ41222707 [reflXM 373314.11(41222707] giJ41222711 IreflXM. 373315.11(41222711] giJ41222713 JreflXM..373316.1 [[41222713] giJ41222716 IreflXM 373317.11(41222716] gi|41222722 IreflXM..373319.1 |[41222722] gi|41148823 [reflXM .373320.1 j[41148823] gi|41057827 IreflXM 373322.11(41057827] gi|41148885 IreflXM .373324.1 [[41148885] giJ41148887 IreflXM..373325.11(41148887] giJ41148889 (reflXM 373326.11(41148889] giJ41148895 |ref|XM 373327.1 |[41148895] gi|41148897] IreflXM..373328.11(41148897] gi|41148899 [reflXM..373329.1 |[41148899] giJ41148905 JreflXM 373331.11(41148905] giJ41148935 IreflXM .373334.1 ([41148935] giJ41151796 (reflXM .373335.1 |[41151796] gi|41151815 IreflXM 373336.1 j[41151815] giJ41151821 IreflXM .373337.11(41151821] gi|41151614 lref|XM 373338.1 ([41151614] giJ41151616 IreflXM .373339.1 [[41151616] giJ41151618 JreflXM..373340.11(41151618] giJ41151620 IreflXM .373341.1 |[41151620] gi|41210573 IreflXM .373343.11(41210573] gi|41151504 [reflXM 373344.1 |[41151504] giJ41151506 IreflXM 373345.11(41151506] giJ41151508 IreflXM 373347.1 |[41151508] gi|41151528 |ref|XM .373348.11(41151528] gi|41151530 |ref|XM..373349.1 ([41151530] gi|41151532 JreflXM .373350.1 [[41151532] gi|41151520 IreflXM .373351.11(41151520] giJ41151522 |ref|XM .373352.11(41151522] giJ41151565] IreflXM. 373353.1 [[41151565]
gi|41151567|ref |XM_373354.11[41151567] giJ41151569|reflXM_373355.1 |[41151569] giJ42662625[ref|XM_373356.2|[42662625] gi|41151573|ref|X _373357.11[41151573] gi|42662588(ref|XM_373358.2([42662588] gi|41210298|ref|XM_373359.1 |[41210298] giJ41210305|ref|XM_373362.1 |[41210305] giJ41151691 (ref|XM_373365.1 [[41151691] gi|41151693|ref|XM_373366.1 |[41151693] gi|41151695|ref|XM_373367.11[41151695] giJ41151697Jref|XM_373368.11[41151697] gi|41151699|reflXM_373369.l i[41151699] giJ41151649Jref|XM_373370.l j[41151649] gi|41151653|ref|XM_373372.11[41151653] giJ41151655(reflXM_373373.1 [[41151655] gi|41151659|ref|XM_373374.1 |[41151659] gi|41210462|ref|XM_373378.11[41210462] giJ41151667|ref|XM_373381.1 j[41151667] gi|41151705|ref|XM_373382.1 [[41151705] gi|41151719|reflXM_373384.1 |[41151719] gi(41151725|ref|XM_373388.1 [[41151725] giJ41151758Jref JXM_373391.1 j[41151758] gi|4115175θ[ref|XM_373395.l j[41151750] gi|41151754Jref|XM_373397.1 ([41151754] gi|41151756|ref|XM_373398.1 |[41151756] giJ41151743|ref|X _373399.1 ([41151743] gi|42661997iref|XM_373407.2([42661997] gi|42659665|ref|XM_373413.2[[42659665] gi(42657614Jref|XM_373419.2[[42657614] gi|42660118[ref|XM_373431.2|[42660118] gi|42661982|reflXM_373433.2|[42661982] gi(42657277[ref|XM_373440.2|[42657277] giJ41149241 [ref|XM_373444.1 [[41149241] gi|41149243Jref|XM_373445.11[41149243] gi|41149245|ref|XM_373446.1 [[41149245] gi[41149255(ref|XM_373447.11[41149255] giJ41149348|ref|XM_373448.1 ([41149348] gi|41149352|ref|XM_373449.1 [[41149352] giJ41149354|ref|XM_373450.11[41149354] gi|42659552|ref|XM_373451.2|[42659552] gi|41149383Iref|XM_373452.11(41149383] giJ41149389[ref|XM_373453.1 ([41149389] giJ41149397Jref|XM_373454.1 ([41149397] gi|42659404Iref|XM_373456.2|[42659404] giJ41149085(ref|XM_373460.11[41149085] gi|41149304|ref|XM_373461.11[41149304] gi|41149314[reflXM_373463.11[41149314] gi[42659519iref|XM_373464.2|[42659519] gi|4265952l [ref|XM_373465.2|[42659521] giJ41149324JreflXM_373466.11[41149324] gi|41149118(ref|XM_373468.11[41149118] gi[41149126JreflXM_373469.1 ([41149126] giJ41149132|ref(xM_373470.11[41149132] gi|41149136|ref|XM_373471.11[41149136] gi|41149144|ref|XM_373472.11[41149144] gi|41149153|ref|XM_373474.11[41149153] giJ41149156iref|XM_373475.l j[41149156]
141149172 | refJjXM 373477.11(41149172] 141149233 IrefJjXM..373478.1j[41149233] 141149411 jrefJjXM 373479.1|[41149411] J41149093 IrefJjXM .373480.1 [[41149093] |42659322 jreflJXM..373481.2j[42659322] |41149594 IreflJXM 373483.11(41149594] (41149596 IrefJjXM 373484.1[[41149596] 141149603 jrefJjXM .373485.1|[41149603] [41149649 jrefJjXM.373486.11(41149649] (41149651 ireflJXM.373487.11(41149651] 142659791 IrefJ|XM .373488.2[[42659791] 141149660 jrefJ|XM 373489.1[[41149660] [41200852 | refJ |XM 373490.1[[41200852] 141200860 irefJlXM .373491.1([41200860] (41200870 lrefJ|XM .373492.1[[41200870] [41200889 jrefJ|XM..373494.1|[41200889] (41200897 jrefJ|XM 373495.1|[41200897] 141200900 JrefJlXM 373496.1|[41200900] [42659836 IrefJjXM .373497.2([42659836] (41200904 ]refJ|XM.373498.1 [[41200904] [41200910 jrefJ|XM .373499.11(41200910] (41149516 |refJjXM.373500.1([41149516] [41200333 IrefJlXM .373502.1|[41200333] (41200338 IreflJXM 373503.11(41200338] [41200362 IreflJXM .373505.1 |[41200362] (41200365 IreflJXM..373506.1[[41200365] [42659693 jrefJ|XM.373507.2([42659693] (42659738 Irefl(XM .373509.21(42659738] 141149571 I refJ |XM.373511.1[[41149571] 141149573 jrefJ|XM .373512.11(41149573] 141149435] IreflJXM.373513.1|[41149435] |41149444 IreflJXM .373515.1|[41149444] 141149461 IreflJXM..373516.1|[41149461] (41149463 jreflJXM.373517.1([41149463] 141149467] IrefJjXM .373518.1|[41149467] 141149471 |refJ|XM .373519.11(41149471] 141149473 IreflJXM .373520.1 |[41149473] 141149857 jrefJ|XM..373521.11(41149857] [41149781 IreflJXM.373522.1([41149781] (41149799 IreflJXM .373524.11(41149799] 141149815 IreflJXM .373525.1|[41149815] [41202053 jrefJ|XM.373527.11(41202053] 141202060 jrefJ]XM .373529.1[[41202060] J42659932 [refJjXM.373530.2|[42659932] 141149752 IreflJXM .373531.1([41149752] |41149765 jrefJ|XM.373533.1 |[41149765] [41201706 IreflJXM.373534.1([41201706] (41201708 jrefJ|XM.373535.1|[41201708] 141201730 IreflJXM .373537.1[[41201730] 141201741 IreflJXM .373538.1([41201741] J41149683 jrefJ|XM..373539.11(41149683] [41149693 jreflJXM..373540.1j[41149693] (41149697 |refJ|XM 373541.11(41149697] 141149705 |refJ|XM 373542.1[[41149705] 141149707 IreflJXM .373543.11(41149707] (42659909 IreflJXM 373544.2([42659909] 141149868 jrefJjXM 373545.1([41149868]
gi|41149872[ref|XM_373546.1 [[41149872] gi|41149878|ref|XM_373547.1 j[41149878] giJ4114988θjref|XM_373548.1 j[41149880] gi|42660092|ref|XM_373549.2J[42660092] gi|41149884|ref|XM_373550.11[41149884] gi|41149833|ref|XM_373553.1 ([41149833] gi|41149841 |ref|XM_373554.1 j[41149841] gi|41149843|ref|XM_373555.lj[41149843] gi|41149771 Iref|XM_373556.11[41149771] gi|41202572|ref|XM_373557.11[41202572] gi|41202576|ref|XM_373558.11[41202576] gi J41202581[reflXM_373559.1 ([41202581] giJ41202591 jref|XM_373560.1 j[41202591] gi|4266015ljref|XM_373561.2|[42660151] giJ41202610IreflXM_373562.lj[41202610] gi|41202613|ref|XM_373563.lj[41202613] gi|41202642[ref|XM_373564.lj[41202642] giJ41202658irefixM_373566.l[[41202658] gi|4120266θjref|XM_373567.l([41202660] gi|41202668|ref|XM_373568.1|[41202668] giJ41202670|ref|XM_373569.11[41202670] gi|41202681 |ref|XM_373570.11[41202681] gi|42660111 |reflXM_373571.2|[42660111] gi|41149904Jref|XM_373572.1 [[41149904] gi|41149906|ref|XM_373573.1 [[41149906] gi|41149908|ref|XM_373574.11[41149908] giJ41149918Iref|XM_373575.li[41149918] giJ41149933|ref|XM_373576.1 ([41149933] giJ41149935Jref|XM_373577.1 j[41149935] giJ41149939Jref|X _373578.lj[41149939] gi|41149951 |ref|XM_373580.11[41149951] giJ41149955Jref|XM_373581.1 [[41149955] gi|41149963|ref|XM_373583.11[41149963] giJ41149969Jref|XM_373585.1|[41149969] gi|41149977|ref|XM_373587.11[41149977] gi|41149981 jref|XM_373588.11[41149981] gi|41203706|ref|XM_373593.1|[41203706] giJ41203716|refjXM_373594.l([41203716] giJ41203719Jref|XM_373595.1|[41203719] giJ41203727|ref|XM_373596.1|[41203727] gi|41203763|ref|XM_373599.1|[41203763] gi|41203766|ref|XM_373600.1 ([41203766] giJ41203769(ref|XM_373601.lj[41203769] gi|41203785|ref|XM_373602.lj[41203785] giJ41203788(ref|XM_373603.1|[41203788] gi|42660347|ref|XM_373604.2J[42660347] gi|41203796|ref|XM_373605.1|[41203796] gi|42660348Jref|XM_373606.2|[42660348] giJ412038 θ[reflXM_373607.1|[41203810] gi|41203815|ref|XM_373608.lj[41203815] gi|41203818|ref|XM_373609.1|[41203818] gi|41203821 |ref|XM_373610.lj[41203821] gi|41203826Jref[XM_373611.1 [[41203826] gi|42660612|ref|XM_373615.2|[42660612] giJ42660635jref|XM_373616.2][42660635] gi|41150186|ref|XM_373617.1|[41150186] gi|41150058|ref|XM_373622.1|[41150058]
gi[41150196|ref[XM_373623.1 [[41150196] giJ42660398|reflXM_373624.2J[42660398] giJ42660434Jref|XM_373625.2J[42660434] gi]41150026|ref|XM_373626.1I(41150026] gi|41204872|ref|XM_373627.1 |[41204872] giJ41204912JrefjXM_373628.lj[41204912] gi[41204932|ref |XM_373630.1 ([41204932] gi|41204939JreflXM_373631.1 ([41204939] gi|42660481|ref|XM_373632.2([42660481] gi|41204956(reflXM_373633.l i[41204956] giJ41204969|ref|XM_373634.l j[41204969] gi|41204976|ref|XM_373635.lj[41204976] giJ42660505JreflXM_373636.2J[42660505] giJ41204983|ref[XM_373637.lj[41204983] giJ42660511 |ref|XM_373638.2J[42660511] gi|41204998|ref|XM_373639.11[41204998] gi|42660646Jref|XM_373641.2J[42660646] gi|41150050|reflXM_373645.1 j[41150050] gi|41150052|ref|XM_373646.1 [[41150052] gi|41150054|ref|XM_373647.1 ([41150054] gi|41205612|ref|XM_373648.1 |[41205612] gii42660586|ref|XM_373649.2J[42660586] giJ41205622iref|XM_373650.lj[41205622] gi|41150215|ref|XM_373653.11(41150215] gi[4266100θiref|XM_373654.2[[42661000] gi|41150491 |ref|XM_373655.11[41150491] gi|41150499 jref JXM_373657.1 j[41150499] giJ41150508(ref|XM_373659.li[41150508] gi|41150319Jref|XM_373660.1 |[41150319] giJ42660860]ref|XM_373662.2i[42660860] giJ41150235Jref|XM_373666.1 |[41150235] gi(41150239|refjXM_373667.li(41150239] giJ41150251 [ref|XM_373668.1 ([41150251] gi|42660728|ref|XM_373669.2|[42660728] giJ41206117Jref|XM_373670.lj[41206117] gi|41206133lref|XM_373671.1 |[41206133] giJ41150389|ref|XM_373672.11[41150389] gi|41150411|ref|XM_373673.1 [[41150411] giJ42660958|reflXM_373674.2J[42660958] giJ42660959|ref|XM_373675.2J[42660959] gii42660973(reflXM_373676.2i[42660973] gi|42660978|ref|XM_373677.2|[42660978] gi|42661022iref|XM_373679.2J[42661022] giJ41150349Jref|XM_373681.li(41150349] giJ41150351 |ref|XM_373682.1 j[41150351] gi|41150286Jref|XM_373683.11[41150286] giJ4115028δjref |XM_373684.11[41150288] gi|42660767|ref|XM_373685.2|[42660767] giJ41150280|ref|XM_373686.l([41150280] giJ41150284Jref|XM_373687.1 [[41150284] gii41206406Jref|XM_373688.1 |[41206406] gi|41206408|ref|XM_373689.1 |[41206408] gi|4120641 ljref[XM_373690.l [[41206411] gi(41206416|ref|XM_373691.1|[41206416] gi|41150361 jref|XM_373692.1|[41150361] giJ42660903|ref|XM_373693.2([42660903] gi|41150373Jref |XM_373694.11[41150373]
141150381 Irefl JXM .373695.1 |[41150381] 141150383 Irefl JXM.373696.11(41150383] (41150385| jref J|XM .373697.1 |[41150385] 141206621 lref J|XM 373698.11(41206621] 141206624 jrefl JXM 373699.1 |[41206624] 141150339 Jref JjXM.373700.11(41150339] |42660886 jrefl JXM 373701.2 |[42660886] (41150299 Jref J|XM 373702.11(41150299] |42660845 Irefl JXM.373703.2 |[42660845] |42660846 jref JjXM .373704.21(42660846] 141150784 Irefl JXM 373705.1 [[41150784] [41150590 jrefl JXM 373707.1 ([41150590] 141150592 jref JjXM .373708.1 |[41150592] (41150603 Irefl JXM 373709.1 [[41150603] [41150624 jrefl JXM..373711.1 |[41150624] 141150636 Irefl JXM 373713.11(41150636] 141150649 Iref JjXM 373714.1 |[41150649] 141150653 jref JjXM..373715.11(41150653] 142661130] Irefl JXM .373717.2 [[42661130] 141150677] Jref JjXM 373718.11(41150677] 142661285 Jref JjXM 373719.2 ([42661285] [42661299 | ref J]XM .373721.21(42661299] 141150720 jref JjXM..373722.1 |[41150720] 141150730 Irefl JXM 373723.11(41150730] 141150732 Irefl JXM .373724.1 |[41150732] |41150818 jref J|XM .373726.1 ([41150818] 141150820 |ref J|XM 373727.1 |[41150820] 141150705 |ref J|XM..373728.11(41150705] [42661149 jref J|XM 373730.2 [[42661149] 142661150 Irefl JXM .373731.21(42661150] J42661159 Irefl JXM..373733.2 ([42661159] 142661163 jref J|XM 373734.2 [[42661163] 141222806 jref J|XM .373735.1 |[41222806] (41222823 Irefl JXM 373737.1 |[41222823] [41222846 jref J|XM .373740.1 ([41222846] (41222854 Irefl JXM 373741.1 |[41222854] 142661218 jref J |XM .373742.2 |[42661218] J41150540 Irefl JXM 373744.11(41150540] 141150542 Irefl JXM .373745.11(41150542] J41150544] Irefl JXM.373746.1 |[41150544] 141150546 jref J|XM .373747.11(41150546] 141150548 Iref JjXM..373748.1 |[41150548] 141150558 jref JjXM .373749.11(41150558] 141150563 Jref JjXM .373750.1 [[41150563] 141150565 jref J]XM.373751.11(41150565] 142661270 jrefl JXM.373753.2 ([42661270] [41150766 Irefl JXM .373754.11(41150766] (41150768 Irefl JXM .373755.1 [[41150768] 141150772 jref J|XM.373756.11(41150772] 142661279 jref JjXM.373757.21(42661279] 141150780 irefl JXM .373758.11(41150780] (41150583 Irefl JXM .373760.1 |[41150583] [41150585 jrefl JXM..373761.11(41150585] (42661307 Irefl JXM..373763.2 ([42661307] J41150838 Irefl JXM..373765.1 |[41150838] J41150846 Irefl JXM..373766.11(41150846] J42661325 Irefj IXM 373767.2 |[42661325]
gi|41150858|ref|XM 373769.11[41150858] gi|41150866|ref|XM_373770.11[41150866] gi|41150868|ref|XM_373771.11[41150868] gi|41150876|ref|XM_373772.11[41150876] gi|41150880|ref|XM_373773.1|[41150880] gi|41150893ireflXM_373774.l[[41150893] giJ41150896Jref|XM_373775.1 ([41150896] gi|41207716|ref|XM_373776.1|[41207716] giJ41150940|ref|XM_373778.11[41150940] giJ41150944Jref|XM_373779.11[41150944] gi|41150946|ref|XM_373780.l([41150946] gi|41150932|ref|XM_373782.11[41150932] gi|4 150914|ref|X _373783.1 J[41150914] gi|41150916JrefjX _373784.l([41150916] giJ42661839 jref (XM_373785.2|[42661839] gi|41151028Jref|XM 373786.1 ([41151028] giJ4115103θjref|XMl373787.1|[41151030] giJ42661867JreflXM_373788.2J[42661867] giJ41151044Jref|XM_373789.1 j[41151044] gi J41151047|ref|XM_373790.1 ([41151047] giJ41151194|ref |XM_373792.1 ([41151194] gi|41151072|ref|XM_373793.1|[41151072] gi|41151108Jref|XM_373795.1 j[41151108] gi|41151115|ref|XM_373796.11[41151115] giJ4115 121 |ref|XM_373797.11[41151121] gi|42662022|ref|XM_373798.2|[42662022] gi[41150958[ref|XM_373799.1|[41150958] gi|41150960|ref|XM_373800.lj[41150960] gi|41150962|ref|XM_373801.1|[41150962] gi|41150964|ref|XM_373802.11[41150964] gi|41150981 |ref|XM_373803.11[41150981 ] gi|42661623Jref|XM_373804.2|[42661623] gi|4266163ljref|XM_373805.2|[42661631] gi]41208726|ref|XM_373808.1|[41208726] giJ41208747Jref|XM_373809.1 ([41208747] giJ42661669|ref|XM_373810.2|[42661669] gi|41208825Jref|XM_373811.1 ([41208825] giJ41208862|ref|XM_373812.lj[41208862] gi|41114913|ref|XM_373814.1|[41114913] giJ41114916|reflXM_373815.1 ([41114916] gi|41114967|ref|XM_373817.1|[41114967] gi|41115071 |ref|XM_373818.1 j[41115071] gi|41107715|ref|XM_373820.1 ([41107715] gi|42655847[reflXM_373821.2J[42655847] gi|41107705|ref|XM_373822.1 [[41107705] gi|41107707|ref|XM_373823.1 [(41107707] gi|41109282|ref|XM_373824.11(41109282] gi|41058214[ref|XM_373825.lj[41058214] gi|41058232Jref|XM_373826.li[41058232] gi|42656066|ref|XM_373827.2|[42656066] gi|41111276|ref|XM_373828.11[41111276] giJ41188259(ref|XM_373831.11[41188259] giJ41107683|ref|XM_373832.1 [[41107683] gi|41107691 [ref|XM_373833.1 j[41107691] gϊ)41109026|ref|XM_373835.11[41109026] gi|41109030|ref|XM_373836.lj[41109030] gi|41109033|ref|XM_373837.11[41109033]
|41116231 Irefl JXM .373838 |[41116231] 141107409 Irefl JXM..373839 1(41107409] (41107411 Irefl JXM 373840 |[41107411] |42655758 Irefl JXM..373843 1(42655758] 141107456 jref JjXM .373844 j[41107456] 141107464] Irefl JXM..373845 |[41107464] I42655786 |ref J|XM .373846 [[42655786] [42655787 |ref J|XM 373847 |[42655787] (41058273 Iref JjXM .373848 1(41058273] J41058277 Iref JjXM..373849 |[41058277] J41058279 jref JjXM..373850 ([41058279] 141188542 | refl JXM 373852 [[41188542] J41103622 iref JjXM..373853 1(41103622] 141103625 jrefl JXM 373854 ([41103625] 141104162 |ref J]XM .373855 1(41104162] J42655583 jrefl JXM 373856 [[42655583] 141057820 Iref JjXM..373857 |[41057820] 141113275 jrefl JXM..373858 ([41113275] 142656026 |ref J|XM 373859 [[42656026] |42655798 jref JjXM 373862 |[42655798] J41115522 |ref JjXM..373864 ([41115522] 142655813 jrefl JXM..373865 [[42655813] 142655710 Irefl JXM 373866 1(42655710] 141106693 |ref JjXM..373867 |[41106693] (42655718 jrefl JXM 373868 ([42655718] 141188793 Irefl JXM .373870 |[41188793] [41188811 Irefl JXM 373871 1(41188811] 141112045] iref J|XM .373872 [[41112045] 141112066 jref J|XM..373873 ([41112066] (41188084 Irefl JXM .373875 1(41188084] 141188088 Irefl JXM .373876 |[41188088] 141188092 Irefl JXM .373877 1(41188092] J41188103 Irefl JXM..373878 |[41188103] J41112850 jref J|XM.373879 ([41112850] 141112860 jref J|XM .373880 |[41112860] 141188450 jref JlXM .373881 1(41188450] J41188458 jref J|XM 373882 |[41188458] 141107517 Irefl JXM .373883 [[41107517] J42655597 jref J|XM..373884 ([42655597] (41058112 |ref JjXM .373885 [[41058112] J42655612 Iref JjXM .373886 1(42655612] [42655912 | refl JXM 373889 j[42655912] J41110098 Irefl JXM .373890 [[41110098] 141110136 Irefl JXM..373893 |[41110136] (41058266 jref J|XM 373894 ([41058266] [41114091 Irefl jXM .373896 j[41114091] (41114103 jref J|XM 373898 1(41114103] [42662258 Irefl jXM .373903 ([42662258] (42662259 Irefl JXM 373904 [[42662259] J41151269 jrefl JXM..373905 1(41151269] 141151271 jrefl JXM 373906 1(41151271] 141151273 Irefl JXM .373907 1(41151273] J42662272 |ref J|XM .373908 j[42662272] [41151279 Irefl JXM .373909 |[41151279] 141151281 |ref J|XM 373910 ([41151281] J41151283 Irefl JXM 373911 |[41151283] 141151295 |ref J|XM 373913 [[41151295]
gi|41151214|refjXM_373914.1 |[41151214] giJ41151223|reflXM_373915.1 ([41151223] gi|41151232|ref|XM_373916.1 [[41151232] gi|41151236|ref|XM_373917.l j[41151236] gi|41151196|ref |XM_373920.1 j[41151196] gi|42662080|ref|XM_373921.2|[42662080] gi|4115120θjrefJXM_373922.1 j[41151200] giJ41151204|ref|XM_373924.1 [[41151204] gi|42662309iref|XM_373925.2|[42662309] gi|4115131θ(ref|XM_373926.1 |[41151310] gi|41151312|ref|XM_373927.1 |[41151312] gi|41151314|ref|XM_373928.1 |[41151314] gi|42662325|ref|XM_373930.2J[42662325] giJ41151326Jref]XM_373931.1 j[41151326] gil42662329|ref|XM_373932.2|[42662329] gi|4115133θjref|XM_373933.11[41151330] giJ4120934l [ref|XM_373936.lj[41209341] gi|41151346|ref|XM_373938.1 |[41151346] gi|42662363Jref|XM_373939.2|[42662363] gi|41151350|ref|XM_373940.l ([41151350] giJ42662365|ref|XM_373941.2|[42662365] gi|41151476|ref|XM_373942.1 j[41151476] gi|41151368Jref)XM_373943.1 ([41151368] giJ41151374|ref|XM_373944.l[[41151374] gil42662398|ref|XM_373945.2|[42662398] gi|41151429Jref|XM_373947.11[41151429] gi|41209810[ref|XM_373949.1|[41209810] gi(41209817Jref|XM_373950.l([41209817] gi[41209831 jref|XM_373951.1 j[41209831] gi|41209834iref|XM_373952.11[41209834] giJ41209837|ref|XM_373953.1 |[41209837] giJ41209844iref|XM_373954.lj[41209844] gi[41209853JreflXM_373955.l [[41209853] gi|41209858|ref[XM_373957.1|[41209858] giJ41209860Jref|XM_373958.l [[41209860] gii4266247θiref|XM_373959.2J[42662470] gi|41209875(ref|XM_373962.11[41209875] gi|41151439JrefixM_373964.lj[41151439] giJ41151441 |ref|XM_373965.1 [[41151441] gi|41151443Jref|XM_373966.11[41151443] giJ41151450|refjXM_373967.l ((41151450] gi|41151452|ref|XM_373968.1|[41151452] giJ41151454|ref(xM_373969.lJt41151454] gi|41151458|ref|XM_373970.1 [[41151458] gi|42662524|ref|XM_373971.2|[42662524] gi|42656378|ref|XM_373972.2|[42656378] gi|41123975|ref|XM_373973.11(41123975] gii41127706|ref|XM_373974.1|[41127706] gi|41123794|ref|XM_373975.11[41123794] giJ41123815Jref|XM_373977.11[41123815] giJ41123819Jref|XM_373978.l i[41123819] gi|41123846Iref|XM_373979.11(41123846] gi|41190370|ref|XM_373980.11[41190370] gi|42656292iref|XM_373981.2J[42656292] gi|41190392[ref|XM_373982.11[41190392] giJ41190395(ref|XM_373983.1 [[41190395] gii41190399iref|XM_373984.l j[41190399]
gi|42656303|ref|XM_373985.2|[42656303] gi|4119041δjref JX _373986.1 j[41190418] gi(41190455|ref|XM_373989.1 |[41190455] gi[41190462|refixM_373990.1 [[41190462] gi|41190472Jref|XM_373991.11(41190472] giJ41190479Jref|XM_373992.1 j[41190479] giJ41124484|reflXM_373995.1 ([41124484] gi J41119844|ref|XM_373997.11[41119844] gi[42656231 |ref|XM_373998.2J[42656231] gi|41119871 [ref|XM_374001.1 ([41119871] giJ42656211 |ref|XM_374002.2[[42656211] giJ41118745Jref |XM_374003.11[41118745] giJ411 874δ(ref |XM_374004.11[41118748] gi|42656213[ref|XM_374005.2J[42656213] gi|41118759|ref|XM_374006.1 ([41118759] gi|41126354(ref|XM_374007.11[41126354] gi|41126357|ref|XM_374008.1 |[41126357] gi|41126366|ref|XM_374009.11(41126366] giJ41126369|ref|XM_374010.11[41126369] giJ41125701 jref|XM_374011.1 [[41125701] gi[41125717|ref|XM_374012.l ([41125717] giJ41125741 (ref|XM_374013.11[41125741] gii41125747|ref|XM_374014.l ([41125747] gi|4112575θ(reflXM_374015.11[41125750] giJ41133988|ref |XM_374016.11[41133988] gi|41134017Jref|XM_374017.l j[41134017] gi|41191405Iref|XM_374019.1 |[41191405] gi(41191410|ref|XM_374020.11[41191410] giJ4119143θ[ref|XM_374021.1 ([41191430] gi|4119144θ(reflXM_374022.1 |[41191440] gi|41191454JreflXM_374023.1 [[41191454] gi|41191480|ref|XM_374025.1 |[41191480] giJ41191492|ref(xM_374026.1 |[41191492] gi|41126918|ref|XM_374028.1 [[41126918] gi|41120019iref[xM_374029.l i[41120019] gi[41144269|ref|XM_374033.11[41144269] gi|41144310|ref|XM_374035.11[41144310] g|J41144328(ref|XM_374036.1 [[41144328] giJ41144338|ref|XM_374037.11[41144338] giJ41144354[reflXM_374038.1 j[41144354] gi|42656645Jref|XM_374042.2|[42656645] giJ41146525|ref|XM_374045.1 ([41146525] gi|41146541 lref[xM_374046.1 [[41146541] gi|42656801 |ref|XM_374047.2|[42656801] gi|42656812(reflXM_374048.2i(42656812] gi|41146560|ref|XM_374049.1 ([41146560] gii42656837[reflXM_374050.2J[42656837] gi|41146573[reflXM_374051.1 [[41146573] gi|41146579[ref|XM_374052.11[41146579] gi|41146587|ref|XM_374053.11[41146587] giJ41146589[ref|XM_374054.1 ([41146589] giJ41146599|ref|XM_374055.1 [[41146599] gi|41146603|ref|XM_374056.11[41146603] gi|41146605[ref|XM_374057.1 ([41146605] gi(41146607|ref|XM_374058.1 |[41146607] gi|41146609(ref|XM_374059.1 [[41146609] gi[42656849ireflXM_374060.2i[42656849]
gi|41146615|ref|XM_374061.11[41146615] giJ4114662δjref|XM_374062.1 j[41146628] giJ41146648(reflXM_374064.11[41146648] gii42656888|ref|XM_374065.2[[42656888] gi|41146652|ref|XM_374066.11[41146652] gii42656889iref[xM_374067.2i[42656889] giJ41194212|ref|XM_374068.l j[41194212] giJ41194215JrefjX _374069.l i[41194215] giJ41144864[ref|XM_374070.11[41144864] gi(41144869[ref|XM_374071.1 [[41144869] gij41146831 (reflXM_374075.1 [[41146831] gi|41146825|ref|XM_374076.11[41146825] gi|41146856ireflXM_374077.l j[41146856] gi|41146861 |ref|XM_374078.11[41146861] giJ41146872Jref|XM_374079.l ([41146872] gi|42657036Jref|XM_374080.2i[42657036] gi|41146898|ref|XM_374081.11[41146898] gi|41146668|ref|XM_374082.1 [[41146668] gi|42656975|ref|XM_374086.2J[42656975] gii41146696|ref|XM_374088.1 ([41146696] gi|41146699(ref|XM_374089.1 [[41146699] gii41146703[ref|XM_374090.1 [[41146703] gi|41146705|ref|XM_374091.1 [[41146705] gi|41146709|ref|XM_374092.1 [[41146709] gi|41146714|ref|XM_374093.1 ([41146714] gi[41146716(reflXM_374094.11[41146716] gi|41146752(ref|XM_374095.1 [[41146752] gi|42657020|ref|XM_374096.2|[42657020] gi[41146756Jref|XM_374097.11[41146756] gi|41146760|ref|XM_374098.11[41146760] gi[42657022Jref|XM_374099.2i[42657022] gi|41146770|ref|XM_374100.1 |[41146770] giJ41146772iref|XM_374101.l ([41146772] gil41146778Jref|XM_374103.l j[41146778] gi|41146780|ref|XM_374104.11[41146780] gi|41146910|ref|XM_374105.1 [[41146910] giJ41146916(ref|XM_374106.1 |[41146916] gi|42657155[ref |XM_374107.2[[42657155] gi|41146921 jref|XM_374108.1 [[41146921] gi|42657163|ref |XM_374109.2|[42657163] gi|41146927|ref|XM_374110.1 [[41146927] gi|41146929Iref|XM_374111.11[41146929] gi|41195514|ref|XM_374112.1 ([41195514] gi|42657181 |ref|XM_374113.2|[42657181] gil42657183|ref|XM_374114.2([42657183] giJ41195521 [ref |XM_374115.1 [[41195521] giJ41195530|ref|XM_374117.1 ([41195530] giJ41147119Jref|XM_374118.11[41147119] gi|41147127|ref|XM_374120.1 ([41147127] gi|41147078Jref|XM_374121.1|[41147078] gi(42658900|ref|XM_374123.2|[42658900] giJ41147175Jref|X _374124.11[41147175] gi[42657380|ref|XM_374125.2|[42657380] gi[41147197|ref|XM_374127.1 |[41147197] gi|41147200|ref|XM_374128.1 [[41147200] giJ41146976|ref|XM_374129.1 ([41146976] gi|41146980|reflXM_374130.11(41146980]
gi|41146982|ref|XM_374131.11[41146982] gi|41147023JrefjX _374132.1 j[41147023] giJ41147027JrefjXM_374133.1 [[41147027] gi|4114703θjref|XM_374134.lj[41147030] gi|41147042|ref|XM_374135.11 [41147042] gi|41147044|ref|XM_374136.l[[41147044] gi|41147052Jref|XM_374137.1|[41147052] gii41147054ireflXM_374138.l[[41147054] giJ41147056[ref|XM_374139.11[41147056] gi|41147058|ref|XM_374140.1 ([41147058] giJ4114706θ[ref|XM_374141.1|[41147060] gi|41147063|reflXM_374142.lj[41147063] gi|41147065Iref|XM_374143.11[41147065] gi|42657239iref|XM_374144.2([42657239] giJ41146997JrefjXM_374145.lj[41146997] giJ41146999[ref|XM_374146.11[41146999] giJ41146931 (reflXM_374147.1 ([41146931] giJ41146946|ref|XM_374148.lj[41146946] gii42657209ireflXM_374149.2i[42657209] giJ4114695θjref|X _374150.11[41146950] gi|42657218[ref|XM_374151.2([42657218] gi|41147237[ref|XM_374152.1|[41147237] giJ4265604ljref|XM_374153.2|[42656041] giJ41147292Jref|XM_374154.1 j[41147292] gi|41147294|ref|XM_374155.1|[41147294] giJ41147296|ref|XM_374156.1|[41147296] gi|41147298JreflXM_374157.1 [[41147298] giJ4114730θ[ref|XM_374158.lj[41147300] giJ41147302JrefjXM_374159.1 j[41147302] giJ41147397|ref|XM_374161.1|[41147397] gi|41197059Jref|XM_374162.1 j[41197059] gi|41197071 |ref[xM_374163.1 ([41197071] gi|4119709θjref|XM_374164.11[41197090] gi|41197095|ref|XM_374165.11[41197095] giJ42657647ireflXM_374167.2J[42657647] gi|41197129|ref|XM_374168.11[41197129] giJ41197134Jref JXM_374169.11[41197134] gi|41197146|ref|XM_374170.1|[41197146] giJ41197149Jref|XM_374171.1[[41197149] gi|41147315|ref|XM_374172.1 ([41147315] gi|41147324JreflXM_374173.1 |[41147324] gi|41147328|ref|XM_374174.l([41147328] gii42657518|reflXM_374175.2[[42657518] giJ4114734θjref|XM_374176.1 [[41147340] gi|41147342Jref|XM_374177.lj[41147342] gi|41147277|ref|XM_374178.lj[41147277] giJ41147284|ref|XM_374179.1|[41147284] gi|41147286|ref|XM_374180.1|[41147286] gi|41196201 |ref|XM_374181.11[41196201] giJ41196235Jref|XM_374183.11[41196235] gi|41196248|ref|XM_374184.1|[41196248] giJ41196250|reflXM_374185.1 [[41196250] gi|41147355|ref|XM_374186.1|[41147355] gi|41147357|ref|XM_374187.lj[41147357] gi|42657547|ref|XM_374188.2J[42657547] giJ41147361 |ref|XM_374189.1 j[41147361] giJ41147363Jref|XM_374190.lj[41147363]
gi|41147365 |ref|XM_ 374191.1 |[41147365] giJ41147373 [ref|XM_ 374192.1 1(41147373] gi|41147381 jrefixM.374193.1 [[41147381] giJ41147351 IreflXM.374194.1 1(41147351] gjJ4 147403 |ref|XM_ .374196.1 [[41147403] g||41147425 JreflXM.374198.1 1(41147425] gi|41147544 (reflXM 374199.1 [[41147544] gi|41222008 IreflXM 374200.1 [[41222008] gi|42657828 IreflXM 374202.2 ([42657828] gi|41222059 [reflXM .374203.1 [[41222059] gi|41222065 JreflXM 374204.1 |[41222065] giJ41147480 IreflXM 374218.1 j[41147480] gfl41147614 |ref|XM_ 374222.1 1(41147614] giJ41147636 IreflXM 374226.1 1(41147636] gi|41147640 |ref|XM_ 374227.1 1(41147640] gi|41199227 [reflXM 374232.1 ([41199227] gi|41147546 JreflXM .374236.1 j[41147546] gi|42657802 |ref|XM_ 374242.2 [[42657802] gϊJ41148489 (reflXM .374243.1 [[41148489] gi]41148493 |ref|XM_ 374244.1 1(41148493] giJ41148501 JreflXM .374245.1 |[41148501] gi|41148585 Iref|XM 374247.1 [[41148585] gi|41148587 IreflXM. 374248.1 ([41148587] gi|42658875 [reflXM. 374249.2 [[42658875] gi|41148597 (reflXM .374250.1 [[41148597] gi|42658853 IreflXM 374251.2 [[42658853] gi]41148539 |ref|XM_ 374252.1 |[41148539] giJ41148543 |ref|XM_ 374253.1 1(41148543] gi|41148549 [reflXM .374254.1 ([41148549] gi|41148551 IreflXM 374255.1 1(41148551] gi|41148553 |ref|XM_ 374256.1 j[41148553] gi]41148555 JreflXM .374257.1 [[41148555] gi|41148428 JreflXM..374258.1 1(41148428] giJ41148430 (reflXM .374259.1 |[41148430] gi|42658766 IreflXM 374260.2 ([42658766] giJ41148436 IreflXM..374261.1 1(41148436] giJ41148448 IreflXM .374262.1 [[41148448] gi|41148634 IreflXM..374263.1 |[41148634] gϊJ41148636 (reflXM 374264.1 [[41148636] gi|41148761 IreflXM 374266.1 [[41148761] gi]41148459 (reflXM .374267.1 1(41148459] gi|41148694 IreflXM 374268.1 [[41148694] giJ41148696 IreflXM .374269.1 |[41148696] gi|42658949 IreflXM .374270.2] 1(42658949] gi|41148715 IreflXM. 374271.1 ([41148715] gi|41148721 [reflXM .374272.1 |[41148721] gi|41148727 IreflXM 374273.1 j[41148727] gi|42658971 IreflXM .374274.2 [[42658971] gi|41148733 IreflXM..374275.1 [[41148733] gi|41148643 (reflXM .374276.1 ([41148643] gi|41148660 |ref|XM .374277.1 [[41148660] gi|41148662 IreflXM..374278.1 1(41148662] giJ41148664 IreflXM .374279.1 |[41148664] gi|41148668 Jref|XM .374280.1 ([41148668] gi|41148670 IreflXM _374281.1 [[41148670] gi|41148673 IreflXM _374282.1 [[41148673] giJ42658843 IreflXM 374283.2 |[42658843]
141148463 |ref J|XM .374284.11(41148463]
141148467 jrefj |XM 374285.1 [[41148467]
141148471 | ref | |XM 374286.1 [[41148471]
J41148473 Irefl JXM .374287.11(41148473]
141148944 Irefl JXM 374289.1 [[41148944]
[41148946 | ref J |XM 374290.11(41148946]
(41148962 iref JlXM..374291.1 ([41148962]
141148964 jref J |XM 374292.1 [[41148964]
141148987 jref jlXM .374293.11(41148987]
J41148989 jref J|XM..374294.1 ([41148989]
J41148792 iref J|XM..374295.1 [[41148792]
[41148921 Irefl (XM 374296.1 ([41148921]
[41149070 Irefl JXM .374297.1 |[41149070]
141149072 jref J|XM..374298.11(41149072]
J41149074 jref J|XM 374299.1 [[41149074]
141149033 jref J|XM..374300.11(41149033]
[41149035 jref J|XM .374301.1 ([41149035]
141149046 Jref jjXM 374302.1 |[41149046]
[41149048 jref JjXM..374303.1 |[41149048]
141149052 jref J|XM..374304.1 ([41149052]
(42659274 Jref J|XM..374305.2 ([42659274]
|42659275 jref JjXM..374306.21(42659275]
[41148821 |ref JjXM..374307.1 ([41148821]
J41057795 Irefl JXM 374308.1 [[41057795]
141057810 |ref J|XM..374309.1 ([41057810]
|42659037 Irefl JXM 374313.2 |[42659037]
141199580 Irefl jXM 374315.1 |[41199580]
141151804 jref JlXM .374317.11(41151804]
141151811 jref j|XM 374318.1 |[41151811]
I42662782 Irefl JXM .374319.21(42662782]
141151597 |ref J|XM..374323.1 |[41151597]
J41210563 irefl JXM 374325.1 ([41210563]
141151510 Irefl JXM .374326.1 |[41151510]
141151516 |ref J]XM .374327.1 [[41151516]
(42662613 jref J]XM..374328.21(42662613]
[42662616 jref J]XM.374329.2 [[42662616]
141151540 [ref] (XM .374330.1 [[41151540]
(41151544 Irefl JXM..374331.1 |[41151544]
[41210271 jref JjXM 374333.11(41210271]
J42662683 Irefl JXM..374336.2 |[42662683]
[41151632 Irefl JXM 374337.1 |[41151632]
[41151639 Irefl JXM .374338.1 j[41151639]
141151643 jref J|XM..374339.1 ([41151643]
|41151645 jref J|XM.374340.11(41151645]
[42662723 Irefl JXM 374341.2 |[42662723]
(41151711 Irefl JXM .374342.1 ([41151711]
[41151713 jref JjXM 374343.11(41151713]
|41149249 Irefl JXM .374349.1 [[41149249]
|41149847j jref J|XM 374352.11(41149847]
141150062 Iref JjXM .374354.1 [[41150062]
|41205023 jrefl JXM .374355.1 |[41205023]
141205031 |ref J|XM .374356.11(41205031]
141150311 jref| |XM..374357.1 |[41150311]
|41150315 Irefl JXM. 374358.1 ([41150315]
(41150902 Irefl JXM. 374359.1 |[41150902]
141150926 Irefl JXM 374361.11(41150926]
141107711 Iref JjXM 374362.1 [[41107711]
gi|41058036|ref|XM_374364.1 |[41058036] giJ4115129l jrefjX _374365.l j[41151291] gi|41146792|ref|XM_374367.11[41146792] gi|41146796 jref |XM_374368.11[41146796] gi|41146798(ref|XM_374369.11[41146798] gi|41147464|ref|XM_374370.1 J[41147464] gi|41148521 jrefjXM_374371.1 ([41148521 ] gi|41148753|ref|XM_374372.1 [[41148753] giJ41151746JreflXM_374377.11[41151746] gi|41151748|ref|XM_374378.11[41151748] gi|41151752Jref|XM_374379.1 |[411517523 gi|41147673|ref|XM_374382.11[41147673] gi|42658179|ref|XM_374386.2[[42658179] gi|41147685|ref|XM_374388.1 [[41147685] gi|41147687|ref|XM_374389.1 |[41147687] gi|41147709Jref|XM_374396.1 ([41147709] gi|42658207|ref|XM_374399.2|[42658207] gii42658214iref|XM_374401.2[[42658214] gi(42658222ireflXM_374404.2|[42658222] gi|42658224|ref|XM_374405.2J[42658224] gi|42658226Jref|XM_374406.2J[42658226] gil41147749Jref|XM_374413.1 ([41147749] gil42658250|ref|XM_374414.2|[42658250] gi|41147771 |ref|XM_374419.11[41147771] giJ41147777|ref|XM_374422.1 j[41147777] gi|42658278|ref|XM_374425.2|[42658278] giJ41147791 jref|XM_374427.1 [[41147791] gi|42658283(ref|XM_374428.2i[42658283] giJ41147797|ref|XM_374430.11[41147797] gi|41147799|ref|XM_374431.11[41147799] gi|41147801 jref|XM_374432.1 j[41147801] giJ41147807[ref[xM_374434.11[41147807] gi|42658312|ref|XM_374435.2J[42658312] giJ41147813|ref|XM_374437.1 |[41147813] giJ41147817|ref|XM_374439.1 l(41147817] gi|42657766|ref|XM_374460.2|[42657766] gi|42657768Jref|XM_374461.2i[42657768] giJ42658456irefjXM_374473.2|[42658456] gi|42658462|ref|XM_374475.2|[42658462] giJ42658478[reflXM_374479.2i[42658478] gi|42658141 |ref|XM_374481.2|[42658141] gi|42658484iref(xM_374483.2([42658484] giJ41147989|ref(xM_374484.1 ([41147989] gi|42658498|reflXM_374487.2[[42658498] gi|42658503(reflXM_374489.2i[42658503] giJ41148003|ref|XM_374490.1 [[41148003] gi|41148005|ref|XM_374491.11[41148005] gi|42658522|ref|XM_374496.2|[42658522] gi[42658525iref|XM_374498.2|[42658525] gi|41148031 |ref|XM_374501.11[41148031] gi|41148067[ref|XM_374502.11[41148067] gi|42658562|ref|XM_374506.2([42658562] giJ41148093JrefjXM_374514.1 |[41148093] gi|41148103|ref|XM_374517.11[41148103] giJ42658163Jref|XM_374518.2J[42658163] gi|41148107|ref|XM_374519.l j[41148107] gi|42658604(ref|XM_374522.2[[42658604]
gi|41148135] JrefJ|XM..374526.11(41148135] gi|42658618 Irefl JXM_374529.2|[42658618] gi|42658619 |ref JjXM .374530.21(42658619] gi|41148163 irefl |XM_374538.1([41148163] giJ41148240 jref JjXM .374579.11(41148240] gi|41148244 Irefl JXM.374581.11(41148244] gi|41148250 Irefl JXM..374583.11(41148250] giJ41148254 Irefl |XM_374584.1 |[41148254] giJ41148256 jref J|XM.374585.1([41148256] gi|41148258 jref J|XM..374586.11(41148258] gi|41148406 irefj JXM
..374589.1|[41148406] gi|42658757 |ref J|XM_374590.2|[42658757] giJ41148412 |ref J|XM.374591.11(41148412] giJ42658761 jref J|XM .374592.2([42658761] giJ41147857 [ref J|XM .374600.1 [[41147857] gi|41147863 jref J|XM.374603.11(41147863] gi|42658279 Irefl JXM.374604.2j[42658279] gi|41147867 Iref JjXM .374605.1[[41147867] giJ41147871 |ref J|XM
..374607.1([41147871] gi|41147873 jref||XM_374608.11(41147873] gi|41148045 jref JjXM..374625.11(41148045] gi|41148049 Irefl (XM_374627.11(41148049] giJ41148051 jref J|XM..374628.1 [[41148051] gi|41148268 Irefl JXM 374636.1 |[41148268] gi|41148270 jref J|XM .374637.1[[41148270] giJ41148272 Irefl JXM 374638.1|[41148272] giJ41148288 jref J|XM..374646.1[[41148288] gi|41148292 Irefl JXM 374648.1|[41148292] gi|42658612 jref J|XM..374650.2([42658612] gi|41148302 Irefl JXM..374653.1 ([41148302] gi|41148306 Irefl JXM 374655.11(41148306] giJ41148310 jrefl JXM .374657.1{[41148310] giJ41148356 jref J|XM..374680.11(41148356] gi|41148360 jrefl JXM 374682.11(41148360] gi|41148362 Irefl JXM .374683.1 [[41148362] gi|41148364 jrefl JXM..374684.1 ([41148364] giJ41148370 Irefl JXM 374687.1 |[41148370] giJ41148374 Irefl JXM..374689.11(41148374] giJ41148384 jref J|XM .374694.1([41148384] giJ41148394 Iref JjXM 374699.11(41148394] gi|42658687 jref J|XM..374702.2|[42658687] gi|41148420 | ref JjXM.374705.1([41148420] g||41148422 |ref J|XM 374706.1|[41148422] giJ41147689 IrefljXM 374710.1 [[41147689] giJ41147691 jrefl JXM 374711.1([41147691] giJ42658199 jref JlXM 374713.2[[42658199] gιJ41147747 jref JjXM 374715.1j[41147747] gi|41147787 |ref J|XM.374721.11(41147787] giJ41147789 jref JjXM .374722.1 ([41147789] gi|41147897 Irefl |XM 374730.1|[41147897] gi|41147939 jref JjXM..374734.11(41147939] gi]41147963 Irefl JXM..374735.1[[41147963] gi|41148007 Irefl IXM..374741.1 |[41148007] giJ41148129 |ref| JXM 374751.11(41148129] gi|41148131 jrefl JXM 374752.11(41148131] giJ41148408 Iref JlXM 374761.1|[41148408] giJ41148418 Iref JjXM 374763.1([41148418]
gi|41148424 refj XM 374764.1 |[41148424] giJ42659447| refl XM 374765.11(42659447] gi|42659452 refj XM .374766.1 |[42659452] gi|42659454 refl XM 374767.1 j[42659454] gi|42659463 refl XM.374768.1 |[42659463] gi|42659465 refl XM 374769.1 |[42659465] gi|42659468 refl XM 374770.1 |[42659468] gi|42659544J refl XM 374779.1 |[42659544] giJ42659549 refl XM 374781.1 |[42659549] gi|42659554 refl XM
,374782.11(42659554] giJ42659565J refl XM 374786.1 |[42659565] giJ42659399 refl XM .374787.1 j[42659399] gi|42659397 refl XM 374792.11(42659397] gi[42659393| refl XM .374799.1 [[42659393] gi(42659313 refl XM .374801.1 ([42659313] gi|42659315] refl XM .374802.1 |[42659315] gi|42659317j refl XM .374803.1 j[4265931 ] gi|42659499] reflXMJ .374807.1 |[42659499] gi|42659505' refl XM
,374809.1 |[42659505] gi|42659507 refl XM 374810.11(42659507] gi|42659517] refl XM 374813.1 |[42659517] gi|42659530 refl XM..374817.11(42659530] gi|42659368 refl XM 374829.11(42659368] gi|42659370 refj XM.374830.11(42659370] gi|42659372 refl XM 374831.11(42659372] gi|42659379 refl XM. 374832.11(42659379] giJ42659429 refl XM 374835.1 [[42659429] gi|42659431 refl XM. 374836.11(42659431] gi|42659420 refl XM..374839.1 j[42659420] gi|42659422 refl XM 374840.1 ([42659422] gi|42659425| refl XM..374842.11(42659425] gi|42659573 refl XM 374844.1 [[42659573] gi[42659323 refl XM .374847.1 ([42659323] gi|42659483 refl XM. 374851.11(42659483] gi|42659487| refl XM .374852.1 |[42659487] gi|42659493 refl XM. 374854.11(42659493] giJ42659495 refl XM 374855.1 |[42659495] gi)42659757 refj XM 374857.1 [[42659757] giJ42659759 refl XM .374858.1 [[42659759] gi|42659763 refl XM..374860.1 [[42659763] gi!42659793 ref] XM 374864.1 |[42659793] gi|42659812| refl XM 374873.1 [[42659812] gi(42659842
; refl XM 374877.1 |[42659842] gi|42659844, refl XM .374879.1 j[42659844] gi|42659846 refl XM..374880.11(42659846] gi|42659851 refl XM 374882.11(42659851] giJ42659655 refl XM..374885.1 j[42659655] gi|42659668 refl XM 374890.11(42659668] gi)42659673 refl XM .374893.1 ([42659673] gi|42659678 refl XM..374898.1 [[42659678] gi|42659687 refl XM 374899.1 |[42659687] gi|42659689 refl XM 374900.1 j[42659689] gi)42659694 refl XM..374902.1 |[42659694] gi|42659718 refl XM..374904.1 j[42659718] gi|42659720 refl XM 374905.1 [[42659720] gi|42659722 refj XM .374906.1 |[42659722] gi(42659726 refl XM 374907.1 |[42659726]
gi|42659730|ref|XM_374909.1|[42659730] gi|42659734[ref[XM_374911.11[42659734] gi|42659736|ref|XM_374912.1|[42659736] gi|42659745|ref|XM_374915.lj[42659745] gi|42659773|ref|XM_374917.lj[42659773] gi[42659775Jref|XM_374919.11[42659775] gi|42659777|refJXM_374920.1|[42659777] gi J42659779|ref|XM_374922.1 ([42659779] gi|42659577|ref|XM_374927.1|[42659577] gi|42659584|ref|XM_374930.1 [[42659584] gi|42659588[ref|XM_374932.1 [[42659588] gi|42659592|ref|XM_374933.1|[42659592] gi|42659603Jref|XM_374936.1|[42659603] gi|42659610)reflXM_374937.11[42659610] giJ42659624ireflXM_374944.li[42659624] gi|42659626|ref|XM_374945.1 [[42659626] gi|42659634Jref|XM_374948.1 ([42659634] gi|42659638|ref|XM_374949.11[42659638] gi|42660082Jref|XM_374952.1|[42660082] gi|42660008(ref|XM_374959.li(42660008] gi|42660022|reflXM_374965.1J[42660022] gi|42660028|ref|XM_374967.1|[42660028] gi|42659922|ref|XM_374972.11[42659922] gi|42659925|refjXM_374973.1|[42659925] gi[42659942|ref |XM_374976.11[42659942] gi|42659951 |ref|XM_374981.11[42659951] giJ42659953|reflXM_374982.1|[42659953] gi|42659959|ref|XM_374983.1 [[42659959] gi|42659962|ref|XM_374985.1|[42659962] gi|42659887|ref|XM_374987.1|[42659887] gi|42659890|reflXM_374989.1|[42659890] giJ42659907JrefjXM_374995.lj[42659907] gi|42659911|ref|XM_374996.1[[42659911] gi|42660086|ref|XM_374997.1|[42660086] gi|42660094|ref|XM_374999.1|[42660094] gi|42660098Jref|XM_375000.11[42660098] gi|42660046|ref|XM_375004.1|[42660046] gi|42660048|ref|XM_375005.1 ([42660048] gi|42660132|ref|XM_375007.1|[42660132] gii42660146irefjXM_375013.li[42660146] gi|42660159|reflXM_375018.1|[42660159] gi[42660175Jref[XM_375023.1 j[42660175] gi|42660186|reflXM_375027.lj[42660186] gi|42660191 |ref|XM_375029.11[42660191] gi[42660196|ref|XM_375031.11[42660196] gi|42660198|reflXM_375032.11[42660198] giJ42660112Jref[XM_375033.1|[42660112] giJ42660120|ref|XM_375035.1|[42660120] gi|42660213|ref|XM_375038.11[42660213] gi|42660219|ref|XM_375039.11[42660219] gi|42660228|ref|XM_375041.11[42660228] giJ42660230[ref(XM_375042.lj[42660230] gi |42660234|reflXM_375045.11[42660234] gi|42660238|ref|XM_375065.1 [[42660238] giJ42660241 |ref[XM_375067.1 ([42660241] gi|42660252|ref|XM_375074.1|[42660252] gi|42660256|reflXM_375075.1 ([42660256]
gi|42660260|ref|XM_375076.11[42660260] gii42660262Jref|XM_375077.l j[42660262] gil42660268[ref|XM_375078.l i[42660268] gi[42660285|ref|XM_375080.1 |[42660285] gi(42660287iref|XM_375081.1 [[42660287] giI42660295|ref|XM_375084.1 I(42660295] gi[42660297iref|XM_375085.l [[42660297] gi|42660301 (reflXM_375086.11[42660301] gi|42660303iref|XM_375087.l ([42660303] gi|42660305(ref|XM_375088.1 |[42660305] gi|42660310|reflXM_375090.1 [[42660310] giJ426603311 ref |XM_375099.1 ([42660331] giJ42660339[ref|XM_375101.1 ([42660339] gi|42660355|ref|XM_375105.11[42660355] gi[42660369(ref|XM_375108.11[42660369] giJ4266061θiref|XM_375145.1 |[42660610] gi|42660614|ref|XM_375147.1 |[42660614] gii42660616JreflXM_375148.1 ([42660616] giJ42660619iref|XM_375150.1 [[42660619] giI42660625|ref|XM_375152.1 |[42660625] gi[42660629iref|XM_375153.1 [[42660629] gi[42660632Jref|XM_375154.1 [[42660632] gil42660637|ref|XM_375157.11(42660637] giI42660666(reflXM_375163.1 ([42660666] gi|42660671 |ref|XM_375165.1 |[42660671] gi(42660677Jref|XM_375170.11[42660677] gi!42660401 |ref|XM_375171.11(42660401] gii42660403[ref|XM_375174.11[42660403] gi|42660405|ref|XM_375176.11[42660405] gii42660409[ref|XM_375178.1 |[42660409] giJ426604111 ref |XM_375179.11 [42660411] gi|42660414Jref|XM_375181.11[42660414] giJ4266041 βjref |XM_375183.1 [[42660418] gi|42660420|ref|XM_375185.1 |[42660420] gi[42660422|ref|XM_375187.1 ([42660422] gi[42660430|reflXM_375190.1 [[42660430] gil42660432|ref|XM_375191.11(42660432] gi|42660442|ref|XM_375196.1 ([42660442] giJ42660451 (ref|XM_375200.1 [[42660451] giI42660454Iref|XM_375203.1 |[42660454] gi[42660470|ref|XM_375207.1 ([42660470] gi|42660473|reflXM_375209.1 |[42660473] giI42660475|ref|XM_375210.11 [42660475] gii42660516[ref|XM_375224.1 ([42660516] gi[42660524ireflXM_375226.l i[42660524] gi|42660527|ref|XM_375228.1 |[42660527] gi|42660529[ref|XM_375230.1 |[42660529] gi|42660644|ref|XM_375235.1 |[42660644] gi|42660650|ref|XM_375239.1 |[42660650] gi(42660652|ref|XM_375240.1 [[42660652] gi(42660656iref|XM_375242.1 l[42660656] gi|42660658iref|XM_375243.l ([42660658] gi[42660553(ref|XM_375246.1 |[42660553] gii42660555[ref|XM_375247.l i[42660555] gi(42660557|reflXM_375248.1 |[42660557] gii42660559(ref|XM_375249.l i[42660559] gi[42660563iref|XM_375252.1 l[42660563]
142660574 Irefl JXM 375260.1 |[42660574] 142660576 Irefl JXM..375261.11(42660576] (42660578 jref J|XM .375263.1 |[42660578] J42660600 jref j |XM .375266.11(42660600] J42660606 |ref J|XM .375268.11(42660606] 142660608 jref jjXM .375269.1 [[42660608] J42660684 Irefl JXM .375272.1 |[42660684] 142660688 jref jjXM .375274.1 ([42660688] 142660690 Irefl JXM 375275.1 ([42660690] [42660692 jref J|XM..375276.1 j[42660692] J42660696 jrefl JXM 375277.1 [[42660696] J42660698 Irefl jXM .375280.1 [[42660698] [42660996 jref J|XM .375282.11(42660996] 142660998 Irefl jXM .375284.1 j[42660998] J42661006 Irefl JXM 375288.1 ([42661006] 142661009 Irefl JXM .375292.1 [[42661009] |42660858 jref J|XM 375298.11(42660858] |42660706 Irefl JXM .375302.1 |[42660706] [42660722 jref j|XM 375305.1 ([42660722] I42660726 |ref J|XM..375306.1 |[42660726] (42660730 jref J|XM..375307.1 [[42660730] [42660732 iref J|XM .375308.1 ([42660732] (42660742 jref J|XM..375313.1 j[42660742] [42660747 jref J|XM..375316.11(42660747] (42660756 Irefl |XM_ .375319.1 |[42660756] [42660758 jref J|XM .375320.11(42660758] (42660930 Irefl |XM .375325.1 |[42660930] [42660946 jref J|XM.375330.1 [[42660946] I42660948 Irefl JXM .375331.1 |[42660948] |42660950 Irefl JXM..375333.11(42660950] J42660954 jref JjXM 375334.1 j[42660954] 142660971 |ref J|XM .375341.1 |[42660971] |42660974 jref JjXM..375344.1 ([42660974] 142660981 |ref J|XM .375349.1 |[42660981] 142661018 Irefl JXM..375351.11(42661018] 142661020 jrefl JXM 375352.1 [[42661020] I42660894 Irefl JXM .375353.1 |[42660894] 142660815 jref JjXM.375355.1 ([42660815] 142660771 Iref J|XM .375357.1 [[42660771] |42660778 I ref J |XM..375358.1 j[42660778] [42660783 Irefl (XM..375359.11(42660783] (42660785 Irefl IXM..375360.1 |[42660785] [42660795 Irefl (XM .375362.1 [[42660795] |42660799 Irefl JXM..375363.1 |[42660799] 142660801 Irefl jXM .375364.1 ([42660801] [42660901 Irefl IXM..375369.1 |[42660901] J42660906 jref J|XM 375373.1 [[42660906] |42660908 Irefl JXM 375375.1 j[42660908] J42660910 Irefl JXM .375376.11(42660910] (42660912 Jref J|XM .375377.1 |[42660912] [42660874 jref J|XM..375378.11(42660874] (42660821 jref J|XM .375379.1 |[42660821] [42660834 Irefl JXM .375383.11(42660834] (42660843 |ref JjXM .375384.1 [[42660843] (42660848 |ref JjXM..375386.11(42660848] [42661283 jref JjXM..375387.1 ([42661283] [42661071 jrefl JXM. 375392.1 |[42661071]
142661077 Irefl JXM 375397.11(42661077] J42661080 jref J|XM 375399.1 |[42661080] 142661092 Irefl JXM 375404.11(42661092] 142661103 Irefl JXM. 375408.1 [[42661103] 142661105 jref J|XM 375410.11(42661105] [42661110 jrefl JXM .375412.11(42661110] [42661131 I ref J]XM. 375418.1 ([42661131] J42661290 Irefl JXM .375423.1 |[42661290] 142661292 I ref J]XM..375424.1 ([42661292] [42661295 jref JjXM. 375426.11(42661295] 142661243 Iref JjXM .375430.11(42661243] (42661249 | refl JXM .375434.11(42661249] [42661253 Irefl JXM .375436.1 |[42661253] 142661260 | ref J|XM .375438.1 JJ42661260] (42661262 jref J|XM. 375439.1 j[42661262] [42661305 |ref J|XM .375443.1 |[42661305] (42661139 | ref J |XM. 375446.1 ([42661139] [42661145 jref ( |XM .375449.1 |[42661145] (42661151 I ref J |XM.375452.1 [[42661151] 142661155 Irefl JXM .375453.11(42661155] 142661165 I refl JXM.375456.1 j[42661165] J42661179 irefl JXM..375469.1 [[42661179] 142661184 Irefl JXM.375471.1 |[42661184] (42661202 jref ( |XM .375475.1 [[42661202] [42661209 Irefl JXM .375478.1 |[42661209] 142661216 jref J|XM.375482.1 ([42661216] 142661222 Irefl |XM .375484.1 |[42661222] 142661227 Irefl JXM.375485.1 ([42661227] [42661044 jref JjXM .375491.1 j[42661044] 142661050 irefl JXM.375492.1 JJ42661050] [42661272 jref J|XM .375494.1 ([42661272] (42661274 Irefl JXM .375495.1 [[42661274] 142661276 Irefl JXM.375496.1 |[42661276] [42661280 jref J|XM..375500.1 ([42661280] (42661060 jref J|XM .375502.1 |[42661060] 142661317 jref J|XM 375511.1 j[42661317] 142661326 Irefl JXM .375514.1 |[42661326] 142661329 Jref J|XM .375516.11(42661329] 142661331 jref J|XM .375517.1 ([42661331] 142661333 Iref JjXM .375520.1 [[42661333] J42661342 jref JlXM .375527.1 ([42661342] 142661395 Irefl JXM.375537.1 [[42661395] 142661415 jref J|XM .375543.1 ([42661415] (42661407 Irefl JXM .375544.1 [[42661407] 142661413 |ref J|XM .375545.1 |[42661413] [42661835 I ref J |XM .375548.1 ([42661835] 142661837 Irefl JXM .375549.1 |[42661837] 142661841 Irefl JXM..375550.11(42661841] 142661845 jref J|XM..375551.1 [[42661845] 142661849 jref J|XM .375552.1 j[42661849] 142661852 Irefl JXM .375553.11(42661852] J42661865 Irefl JXM .375557.1 j[42661865] J42661870 Irefl JXM .375558.1 j[42661870] J42661873 Irefl JXM..375559.1 [[42661873] 142661876 Irefl JXM..375560.11(42661876] [42661954 Irefl JXM .375563.1 |[42661954] 142661967 jref JjXM 375568.11(42661967]
gi|42661972|ref|XM _375569.1 |[42661972] gi|42661977|reflXM _375575.1 1(42661977] gi|42662005|ref|XM _375583.1 ([42662005] gi|42662014|ref|XM .375589.1 |[42662014] gi|42662016Jref|XM .375590.1 1(42662016] gi|42661561 [reflXM .375593.1 [[42661561] gi|42661566|ref|XM .375594.1 [[42661566] gi[42661576(ref|XM .375599.1 ([42661576] gi|42661589|ref|XM .375602.1 1(42661589] gi|42661591 IreflXM .375603.1 [[42661591] gi[42661593(reflXM 375604.1 |[42661593] giJ42661602|ref|XM..375606.1 [[42661602] giJ42661612Jref|XM .375608.1 [[42661612] gi[42661614(reflXM 375609.1 j[42661614] giJ42661629Jref|XM .375614.1 |[42661629] gi|42661643Iref|XM .375618.1 j[42661643] gi[42661647|ref|XM .375619.1 |[42661647] gi|42661658|ref|XM..375629.1 j[42661658] giJ42661663JreflXM 375631.1 |[42661663] giJ42661665[reflXM .375632.1 [[42661665] gi|42661667|ref|XM 375633.1 j[42661667] giJ42661670(refjXM .375634.1 [[42661670] gi|4266169θ[ref|XM .375638.1 [[42661690] gi|42661692|ref|XM 375639.1 |[42661692] gi(42661694JrefjXM .375640.1 1(42661694] gi|42661704|ref|XM 375646.1 1(42661704] gi[42661710|ref|XM .375651.1 j[42661710] gi|42661714|ref|XM 375654.1 j[42661714] gi|42661716|ref|XM .375655.1 |[42661716] gi|42661719Jref|XM .375656.1 1(42661719] gi|42661722|ref|XM 375658.1 1(42661722] gi|42661728Jref|XM 375660.1 |[42661728] gi|42661731 |ref|XM .375663.1 1(42661731] giJ42661734|ref|XM 375664.1 [[42661734] gi|42661737|reflXM .375665.1 1(42661737] gii42656035|ref|XM_ .375667.1 j[42656035] giJ42656037Jref|XM .375668.1 1(42656037] giJ42656039(ref|XM .375669.1 |[42656039] gi|42656043lref|XM_ .375670.1 1(42656043] gi|42656045|ref|XM_ .375671.1 ([42656045] gi[4265606θ(reflXM_ .375678.1 |[42656060] gi[4265584θiref|XM_ 375681.1 |[42655840] gi|42655842|ref|XM_ 375682.1 j[42655842] gi|42655621 |ref|XM_ .375684.1 [[42655621] gi[42655623JrefjXM_ 375685.1 1(42655623] gii4265563θirefjXM_ .375687.1 j[42655630] gi|42656004[ref|XM_ 375688.1 ([42656004] giJ42656007|reflXM_ .375690.1 j[42656007] gi|42655929|ref|XM_ 375695.1 1(42655929] gi[42655931 |ref|XM_ .375696.1 j[42655931] gi|42655933|reflXM_ .375697.1 ([42655933] giJ42655935|ref|XM_ .375698.1 j[42655935] giJ42656024(ref|XM_ .375700.1 [[42656024] gi|42655832|ref|XM_ .375707.1 [[42655832] gi|42655885(ref|XM_ .375712.1 j[42655885] gi(42655888Iref|XM_ .:375713.1 1(42655888] gi|42655890|ref|XM_: 375714.1 |[42655890]
gi|42655733|reflXM_375718.1 |[42655733] giJ42655739iref[xM_375720.1 |[42655739] gi[42655761 |reflXM_375726.1 ([42655761] gi|42655770JreflXM_375729.1 |[42655770] gi|42655775|ref|XM_375732.1 |[42655775] giJ42655781 [reflXM_375737.11[42655781] gi[42655783irefjXM_375738.l [[42655783] gi|42655656|ref|XM_375744.l j[42655656] gi[42655967(ref|XM_375746.1 [[42655967] gi|42655969iref|XM_375747.l j[42655969] gi|4265567θ(ref|XM_375753.11[42655670] gi|42655672[refjXM_375754.1 ([42655672] gi|42655676iref|XM_375756.1 |[42655676] gi|42655681 |ref|XM_375761.1 [[42655681] gi|42655683|ref|XM_375762.1 [[42655683] gi[42655586iref|XM_375770.l j[42655586] giJ42655646|ref|XM_375774.1 |[42655646] gi|42656027|ref|XM_375775.1 |[42656027] gii42656072|ref|XM_375779.l j[42656072] giJ42655801 (reflXM_375783.1 j[42655801] giJ42655805(ref|XM_375785.l j[42655805] gi|42655720|refjXM_375802.l ([42655720] gi|42655722(ref|XM_375803.1 |[42655722] giJ42656116[reflXM_375806.1 [[42656116] gi|42656118(ref|XM_375809.11[42656118] giJ42656120|ref|XM_375810.l i[42656120] gi|42656122|ref|XM_375811.1 |[42656122] gii42656124|ref|XM_375812.l j[42656124] giJ4265613θiref|XM_375814.l i(42656130] gi|42656138|ref|XM_375816.1 |[42656138] gi|42655876(ref|XM_375821.1 [[42655876] gi|42655959|ref|XM_375825.1 |[42655959] gi[42655825iref|XM_375833.1 [[42655825] gi|42655592|ref|XM_375834.1 |[42655592] gi(42655605Jref|XM_375837.11[42655605] gi|42655607(ref|XM_375838.1 |[42655607] giJ42655614|ref(xM_375841.1 ([42655614] giJ42655616|ref|XM_375842.1 |[42655616] giJ42655618|ref|XM_375843.11[42655618] gi|4265591θ[refixM_375845.l i[42655910] gi|42655913[ref|XM_375846.1 |[42655913] gi(42655918iref|XM_375848.1 ([42655918] gi|42655920|ref|XM_375849.1 |[42655920] gii42655923[reflXM_375850.l i[42655923] gi(42655925|ref|XM_375851.1 [[42655925] giJ42655983ireflXM_375853.1 |[42655983] gii42655989Jref|XM_375856.1 |[42655989] gii42655729|ref|XM_375863.l j[42655729] gi|42656146|ref|XM_375865.l ([42656146] gi|42656080|reflXM_375869.1 ([42656080] giJ42656088|ref|XM_375873.1 J[42656088] gii42656092iref|XM_375875.l ([42656092] gi|42656094|ref|XM_375876.1 |[42656094] gi|42656096[ref|XM_375882.1 |[42656096] giJ4265610l [ref|XM_375885.11[42656101] gi|4265615l (ref|XM_375887.l i[42656151] gi|42656157|ref|XM_375897.11[42656157]
gi|42656183|ref|XM_375899.1 |[42656183] gi J42662248 jref |XM_375902.1 j[42662248] giJ4266226l(ref|XM_375904.1 ([42662261] gi[4266228θ[ref|XM_375911.11[42662280] giJ42662282|ref|XM_375912.l j[42662282] gii4266229ljref|XM_375914.1 ([42662291] giJ42662221 jref|XM_375917.1 j[42662221] giJ42662241 jref|XM_375922.1 [[42662241] gi|42662088[reflXM_375925.1 |[42662088] gi|42662092Jref|XM_375928.1 |[42662092] giI42662303ireflXM_375929.lj[42662303] gi|42662305(ref|XM_375930.11[42662305] gi|42662307Jref|XM_375931.11[42662307] gi|42662311 |ref|XM_375934.11[42662311] giJ42662313Jref|XM_375935.1 |[42662313] gi|42662315|ref|XM_375936.1 [[42662315] gi(42662333|refjXM_375941.1 ([42662333] gi|42662375|ref|XM_375946.1 [[42662375] gi|42662531 |ref|XM_375948.1|[42662531] gii42662383JreflXM_375951.1 [[42662383] giJ42662390|ref|XM_375953.1 |[42662390] gii42662392irefixM_375954.l j[42662392] giJ42662396|ref|XM_375955.l j[42662396] gi|42662399Iref|XM_375958.lj[42662399] gi|42662417[ref|XM_375963.1 ([42662417] giJ42662419[ref|XM_375964.11[42662419] giJ42662421 |ref|XM_375965.1 ([42662421] gi|42662423|ref|XM_375966.11[42662423] gii42662428Jref|XM_375993.lj[42662428] giJ42662431 |reflXM_375996.1 [[42662431] giJ42662433|reflXM_375997.1 |[42662433] gi[42662448JreflXM_376001.1 j[42662448] gi|42662456|ref|XM_376003.1 ([42662456] gi[42662461 jref|XMJ376007.1 j[42662461] gi[42662463Jref[xM_376008.l i[42662463] giJ42662472|ref|XM_376010.11[42662472] gi|42662474|ref|XM_376013.1 [[42662474] gi[4266250θireflXM_376018.1 ([42662500] gi|42662503|ref|XM_376019.1|[42662503] giJ42662505|ref|XM_376020.l[[42662505] gi(42662512[ref|XM 376021.1 |[42662512] gi|42662520|ref|XMl376022.1 |[42662520] giJ42662522JreflXM_376023.li[42662522] gi|42662529|ref|XM_376024.11[42662529] gi|42656493 jref|XM_376031.1 j[42656493] gi[42656495iref|XM_376032.l [[42656495] gi|42656502|ref|XM_376033.1 |[42656502] giJ42656509|ref|XM_376034.1|(42656509] gi J42656270|ref JXM_376043.1 ([42656270] gi|42656274|ref|XM_376044.1|[42656274] gii4265629θ[ref|XM_376048.l i[42656290] gii42656293|ref|XM_376049.l ([42656293] gi|42656301 |ref[XM_376051.11[42656301] gi|42656307|ref|XM_376056.11[42656307] gi|42656322lref|XM_376059.l([42656322] gi|42656329|ref|XM_376060.1 |[42656329] gi|42656335Jref|XM_376062.1 |[42656335]
gi|42656342|reflXM_376068.1|[42656342] giI42656349(ref|XM_376072.1|[42656349] gi|42656351 |ref|XM_376073.11[42656351] gi[42656353iref|XM_376074.1[[42656353] gi|42656380|ref[XM_376094.1|[42656380] gi|42656385)ref|XM_376097.1|[42656385] gi|42656387|ref|XM_376099.1|[42656387] gi|42656389|ref|XM_376100.1 j[42656389] gi|42656391(ref|XM_376101.1[[42656391] giJ42656221 jref|XM_376106.1 ([42656221] gi|42656234|ref|XM_376108.11[42656234] gii42656244iref|XM_376111.1 j[42656244] gi|42656246|ref|XM_376112.11[42656246] gii42656405(refJXM_376117.li[42656405] gi|42656627(ref|XM_376118.11[42656627] giJ42656204|ref|XM_376121.11[42656204] gi|42656463|ref|XM_376125.1|[42656463] gi|42656465|ref|XM_376126.1|[42656465] gi|42656467|refJXM_376127.1|[42656467] gi|42656428|ref|XM_376130.1|[42656428] gi|42656437)ref|XM_376139.11[42656437] gi|42656440|ref|XM_376141.11[42656440] gi|42656442|ref|XM_376142.11[42656442] giJ42656446|ref[XM_376144.1[[42656446] gi|42656523|ref|XM_376148.1|[42656523] gi|42656528|ref|XM_376150.11[42656528] gi|426565311 ref |XM_376154.1 j[42656531] gi|42656542|ref|XM
„376158.11[42656542] gi|42656547|ref|XM_376160.l([42656547] gi[42656552|ref|XM_376161.1([42656552] gi|42656554|ref|XM_376162.11[42656554] gi|42656558Jref|XM_376165.lj[42656558] gi|42656567|ref|XM_376171.1 [[42656567] gi|42656572Jref|XM_376172.1|[42656572] gi[42656587|ref|XM_376178.11[42656587] gi|42656593|ref|XM_376179.11[42656593] giJ42656595|ref|XM_376180.11[42656595] gi|42656477|ref|XM_376185.11[42656477] giJ42656479|ref|XM_376186.11[42656479] gi|42656489|ref|XM_376189.1|[42656489] gi|42656256|ref|XM_376190.11[42656256] gi(42656258[ref|XM_376191.1 [[42656258] gi|42656617|ref|XM_376193.11(42656617] gi|42656418|refJXM_376195.lj[42656418] gi|42656633|ref|XM_376200.1 [[42656633] gi[42656662|ref|XM_376201.11[42656662] gi|42656673Jref|XM_376203.1|[42656673] gi|42656680|ref JXM_376206.1 [[42656680] gi|42656682|ref|XM_376207.1([42656682] gi|42656688iref|XM_376209.1|[42656688] gi|42656696|ref[XM_376212.1|[42656696] gi[42656726[ref(XM_376225.1|[42656726] gi|42656730|ref|XM_376227.1|[42656730] gi|42656736Jref[XM_376232.1[[42656736] gi|42656738(ref|XM_376233.1 ([42656738] gi|42656742|ref|XM_376238.1|[42656742] gι'J42656744 jref|XM_376239.11[42656744]
giJ42656749 IreflXM.376241.1 |[42656749] gi|42656635 (reflXM 376243.11(42656635] gi(42656647 IreflXM 376247.1 |[42656647] gi|42656649 IreflXM 376248.1 ([42656649] gi|42656651 IreflXM 376249.1 [[42656651] gi|42656774 IreflXM 376254.1 [[42656774] gi|42656779 IreflXM 376256.1 JJ42656779] gi|42656794 IreflXM .376257.1 |[42656794] gi|42656796 IreflXM .376258.1 ([42656796] giJ42656823 IreflXM 376267.1 [[42656823] gi|42656831 |ref|XM 376268.1 |[42656831] gi(42656843 IreflXM .376269.11(42656843] gi[42656860 IreflXM .376278.1 ([42656860] giJ42656864 IreflXM .376280.1 |[42656864] gi|42656866 |ref|XM 376281.1 [[42656866] gi|42656874 [reflXM 376284.1 ([42656874] gi|42656878 IreflXM .376287.1 [[42656878] gi[42656892 IreflXM 376290.11(42656892] gi(42656760 IreflXM. 376292.1 j[42656760] gi|42657047 JreflXM 376299.1 |[42657047] gi[42657069 lref|XM 376300.11(42657069] gi(42657060 [reflXM .376301.11(42657060] gi|42657062 IreflXM. 376303.1 |[42657062] gi|42657093 IreflXM 376305.11(42657093] gi[42657095 IreflXM 376306.1 [[42657095] gi(42657099 |ref|XM .376307.1 j[42657099] giJ42657114 )ref|XM 376309.1 j[42657114] gi|42657116 JreflXM. 376310.11(42657116] giJ42657119 (reflXM .376312.11(42657119] gi|42656918 IreflXM. 376314.1 ([42656918] gi|42657088 IreflXM..376317.1 [[42657088] gi[42657038 [reflXM .376318.1 ([42657038] gi(42657012 Jref|XM 376320.1 ([42657012] gi[42657137 IreflXM..376322.1 |[42657137] gi|42657139 IreflXM .376323.1 ([42657139] gi|42657141 IreflXM. 376324.1 |[42657141] gi|42657075 IreflXM..376325.11(42657075] gi|42656927 JreflXM 376327.1 [[42656927] gi|42656929 IreflXM..376328.1 ([42656929] gi|42656933 IreflXM .376331.1 [[42656933] gi|42656937 IreflXM 376333.1 [[42656937] giJ42656939 IreflXM .376334.11(42656939] gi|42656946 JreflXM 376338.1 [[42656946] gi[42656952 IreflXM..376339.1 |[42656952] gi|42656959 JreflXM 376342.1 ([42656959] giJ42656979 IreflXM 376347.1 ([42656979] gi|42656989 IreflXM .376348.1 [[42656989] gi(42656995 IreflXM 376349.1 [[42656995] giJ42656997 IreflXM .376350.11(42656997] giJ42657014 (reflXM .376353.1 [[42657014] giJ42657016 IreflXM 376354.1 ([42657016] gi|42657018 IreflXM 376355.1 ([42657018] gi|42657147 IreflXM 376364.1 ([42657147] giJ42657149 IreflXM 376365.1 [[42657149] giJ42657151 IreflXM .376366.11(42657151] gi|42657161 IreflXM .376368.1 |[42657161] giI42657172 IreflXM 376370.11(42657172]
J42657177 I ref J |XM 376371.1 |[42657177] 142657179 Irefl JXM .376372.11(42657179] I42657328 |ref J|XM 376376.1 [[42657328] J42657336 |ref J|XM 376379.1 |[42657336] [42657306 jrefl JXM .376383.1 ([42657306] (42657312 Irefl JXM .376386.1 |[42657312] 142657314 |ref J|XM .376387.11(42657314] [42657426 |ref J|XM..376389.1 [[42657426] |42657430 irefl JXM..376391.1 ([42657430] |42657295 |ref J|XM 376394.1 |[42657295] |42657297 jrefl JXM 376395.11(42657297] (42657432 Irefl JXM 376397.1 [[42657432] I42657377 |ref J|XM 376403.1 ([42657377] |42657383 jrefl JXM 376405.1 |[42657383] [42657229 |ref J|XM .376412.1 [[42657229] (42657231 jref j|XM. 376413.1 |[42657231] [42657264 Irefl (XM..376416.1 ([42657264] J42657268 jref J|XM 376419.1 j[42657268] [42657271 jref JXM..376420.1 JJ42657271] |42657275 jref j|XM .376423.11(42657275] |42657279 Irefl JXM .376427.1 [[42657279] J42657281 Irefl JXM .376428.1 ([42657281] |42657236 Irefl JXM. 376430.11(42657236] J42657246 Irefl JXM .376433.1 [[42657246] J42657252 jrefl JXM 376436.1 |[42657252] J42657195 irefl JXM .376440.1 ([42657195] (42657207] |ref J|XM. 376443.11(42657207] J42657210 Iref JjXM .376444.1 |[42657210] 142657216 Jrefj JXM .376447.1 [[42657216] (42657403 Irefl JXM..376453.1 j[42657403] |42657405 jrefl JXM. 376454.11(42657405] [42657414 Irefl JXM .376458.1 |[42657414] |42657496 Irefl JXM .376461.1 ([42657496] (42657508 jref JlXM 376463.1 j[42657508] 142657512 I ref J|XM .376464.1 |[42657512] |42657577 Irefl JXM .376469.11(42657577] I42657582 I ref j |XM .376471.1 j[42657582] |42657584 I ref J |XM 376472.1 |[42657584] [42657587 Irefl JXM .376473.1 j[42657587] (42657589 I ref J |XM 376474.1 |[42657589] [42657610 Irefl JXM .376479.1 [[42657610] J42657612 Jref JlXM 376480.1 |[42657612] [42657618 jref J |XM .376486.1 ([42657618] (42657620 Irefl JXM..376487.1 [[42657620] [42657622 jref J|XM 376488.11(42657622] (42657624 Irefl JXM .376491.1 ([42657624] [42657639 Irefl JXM 376498.11(42657639] J42657641 jref J|XM 376499.1 [[42657641] J42657652 jref JjXM..376503.1 ([42657652] J42657657 |ref JjXM 376505.1 [[42657657] I42657663 jref JjXM 376508.1 ([42657663] J42657516 Iref JjXM 376516.1 [[42657516] J42657523 Iref JjXM .376518.11(42657523] J42657528 Irefl JXM .376519.11(42657528] J42657439 Irefl JXM 376522.1 ([42657439] J42657442 Irefl JXM 376525.1 |[42657442] J42657446 Irefl JXM. 376527.1 j[42657446]
gi|42657453|ref|XM_376532.1 |[42657453] gi J42657455Jref |XM_376533.1 j[42657455] gi|42657458|reflXM_376535.lj[42657458] gi|42657465(ref|XM_376536.1 |[42657465] gi|42657469|ref|XM_376537.1 |[42657469] giJ42657481 (ref|XM_376540.11[42657481] giJ42657486[ref JXM_376541.1 ([42657486] gii42657493|ref|XM_376547.1 |[42657493] gii42657542(ref|XM_376549.11[42657542] gi|42657544|ref|XM_376550.1 |[42657544] gi|42657561 (ref|XM_376554.11[42657561] gi|42657531 (ref|XM_376555.1 j[42657531] gii42657538Jref|XM_376556.l j[42657538] giJ4265754θiref[xM_376557.1 |[42657540] gi|42657671 |ref|XM_376558.11[42657671] gi|42657675iref|XM_376560.1 |[42657675] giJ42657804Jref|XM_376564.11[42657804] gi|42657814|ref|XM_376565.1)[42657814] gii42657816|ref|XM_376566.11[42657816] gi|42657818Jref|XM_376567.1 |[42657818] gi|42657822Jref|XM_376568.1 |[42657822] giJ42657826[ref JXM_376569.11[42657826] gi|42657830|ref|XM_376571.11[42657830] gi|42657834Jref|XM_376573.l j[42657834] giJ42657837|refjXM_376575.l [[42657837] gi|42657842|ref|XM_376576.1 |[42657842] gi|42657844|ref|XM_376577.1 [[42657844] gi(42657846(ref|XM_376578.1 j[42657846] gi|42657861 |ref|XM_376585.1|[42657861] gii42657863[ref|XM_376586.l i[42657863] gi|42657869|reflXM_376587.1 |[42657869] giJ42657873iref|XM_376588.l j[42657873] gi|42657881|refJXM_376589.1 |[42657881] gi|42657883|ref|XM_376590.1 |[42657883] gi[42657887|ref|XM_376591.1 j[42657887] gi|42657891 |ref|XM_376593.11[42657891] giJ42657895(ref|XM_376595.1 |[42657895] gi|42657900|ref |XM_376597.1 [[42657900] giJ42657904Jref|XM_376598.lj[42657904] giJ42657906|ref|XM_376600.l ([42657906] gi|42657908|ref|XM_376602.1 |[42657908] gil42657912|ref|XM_376604.1 |[42657912] giJ42657914|ref|XM_376605.11[42657914]
■ giJ42657918|ref|XM_376607.11[42657918] giJ42657687Jref]XM_376609.lj[42657687] gi|42657689|ref|XM_376610.11(42657689] gi|42657691 jref|XM_376611.11[42657691] gi|42657693|ref|XM_376612.1|[42657693] gi|42657695|ref|XM_376613.11[42657695] gi|42657685|ref|XM_376614.1 |[42657685] gi|42657726|ref|XM_376615.11[42657726] gi|42657732|reflXM_376616.l i[42657732] gi|42657735|ref|XM_376617.11[42657735] giJ42657737|ref|XM_376618.1 [[42657737] gi|42657739|ref|XM_376619.11[42657739] gi(42657748|ref|XM_376621.11[42657748] gi|42657750|ref|XM_376622.l j[42657750]
J42657752 Irefl JXM .376623.11(42657752] [42657770 jrefl JXM 376625.1 ([42657770] 142657772 I ref J |XM .376626.11(42657772] 142657777 Irefl JXM 376628.1 ([42657777] 142657779 Irefl JXM..376629.11(42657779] (42657781 Irefl JXM 376630.1 j[42657781] I42657783 Irefl JXM .376631.1 j[42657783] I42657788 Irefl JXM .376636.1 |[42657788] J42658015 jref jlXM 376638.1 j[42658015] 142658019 Irefl JXM .376639.1 |[42658019] 142658021 Irefl JXM 376640.11(42658021] |42658023 jref J|XM..376642.1 IJ42658023] |42658026 Irefl (XM 376643.1 j[42658026] |42658038 Irefl JXM..376647.1 ([42658038] (42658046 jref J|XM..376648.11(42658046] [42658048 |ref J|XM 376649.1 [[42658048] I42658050 Irefl JXM .376651.1 ([42658050] (42658054 jref J|XM..376652.1 |[42658054] [42658056 Irefl (XM .376653.11(42658056] (42658061 Irefl JXM..376655.1 |[42658061] [42658063 jref J|XM 376656.1 [[42658063] (42658065 Irefl JXM 376657.1 ([42658065] |42658067 jrefl JXM..376658.1 |[42658067] I42658079 Irefl JXM 376663.1 [[42658079] |42658083 Iref JjXM..376664.1 ([42658083] J42658090 jref JjXM..376665.1 [[42658090] 142658099 Iref JjXM .376668.1 ([42658099] 142658101 jrefl JXM..376670.11(42658101] J42658108] Irefl JXM 376671.1 [[42658108] 142658112 Irefl JXM .376672.1 ([42658112] 142658126] jrefl JXM .376677.1 |[42658126] [42658129] I ref J]XM .376679.1 [[42658129] |42657947] I ref J]XM 376680.11(42657947] J42657949 Irefl JXM..376681.1 ([42657949] J42657953 jrefl JXM 376683.1 [[42657953] [42657956 jref] JXM..376684.1 ([42657956] J42657959 jrefl JXM..376707.1 j[42657959] |42657963 Jref JXM .376712.1 [[42657963] |42657965 jrefl JXM..376713.1 |[42657965] I42657967 jref JjXM. 376715.1 j [42657967] |42657969 Irefl JXM .376716.1 ([42657969] (42657972 Irefl JXM..376717.1 ([42657972] [42657974 jref J|XM..376718.1 |[42657974] (42657976 I ref J|XM .376719.1 ([42657976] |42657978 Irefl JXM .376720.1 [[42657978] J42657986 jrefl JXM..376722.1 j[42657986] |42657990 jrefl JXM. 376724.1 [[42657990] |42657992 [ref J|XM .376725.11(42657992] [42657711 Irefl JXM .376727.1 j[42657711] (42657713 jref J|XM..376728.1 ([42657713] [42657719 jref J|XM..376730.1 [[42657719] |42658805 Iref JjXM 376736.11(42658805] J42658818 I ref J]XM 376741.1 |[42658818] J42658879 I ref J]XM 376746.1 ([42658879] [42658887 jref J]XM .376750.1 j[42658887] 142658851 I ref J]XM 376754.11(42658851] J42658855 jrefl JXM 376756.11(42658855]
gi[42658861 |ref|XM_376757.1 [[42658861] gi|42658769|ref|XM_376761.11[42658769] gi|42658773|ref|XM_376763.lj[42658773] gii42658776iref|XM_376764.1J[42658776] gi|42658946|ref JXM_376771.11[42658946] gi[4265895θiref|XM_376774.l[[42658950] gi|42658956|reflXM_376776.1 [[42658956] giJ42658973|ref|XM_376780.lj[42658973] gi[42658975|reflXM_376781.1 [[42658975] gi|42658893|reflXM_376783.li[42658893] gi|42658905|ref|XM_376784.1|[42658905] gi|42658907|ref|XM_376785.11[42658907] gi|42658909|ref|XM_376786.1|[42658909] giJ42658911 [ref|XM_376787.11[42658911] gi|42658923[ref|XM_376791.1 ([42658923] gi|42658929|ref|XM_376793.1|[42658929] gii42658835iref|XM_376795.1 j[42658835] gi J42658837Jref|XM_376797.11[42658837] gi|42658782|ref|XM_376800.lj[42658782] gi[42658785iref[xM_376801.1 ([42658785] gi|42658896|reflXM_376809.1|[42658896] gi|42658898iref(XM_376810.1|[42658898] gi(42659144|reflXM_376814.lj[42659144] gi|42659167|ref|XM_376819.11[42659167] gi|42659172[ref|XM_376821.1 [[42659172] gii42659176|ref|XM_376822.11[42659176] gi[42659178[reflXM_376824.lj[42659178] gi|42659188|ref|XM_376829.1|[42659188] giJ42659193JreflXM_376830.1|[42659193] gi|42659207|ref|XM_376833.l[[42659207] gi|42659059|ref|XM_376838.1|[42659059] giJ42659063[ref|XM_376840.1|[42659063] gi|42659065|ref|XM_376841.11[42659065] gii42659067iref|XM_376843.lj[42659067] gi|42659072ireflXM_376846.l[[42659072] gii42659119|ref|XM_376847.1 ([42659119] gi|42659074|ref|XM_376848.1|[42659074] gi|42659076Jref|XM_376850.1 ([42659076] gi J4265908θ[ref |XM_376852.11[42659080] gi|42659084|ref|XM_376855.1|[42659084] gii42659090|ref|XM_376858.l([42659090] giJ42659093(ref|XM_376861.11[42659093] gi|42659095Jref|XM_376863.11[42659095] giJ42659103JreflXM_376866.1 [[42659103] gi|42659105|ref|XM_376869.1|[42659105] giJ42659115Jref|X _376872.1 [[42659115] gi|42659012|ref|XM_376874.1 |[42659012] gi|42659017JreflXM_376876.11[42659017] giJ42659021 |ref|XM_376880.1 j[42659021] gi|42659027|ref|XM_376885.lj[42659027] gii42659033(ref|XM_376888.li[42659033] gi|42659297iref|XM_376890.1|[42659297] gi|42659304|refjXM_376892.1|[42659304] giJ42659228Jref|XM_376895.1 j[42659228] gi|42659233|ref|XM_376898.lj[42659233] gi[42659235|ref|XM_376899.l([42659235] gii42659238Jref|XM_376900.11[42659238]
gi|42659242 I ref J |XM 376902.1 IJ42659242] gi(42659248 jref JjXM 376903.1 ([42659248] giJ42659252 Irefl JXM 376905.1 |[42659252] gi|42659258 jref J|XM. 376909.11(42659258] gi|42659054 Irefl JXM 376917.1 |[42659054] gi(42659057 Irefl JXM 376921.1 [[42659057] giJ42658996 jref J|XM 376924.1 |[42658996] giJ42659039 Irefl JXM 376925.1 ([42659039] gi|42659048 Irefl JXM .376930.1 |[42659048] gi|42659050 jrefl JXM .376931.1 ([42659050] gi|42659123 Irefl JXM 376939.1 j[42659123] giJ42659134 jref J|XM .376947.11(42659134] gi|42659138 Irefl JXM 376949.1 |[42659138] gi(42659140 jrefl JXM. 376950.1 [[42659140] gi|42662769 Irefl JXM..376960.11(42662769] giJ42662771 Irefl JXM .376965.1 |[42662771] giJ42662775 Irefl JXM 376968.11(42662775] gi|42662712 Irefl JXM..376978.1 |[42662712] giJ42662658 |ref JjXM .376981.1 ([42662658] gi(42662730 jref J|XM..376986.1 |[42662730] gi|42662539 |ref J|XM .376989.11(42662539] giJ42662565 irefl JXM .377000.1 [[42662565] gi|42662567 jref J|XM 377002.1 |[42662567] gi|42662560 jref J|XM..377012.11(42662560] gi|42662611 Irefl JXM..377014.1 |[42662611] gi[42662619 Irefl (XM .377018.11(42662619] gi|42662621 |ref J|XM 377019.1 ([42662621] giJ42662576 Iref JjXM..377024.1 [[42662576] giJ42662578 jref JjXM 377025.1 IJ42662578] gi|42662580 [ref J|XM .377026.11(42662580] giJ42662582 jrefl JXM. 377027.1 ([42662582] gi|42662584 Irefl JXM .377028.1 [[42662584] giJ42662591 jref| |XM 377031.1 |[42662591] gi (42662593 irefl JXM .377032.1 ([42662593] gi|42662598 jrefj |XM 377033.1 |[42662598] gi|42662602 Irefl JXM..377034.1 [[42662602] gi(42662678 Irefl JXM 377041.1 |[42662678] gi|42662638 Irefl JXM .377053.1 |[42662638] gi[42662641 jref J|XM..377060.1 ([42662641] gi|42662643 Irefl JXM 377062.11(42662643] giJ42662704 Irefl JXM .377071.1 [[42662704] gi[42662695 jref J|XM..377072.1 ([42662695] gi(42662697 Irefl JXM 377073.1 [[42662697] gi|42662721 Irefl JXM .377076.1 |[42662721] gi|42662760 jref J]XM..377087.1 j[42662760] gi|42662746 Irefl JXM .377097.11(42662746] gi[42662748 Irefl JXM 377098.1 |[42662748] gi|42662750 jref J|XM..377102.1 ([42662750] gi|42662752 jref JjXM..377104.1 |[42662752] gi|42659567 Irefl JXM 377109.1 ([42659567] gi|42659569 Irefl JXM 377110.1 [[42659569] gi|42659412 Irefl JXM 377115.1 |[42659412] giJ42659532] jref J]XM..377117.11(42659532] gi|42659536 jrefl JXM..377122.1 [[42659536] giJ42659391 jref JjXM..377129.1 j[42659391] gi(42659435 jref JjXM..377133.1 j[42659435] gi(42659476 jref JjXM. 377136.11(42659476]
gi|42659478|ref|XM_377137.11[42659478] gi|4265948θ[ref|XM_377140.1 ([42659480] gi|42659489|ref|XM_377142.1 |[42659489] gi|42659491 |ref|XM_377143.11[42659491] gi|42659855|ref|XM_377147.1|[42659855] gi|42659861 [ref|XM_377154.1 [[42659861] gi|42659863iref|XM_377155.1 ([42659863] gi|42659865|ref|XM_377156.11[42659865] gi|42659869|ref|XM_377158.11[42659869] gi)42659871|ref|XM_377159.11[42659871] gil42659875(ref|XM_377179.11[42659875] gi(42659877(ref|XM_377185.1 [[42659877] gi|42659883|ref|XM_377189.11[42659883] gi|42659700|ref|XM_377200.11[42659700] gi|42659708[ref|XM_377218.1 ([42659708] gi|42659712|ref|XM_377222.11[42659712] gi|42659753[ref|XM_377230.1|[42659753] gi[42659755ireflXM_377231.1 ([42659755] gi|42659646|ref|XM_377240.1|[42659646] gi|42660042|ref|XM_377259.1|[42660042] gi|42659988ireflXM_377262.1|[42659988] gil42659992]ref|XM_377265.1|[42659992] gil42659916[ref|XM_377278.1|[42659916] gii42660107JrefixM_377283.11[42660107] gi|42660109|ref|XM_377285.11[42660109] gi|42660069|ref|XM_377287.1|[42660069] gi[42660205iref|XM_377296.li[42660205] gi[42660221 |ref|XM_377305.1 ([42660221] gi|42660226|ref|XM_377306.1 |[42660226] gi|42660386|ref|XM_377337.11[42660386] gi|42660388Jref|XM_377338.li[42660388] gi|42660390|ref|XM_377343.1|[42660390] gi|42660392|ref|XM_377355.1|[42660392] gi[42660673irefixM_377369.1 |[42660673] gi|42660531 |ref|XM_377374.11[42660531] gii42660533(ref|XM_377376.1|[42660533] gi|42660535|reflXM_377377.1j[42660535] gi|42660539|ref[xM_377383.1|[42660539] gi|42660541 |ref|XM_377388.11[42660541] gi|42660543|ref|XM_377390.1 |[42660543] gi(4266066θiref|XM_377394.11[42660660] gi|42660590|ref|XM_377395.1|[42660590] gi|42661012|ref|XM_377407.l[[42661012] gi[42661014iref[xM_377408.1 ([42661014] gi|42661016|ref|XM_377412.1 |[42661016] gi|42660760|ref|XM_377414.1|[42660760] gil42660762|ref|XM_377416.1|[42660762] gii42660983Jref|XM_377420.l([42660983] gi|42660985lref[XM_377423.l([42660985] gii42660987iref|XM_377424.1|[42660987] gi|42660989lref|XM_377425.1|[42660989] gi|42660991 lrefJXM_377426.11[42660991] gi[42660993iref|XM_377429.li[42660993] gi|42660811 |ref|XM_377439.11[42660811] giJ42660916[ref|XM_377444.11[42660916] gi|42660918|ref|XM_377445.11[42660918] gil42660920|ref|XM_377446.1|[42660920]
gi|42660922 reflXM.377447.1 [[42660922] gi[42660852 reflXM.377451.1 |[42660852] gi|42661133 reflXM.377455.11(42661133] gi|42661135 reflXM.377457.11(42661135] giJ42661264 reflXM.377464.1 j [42661264] giJ42661229 reflXM 377475.1 |[42661229] giJ42661231 reflXM 377476.1 ([42661231] gi|42661233 reflXM .377477.1 ([42661233] gi|42661235| reflXM 377480.11(42661235] gi|42661364 reflXM .377488.1 [[42661364] gi|42661370 reflXM 377496.11(42661370] giJ42661372 reflXM..377498.1 ([42661372] gi|42661879 reflXM 377500.1 j[42661879] gi|42661883 reflXM..377506.1 |[42661883] gi|42662032 reflXM .377511.1 [[42662032] gi(42662034 reflXM .377512.1 ([42662034] gi[42662039 reflXM 377514.1 IJ42662039] gi|42662041 reflXM..377515.11(42662041] gi|42662043 reflXM .377516.1 ([42662043] giJ42662047 reflXM 377521.1 [[42662047] giJ42662053 reflXM 377522.1 ([42662053] gi|42661739 reflXM 377527.1 [[42661739] gi|42661743 reflXM .377529.11(42661743] giJ42661752| reflXM 377533.1 |[42661752] gi|42661754 reflXM .377537.11(42661754] giJ42661756: reflXM .377538.1 J[42661756] gi|42661761 reflXM. 377553.11(42661761] gi|42661763 reflXM. 377554.1 |[42661763] gi|42656051 reflXM 377555.1 ([42656051] gi|42655850 reflXM 377556.1 [[42655850] gi[42655906 reflXM 377558.1 ([42655906] gi(42655642 reflXM 377563.11(42655642] gi|42656018 reflXM..377565.1 |[42656018] gi[42656020 reflXM 377566.1 j[42656020] gi|42656022 reflXM..377568.1 |[42656022] giJ42656015 reflXM .377570.1 ([42656015] gi(42655937 reflXM 377577.1 |[42655937] gi|42655836 reflXM..377579.1 |[42655836] giJ42655838 reflXM..377580.11(42655838] gi|42655900 reflXM 377583.1 [[42655900] gi|42655892 reflXM 377585.1 |[42655892] gi|42655894 reflXM..377586.11(42655894] gi|42655658 reflXM .377593.1 |[42655658] gi|42655975 reflXM .377594.1 ([42655975] gi(42655679 reflXM .377595.1 |[42655679] giJ42655981 | reflXM .377597.1 [[42655981] gil42656070 reflXM 377599.1 |[42656070] gi|42656076 reflXM .377600.11(42656076] gi(42655807 reflXM..377601.1 JJ42655807] gi|42656144 reflXM .377611.1 ([42656144] giJ42655951 reflXM .377613.1 ([42655951] gi|42655881 reflXM .377618.1 |[42655881] gi(42655964 reflXM 377620.1 ([42655964] gi[42655927 reflXM .377630.1 ([42655927] giJ42656000 reflXM .377631.1 |[42656000] gi|42656002 reflXM 377633.1 [[42656002] gi|42655700 reflXM. 377635.1 |[42655700]
gi|42655706|ref[XM_377643.1|[42655706] gi|42655708 jref|XM_377644.11[42655708] giJ4265603ljrefjXM_377645.11[42656031] giJ42656084iref|XM_377649.1 ([42656084] gi|42656161 |ref|XM_377655.11[42656161] gi|42656163(ref|XM_377657.1 |[42656163] giJ426561
βδjref JXM_377658.1 ([42656165] giJ42656167|ref|XM_377659.1 ([42656167] gi|42656169(ref|XM_377660.11[42656169] gi|42656171 |ref|XM_377661.11[42656171] giJ42656173JreflXM_377662.l i[42656173] gi|42656175|ref|XM_377663.11[42656175] gi|42656177|ref|XM_377664.11[42656177] gi|42656179Jref|XM_377665.11[42656179] gi|42656181 jref|XM_377666.11[42656181] gi|42662286Jref|XM_377668.1 |[42662286] gi|42662299JreflXM_377675.1 ([42662299] giJ42662337JrefjXM_377687.l j[42662337] gi|42662368Jref|XM_377690.1 |[42662368] gi|4266237θ(refjXM_377691.1 ([42662370] gi|42662405|ref|XM_377694.1 |[42662405] gi[42662407JrefixM_377695.11[42662407] gij42662409|ref|XM_377696.11[42662409] giJ42662484Jref|XM_377700.l j[42662484] gil42662488(ref|XM_377713.11[42662488] gi|42662492|ref|XM_377715.11[42662492] giJ42662494iref|XM_377716.l [[42662494] gi|42662496Jref|XM_377717.11[42662496] giJ42662498|ref|XM_377720.l j[42662498] gi|42662516|ref|XM_377721.11[42662516] gi|42656370|ref|XM_377725.1 |[42656370] gi|42656515Jref|XM_377728.l [[42656515] gi|42656198|ref)XM_377732.1 [[42656198] gi|42656363|reflXM_377741.1 j[42656363] gi|42656365[ref|XM_377742.1 |[42656365] gi|42656414|ref|XM_377751.1 ([42656414] gi(4265640θjref|XM_377754.l ([42656400] giJ4265625θiref|XM_377755.1 ([42656250] gi|42656252JreflXM_377756.l j[42656252] gii42656409Jref|XM_377760.1 l[42656409] giJ42656629[ref|XM_377761.1 ([42656629] gi|42656474|ref|XM_377766.1 |[42656474] gi|42656452[ref|XM_377768.l ([42656452] gi|42656454|ref)XM_377771.1 [[42656454] gi]42656598|ref|XM_377774.1 |[42656598] giJ4265660θ(ref|XM_377776.1 [[42656600] gi|42656602|ref|XM_377778.1 |[42656602] gi|42656607|ref|XM_377783.1|[42656607] gi|42656609Jref [XM_377786.1 ([42656609] giJ42656753|reflXM_377797.1 |[42656753] gi|42656755[ref|XM_377803.11[42656755] gi|42656898|ref|XM_377809.1 |[42656898] gi|42656900(ref|XM_377811.11[42656900] gi|42656902(ref|XM_377815.1 ([42656902] gi]42656906|ref|XM_377818.11[42656906] gi|4265691θ(ref|XM_377820.11(42656910] gi|42656912|ref|XM_377823.1 |[42656912]
gi|42656769|ref|XM_377824.1 |[42656769] gi|42657056iref|XM_377828.l [[42657056] gi(42657073[ref|XM_377829.1 ([42657073] gi|42657121 |ref|XM_377830.1 [[42657121] gi|42657123|ref|XM_377831.1 H42657123] gi|42657125|ref|XM_377832.1 ([42657125] gi(42657127|ref|XM_377834.11[42657127] gi|42657129|ref|XM_377835.1 |[42657129] gi|4265713l (ref|XM_377836.l i[42657131] gi(42657133iref|XM_377837.1 ([42657133] gi|42657041 jreflXM_377841.11[42657041] gi[42656967iref|XM_377845.1 [[42656967] gi|42656969|ref|XM_377847.1 |[42656969] gi|4265697l jref|XM_377849.1 [[42656971] gi|42657034|ref|XM_377861.11[42657034] gi|42657349|ref|XM_377875.1 |[42657349] giJ42657351 (ref|XM_377877.1 [[42657351] gi|42657353|ref|XM_377878.1 |[42657353] gi|42657355|ref|XM_377879.1 |[42657355] giJ42657357iref|XM_377880.1 |[42657357] gi|42657359[ref|XM_377881.1 [[42657359] gi[42657361 |ref|XM_377882.11[42657361] gi[42657363iref|XM_377883.l i[42657363] gii42657365iref|XM_377884.l j[42657365] gi|42657367|ref|XM_377885.1 |[42657367] gi|42657369Jref|XM_377886.l ([42657369] gi|42657371 |ref|XM_377887.11[42657371] gii42657373[ref|XM_377889.l i[42657373] gi|42657316[ref|XM_377896.1 [[42657316] gi[42657396|ref|XM_377904.l ([42657396] gi[42657289|ref|XM_377907.1 |[42657289] gi|4265729l (ref|XM_377910.1 [[42657291] gi|4265726θ(ref|XM_377911.1 [[42657260] gi|42657262|ref|XM_377912.1 |[42657262] gi|42657219(ref|XM_377918.1 [[42657219] gi|42657221 j ref |XM_377919.11 [42657221] gi|42657223|ref|XM_377920.1 |[42657223] gi|42657424[ref|XM_377924.l i[42657424] giJ42657921 |ref|XM_377925.1 [[42657921] gi|42657923|ref|XM_377926.1 J[42657923] gi|42657925|ref|XM_377927.11[42657925] gi|42657927|ref|XM_377928.l ([42657927] gi|42657929iref|XM_377929.l [[42657929] gi|42657933|ref|XM_377931.11[42657933] gi|42657935[ref|XM_377932.l i[42657935] gi|42657937|ref|XM_377933.1 |[42657937] giJ42657939iref|XM_377934.l [[42657939] gi|42657941 |ref|XM_377935.11[42657941] gi|42657943|ref|XM_377938.l ([42657943] gi|42657790|ref|XM_377941.11[42657790] gi|42657792[reflXM_377942.l i[42657792] gi(42657794JrefixM_377943.l [[42657794] gii42657796|ref|XM_377944.1 |[42657796] gi|42657798|ref|XM_377945.1 |[42657798] gii42657800|ref|XM_ 377946.1 |[42657800] gi|42658133|ref|XM_377947.11[42658133] gi|42658135(ref |XM_377949.1 [[42658135]
gi|42658137 |refJ|XM 377950.11(42658137] gi[42658139 Irefl JXM.377951.1|[42658139] giJ42658143 Irefl JXM 377955.11(42658143] gi|42658145 Irefl JXM 377956.1|[42658145] gi|42658147 |ref J|XM 377957.11(42658147] gi[42658149 jrefl JXM 377958.1 [[42658149] gi|42658151 Irefl JXM 377959.1 ([42658151] gi(42658153 I refl JXM..377961.11(42658153] gi|42658155 J|ref JlXM .377962.1|[42658155] gi[42658157 Irefl JXM.377964.11(42658157] gi(42658159 Irefl JXM .377969.1|[42658159] gi|42658161 Irefl JXM..377970.11(42658161] gi|42658165 jref (jXM .377972.1 [[42658165] gi[42658167 Jref JjXM.377973.1 ([42658167] gi|42658169 Irefl JXM .377974.11(42658169] gi(42658001 Irefl JXM 377976.1[[42658001] gi[42658003 jrefl JXM..377995.1[[42658003] giJ42658005 jref J|XM..377997.1|[42658005] giJ42658007 Irefl JXM 377998.11(42658007] giJ42658009 Irefl JXM 377999.1|[42658009] gil42658011 jref J|XM..378001.11(42658011] gi|42658013 jref J|XM..378002.1 |[42658013] gi|42657724 Iref JjXM 378003.1 [[42657724] gi[42658822 Irefl JXM .378007.1 ([42658822] gi(42658824 jrefj JXM..378008.1 [[42658824] gi[42658826 Irefl JXM 378009.11(42658826] gi|42658828 |ref J|XM..378010.1|[42658828] giJ42658830 jref JjXM .378014.11(42658830] gi|42658832 Irefl JXM 378015.1[[42658832] giJ42658890 jref J|XM..378018.1JJ42658890] gi|42658978 iref| JXM 378028.1J[42658978] gi[42658980 Irefl JXM..378031.11(42658980] gi[42658982 jrefj|XM 378033.1|[42658982] gi(42658984 Irefl JXM .378035.1([42658984] gi|42658986 jrefj|XM 378036.1|[42658986] giJ42658787 Irefl JXM..378043.1([42658787] giJ42658789 Irefl JXM 378044.1|[42658789] giJ42658870 IrefljXM .378046.1 ([42658870] giJ42659216 jrefl JXM..378052.1 [[42659216] giJ42659218 Irefl JXM .378054.11(42659218] gi[42659061 Irefl JXM .378062.1([42659061] gi|42659069 Irefl JXM .378063.1([42659069] giJ42659078 jrefl JXM..378064.1 ([42659078] gi|42659107 Iref JjXM 378067.1 [[42659107] gi(42659289 |ref J|XM 378078.1 ([42659289] giJ42659293 Irefl JXM .378080.1[[42659293] gi(42659004| jref J|XM..378087.1[[42659004] giJ42659006 Irefl JXM 378089.1 |[42659006] gi(42659008 Irefl JXM .378090.11(42659008] giJ42662790 jref J|XM..378102.1([42662790] gi(42662710 |ref J|XM 378103.11(42662710] gi|42662569 Irefl JXM 378113.1 ([42662569] giJ42662571 jref J]XM..378116.11(42662571] gi|42662562 Irefl JXM..378118.1|[42662562] gi(42662636 Irefl JXM..378123.1([42662636] giJ42662732 Irefl JXM..378124.11(42662732] giJ42662606 |ref (|XM.378125.1 |[42662606]
gi|42662608|ref[XM_378128.1|[42662608] giJ42662691|ref|XM_378137.l j[42662691] giJ42662647|ref|XM_378143.1 |[42662647] gii42662726Jref|XM_378152.1 ([42662726] gi|42662758|ref|XM_378155.11 [42662758] gi|42662762[ref|XM_378156.l j[42662762] gi|42662742Jref JXM_378158.1 j[42662742] gi|42660547|ref|XM_378172.l j[42660547] gi|42660596(ref|XM_378173.11[42660596] gi|42660924|ref|XM_378175.11[42660924] gi[42661237|reflXM_378177.lj[42661237] gi|42661554|ref|XM_378178.1|[42661554] giJ42661950(ref JXM_378180.1 ([42661950] giJ42655731 [ref|XM_378181.11[42655731] gi|42655816|ref |XM_378183.11(42655816] gii42655852Jref|XM_378184.l i[42655852] gi|42655856|reflXM_378185.1|[42655856] giJ42655908Jref|XM_378186.1 j[42655908] gi|42655939|ref|XM_378187.l j[42655939] gi|42662411 j ref |XM_378189.11[42662411] gi|42656262|ref|XM_378190.1 ([42656262] gi|42656517|ref[XM_378191.1 [[42656517] giJ42656973Jref|XM_378192.11[42656973] gi|42657058|ref|XM_378193.11[42657058] gi|42657299|ref|XM_378194.11[42657299] gi|4265757θjref|XM_378195.1 |[42657570] gi|42657945|ref|XM_378196.1|[42657945] gi|4265878θ[ref|XM_378197.1 [[42658780] giJ42659220|ref|XM_378199.11(42659220] gi(42659437iref|XM_378200.1 |[42659437] gi|42659439|ref|XM_378201.11[42659439] gi(42659441 [ref|XM_378202.1 j[42659441] gil42659443 jref|XM_378203.11[42659443] gi|42659445Jref|XM_378207.1|[42659445] gii4265945θiref|XM_378208.1 |[42659450] gi|42659459|ref|XM_378210.1 |[42659459] giJ42659461 [ref[XM_378211.1 ([42659461] giJ42659539|ref|XM_378215.1 |[42659539] gi|42659558Jref|XM_378218.11[42659558] gi[4265956θiref|XM_378219.l [[42659560] gi|42659410|ref|XM_378223.1 ([42659410] gi|42659427|ref|XM_378224.l j[42659427J gi|42659320|ref|XM_378226.11[42659320] gi|42659416Jref|XM_378227.l j[42659416] giJ42659497JreflXM_378228.lj[42659497] gi|42659502|ref|XM_378230.l ([42659502] gi|42659511 |ref|XM_378232.11[42659511] gii42659522Jref|XM_378235.l i[42659522] gi J42659524(ref [XM_378236.1 j[42659524] gi|42659526JreflXM_378238.1 |[42659526] giJ42659336|ref|XM_378239.1 ([42659336] gi|42659345(ref|XM_378240.1 |[42659345] gi|42659361 |ref|XM_378247.11[42659361] gi|42659377Jref|XM_378250.11[42659377] gi|42659381 |ref|XM_378251.lj[42659381] gi|42659332|ref|XM_378255.1 |[42659332] gi|42659330|ref|XM_378257.l j[42659330]
gi|42659325|ref|XM_378259.1|[42659325] gi|42659327|ref|XM_378260.1 [[42659327] gi|42659485Jref|XM_378261.1 j[42659485] gi(42659761 |ref|XM_378262.1 ([42659761] gii42659789|ref|XM_378266.1 ([42659789] giJ42659804(ref[XM_378271.11[42659804] gi|42659806|reflXM_378272.1 j[42659806] giJ42659808Jref|XM_378273.lj[42659808] gi|42659814|ref|XM_378276.l[[42659814] gi|42659818Jref|XM_378277.lj[42659818] gi|42659822|ref|XM_378279.lj[42659822] gi(42659824Jref |XM_378280.1 ([42659824] gi[42659833irefixM_378283.lj[42659833] gi|42659838irefixM_378286.l[[42659838] gi|42659840ireflXM_378288.1|[42659840] gi|42659685|ref|XM_378297.lj[42659685] gi|42659769|ref|XM_378299.11[42659769] gi|42659716|ref|XM_378300.11[42659716] gi|42659724|refJXM_378301.1 [[42659724] giJ42659741 jref|XM_378303.1 [[42659741] gi|42659743Jref|XM_378304.1|[42659743] gi|42659749|ref|XM_378305.1|[42659749] gi(42659579|refjXM_378308.1 ([42659579] giJ42659594|ref]XM_378309.1|[42659594] gi|42659596Jref|XM_378311.1 [[42659596] gi|42659598|ref|XM_378312.11[42659598] giJ42659606|ref|XM_378313.l[[42659606] gi(42659608|ref[XM_378314.1|[42659608] gi|42659614Jref|XM_378316.lj[42659614] gi|42659619|ref|XM_378317.11[42659619] gil42659629|ref|XM_378320.11[42659629] gij4265963ljref|XM_378321.1|[42659631] gi|42659636|ref|XM_378325.11[42659636] gi|42659640JrefjXM_378326.1|[42659640] gi|42659644|ref|XM_378327.1|[42659644] gi|42660072JreflXM_378328.11[42660072] gii42660074[refjXM_378329.1l[42660074] gi|42660076|ref|XM_378330.1|[42660076] giJ42660078Jref|XM_378331.11[42660078] gi|42660080|ref|XM_378332.1 |[42660080] gi|42659999|ref|XM_378336.1|[42659999] gi|42660011 |ref|XM_378339.1 ([42660011] gi|42660015iref|XM_378340.1|[42660015] gi|42660017|reflXM_378342.1 [[42660017] gi|42660019|ref|XM_378343.lj[42660019] giI42660030|ref|XM_378344.l j(42660030] giJ42660032|reflXM_378346.li[42660032] gii42660037|reflXM_378349.li[42660037] giJ42659928|ref|XM_378350.11[42659928] giJ42659933|ref|XM_378353.l([42659933] gi(42659935[ref|XM_378354.1 ([42659935] gi|42659944Jref|XM_378355.li[42659944] gi|42659948Jref[xM_378356.1|[42659948] gi|42659955|ref|XM_378358.11[42659955] giJ42659957|ref|XM_378360.1|[42659957] gi|42659972Jref|XM_378362.11[42659972] gi|42659974|ref|XM_378363.11[42659974]
giJ42659979|ref|XM_378365.1|[42659979] gi[42659981|ref|XM_378366.1|[42659981] gi|42659984Jref|XM_378367.lj[42659984] gi[42659986|ref|XM_378368.1|[42659986] giJ42659892|ref |XM_378371.1 j[42659892] gi|42659894|ref|XM_378372.1|[42659894] giJ42659896(ref(xM_378374.li[42659896] gi|42659903|ref|XM_378379.1|[42659903] giJ42659914|refjXM_378381.lj[42659914] giJ4266010θirefixM_378388.li(42660100] gi|42660055|ref|XM_378389.1 |[42660055] giJ42660057Jref|XM_378390.l[[42660057] giJ4266006θjref |XM_378392.1 j[42660060] giJ42660063Jref|XM_378393.l([42660063] gi(42660065|ref|XM_378394.lj[42660065] gi|42659994|ref|XM_378398.1|[42659994] gi|42660137|ref|XM_378399.1|[42660137] gi|42660143|ref|XM_378404.1|[42660143] giJ42660149|ref|XM_378405.lj[42660149] gi(42660152[ref|XM_378407.1|[42660152] gi|42660164|ref|XM_378411.1|[42660164] giJ42660166irefixM_378412.li[42660166] giJ42660169|ref|XM_378413.1 ([42660169] giJ42660173|refJXM_378414.lj[42660173] gi(42660178[ref|XM_378416.1 ([42660178] gi|42660188|ref|XM_378419.1|[42660188] giJ42660194iref|XM_378421.lj[42660194] giJ42660114|ref|XM_378425.1|[42660114] gi|42660116|reflXM_378428.lj[42660116] giJ42660122Jref|XM_378430.11[42660122] gi|42660127|ref|XM_378431.11[42660127] gi|42660207|ref|XM_378434.1|[42660207] giJ42660209(ref|XM_378436.li[42660209] giJ42660211|ref|XM_378437.1|[42660211] gi|42660217|ref|XM_378439.lj[42660217] giJ42660224Jref|XM_378441.11[42660224] giJ42660249Jref|XM_378449.l([42660249] gi|42660258|ref|XM_378452.1|[42660258] giJ42660264Jref|XM_378453.lj[42660264] gi|42660266(ref|XM_378454.l[[42660266] gi|42660274|ref|XM_378455.1|[42660274] gi|42660276lref|XM_378456.1 |[42660276] giJ42660279JrefixM_378457.lj[42660279] gi J42660289Jref JXM_378460.1 ([42660289] giJ42660299[refjXM_378462.lj[42660299] gi|42660313|ref|XM_378465.l([42660313] gi|42660325|ref|XM_378467.1 |[42660325] gi|42660329JrefjXM_378470.lj[42660329] gi(42660334[ref|XM_378472.11[42660334] gi|42660336|ref|XM_378473.1|[42660336] gi|42660343|ref|XM_378476.1|[42660343] gi(42660345|ref|XM_378477.11[42660345] gi|42660350[ref|XM_378482.1([42660350] gi|42660360|ref|XM_378487.1|[42660360] gi|42660365|ref|XM_378488.lj[42660365] gi|42660367|ref|XM_378489.1|[42660367] gi|42660373[ref|XM_378490.1|[42660373]
gi|42660379|ref|XM_378491.11[42660379] giJ42660621|ref[XM_378493.1[[42660621] gi|42660627|ref|XM_378494.1|[42660627] gi|42660640|ref|XM_378496.1|[42660640] gi|42660416|ref|XM_378506.1|[42660416] gi|42660428|ref|XM_378507.1|[42660428] gi|42660445Jref|XM_378511.1 ([42660445] gi|42660448|refjXM_378512.1|[42660448] gi|42660461 |ref|XM_378514.11[42660461] gi|42660463|ref|XM_378515.11[42660463] gi|42660466|ref[XM_378516.11[42660466] gi|42660468|ref|XM_378517.1 [[42660468] gi(42660484Jref |XM_378522.11[42660484] gi|42660489|ref|XM_378523.1][42660489] gi|42660495 jref |XM_378525.1 ([42660495] gi J42660497 jref |XM_378526.11[42660497] gi|4266050l(refJXM_378528.11[42660501] gi|42660503|ref|XM_378529.11[42660503] gi|42660507|ref|XM_378532.1|[42660507] giJ42660518(ref(XM_378535.l([42660518] gi|42660549[ref|XM_378538.1|[42660549] giI42660569Jref[XM_378542.li[42660569] gi|42660580|ref|XM_378544.1|[42660580] gi|42660582|ref|XM_378545.1|[42660582] gi|42660584|ref|XM_378546.lj[42660584] gi[42660588|ref|XM_378549.11[42660588] gi|42660602|ref|XM_378550.lj[42660602] giJ42660604|ref|XM_378551.1|[42660604] gi|42660694|ref|XM_378553.1|[42660694] gi|42660854|ref|XM_378558.1|[42660854] gi|42660870|ref |XM_378562.11[42660870] gi|42660708|ref|XM_378564.lj[42660708] giJ4266072θiref|XM_378567.11[42660720] gi|42660745|ref[XM_378571.11[42660745] giJ4266075lirefjXM_378573.1 ([42660751] gi|42660932|ref|XM_378576.11[42660932] gi|42660934|refjXM_378577.1|[42660934] gi[42660936|ref|XM_378578.l([42660936] gi|42660952|ref|XM_378579.1|[42660952] gi|42660961 |ref|XM_378582.11[42660961] gi|42660888 jref|XM_378586.11[42660888] gi|42660890|ref|XM_378588.lj[42660890] gi|42660892|ref|XM_378589.1|[42660892] gi|42660897|ref|XM_378590.1|[42660897] gi(42660813iref(XM_378592.lj[42660813] gi|42660817|ref|XM_378594.11[42660817] gi)42660819Jref|XM_378595.1|[42660819] gi|42660768|ref|XM_378599.1 [[42660768] gi|42660776|ref|XM_378601.11[42660776] gi|42660781 |ref|XM_378606.11[42660781] gi|42660787|ref |XM_378607.11[42660787] gi|42660797|ref|XM_378608.1|[42660797] giJ42660803|ref|XM_378609.1 ([42660803] gi|42660808|ref|XM_378610.11[42660808] gi|42660914Jref|XM_378617.lj[42660914] gi|42660876[ref|XM_378620.11[42660876] gi|42660878JrefJXM_378621.1 [[42660878]
gi|42660880|ref|XM_378623.11[42660880] gi J42660882[reflXM_378625.1 j[42660882] giJ42660884JrefjXM_378626.l[[42660884] gi|42660823JreflXM_378628.1|[42660823] gi|42660828|ref|XM_378629.1|[42660828] gii4266083θ[reflXM_378630.1([42660830] gi|42660832JreflXM_378631.lj[42660832] giJ42660836JrefjXM_378632.lj[42660836] gi|42660838Jref|XM_378633.l[[42660838] gi|42660840|ref|XM_378634.1|[42660840] gi(4266085θ[ref[XM_378637.l([42660850] gi|4266130θjref|XM_378639.li[42661300] gι
'J42661065Jref|XM_378642.1 j[42661065] giJ42661069|ref|XM_378643.1 ([42661069] gi|42661084|ref|XM_378646.1 j[42661084] gi|42661094Jref|XM_378648.1|[42661094] gi|4266110ljref[XM_378649.1|[42661101] giJ42661107|ref|XM_378650.11(42661107] gi|42661112|ref|XM_378653.1 ([42661112] giJ4266112θ[ref(XM_378655.11[42661120] gi|42661123|ref|XM_378656.11[42661123] gii42661286JrefjXM_378660.li[42661286] gi|42661297Jref|XM_378661.1|[42661297] giJ42661241|ref|XM_378664.lj[42661241] gi[4266125l(ref|XM_378667.1 [[42661251] gi|42661258|ref|XM_378668.1|[42661258] gi|42661153|ref|XM_378675.11[42661153] giJ42661157Jref(XM_378676.11 [42661157] gi|42661167|ref|XM_378678.1 ([42661167] gii42661175Jref|XM_378680.lj[42661175] gi|42661181|ref|XM_378682.1|[42661181] gi|42661187[ref[XM_378683.1|[42661187] gι
'J42661189JreflXM_378684.1 ([42661189] gi|4266119ljref|XM_378685.1 [[42661191] gi|42661193|ref|XM_378686.11[42661193] gi|42661198|ref|XM_378687.11[42661198] gi|42661200Jref|XM_378688.1|[42661200] gi|42661211|reflXM_378689.11(42661211] gi|42661213Jref|XM_378692.lj[42661213] giJ42661220|ref|XM_378693.l[[42661220] gil42661225|ref|XM_378694.11[42661225] gi|42661023|ref|XM_378695.1J[42661023] giJ42661027|reflXM_378698.1|[42661027] gi|42661029|ref|XM_378700.11[42661029] gi|4266103ljrefjXM_378701.lj[42661031] gi(42661033|ref|XM_378703.1|[42661033] gi|42661035|ref|XM_378705.1|[42661035] gi|42661042Jref|XM_378706.li[42661042] gi|42661046|ref|XM_378708.11[42661046] giJ42661048|refjXM_378709.lj[42661048] gi[42661057|ref|XM_378712.1|[42661057] gi|42661268|ref|XM_378713.1|[42661268] gi(42661308(ref|XM_378723.li[42661308] gi|42661314|ref|XM_378724.1|[42661314] gij42661319|ref|XM_378727.1|[42661319] gi|42661321|ref|XM_378728.11[42661321] gi|42661335|ref|XM_378730.11[42661335]
|42661340 | refl JXM 378734.11(42661340] J42661344 | refl jXM .378735.1 j[42661344] 142661348 |ref J|XM 378738.11(42661348] 142661360 | refl JXM..378741.1 j[42661360] 142661376 |ref j|XM 378742.11(42661376] J42661380 | refl JXM .378743.1 |[42661380] J42661387 |ref j|XM .378745.11(42661387] 142661389 |ref J|XM 378746.1 [[42661389] J42661401 | refl JXM 378747.1 [[42661401] 142661417 | ref J|XM .378750.1 |[42661417] (42661419 |ref J|XM 378751.11(42661419] 142661421 |ref J|XM .378753.1 |[42661421] J42661423 | refl JXM 378754.11(42661423] (42661427 |ref J|XM .378755.1 |[42661427] 142661405 jref JjXM .378756.11(42661405] 142661409 |ref J|XM. 378757.1 |[42661409] 142661411 | refl JXM. 378758.11(42661411] J42661366 Iref JjXM .378760.1 |[42661366] 142661368 |ref J|XM .378763.11(42661368] 142661847 | ref J|XM .378765.1 |[42661847] J42661854 | ref J|XM .378766.11(42661854] 142661859 | ref J |XM .378767.1 |[42661859] 142661861 | ref J|XM .378769.1 [[42661861] (42661863 | ref J|XM .378770.11(42661863] [42661952 | ref J|XM. 378776.11(42661952] (42661956 |ref J|XM .378777.1 |[42661956] 142661991 | ref J|XM..378780.1 ([42661991] 142662010 jrefl JXM 378783.1 [[42662010] [42662020 |ref J|XM .378784.1 |[42662020] 142661557 |ref J|XM. 378786.11(42661557] [42661559 jref J|XM .378787.1 |[42661559] J42661564] | ref J|XM 378791.11(42661564] J42661568 jref j |XM .378793.1 |[42661568] 142661581 | ref J|XM .378794.1 ([42661581] J42661584 jref JjXM .378795.1 |[42661584] |42661586 | ref J|XM 378796.1 [[42661586] [42661599 |ref J|XM .378798.1 |[42661599] 142661604 |ref J|XM..378799.11(42661604] 142661616 | ref J|XM 378800.1 ([42661616] J42661618 jref JjXM 378801.1 [[42661618] (42661634] jref JjXM .378804.11(42661634] 142661636 | ref J|XM .378805.1 |[42661636] 142661638 ( ref J |XM 378806.1 j[42661638] 142661645] | ref J|XM 378807.1 j[42661645] 142661656 jref J|XM .378810.11(42661656] 142661674 | ref j|XM 378812.1 |[42661674] (42656033 jrefl JXM .378815.1 ([42656033] [42655829 |ref J|XM .378820.1 |[42655829] J42656047 |ref J|XM .378822.1 [[42656047] [42656053 |ref J|XM 378823.1 |[42656053] |42656055 |ref J|XM 378824.1 ([42656055] (42656057 | ref J |XM .378825.11(42656057] [42655848 |ref J|XM 378828.1 [[42655848] (42655902 |ref J|XM 378831.11(42655902] |42655904 |ref J|XM .378832.1 |[42655904] (42655626 | refl JXM. 378835.11(42655626] [42656011 jrefl JXM. 378837.1 ([42656011]
|42656064 |ref J|XM 378838.1 |[42656064] 142655685 [ref J]XM 378840.11(42655685] 142655687 Irefl JXM..378841.1 |[42655687] 142655689 [ref J]XM 378842.1 [[42655689] 142655693 Irefl JXM 378843.1 |[42655693] (42655896 Irefl JXM .378848.11(42655896] [42655883 Irefl JXM .378852.1 ([42655883] (42655735 |ref J|XM .378855.1 [[42655735] [42655741 | ref J]XM 378858.11(42655741] I42655745 |ref J]XM..378859.1 ([42655745] (42655748 Iref JjXM .378860.1 |[42655748] [42655751 I ref J]XM..378861.11(42655751] |42655756 Irefl JXM .378862.1 [[42655756] (42655777 jref J]XM..378865.11(42655777] |42655779 Irefl JXM 378866.1 ([42655779] |42655654 jref J]XM..378873.1 [[42655654] J42655971 JXM .378874.1 ([42655971] (42655973 Irefl JXM 378876.1 |[42655973] J42655664 [ref J]XM..378877.1 [[42655664] |42655666 jref J]XM .378879.1 |[42655666] |42655668 Irefl JXM .378880.1 ([42655668] |42655578 I ref J]XM .378883.1 [[42655578] (42655977 Irefl JXM 378886.1 ([42655977] [42655644 Irefl JXM .378889.1 [[42655644] 142656103 jref J]XM. 378890.1 ([42656103] (42656105 jref J]XM .378891.1 [[42656105] [42656068 Irefl JXM .378892.1 ([42656068] |42655793 | refl JXM..378893.1 |[42655793] (42655795 jref J]XM..378894.1 [[42655795] [42656110 Irefl JXM .378897.1 |[42656110] 142656112 Iref JjXM .378898.11(42656112] (42655809 | ref J |XM 378899.1 |[42655809] [42655724 I ref J|XM. 378901.11(42655724] J42656126 jref J|XM .378903.1 ([42656126] 142656132 | ref J |XM .378905.1 ([42656132] J42655942 iref JlXM..378908.1 |[42655942] |42655944 | ref J |XM .378909.1 [[42655944] |42655946 Irefl JXM .378910.1 IJ42655946] |42655949 jref JjXM .378912.11(42655949] 142655861 Iref JjXM .378914.1 JJ42655861] |42655865 Iref JjXM .378917.1 j[42655865] |42655867 Iref JjXM 378918.1 |[42655867] (42655869 jref J|XM 378919.1 j[42655869] [42655871 jref J]XM .378921.1 ([42655871] |42655879 Irefl JXM..378923.11(42655879] (42655957 [ref |XM .378925.1 |[42655957] I42655820 jref J|XM 378930.1 [[42655820] J42655599 Irefl JXM .378933.1 [[42655599] J42655601 Irefl JXM..378934.11(42655601] [42655986 jref J|XM .378941.1 ([42655986] (42655994 I ref J|XM 378945.1 ([42655994] I42655996 jref J|XM .378946.1 |[42655996] [42655998 jref JlXM .378947.1 [[42655998] (42656148 Jref JlXM .378949.1 |[42656148] [42656086 Jref J|XM..378950.1 j [42656086] I4265609O Irefl (XM..378951.1 | [42656090] J42656185 Irefj JXM. 378954.11(42656185]
142656187 Irefl JXM 378955.11(42656187] |42662254 Irefl JXM .378956.1 j[42662254] (42662256 Irefl JXM 378957.1 ([42662256] |42662267 |ref J|XM 378961.11(42662267] [42662269 |ref J|XM 378964.1 j[42662269] |42662273 |ref J|XM .378969.1 |[42662273] |42662276 Iref JjXM. 378970.11(42662276] (42662278 Irefl JXM 378971.1 ([42662278] I42662284 |ref J|XM 378973.1 [[42662284] [42662288 Iref JjXM 378974.1 ([42662288] (42662295 Iref JjXM .378976.11(42662295] [42662213 |ref J|XM .378977.1 |[42662213] (42662217 Iref JjXM .378978.11(42662217] [42662219 |ref J|XM 378980.1 |[42662219] (42662225 jref JjXM. 378981.1 j[42662225] [42662227 Irefl JXM 378982.11(42662227] J42662229 jref J|XM 378983.1 [[42662229] 142662231 jref J|XM. 378985.1 j[42662231] |42662244] lref J|XM 378988.1 |[42662244] [42662301 Irefl JXM .378989.11(42662301] 142662081 Irefl JXM..378992.1 ([42662081] [42662083 |ref J|XM 378993.11(42662083] (42662086 Irefl [XM .378994.1 [[42662086] 142662318 Irefl JXM..379002.1 j[42662318] J42662320 jrefl JXM 379003.1 JJ42662320] J42662323 Irefl JXM..379004.11(42662323] [42662326 irefl JXM 379006.1 j[42662326] (42662331 Irefl JXM .379009.11(42662331] J42662335 Irefl |XM_ 379011.1 |[42662335] |42662373 jref J|XM .379012.11(42662373] J42662339 Iref J|XM .379014.1 [[42662339] 142662341 Irefl JXM..379015.1 [[42662341] (42662343 Jref J|XM. 379016.1 |[42662343] |42662345 jref JjXM .379017.11(42662345] (42662347] Jref j|XM .379018.1 ([42662347] (42662349 Irefl |XM_ 379020.1 [[42662349] J42662351 jref jjXM .379021.11(42662351] |42662353 jref j|XM .379022.1 ([42662353] I42662355 Jref j|XM 379023.11(42662355] J42662361 jref| |XM_ 379025.1 j[42662361] I42662377] Irefl JXM .379029.1 |[42662377] J42662379 jref J|XM. 379030.1 j[42662379] J42662385 ]ref J|XM .379032.11(42662385] 142662415 Irefl JXM 379036.11(42662415] [42662440 jref J|XM 379040.1 | [42662440] I42662444 Irefl JXM .379041.11(42662444] [42662450 jref J|XM 379044.1 j[42662450] (42662454 jref J |XM .379046.1 ([42662454] [42662466 Irefl JXM .379052.1 |[42662466] (42662476 jrefl |XM_ .379054.11(42662476] [42662480 Irefl JXM..379055.1 ([42662480] (42662510 Irefl JXM .379060.1 [[42662510] [42662514 jrefl JXM .379064.1 ([42662514] |42662518 jrefl JXM. 379065.1 [[42662518] (42656372 |ref J|XM..379068.1 ([42656372] [42656374 |ref J|XM..379069.1 |[42656374] [42656491 Iref JjXM. 379072.1 [[42656491]
142656498 | ref j |XM 379073.11(42656498] 142656500 I ref J|XM 379074.1 |[42656500] (42656504 | ref j|XM 379075.11(42656504] 142656513 |ref J|XM .379077.1 |[42656513] [42656189 I ref j|XM 379078.11(42656189] 142656192 jref J|XM..379079.1 JJ42656192] (42656195 I ref J|XM 379080.11(42656195] I42656282 I ref J|XM..379085.11(42656282] |42656284 jref J|XM 379086.11(42656284] |42656299 I ref j|XM 379089.1 ([42656299] [42656305 | ref J|XM..379094.11(42656305] (42656311 Irefl JXM 379096.1 |[42656311] (42656313 |ref J|XM 379097.11(42656313] 142656315 Irefl JXM .379098.1 ([42656315] (42656320 jrefl JXM 379099.1 JJ42656320] [42656324 I ref J |XM .379100.1 IJ42656324] (42656327 | ref J |XM..379101.1 j[42656327] |42656333 I ref | JXM 379102.11(42656333] [42656345 I ref J|XM .379106.1 j[42656345] |42656359 | ref J |XM..379108.1 |[42656359] (42656411 jref (lXM 379109.11(42656411] [42656395 jref J|XM..379111.1 [[42656395] 142656217 jref JlXM 379112.1 [[42656217] (42656225 jref J|XM .379113.11(42656225] J42656227 jref J|XM..379114.1 ([42656227] |42656238 Iref JXM 379117.11(42656238] (42656240 | ref J|XM..379118.1 |[42656240] [42656242 jref |XM..379119.1 [[42656242] |42656407 jrefl JXM 379121.1 ([42656407] J42656202 |ref JXM .379122.1 [[42656202] J42656206 | ref J]XM .379123.1 |[42656206] J42656459 jref J]XM. 379131.11(42656459] 142656461 jrefl JXM .379133.1 ([42656461] |42656470 I ref J|XM .379135.11(42656470] J42656422 |ref JjXM .379136.1 [[42656422] (42656450 Iref JjXM..379141.11(42656450] |42656533 Iref JjXM .379145.1 |[42656533] J42656535 I refl JXM..379146.11(42656535] |42656537 jrefl JXM .379147.1 ([42656537] |42656544 I ref J|XM .379149.11(42656544] |42656574 jref J]XM 379154.1 [[42656574] (42656581 jref J]XM .379156.1 ([42656581] |42656589 | ref JXM..379158.1 |[42656589] [42656591 jref J|XM 379159.1 [[42656591] (42656481 I ref J]XM .379161.1 |[42656481] |42656483 | ref J |XM 379163.11(42656483] [42656487 Jref J|XM 379164.1 ([42656487] (42656260 jref J|XM..379166.1 j[42656260] [42656612 jref J |XM..379167.1 |[42656612] 142656619 Irefl (XM 379168.1 ([42656619] J42656416 I ref j|XM .379169.1 j[42656416] [42656631 Irefl JXM .379170.11(42656631] J42656660 |ref J|XM .379171.1 |[42656660] J42656664 jrefl JXM .379172.1 [[42656664] |42656666 jrefl JXM .379173.1 ([42656666] |42656668 |ref J|XM .379174.1 [[42656668] J42656670 jref JjXM 379175.1 |[42656670]
giJ42656675 refl JXM. 379177. 11(42656675] gi|42656686 ref J|XM 379179 I [[42656686] giJ42656698 refl JXM. 379180. 11[42656698] gi|42656700 ref J|XM 379181 . l |[42656700] gi|42656702 refl JXM 379182. l |[42656702] gi(42656704 refl JXM 379183. 11[42656704] gi|42656706 refl JXM 379184 I [[42656706] gi|42656719 refl JXM 379189 l |[42656719] gi[42656724 refl JXM 379190. 11[42656724] gi(42656732 ref JjXM. 379191 . 11[42656732] gi[42656734 refl JXM 379192. I ([42656734] gi|42656740 ref J|XM
"379194. 11[42656740] gi(42656747 ref JjXM. 379195. I ([42656747] giJ42656751 ref J|XM 379196. I [[42656751] giJ42656637 ref J|XM
"379197. l |[42656637] giJ42656639 ref j|XM .379198. 11(42656639] gi|42656656 ref JjXM. 379200. I [[42656656] gi|42656777 refl JXM 379201 . 11[42656777] gi|42656781 refl JXM. 379203. 1 |[42656781] gi|42656783 refl JXM .379204. 11[42656783] gi[42656787 ref J|XM
"379205. I [[42656787] gi(42656792 refl JXM..379206. 11(42656792] gi|42656799 ref J|XM 379207. I ([42656799] giJ42656810 ref J|XM..379210. l |[42656810] gi|42656819 ref J|XM 379213. l |[42656819] gi(42656826 ref J|XM. 379214. I [[42656826] giJ42656828 ref J|XM 379215. I I [42656828] gi|42656854 ref J|XM..379228. 11(42656854] gi|42656858 ref J|XM 379229. 11[42656858] giJ42656862 ref J|XM..379230. 11[42656862] gi(42656868 ref J|XM .379231 . I [[42656868] giJ42656872 ref J|XM..379233. l |[42656872] gi|42656881 ref J|XM .379234. 11[42656881] gi|42656883 ref J|XM..379235. 11[42656883] gi|42656890 ref JjXM..379240. 11(42656890] giJ42656771 refl JXM..379243. l |[42656771] giJ42656758 refl JXM 379244. 11[42656758] gi|42656766 refl JXM .379247. 11[42656766] giJ42657043 ref J|XM .379248. 11[42657043] gi|42657045 ref J|XM .379249. I ([42657045] gi|42657052 ref JjXM .379250. l |[42657052] gi|42657071 refl jXM .379252. 1 |[42657071] giJ42657065 refl JXM 379254. I j[42657065] giJ42657067 refl JXM .379255. I [[42657067] gi|42657091 refl JXM .379256. 11[42657091] gi|42657105 refl JXM .379258. 11[42657105] giJ42657107] ref J|XM .379260. 11[42657107] gi|42656914 ref JjXM .379262. 11[42656914] gi[42656920 ref J|XM .379263. 11[42656920] gi|42656922 ref JjXM..379264. 11[42656922] gi(42657079 refl JXM 379265. I ([42657079] gi|42657005 refl JXM .379267. 11[42657005] giJ42657007 ref J|XM 379268. 11[42657007] gi|42657135 ref JjXM 379270. I [[42657135] gi|42656931 ref J|XM. 379271 . 11[42656931] giJ42656944 refl JXM 379273. 11[42656944] gi(42656948 ref JlXM. 379274 I j[42656948]
|42656957 Iref JlXM .379275.11(42656957] [42656964 Irefl JXM .379276.11(42656964] 142656977 Irefl JXM. 379278.11(42656977] J42656982 Irefl JXM 379280.1 |[42656982] 142656991 Irefl JXM..379287.1 |[42656991] J42656993 |ref J|XM .379288.1 ([42656993] |42657024 Irefl JXM..379294.1 |[42657024] I42657027 Irefl JXM .379295.1 [[42657027] (42657029 Irefl JXM..379298.1 IJ42657029] I42657032 Iref JjXM .379299.1 ([42657032] 142657153 Jref JlXM 379303.1 |[42657153] 142657158 Irefl JXM .379306.1 [[42657158] [42657165 iref J|XM..379309.1 |[42657165] J42657175 Irefl JXM 379313.1 ([42657175] 142657185 |ref J|XM .379317.1 |[42657185] 142657187 |ref J|XM .379318.11(42657187] (42657189 Irefl JXM .379320.1 j[42657189] |42657320 Irefl JXM .379321.11(42657320] I42657322 |ref J|XM .379322.1 ([42657322] I42657324 Irefl JXM .379323.1 ([42657324] I42657326 Irefl JXM. 379324.1 |[42657326] I42657330 jrefl JXM 379325.1 [[42657330] [42657332 Irefl JXM .379326.1 ([42657332] (42657334 Iref JjXM..379327.1 [[42657334] [42657338 | ref J |XM 379328.1 |[42657338] |42657343 Irefl JXM .379331.11(42657343] (42657345 Jref JjXM. 379332.1 |[42657345] I42657347 jref (jXM .379333.1 ([42657347] 142657301 |ref JjXM .379334.1 |[42657301] J42657304 iref JlXM..379336.1 ([42657304] I42657387 jrefl JXM 379339.11(42657387] (42657392 Irefl JXM..379340.1 [[42657392] [42657225 |ref J|XM..379343.1 ([42657225] |42657233 jrefl JXM .379347.1 |[42657233] (42657284 |ref J|XM .379355.1 [[42657284] [42657286 |ref J|XM .379359.1 ([42657286] (42657250 Jref J|XM..379363.11(42657250] [42657254 Irefl JXM. 379364.1 |[42657254] |42657256 Jref J|XM .379366.11(42657256] (42657258 |ref J|XM .379368.1 j[42657258] |42657192 | ref J |XM. 379371.11(42657192] [42657197 |ref J|XM .379372.1 ([42657197] (42657205 |ref J|XM..379373.11(42657205] [42657212 |ref J|XM .379377.1 [[42657212] (42657214 Irefl JXM .379378.1 ([42657214] I42657400 Irefl JXM..379380.1 |[42657400] |42657407 Irefl JXM .379381.1 |[42657407] [42657410 Irefl JXM .379382.1 [[42657410] J42657498 Irefl JXM .379384.11(42657498] I42657500 Irefl JXM..379386.1 ([42657500] |42657502 jref JjXM .379391.11(42657502] |42657504 Irefl JXM..379392.1 |[42657504] J42657510 jrefj JXM..379393.1 [[42657510] |42657572 Irefl JXM..379395.1 IJ42657572] |42657575 Irefl JXM..379396.1 [[42657575] J42657580 Irefl JXM .379398.1 [[42657580] J42657593 jref JjXM. 379401.11(42657593]
gi|42657600|ref|XM_379402.1|[42657600] giJ42657605(ref|XM_379403.lj[42657605] gi|42657607|ref |XM_379404.11[42657607] gi|42657626|ref|XM_379406.1|[42657626] gi|42657628|ref|XM_379408.1|[42657628] gi|42657630|ref|XM_379409.11[42657630] giJ42657632JrefJXM_379410.li[42657632] gi|42657636|ref|XM_379411.11[42657636] gi|42657645|ref|XM_379413.11[42657645] gi|42657661|ref|XM_379417.1|[42657661] gi|42657520|ref|XM_379424.1 [[42657520] giJ42657448|refjXM_379430.1|[42657448] gi|42657460|ref|XM_379432.11[42657460] gi|42657462|ref|XM_379433.1|[42657462] gi|42657467|ref|XM_379434.1|[42657467] gi|42657471|ref|XM_379435.1|[42657471] gi|42657475|ref|XM_379436.1|[42657475] gi J42657477|ref|XM_379437.1 ([42657477] gi|42657479|ref|XM_379438.1|[42657479] giJ42657484|ref|XM_379439.1 j[42657484] gi|42657490|ref|XM_379441.11[42657490] giJ42657550lreflXM_379450.1J[42657550] gi|42657553|ref|XM_379452.11[42657553] gi|42657555|ref|XM_379453.1|[42657555] giJ42657559[refixM_379454.1 ([42657559] gi|42657533|ref|XM_379456.1|[42657533] giJ42657536Jref|XM_379458.lj[42657536] gi|42657565(ref jXM_379459.1 ([42657565] gi|42657567|ref|XM_379460.1|[42657567] gi|42657667JreflXM_379461.11[42657667] gi|42657669|ref|XM_379462.11[42657669] gii42657673|refJXM_379463.lj[42657673] gi|42657806|ref|XM_379467.1|[42657806] gi|42657810|ref|XM_379469.1J[42657810] gi|42657820|ref|XM_379471.11[42657820] gi|42657824|ref|XM_379472.1|[42657824] giJ42657852Jref|XM_379474.1|[42657852] gi|42657857|ref|XM_379476.11[42657857] gi|42657865Jref|XM_379477.1|[42657865] gi|42657867[ref(XM_379478.1|[42657867] giJ42657871 |ref|XM_379479.11[42657871] giJ42657875|ref|XM_379480.lj[42657875] giJ42657877|ref|XM_379481.1 ([42657877] gi|42657879|ref|XM_379482.1|[42657879] giJ42657885irefjXM_379483.1|[42657885] gi|42657893|ref|XM_379484.1|[42657893] gi!42657902[ref|XM_379485.1|[42657902] gii42657910|ref|XM_379486.1|[42657910] gi|42657916|ref|XM_379487.1|[42657916] gi|42657699|ref|XM_379488.11[42657699] gi|42657701 |ref|XM_379489.11[42657701] gi(42657703[ref|XM_379490.1|[42657703] gi|42657681|ref|XM_379491.11[42657681] gi|42657683(ref|XM_379492.lj[42657683] giJ42657728 jreflXM_379493.11[42657728] gi|42657730|reflXM_379494.1|[42657730] giJ42657742Iref|XM_379495.1|[42657742]
142657746 refl XM. 379496.11 [42657746] J42657754] refl XM 379498, 1 |[42657754] 14265775 reflXM .379499.1 1[42657757] (42657759 refl IXM 379500.1 [[42657759] J42657762] refl XM .379501 , 1 J [42657762] |42657764 refl JXM..379502.11[42657764] 142657774] ref XM 379503, 1 |[42657774] J4265801 refl jXM 379504.1 |[42658017] |42658034 refl |XM .379506.1 JJ42658034] [42658042] refl [XM 379507, 11[42658042] |42658044 refl JX 379508, 1 |[42658044] |42658052] refl XM 379510, 1 1(42658052] J42658059 refl XM 379511.1 1(42658059] |42658076 refl IXM .379512, 1 1(42658076] [42658081 refl XM 379513, 1 1[42658081] |42658085 refl XM. 379514, 1 1[42658085] |42658088] refl XM .379515, 1 1[42658088] J42658093 refl XM 379516, 1 1[42658093] I42658095 refl XM 379517. I J[42658095] J42658097 refl XM 379518.1 JJ42658097] (42658110 refj XM .379520.1 |[42658110] 142658115] refl XM 379521 , 1 |[42658115] 142658122 refl XM 379522, I I [42658122] J42658124] refj XM .379523, 1 1[42658124] J42658131 reflXM 379524, 1 1[42658131] 142657951 refl XM 379526, 1 [[42657951] I42657980] refj XM .379527, 1 1[42657980] |42657982 refl XM .379528, 1 1[42657982] |42657984 refl IXM .379529.1 1[42657984] J42657988 refl XM .379530, 1 1[42657988] |42657994 refl XM 379531, 1 1[42657994] |42657996 reflXM .379532, 1 1[42657996] J42657705 refl XM .379533, 1 [[42657705] I42657709] refl !XM 379534.1 |[42657709] |42657715i refl XM..379535.1 [[42657715] 142657722 refl IXM .379536.1 1[42657722] 142657707 reflXM .379537, 1 1[42657707] 142658791 refl XM 379539, 1 |[42658791] I42658793 refl XM 379540.1 |[42658793] I42658795 refl XM .379541, 1 1[42658795] 142658801 refl XM .379543, 1 1[42658801] I42658803 refl XM. 379545, 1 1[42658803] |42658807 refl XM .379547, 1 j [42658807] 142658811 ref] XM .379548, 1 |[42658811] (42658820 refl XM .379550 1 ([42658820] J42658872 refl XM 379551, 1 1[42658872] I42658877 ref] XM 379552 1 1[42658877] J42658882 refl JXM .379553, 1 1[42658882] |42658884| refl XM .379554, 1 1[42658884] J42658857
1 refl XM 379559 1 1[42658857] J42658863 refj [XM 379562 1 1[42658863] |42658902 refl XM .379573 1 1[42658902] 142658952 refl XM .379582 1 1[42658952] |42658960, refl XM .379583 1 1(42658960] J42658962 refl XM .379584 1 1[42658962] 142658964, refj XM .379586 1 1[42658964] 142658966 refl XM 379587 ,1 1[42658966]
gi|42658913 reflXM 379592.11(42658913] gi|42658915 reflXM 379594.1 |[42658915] gi[42658921 reflXM 379595.1 ([42658921] gi|42658925 reflXM 379596.1 |[42658925] gi|42658927| reflXM 379597.1 j[42658927] gi|42658933 reflXM 379601.1 |[42658933] gi|42658937 reflXM 379603.1 [[42658937] gi(42658939 reflXM 379605.1 ([42658939] giJ42658845 reflXM 379608.1 [[42658845] gi(42658847 reflXM 379609.1 ([42658847] gi|42658849 reflXM .379610.1 [[42658849] gi|42659148 reflXM 379617.11(42659148] giJ42659150 reflXM 379618.1 ([42659150] giJ42659152 reflXM 379619.11(42659152] giJ42659154 reflXM .379622.1 j[42659154] giJ42659164 reflXM 379623.1 |[42659164] gi|42659169 reflXM .379625.11(42659169] giJ42659174 reflXM .379627.1 |[42659174] gi|42659180 reflXM .379628.1 j[42659180] gi|42659182 reflXM..379629.1 |[42659182] giJ42659195 reflXM .379630.1 [[42659195] giJ42659199 reflXM .379632.1 |[42659199] gi|42659201 reflXM..379634.1 ([42659201] giJ42659210 reflXM 379635.1 j[42659210] giJ42659214 reflXM .379636.11(42659214] giJ42659121 reflXM .379637.1 |[42659121] giJ42659088 reflXM..379638.1 [[42659088] gi(42659097 reflXM 379639.1 [[42659097] giJ42659099 reflXM..379640.1 j[42659099] giJ42659109| reflXM..379641.1 |[42659109] gi|42659113| reflXM..379642.11(42659113] gi|42659010| reflXM .379643.1 |[42659010] gi|42659019| reflXM .379644.11(42659019] gi|42659031 reflXM..379645.1 j[42659031] gi|42659035 reflXM .379647.11(42659035] giJ42659132| reflXM 379648.1 ([42659132] gi|42659299| reflXM .379650.11(42659299] gi|42659302 reflXM .379651.1 ([42659302] gi|42659308 reflXM 379655.1 |[42659308] gi|42659222 reflXM..379656.1 [[42659222] gi|42659225 reflXM 379657.1 ([42659225] giJ42659231 reflXM .379660.11(42659231] giJ42659244 reflXM 379664.1 |[42659244] gi|42659246 reflXM..379665.1 [[42659246] gi|42659254 reflXM 379667.11(42659254] gi(42659267 reflXM .379668.1 ([42659267] gi[42659277 reflXM .379671.1 [[42659277] giJ42659279 ref|XM..379672.1 j[42659279] gi|42658994 reflXM 379676.11(42658994] gi[42658998 reflXM 379677.11(42658998] gi|42659000 reflXM .379678.1 ([42659000] gi|42659002 reflXM. 379680.11(42659002] giJ42659043 reflXM..379682.1 |[42659043] giJ42659142 reflXM..379684.11(42659142] gi|42662773 reflXM..379686.1 [[42662773] gi|42662779 reflXM .379688.11(42662779] gi(42662783 reflXM 379690.1 |[42662783]
gi|42662785|ref|XM_379691.11[42662785] giJ42662787|ref[XM_379692.1([42662787] gi|42662706(ref|XM_379693.1|[426627O6] giJ42662693|ref|XM_379694.lj[42662693] gi|42662649JrefixM_379695.l[[42662649] gi|42662651 jref|XM_379696.11[42662651] giJ42662662JreflXM_379697.lj[42662662] gi|42662676|reflXM_379699.1 [[42662676] gi|42662728|ref|XM_379700.1|[42662728] gi|42662537Jref|XM_379702.li[42662537] gi|42662541|ref|XM_379703.1|[42662541] gi|42662543Jref|XM_379704.1[[42662543] gi|42662551 |ref|XM_379705.11[42662551] gi[42662623|ref|XM_379714.1|[42662623] gi|42662630|ref|XM_379715.1|[42662630] gi|42662634|ref |XM_379716.11[42662634] giJ42662586[ref(XM_379717.lj[42662586] gi|42662699|ref|XM_379720.1|[42662699J gi|42662533|ref[XM_379721.11[42662533] giJ42662714Jref|XM_379722.1 ([42662714] gi J42662716|ref|XM_379723.11[42662716] gi|42662685|ref|XM_379728.1)[42662685] gi|42662702ireflXM_379730.l([426627O2] gi|42662754|ref|XM_379735.1|[42662754] gi|42662764Jref(XM_379736.li[42662764] gi(42662737|ref[XM_379738.1|[42662737] gi|42662739|ref)XM_379739.lj[42662739] giJ4265975l(ref|XM_379741.1 [[42659751] gi|42661161|reflXM_379749.1|[42661161] gi(42662708iref[XM_379760.lj[426627O8] gi|42662756|ref|XM_379761.11[42662756] gi|42658173Jref|XM_379766.11[426581 73] gi[42658175|reflXM_379767.11[42658175] gi|42658183|ref|XM_379771.11[426581 83] giJ42658185Jref|XM_379772.1 [[426581 85] gi|42658187|reflXM_379773.11[426581 87] gi|42658189iref|XM_379774.lj[426581 89] gi|42658193|ref|XM_379775.1 [[426581 93] gi|42658197|ref|XM_379776.11[426581 97] gi|42658201 |ref|XM_379777.11[42658201] gi|42658203|ref|XM_379780.1 [[42658203] gi|42658205Jref|XM_379781.11[42658205] gi|42658210|ref|XM_379783.1|[42658210] gi|42658212|ref|XM_379784.1|[42658212] gi|42658216Jref|XM_379786.li[42658216] gi|42658218Jref|XM_379787.1|[42658218] gi|42658220|ref|XM_379788.lj[42658220] gi[42658230|ref|XM_379792.1 ([42658230] gi|42658232|ref|XM_379793.1|[42658232] gi|42658234|ref|XM_379794.1|[42658234] gi|42658238|ref|XM_379796.1|[42658238] gi|42658240|ref|XM_379797.lj[42658240] giJ42658242|ref|XM_379798.1 ([42658242] gi|42658253|ref|XM_379800.1|[42658253] gi|42658261|ref|XM_379801.1 ([42658261] gi|42658263|ref|XM_379802.11[42658263] giJ42658267Jref|XM_379803.1|[42658267]
142658270 |ref J|XM 379806.11(42658270] J42658272] jref JjXM .379807.1 ([42658272] 142658276 Irefl JXM 379809.1 j[42658276] 142658281 Jref J|XM 379812.11(42658281] J42658287 Irefl JXM .379815.1 j[42658287] 142658291 Irefl JXM 379816.1 |[42658291] |42658293 Irefl JXM 379817.1 ([42658293] J42658295 iref J|XM .379818.11(42658295] [42658297 Irefl JXM
" .379819.1 |[42658297] |42658299 Irefl JXM 379820.11(42658299] [42658303 Irefl JXM 379824.1 |[42658303] |42658305 Irefl JXM 379825.1 ([42658305] J42658310 |ref J|XM 379827.1 ([42658310] 142658313] jref J|XM .379830.1 |[42658313] 142658315 Irefl JXM 379831.11(42658315] 142658317 Irefl JXM .379832.1 j[42658317] (42658319 [ref| JXM 379834.1 |[42658319] 142658321 Irefl JXM
" .379835.1 ([42658321] J42658323 Irefl JXM .379836.1 [[42658323] |42658328 jref J|XM .379838.1 |[42658328] [42658334 J]refl JXM 379839.11(42658334] [42658336 lref J|XM .379840.1 ([42658336] |42658338 |ref J|XM 379841.1 |[42658338] [42658344 Irefl JXM .379842.1 ([42658344] J42658374 Irefl JXM 379843.1 |[42658374] I42658380 Iref JlXM .379844.1 ([42658380] J42658382 Irefl JXM .379845.11(42658382] |42658384 Irefl JXM 379846.1 j[42658384] |42658386 jref JjXM 379847.1 ([42658386] [42658388 Irefl JXM .379848.1 |[42658388] J42658390 Irefl JXM .379849.1 ([42658390] [42658394 Iref JjXM .379850.1 ([42658394] J42658396 jref J]XM 379851.1 j[42658396] |42658398 jref J|XM 379852.11(42658398] J42658400 Irefl JXM .379853.1 j [42658400] J42658404 jref J|XM .379854.1 ([42658404] |42658406 Iref JjXM .379855.1 |[42658406] J42658408 Irefl JXM 379856.1 [[42658408] J42658416 |ref J]XM 379857.1 |[42658416] |42658422 Irefl JXM 379858.1 |[42658422] J42658424 Irefl JXM .379859.1 j [42658424] I42658426 Irefl JXM .379860.1 | [42658426] J42658430 Irefl JXM 379861.1 [[42658430] (42658432 Iref JjXM .379862.1 j [42658432] |42658434| jref J|XM .379863.1 | [42658434] |42658436 jref JjXM 379864.1 | [42658436] J42658438 Irefl JXM .379865.1 | [42658438] J42658440 jref J|XM .379866.1 | [42658440] I42658458 Irefl JXM 379868.1 j [42658458] J42658460] jref J|XM .379869.1 | [42658460] J42658464 jref J|XM .379871.1 | [42658464] I42658466 [ref JjXM .379872.1 | [42658466] J42658470! Irefl JXM 379874.1 |[42658470] J42658472 Iref JjXM .379875.1 |[42658472] |42658474| Irefl JXM .379876.1 |[42658474] |42658476 [ref J]XM .379877.11(42658476] |42658482 Irefl JXM 379879.1 |[42658482]
M 379881.11(42658486] M 379883.1 |[42658490] M 379884.11(42658492] M 379885.1 j[42658494] M .379887.11(42658501] M 379891.11(42658504] M .379892.11(42658508] M 379893.11(42658510] M .379894.1 |[42658514] M 379895.11(42658516] M 379896.1 |[42658518] M 379897.1 j[42658520] M .379899.1 |[42658523] M 379901.1 |[42658526] M 379904.11(42658556] M..379905.11(42658558] M..379906.11(42658560] M..379908.11(42658566] M 379909.1 |[42658568] M 379910.1 I[42658570] M..379911.11(42658574] M 379913.1 |[42658578] M .379914.1 j[42658584] M..379917.1 I[42658588] M 379919.1 I[42658598] M .379920.1 I[42658600] M..379921.1 I[42658602] M 379923.1 |[42658606] M .379927.11(42658614] M..379931.11(42658623] M.379932.11(42658625] M 379933.11(42658627] M .379934.1 |[42658631] M .379935.1 [[42658633] M.379936.1 ([42658635] M .379938.1 |[42658639] M..379939.1 |[42658641] M.379940.1 |[42658643] M .379954.11(42658647] M 379958.1 |[42658651] M .379959.11(42658653] M .379962.11(42658655] M.379964.11(42658657] M .379965.1 I[42658659] M 379966.1 |[42658661] M .379967.11(42658663] M 379968.11(42658665] M .379970.11(42658671] M .379974.11(42658675] M .379975.11(42658677] M 379976.1 j[42658683] M .379977.11(42658685] M .379979.11(42658751] M .379980.11(42658753] M 379983.11(42658759] M 379986.11(42658346]

M.379987.11(42658348]
gi|42658350|ref|XM_379988.1 l(42658350] gi[42658352|ref|XM_379989.1 |[42658352] giJ42658354[ref|XM_379990.l [[42658354] gi(42658356iref(xM_379991.11[42658356] gi[42658358|ref|XM_379993.1 [[42658358] gi|42658360|ref|XM_379994.1 |[42658360] gi|42658362Jref|XM_379995.l [[42658362] giJ42658364|ref|XM_379996.1 |[42658364] gi|42658366[reflXM_379997.l i[42658366] gi|42658368|ref|XM_379998.1 |[42658368] gii4265837θiref|XM_379999.l ([42658370] gii42658372|ref(xM_380002.l i[42658372] gi|42658442(ref|XM_380005.1 [[42658442] gi|42658444|ref|XM_380006.1 |[42658444] gi|42658446iref[xM_380007.l i[42658446] gi|42658448[ref|XM_380008.1 |[42658448] giJ42658450|reflXM_380009.1 ([42658450] gi(42658452[ref|XM_380010.1 ([42658452] gi(42658454|ref|XM_380011.11[42658454] gi[4265853θireflXM_380012.1 ([42658530] gii42658532Jref|XM_380013.1 |[42658532] gi|42658534|ref|XM_380014.l [[42658534] gi|42658536|ref|XM_380015.1 |[42658536] gi(42658538[ref|XM_380018.1 ([42658538] giJ4265854θirefixM_380019.l ([42658540] gi|42658542|ref|XM_380020.1 |[42658542] gi[42658544|reflXM_380021.11[42658544] gi[42658546|ref|XM_380022.l ([42658546] gi[42658548iref|XM_380024.l i[42658548] gi|42658550JreflXM_380025.l i[42658550] gi|42658552(ref|XM_380026.1 |[42658552] giJ42658691 [ref|XM_380028.11[42658691] gil42658693(ref|XM_380031.1 [[42658693] gi|42658695|ref|XM_380032.l [[42658695] gi[42658697|reflXM_380033.l i[42658697] gi(42658699|ref|XM_380034.1 |[42658699] giJ42658701 (ref|XM_380036.1 [[42658701] gi|42658703|ref|XM_380038.11[42658703] gi(42658705|ref|XM_380039.1 |[42658705] gi|42658707[ref|XM_380040.l [[42658707] gi|42658709|reflXM_380042.11[42658709] gi|42658711 |ref|XM_380044.1 [[42658711] gi(42658713iref|XM_380045.1 |[42658713] gi(42658715|ref|XM_380047.1 [[42658715] gi|42658717|ref|XM_380048.1 [[42658717] gi|42658719|ref|XM_380055.1 |[42658719] giJ42658721 [ref|XM_380056.11[42658721] gi|42658723|ref|XM_380057.l ([42658723] gi[42658725ireflXM_380059.1 [[42658725] gi|42658727|ref|XM_380063.1 |[42658727] gi|42658729(ref|XM_380067.11[42658729] gi|42658731 |ref|XM_380068.1 j[42658731] gi|42658733(ref|XM_380069.1 |[42658733] gi|42658735[ref|XM_380070.1 |[42658735] gi|42658737Jref|XM_380072.l j[42658737] gi(42658739|ref|XM_380073.l ([42658739] giJ42658741 (ref|XM_380074.1 [[42658741]
giI42658743 Irefl JXM 380076.11(42658743] gi|42658745] Irefl jXM 380077.1 |[42658745] giJ42658764 Irefl JXM 380078.1 ([42658764] giJ42658554] iref JlXM 380081.1 |[42658554] gi|42658177 jref J|XM 380082.11(42658177] giJ42658181 Irefl JXM 380085.1 ([42658181] gi|42658191 jrefl JXM .380087.1 ([42658191] giJ42658195 Irefl JXM..380088.1 ([42658195] gi|42658228 jrefl |XM..380089.1 [[42658228] gi|42658236 Irefl JXM .380091.1 IJ42658236] giJ42658244 Irefl jXM .380092.1 j[42658244] giJ42658246 jref JjXM .380093.11(42658246] gi|42658248 iref JjXM .380094.1 j[42658248] gi|42658251 Iref JjXM .380095.1 [[42658251] giJ42658255 Irefl JXM .380096.1 ([42658255] giJ42658257 Irefl JXM..380097.1 JJ42658257] gi|42658259 jref J|XM .380098.11(42658259] gi|42658265 Irefl JXM .380099.1 j [42658265] gi(42658274 Irefl JXM .380100.11(42658274] gi|42658285 Irefl JXM .380103.1 [[42658285] giJ42658289 Irefl JXM .380104.1 ([42658289] giJ42658301 Irefl JXM.380105.1 [[42658301] giJ42658308 Irefl JXM 380106.11(42658308] gi|42658326 jref jlXM .380107.1 ([42658326] gi[42658330 I ref J |XM 380108.1 JJ42658330] giJ42658332 jref JjXM .380109.1 |[42658332] gi|42658340 |ref JjXM.380110.1 [[42658340] gi(42658342 |ref JjXM .380111.1 ([42658342] gi|42658376 jref JlXM..380112.1 [[42658376] gi|42658378 I ref J |XM.380113.11(42658378] gi|42658392 Irefl JXM .380114.1 |[42658392] gi|42658402 Irefl JXM .380115.11(42658402] gi|42658410 Irefl JXM.380117.1 |[42658410] gi|42658412 jref J|XM .380118.1 |[42658412] giJ42658414 Irefl JXM..380119.11(42658414] gi|42658418 Irefl jXM .380120.1 j[42658418] giJ42658420 Irefl JXM.380121.1 j[42658420] gi|42658428 Irefl (XM 380122.1 |[42658428] gi|42658468 irefl JXM .380125.1 |[42658468] giJ42658480 jrefl JXM.380126.11(42658480] gi|42658488 Irefl JXM..380127.11(42658488] giJ42658496 jref JjXM..380128.11(42658496] gi|42658499 iref JjXM .380129.1 |[42658499] gi(42658506 jref JjXM .380131.1 ([42658506] gi|42658512 Iref JjXM .380133.11(42658512] giJ42658528 Iref JjXM 380134.1 |[42658528] gi(42658564 |ref JjXM..380135.11(42658564] gi|42658572 |ref JjXM 380136.11(42658572] giJ42658576 |ref J]XM..380137.1 [[42658576] giJ42658580 jref J]XM .380138.1 ([42658580] giJ42658582 Irefl JXM .380139.1 [[42658582] gi|42658590 jref J|XM .380140.11(42658590] gi|42658592 |ref J]XM..380141.1 |[42658592] giJ42658594 jref J]XM .380142.1 ([42658594] giJ42658596 jref J]XM.380143.11(42658596] giJ42658610 Iref JjXM.380146.1 |[42658610] giJ42658616 jref JjXM.380147.1 |[42658616]
142658621 |ref|XM_380148.1|[42658621]
|42658629JrefjXM_380149.1|[42658629]
J42658637Jref|XM_380150.1 [[42658637]
|42658645|ref|XM_380151.11[42658645]
|42658667|ref|XM_380152.1 |[42658667]
|42658669|ref|XM_380153.1|[42658669]
J42658673Jref|XM_380154.1 ([42658673]
J42658679Jref|XM_380155.1 [[42658679]
142658681 |ref|XM_380156.1 ([42658681]
|42658689|ref|XM_380157.1|[42658689]
|42658747|ref|XM_380158.1|[42658747]
|42658749|reflXM_380159.lj[42658749]
|42658755|ref|XM_380160.1 j[42658755]
|42658762|ref|XM_380162.1 |[42658762] l42656082|ref|XM_380170.1 |[42656082]
J42657167[ref|XM_380171.1|[42657167]
|42657375 jref|XM_380173.1 [[42657375]
|42656614|reflXM_380174.1|[42656614]
142657318|ref|XM_380175.11[42657318]
J42655791 |ref|XM_380176.11[42655791]
|42406218Jref|NC_000001.7|NC_000001[42406218]
J42406219|refjNC_000002.8JNC_000002[42406219]
|4240622θjref|NC_000003.8JNCJ)00003[42406220]
J42406221 |ref|NC_000004.8|NC_000004[42406221]
J42406224Jref|NC__000005.7|NC__000005[42406224]
|42406225|ref|NC_000006.8(NC_000006[42406225I i4240629ljref|NC_000007.10|NC_000007[42406291]
J42406292(ref|NC_000008.8|NC_000008[42406292]
J42406293|reflNC_000009.8JNC_000009[42406293]
|42406297|ref|NC_000010.7|NC_000010[42406297]
|42406298|ref|NC_000011.7|NC_000011 [42406298]
|42406299[refiNC_000012.8(NC_000012[42406299]
(42406300|ref|NC_000013.8|NC_000013[424063O0]
|4240630ljrefJNC_000014.6JNC_000014[424063O1]
(42406302|ref|NC_000015.7|NC_000015[424063O2]
|42406303|ref|NC_000016.7iNC_000016[424063O3]
J42406304(ref|NC_000017.8|NC_000017(42406304]
[42406305|ref|NC_000018.7|NC_000018[424063O5]
J42406306|ref|NC_000019.8|NC_000019[424063O6] i42406307|ref|NC_000020.8(NC_000020[424063O7]
|42406308|reflNC_000021.6|NC_000021 [42406308]
|42406309|ref|NC_000022.7|NC_000022[424063O9]
|42406310[ref|NC_000023.7|NC_000023[42406310]
J42406311 |ref|NC_000024.6|NC_000024[42406311]
|17981852[ref|NC_001807.41(17981852]
|18860922(refJNG_000002.1|[18860922]
|21536445|ref|NG_000004.2|[21536445]
|14523048|ref|NG_000006.lj[14523048]
J28380636|refJNG_000007.3J[28380636]
[24497632|ref|NG_000008.5([24497632]
|18254505Jref|NG_000009.1 ([18254505]
|50284693|ref|NG_000012.2[[50284693]
|21464097|ref|NG_000013.2[[21464097]
[l4861872Jref|NG_000016.l j[14861872] i28558729Jref|NG_000017.2|[28558729]
|15208635Jref|NG_000018.1[[15208635]
J21592961 |refJNG_000019.3|[21592961]
|21464096 I ref J |NG 000827.11(21464096] [19743567 J]ref JlNG .000833.1 i(19743567] 119718803 iref JlNG .000834.1 |[19718803] J19718802 jref JjNG .000837.1 j[19718802] j19913448 Irefl JNG .000839.1 |[19913448] 120128052 Irefl JNG .000840.1 i(20128052] 120128053 Irefl JNG 000841.1 |[20128053] 120128054 Irefl JNG .000842.1 [[20128054] |20128055| |ref J|NG .000843.1 |[20128055] 131621294 jref J|NG 000845.2 [[31621294] 120128057 jref JjNG .000846.1 [[20128057] (20128058 iref J|NG .000847.1 [[20128058] J20128059 Irefl JNG 000848.1 |[20128059] (24046334 iref JlNG .000849.11(24046334] 131581526 iref JlNG .000850.21(31581526] |20270403 jref J|NG .000851.1 j[20270403] I24046336 iref JlNG .000852.1 [[24046336] 120128060 Irefl JNG 000853.1 [[20128060] J20128463 Irefl JNG .000854.1 [[20128463] |20270405 Irefl JNG .000858.1 |[20270405] 122165413 jref J|NG .000859.2 j[22165413] 120128062 Irefl JNG .000861.1 |[20128062] [20128063 I ref J |NG .000862.1 [[20128063] I24046337 Irefl JNG 000863.1 |[24046337] 132480612 Irefl JNG .000865.31(32480612] J29789590 jref J|NG .000866.1 j[29789590] J20128065 Irefl JNG 000867.1 |[20128065] (20128066 Irefl JNG .000868.1 [[20128066] |20270406 Irefl JNG .000869.1 |[20270406] [20128067 jref J |NG 000870.1 [[20128067] 120128068 Irefl JNG..000871.1 |[20128068] J20270402 jref J|NG 000872.1 [[20270402] J20128069 Irefl JNG .000873.1 |[20128069] J20128070 iref JlNG .000874.1 [[20128070] |20270408 jref J|NG 000877.1 |[20270408] (20128071 jref J|NG .000878.1 ([20128071] |20270409 Iref JjNG .000880.1 j[20270409] [20270410 Irefl JNG 000881.1 |[20270410] 120270411 jref JjNG .000882.11(20270411] J29789730 Irefl JNG 000883.1 ([29789730] J20128072 | refl JNG .000884.11(20128072] J24046339 Jref JlNG .000885.1 |[24046339] J22902139 Irefl JNG 000886.2 [[22902139] J22902140 jref J|NG .000887.2 |[22902140] J20270414 Irefl JNG 000889.1 ([20270414] 120128073 jref JjNG .000890.1 [[20128073] |24046340 Irefl JNG 000891.1 [[24046340] (20128074 Irefl JNG 000892.1 ((20128074] 120128075 Irefl JNG 000893.1 |[20128075] 120128076 jref J|NG 000894.1 |[20128076] J24046341 jref J|NG 000895.1 ([24046341] 120128077 jref J|NG 000896.1 [[20128077] J27502364 jrefl JNG 000897.2 ([27502364] 120128078 Irefl JNG 000898.1 j[20128078] 120128079 Irefl JNG 000899.1 |[20128079] 120270416 Irefl JNG 000900.11(20270416] [20128081 Iref JjNG 000901.11(20128081]
120128080 |ref J|NG .000903.11(20128080] (20270417 |ref JNG .000904.1 [[20270417] J20128082 Irefl JNG 000906.1 1(20128082 J20128083 |ref JjNG .000907.1 j[20128083] J20128084 Irefl JNG .000908.1 j[20128084] J20128085 |ref JjNG .000909.1 1(20128085] (20128086 Irefl JNG 000910.1 |[20128086] J20128087 jref J]NG .000911.1 ([20128087] (20270418 | ref J |NG 000912.1 ([20270418 J20128088 Irefl JNG .000913.1 ([20128088] J20128090 |ref JjNG 000914.1 ([20128090] J20128091 iref J|NG .000915.1 1(20128091 [20128092 jref JjNG 000916.1 |[20128092] J20128093 iref J|NG .000917.1 |[20128093] |28882058 Irefl JNG .000919.2 ([28882058 J20128095 Irefl JNG .000920.1 |[20128095] 120128096 Irefl JNG .000921.1j[20128096] [20128097 j]ref J|NG 000922.1 |[20128097 [20128098 Jref J|NG .000923.1 ([20128098] J20128099 | ref J |NG .000924.1 ([20128099] (20128100 Irefl JNG .000925.1 ([20128100 120128101 jref JjNG .000927.1 1(20128101 120128102 Irefl JNG .000929.1 1(20128102 120128103 | ref J |NG .000930.1 |[20128103] 120270419 Irefl JNG 000933.1 ([20270419 (34330152] Irefl JNG .000936.2 |[34330152] (20128105] jref J|NG 000938.1 ([20128105] 120128106 iref JlNG .000939.1 |[20128106' (20128107] jref J|NG 000940.1 1(20128107] 120128108 Irefl JNG .000941.1 ([20128108' (20128109 jrefl JNG .000942.1 1(20128109 120128110 Irefl JNG 000943.1 ([20128110] 120270421 Irefl JNG .000944.1 1(20270421 (20270422] jrefl JNG 000945.1 |[20270422] (20128111 jrefl JNG .000946.1 [[20128111 |20270423 Irefl JNG .000948.1 [[20270423] (20128112 Irefl JNG .000949.1 1(20128112] [20270424 jrefl JNG .000950.1 ([20270424] |20270425 Irefl JNG .000951.1 j[20270425] [20128113 Irefl JNG .000952.1 ([20128113 J20128114 jref J|NG 000953.1 |[20128114] 134330150 Jref J|NG .000954.2 j[34330150] (29837646 Irefl JNG 000955.2 ([29837646] |20270427 Jref J|NG .000956.1 [[20270427 (20128116 Irefl JNG .000957.1 |[20128116] J20128117 jref JjNG .000958.1 j[20128117] |20270428 |ref J|NG 000959.1 [[20270428] J20128118 jref J|NG .000960.1 1(20128118] J20270429 Irefl JNG 000961.1 [[20270429] J20128119 | ref J |NG 000962.1 ([20128119] (20128120 jref JjNG .000963.1 [[20128120] [20128121 Irefl JNG .000964.1 |[20128121 (24046342 Irefl JNG 000965.1 ([24046342] 120128122 Irefl JNG 000966.1 j[2012812Z [20270430 Irefl JNG 000967.1 [[20270430] (20270431 Irefl JNG 000968.1 |[20270431 |20128123 Irefl JNG 000969.1 j[20128123
gi|20270432|ref|NG_000970.1l[20270432] gi|20128124Jref|NG_000971.1|[20128124] gi[20128125Jref|NG_000972.1|[20128125] gi|20270433|reflNG_000973.1|[20270433] gii20128126ireflNG_000974.l([20128126] gi[20128127iref|NG_000975.l([20128127] gi[28866967|ref|NG_000977.2|[28866967] giJ20128129JrefJNG_000978.lj[20128129] gi|20128130|ref|NG_000979.11[20128130] gi|28316813|ref|NG_000980.2|[28316813] gi[20270434|ref[NG_000981.1 ([20270434] gii20270435iref|NG_000982.l[[20270435] gi(20128132[ref[NG_000983.li[20128132] giJ20270436|reflNG_000984.1|[20270436] giJ20128133Jref|NG_000986.l([20128133] gi|20128466[ref|NG_000987.l([20128466] gi(20128134[refiNG_000988.li[20128134] gi(20270629iref|NG_000989.li[20270629] gi|20128135|ref|NG_000990.l([20128135] gi|20128136Jref|NG_000991.11[20128136] giJ20128137)ref|NG_000994.11(20128137] gi[20270437[ref|NG_000995.li[20270437] gi(20128138ireflNG_000996.1|[20128138] giJ20128139|ref|NG_000997.lj[20128139] gi|20128140|ref|NG_000999.1|[20128140] gi|20128141lref|NG_001000.1|[20128141] gi|20128142|ref|NG_001001.lj[20128142] giJ20128143[ref|NG_001002.lj[20128143] gi|20270438|ref|NG_001004.1 j[20270438] gi(20270439|ref|NG_001005.11[20270439] gi(20128144Jref|NG_001006.l[[20128144] giJ20128145|ref|NG_001007.1 ([20128145] gi[20270440|ref|NG_001008.1 [[20270440] giJ2027044l (ref|NG_001009.1 [[20270441] gi|20128146irefJNG_001010.1|[20128146] gi|20128147Jref|NG_001012.lj[20128147] gi[20270442lreflNG_001013.1 ([20270442] gi[21729868|ref|NG_001014.l[[21729868] gi(20270443|ref|NG_001016.1 [[20270443] gi|20128149|ref|NG_001017.1|[20128149] gi|4892799θ[ref|NG_001019.3|[48927990] gi[20128462|ref(NG_001020.1j[20128462] gi(2012815liref|NG_001021.1|[20128151] gi|20128152(refiNG_001022.1|[20128152] giJ20128153|ref|NG_001023.l[[20128153] gi|20128154irefJNG_001024.1|[20128154] gi|20128155|ref|NG_001025.1J[20128155] giJ20128156|reflNG_001026.1 ([20128156] gi|20128157|refJNG_001027.li[20128157] gi|20128158Jref|NG_001028.1|(20128158] gi|20270444Jref|NG_001029.11[20270444] gi(20270445iref|NG_001030.1 ([20270445] giJ20 28461 [reflNG_001031.1 [[20128461] giJ20128159|ref|NG_001032.l([20128159] gi|20270446(ref|NG_001033.1 ([20270446] gi|2012816θjref|NG_001035.lj[20128160] giJ2012816ljref(NG_001036.1|[20128161]
gi|20128464 Irefl JNG 001037.11(20128464] giJ20270447 Irefl JNG 001038.1 |[20270447] gi(20270448 jref JjNG 001040.1 j[20270448] giJ20128162 Irefl JNG 001041.1 |[20128162] giJ20128163 jref J|NG .001043.1 j[20128163] giJ20128164 Irefl JNG 001045.1 ([20128164] gi|20128165 jref J|NG .001046.1 |[20128165] gi|20128166 jref J|NG 001047.11(20128166] giJ20128167 Irefl JNG 001048.1 ([20128167] gi|20128465 jrefl JNG 001049.1 |[20128465] gi|20128168 Irefl JNG .001050.1 [[20128168] gi[20270449 Jref J|NG 001051.1 |[20270449] gi|20128169 jref J|NG 001052.11(20128169] giJ20128170 |ref J|NG 001053.1 |[20128170] gi|20270450 Irefl JNG 001054.1 ([20270450] giJ20128171 jref J|NG 001055.1 [[20128171] gi|20128172 jref JlNG 001056.11(20128172] gi|20128173 Irefl JNG .001057.1 ([20128173] gi|20128174 jref J|NG 001058.1 j[20128174] giJ20128467 Irefl JNG .001060.11(20128467] gi[20270451 Irefl JNG 001061.1 [[20270451] giJ20270452 jrefl JNG .001062.11(20270452] gi|20270453 Irefl JNG 001063.1 |[20270453] gi|20270454 Irefl JNG 001064.1 ([20270454] gi[20270455 jref JjNG .001065.1 [(20270455] giJ42415476 jref JlNG 001066.1 [[42415476] gi(20270626 Irefl JNG .001067.11(20270626] gi[20270456 Irefl JNG .001068.1 ([20270456] gi|20270457 Irefl JNG 001069.11(20270457] giJ20270458 jref JjNG .001070.1 [[20270458] giJ20270459 Irefl JNG 001071.1 |[20270459] gi|29789827 jrefl JNG .001073.11(29789827] gi|29789826 [refl JNG 001074.1 ([29789826] gi(20270461 Irefl JNG 001075.1 |[20270461] gi|29789822 jref J|NG .001076.1 [[29789822] gi[20270462 jref JjNG .001077.1 [[20270462] gi(20270463 Irefl JNG 001078.11(20270463] giJ29789823 Irefl JNG .001080.1 |[29789823] gi|20270464 Irefl JNG .001081.1 [[20270464] gi|20270465 jref J|NG 001082.1 ([20270465] gi(20270466 Irefl JNG .001083.11(20270466] gi|20270467 | ref J |NG 001084.1 ([20270467] gil20270468 Irefl JNG .001085.1 [[20270468] gi|20270469 jrefl JNG .001086.1 j[20270469] gi[29789502 jref JjNG .001087.1 [[29789502] gi|29789809 Irefl JNG .001088.11(29789809] giJ20270470 jref J|NG 001089.1 |[20270470] gi|20270471 iref J|NG .001090.11(20270471] gi[29789807 jrefl JNG .001091.1 [[29789807] gi(20270472 Irefl JNG 001092.11(20270472] gi[20270473 I ref J|NG .001093.1 |[20270473] gi|42543997| I ref J |NG .001094.11(42543997] giJ20270474| jref JjNG .001095.11(20270474] gi|20270627| Irefl JNG .001096.1 |[20270627] gi|20270475 Irefl |NG 001097.11(20270475] gi(20270476 jref J|NG .001098.1 [[20270476] gi(20270477 jref JjNG 001099.1 ([20270477]
120270478 Irefl JNG .001100.11(20270478]
120270479 I ref JNG .001101.1 |[20270479]
120270480 Irefl JNG 001102.1 j[20270480]
J20270481 jref J|NG .001103.1 [[20270481]
|20270482 Irefl JNG 001104.1 ([20270482]
|20270483 jref J|NG .001105.11(20270483]
|20270484 Irefl JNG .001106.1 [[20270484]
I42543998 jref JlNG .001107.11(42543998]
|20270485 jref JlNG .001108.1 |[20270485]
|20270486 jref J|NG 001109.1 ([20270486]
(29789797] |ref J|NG .001110.1 j[29789797]
[29789779 jref J|NG 001112.1 [[29789779]
(29789778 jref J|NG 001113.1 |[29789778]
[20270488 Irefl JNG .001114.1 j[20270488]
(29789768 Irefl JNG 001115.1 |[29789768]
|29789757 jref J|NG .001116.1 j[29789757]
[29789756 jref J|NG 001117.1 |[29789756]
I29789755 I ref J |NG .001118.11(29789755]
(20270489 jref J|NG..001119.1 |[20270489]
|20270490 Irefl JNG .001120.1 ([20270490]
[29423721 irefl JNG 001121.2 ([29423721]
J20270492 I refl JNG .001122.1 ([20270492]
[20270493 Irefl JNG .001123.1 j[20270493]
[29789748 Irefl JNG .001124.1 [[29789748]
120270494] jrefl JNG .001125.1 ([20270494]
I29789745 jrefl JNG 001126.1 [[29789745]
|29789746 Irefl JNG .001127.1 j[29789746]
[42415477 jref J|NG..001128.11(42415477]
142415478 Irefl JNG 001129.1 j[42415478]
J20270495 jref JlNG .001130.1 [[20270495]
[20270496 jref J|NG..001131.1 j[20270496]
(20270497 Irefl JNG 001132.1 j[20270497]
[29789741 Irefl JNG .001133.11(29789741]
(20270498 jref J |NG 001134.1 [[20270498]
|20270499 jref JlNG 001135.1 [[20270499]
I20270500 jref JlNG .001136.1 j[20270500]
[29789732 jref J|NG 001141.1 ([29789732]
|29789729 Irefl JNG .001142.1 j[29789729]
129789719 jref JjNG..001144.1 |[29789719]
J29789720 Irefl JNG .001145.1 ([29789720]
J29789717 Irefl JNG .001146.1 [[29789717]
J29789718 |ref J|NG..001147.11(29789718]
J29789715 Irefl JNG..001148.1 ([29789715]
J20270501 Irefl (NG .001149.11(20270501]
J20270502 Irefl JNG .001150.11(20270502]
I29789587] |ref J|NG .001151.1 j[29789587]
(20270503 jref JjNG..001152.1 |[20270503]
J20270504 Irefl (NG .001153.11(20270504]
J20270506 jrefl JNG .001155.1 |[20270506]
I20270507] jrefl JNG .001156.1 |[20270507]
J20270508 jref J|NG .001157.1 ([20270508]
|20270509 jref J|NG. 001158.1 [[20270509]
J20270510 Jref J|NG 001159.1 [[20270510]
[29789647 Irefl JNG. 001160.1 |[29789647]
(20270511 Irefl JNG 001161.1 |[20270511]
[29789631 Irefl JNG 001162.11(29789631]
J29789629 Iref JjNG 001163.1 [[29789629]
G .001164.1 |[29789626] G 001165.1 j[29789627] G .001166.1 |[29789624] G 001167.1 j[29789625] G 001169.1 |[20270512] G .001170.11(29789608] G 001171.1 j[29789605] G .001172.11(20270513] G 001173.1 j[20270514] G 001174.1 i(20270515] G 001175.1 |[20270516] G .001176.11(20270517] G 001177.1 [[29789604] G .001178.1 |[20270518] G .001179.1 JJ42538974] G .001180.2 |[32964834] G..001181.1 j[20270519] G .001182.1 |[20270520] G .001183.11(20270521] G 001184.1 j[20270522] G .001185.1 i(29789582] G .001186.1 |[29789578] G 001187.11(29789575] G .001188.1 j[20270523] G 001189.11(20270524] G .001190.1 j[20270525] G 001191.11(20270526] G..001192.1 j[29789536] G 001193.1 i(29789535] G .001194.11(20270527] G..001195.1 j[20270528] G 001196.2] 1(29423719] G .001197.1 j[20270530] G .001198.11(29789766] G.001199.1 j[20270531] G .001200.1 j[20270532] G .001201.1 j[29789490] G .001202.1 |[20270533] G 001204.1 |[20270630] G .001205.1 j[20270631] G .001206.1 |[20270534] G .001207.1 i(29789486] G .001208.1 |[29789483] G .001209.1 i(20270535] G .001210.1 j[29789479] G 001211.1 i(20270536] G .001212.1 |[28866961] G .001213.11(28849911] G .001214.1 |[29789469] G .001215.2 j[33342274] G .001216.1 |[29789830] G .001217.1 j[20270538] G..001218.1 i[42409515] G .001219.1 i(20270539] G .001220.1 i[20270540] G..001221.1 j[20270541]

G. 001222.1 |[29789644]
gi[20270542|ref(NG_001223.1 ([20270542] giJ20270543Jref|NG_001224.l j[20270543] giJ29789742JreflNG_001226.1 j[29789742] gi[29789796JreflNG_001228.1 [[29789796] gi|20270545|ref|NG_001229.1 |[20270545] giJ20270546Jref|NG_001230.1 |[20270546] gii20270547JreflNG_001231.1 |[20270547] gi|20270548Jref|NG_001232.1 |[20270548] giJ20270549Jref|NG_001233.1 [[20270549] gi|2027055θjref|NG_001234.1 J[20270550] gi|20270551 |ref|NG_001235.11[20270551] giJ20270552JreflNG_001236.1 J[20270552] gi(20270553Jref|NG_001237.1 |[20270553] gi|20270554|ref|NG )01238.11[20270554] giJ20270555Jref|NG_001239.1 ([20270555] gi|20270556|ref|NG_001240.11[20270556] gi|20270557|ref|NG_001241.11[20270557] gi|20270558Jref|NG_001242.l j[20270558] gi|20270559|ref|NG_001243.l j[20270559] gi|2027056θjref|NG_001244.l [[20270560] gi|29789744|ref|NG_001245.1 |[29789744] gi|20270561|ref|NG_001246.1 [[20270561] giJ20270562JreflNG_001247.l([20270562] giJ29789572|reflNG_001248.11[29789572] giJ29789571 |ref|NG_001249.1 [[29789571] gi|29789574lref|NG_001250.11[29789574] giJ29789573JreflNG_001251.lj[29789573] gi|29789576[ref|NG_001252.1|[29789576] giJ29789577Jref|NG_001253.l j[29789577] giJ2978958θ(ref|NG_001254.1 |[29789580] giJ29789579|ref|NG_001255.1 j[29789579] gi|29789581|ref|NG_001256.1 [[29789581] gi[29789584Jref(NG_001257.1 |[29789584] gi|29789583Jref|NG_001258.1 |[29789583] gi|20270563|ref]NG_001259.l ([20270563] gi|20270564JreflNG_001260.11[20270564] gi|20270565|ref|NG_001261.11[20270565] giJ20270566[ref|NG_001262.li[20270566] gi|20270567|ref|NG_001263.11[20270567] giJ20270568Jref|NG_001264.1 |[20270568] gi|29789831 |ref|NG_001265.11[29789831] gi|20270569|ref|NG_001266.11[20270569] gii2027057θiref|NG_001267.l j[20270570] giJ20270571|ref|NG_001268.l ([20270571] gi|29789833[reflNG_001269.11[29789833] gi|29789835|ref|NG_001270.1 ([29789835] gi|20270572|ref|NG j01271.1 |[20270572] gii20270573JreflNG_001272.l i[20270573] gi|20270574|ref|NG_001273.11[20270574] gi|20270575Jref|NG_001274.l j[20270575] giJ20270576|ref|NG_001275.l j[20270576] gi|20270577|ref|NG_001276.11[20270577] giJ20270578iref|NG_001277.l ([20270578] giJ20270579Jref[NG_001278.1 [[20270579] gi|2027058ljref|NG_001279.1 |[20270581] gi|20270582Jref|NG_001280.l [[20270582] gi(20270583JrefJNG_001281.1 |[20270583]
gi|20270584|ref|NG_001282.1[[20270584] gi[29423722Jref|NG_001286.2|[29423722] giJ2027058θjrefJNG_001287.1 j[20270580] gi|20270589(ref|NG )01288.1|[20270589] gi|20270590|ref|NG_001289.l[[20270590] gii29789599Jref[NG_001290.li[29789599] gi)29789585|ref|NG_001291.11[29789585] giJ29789588Jref|NG_001292.11[29789588] gi|2027059l(ref|NG_001293.1 [[20270591] gi|20270592|ref|NG_001294.1|[20270592] giJ20270593lref|NG_001295.l([20270593] gi|20270594|refJNG_001296.l([20270594] gi]20270595|ref|NG_001297.1|[20270595] giJ20270596|refiNG_001298.l[[20270596] gi|20270597ireflNG_001299.1|[20270597] gi|20270598|ref|NG_001300.lj[20270598] gi|20270599|ref[NG_001301.11[20270599] giJ29789542Jref|NG_001302.1 j[29789542] gi|20270600|ref|NG_001303.11[20270600] gi|2027060l[ref|NG_001305.11[20270601] gi|20270602|ref|NG_001306.11[20270602] gi|20270603irefJNG_001307.lj[20270603] gi[20270604(ref|NG_001308.1 ([20270604] giJ20270605JrefJNG_001309.lj[20270605] giJ20270606Jref[NG_001311.1 j[20270606] gi|20270608|ref|NG_001313.1 |[20270608] gi(29789694iref[NG_001314.li[29789694] gi(20270609ireflNG_001315.li[20270609] gi|20270610|ref|NG_001316.11[20270610] gi|20270611|ref|NG_001317.11[20270611] gi|20270612|ref|NG_001318.lj[20270612] giJ20270613|ref|NG_001319.1|[20270613] gi(20270615ireflNG_001321.11[20270615] gi|20270616|ref|NG_001322.1 j [20270616] giJ20270617|reflNG_001323.1 [[20270617] gi|20270618|ref|NG )01324.11[20270618] gi[20270619iref|NG_001325.lj[20270619] gi|20270620jrefjNG_001326.11[20270620] gi|20270622JreflNG_001328.1 j[20270622] gi|20270623Jref|NG_001329.11[20270623] giJ20270624JrefJNG_001330.1 ([20270624] gi|20270625|ref|NG_001331.11[20270625] giJ2136312ljref(NG_001332.1([21363121] gi|21536269JreflNG_001333.lj(21536269] gi|21463739Jref|NG_001334.1 ][21463739] giJ2458669ljref|NG_001335.lj[24586691] gi|28436398|ref|NG_001336.2|[28436398] gi|22143966Jref|NG_001337.1|[22143966] gi(24046436(ref|NG_001526.1|[24046436] giJ24046437ireflNG_001528.1 J[24046437] gi[24046438|reflNG_001529.1 [[24046438] gi|24046439|ref|NG_001531.1 |[24046439] gi|24046440|ref|NG_001532.1|[24046440] giJ2404644ljrefJNG_001533.lj[24046441] giJ24046442JrefJNG_001534.11[24046442] gi|24046443lref|NG_001535.1 ([24046443] gi|24046444|reflNG_001537.1 j[24046444]
gi|24046445 Irefl JNG_ 001538.1 |[24046445] gi|24046446 Irefl |NG_ .001539.11(24046446] gi[24046447 Irefl JNG.001540.11(24046447] gi(24046448 Irefl JNG .001541.1 j[24046448] gi[24046449 Irefj |NG_ 001542.11(24046449] gi(24046450 jref JlNG .001543.1 j[24046450] gil24046451 Irefl JNG..001544.11(24046451] gi|24046452 jref JNG.001545.1 ([24046452] gi|24046453 Irefl |NG_ 001546.1 [[24046453] gi[24046454 Irefl JNG .001547.1 |[24046454] gi(24046455 Irefl JNG .001548.1 j[24046455] giJ29789697 Irefl |NG 001549.1 [[29789697] gi|24046456 jrefl JNG..001550.1 j[24046456] gi(24046457 Irefl JNG_ 001551.1 |[24046457] gi|24046458 jref J|NG .001552.1 [[24046458] gi|24046459 Irefl JNG 001553.11(24046459] gi[24046460 Irefl JNG 001554.1 [[24046460] gi|24046461 Irefl JNG .001555.11(24046461] gi(24046462 jref JlNG .001556.1 [[24046462] gi|24046463 |ref J|NG .001557.11(24046463] gi|24046464 jref JjNG .001558.1 [[24046464] gi|24046465 Irefl JNG 001559.1 [[24046465] gi|24046466 Iref JjNG .001560.11(24046466] gi|24046467 Irefl JNG 001561.1 j[24046467] gi|24046468 jref JlNG .001562.11(24046468] gi[24046469 Irefl JNG .001563.1 [[24046469] gi|24046470 jref J|NG .001564.1 [[24046470] gi|24046472 Irefl jNG 001566.11(24046472] gi|24046473 jref J|NG..001567.1 [[24046473] gi(24046474 iref (|NG.001568.1 j[24046474] giJ24046475 |ref JjNG 001569.1 j[24046475] gi|24371385 |ref J|NG .001571.11(24371385] gi|24046476 jref J|NG .001572.1 ([24046476] gi|24046477 jref JlNG 001573.1 |[24046477] gi|24046478 jref JlNG .001574.1 |[24046478] gi|24046479 jref JjNG .001576.1 [[24046479] giJ24046480 |ref JNG 001577.1 |[24046480] gi|24046481 Irefl JNG .001578.1 j[24046481] gi|24046482 jref J |NG 001579.1 ([24046482] giJ24046483 Irefl JNG 001580.11(24046483] gi|24046484 Irefl JNG..001581.1 j[24046484] gi|47059202 Irefl JNG .001582.2 [[47059202] gi(24046486 Irefl JNG 001583.11(24046486] gi|24046487 jref |NG .001584.1 [[24046487] gi|24046488 Irefl JNG 001585.1 [[24046488] gi|24046489 jref J|NG .001586.1 j[24046489] giJ24371387 Irefl JNG .001587.11(24371387] giJ24046490 Irefl JNG .001588.1 [[24046490] gi|24046491 Irefl JNG .001589.11(24046491] gi[24046492 jref J|NG .001590.1 |[24046492] gi(24046493 jrefl JNG .001591.1 ([24046493] gi|29611657 |ref JjNG .001592.2 |[29611657] giJ24047046 Irefl JNG .002151.1 ([24047046] gi|24047047 Irefl JNG 002153.1 [[24047047] gi|24047048 ]ref J|NG .002154.11(24047048] gi(46047489 Irefl (NG .002156.2 [[46047489] gi|24047050 Iref JjNG. 002158.1 [[24047050]
124047051 I ref J |NG 002160.1 |[24047051] [24047052 I ref j |NG 002162.1 j[24047052] I24047053 iref JlNG .002164.1 |[24047053] |24047054 Irefl jNG ~002166.1 ([24047054] (46047441 jref J|NG 002168.2 [[46047441] |24047056 Irefl jNG 002170.1 |[24047056] [24047057 jref J |NG 002172.11(24047057] |24047058 iref jlNG 002173.1 [[24047058] J24047059 Irefl JNG 002175.11(24047059] |24047060 jref j|NG .002177.1 |[24047060] J24047061 jref J|NG 002179.11(24047061] J24047062 |ref J|NG 002181.11(24047062] J48597071 Irefl JNG .002183.2 j[48597071] 142415480 Irefl JNG 002184.1 [[42415480] [24047064 iref J|NG .002185.1 j[24047064] |24047066 jref jjNG 002189.1 |[24047066] |24047067 Irefl JNG..002191.11(24047067] J24047069 jrefj JNG 002195.1 ([24047069] 141281755 Irefl JNG .002196.1 ([41281755] J24047071 Irefl jNG .002199.1 ([24047071] |48597074 jref J|NG 002200.21(48597074] |48525352 iref JlNG .002201.2 |[48525352] J24047074 Irefl JNG .002202.11(24047074] |24047075 jref | JNG .002203.1 j[24047075] J24047076 iref JlNG .002204.11(24047076] [50055022 jref J |NG .002205.1 |[50055022] J48597072 I ref J |NG 002207.1 j[48597072] |24047078 iref JlNG .002211.1 |[24047078] J24047079 jref J|NG .002212.11(24047079] 124047081 iref JlNG .002215.1 |[24047081] J24047082 jref J|NG 002216.1 [[24047082] J24047084 Irefl JNG 002218.1 ([24047084] J24047085 Iref JjNG .002219.1 |[24047085] J24047086 Irefl JNG .002220.1 [[24047086] J24047087 Irefl JNG 002221.1 ([24047087] [24047090 jref JjNG 002225.1 |[24047090] J24047092 Iref JjNG .002229.1 |[24047092] J24047093 jref J|NG .002232.1 [[24047093] J48597080 jref JjNG .002233.4 j[48597080] J29423720 Irefl JNG .002234.21(29423720] I39777575 Irefl JNG .002235.2 |[39777575] |24047097 Irefl JNG .002236.1 [[24047097] |24047098 jref ( |NG 002238.1 |[24047098] (41281757 Irefl JNG .002239.11(41281757] |39777578 Irefl JNG..002240.2 |[39777578] [48597073 jref J|NG .002241.2 [[48597073] 124047101 jref J|NG 002242.1 |[24047101] (24047102 jrefl JNG 002243.1 ([24047102] 150055011 Irefl JNG .002246.1 |[50055011] 141281759 jrefl JNG .002247.1 [[41281759] [48525353 Irefl JNG .002250.2 ([48525353] (50055031 jrefl JNG 002251.11(50055031] J24047104 Irefl JNG .002252.1 [[24047104] (41281761 Irefl JNG 002253.1 [[41281761] J24047105 jref J|NG .002254.1 [[24047105] |24047106 Irefl JNG .002255.1 |[24047106] J24047107 jref JjNG 002256.1 j[24047107]
148597078 I ref J |NG 002257.2 |[48597078] 148597075 jref J|NG 002258.2 |[48597075] [24047110 iref JlNG 002259.11(24047110] 124047111 jref J|NG 002260.11(24047111] J24047112 jrefl JNG 002261.11(24047112] J24047113 jref J |NG 002262.1 |[24047113] [24047114 jref JjNG 002263.1 [[24047114] 124047115 |ref J|NG 002264.1 |[24047115] [39777576 Irefl JNG 002265.21(39777576] 124047117 |ref JjNG 002266.1 |[24047117] 124047118 Irefl JNG .002267.1 ([24047118] (24047119 |ref jjNG .002268.1 |[24047119] [24047120 jref J|NG .002269.11(24047120] 124047121 Irefl JNG 002270.1 j[24047121] (24047122 Irefl JNG 002271.1 ([24047122] 124047123 Irefl JNG..002272.1 ([24047123] 124047124 Irefl jNG 002273.1 |[24047124] |39777574 Irefl JNG 002274.2 [[39777574] J41281763 jref jlNG 002275.11(41281763] |46048461 Irefl JNG .002277.2 |[46048461] J48597079 Irefl JNG .002278.2 [[48597079] J29789862 Irefl JNG 002279.1 ([29789862] J29789864 Irefl jNG .002281.1 [[29789864] J29789614 jref J|NG .002282.1 |[29789614] J24047129 Irefl JNG 002284.11(24047129] 124047130 Irefl JNG .002285.1 |[24047130] J24047131 jrefl JNG 002286.11(24047131] 124047132 Iref JjNG .002287.1 ([24047132] (24047133 jref JjNG .002288.1 ([24047133] 124047134 Irefl JNG 002289.1 [[24047134] J24047135 jref J |NG .002290.11(24047135] J24047136 Irefl JNG 002291.1 |[24047136] [24047142 jref JlNG .002298.1 ([24047142] J24047143 jrefl JNG 002299.1 j[24047143] 141281773 jrefl JNG .002302.11(41281773] I46047472J jrefl JNG .002305.2 |[46047472] 124047149 Irefl JNG .002306.1 ([24047149] J46048473 Irefl JNG 002307.21(46048473] J24047154J jrefl JNG .002311.1 |[24047154] J45504398
1 Irefl JNG 002315.21(45504398] J24047159 Irefl JNG 002316.1 |[24047159] J24047160 jref J|NG .002317.11(24047160] J24047162 Irefl JNG..002319.1 ([24047162] 124371388 jref J|NG .002320.1 [[24371388] 124047163 Irefl jNG .002321.1 |[24047163] J24047164 I ref J |NG .002322.1 j[24047164] (24047165 |ref J|NG .002323.1 |[24047165] [24047166 Iref JjNG 002324.11(24047166] J24047168 jref JjNG .002325.1 j[24047168] J29789776 jrefl JNG .002326.1 |[29789776] J29789869 |ref J|NG .002327.1 |[29789869] J24047169 | ref J|NG .002328.11(24047169] |24047167| jref JjNG .002329.1 ([24047167] 124371379 Irefl JNG .002331.11(24371379] 124047170 jref JlNG .002332.1 [[24047170] J24047171 Jref J|NG .002333.11(24047171] |29789868 jref JjNG 002334.1 |[29789868]
gi|41281775|ref|NG_002335.11[41281775] giJ29789866|ref|NG_002336.l([29789866] gi|240471 2|ref|NG_002337.11[24047172] gi|29789867|ref|NG_002338.lj[29789867] gi|29789865|refjNG_002339.1 [[29789865] gi|24047173jref|NG_002340.1|[24047173] giJ24047174irefJNG_002341.1 ([240471 4] gi|24047175|refJNG_002342.1 j[24047175] gi|29789863|ref|NG_002343.lj[29789863] gi|24047177Jref|NG_002348.lj[24047177] gi|34328081|ref|NG_002349.2|[34328081] gi|24047179Jref|NG_002350.1 j[24047179] gi|2404718θ(ref|NG_002351.11[24047180] gi|24047181|ref|NG_002352.1|[24047181] giJ24047182[reflNG_002353.1 ([24047182] gi|24047183|ref|NG_002354.1|[24047183] gi[24047184|ref(NG_002355.1 [[24047184] gi|48597077|ref|NG_002356.2|[48597077] gi|24047186|ref|NG_002357.1|[24047186] giJ24047187|reflNG_002360.1 ([24047187] gi|24047188|ref|NG_002361.1|[24047188] giJ24047189|reflNG_002363.lj[24047189] gi|24047190|ref|NG_002364.11[24047190] gi|24047191|ref|NG_002365.1|[24047191] giJ24047192JrefJNG_002366.1 ([24047192] gi|24047193|ref|NG_002367.1 [[24047193] gi[24047194|ref[NG_002368.1([24047194] gi|24047195|ref|NG_002369.11[24047195] gi|24047196|ref|NG_002370.lj[24047196] gi|24047197|ref|NG_002371.11[24047197] gi|24432180|ref|NG_002372.1|[24432180] giJ24047198Jref(NG_002373.li[24047198] gi|24047199|ref|NG_002374.11[24047199] gi|24047200|ref|NG_002375.1|[24047200] gi|24047201 jref|NG_002376.11[24047201] gi|24047203|ref[NG_002378.11[24047203] giJ24047204Jref|NG_002379.lj[24047204] gi|24047205|ref|NG )02380.1|[24047205] gi|24047206|ref|NG_002381.1 ([24047206] gi(34330153(ref|NG_002382.2|[34330153] gi|24047208|ref|NG_002383.1|[24047208] gi|29789622|reflNG_002384.l([29789622] gi|29789700|reflNG_002385.1|[29789700] gi|29789688|ref|NG_002387.1|[29789688] giJ29789689|ref|NG_002388.1 [[29789689] gi|29789686|ref(NG_002389.1|[29789686] giJ29124352|ref|NG_002392.2J[29124352] gi|29789829|ref[NG_002393.1|[29789829] gi|29789687|ref|NG_002394.1|[29789687] gi|29789684Jref|NG_002395.1 [[29789684] gi|50284694|ref|NG_002397.2|[50284694] gi|50295102(ref|NG_002398.2|[50295102] gi|29789489|ref|NG_002399.11[29789489] gi|29135348|ref|NG_002400.lj[29135348J gi|29135351 |ref|NG_002401.1 [[29135351] gi|29135349|ref|NG_002402.1|[29135349] giJ29135354Jref|NG_002403.l[[29135354]
129789521 IreflJNG .002404 |[29789521]
[29789586 Irefl JNG 002405 j[29789586]
(29789613 Irefl JNG .002406 |[29789613]
[29789615 jrefl JNG .002408 i(29789615]
|48525354 |ref J|NG 002409 i[48525354]
J29789685 IrefljNG 002410 1(29789685]
|29789500 Irefl JNG .002411 j[29789500]
J29789683 jref J|NG 002412 1(29789683]
|29789589| Irefl JNG .002415 j[29789589]
J29789649J Irefl JNG 002416 1(29789649]
|29789680 Irefl JNG .002417 |[29789680]
(29789691 jref JjNG .002418 j[29789691]
[29789771 Iref JjNG .002419 P9789771]
(29789806 jref J|NG .002423 1(29789806]
142415481 Irefl JNG 002425 JJ42415481]
142415482 jref J|NG .002426 j[42415482]
[42415483 Irefl JNG .002427 1(42415483]
142415485 jref J|NG .002428 1(42415485]
142415484 |ref J|NG .002429 j[42415484]
J29789650 irefl JNG .002430 j[29789650]
129789681 jrefl JNG .002431 1(29789681]
128212469 Irefl JNG 002432 ([28212469]
(28212470 jref J|NG. .002433 |[28212470]
J29789609 I ref J |NG 002434 1(29789609]
J29789794 Irefl JNG .002437 [[29789794]
[29789701 Irefl JNG .002438 1(29789701]
J29789699J |ref J|NG .002440 [[29789699]
J29789661 jref JjNG .002448 ([29789661]
J29789657 Irefl JNG .002449 [[29789657]
[29789676 jref JjNG 002450 ([29789676]
(29789659 jref JjNG .002451 1(29789659]
|29789656 jref J|NG 002452 |[29789656]
[29789658 Irefl JNG .002453 j[29789658]
J29789660 Irefl JNG .002454 1(29789660]
J28603821 |ref J|NG .002456 1(28603821]
J28603822 jrefl JNG .002457 ([28603822]
(28571461 Irefl JNG .002458 |[28571461]
[29789475 | refl JNG .002459 [[29789475]
(29789725 jrefl JNG .002460. 1(29789725]
[29789677 Irefl |NG .002461 |[29789677]
129789812 jrefl JNG 002462 1(29789812]
J29789674 jref JjNG .002463 1(29789674]
[29789499 jref J |NG 002464. ([29789499]
J29789808 |refj|NG .002465 1(29789808]
[29789828 | ref J |NG .002467 |[29789828]
(29789690 jrefjlNG 002468 j[29789690]
[29789810 Irefl JNG 002469 1(29789810]
(29789811 jrefj |NG .002470 |[29789811]
I29789675 Irefl JNG .002471 ([29789675]
J29789803 jrefjlNG .002472. [[29789803]
|29789804 Irefl JNG. .002473. ([29789804]
(29789805 jref JjNG. .002474. |[29789805]
[29789802 jref JjNG 002475. 1(29789802]
J29789800 Irefl JNG .002476 |[29789800]
J29789801 Irefl JNG .002477 1(29789801]
J29789825J jrefjjNG .002478 |[29789825]
J29789798J Irefl JNG 002479 |[29789798]
gi|29789777|ref|NG_002480.1|[29789777] giJ28372978(refiNG_002481.1 [[28372978] giJ29789557Jref|NG_002482.lj[29789557] gi|29789606(ref|NG_002483.li[29789606] giJ29789603|ref[NG_002484.1 |[29789603] gii29789753iref(NG_002485.li[29789753] gi|29789703 jref|NG_002486.1 ([29789703] gi|29789555Jref|NG_002487.1|[29789555] gi|2978956θjref]NG_002488.l([29789560] gi|29789706[ref|NG_002489.1|[29789706] gii29789482|ref|NG_002490.lj[29789482] giJ29789553|ref|NG_002491.11[29789553] giJ29789824(ref|NG_002492.lj[29789824] gi|29789773|ref|NG_002493.1|[29789773] gi|29789774Jref|NG_002494.lj[29789774] gi|29789765|ref|NG_002495.1|[29789765] giJ29789772Jref[NG_002496.li[29789772] gi(29789819JrefJNG_002497.11[29789819] giJ29789769|ref|NG_002498.1|[29789769] giJ2978977θjref|NG_002499.1l[29789770] gii29789820|ref|NG_002500.1|[29789820] gi[29789474[ref|NG_002501.1 j[29789474] gi|29789473[ref[NG_002502.1|[29789473] giJ29789480 jref |NG_002503.1|[29789480] giJ29789472iref|NG_002504.1|[29789472] gi|29789634|ref|NG_002505.lj[29789634] giJ42415486|ref|NG_002506.li[42415486] gi|29789704Jref|NG_002507.l([29789704] gi|29789727|ref|NG_002508.1J[29789727] gi|29789728|ref|NG_002509.lj[29789728] gi|29789726Jref|NG_002510.11[29789726] gi|29789723|refJNG_002511.1 ([29789723] gi|29789724Jref|NG_002512.lj[29789724] gi|29789570Jref|NG_002513.lj[29789570] gi|2978982ljref|NG_002514.1 ([29789821] giJ29789818iref|NG_002515.11[29789818] gi|29789813|ref|NG_002516.11[29789813] gi(29789747Jref|NG_002517.11[29789747] giJ29789567|ref|NG_002518.lj[29789567] gi|29789559|ref|NG_002519.1|[29789559] gi|29789558(ref|NG_002520.l[[29789558] gi|29789556Jref JNG_002521.1 ([29789556] gi(29789523iref|NG_002522.lj[29789523] giJ29789525Jref(NG_002523.1 ([29789525] gi[29789484irefJNG_002524.1|[29789484] gi(29789549iref[NG_002525.li[29789549] gi|29789534|ref|NG_002526.1 |[29789534] gi|29789548Jref|NG_002527.li[29789548] gi|29789481 |reflNG_002528.1 j[29789481] gi|29789487|ref|NG_002529.1|[29789487] gi|29789497|ref|NG_002530.11[29789497] gi[29789498|ref|NG_002531.11[29789498] gi|29789501 [ref|NG_002532.1 j[29789501] giJ29789503Jref|NG_002533.lj[29789503] gi|29789505|ref|NG_002534.1|[29789505] gi(29789568|ref|NG_002535.1|[29789568] gi|29789504Jref|NG_002537.1|[29789504]
gi|29789565|ref|NG_002538.1J[29789565] giJ29789566iref(NG_002539.lj[29789566] giJ29789563|ref|NG_002540.1 ([29789563] gi|29789564jref|NG_002541.11[29789564] giJ29789561 [ref|NG_002542.1 [[29789561] gi|29789562|ref|NG_002543.11[29789562] giJ29789554JrefJNG_002544.lj[29789554] gi|29789551 (ref|NG_002545.11[29789551] gi|29789552|ref|NG_002546.1|[29789552] gι'J29789767Jref |NG_002547.1 J[29789767] gi|29789722|ref|NG_002548.11[29789722] gi|29789550|ref|NG_002549.1|[29789550] gi|29789547|ref|NG_002550.11[29789547] gi)29789545|ref|NG_002551.11[29789545] gii29789716iref(NG_002552.1|[29789716] gi|29789546|ref|NG_002553.1|[29789546] gi|29789633|ref|NG_002554.1|[29789633] gi|29789543 jref JNG_002555.11[29789543] gi|29789544|ref|NG_002556.l([29789544] gi|29789654|ref|NG_002557.1|[29789654] gi|29789541 jref|NG_002558.11[29789541] gi(29789539|ref|NG_002559.1[[29789539] gi|29789540|ref|NG_002560.1 ([29789540] gi|29789537|ref|NG_002561.1 J[29789537] giJ29789538|ref|NG_002562.1|[29789538] gi|29789533Jref|NG_002563.1|[29789533] gi|29789530Jref|NG_002564.lj[29789530] gi[29789600|ref|NG_002565.lj[29789600] gi|29789531jref|NG_002566.11[29789531] giJ29789528[refJNG_002567.lj[29789528] gi|29789618|ref|NG_002568.1|[29789618] giJ29789529|ref|NG_002570.lj[29789529] gi|29789526|ref|NG_002571.1 ([29789526] gi|29789527|ref|NG )02572.1|[29789527] gi|29789524|ref[NG_002573.11[29789524] gi]29789520|ref|NG_002574.1|[29789520] gi|29789532JreflNG_002575.lj[29789532] gi|29789522Jref|NG_002576.11[29789522] gi|29789465|ref|NG_002577.1|[29789465] giJ29789464(ref[NG_002578.1 [[29789464] gi|29789461|ref|NG_002579.11[29789461] gi(29789463|ref|NG_002580.1|[29789463] gi(29789696|ref|NG_002581.11[29789696] gi|29789466|ref|NG_002582.1|[29789466] giJ29789462irefJNG_002583.l[[29789462] gi|29789460|ref |NG_002584.11[29789460] giJ49615102(refJNG_002585.1|[49615102] giJ29789468|ref|NG_002586.1|[29789468] gi|49615103|ref|NG_002587.11[49615103] gi|29789655|ref|NG_002588.l[[29789655] gi|29789653|ref|NG_002589.1|[29789653] giJ29789651|refJNG_002590.lj[29789651] giJ29789652|reflNG_002591.1 ([29789652] gi|29789665Jref|NG )02592.1|[29789665] gi|29789646|ref|NG_002593.1|[29789646] gi|29789635|ref|NG_002594.1|[29789635] gi[29789632|ref|NG_002595.1|[29789632]
129789630 Irefl JNG .002596.1 |[29789630]
J29789628 Irefl JNG 002597.1 j[29789628]
(29789623 jrefl JNG .002598.1 j[29789623]
129789621 Irefl JNG 002599.11(29789621]
J46358430 jrefl JNG 002601.1 i[46358430]
J29789619 Irefl JNG .002603.11(29789619]
[29789612 Irefl JNG 002604.1 j[29789612]
J29789610 jref JjNG .002605.1 ([29789610]
I29789607 J]refl JNG 002606.1 |[29789607]
J29789601 | ref j |NG .002607.1 ([29789601]
(29789602 jref JlNG .002608.1 j[29789602]
I29789592 jref jlNG 002609.11(29789592]
[29789569 jref J|NG .002610.1 [[29789569]
(29789678 Irefl jNG 002611.11(29789678]
[29789758 jref J|NG .002612.1 |[29789758]
J29789759 Irefl jNG .002613.1 ([29789759]
[29789734 Irefl JNG 002616.1 |[29789734]
J29789733 Irefl JNG .002617.1 [[29789733]
129789731 Irefl JNG..002618.1 |[29789731]
|29789735 Irefl JNG 002619.1 ([29789735]
|29789736 iref J|NG .002620.1 |[29789736]
|29789738 Iref JjNG .002621.1 |[29789738]
|29789679 jref JjNG .002622.1 [[29789679]
128571462 jref JlNG .002623.1 |[28571462]
J28827827 jref J|NG .002624.1 j[28827827]
J28827828 Irefl JNG 002625.1 j[28827828]
J47059115 Irefl JNG .002626.1 [[47059115]
|29789672 Irefl JNG .002627.1 ([29789672]
J29789673 jrefl JNG .002628.1 [[29789673]
|29789670 iref J|NG .002629.1 |[29789670]
J29789671 Irefl JNG 002630.1 [[29789671]
|29789668 jref J|NG .002631.1 j[29789668]
I29789669 Irefl JNG.002632.11(29789669]
|28626502 | ref J |NG .002633.1 |[28626502]
(28629869 Irefl JNG .002634.1 ([28629869]
J28866958 Irefl JNG.002636.1 [[28866958]
[28849908 Irefl JNG .002637.11(28849908]
|28866962 jref JjNG .002638.1 ([28866962]
[50979291 |ref JNG .002639.2 [[50979291]
(32480613 Irefl JNG 002640.1 |[32480613]
J28882050 Irefl JNG .002641.1 j[28882050]
128912917 Irefl JNG .002642.1 [[28912917]
147059116 iref J|NG .002645.11(47059116]
(28973788 jref JjNG .002646.1 j[28973788]
J28973793 Irefl jNG .002647.1 [[28973793]
[32480616 Irefl JNG 002648.1 [[32480616]
132480614 jref J |NG .002649.1 |[32480614]
[32480618 Irefl JNG .002650.1 [[32480618]
(32480615 jref j|NG .002651.1 [[32480615]
[32480620 jref JlNG .002652.11(32480620]
132480617 jref j|NG .002653.1 |[32480617]
(28976149 jref J|NG .002655.1 ([28976149]
J28976152 Irefl jNG .002656.1 |[28976152]
[28976150 Irefl JNG .002657.11(28976150]
(28976155 Iref J|NG .002658.1 |[28976155]
[47059119 Irefl jNG .002659.1 ([47059119]
J28976151 Iref JjNG 002660.1 ([28976151]
gi|28976157|ref|NG. 002661.1 |[28976157] giJ3248062l jrefJNG 002662.11[32480621] gi|32480619Jref[NG 1_002663.1 J[32480619] gi|32480623|ref|NG i._002665.1 ([32480623] gii32480622|refJNG 002666.11[32480622] gi|31711995|ref|NG i._002667.2J[31711995] gi(28976162iref(NG 002668.1 [[28976162] gi|28976158Jref|NG 002669.1 j[28976158] giJ45361708|ref|NG *_002670.2|[45361708] giJ28976161 |ref|NG 002671.1 j[28976161] gi|31711993|ref|NG _002672.2|[31711993] gi|28976164|ref|NG _002673.1 [[28976164] giJ32480624JreflNG. .002674.11[32480624] giJ32480625Jref|NG _002675.11(32480625] gi(32480629Jref[NG 002676.1 |[32480629] gi|29150256|ref|NG .002679.1 [[29150256] gi(29336064Jref|NG .002680.1 [[29336064] gi|29336072|ref|NG .002681.1 [[29336072] gi|32480626Jref|NG .002682.1 j[32480626] gi[32480632|ref|NG .002683.11[32480632] giJ29336067|ref|NG .002684.11[29336067] gi|29336075|ref|NG 002685.1 |[29336075] gi|29171299Jref|NG .002687.11[29171299] gi|29171302|ref|NG 002688.1 j[29171302] gi|29171308|ref|NG
" _002689.1 |[29171308] gi|30316443Jref|NG _002690.1 |[30316443] giJ29383684|refiNG 002691.1 |[29383684] gi|29366831|ref|NG .002692.11[29366831] giJ29366836|reflNG .002693.1 j[29366836] gi|29366839(ref|NG. .002694.1 [[29366839] gi|29423716(ref|NG .002696.1 |[29423716] gi(29423717JrefJNG .002697.1 ([29423717] giJ29423718|ref|NG .002698.1 ([29423718] gi|29469072|ref|NG
_ .002699.1 ([29469072] gi|41281879|refJNG _002700.l([41281879] giJ4128190θ(ref|NG~ 002701.1 J[41281900] giJ41281903|ref|NG 002702.1 |[41281903] giJ30242643(ref|NG~ 002703.1 j[30242643] gi|30242644|ref|NG~ 002705.1 |[30242644] giJ30242646[ref|NG 002707.1 [[30242646] gi|3024265θ[ref|NG~ 002709.11[30242650] gii30242655(ref|NG_ 002711.1 |[30242655] gi|30242641 [reflNG .002713.1 [[30242641] gi|29824428[ref(NG 002716.1 J[29824428] gi|29725628|ref|NG_ 002717.1 j[29725628] gi|29725625|ref|NG_ .002718.11[29725625] gi|29725629|ref[NG .002719.11[29725629] giJ2972563θjref|NGl 002720.1 ([29725630] giJ2972563lireflNG_ .002721.11[29725631] giJ29837651|reflNG_ 002723.1 j[29837651] giJ30089684Jref|NG_ .002724.11(30089684] gi|29837660|ref|NG_ .002725.11(29837660] gi|29837663|ref|NG_ .002726.1 j[29837663] gi|30089698|ref|NG_ 002727.1 J[30089698] giJ30089699|ref|NG_ 002728.11[30089699] giJ30089700|ref|NG_ 002729.1 j[30089700] gi|30089701 |reflNG_ 002731.11[30089701]
gi|30089702 |ref J|NG 002733.11(30089702] gi|30172549 irefl JNG 002735.1 |[30172549] giJ30172550 Irefl JNG .002736.1 j[30172550] gi|30172551 Irefl JNG 002737.1 [[30172551] giJ30172552 |ref J|NG. 002738.1 |[30172552] giJ30172553 | ref | NG .002739.1 [[30172553] giJ30172554 iref| NG 002740.11(30172554] gi|30172555 jrefl JNG .002741.1 |[30172555] gi|30172556 Irefl JNG 002742.1 j[30172556] gi|30172557 jref] NG .002743.1 [[30172557] giJ30172558 jref J|NG .002744.1 [[30172558] gi|30172559 iref JlNG .002745.1 ([30172559] gi|30387634 jrefl JNG .002746.1 [[30387634] gi[30387635 jrefl JNG..002747.11(30387635] gi(30387636 Irefl NG..002748.1 j[30387636] gi[30425556 I ref | NG .002749.11(30425556] gi|30425554 Irefl JNG .002750.1 [[30425554] gi(30425555 Irefl JNG .002752.1 ([30425555] gi[30425558 jrefl JNG .002753.1 [[30425558] gi(30425559 Iref JjNG .002754.1 |[30425559] gi[30520308 jref J|NG 002761.11(30520308] gi|32996738 jref J|NG .002762.11(32996738] gi|39841019 Irefl JNG..002763.2 ([39841019] gi|30520304 irefl (NG .002764.1 |[30520304] gi(32996739 jrefl JNG .002765.11(32996739] gi[30520307 Irefl JNG .002766.1 ([30520307] gi(30520323 |ref J|NG .002767.1 j[30520323] gi|32996740 Irefl JNG .002768.1 [[32996740] giJ30520344 Irefl JNG 002769.11(30520344] giJ30520355 Irefl JNG..002770.1 ([30520355] gi|30578394 jref JjNG .002771.11(30578394] gi[30578387 Irefl JNG .002772.1 j[30578387] gi|32189419 jref JlNG..002773.1 ([32189419] giJ30911114 I ref J |NG 002775.1 |[30911114] gi|30911110 Irefl JNG .002776.1 [[30911110] gi|30911116 Irefl JNG .002777.1 |[30911116] giJ30911118 Irefl JNG .002778.11(30911118] gi|30911115 Irefl JNG..002779.1 ([30911115] giJ30911119 jref JlNG .002780.1 |[30911119] giJ30911117 Iref JjNG 002781.11(30911117] gi|30911120 jref JlNG..002782.1 |[30911120] gi|32189416 Irefl NG .002785.1 ([32189416] giJ31126969 Irefl NG .002786.11(31126969] gi|31126964 Irefl JNG .002787.11(31126964] giJ31340733 Irefl NG .002788.11(31340733] giJ31791043 Irefl NG .002790.1 [[31791043] giJ32171234 iref JlNG .002791.11(32171234] gi|32171197 Irefl JNG 002792.1 |[32171197] gi|32307633 Irefl NG .002793.11(32307633] giJ32996710 irefl NG .002795.11(32996710] gi|47575700 Irefl JNG .002796.2 ([47575700] giJ32996711 Iref JjNG .002797.1 |[32996711] giJ32996712 Iref JjNG 002798.11(32996712] gi|32490568 |ref J|NG..002799.1 |[32490568] giJ32996713] jref JjNG. 002800.1 |[32996713] gii32563558 jref J|NG. 002801.1 [[32563558] giJ32563560 iref (jNG. 002802.1 j[32563560]
gi|32563559|refJ|NG 002803.11(32563559] giJ32563563[ref J|NG .002804.1 |[32563563] gi|32698797Jref| |NG_002805.1 ([32698797] giJ32698793Jref JjNG_002806.1 |[32698793] gi|32698799Jref| |NG_002807.1 1(32698799] giJ32698796Jref| |NG_002808.1 1(32698796] gi|3269880l [ref| |NG_002809.1 1(32698801] giJ32698798Jref JjNG 002810.1 |[32698798] gi]32698803Jref J|NG 002811.1 [[32698803] gi|3269880θ[ref| |NG_.002812.1 |[32698800] giJ32698805Jref JjNG 002813.1 1(32698805] giJ32698802(refj |NG_002814.1 |[32698802] giJ32698804Jref| |NG_002815.1 ([32698804] giJ3269881θ[ref J|NG_002816.1 |[32698810] gi|32698806Jrefj |NG_002817.1j[32698806] gi|32698813|ref J|NG 002818.1 |[32698813] giJ32698809Jref |NG_.002819.1 1(32698809] gi|32698811 |ref J|NG .002821.1 [[32698811] gi[32698812Jref J|NG.002822.1 [[32698812] gi|32698816|ref JjNG..002823.1 ([32698816] gi|32698817|ref J|NG 002824.1 |[32698817] giJ32698818Jref J|NG.JD02825.1j[32698818] giJ32698819|ref J|NG.002826.1j[32698819] giJ32698833Jref JjNG .002827.1 ([32698833] giJ32698842Jref J|NG 002828.1 ([32698842] giJ32698843(ref J|NG 002829.1 ([32698843] giJ32698862(ref JiNG .002830.1j[32698862] gi|32698999Jrefj(NG 002831.1 |[32698999] giJ32699024Jref J|NG .002832.1 [[32699024] gi|32699031 |ref J|NG .002833.1 |[32699031] gi|32699039|ref J|NG 002834.1j[32699039] giJ32699034Jref J|NG .002835.1 |[32699034] giJ3269904θjref J]NG 002836.1 [[32699040] giJ32699043Jref J|NG 002837.1 ([32699043] giJ32699046(ref J|NG .002838.1 [[32699046] gi|32699077Jref J|NG 002839.1 1(32699077] giJ32699078|ref J|NG .002840.1 ([32699078] gi|32699079|ref J|NG 002841.1 1(32699079] gi[32699080|ref J|NG .002842.1 ([32699080] giJ3269908 jref J|NG ~002843.1 |[32699081] gi|32699082|ref J|NG 002844.1 1(32699082] giJ32699083Jref J|NG.002845.1j[32699083] gi|32699084Jref JjNG.002846.1 1(32699084] giJ32699085Jref J|NG.002847.1 [[32699085] giJ32699086Jref J|NG.002848.1 |[32699086] giJ32699087Jref J|NG.002849.1 [[32699087] gi|32699088|ref J(NG.002850.1 |[32699088] gi(32699089Jref J|NG.002851.1 [[32699089] gi|3269909θjref J|NG.002852.1 [[32699090] gi|3269909l[ref J|NG.002853.1 [[32699091] giJ32699092Jref J]NG_002854.1 [[32699092] giJ32699095Jref J|NG_002855.1 1(32699095] gi|32699096|ref J|NG._002856.1 [[32699096] giJ32699097Jref J|NG_002857.1 1(32699097] giJ32699098[ref J|NG_002858.1 [[32699098] gi|32699099|ref JjNG 002859.1 |[32699099] giJ3269910θjref J|NG 002860.1 1(32699100]
G.002861.11(32699101] G .002862.1|[32699102] G 002863.11(32699103] G 002864.1|[32699104] G..002865.11(32699105] G 002866.1([32699106] G..002867.11(32699107] G 002868.1[[32699108] G..002869.11(32699109] G 002870.1|[32699110] G 002871.11(32699111] G..002872.1 |[32699112] G 002873.11(32699113] G..002874.1 |[32699114] G 002875.11(32699115] G..002876.1|[32699116] G 002877.1j[32699118] G..002878.1j[32699119] G 002879.1|[32699120] G .002880.1[[32699121] G .002881.1 [[32699122] G 002882.11(32699123] G .002883.1 j[32699124] G..002884.1 j[32699125] G 002885.11(32699126] G .002886.1 ([32699127] G 002887.11(32699128] G .002888.11(32699129] G.002889.1 [[32699130] G .002890.1 ([32699131] G..002891.1 |[32699132] G.002892.1 [[32699133] G .002893.1 |[32699134] G .002894.1([32699135] G..002895.11(32699136] G.002896.1 [[32699137] G .002897.11(32699138] G.002898.11(32699139] G .002899.1 ([32699140] G.002900.1 [[32699141] G .002901.11(32880204] G 002902.1 ([32880210] G .002903.1 [[32880209] G 002904.11(32880217] G .002905.1 |[33186879] G..002906.1 |[33186880] G .002907.1 [[33186890] G .002908.1j[33186885] G .002909.11(33186892] G..002910.11(33186891] G..002911.1[[33186899] G 002912.11(33186893] G .002913.1 ([33186902] G .002914.11(33186898] G 002915.1 [[33186903] G..002916.1 |[33300640]

G 002917.11(33342269]
G 002918. |[33342270] G .002919. ([33342271] G 002920. |[33342272] G .002921. [[33342273] G 002922. ([33438578] G .002923. |[33354258] G 002924. |[33354259] G .002926. 1(33354260] G .002927. 1(33354261] G 002928. [[33354262] G .002929. 1(33354263] G .002930. |[33354264] G 002931. ([50055034] G .002932. |[33386681] G 002933. ([33386679] G .002934. j[33386684] G .002935. 1(33386680] G 002936. j[33386686] G .002937. 1(33386688] G 002938. j[33386685] G .002939. 1(33386689] G .002940. [[33386687] G 002941. ([33413415] G .002942. |[33413416] G .002943. 1(33413419] G 002944. 1(33413420] G 002945. |[33413422] G .002946. 1(33413421] G 002947. |[33413426] G .002948. j[33413423] G .002949. |[33413427] G 002950. 1(33457309] G .002951. 1(33457304] G 002952. |[33457312] G .002953. 1(33563318] G .002954. 1(33563320] G .002955. ([33563317] G .002956. 1(33563322] G .002957. ([33563319] G 002958. 1(33563324] G .002959. [[33563321] G .002960. |[33563326] G .002961. 1(33563323] G 002962. ([33563328] G .002963. |[33563325] G..002964. [[33563330] G .002965. ([33563327] G .002966. |[33563329] G..002967. 1(33563334] G..002968. [[33563331] G .002969. ([33563336] G .002970, [[33563333] G..002971 , |[33563338] G .002972, ([33563335] G 002973, j[33563337] G 002974, ][33563341]

G 002975, 1(33563342]
G 002976.11(33563343
' G 002977.11(33563344] G .002978.1([33563345] G 002979.1[[33563346] G .002980.1([33563347 G 002981.11(33563348] G .002982.1[[33563349 G .002983.11(33563350] G .002984.11(33563351 G 002985.1([33563352 G .002986.1 [[33563353] G..002987.1 ([33563356] G .002988.1|[33563357] G .002989.1[[33563358] G .002990.11(33563359
' G .002991.1([33563360 G 002992.1|[33563361 G .002993.1[[33563362] G 002994.1([33563363 G .002995.1[[33563364] G 002996.1 ([33589840 G .002997.1 |[33589837] G 002998.11(33589841 G .002999.1 [[33589839] G 003006.11(34303967 G .003008.1|[34098955] G 003009.11(34304063 G .003010.1[[34304009] G .003011.1[[34304012
" G 003012.11(34304013 G .003013.1|[34304014] G.003014.1([34304015
' G .003015.11(34304016 G .003016.1 |[34304017] G .003017.1 [[34328083] G 003018.1 |[34328088 G .003019.11(34328084
' G .003020.1|[34328090] G .003021.1([34328087] G 003022.1|[34328093 G .003023.11(34328089 G 003024.11(34328096] G .003025.1j[34395920] G 003026.1([34419619] G .003027.11(34419623] G 003028.1 ([34419620] G .003029.1 [[34419626 G 003030.1 |[34447214] G .003031.11(34447212] G .003032.1|[34447213 G .003033.1([34447219 G .003034.1|[34447215 G .003035.11(34447221 G 003036.1|[34447218] G .003037.11(34447220 G 003038.1[[34447225]
G 003039.11(34482048
'
gi|34536817|ref|NG_003040.1|[34536817] giJ34594662|ref|NG_003041.1 ([34594662] gi|34594667Jref|NG_003042.1 ([34594667] giJ34594665|ref|NG_003043.1|[34594665] giJ35215289[ref|NG_003044.li[35215289] gi|35215290|ref|NG_003045.1|[35215290] gij37620190|refJNG_003061.1J[37620190] gi|37620207|ref|NG_003062.11[37620207] gi|37620208|ref|NG_003063.1|[37620208] gi|37674243|ref|NG_003064.1|[37674243] giJ37693501 |ref|NG_003070.11[37693501] gi|38604065|ref|NG
„003072.1|[38604065] giJ37700235|ref|NG_003073.1 [[37700235] gi|37700242|ref|NG_003074.1)[37700242] giJ37700247(ref|NG_003075.1|[37700247] gi|37700250|ref|NG_003076.1|[37700250] gi|37700257|ref|NG_003077.1|[37700257] gi|37700258|ref|NG_003078.1|[37700258] gi|37700259|ref|NG_003079.1l(37700259] gi|37700260|ref|NG_003080.l[[37700260] gi|37700261 |ref|NG_003081.11[37700261] gil37700262|reflNG_003082.1J[37700262] gi|37700263[ref|NG_003083.1|[37700263] gi|37700264|ref|NG_003084.1|[37700264] gi(37700265|ref|NG_003085.1[[37700265] gi|37700266|ref[NG_003086.1|[37700266] giJ37700267[ref(NG 003087.1 j[37700267] gi|37700268|ref|NG )03088.11[37700268] giJ37700269|ref|NG_003089.1|[37700269] gi[37700270|ref|NG_003090.1|[37700270] gi|37700271|ref|NG_003091.1|[37700271] gi|37700272|reflNG_003092.lj[37700272] gi|37700273|ref|NG_003093.1|[37700273] gi|37700277|ref|NG_003094.1J[37700277] gi|37700275iref|NG_003095.l([37700275] giJ37700278|ref|NG_003096.1|[37700278] gi|37700276|ref|NG_003097.lj[37700276] gi|37700279|ref|NG_003098.1|[37700279] gi|37700280|ref|NG_003099.1|[37700280] gi|38570424|ref|NG_003100.1|[38570424] gi|38016168|ref|NG_003101.11[38016168] gi|38016169|ref|NG_003102.1|[38016169] gi|38016170|ref|NG_003103.1|[38016170] gi|38570426|ref|NG_003104.1|[38570426] gi|38016185Jref|NG_003105.1 ([38016185] gi|38016181|ref|NG_003106.1|[38016181] gi|38194230|ref|NG_003107.1 ([38194230] gi|38257142|ref|NG_003108.1|[38257142] gi|38257149|ref|NG_003109.11[38257149] gi]38570425|refJNG_003110.l[[38570425] gi|38348441|ref|NG_003111.11[38348441] gi|38371733(ref[NG_003114.1[[38371733] gi|38371741 |ref|NG_003115.1 ([3837 741] gi|38371740|ref|NG_003116.1 [[38371740] gi|38371746|ref|NG_003117.1 [[3837 746] gi|38371742|ref|NG_003118.11[38371742] giJ38371748|ref|NG_003119.1J[38371748]
138371745 Irefl JNG 003120 |[38371745] J38371749 Irefl JNG .003121 1(38371749] 138371747 Irefl jNG
"003122 1(38371747] 138371751 Irefl JNG .003125 1(38371751] J38371750 iref j|NG
"003126 1(38371750] J38371752 Irefl JNG 003127 ([38371752] I47894453 Iref JjNG
~003128, |[47894453] [38371756 jrefl JNG 003129 [[38371756] 138488761 Iref JjNG 003130 |[38488761] |38488760 jref J|NG 003131, [[38488760] (38488762 Irefl JNG 003132, |[38488762] |38488769 | ref J |NG .003133, 1(38488769] |38488765 Irefl JNG
"003134, |[38488765] 138488771 Irefl JNG .003135, 1(38488771] [38488768 jrefl JNG .003136, |[38488768] |38488773 Irefl JNG 003137, [[38488773] I38488770 Irefl JNG 003138, ([38488770] |38488775 |ref J|NG..003139, [[38488775] |38488772 jref J|NG..003140, ([38488772] J38488777 jref J|NG 003141, j[38488777] |38488774 jref JlNG 003142. ([38488774] |38488779 jref J|NG 003143. |[38488779] [38488776 Irefl jNG .003144. |[38488776] J38488780 jref J|NG 003145. ([38488780] I38488778 Irefl JNG .003146 ([38488778] I38488782 jrefl JNG 003147 |[38488782] (38488781 Iref JjNG 003148 j[38488781] |38488783 Iref JjNG .003149 1(38488783] [38488784 Iref JjNG 003150. |[38488784] |38488786 jrefl JNG .003151 [[38488786] (38488785 Jref JlNG 003152 |[38488785] I38488787 ]ref J|NG 003153 j[38488787] [38502319 Irefl JNG .003154 1(38502319] J39573710 Irefl JNG .003155. |[39573710] J39573711 Irefl JNG 003156 j[39573711] 139573719 Irefl JNG .003157. 1(39573719] 139573721 jrefl JNG 003158 [[39573721] J39573722 irefl JNG .003159. ([39573722] |39573720 Irefl JNG .003160 [[39573720] 140217618 irefl JNG .003162. ([40217618] 140217619 Irefl JNG..003163. 1(40217619] (40353208 |ref J|NG .003164 |[40353208] 140217620 iref J|NG 003165 [[40217620] [40217623 Irefl JNG .003166. j[40217623] 140217624 jref J|NG .003167 |[40217624] 140217631 Irefl JNG .003168. 1(40217631] 140217632 |ref J|NG .003169 |[40217632] J40217633 Irefl JNG.003170. |[40217633] 140217634 jref J|NG .003171. 1(40217634] |40217635 jref ( |NG .003172 [[40217635] 140217636 Irefl JNG .003173 1(40217636] |40806230 JrefJlNG .003180 1(40806230] J41349935 IreflJNG .003183 |[41349935] J41680696 IreflJNG .003186 1(41680696] [41680699 jrefJ|NG .003187 |[41680699] (47894442 IreflJNG .003188 [[47894442] 142761477 jrefJjNG 003189 |[42761477]
G.003190. 1(45120107] G .003193. IJ45504397] G 003194. 1(45504395] G .003195. |[45504396] G 003196. 1(48525360] G .003197. |[45504378] G 003198. ([49457787] G .003200. 1(45592950] G .003201. [[45592947] G.003215. [[46047452] G .003216. [[46047437] G.003217. [[46047457] G .003218. [[46047450] G .003219. |[46047461] G .003221. j[46047469] G .003222. 1(46047477] G .003230, [[46048465] G 003253, ([46048483] G .003254, 1(46243669] G .003255, [[46389547] G .003256, j[46395481] G.003258, |[46409255] G .003259, 1(46409256] G .004075, ([47458033] G..004077, j[47458034] G .004085, |[47777326] G .004086, j[48525355] G 004087, [[48525356] G .004088. 1(49615106] G..004089. |[47777331] G.004091. 1(47777338] G .004092. 1(47777341] G.004093. [[47777342] G .004095. 1(47827226] G .004096. ([47827223] G .004097. [[47827227] G.004098. |[47827228] G .004099. ([47827229] G .004100. |[47827230] G.004101. [[47827231] G .004102. |[47827232] G .004103. ([47827233] G.004109. |[48427665] G 004110. [[48475056] G..004111. 1(48475053] G 004112. j[49487492] G .004113. 1(49487494] G 004115. [[48597004] G .004116, [[48597002] G 004122, |[48597025] G 004123, 1(48597022] G 004124, ([48597024] G 004125, 1(48597029] G 004126, [[48597026] G 004127, |[48597031] G .004128, ([48597028]

G 004129, |[48597032]
gi|48597030|ref|NG_004130.11[48597030] giJ48597033|refJNG_004131.1 j(48597033] gi|48597034|ref|NG_004132.1|[48597034] gi|48597036lref|NG_004133.1|[48597036] giJ48597037(ref|NG_004134.1 ([48597037] gi[48597038|ref|NG_004135.1J[48597038] giJ4859704θjrefJNG_004136.1|[48597040] gi|48597041 [ref|NG_004137.11[48597041] gi|48597043|ref|NG_004138.1|[48597043] giJ48597039JrefJNG_004139.lj[48597039] gi|48597045|ref|NG_004140.1|[48597045] gi|48597042|ref|NG_004141.11[48597042] giJ48597047|ref|NG_004142.1 ([48597047] gi|48597044|ref|NG_004143.1|[48597044] giJ48597049irefJNG_004144.li[48597049] gi|48597046|ref[NG_004145.1|[48597046] gi|48597050|ref(NG_004146.1|[48597050] gi|48597048|ref|NG_004147.1 ([48597048] gi|48597051)reflNG_004148.1|[48597051] gi|48597052|ref|NG_004149.1|[48597052] gij48597053|ref|NG_004150.11(48597053] giJ48597054JreflNG_004151.1 [[48597054] gi|48597055|ref|NG_004152.11[48597055] giJ48597056|ref|NG_004153.11[48597056] giJ48597059Jref|NG_004154.lj[48597059] gi)48597057|ref)NG_004155.11[48597057] giJ48597060JreflNG_004156.l[[48597060] gi|48597058|ref|NG_004157.1j[48597058] gi|48597061|ref|NG_004158.1 ([48597061] gi|48597062Jref|NG_004159.1 ([48597062] gi|48597063|ref|NG_004160.1 ([48597063] gi(48597064|ref[NG_004161.1|[48597064] gi|48597065|ref|NG_004162.1|[48597065] gi|48597066|ref|NG_004163.1|[48597066] gi|48597067|ref|NG_004164.1|[48597067] gi|48597068|ref|NG_004165.11[48597068] gi(48597069Jref|NG_004166.1|[48597069] gi j48597070|ref jNG_004167.1 ([48597070] gi|48717107|ref|NG_004168.1|[48717107] giJ48717108(ref|NG_004169.l([48717108] gi|48717109|ref(NG_004170.1 [[48717109] giJ4871711θjref|NG X)4171.1|[48717110] gi|48717111 |ref|NG_004172.1 ([48717111] gi|48717112|ref|NG_004173.1|[48717112] gi|48717113irefJNG_004174.li[48717113] gi[48717114|ref|NG_004175.11[48717114] giJ48717115|ref|NG_004176.1|[48717115] gi|48717116|reflNG_004177.lj[48717116] gi|48717117|ref|NG_004178.11[48717117] gi|48717118|ref|NG_004179.1 ([48717118] gi|48717119|ref|NG_004180.1)[48717119] giJ48717120Jref|NG_004181.1|[48717120] gi[48717121|reflNG_004182.1|[48717121] gi|48717122|ref|NG_004183.1|[48717122] gi|48717123[reflNG_004184.1|[48717123] gi|48717124|ref|NG_004185.1|[48717124] gi|48717125JrefJNG_004186.lj[48717125]
gi|48717127|ref|NG_004188.1 |[48717127] gi|48717128|reflNG_004189.1 [[48717128] gi|48717129|ref|NG_004190.l [[48717129] giJ4871713θjrefJNG_004191.l j[48717130] giJ4871713l jref|NG_004192.l j[48717131] gi[48717132lrefJNG_004193.l j[48717132] gi(48717133iref|NG_004194.l ([48717133] gi|48717134|ref|NG_004195.11[48717134] gi|48717135Jref|NG_004196.l [[48717135] gi(48717136JrefjNG_004197.1 J[48717136] giJ48717137|ref |NG_004198.11[48717137] gi[48717138Jref |NG_004199.1 j[48717138] gi|48717139JreflNG_004200.l i[48717139] gi|4871714θjref|NG_004201.11[48717140] gi[4871714l (ref|NG_004202.1 [[48717141] gi|48717142|ref|NG_004203.11[48717142] gi|48717143iref|NG_004204.1 |[48717143] gi|48717144Jref|NG_004205.l j[48717144] gi|48717145|ref|NG_004206.11[48717145] gi[48717146iref(NG_004207.11[48717146] gi|48717147|ref|NG_004208.1 |[48717147] gi|48717148iref|NG_004209.l j[48717148] giJ48717149[refJNG_004210.l j[48717149] gi|48717150|ref|NG_004211.1 [[48717150] giJ4871715l jref]NG_004212.l [[48717151] gi|48717152|ref|NG_004213.11[48717152] giJ48717153|ref|NG_004214.1 ([48717153] gi|48717154Jref|NG_004215.1 [[48717154] gij48717155Jref|NG_004216.l i(48717155] gi|48717156|ref|NG_004217.11[48717156] giJ48717157|reflNG_004218.11[48717157] gi|48717158|ref|NG_004219.1 ([48717158] gi|48717159(ref|NG_004220.1 ([48717159] gi|48717160|ref|NG_004221.11[48717160] gi|48717161 |ref|NG_004222.1 ([48717161] gi|48717162[reflNG_004223.1 ([48717162] gi|48717163|ref|NG_004224.11[48717163] giJ48717164|ref|NG_004225.1 [[48717164] gi|48717165[ref|NG_004226.11[48717165] giJ48717166|ref|NG_004227.l ([48717166] gi[48717167Jref|NG_004228.l i(48717167] gi(48717168(ref|NG_004229.1 |[48717168] giJ48717169[ref|NG_004230.1 j[48717169] gi|48717170|ref|NG_004231.11[48717170] gi[4871717l jreflNG_004232.l j[48717171] gi|48717172|ref|NG_004233.11(48717172] giJ48717173|ref|NG_004234.1 [[48717173] gi|48717174|ref|NG_004235.11[487171 4] gi|48717175|ref|NG_004236.1 |[48717175] gi|48717176Jref|NG_004237.l j[48717176] giJ48717177[ref|NG_004238.1 ([48717177] gi|48717178|ref|NG_004239.1 |[48717178] gil48717179|ref|NG_004240.1 |[48717179] gi|48717180|ref|NG_004241.11[48717180] giJ48717181 iref|NG_004242.l j[48717181] gi|48717182[ref|NG_004243.l j[48717182] gi[48717183(reflNG_004244.11[48717183]
gi|48717184|refJ|NG_.004245.1 [48717184] giJ48717185Jrefj |NG_004246.1 [48717185]' gi|48717186|ref| |NG_004247.1 [48717186] giJ48717187jref( |NG_004248.1 [48717187] gi|48717188[ref J|NG_004249.1 [48717188] giJ48717189|ref| |NG_004250.1 [48717189] gi|4871719θ(ref| |NG_004251.1 [48717190] gi|48717191 |ref| |NG_004252.1 [48717191] giJ48717192|refl |NG_004253.1 [48717192] gi|48717193Jref JNG 004254.1 [48717193] giJ48717194|ref| |NG_.004255.1 [48717194] giJ48717195Jref JjNG_004256.1 [48717195] gi|48717196|ref| |NG_004257.1 [48717196] giJ48717197lref J|NG .004258.1 [48717197] gi|48717198Jrefj |NG_004259.1 [48717198] gi|48717199|ref J|NG.004260.1 [48717199] gi|48717200|ref JjNG.004261.1 [48717200] gi|48717201 (refl JNG.004262.1 [48717201] gi|48717202(ref J|NG.004263.1 [48717202] giJ48717203Jref J|NG .004264.1 [48717203] gi|48717204|ref J|NG 004265.1 [48717204] gi[48717205Jref J|NG 004266.1 [48717205] gi|48717206|ref JjNG 004267.1 [48717206] giJ48717207|ref J|NG 004268.1 [48717207] giJ48717208Jref JjNG 004269.1 [48717208] gi|48717209Jref J|NG 004270.1 [48717209] gi|48717210|ref J|NG 004271.1 [48717210] giJ48717211 |ref J|NG 004272.1 [48717211] giJ48717212Jref J|NG .004273.1 [48717212] gi|48717213Jref J|NG 004274.1 [48717213] gi|48717214|ref J|NG..004275.1 [48717214] gi[48717215Jref J|NG .004276.1 [48717215] giJ48717216Jref| |NG .004277.1 [48717216] gi|48717217Jref J|NG 004278.1 [48717217] gi(48717218Jref J|NG .004279.1 [48717218] gi|48717219|refj|NG .004280.1 [48717219] giJ48717220|ref J|NG .004281.1 [48717220] gi|48717221 |ref J|NG .004282.1 [48717221] gi|48717222|ref JjNG 004283.1 [48717222] giJ48717223Jref J|NG .004284.1 [48717223] giJ48717224Jref J|NG 004285.1 [48717224] gil48717227|ref J|NG 004286.1 [48717227] giJ48717228Jref J|NG.004287.1 [48717228] gi|48717229|ref J|NG 004288.1 [48717229] gi|48717232|ref J|NG.004289.1 [48717232] gi|48717237|ref J|NG.004290.1 [48717237] gi|48717242Jref J|NG.004291.1 [48717242] giJ48717245Jref JjNG.004292.1 [48717245] gi|48717246|ref J|NG.004293.1 [48717246] giJ48717247Jref J|NG.004294.1 [48717247] gi|48717250|ref J]NG.004295.1 [48717250] gi|48717251 |ref J|NG.004296.1 [48717251] gi(48717252Jref J|NG_004297.1 [48717252] gi|48717253Jref J|NG._004298.1 [48717253] gi|48717254|ref JjNG._004299.1 [48717254] gi|48717255|ref J|NG_004300.1 [48717255] gi|48717256|ref J]NG 004301.1 [48717256]
gi|48717257|ref|NG_004302.1|[48717257] giJ48717258[ref|NG_004303.lj[48717258] giJ48717259|ref|NG_004304.11[48717259] giJ48717260|ref|NG_004305.11[48717260] gi|48717266|ref|NG_004307.1|[48717266] gi|48717267|ref|NG_004308.1 ([48717267] gi|48717271(ref|NG_004309.1|[48717271] giJ48717273|ref|NG_004310.11[48717273] gii4871727θ(ref|NG_004311.1 [[48717270] gi|48717275Jref|NG_004312.1|[48717275] gii48717272|ref|NG_004313.1|[48717272] gi|48717274|ref|NG_004314.1|[48717274] gi|48717276Jref[NG_004315.11[48717276] gi|48717287|ref|NG_004317.1|[48717287] gi|48717283|ref|NG_004318.11[48717283] gi|48717289irefJNG_004319.li[48717289] giJ48717286[ref|NG_004320.li(48717286] giJ48717291|ref|NG_004321.lj[48717291] giJ48717288JreflNG_004322.11(48717288] gi|48717293|ref|NG_004323.lj[48717293] gi|48717290|ref|NG_004324.11[48717290] gi|48717295(refJNG_004325.1 [[48717295] giJ48717292lref)NG_004326.lj[48717292] gi|48717297|ref|NG_004327.1|[48717297] gi|48717294|reflNG_004328.11[48717294] gi|48717299|ref|NG_004329.11[4871 299] giJ48717296|ref|NG_004330.11[48717296] gi|48717301 jref|NG_004331.1 [[48717301] gi|48717298|ref[NG_004332.1 j[48717298] giJ48717303irefJNG_004333.lj(48717303] gi|48717300|ref|NG_004334.11[48717300] gi|48717305Jref|NG_004335.1 [[48717305] gi|48717302JrefJNG_004336.lj[48717302] gi|48717307|ref|NG_004337.1|[48717307] gi|48717304Jref|NG_004338.1 [[48717304] gi|48717309|ref|NG_004339.11[48717309] gi(48717306|ref|NG_004340.lj[48717306] giJ4871731 liref|NG_004341.l([48717311] gi|48717308|ref|NG_004342.1|[48717308] giJ48717313Jref|NG_004343.1 j[48717313] gi|48717310|ref|NG_004344.11[48717310] gi|48717315|ref|NG_004345.1|[48717315] gi|48717312(ref|NG_004346.1|[48717312] gi|48717317|reflNG_004347.lj[48717317] gi|48717314Jref|NG_004348.1][48717314] giJ487 7319|ref|NG_004349.1 [[48717319] gi|48717316|refJNG_004350.11[48717316] gi|48717321 |ref|NG_004351.11[48717321] gi|48717318Jref|NG_004352.lj[48717318] gi|48717325|ref|NG_004353.1|[48717325] gi[4871732θiref|NG_004354.1 ([48717320] gi|50054346irefJNG_004355.2i[50054346] gi(48717330[ref[NG_004356.1([48717330] gij48717326|refjNG_004357.1|[48717326] gi]48717332Jref[NG_004358.lj[48717332] gi|48717329Jref |NG_004359.1 [[48717329] gi|48717334Jref|NG_004360.11[48717334]
gi|48717331 |refJ|NG .004361 1(48717331] gi|48717333 |refJjNG 004362 j[48717333] gi|48717339 irefJ|NG .004363 |[48717339] gi|48717335 jrefjjNG .004364 j[48717335] giJ48717341 IreflJNG.004365 1(48717341] gi|48717338 IreflJNG.004366 j[48717338] gi|48717343 |refJ|NG .004367 |[48717343] gi(48717340 jrefJjNG 004368 1(48717340] giJ48717345 IreflJNG .004369 1(48717345] giJ48717342| |refJ|NG 004370 j[48717342] giJ48717347| IrefJjNG .004371 |[48717347] giJ48717344 irefJ|NG .004372 i(48717344] gi|48717349 IreflJNG 004373 j[48717349] giJ48717346 [refJ|NG .004374 i(48717346] gi|48717351 IreflJNG 004375 j[48717351] giJ48717348 IreflJNG .004376 |[48717348] gi|48717353 IreflJNG .004377 1(48717353] giJ48717350 I reflJNG .004378 |[48717350] giJ48717355 jrefJjNG .004379 1(48717355] giJ4871 352 IreflJNG..004380 |[48717352] giJ48717354 IreflJNG..004381 i(48717354] gi|48717356 IreflJNG..004382 i(48717356] giJ50055017 irefJ|NG .004383 1(50055017] giJ48717362 [refJ|NG .004384 1(48717362] giJ48717397 IreflJNG.004385 |[48717397] gi(48717424 jreflJNG .004386 j[48717424] giJ48717427 IreflJNG .004387 |[48717427] gi|48717428 | refJ |NG .004388 j[48717428] gi|48717429 irefJlNG 004389 j[48717429] gi|48717430 jreflJNG .004390 j[48717430] gi|48717431 IreflJNG
.004391 ([48717431] giJ48717432 jreflJNG .004392 |[48717432] giJ48717433 IrefljNG .004393 1(48717433] giJ48717434 jrefJ|NG .004394 ([48717434] gi|48717435 IreflJNG..004395 |[48717435] giJ48717436 I reflJNG.004396 1(48717436] gi|48717439 jreflJNG .004397 1(48717439] gi|48717437 IreflJNG .004398 [[48717437] giJ48717441 IrefJjNG.004399 |[48717441] giJ48717438 jreflJNG .004400 j[48717438] giJ48717443 jrefjJNG .004401 |[48717443] gi|48717440 jrefJjNG.004402 1(48717440] gi|48717445 jrefJjNG .004403 |[48717445] gi|48717442 IrefJjNG 004404 ([48717442] gi|48717449 jrefJjNG.004405 |[48717449] giJ48717444 jrefJjNG .004406 1(48717444] gi|48717451 jrefJjNG .004407 |[48717451] gi|48717448 jrefJ]NG 004408 j[48717448] gi|48717453 IrefJjNG.004409 |[48717453] gi(48717450 jreflJNG .004410 1(48717450] gi|48717455 jrefJjNG .004411 |[48717455] gi|48717452 IrefJjNG 004412 j[48717452] gi|48717456 I refJ|NG .004413 |[48717456] gi|48717454 IreflJNG 004414 1(48717454] gi|48717457 jrefJ|NG 004415 |[48717457] gi|48717458 jrefJ|NG..004416 [[48717458] gi|48717459 jrefJjNG.004417 j[48717459]
j|ref |NG_004418.11[48717460] I |ref|NG_004419.11[48717461] .jreflNG_004420.1 ([48717462] j(ref|NG_004421.11[48717465] 3|ref |NG_004422.1 [[48717463] 3|ref |NG_004423.11[48717466] l[ref(NG_004424.1 j[48717464] πref|NG_004425.11[48717467] 3(ref[NG_004426.1 ([48717468] 3|ref|NG_004427.11[48717469] )|ref|NG_004428.1 [[48717470] I |ref|NG_004429.11[48717471] -|ref|NG_004430.1 j[48717472] 3|ref|NG_004431.1 [[48717473] l[ref (NG_004432.1 j[48717474] 5|ref|NG_004433.11[48717475] 3|ref|NG_004434.11[48717476] πref|NG_004435.11[48717477] 3|ref|NG_004436.1 [[48717478] 3JrefJNG_004437.11[48717479] 3|ref|NG_004438.11[48717480] I jref|NG_004439.1 [[48717481] >[ref|NG_004440.1 ([48717482] 3|ref|NG_004441.11(48717483] 3|ref|NG_004443.1 ([48717486] |ref|NG_004444.11(48717490] ηreflNG_004445.1 ([48717487] 3[ref|NG_004446.11[48762516] πref|NG_004625.1 ([49227707] 3|ref|NG_004626.11[49227716] 5|ref|NG_004627.1 [[49227735] 3|ref|NG_004628.11[49227779] -|ref|NG_004629.1 ([49227822] πref|NG_004630.11[49227837] I |ref|NG_004631.11[49227451] 3|ref|NG_004632.11[49227006] 3|ref|NG_004633.11[49227863] t-|ref|NG_004634.11[49226814] 3(refJNG_004635.1 ([49226776] -|ref|NG_004638.11[50054452] t[ref|NG_004639.1 j[50054484] I |ref|NG_004652.11[50080191] l|ref|NG_004656.1 ([50263034] 3|ref|NG_004658.11[50345829] j[ref|NG_004662.1 ([50657353] jreflNG_004663.11[50657350] r|ref|NG_004666.11[51011127] 5|ref|NG_004670.11[51092275] ljref|NT_004321.15|Hs1_4478[29791384] 3|ref|NT_004350.16[Hs1_4507[37547298] .(refl NT_004433.16JHs1_4590[37547322] ηreflNT_004434.16|Hs1_4591[37547377] 3|ref|NT_004487.16|Hs1_4644[37547538] 1|ref|NT_004511.16|Hs1_4668[37548184] 3Jref|NT_004538.15[Hs1_4695[37548196] DirefJNT_004547.16JHs1_4704[37548319] πreflNT_004559.11 JHs1_4716(37548557]
|37548632|ref|NT_004610.16|Hs1_4767[37548632]
J37548668(ref|NT_004668.16|Hs1_4825[37548668]
|29793399|ref|NT_004671.15|Hs1_4828[29793399] i37548902iref|NT_004686.16[Hs1_4843[37548902]
|37549274(ref|NT_004754.15JHs1_4911(37549274]
|37549514|ref|NT_004836.15|Hs1_4993[37549514]
(3754954l iref|NT_004873.15(Hs1_5030[37549541]
|37547378Jref|NT_005058.14|Hs2_5215(37547378]
[29792066|ref|NT_005079.12|Hs2_5236[29792066]
|37547496|ref|NT_005120.14|Hs2_5277[37547496]
(37547741 |ref|NT_005334.14JHs2_5491 [37547741]
(37551287ireflNT_005403.14iHs2_5560[37551287]
(37549577[ref|NT_005416.11 JHs2_5573[37549577]
|29789894|ref|NT_005535.15|Hs3_5692[29789894] i37550867iref|NT_005612.14JHs3_5769[37550867]
137540616(ref|NT_006051.16JHs4_6208[37540616]
J37540627(ref|NT_006081.16|Hs4_6238[37540627]
J29791409Jref|NT_006216.14|Hs4_6373[29791409]
|29791548|ref|NT_006238.1θ(Hs4_6395[29791548]
J37540649(ref|NT_006307.15iHs4_6464[37540649]
|29792366(ref|NT_006316.15|Hs4_6473[29792366] i29798426(ref|NT_006431.13JHs5_6588[29798426]
|37550355Jref|NT_006576.14JHs5_6733[37550355] i29799535iref|NT_006713.13JHs5_6870[29799535]
[29740881 iref[NT_007299.12(Hs6_7456[29740881]
|37551434|ref|NT_007302.12|Hs6_7459[37551434]
J29803241 |ref|NT_007422.12JHs6_7579[29803241]
J27484631 (ref|NT_007583.11 |Hs6_7740[27484631]
|29804415|ref|NT_007592.13JHs6_7749[29804415]
J29794921 [reflNT_007741.12|Hs7_7898[29794921]
[29795292Jref|NT_007758.1θ[Hs7_7915[29795292] i3753847θirefiNT_007819.14JHs7_7976[37538470]
|37538573(ref|NT_007914.13|Hs7_8071[37538573]
J37538784ireflNT_007933.13|Hs7_8090[37538784]
|29795347|ref|NT_007995.13|Hs8_8152[29795347] i37555983iref|NT_008046.14[Hs8_8203[37555983]
|37555994|ref|NT_008127.15|Hs8_8284[37555994]
(37556103|reflNT_008183.17|Hs8_8340[37556103] i37556153Jref|NT_008251.14[Hs8_8408[37556153]
|37540361 |ref|NT_008413.16|Hs9_8570[37540361]
J3754059θiref|NT_008470.16[Hs9_8627[37540590]
|29734309Jref|NT_008476.14|Hs9_8633[29734309] i29795374(reflNT_008554.15JHs9_8711 [29795374]
J37551286JrefJNT_008583.16|Hs10_8740[37551286]
[37551352|ref|NT_008705.15|Hs10_8862[37551352]
J37551413[ref|NT_008818.15|Hs10_8975[37551413] l37541484|ref|NT_008984.16|Hs11_9141 [37541484]
|37541814JrefJNT_009237.16iHs11_9394[37541814]
[29804225|ref|NT_009487.15|Hs12_9644[29804225]
|37543832(ref|NT_009714.16JHs12_9871[37543832]
(37543915|ref|NT_009755.16|Hs12_9912[37543915]
|2980520θ[ref|NT_009759.15|Hs12_9916(29805200]
|37544143|ref|NT_009775.14JHs12_9932[37544143]
|37544901 |ref|NT_009952.14|Hs13_10109[37544901]
|37540936(reflNT_010194.16[Hs15_10351[37540936]
(29802090|ref|NT_010274.15[Hs15_10431 [29802090]
|37540985(ref|NT_010280.16JHs15_10437(37540985]
|37541485|ref[NT_010393.14|Hs16_10550[37541485] J37541544JrefJNT_010498.14JHs16_10655[37541544] 137541553Jref|NT_010505.14|Hs16_10662(37541553] 137541576|ref|NT_010542.14[HS16_10699[37541576] 129809129|ref|NT_010552.13|Hs16_10709(29809129] |3754259l(ref|NT_010641.14(Hs17_10798[37542591] (37542676|ref|NT_010663.14|Hs17_10820(37542676] |3754350θ(ref|NT_010718.14|Hs17_10875[37543500] |37543812|ref|NT_010755.14|Hs17_10912[37543812] 137544107|ref |NT_010783.14|Hs17_10940(37544107] |37544509Jref|NT_010799.14|Hs17_10956[37544509] |3754515θjref|NT _010859.13|Hs18_11016(37545150] |37545286|ref|NT_010879.14|Hs18_11036(37545286] J29823167Jref|NT_010966.13|Hs18_11123(29823167] |29800594|ref|NT_011109.15|Hs19_11266(29800594] |37552371|ref|NT_011255.14|Hs19_11412(37552371] J29801560Jref[NT_011295.10JHs19_11452(29801560] )20558635|ref|NT_011333.5|Hs20_11490(20558635] [29805814JrefJNT .011362.8JHs20_11519(29805814] J27501067|ref|NT_011387.8|Hs20_11544(27501067] |37558541 |ref|NT_011512.9|Hs21_11669(37558541] |29806340[ref|NT_011515.10|Hs21_11672[29806340] 114776721 |ref|NT_011516.5|Hs22_11673(14776721] J29806588JrefJNT_011519.10|Hs22_11676(29806588] |29807292|ref|NT_011520.9|Hs22_11677(29807292] |29807329|ref|NT_011521.2|Hs22_11678(29807329] J29807376(refJNT_011522.4|Hs22_11679[29807376] [29807498(ref[NT_011523.9|Hs22_11680[29807498] (29807536[ref|NT_011525.5JHs22_11682(29807536] J29807563Jref|NT_011526.5[HS22_11683(29807563] J37546122Jref |NT_011565.6JHsX_11722(37546122] J37546162|reflNT_011568.13|HsX_11725(37546162] 137546193|ref|NT_011630.14|HsX_11787(37546193] 137546211 |ref|NT_011638.12|HsX_11795(37546211] J37546266Jref|NT_011651.14[HsX_11808(37546266] |37546327|ref|NT_011669.14|HsX_11826(37546327] |37546334|ref|NT_011681.13|HsX_11838(37546334] [37546345|refJNT_011726.11 JHsX_11883(37546345] J37546406|ref|NT_011757.13|HsX_11914(37546406] |37546455[refJNT_011786.14JHsX_11943(37546455] |37546625|ref|NT_011875.10|HsY_12032(37546625] |37546636|ref|NT_011878.8|HsY_12035[37546636] i37546655JrefJNT _011896.8JHsY_12053[37546655] |37546676|ref|NT_011903.10|HsY_12060(37546676] J29789874Jref|NT_015926.13JHs2_16082(29789874] |37539484|ref|NT_016297.15|Hs4_16453(37539484] 137539910|ref|NT_016354.16|Hs4_16510(37539910] J37540362|reflNT_016606.16|Hs4_16762[37540362] J29737804Jref JNT.017696.14|Hs10_17852(29737804] |37550477|ref|NT_017795.16|Hs10_17951 [37550477] jl4779868|refJNT_019197.3|Hs22_19353[14779868] J37539512|ref |NT_019273.16|Hs1_19429(37539512] |29731701 |ref|NT_019501.12|Hs9_19657[29731701] [29802923iref[NT .019546.15JHs12_19702(29802923] J29809209Jref JNT_019609.11 JHs16_19765(29809209] |37546460|refJNT_019686.8|HsX 9842(37546460] J37539616|refJNT_021877.16JHs1_22033[37539616]
137539904 ref|NT_021937.16|Hs1_22093[37539904] J37539921 refiNT_021973.16[Hs1_22129[37539921] I29728076] ref|NT_022052.2|Hs1_22208[29728076] I29728085' ref|NT_022071.12|Hs1_22227[29728085] I29789875J ref|NT_022135.13|Hs2_22291[29789875] |37539622| ref[NT_022139.12(Hs2_22295[37539622] I29789878 ref|NT_022171.13JHs2_22327[29789878] J29789879] ref|NT_022173.11 |Hs2_22329[29789879] 137547123 ref|NT_022184.13JHs2_22340[37547123] 137547133 ref|NT_022221.1 O(HS2_22377[37547133] 137547140 ref|NT_022270.12|Hs2_22426[37547140] 137547164 ref|NT_022327.13|Hs2_22483[37547164] 137549731 ref(NT_022459.13JHs3_22615[37549731] 137550163 ref|NT_022517.16|Hs3_22673[37550163] J37540381 ref|NT_022778.14|Hs4_22934[37540381] |37540430 ref|NT_022792.16|Hs4_22948[37540430] I29789903 ref|NT_022794.9|Hs4_22950[29789903] I29789926 ref|NT_022853.14|Hs4_23009[29789926] J29796148] ref|NT_023089.13|Hs5_23245[29796148] J29796698 ref|NT_023133.11 |Hs5_23289[29796698] I37549963 ref|NT_023148.12(Hs5_23304[37549963] J37538129 ref|NT_023603.5|Hs7_23759[37538129] |29794492] ref|NT_023629.12|Hs7_23785[29794492] J37552400 ref|NT_023666.16|Hs8_23822[37552400] I37552409 ref|NT_023678.15[Hs8_23834[37552409] J37552441 ref|NT_023684.16|Hs8_23840[37552441] [37552484 ref|NT_023736.16|Hs8_23892[37552484] J37539567 ref)NT_023929.16|Hs9_24085[37539567] J37539568 ref|NT_023932.16JHs9_24088[37539568] J37539725 ref|NT_023935.16|Hs9_24091 [37539725] J29793214 ref|NT_024000.15|Hs9_24156[29793214] |29737906i ref|NT_024040.14|Hs10_24196[29737906] 117452488 ref|NT_024133.4|Hs10_24289[17452488] J29802945| ref)NT_024477.12|Hs12_24633[29802945] J37544912 ref|NT_024498.12|Hs13_24654[37544912] J37546841 ref|NT_024524.13|Hs13_24680[37546841] I29809252 ref|NT_024773.11 |Hs16_24929[29809252] 137541871 ref|NT_024797.14|Hs16_24953[37541871] (29809804 ref|NT_024812.10JHs16_24968[29809804] 137544515 ref|NT_024862.13|Hs17_25018[37544515] J37544588 ref|NT_024871.11 |Hs17_25027[37544588] |37544595 ref|NT_024972.7|Hs17_25128(37544595] [29823169 ref|NT_025004.13|Hs18_25160(29823169] J29823171 ref [NT_025028.13JHs18_25184(29823171] j 14772189 refJNT_025215.4|Hs20_25371 [14772189] |37546463 ref|NT_025302.12|HsX_25458[37546463] J37546468 refJNT_025307.14|HsX_25463[37546468] I37546473 ref|NT_025319.14|HsX_25475[37546473] J37546677 refJNT_025441.2|HsY_25597[37546677] [37551196 ref|NT_025741.13|Hs6_25897[37551196] J37550487 ref|NT_025835.11 |Hs10_25991 [37550487] |37546492 ref|NT_025965.12|HsX_26121 [37546492] |13626247 ref|NT_025975.2|HsY_26131[13626247] J29736559: ref|NT_026437.10|Hs14_26604[29736559] [29802188 ref|NT_026446.12JHs15_26613[29802188] J37540182 ref|NT_026943.13|Hs1_27103[37540182] J29791404 ref|NT_026970.9|Hs2_27130[29791404]
137545858 ref|NT_027140.6|Hs13_27300[37545858]
137546771 ref|NT_028050.13|Hs1_28209[37546771]
J37552491 ref)NT_028251.11 JHs8_28410(37552491]
117458490 reflNT_028392.4|Hs20_28551 [17458490]
I29807603 ref|NT_028395.2|Hs22_28554[29807603]
J29804531 reflNT .028405.9JHsX_28564[29804531]
J37546501 ref|NT_028413.7|HsX_28572[37546501]
J37550092 ref|NT_029289.10|Hs5_29448[37550092]
I29803948 ref|NT_029419.10JHs12_29578[29803948]
122067441 ref|NT_029490.3|Hs21_29649[22067441]
J37546778 refJNT _029860.11JHs1_30115(37546778]
J37550265 reflNT_029928.11 JHs3_30183(37550265]
J29732996 ref|NT_029998.6|Hs7_30253[29732996]
|29794552 ref(NT_030008.6JHs7_30263[29794552]
|27480958 ref|NT„030058.3(Hs9_30313[27480958]
J37550928 ref|NT_030059.11|Hs10_30314[37550928]
[16166615 refJNT_030187.1(Hs21_30442[16166615]
|37559293 ref|NT_030188.3JHs21_30443[37559293]
|37546789 ref|NT_030584.10|Hs1_30840[37546789]
J37552654 ref|NT_030737.8|Hs8_30993[37552654]
|37550940 ref|NT_030772.9|Hs10_31028(37550940]
J37544716ι ref|NT_030843.7|Hs17_31099[37544716]
117458602 ref|NT_030871.lJHs20_31127(17458602]
J29807617 ref|NT_030872.2|Hs22_31128(29807617]
J29791369 ref|NT_031730.8JHs1_31901[29791369]
J29738873 ref|NT_031847.7|Hs10_32018[29738873]
J37546839 ref|NT_032962.5JHs1_33138[37546839]
J37546840 ref)NT_032968.6|Hs1_33144[37546840]
137547124 ref|NT _032977.6|Hs1_33153[37547124]
129791412 ref|NT_032994.5JHs2_33170[29791412]
|37547233 ref|NT_033000.6|Hs2_33176[37547233]
J29801791 ref[NT_033172.4JHs6_33348[29801791]
J29738878 reflNT_033224.4JHs10_33400[29738878]
J37546506 refJNT_033330.6|HsX_33506[37546506]
J37540935 reflNT_033899.6JHs11_34054[37540935]
137541191 ref|NT_033903.6|Hs11_34058[37541191]
[37541321 reflNT_033927.6(Hs11_34082[37541321]
J27483040 reflNT_033948.3|Hs6_34103[27483040]
137538184 ref|NT .033968.5JHs7_34123[37538184]
J37550981 ref|NT_033985.6JHs10_34140[37550981]
129791378 ref|NT_034398.4(Hs1_34560[29791378]
|27478327 reflNT_034400.2|Hs1_34562[27478327]
137547157: ref|NT.034401.5|Hs1_34563[37547157]
129729791 ref|NT_034403.3JHs1_34565[29729791]
137547201 ref|NT_034410.5|Hs1_34572[37547201]
J29791381 ref|NT_034471.3|Hs1_34633[29791381]
|29791436 reflNT_034485.4|Hs2_34647[29791436]
129791451 ref|NT_034508.2|Hs2_34670[29791451]
I37550270 ref(NT_034772.5|Hs5_34934[37550270]
J29797964 ref|NT_034819.3JHs5_34981[29797964]
J37551285 ref|NT_034880.3|Hs6_35042[37551285]
137538187 ref|NT_034885.3JHs7_35047[37538187]
J29732427[ ref|NT_035014.3[Hs9_35176[29732427]
J27482319 ref|NT_035036.2|Hs10_35198(27482319]
J37550994] ref|NT_035040.4JHs10_35202[37550994]
J29806293J ref|NT_035086.3(Hs11_35248[29806293]
137541345 refl NT.035113.5| Hs11_35275[37541345]
129806334 ef|NT_035158.2|Hs11_35320[29806334] (29803986 ef|NT_035243.4|Hs12_35405[29803986] 137541078 ef|NT_035325.5|Hs15_35487[37541078] J29809860 ef|NT_035363.3|Hs16_35525[29809860] 129799518 ef|NT_035414.4|Hs17_35576[29799518] J22063824 eflNT_035608.l JHs20_35770[22063824] J29791382 ef|NT_037485.3[Hs1_37489[29791382] 129791461 ef|NT_037540.2JHs2_37544[29791461] J29789976 efJNT_037622.3(Hs4_37626[29789976] |37540599 ef|NT_037623.4|Hs4_37627[37540599] [29729468 ef|NT_037645.2(Hs4_37649[29729468] |37574696 ef|NT_037704.4|Hs8_37708[37574696] J27482375 ef|NT_037750.1 [Hs10_37754[27482375] J37551011 ef|NT_037753.4|Hs10_37757[37551011] J37541180 ef|NT_037852.4JHs15_37856[37541180] J29810036 ef|NT_037887.3|Hs16_37891 [29810036] 129794124 ef|NT_077382.2|Hs1_77431 [29794124] (37549544 ef|NT_077383.3JHs1_77432[37549544] [29733654 ef|NT_077384.1 |Hs1_77433[29733654] 129794331 ef|NT_077387.2|Hs1_77436[29794331] |37549576 ef|NT_077388.3|Hs1_77437[37549576] (29794365 ef|NT_077389.2|Hs1_77438[29794365] |29794380 ef|NT_077390.2|Hs1_77439[29794380] [29794392 ef|NT_077402.1 |Hs1_77451 [29794392] [29794254 ef|NT_077407.2|Hs2_77456[29794254] J29792454 ef|NT_077444.2|Hs4_77493[29792454] J37550440 eflNT_077451.3|Hs5_77500[37550440] I29733246 ef|NT_077528.1 |Hs7_77577[29733246] [37552698 ef|NT_077531.3|Hs8_77580[37552698] 137551026 ef|NT_077567.3(Hs10_77616[37551026] (29800963 ef|NT_077569.2|Hs10_77618(29800963] [29739944 ef|NT_077570.1 |Hs10_77619(29739944] J29739967 ef[NT_077571.1 JHs10_77620[29739967] J29799665 ef|NT_077627.2|Hs13_77676[29799665] J29802368 ef|NT_077631.1 JHsl 5_77680[29802368] J37541217 ef|NT_077661.2|Hs15_77710(37541217] 129810067 ef|NT_077677.2JHs16_77726[29810067] (37574721 ef|NT_077812.2|Hs19_77861[37574721] |37546537 ef|NT_077819.3|HsX_77868[37546537] J29794400, ef|NT_077911.1 |Hs1_77980[29794400] |29794409| ef|NT_077912.l (Hs1_77981[29794409] (37549614 efJNT_077913.2|Hs1_77982[37549614] [37549622 eflNT_077914.2(Hs1_77983[37549622] [29794459 ef|NT_077915.1 |Hs1_77984[29794459] J37549624 ef|NT_077919.2(Hs1_77988[37549624] |37549633 eflNT_077920.2[Hs1_77989[37549633] J29794539 ef|NT_077921.1 JHs1_77990[29794539] J37549636 ef|NT_077922.2(Hs1_77991 [37549636] |29794679 ef|NT_077930.l [Hs1_77999[29794679] (37549641 ef|NT_077931.2JHs1_78000[37549641] J37549644 ef|NT_077932.2|Hs1_78001[37549644] |29794723 ef|NT_077933.1|Hs1_78002[29794723] J37549653 ef | NT_077936.2| Hs1 _78005[37549653] (29794872 ef|NT_077939.l (Hs1_78008[29794872] J29794900 eflNT_077941.11HS1_78010[29794900] J29794939 ef|NT_077944.1 |Hs1_78013[29794939] J29794950 ef|NT_077945.1 |Hs1_78014[29794950]
|29794965|ref|NT_077946.1|Hs1_78015[29794965] i29795012iref|NT_077949.l [Hs1_78018[29795012] |29795183|ref|NT_077962.1 |Hs1_78031[29795183] i37549666iref|NT_077964.2(Hs1_78033[37549666] 129795221 |ref|NT_077966.1 |Hs1_78035[29795221] J37549729iref[NT_077967.2[Hs1_78036[37549729] 129795291 |ref|NT_077970.1 |Hs1_78039[29795291] i37549730JrefiNT_077988.2JHs1_78057[37549730] |29794265|ref|NT_077995.1 |Hs2_78064[29794265] J29794307|reflNT_077999.1 |Hs2_78068[29794307] i29794317irefiNT_078000.l JHs2_78069[29794317] J29794350|reflNT_078003.l iHs2_78072[29794350] |29792480|ref|NT_078010.1 |Hs4_78079[29792480] [2979250θireflNT_078012.l iHs4_78081[29792500] |37540684|ref|NT_078016.2|Hs4_78085[37540684] [29799691 (ref|NT_078018.1 JHs5_78087[29799691] i29799708iref|NT_078019.l [Hs5_78088[29799708] J37552304Jref|NT_078020.2iHs6_78089[37552304] |37552305|ref|NT_078023.2|Hs6_78092[37552305] |29794804|ref|NT_078033.1 |Hs7_78102(29794804] i37552703|ref|NT_078037.2|Hs8_78106[37552703] |29795147|reflNT_078038.1 |Hs8_78107[29795147] |29793286|ref|NT_078040.1 |Hs9_78109(29793286] i29793299|ref|NT_078041.1 JHs9_78110(29793299] i37539905iref(NT_078042.2iHs9_78111 [37539905] 137539913|ref|NT_078043.2|Hs9_78112(37539913] |29793356(ref|NT_078044.1 JHs9_78113(29793356] [29793369iref[NT_078045.1 |Hs9_78114(29793369] [3753992θiref|NT_078046.2JHs9_78115(37539920] (37539922|ref|NT_078047.2|Hs9_78116(37539922] |37539923[ref|NT_078048.2|Hs9_78117(37539923] 129793419|ref|NT_078049.l JHs9_78118(29793419] J37539926|reflNT_078051.2JHs9_78120[37539926] |37539927|refJNT_078052.2[Hs9_78121 [37539927] J37539930JrefiNT_078053.2JHs9_78122[37539930] |37539933|reflNT_078055.2[Hs9_78124[37539933] i29793548JrefiNT_078057.l JHs9_78126(29793548] i29793568iref|NT_078058.l (Hs9_78127(29793568] J37539942|ref|NT_078059.2JHs9_78128[37539942] (29793603[ref|NT_078061.1 |Hs9_78130(29793603] i2979361θ[ref|NT_078062.l iHs9_78131 [29793610] i37539951 lreflNT_078063.2[Hs9_78132[37539951] i29793633|ref|NT_078064.1 |Hs9_78133[29793633] |37539960|ref|NT_078065.2|Hs9_78134[37539960] i37539963|ref|NT_078066.2(Hs9_78135[37539963] |37539964iref(NT_078067.2JHs9_78136[37539964] |29793707|ref|NT_078068.1 |Hs9_78137[29793707] |29793717|ref|NT_078069.l (Hs9_78138[29793717] |29793738[ref|NT_078070.1 |Hs9_78139(29793738] 137539981 |ref|NT_078071.2JHs9_78140[37539981] J29793761 |ref|NT_078072.1 |Hs9_78141 [29793761] |29793793ireflNT_078074.l iHs9_78143[29793793] |29793806|ref|NT_078075.1 |Hs9_78144[29793806] (37539982iref|NT_078076.2(Hs9_78145[37539982] i37539983(ref|NT_078077.2JHs9_78146[37539983] |37539988JreflNT_078078.2|Hs9_78147[37539988] i29793857[refJNT_078079.l JHs9_78148[29793857]
|37539999|ref|NT_078081.2|Hs9_78150[37539999]
137540010|ref|NT_078083.2|Hs9_78152(37540010]
J37551062|ref JNT_078087.2|Hs10_78156(37551062] i37541352|ref|NT_078088.2|Hs11_78157(37541352]
|37545863ireflNT_078092.2JHs13_78161 [37545863] i29802447(ref|NT_078094.1 |Hs15_78163(29802447] i29802469iref[NT_078095.liHs15_78164[29802469]
|37541233(ref|NT_078096.2lHs15_78165[37541233] i37542234[reflNT_078099.2iHs16_78168[37542234]
|29799562Jref|NT_078100.1 |Hs17_78169(29799562]
J29824109[ref|NT_078102.1 JHs17_78171 [29824109]
|29801587Jref|NT_078103.1|Hs19_78172[29801587]
|29805042ireflNT_078109.liHsX_78178[29805042]
|37546556Jref|NT_078110.2[HSX_78179(37546556]
J37546559|ref|NT_078114.2JHsX_78183[37546559]
|37546560|ref|NT_078115.2JHsX_78184[37546560] i37546563|ref|NT_078116.2(HsX_78185[37546563]
|37546564|ref|NT_078118.2|HsX_78187[37546564]
|3754974θ[ref|NT_079482.1|Hs1_79547[37549740]
J37549741 |ref|NT_079483.1 |Hs1_79548[37549741] i37549803iref|NT_079484.l [Hs1_79549[37549803]
|37549806|ref|NT_079485.1|Hs1_79550[37549806]
J37549811 (ref|NT_079486.1 [HS1_79551 [37549811]
J37549812Jref|NT_079487.l[Hs1_79552[37549812]
137549815ireflNT_079488.liHs1_79553[37549815] i37549816iref|NT_079489.1|Hs1_79554[37549816] i37549817|ref|NT_079490.l[Hs1_79555[37549817]
|3754982θ(ref|NT_079491.1 JHs1_79556[37549820]
(37549821 |reflNT_079492.1 JHs1_79557[37549821]
[37549822(ref|NT_079493.1|Hs1_79558[37549822] i37549823iref|NT_079494.lJHs1_79559[37549823]
J37549826ireflNT_079495.liHs1_79560[37549826]
|37549829|ref|NT_079496.1 (HS1_79561 [37549829]
|37549836Jref|NT_079497.1|Hs1_79562[37549836]
J3754986liref|NT_079498.l[Hs1_79563[37549861]
|37549864iref|NT_079499.l[Hs1_79564[37549864] i37549865irefJNT_079500.lJHs1_79565[37549865]
|37549866(ref|NT_079501.1 JHs1_79566[37549866] i37549602[ref|NT_079502.1 [Hs2_79567[37549602] i37549605ireflNT_079503.l iHs2_79568[37549605]
|37549608|ref|NT_079504.1|Hs2_79569[37549608]
(37549617[ref(NT_079505.1 [HS2_79570[37549617]
|37550890|ref[NT_079506.1 |Hs3_79571 [37550890] i37550893iref|NT_079507.lJHs3_79572[37550893]
|37550894|ref|NT_079508.l(Hs3_79573[37550894]
J37550895JreflNT_079509.liHs3_79574[37550895]
|37540685|ref|NT_079510.1 |Hs4_79575[37540685]
|37540686|ref|NT_079511.1 |Hs4_79576[37540686]
J37540687|reflNT_079512.1 |Hs4_79577[37540687]
[37550453(ref|NT_079513.1 [HS5_79578[37550453]
|37538270|ref|NT_079514.1|Hs7_79579[37538270]
|37538277|ref|NT_079515.1 |Hs7_79580[37538277]
J37538282|ref|NT_079516.1 |Hs7_79581 [37538282]
[37552706|ref |NT_079517.1 |Hs8_79582[37552706]
[37552707|ref|NT_079518.l[Hs8_79583[37552707]
[37552708|ref|NT_079519.1 [HS8_79584[37552708] i37552709iref|NT_079520.1|Hs8_79585[37552709]
137552710|ref|NT_079521.1 |Hs8_79586[37552710] J37552711 |ref|NT_079522.1 |Hs8_79587[37552711] J37552712iref|NT_079523.l JHs8_79588[37552712] |37552713iref|NT_079524.1 |Hs8_79589[37552713] |37552714ireflNT_079525.lJHs8_79590[37552714] 137552721 |ref|NT_079526.1 |Hs8_79591 [37552721] |37552722(ref|NT_079527.l JHs8_79592[37552722] [3754001 liref|NT_079528.l JHs9_79593[37540011] i37540016ireflNT_079529.l JHs9_79594[37540016] i37540017[ref|NT_079530.l [Hs9_79595[37540017] |37540020|ref|NT_079531.1 |Hs9_79596[37540020] J37540021 |ref|NT_079532.1 [HS9_79597[37540021] [37540026iref|NT_079533.l [Hs9_79598[37540026] (37540031 [ref|NT_079534.1 |Hs9_79599[37540031] |37540057(ref|NT_079535.l JHs9_79600[37540057] i37540083ireflNT_079536.1 JHs9_79601 [37540083] 137540119|ref|NT _079537.1 |Hs9_79602[37540119] |37540152Jref|NT_079538.l (Hs9_79603[37540152] |37540155|ref|NT_079539.1|Hs9_79604[37540155] J37551063iref|NT_079540.l JHs10_79605[37551063] |37551066(ref|NT_079541.l iHs10_79606[37551066] |37551071 |ref|NT_079542.1 |Hs10_79607[37551071] J37551076|ref|NT_079543.1 JHsl 0_79608[37551076] (37551079[ref|NT_079544.1 |Hs10_79609[37551079] [37551082Jref|NT_079545.1 JHsl 0_79610[37551082] J37541240|ref|NT_079546.l [Hs15_79611(37541240] i37541243|ref[NT_079547.l iHs15_79612[37541243] i37541254|ref[NT_079548.1 |Hs15_79613[37541254] J37541257iref|NT_079549.l JHs15_79614[37541257] J37541258iref|NT_079550.1 |Hs15_79615[37541258] [37541259irefiNT_079551.l (Hs15_79616[37541259] i37541260|refJNT_079552.l [Hs15_79617[37541260] (37541261 jref|NT_079553.1 |Hs15_79618[37541261] i37541262iref|NT_079554.l (Hs15_79619[37541262] J37544921 |ref|NT_079555.1 |Hs17_79620[37544921] 137544931 |ref|NT_079556.1 |Hs17_79621 [37544931] (37544936JreflNT_079557.1 |Hs17_79622[37544936] |37544945|ref(NT_079558.1 |Hs17_79623[37544945] (37544957[ref|NT_079559.1 |Hs17_79624[37544957] |37544965|ref|NT_079560.1 |Hs17_79625[37544965] [37544968|ref|NT_079561.1 JHsl 7_79626[37544968] J37544969[reflNT_079562.1 |Hs17_79627[37544969] (37544972|ref|NT_079563.1 |Hs17_79628[37544972] |37544979[reflNT_079564.1 JHsl 7_79629[37544979] i37544982[ref|NT_079565.1 |Hs17_79630[37544982] |37544984[reflNT_079566.1 |Hs17_79631 [37544984] J37544986|reflNT_079567.1 JHsl 7_79632[37544986] i37544996[ref|NT_079568.1|Hs17_79633[37544996] (37544998|ref|NT_079569.1 JHsl 7_79634[37544998] i3754500θireflNT_079570.1 |Hs17_79635[37545000] |37545007|ref|NT_079571.1 |Hs17_79636[37545007] i37545883(ref|NT_079572.1 JHsl 8_79637[37545883] i37546587[ref|NT_079573.1 |HsX_79638[37546587] |37546602|ref|NT_079574.1 |HsX_79639[37546602] |37546603(ref|NT_079575.1 |HsX_79640[37546603] |37546604[ref|NT_079576.1 |HsX_79641 [37546604] J37546605|ref|NT_079577.1 |HsX_79642[37546605]
|37546608|ref|NT_079578.1|HsX_79643[37546608]
|37546609|ref|NT_079579.1 |HsX_79644[37546609]
13754661 θjref|NT_079580.1 |HsX_79645[37546610]
J37546680|ref|NT_079581.1 |HsY_79646[37546680] l37546685(ref|NT_079582.1lHsY_79647[37546685]
|3754669θ(ref|NT_079583.1|HsY_79648[37546690]
J37546691 (ref|NT_079584.1 JHsY_79649[37546691] i37546692(ref|NT_079585.1 |HsY_79650[37546692]
|37546695|ref|NT_079586.1 |HsUn_79651 [37546695]
|37546702lreflNT_079587.l(HsUn_79652[37546702]
|37546703!ref|NT_079588.1 |HsUn__79653[37546703] i37546704(ref|NT_079589.l (HsUn_79654[37546704]
|3753813θiref|NT_079590.l [Hs7b_79655[37538130]
137538791 |ref|NT_079591.1 |Hs7b_79656[37538791]
|37538986jref|NT_079592.1 |Hs7b_79657[37538986]
(37539049iref|NT_079593.l[Hs7b_79658[37539049]
(37539058|ref|NT_079594.1|Hs7b_79659[37539058]
J37539165Jref|NT_079595.l[Hs7b_79660[37539165]
|37539429|ref|NT_079596.1 |Hs7b_79661 [37539429]
|37539444(ref|NT_079597.1|Hs7b_79662[37539444]
J37546707 jref [NT.079614.1 JHsUn_79741 [37546707]
|37546708|ref|NT_079615.1|HsUn_79742[37546708]
J37546709|ref|NT_079616.l JHsUn_79743[37546709]
|37546710|ref|NT_079617.1|HsUn_79744[37546710]
137546715|ref|NT_079618.1 |HsUn_79745[37546715]
J37546718|ref|NT_079619.1 JHsUn_79746[37546718]
[37546719|ref|NT_079620.liHsUn_79747[37546719]
|37546722Jref[NT_079621.1 |HsUn_79748[37546722] i37546723|reflNT_079622.l JHsUn_79749[37546723]
|37546730|ref|NT_079623.1|HsUn_79750[37546730]
|37546733]reflNT_079624.1 JHsUn_79751 [37546733]
(37546738(ref|NT_079625.liHsUn_79752[37546738]
|42655516|ref|NT_086323.2|ENr111(42655516]
|42655538|ref|NT_086324.2JENr112(42655538]
|42633566|ref|NT_086325.1 |ENr113(42633566]
]42655573lreflNT_086326.2]ENr114(42655573]
(42655539[refiNT_086327.3(ENr115[42655539]
[42655540|ref|NT_086328.2(ENr121 [42655540]
142655541 jref|NT_086329.2|ENr122(42655541]
142633571 |ref|NT_086330.1 |ENr123(42633571]
|42655542(ref|NT_086331.3[ENr124[42655542]
J42633573|ref|NT_086332.lJENr131 [42633573] i42633574[ref|NT_086333.l[ENr132(42633574] i42655574|ref[NT_086334.3[ENr133(42655574]
|42655544|ref|NT_086335.2|ENr134(42655544]
|42655545|ref|NT_086336.2|ENr211 [42655545] i42655546lref|NT_086337.2[ENr212[42655546]
|42633579|ref [NT_086338.1 |ENr213[42633579]
(42655547|ref|NT_086339.2|ENr214[42655547]
|42655548Jref|NT_086340.3|ENr221[42655548]
|42655520|ref|NT_086341.2JENr222[42655520]
|42655521|ref|NT_086342.2lENr223[42655521]
|42633584|ref|NT_086343.1|ENr224[42633584]
J42655549|ref|NT_086344.3|ENr231[42655549]
|42655523|ref|NT_086345.2|ENr232[42655523]
J42655550|ref|NT_086346.3JENr233[42655550]
]42655525|ref|NT _086347.2|ENr234[42655525]
|42655551 |ref|NT_086348.2|ENr311 [42655551]
|42655552|ref|NT_086349.2|ENr312[42655552] i42655553JrefINT_086350.2iENr313[42655553]
J42655554|refINT_086351.2|ENr314[42655554]
|42655555|ref|NT_086352.3(ENr321[42655555]
J42655556|ref|NT_086353.3(ENr322[42655556]
J42655528|ref|NT_086354.2|ENr323[42655528] i42655557iref|NT_086355.2JENr324[42655557]
[42655529iref|NT_086356.2[ENr331[42655529]
|42655559iref|NT_086357.2(ENm001[42655559]
(42633603|ref|NT_086358.1|ENm002[42633603]
|4265556θiref|NT_086359.2|ENm003[42655560]
[42655561|ref|NT_086360.2|ENm004[42655561]
|42655562|ref|NT_086361.2|ENm005[42655562]
(42655576|ref|NT_086362.4[ENm006[42655576]
|42655564|ref|NT_086363.3|ENm007[42655564]
(42655565[ref|NT_086364.3|ENm008[42655565]
|42655566|ref|NT_086365.3|ENm009[42655566]
J42633611 [ref|NT_086366.1 ]ENm010(42633611]
J42655577Jref|NT_086367.4|ENm011(42655577]
|42655568[ref|NT_086368.3|ENm012[42655568]
[42655569|ref|NT_086369.2(ENm013[42655569]
[42655570lref|NT_086370.2(ENm014[42655570]
|42633616|ref|NT_086371.1|ENm015[42633616]
142655571 [ref|NT_086372.2|ENm016(42655571]
|42655572[ref|NT_086373.2|ENm017[42655572] i42655575[ref|NT_086374.4|ENr332[42655575]
|42633599iref|NT_086375.l [ENr333[42633599]
(42655531|ref|NT_086376.2[ENr334[42655531]
142633601 |ref|NT_086377.1 |ENr335[42633601]
Table II Non-limiting examples of Stabilization Chemistries for chemically modified siNA constructs
CAP = any terminal cap, see for example Figure 10. All Stab 00-22 chemistries can comprise 3 '-terminal thymidine (TT) residues All Stab 00-22 chemistries typically comprise about 21 nucleotides, but can vary as described herein. S = sense strand AS = antisense strand *Stab 23 has a single ribonucleotide adjacent to 3'-CAP *Stab 24 and Stab 28 have a single ribonucleotide at 5 '-terminus *Stab 25. Stab 26. and Stab 27 have three ribonucleotides at 5 '-terminus *Stab 29. Stab 30. and Stab 31. any purine at first three nucleotide positions from 5'-terminus are ribonucleotides p = phosphorothioate linkage
Table III A. 2.5 μmol Synthesis Cycle ABI 394 Instrument
B. 0.2 μmol Synthesis Cycle ABI 394 Instrument
Wait time does not include contact time during delivery.
Tandem synthesis utilizes double coupling of linker molecule




















